Abstract
To gain a more complete understanding of hepatitis C virus (HCV) entry, we initially assessed the rate at which HCV initiates productive attachment/infection in vitro and discovered it to be slower than most viruses. Since HCV, including cell culture-derived HCV (HCVcc), exhibits a broad density profile (1.01 – 1.16 g/ml), we hypothesized that the varying densities of the HCVcc particles present in the inoculum may be responsible for this prolonged entry phenotype. To test this hypothesis, we show that during infection, particles of high-density disappeared from the viral inoculum sooner and initiated productive infection faster than virions of low-density. Moreover, we could alter the rate of attachment/infection initiation by increasing or decreasing the density of the cell culture medium. Together, these findings demonstrate that the relationship between the density of HCVcc and the density of the extracellular milieu can significantly impact the rate at which HCVcc productively interacts with target cells in vitro.
Keywords: Hepatitis C virus, entry, density, infection initiation
Introduction
Hepatitis C virus (HCV), a positive-strand RNA flavivirus, is a major cause of liver disease worldwide, including cirrhosis and hepatocellular carcinoma (Poynard et al., 2000; Poynard et al., 2003). Since the discovery of HCV in 1989 as the causative agent of “non-A non-B hepatitis” (Choo et al., 1989), a major obstacle impeding HCV research has been the lack of robust cell culture and small animal infection models. To circumvent these limitations, alternate systems based on selectable replication-competent HCV RNAs, known as replicons (Blight, Kolykhalov, and Rice, 2000; Blight et al., 2003; Ikeda et al., 2002; Lohmann et al., 1999), and pseudotyped particles (HCVpp) (Bartosch, Dubuisson, and Cosset, 2003) were developed and have provided significant insight into HCV replication and entry, respectively. These systems were limited in that they did not recapitulate the entire viral lifecycle; however, with the 2005 establishment of the HCV infectious cell culture system (Lindenbach et al., 2005; Wakita et al., 2005; Zhong et al., 2005), based on a genotype 2a HCV consensus clone (JFH-1) (Kato et al., 2003; Kato et al., 2001) that can replicate and produce infectious HCV in cell culture (HCVcc), we now possess the means to examine the entire viral lifecycle in detail and possibly learn how to exploit key steps as potential drug targets. In particular, HCV entry represents a “promising multi-faceted opportunity for drug discovery” [reviewed in (Meanwell, 2006)], but a deeper understanding of the process is needed to facilitate such endeavors. The HCV particle itself consists of a positive-strand RNA genome surrounded by a nucleocapsid and a lipid bilayer containing two viral envelope glycoproteins, E1 (residues 192–383) and E2 (residues 384–746), which form a non-covalent heterodimer complex on the surface of the viral particle believed to mediate host cell receptor binding (Moradpour, Penin, and Rice, 2007). HCV particles in patient serum and in vitro exist as low-density lipoprotein (LDL)�virus complexes (Andre et al., 2002; Gastaminza et al., 2008; Gastaminza, Kapadia, and Chisari, 2006). They are associated with triglycerides, β-lipoproteins (very-low-density lipoproteins [VLDL] and LDL) (Gastaminza et al., 2008; Nielsen et al., 2006; Thomssen et al., 1992; Thomssen, Bonk, and Thiele, 1993), cholesterol and sphingomyelin (Aizaki et al., 2008; Kapadia et al., 2007), all of which contribute to the unique broad buoyant density profile (Andre et al., 2002; Gastaminza et al., 2008; Gastaminza, Kapadia, and Chisari, 2006; Lindenbach et al., 2005; Pumeechockchai et al., 2002; Zhong et al., 2005) of the virus and appear to play a critical role in viral entry and fusion (Aizaki et al., 2008; Haid, Pietschmann, and Pecheur, 2009; Kapadia et al., 2007; Lavillette et al., 2006). In patients, serum HCV (HCVser) has a buoyant density profile of ~1.03 – 1.13 g/ml, with the majority of HCV RNA detectable at 1.08 g/ml and below (Andre et al., 2002). The density profile of HCVcc shows a similar broad HCV RNA distribution from 1.01 to 1.16 g/ml, but exhibits an additional peak of relatively non-infectious RNA at ~1.12 g/ml. Like HCVser (Bradley et al., 1991), HCVcc particles of low-density display the highest specific infectivity (Haid, Pietschmann, and Pecheur, 2009; Lindenbach et al., 2005; Zhong et al., 2006) consistent with lipoproteins being important for HCV infectivity.
In vitro, four cell surface receptors, the tetraspanin protein CD81 (Bartosch, Dubuisson, and Cosset, 2003; Hsu et al., 2003; Pileri et al., 1998; Wunschmann et al., 2000; Zhong et al., 2005), the scavenger receptor class B member I (Bartosch, Dubuisson, and Cosset, 2003; Grove et al., 2007; Kapadia et al., 2007; Scarselli et al., 2002; Zeisel et al., 2007), and the tight junction proteins claudin-1 (Evans et al., 2007) and occludin (Benedicto et al., 2009; Liu et al., 2009; Ploss et al., 2009) have been shown to be required for HCVcc entry. In addition, the low-density lipoprotein receptor (Agnello et al., 1999; Molina et al., 2007; Monazahian et al., 1999; Wunschmann et al., 2000), asialoglycoprotein receptor (Saunier et al., 2003), glycosaminoglycans (heparan sulfate) (Barth et al., 2003; Barth et al., 2006; Bartosch, Dubuisson, and Cosset, 2003) and protocaherin β5 (Wong-Staal et al., 2008) have been implicated. Following a relatively undefined multi-step binding process, HCV enters the cell via clathrin-mediated endocytosis (Blanchard et al., 2006; Meertens, Bertaux, and Dragic, 2006) and fusion between the virion envelope and the endosomal membrane occurs in the acidified endosomal compartment (Bartosch et al., 2003; Haid, Pietschmann, and Pecheur, 2009; Hsu et al., 2003; Koutsoudakis et al., 2006; Lavillette et al., 2006; Tscherne et al., 2006) resulting in nucleocapsid release into the cytoplasm.
To expand our working knowledge of HCV entry we sought to assess the rate at which HCVcc productively attaches/interacts with target cells (i.e. infection initiation) in vitro and identify the factors that influence the kinetics observed. Here we show that the rate of HCVcc attachment/infection initiation in vitro is relatively slow and, more importantly, that the kinetics are largely determined by the density of the viral particle relative to the extracellular environment. Our study highlights the importance of understanding how the static nature of traditional cell culture may artificially impact the rate of HCV infection initiation in vitro and the need to carefully consider experimental HCVcc infection conditions to accurately interpret and study viral entry in this now widely used experimental model system.
Materials and Methods
Cells
Huh7 cells [also known as Huh7/scr (Gastaminza, Kapadia, and Chisari, 2006; Zhong et al., 2006) and Huh7-1 (Sainz, Barretto, and Uprichard, 2009) cells)] were obtained from Dr. Francis Chisari (The Scripps Research Institute, La Jolla, CA) (Zhong et al., 2005), and were cultured in complete Dulbecco’s modified Eagle’s medium (cDMEM) (Hyclone, Logan, UT) supplemented with 10% fetal bovine serum (FBS) (Hyclone), 100 units/ml penicillin, 100 mg/ml streptomycin, and 2 mM L-glutamine (Gibco Invitrogen, Carlsbad, CA).
Cell culture propagated HCV (HCVcc)
The genotype 2a JFH-1 plasmid (pJFH-1) has been previously described (Wakita et al., 2005). The JFH-1G451R expression plasmid (pJFH-1G451R), which contains a glycine-to-arginine mutation at amino acid residue 451 in the E2 glycoprotein (Zhong et al., 2006), was generated by site-directed mutagenesis of pJFH-1 using a complementary pair of oligomers of sequence 5’-CAA CTC TTC AAG GTG TCC TGA GAG GTT GGC C-3’ and the QuickChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA), as per the manufacturer’s instructions.
Viral stocks of wild-type (wt) JFH-1 (HCVccwt) or JFH-1G451R (HCVccG451R) were generated as previously described (Sainz, Barretto, and Uprichard, 2009). Supernatants from infected cells were collected and centrifuged at 4,000 rpm for 5 minutes to remove cellular debris, infectivity titers were determined as described below and when indicated, concentrated 10-fold by centrifugal filteration (Amicon Ultra; Millipore, Billerica, MA).
48 hour foci formation assay
In order to achieve a cell confluence of 80% at the time of inoculation, ~5,000 Huh7 cells were seeded in each well of a 96-well plate (Corning, Lowell, MA). Exactly 24 hours post seeding, wells were inoculated with ~100 focus forming units (FFU) of HCVccwt or HCVccG451R in a total volume of 100 µl cDMEM containing 1, 10 or 20% FBS. At indicated times post infection (p.i.), the inoculum was removed from triplicate wells, cells were washed twice with pre-warmed (i.e. 37°C) 1X phosphate buffered saline (PBS), and monolayers overlaid with 200 µl of pre-warmed cDMEM containing a final concentration of 0.25% methylcellulose (vol/vol) (Fluka BioChemika, Switzerland). Forty-eight hours p.i., cells were fixed in 4% paraformaldehyde (Sigma, St. Louis, MO), and HCV-specific foci detected by immunohistochemical staining using a mouse monoclonal anti-NS5A antibody E910 (Lindenbach et al., 2005) as described in (Sainz, Barretto, and Uprichard, 2009).
Density gradient ultracentrifugation
Iodixanol (OptiPrep Solution, Axis-Shield, Norton, MA) density gradients were prepared in Beckman UltraClear 14 by 89 mm ultracentrifugation tubes as a step gradient of 10–50% iodixanol in TNE (150mM NaCl, 10mM Tris-HCl and 2mM EDTA, pH 8). One to 1.5 ml of viral sample was layered on top of the gradient and separated by ultracentrifugation in a SW41Ti (Beckman Coulter, Inc. Fullerton, CA) rotor at 125,000 × g for 16 hours at 4°C. One ml gradient factions were collected from the top of the gradient and titrated for virus infectivity or analyzed for HCV RNA as described below. Fraction densities were determined by measuring the mass of 50µl aliquots of each sample.
Infectivity titration assay
Culture supernatants or iodixanol gradient fractions (diluted 1:2 with cDMEM) were serially diluted 10-fold and used to infect duplicate 96-well Huh7 cultures. At 24h p.i., cultures were overlaid with cDMEM to a final concentration of 0.25% methylcellulose (Fluka BioChemika) to limit long distance extracellular spread and resulting secondary satellite foci formation. Seventy-two hours p.i., cells were fixed in 4% paraformaldehyde (Sigma), and immunohistochemically stained for HCV NS5A using a mouse monoclonal anti-NS5A antibody E910 (Lindenbach et al., 2005). Viral titers are expressed as FFU/ml, determined by the average NS5A-positive foci number detected at the highest HCV-positive dilution, foci being defined as discrete areas of HCV-positive cells.
HCV infection kinetics
Huh7 cells were seeded at 4×103 cells in each well of a 96-well plate (Corning). Twenty four hours post-seeding, cells were infected with HCVccwt at a multiplicity of infection (MOI) of 0.01 FFU/cell in a total volume of 100 µl cDMEM containing 1, 10 or 20% FBS. At indicated times p.i., total cellular RNA was harvested from triplicate wells for reverse transcription followed by real-time quantitative PCR (RTqPCR) analysis.
RNA isolation and RTqPCR
Total cellular RNA was isolated using a 1X Nucleic Acid Purification Lysis Solution (Applied Biosystems, Foster City, CA) and purified using an ABI PRISM™ 6100 Nucleic Acid PrepStation (Applied Biosystems), as per the manufacturer’s instructions. Total RNA from gradient fractions was isolated by the guanidine thiocyanate (GTC) method using 1.6X GTC containing 2 µg of murine liver RNA and following standard protocols (Chomczynski and Sacchi, 1987). One µg of RNA was used for cDNA synthesis using TaqMan reverse transcription reagents (Applied Biosystems), followed by SYBR green RTqPCR using an Applied Biosystems 7300 real-time thermocycler as previously described (Choi et al., 2009). HCV transcript levels were determined relative to a standard curve of serially diluted JFH-1 plasmid and normalized to GAPDH. The PCR primers used to detect GAPDH and HCV were: human GAPDH (NMX002046) 5'-GAAGGTGAAGGTCGGAGTC-3' (sense) and 5'-GAAGATGGTGATGGGATTTC-3' (anti-sense), murine GAPDH (M32599) 5'-TCTGGAAAGCTGTGGCGTG-3' (sense) and 5'-CCAGTGAGCTTCCCGTTCAG-3' (antisense) and JFH-1 HCV (AB047639) 5'-TCTGCGGAACCGGTGAGTA-3' (sense) and 5'-TCAGGCAGTACCACAAGGC-3' (anti-sense).
Pseudotyped retroviral particle production and infections
Pseudotyped viruses were produced as previously described (Sainz, Barretto, and Uprichard, 2009). Briefly, DNA encoding HCV E1/E2 (JFH-1 or H77) or VSV G glycoproteins were co-transfected along with the Env-deficient HIV vector carrying a luciferase reporter gene (pNL4-3-Luc-R−-E−) into 293T producer cells. The supernatants containing the pseudotyped viruses were collected 48 h post transfection and filtered through a 0.45 µm-pore-size filter (BD Biosciences, San Jose, CA), aliquoted, frozen and subsequently titered using the QuickTiter Lentivirus Titer Kit (Cell Biolabs, Inc., San Diego, CA) according to the manufacturer’s instructions.
For experimental HCVpp infections, Huh7 cells were seeded at 4×103 cells in 96-well plates (Corning). Twenty-four hours post seeding, cells were infected with equal titers of pseudotyped virus and at indicated time points the inoculums were removed, cells were washed twice with 1X PBS, and overlaid with 100 µl cDMEM. Seventy-two hours p.i., all cultures were lysed in 20µl of lysis reagent and luciferase activity was measured with a luciferase assay kit (Promega, Madison, WI) and a FLUOstar Optima microplate reader (BMG Labtechnologis Inc, Durham, NC) according to manufacturer’s protocol.
Statistical Analysis
To compare foci number obtained among the different viruses or medium conditions at each time point, we used the Kruskal-Wallis Test and subgroups analysis was performed using the Mann-Whitney Test. To determine whether foci increase slopes differ among viruses or medium conditions, we used a linear regression model (S-PLUS, V.8, Seattle, WA). The slope of this linear-regression line represents the best estimate of foci increase rate. In addition, we included a standard error (SE) interval to indicate the reliability of each estimate. The analysis includes p value (two tailed), testing the null hypothesis that the slope is not different than zero; if the p value is higher than 5%, we conclude that the slope is not different than zero. In all cases, a two-sided P-value of ≤0.05 was considered significant.
Results
HCVcc infection initiation kinetics
To gain insight into the early steps of HCV infection, we assessed HCVcc foci formation after increasing viral inoculation periods on Huh7 cells as a measure of the rate at which HCV virions attach to target cells to initiate a productive infection in vitro. We utilized a 48 hour end-point focus forming assay in which naïve Huh7 cells were inoculated with ~100 FFU of JFH-1 HCVccwt. To determine the number of productive “infection initiation” events (i.e. productive interactions with target cells) that had occurred at specific time points p.i., the inoculum was removed from triplicate wells at frequent time intervals p.i. (e.g. 30 minutes or hourly), cells were washed with 1X PBS, cDMEM containing 0.25% methyl cellulose [an inert substance routinely used in virus titration assays to minimize secondary spread (Cooper, 1967)] was added and the infection was allowed to proceed for a total of 48 hours prior, such that discrete HCV-specific foci (consisting of 4–8 HCV NS5A-positive Huh7 cells grouped as one focal point) could be detected by standard immunohistochemical staining as described in Materials and Methods.
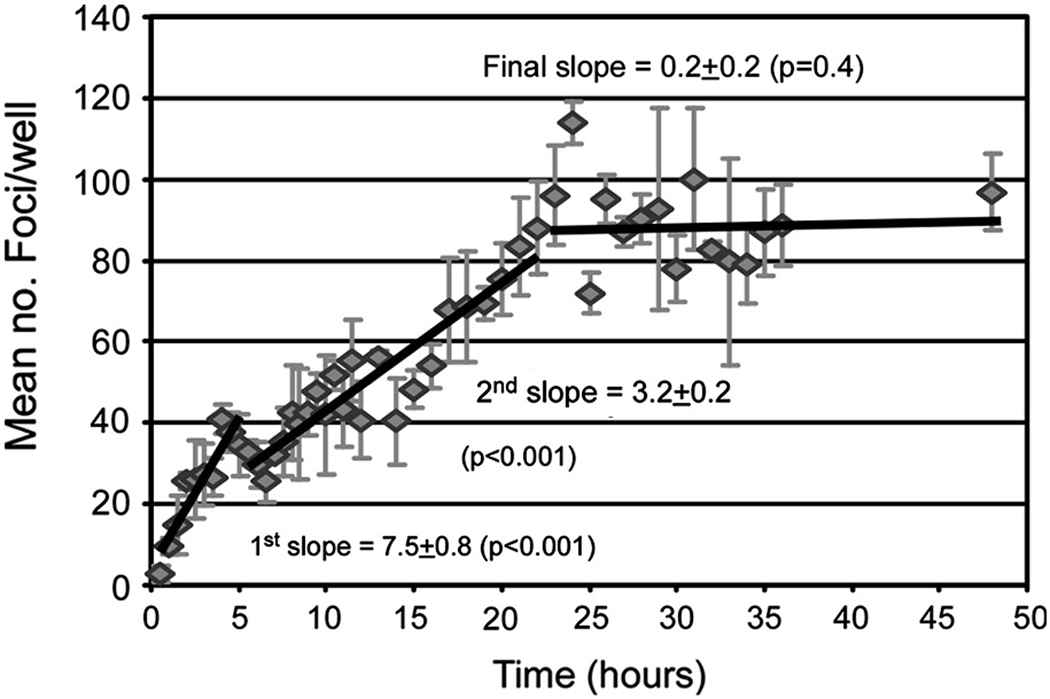
The data in Fig. 1 (representative of four separate experiments, Supplemental Fig. 1) illustrates the mean number of foci achieved at each time point over the course of a 48 hour infection period. Contrary to HCVpp (Supplemental Fig. 2) in which 100% productive infection is initiated within 6 hours and other viral systems (Bagai and Lamb, 1995; Hung et al., 1999; Kartenbeck, Stukenbrok, and Helenius, 1989) in which 100% infection occurs within several hours, our results indicated that HCVcc infection initiation events occurred over an extended window of time. We observed a rapid increase in foci formation (1st slope = 7.5 ± standard error (SE) 0.8 foci/hr) during the first ~6 hours of inoculation (Fig. 1); however, from ~6 to 22 hours post inoculation HCVcc foci formation continued to increase, albeit at a slower rate (2nd slope = 3.2 ± 0.2 foci/hr). This was followed by a plateau (i.e., foci slope = 0.19±0.23 which is not significantly (p=0.42) different than zero; Fig. 1). The same pattern was observed in the three additional experiments shown in Supplemental Fig. 1.
Fig. 1. Multi-phasic time course of HCVcc infection initiation in Huh7 cells.
Huh7 cells were plated in 96-well plates and cultures infected with ~ 100 FFU of HCVccwt. At indicated time points p.i., inoculum was removed from triplicate wells, cells were washed twice with 1X PBS and overlaid with 10% cDMEM containing 0.25% methylcellulose. Forty-eight hours p.i., HCV foci were visualized by HRP-based immunocytochemistry staining using an anti-NS5A antibody, quantified and are expressed as mean number (no.) of foci detected/well ± standard deviation (stdev) for triplicate samples. The slope of foci increase for each of the three phases of HCVcc foci formation were calculated (solid lines). p≤0.05 represents foci slope different than zero.
High density HCVcc particles disappear from viral inoculum sooner than low density HCVcc particles
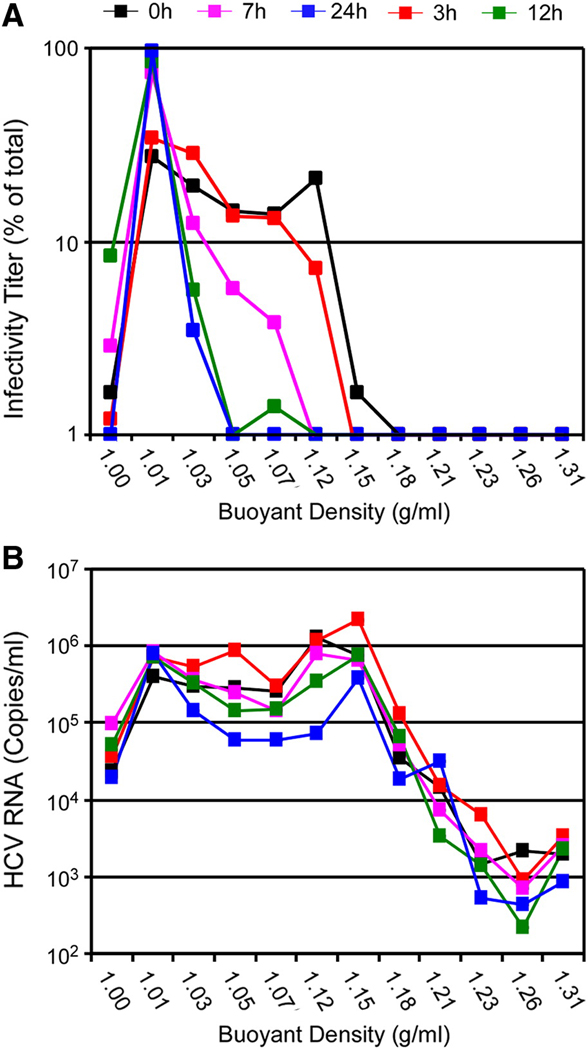
In addition to the prolonged rate of infection initiation observed in Fig. 1, HCVcc foci formation appeared biphasic followed by a plateau (termed here multi-phasic), suggesting that the viral inoculum may contain a heterogeneous population of virions that attach/interact with target cells at different rates. Since HCVser (Andre et al., 2002) and HCVcc (Supplemental Fig. 3A) both exhibit broad density profiles (1.01 – 1.16 g/ml), we hypothesized that the multi-phasic kinetics of infection initiation observed might be due to the varying densities of the HCVcc particles present in the inoculum. To test this hypothesis, we repeated the 48 h foci assay described above, but in parallel infected larger cultures so that sufficient material would be available to assess the density profile of the HCVcc particles remaining in the inoculum over time. For these scaled up infections, 6-well plate cultures were infected with HCVccwt at an MOI of 1.0 FFU/cell and at indicated times p.i. the viral inoculum was removed and stored at −80°C. To determine the buoyant density profile of the infectious particles remaining in the inoculum after 3, 7, 12 and 24 hours incubation with the cells, samples were thawed, overlaid onto discontinuous iodixanol gradients (10 to 50% in TNE) and individual fractions were titrated for virus infectivity and HCV RNA per fraction determined by RTqPCR as described in Materials and Methods. The data demonstrate that over time HCVcc infectious virions disappeared from the viral inoculum (Fig. 2A) in a density-dependent manner, with virions of higher densities (> 1.07 g/ml) no longer detectable by 7 h p.i., which is the approximately the time in which foci formation rate slowed (Fig. 1). In contrast, particle stability, as measured by RNAse resistant HCV RNA levels present in the fraction (Kim, Ha, and Park, 2006), did not significantly fluctuate from 0 to 12 h p.i. (p > 0.05) (Fig. 2B). Only at 24 h p.i. was there a significant (p < 0.01) decrease (~ 1 log) in HCV RNA levels as compared to input levels (Fig. 2B). Taken together, these data suggested that that the disappearance of HCVcc virions of high density from the viral inoculum during the first 12 h of inoculation was not due to loss of particle integrity, but rather a consequence of virions attaching to/interacting with target cells. Thus, the rate at which HCVcc virions initiate productive infection in vitro is dependent on the density of the virion, with more dense particles becoming associated with the Huh7 cells during the first, more rapid foci formation phase.
Fig. 2. HCV particles of higher density disappear/loose infectivity faster than particles of lower density after inoculation into cell cultures.
Cultures of Huh7 cells seeded in 6-well plates, were infected in parallel with HCVccwt at a MOI of 1.0 FFU/cell in a total inoculum volume of 1 ml and at indicated time points p.i., medium was removed from inoculated cultures and frozen at −80°C. Inoculum collected at 0, 3, 7, 12 and 24 h p.i. were thawed and fractionated by discontinuous iodixanol gradient centrifugation (10 to 50% in TNE). Individual fractions were collected, titrated for virus infectivity and HCV RNA levels determined by RTqPCR analysis. (A) Titers are presented as a percentage of the total sum of infectious virus present in all fractions. (B) HCV RNA levels are presented as viral genome copies/ml present in all fractions at each time point.
The rate of HCVcc infection initiation in vitro is density-dependent
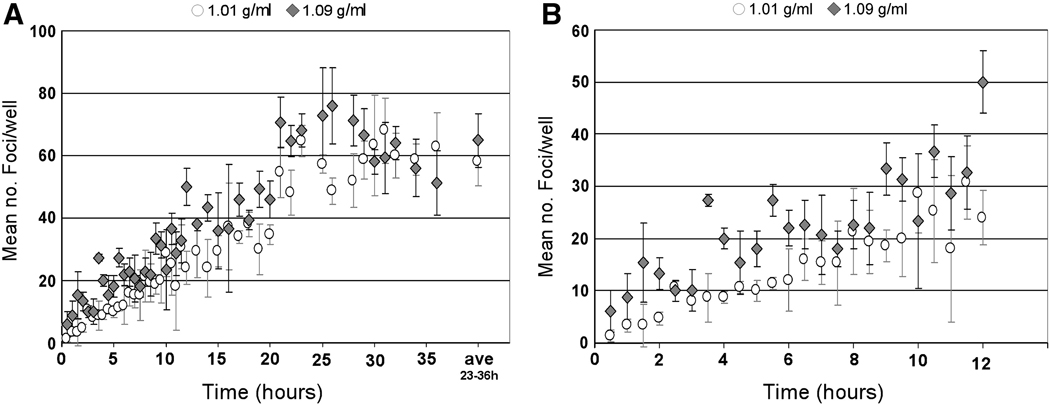
To more directly test this hypothesis, we separated HCVcc particles of different densities and measured the relative rate at which these different density particles initiated foci formation in vitro. For this analysis, 1.2 ml of a concentrated 1×106 FFU/ml HCVccwt stock was separated by ultracentrifugation and fractions were collected and analyzed for HCV RNA and infectivity (data not shown). A 48 hour focus forming assay, as detailed in Fig. 1, was then performed with the 1.01 g/ml and 1.09 g/ml density fractions to determine the rate at which HCVcc virions within two different densities fractions initiated productive foci formation (Fig. 3A). Consistent with particles of higher density attaching to and/or interacting with target cells sooner than particles of lower density, the rate of foci formation calculated in cultures infected with 100 FFU of HCVcc from the 1.09 g/ml density fraction was significantly higher (p=0.0002) when compared to the number of foci achieved in cultures infected with HCVcc from the 1.01 g/ml density fraction (Fig. 3A). This difference in infection initiation kinetics was particularly evident during the first 12 hours p.i. (Fig. 3B). Interestingly, the rate of foci formation was not multi-phasic as observed for HCVccwt (Fig. 1), but rather single-phasic from 0.5 to 24 h p.i. for HCVcc from both the 1.09 g/ml and 1.01 g/ml density fractions (slopes of 1.30±0.05 and 1.04±0.04 foci/hr, respectively). These single-phase increase slopes (rather than the biphasic increase; e.g., Fig. 1) are likely a direct result of using a more homogenous viral inoculum of similar density virions as opposed to using an unpurified inoculum containing heterogeneous broad-density virions (e.g. HCVccwt).
Fig. 3. HCV particles of higher density exhibit faster infection initiation kinetics.
Huh7 cells were plated in 96-well plates, infected with ~ 100 FFU of 1.01 g/ml (open circles) or 1.09 g/ml (grey diamonds) density fractions of HCVccwt in 1% cDMEM. At indicated time points p.i., inoculum was removed from triplicate wells, cells were washed twice with 1X PBS and overlaid with 10% cDMEM containing 0.25% methylcellulose. Forty-eight hours p.i., HCV foci were visualized by HRP-based immunocytochemistry staining using an anti-NS5A antibody, quantified and are expressed as mean number (no.) of foci detected/well ± stdev for triplicate samples. Time course of infection initiation is shown over (A) 36 hours (Additionally, the average number of foci obtained after maximal foci formation was observed (23–36 h p.i.) is shown.) and (B) 12 hours.
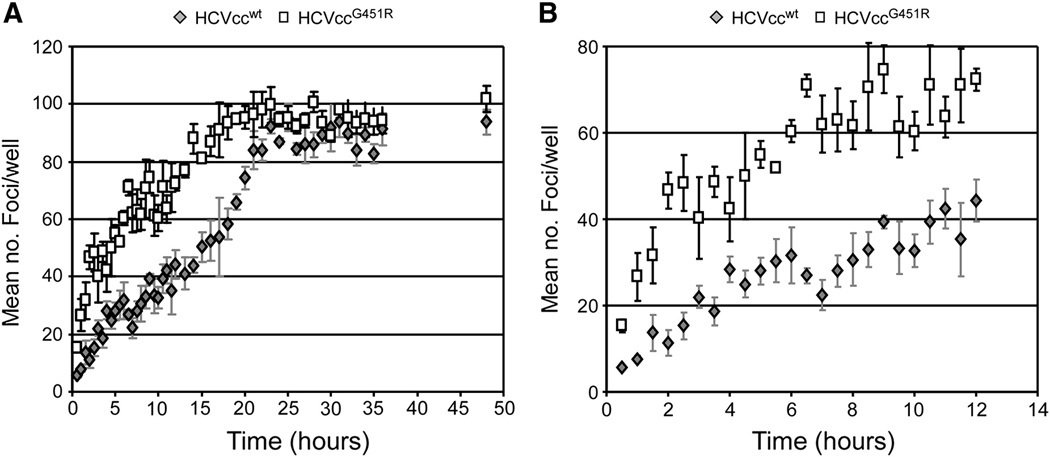
In addition to analyzing fractions of HCVccwt of different densities, we also assessed the infection initiation rate of the mutant virus JFH-1G451R(HCVccG451R) (Zhong et al., 2006), which has been shown to have enhanced infectivity in vitro as well as a distinct density profile as compared to HCVccwt. Specifically, Zhong et al. showed that a glycine-to-arginine mutation at position 451 (G451R) in the viral E2 glycoprotein resulted in production of HCVcc having a narrower density range with a higher average mean density (Zhong et al., 2006). After introducing this mutation into the JFH-1 coding sequence and verifying that we observed a higher mean density profile and more robust HCVccG451R infectivity kinetics, similar to that described by Zhong et al. (Zhong et al., 2006) (Supplemental Figs. 3B–C), we tested whether HCVccG451R exhibited faster infection initiation kinetics than HCVccwt. We found that when comparing HCVccwt with HCVccG451R first phase increase (0 to 6 hrs p.i.) was 4.85±0.36, versus 6.25±0.76 foci/hr, respectively, and 2.98±0.18, versus 2.64±0.21 foci/hr, respectively, during the second/slower phase (6 to 20 hrs p.i.). Foci number accumulation slowed significantly for both viruses 20 hrs p.i. onwards with slopes of 0.24±0.08 foci/hr (HCVccwt) versus 0.22±0.08 foci/hr (HCVccG451R). By comparing the slopes we found as expected that the kinetics of HCVcc foci formation between the two viruses were not significantly (p>0.1) different. However importantly, HCVccG451R foci levels were always significantly (p<0.05) higher than HCVccwt foci levels. Thus, consistent with particles of higher density initiating productive infection sooner than particles of lower density, the results demonstrate that higher a percentage of HCVccG451R attaching to and/or interacting with target cells sooner than HCVccwt (Fig. 4), a characteristics that likely at least in part explains the enhanced infectivity exhibited by this mutant virus.
Fig. 4. Mutations that alter HCV particle density affect viral infection initiation kinetics.
Huh7 cells were plated in 96-well plates, infected with ~ 100 FFU of HCVccwt (grey diamonds) or HCVccG451R (open squares) and at indicated time points p.i., inoculum was removed from triplicate wells, cells were washed twice with 1X PBS and overlaid with 10% cDMEM containing 0.25% methylcellulose. Forty-eight hours p.i., HCV foci were visualized by HRP-based immunocytochemistry staining using an anti-NS5A antibody and are expressed as mean number (no.) of foci detected/well ± stdev for triplicate samples. Linear trend lines are depicted to help illustrate the underlying difference between the two viruses.
The rate of HCVcc infection initiation can be altered by changing the density of the culture medium
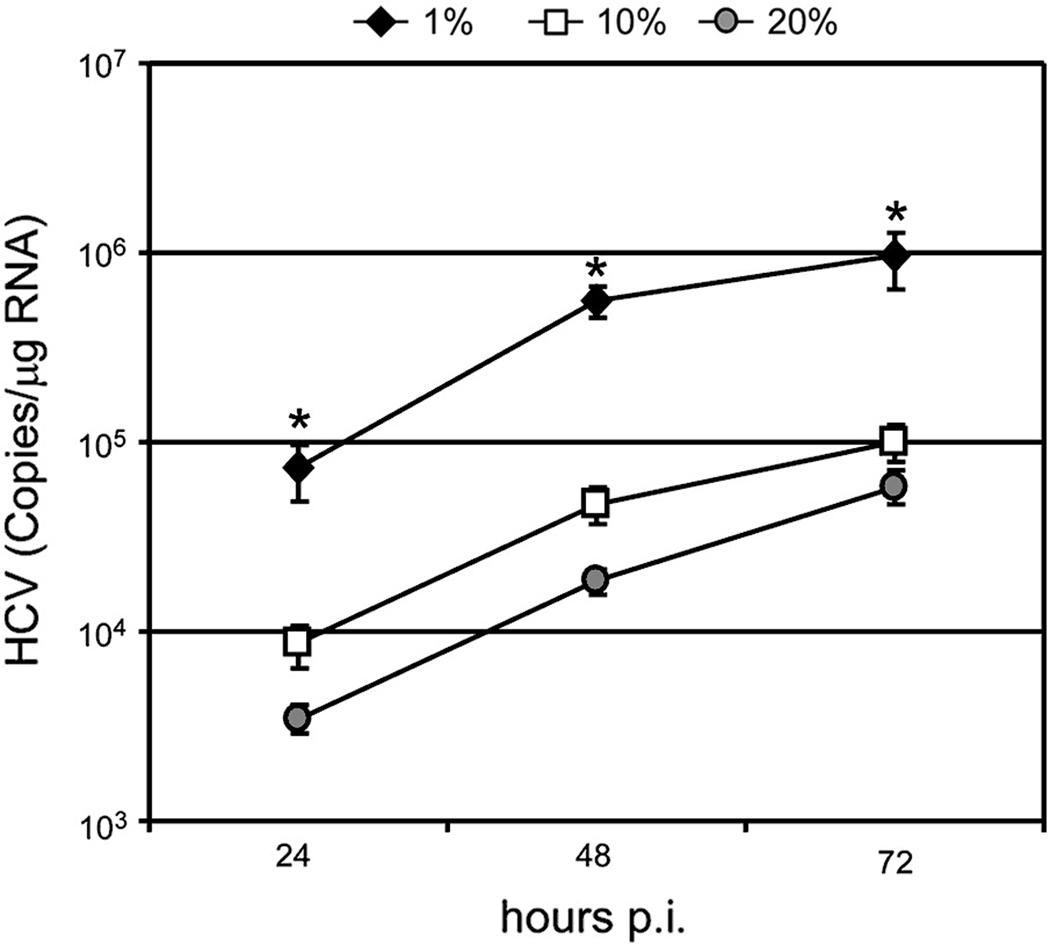
Because the density of complete DMEM supplemented with 10% FBS is ~1.05 g/ml and the majority of infectious HCVccwt particles have a density between 1.01 and 1.09 g/ml, we reasoned that the ability of some infectious HCVcc virions to reach their target cells in vitro is hindered because ~60% of infectious particles are less dense than the density of the standard culture medium and would thus tend to “float” rather than sink to the cell monolayer like most other viruses, whose densities are greater than 1.11 g/ml (Burtonboy et al., 1993; Hirano, Hino, and Fujiwara, 1978; Huang et al., 2004; Kokorev et al., 1976; Macnaughton and Davies, 1980; Niiyama et al., 1975; Smith et al., 1970; Weiss et al., 1996). To test the hypothesis that the relative density of HCVcc particles versus the culture medium influences the kinetics of infection in vitro, we first analyzed HCVcc RNA kinetics following infection in cDMEM supplemented with 1, 10 and 20% FBS, which correspond to media densities of 1.02, 1.05 and 1.07 g/ml, respectively (determined by calculation and empirical measurement of density). As shown in Fig. 5, the kinetics of HCVcc RNA amplification differed based on the density of the extracellular medium. Twenty four hours p.i., the mean intracellular HCV RNA level was 8-fold higher in cultures in which HCVcc infection was performed in the presence of 1% FBS as compared to 10% FBS (p=0.05). Likewise, a mean 3-fold reduction in intracellular HCV RNA was detected in cultures in which HCVcc infection was performed in the presence of 20% FBS as compared to 10% FBS (p=0.05). The differences observed at 48 and 72 h p.i. were also significantly different (p=0.05) (Fig. 5). To specifically assess infection initiation kinetics, we performed a foci formation time course assay with HCVccwt (Fig. 6A–B) and HCVccG451R (Fig. 6C–D) in the presence of increasing concentrations of FBS. We observed that the rate of HCVccwt foci initiation was multi-phasic (biphasic followed by plateau) in medium containing 1%, 10% or 20% FBS (Fig. 6A). In medium containing 1%, 10% or 20% FBS the first phase (~ 0 to 6 hrs p.i.) foci formation rate decreased from 4.69±0.92, 3.06±0.45 and_1.01±0.27 foci/hr, respectively, and the second phase (~ 6 to 20 hr p.i.) foci formation rate decreased from 2.96±0.41, 1.81±0.15 and 0.47±0.11 foci/hr, respectively. By comparing the slopes we found that the kinetics of HCVccwt foci formation among the different medias were significantly different (p <0.01) except for the first phase increase in media containing 1% (1.02 g/ml) and 10% (1.05 g/ml) FBS (p = 0.12). Taken together, these data demonstrate that the rate of HCVccwt foci initiation was enhanced when the density of the medium was lowered to 1.02 g/ml and significantly inhibited when increased to 1.07 g/ml (Figs. 6A–B).
Fig. 5. Reducing the density of the extracellular environment affects HCV infection initiation kinetics.
Huh7 cells were plated in 96-well plates, infected with HCVccwt at a MOI of 0.1 FFU/cell in cDMEM containing 1% (blue diamonds), 10% (red squares) or 20% (green circles) FBS. At indicated time points p.i., inoculum was removed from triplicate wells, intracellular RNA was collected and HCV RNA and cellular GAPDH RNA were quantified by RT-qPCR. HCV RNA is expressed as HCV copies/µg total cellular RNA. Results are graphed as mean ± stdev (n=3). * p-value = 0.05.
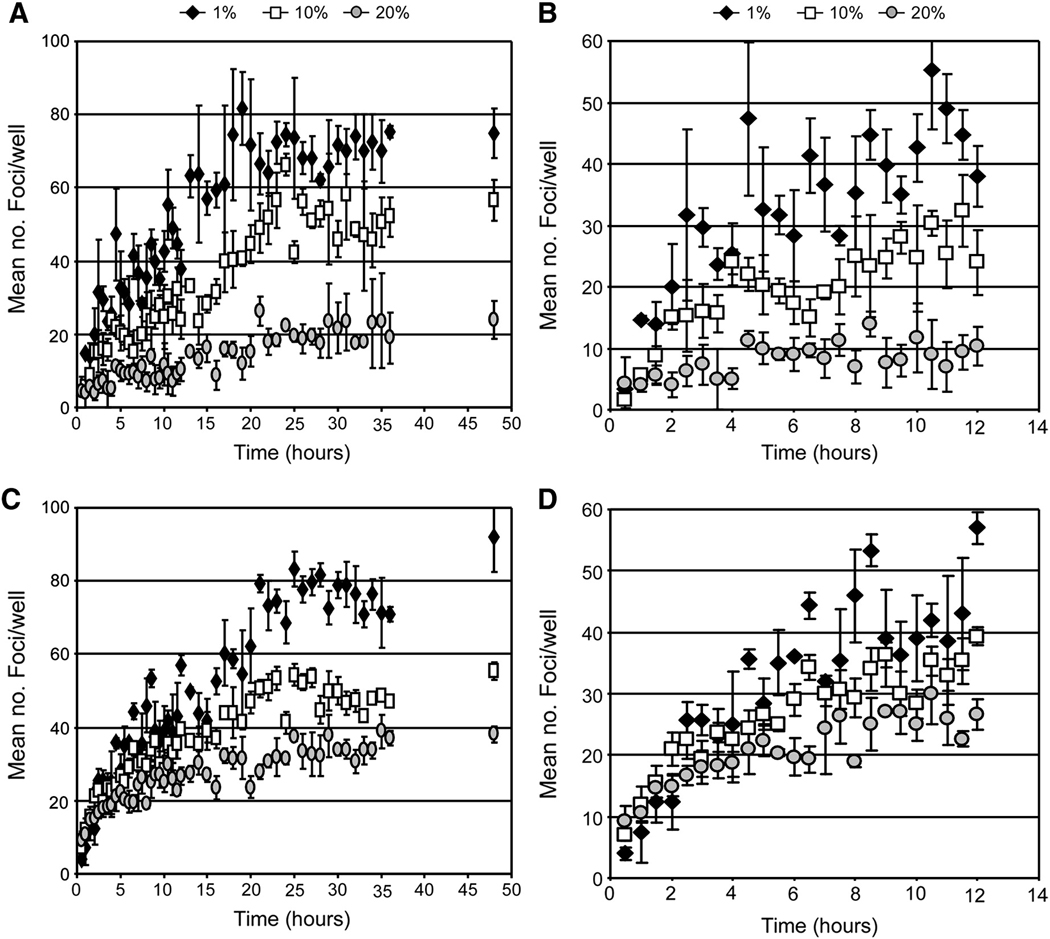
Fig. 6. Reducing the density of the extracellular environment affects the rate of HCV infection initiation.
Huh7 cells were plated in wells of a 96-well plate, infected with ~ 100 FFU of (A–B) HCVccwt or (C–D) HCVccG451R in cDMEM containing 1% (black diamonds), 10% (open squares) or 20% (grey circles) FBS. At indicated time points p.i., inoculum was removed from triplicate wells, cells were washed twice with 1X PBS and overlaid with 10% cDMEM containing 0.25% methylcellulose. Forty-eight hours p.i., HCV foci were visualized by HRP-based immunocytochemistry using an anti-NS5A antibody, quantified and expressed as a mean number (no.) of foci detected/well ± stdev for triplicate samples. Time course of infection initiation is shown over (A, C) 24 hours and (B, D) 12 hours.
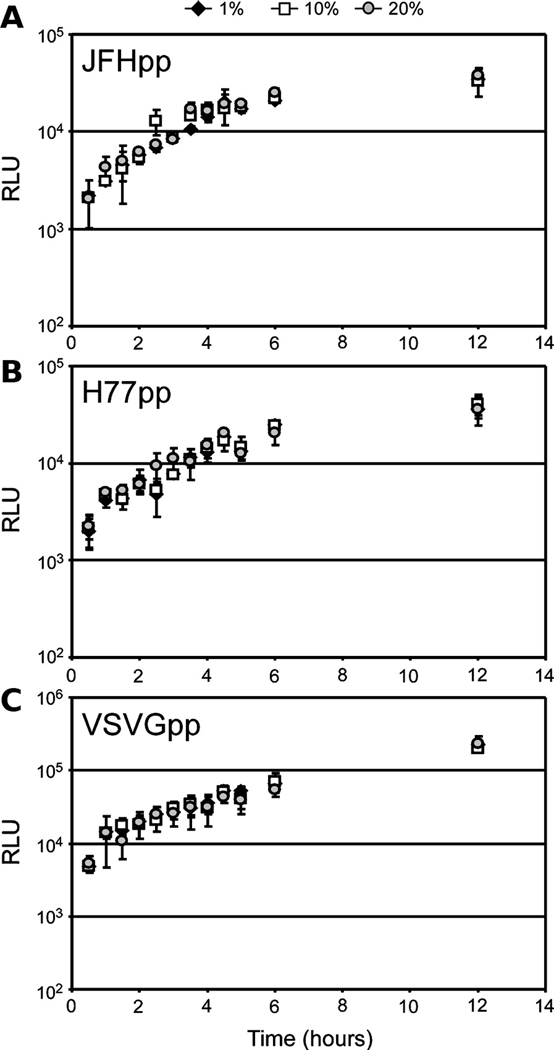
The rate of HCVccG451R foci initiation was also multi-phasic in medium containing 1%, 10% or 20% FBS (Fig. 6C). In medium containing 1%, 10% or 20% FBS, the first phase (~ 0 to 6 hr p.i.) foci formation rate decreased 5.77±0.50, 3.20±0.32 and 2.03±0.25 foci/hr, respectively, and 1.61±0.24, 1.08±0.11 and 0.53±0.13 foci/hr, respectively, during the second phase (~ 6 to 20 hr p.i,). By comparing the foci formation slopes among the medium concentrations we found that all were significantly different from each other (p≤0.05); however, this effect was not as visually prominent as compared to HCVccwt (Fig. 6B vs 6D). Particularly during the first 12 h p.i. with HCVccG451R, when the majority of highly infectious 1.07–1.09 g/ml HCVccG451R particles would be expected to rapidly “sink” to the surface of the cell monolayer in DMEM supplemented with either 1% FBS (1.02 g/ml), 10% FBS (1.05 g/ml), or 20% FBS (1.07 g/ml), similar infection initiation kinetics with HCVccG451R inoculated in the different serum concentrations was observed (Fig. 6D). However, by 12 h p.i., the kinetics of HCVccG451R foci formation more closely paralleled that of HCVccwt (Fig. 6A vs Fig. 6C), which is consistent with the fact that the HCVccG451R stock did contain some lighter particles (Supplemental Fig. 3B) that would be expected to be sensitive to the density of the extracellular environment. Recognizing that FBS is not an inert substance and when present at high concentrations (e.g. 20%) FBS could have a negative impact on the cells and/or virus, we attempted to alter the density of the culture medium with inert substances such as iodixanol, sucrose and glycerol, but, the concentrations of these reagents required to achieve final densities of 1.05 and 1.07 g/ml were cytotoxic and therefore we were unable to alter the density of the medium by these means (data not shown). As an alternative approach, to determine if the effects we had observed were due to some non-specific effect of the FBS, we tested whether concentrations of FBS from 1 to 20% had similar effects on other viral infections. Specifically, we assessed pseudoparticle entry in the presence of increasing concentrations of FBS, and observed that the rates of HCVpp (JFH-1 and H77) and VSVGpp entry were unaffected at any of the FBS concentrations tested (Fig. 7). This would imply that FBS present in the culture medium, at concentration from 1 to 20%, has no indirect negative affects on the cells or E1/E2 bearing viral particles. Lastly, we tested whether these same concentrations of FBS from 1 to 20% had any effect on HCV replication post attachment/entry. For these experiments, Huh7 cells were inoculated with HCVccwt at an MOI of 1.5 FFU/cell for 12 h and subsequently cultured in the presence of increasing concentrations of FBS. Cellular HCV RNA levels were assessed by RT-qPCR to assess amplification of HCV RNA from 12 to 24 hours. As expected, HCV RNA replication levels were not affected at any of the FBS concentrations tested (Supplemental Fig. 4). Thus, the affects observed on HCVcc entry (Fig. 6) are instead, as hypothesized, likely density-related.
Fig. 7. The density of the extracellular environment does not affect HCVpp or VSVGpp entry.
Huh7 cells were plated in 96-well plates and infected with equal titer amounts of (A) JFHpp, (B) H77pp or (C) VSVGpp in cDMEM containing 1% (filled diamonds), 10% (open squares) or 20% (shaded circles) FBS. At indicated time points p.i., inoculum was removed from triplicate wells, cells were washed twice with 1X PBS and overlaid with 10% cDMEM. Seventy-two hours p.i., wells were lysed and infection quantified by relative light units (RLU) ± stdev (n=3).
Discussion
HCV entry represents a key step in the viral lifecycle and a promising target for the development of novel therapeutics to prevent HCV infection and spread. Although prior work using HCVpp has identified essential HCV receptors and demonstrated that post-binding internalization of E1/E2 containing particles is relatively slow (i.e. 300 minutes to achieve 100% entry) (Dreux et al., 2006; Meertens, Bertaux, and Dragic, 2006), we are still far from fully understanding all the steps of this complex and dynamic process particularly as they occur during authentic HCV infection. Aiming to gain a better appreciation of infectious HCVcc entry in vitro, we assessed the rate of HCVcc attachment/interaction with target cells (i.e. “infection initiation”) and observed that not only was HCVcc infection initiation slower than reported for other viruses, but a novel multi-phasic, density-dependent phenotype was observed, as detailed below.
Kinetics of HCVcc infection initiation
Although exceptions to the rule do exist (Culp and Christensen, 2004), viral entry is generally a rapid and highly efficient event. For example, SV40 infection of CV-1 cells has been shown to occur with 20–30 minutes (Kartenbeck, Stukenbrok, and Helenius, 1989), while Dengue virus infection has been shown to occur within 2 hours in vitro (Hung et al., 1999). Thus, it was surprising to find that under the experimental conditions described herein, HCVcc required up to 24 hours for the maximum number of infection initiation event to occur (Fig. 1), suggesting that productive binding of HCVcc particles to the host cells is not synchronous in vitro, but rather individual binding events occur over a significant period of time after inoculation of the virus into the culture.
Since the rate at which viral particles come into contact with their target cells in vitro can be affected by specific cell culture conditions (e.g. inoculation volume and virus-to-cell ratio) as well as extraneous cell-culture manipulations (e.g. culture rocking/shaking and temperature changes), extreme care was taken to ensure that standardized infection conditions were rigorously followed and that the infected cultures were handled minimally and in precisely the same manner. Additionally, when relevant, HCVpp and VSVGpp kinetics were analyzed as controls for manipulation-specific effects. For example, because the kinetic experiments described required a significant amount of culture handling as detailed in Materials and Methods, cDMEM was supplemented with HEPES buffer to ensure proper buffering, plates were handled for no more than 2 minutes outside of the incubator and cultures were always kept at 37°C by pre-warming all cell culture reagents used for washes and overlays. Despite taking excess precautions, however, we of course cannot definitively rule out the possibility that mere shifts of O2/CO2 might have potentially affected cellular conditions and inadvertently skewed our entry kinetics towards a slower rate than what would be observed in the absence of such extraneous manipulations. Importantly, however, identical manipulations during HCVpp and VSVGpp infection did not result in analogous prolonged infection kinetics (Supplemental Fig. 2 and Supplemental Fig. 5) indicating that this phenotype was HCVcc specific.
To ensure that our foci counts were reflective of only initial infection events and did not include secondary satellite foci, an overlay was added to all cultures post infection, even though studies from our laboratory demonstrate that following low multiplicity of infection de novo infectious HCVcc production does not occur until ~48 h p.i. (data not shown). In selecting an overlay, we chose methylcellulose as this inert substance has been routinely used in numerous virus titration assays. Although a standard virological overlay (Cooper, 1967), we nonetheless tested whether the addition of methylcellulose post infection could affect viral entry kinetics and. found no significant difference in foci numbers at any time point regardless of methylcellulose addition (data not shown). Likewise, using the unrelated VSVGpp virus, we show in Supplemental Fig. 5 that the addition of methycellulose to infected cultures post infection had no affect (p value > 0.05) on VSVGpp kinetics. Thus, the observed prolonged HCVcc infection initiation kinetics observed was not indirectly due to de novo HCVcc production or the presence of methylcellulose in the culture media.
Together with the fact that four independent experiments performed by multiple individuals yielded identical prolonged multi-phasic HCVcc infection initiation kinetics (Fig. 1 and Supplemental Fig. 1) and that introduced variables (e.g. particle and/or media density) had reproducible effects on infection initiation kinetics (Figs. 3–6), our results as a whole suggest that the cell culture manipulations performed, assay conditions utilized and experimental parameters followed were not responsible for the unique HCVcc infection kinetics observed but rather, HCVcc entry is indeed a slow and prolonged process.
HCVcc particle density is a determinant of viral infection initiation kinetics in vitro
Of particular interest was the fact that in addition to HCVcc attachment to target cells occurring over a large window of time, the rate of this infection initiation was not linear as is routinely observed for other viral systems, including HCVpp (Supplemental Fig. 2). Rather, the rate at which HCVcc initiated productive infection of Huh7 cells appeared multi-phasic (Fig. 1 and Supplemental Fig. 1). We reasoned that since i) the same prolonged, multi-phasic pattern was not recapitulated by HCVpp (Supplemental Fig. 2), which is produced in lipoprotein-deficient 293T embryonic kidney cells (Farquhar and McKeating, 2008), and ii) viral association with lipoproteins contributes to the broad density profile of HCV (Andre et al., 2002; Gastaminza, Kapadia, and Chisari, 2006; Zhong et al., 2005) (Supplemental Fig. 3A), which is unlike the majority of other viruses that exhibit uniform density (Burtonboy et al., 1993; Hirano, Hino, and Fujiwara, 1978; Huang et al., 2004; Kokorev et al., 1976; Macnaughton and Davies, 1980; Niiyama et al., 1975; Smith et al., 1970; Weiss et al., 1996), that the prolonged and multi-phasic rate of infection initiation observed (Fig. 1) might be directly due to HCVcc virions of differing density existing in the viral inoculum.
By analyzing the density of HCVcc virions remaining in the culture medium following viral inoculation, we initially found that infectious HCVcc particles of particular buoyant densities disappeared from the viral inoculum at different rates (Fig. 2). Interestingly, however, if the HCV particles were disappearing due to productive attachment/interaction to target cells, we would have initially predicted the loss of less dense particles first since low-density HCV particles have been shown to have higher specific infectivity than high-density particles (Lindenbach et al., 2005; Zhong et al., 2006). Instead, we observed the loss of denser HCV particles before that of less dense particles such that by 7 h p.i., essentially no particles of densities greater than 1.07 g/ml were detectable in the viral inoculum (Fig. 2B). Thus, we concluded that the high-density particles must have either i) become associated with the cells prior to low-density particles, ii) were significantly less stable than the low-density particles, or iii) that the disappearance of high-density particles was a consequence of denser particles picking up lipoproteins present in the medium during infection and thus becoming less dense over time.
Although all three conclusions are plausible and may contribute to the observed phenotype, our data most directly support that high-density particles productively interact with cells faster than low-density particles (i.e. conclusion “i"). We tested this hypothesis by fractionating HCVcc virions by density gradient ultracentrifugation and directly assessing the rate at which low- versus high-density HCV particles initiated productive infection in vitro (Fig. 3). Consistent with idea that denser HCV particles attach and interact with target cells first (as measured by HCV foci formation), HCVcc within the 1.09 g/ml density fraction initiated productive foci formation more efficiently than the virus within the 1.01 g/ml density fraction (Fig. 3B). Further supporting our hypothesis that the multi-phasic nature of HCVccwt foci formation (Fig. 1) is due to the HCVcc virions of differing density existing in the viral inoculum, a linear increase in foci formation was observed for HCVcc in both the 1.01 g/ml and 1.09 g/ml density factions (Fig. 3A). These more linear kinetics are likely a direct result of using an inoculum containing more homogenous virions of similar density as opposed to using an inoculum containing heterogeneous virions with a broad range of densities (e.g. HCVccwt).
Importantly, analysis of particle stability over time (as determined by measuring RNase resistant HCV RNA) indicated that high-density HCV particles were as stable as low-density particles over the course of infection (Fig. 2B), suggesting that the immediate loss of high density particles from the inoculum during infection is not a consequence of particle stability (i.e. conclusion “ii”), but rather a result of these particles interacting with cells prior to virions of lower density. Lastly, since both the 1.01 g/ml and 1.09 g/ml HCVcc virions were produced and subsequent experimental infections were performed in identical cDMEM supplemented with only 1% FBS it is unlikely that the HCV particles rapidly became lipidated by FBS components during the course of this analysis (i.e. conclusion “iii”). Hence, take together, our data supports conclusion i, in which the different infection initiation rates observed with the 1.01 and 1.09 g/ml HCV particles (Fig. 3B) represents a density-associated HCV particle characteristic independent of particle decay or lipidation.
Notably our second method of testing whether particles of high-density interact with target cells sooner in vitro, also confirmed this conclusion as the HCVccG451R adaptive mutant, which has been shown to exhibit a higher mean density (1.07–1.09 g/ml) than HCVccwt, exhibited faster infection initiation kinetics than HCVccwt (Figs 4 and 6). Thus together these data support the conclusion that the density of the HCV particle predominantly influences the rate at which the virus attaches/interacts with target cells in vitro.
Density of the cell culture medium is a major determinant of HCV infection initiation kinetics in vitro
The observation that HCV particles of higher density exhibit faster infection initiation kinetics in vitro despite the fact that less dense HCV particles have a higher intrinsic specific infectivity (Haid, Pietschmann, and Pecheur, 2009; Lindenbach et al., 2005; Zhong et al., 2006), suggested to us that the in vitro cell culture environment might be artificially influencing the rate at which HCVcc interacts with target cells. Specifically, since we measured cell culture medium supplemented with 10% FBS to have a density of 1.05 g/ml and the majority of infectious HCVcc has a density of ≤ 1.09 g/ml, one would assume that the density of the medium might impede the some of the more highly infectious HCVcc virions from reaching the cell monolayer. To test this hypothesis, we decreased and increased the density of the extracellular environment by altering the percentage of serum in the medium and assessed whether HCVcc infection initiation was enhanced or impaired, respectively (Figs. 5 and 6). As predicted, reducing serum to 1% (i.e. reducing medium density to 1.02 g/ml) enhanced infection initiation kinetics, suggesting that when the medium was less dense (1% FBS), HCVcc virions of a broader range of densities could more readily sink to the cell monolayer essentially increasing the rate at which virion-cell interactions occurred. In contrast, when the medium density was increased to 1.07 g/ml (20% FBS) fewer HCVcc particles (i.e. only those with a density ≥ 1.07 g/ml) could rapidly sink to interact with target cells. The observed phenotypes were HCVcc-specific and not due to any indirect negative affects of FBS, as neither HCV RNA replication post entry nor HCVpp (JFH-1 and H77) or VSVGpp entry kinetics were affected by the varying concentration of serum present in the media (Supplemental Fig. 4 and Fig. 7, respectively). Taken together, these observations would suggest that the kinetics of HCVcc infection initiation in vitro is directly affected by the density of the particle relative to the density of the extracellular milieu.
Selection pressures that drive HCV cell culture adaptive mutations
Because HCV particles of high-density are able to more efficiently sink to the cell monolayer and interact with target cells faster than low-density particles in vitro, it is likely that the higher average buoyant density of the HCVccG451R mutant drove the selection of this adaptive mutation during long term propagation in vitro and in turn explains its enhanced in vitro infectivity. Notably however, the loss of viral-associated lipoproteins would not necessarily be advantageous in vivo as such associations are believed to provide particle stability (Burlone and Budkowska, 2009), confer a mechanism of escape from the humoral immune response (Andre et al., 2002) and enhance in vivo infectivity (Andreo et al., 2007; Burlone and Budkowska, 2009; Lindenbach et al., 2006). The latter is supported by the fact that virus recovered from mice and chimpanzees infected with HCVcc have a higher specific activity and are of lower buoyant density than the virus produced in cell culture (Lindenbach et al., 2006). Therefore, while in vitro adaptive mutations conferring HCV particles with higher buoyant density, such as the G451R mutation, provide useful research tools (Grove et al., 2008), their in vivo fitness may be compromised as is often the case with tissue-culture adapted viruses.
Conclusions
We show that in vitro HCVcc attachment/interaction with target cells is a slow process, affected by the density of both the viral particle and the extracellular culture medium. During infection in vitro, HCVcc particles must sink through the medium to efficiently reach their target cells, therefore viral particles with buoyant densities higher than the culture medium are able to reach the cells sooner than particles with a buoyant density lower than the culture medium. Although this phenotype is an in vitro phenomenon, it nonetheless reveals the importance of a previously unrecognized variable (i.e density of the culture medium) inherent in the HCV infectious cell culture system. While it is generally accepted that in vivo infection conditions cannot be perfectly modeled in vitro, this study highlights that extreme care must be taken to at least ensure that the in vitro conditions themselves do not limit infection or result in misinterpretation of viral life cycle parameters (e.g. infection initiation kinetics). Whether this in vitro restriction has previously impeded our ability to efficiently culture other HCV genotypes in cell culture (Pietschmann et al., 2009; Yi et al., 2006) remains to be determined; however, the observations presented herein certainly suggest that cell culture practices, such as infections in low-serum medium, might enhance our ability to propagate different HCV clones in vitro.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Charlie Rice of The Rockefeller Institute for donating the mouse monoclonal anti-NS5A antibody (E910) and Dr. Takaji Wakita of the National Institute of Infectious Diseases in Japan for providing the pJFH-1 plasmid. In addition, we thank Danyelle Martin and Drs. Bruno Sainz, Jr., Katharina B. Rothwangl and Naina Barretto for technical assistance.
This work was supported by the National Institutes of Health Public Health Service Grants R01-AI070827 (SLU), R03-AI085226 (SLU) and R21-DK070551 (RFG), Research Scholar Grant RSG-09-076-01 from the American Cancer Society (SLU) and the UIC Walter Payton Center GUILD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1999;96(22):12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizaki H, Morikawa K, Fukasawa M, Hara H, Inoue Y, Tani H, Saito K, Nishijima M, Hanada K, Matsuura Y, Lai MM, Miyamura T, Wakita T, Suzuki T. Critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J Virol. 2008;82(12):5715–5724. doi: 10.1128/JVI.02530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76(14):6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreo U, Maillard P, Kalinina O, Walic M, Meurs E, Martinot M, Marcellin P, Budkowska A. Lipoprotein lipase mediates hepatitis C virus (HCV) cell entry and inhibits HCV infection. Cell Microbiol. 2007;9(10):2445–2456. doi: 10.1111/j.1462-5822.2007.00972.x. [DOI] [PubMed] [Google Scholar]
- Bagai S, Lamb RA. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J Virol. 1995;69(11):6712–6719. doi: 10.1128/jvi.69.11.6712-6719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H, Schafer C, Adah MI, Zhang F, Linhardt RJ, Toyoda H, Kinoshita-Toyoda A, Toida T, Van Kuppevelt TH, Depla E, Von Weizsacker F, Blum HE, Baumert TF. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J Biol Chem. 2003;278(42):41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- Barth H, Schnober EK, Zhang F, Linhardt RJ, Depla E, Boson B, Cosset FL, Patel AH, Blum HE, Baumert TF. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J Virol. 2006;80(21):10579–10590. doi: 10.1128/JVI.00941-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J Exp Med. 2003;197(5):633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosch B, Vitelli A, Granier C, Goujon C, Dubuisson J, Pascale S, Scarselli E, Cortese R, Nicosia A, Cosset FL. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J Biol Chem. 2003;278(43):41624–41630. doi: 10.1074/jbc.M305289200. [DOI] [PubMed] [Google Scholar]
- Benedicto I, Molina-Jimenez F, Bartosch B, Cosset FL, Lavillette D, Prieto J, Moreno-Otero R, Valenzuela-Fernandez A, Aldabe R, Lopez-Cabrera M, Majano PL. The tight junction-associated protein occludin is required for a postbinding step in hepatitis C virus entry and infection. J Virol. 2009;83(16):8012–8020. doi: 10.1128/JVI.00038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard E, Belouzard S, Goueslain L, Wakita T, Dubuisson J, Wychowski C, Rouille Y. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J Virol. 2006;80(14):6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290(5498):1972–1975. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- Blight KJ, McKeating JA, Marcotrigiano J, Rice CM. Efficient Replication of Hepatitis C Virus Genotype 1a RNAs in Cell Culture. J Virol. 2003;77(5):3181–3190. doi: 10.1128/JVI.77.5.3181-3190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, McCaustland K, Krawczynski K, Spelbring J, Humphrey C, Cook EH. Hepatitis C virus: buoyant density of the factor VIII-derived isolate in sucrose. J Med Virol. 1991;34(3):206–208. doi: 10.1002/jmv.1890340315. [DOI] [PubMed] [Google Scholar]
- Burlone ME, Budkowska A. Hepatitis C virus cell entry: role of lipoproteins and cellular receptors. J Gen Virol. 2009;90(Pt 5):1055–1070. doi: 10.1099/vir.0.008300-0. [DOI] [PubMed] [Google Scholar]
- Burtonboy G, Delferriere N, Mousset B, Heusterspreute M. Isolation of a C-type retrovirus from an HIV infected cell line. Arch Virol. 1993;130(3–4):289–300. doi: 10.1007/BF01309661. [DOI] [PubMed] [Google Scholar]
- Choi S, Sainz B, Jr, Corcoran P, Uprichard SL, Jeong H. Characterization of increased drug metabolism activity in dimethyl sulfoxide (DMSO)-treated Huh7 hepatoma cells. Xenobiotica. 2009;39(3):205–217. doi: 10.1080/00498250802613620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Choo Q-L, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Cooper PD. The plaque assay of animal viruses. In: Maramorosch K, Koprowski H, editors. Methods in Virology. Vol. III. New York and London: Academic Press; 1967. pp. 243–311. [Google Scholar]
- Culp TD, Christensen ND. Kinetics of in vitro adsorption and entry of papillomavirus virions. Virology. 2004;319(1):152–161. doi: 10.1016/j.virol.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Dreux M, Pietschmann T, Granier C, Voisset C, Ricard-Blum S, Mangeot PE, Keck Z, Foung S, Vu-Dac N, Dubuisson J, Bartenschlager R, Lavillette D, Cosset FL. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J Biol Chem. 2006;281(27):18285–18295. doi: 10.1074/jbc.M602706200. [DOI] [PubMed] [Google Scholar]
- Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446(7137):801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- Farquhar MJ, McKeating JA. Primary hepatocytes as targets for hepatitis C virus replication. J Viral Hepat. 2008;15(12):849–854. doi: 10.1111/j.1365-2893.2008.01051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82(5):2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastaminza P, Kapadia SB, Chisari FV. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J Virol. 2006;80(22):11074–11081. doi: 10.1128/JVI.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J, Huby T, Stamataki Z, Vanwolleghem T, Meuleman P, Farquhar M, Schwarz A, Moreau M, Owen JS, Leroux-Roels G, Balfe P, McKeating JA. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J Virol. 2007;81(7):3162–3169. doi: 10.1128/JVI.02356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J, Nielsen S, Zhong J, Bassendine MF, Drummer HE, Balfe P, McKeating JA. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J Virol. 2008;82(24):12020–12029. doi: 10.1128/JVI.01569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haid S, Pietschmann T, Pecheur EI. Low pH-dependent Hepatitis C Virus Membrane Fusion Depends on E2 Integrity, Target Lipid Composition, and Density of Virus Particles. J Biol Chem. 2009;284(26):17657–17667. doi: 10.1074/jbc.M109.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N, Hino S, Fujiwara K. Physico-chemical properties of mouse hepatitis virus (MHV-2) grown on DBT cell culture. Microbiol Immunol. 1978;22(7):377–390. doi: 10.1111/j.1348-0421.1978.tb00384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M, Zhang J, Flint M, Logvinoff C, Cheng-Mayer C, Rice CM, McKeating JA. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci U S A. 2003;100(12):7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yang ZY, Kong WP, Nabel GJ. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J Virol. 2004;78(22):12557–12565. doi: 10.1128/JVI.78.22.12557-12565.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung SL, Lee PL, Chen HW, Chen LK, Kao CL, King CC. Analysis of the steps involved in Dengue virus entry into host cells. Virology. 1999;257(1):156–167. doi: 10.1006/viro.1999.9633. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Yi M, Li K, Lemon SM. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J Virol. 2002;76(6):2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia SB, Barth H, Baumert T, McKeating JA, Chisari FV. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J Virol. 2007;81(1):374–383. doi: 10.1128/JVI.01134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartenbeck J, Stukenbrok H, Helenius A. Endocytosis of simian virus 40 into the endoplasmic reticulum. J Cell Biol. 1989;109(6 Pt 1):2721–2729. doi: 10.1083/jcb.109.6.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003;125(6):1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, Nagayama K, Tanaka T, Wakita T. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J Med Virol. 2001;64(3):334–339. doi: 10.1002/jmv.1055. [DOI] [PubMed] [Google Scholar]
- Kim M, Ha Y, Park HJ. Structural requirements for assembly and homotypic interactions of the hepatitis C virus core protein. Virus Res. 2006;122(1–2):137–143. doi: 10.1016/j.virusres.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Kokorev VS, Podoplekin VD, Pereverzev VD, Fedotova TT. Buoyant density of some togaviruses in sucrose density gradient and capacity of their haemagglutinin fractions to interact with antibody. Acta Virol. 1976;20(5):353–360. [PubMed] [Google Scholar]
- Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol. 2006;80(11):5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavillette D, Bartosch B, Nourrisson D, Verney G, Cosset FL, Penin F, Pecheur EI. Hepatitis C virus glycoproteins mediate low pH-dependent membrane fusion with liposomes. J Biol Chem. 2006;281(7):3909–3917. doi: 10.1074/jbc.M509747200. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G, Rice CM. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J Virol. 2009;83(4):2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285(5424):110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Macnaughton MR, Davies HA. Two particle types of avian infectious bronchitis virus. J Gen Virol. 1980;47(2):365–372. doi: 10.1099/0022-1317-47-2-365. [DOI] [PubMed] [Google Scholar]
- Meanwell NA. Hepatitis C virus entry: an intriguing challenge for drug discovery. Curr Opin Investig Drugs. 2006;7(8):727–732. [PubMed] [Google Scholar]
- Meertens L, Bertaux C, Dragic T. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrincoated vesicles. J Virol. 2006;80(23):11571–11578. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P, Smolarsky M, Funaro A, Malavasi F, Larrey D, Coste J, Fabre JM, Sa-Cunha A, Maurel P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46(3):411–419. doi: 10.1016/j.jhep.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Monazahian M, Bohme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J.Med.Virol. 1999;57(3):223–229. doi: 10.1002/(sici)1096-9071(199903)57:3<223::aid-jmv2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5(6):453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J Virol. 2006;80(5):2418–2428. doi: 10.1128/JVI.80.5.2418-2428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niiyama Y, Igarashi K, Tsukamoto K, Kurokawa T, Sugino Y. Biochemical studies on bovine adenovirus type 3. I. Purification and properties. J Virol. 1975;16(3):621–633. doi: 10.1128/jvi.16.3.621-633.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietschmann T, Zayas M, Meuleman P, Long G, Appel N, Koutsoudakis G, Kallis S, Leroux-Roels G, Lohmann V, Bartenschlager R. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog. 2009;5(6):e1000475. doi: 10.1371/journal.ppat.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282(5390):938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457(7231):882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poynard T, Ratziu V, Benhamou Y, Opolon P, Cacoub P, Bedossa P. Natural history of HCV infection. Baillieres Best Pract Res Clin Gastroenterol. 2000;14(2):211–228. doi: 10.1053/bega.1999.0071. [DOI] [PubMed] [Google Scholar]
- Poynard T, Yuen MF, Ratziu V, Lai CL. Viral hepatitis C. Lancet. 2003;362(9401):2095–2100. doi: 10.1016/s0140-6736(03)15109-4. [DOI] [PubMed] [Google Scholar]
- Pumeechockchai W, Bevitt D, Agarwal K, Petropoulou T, Langer BC, Belohradsky B, Bassendine MF, Toms GL. Hepatitis C virus particles of different density in the blood of chronically infected immunocompetent and immunodeficient patients: Implications for virus clearance by antibody. J Med Virol. 2002;68(3):335–342. doi: 10.1002/jmv.10208. [DOI] [PubMed] [Google Scholar]
- Sainz B, Jr, Barretto N, Uprichard SL. Hepatitis C Virus infection in phenotypically distinct Huh7 cell lines. PLoS ONE. 2009;4(8):e6561. doi: 10.1371/journal.pone.0006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunier B, Triyatni M, Ulianich L, Maruvada P, Yen P, Kohn LD. Role of the asialoglycoprotein receptor in binding and entry of hepatitis C virus structural proteins in cultured human hepatocytes. J Virol. 2003;77(1):546–559. doi: 10.1128/JVI.77.1.546-559.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. Embo J. 2002;21(19):5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TJ, Brandt WE, Swanson JL, McCown JM, Buescher EL. Physical and biological properties of dengue-2 virus and associated antigens. J Virol. 1970;5(4):524–532. doi: 10.1128/jvi.5.4.524-532.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomssen R, Bonk S, Propfe C, Heermann K-H, Kochel HG, Uy A. Association of hepatitis C virus in human sera with B-lipoprotein. Med.Microbiol.Immunol. 1992;181:293–300. doi: 10.1007/BF00198849. [DOI] [PubMed] [Google Scholar]
- Thomssen R, Bonk S, Thiele A. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med.Microbiol.Immunol.(Berl) 1993;182(6):329–334. doi: 10.1007/BF00191948. [DOI] [PubMed] [Google Scholar]
- Tscherne DM, Jones CT, Evans MJ, Lindenbach BD, McKeating JA, Rice CM. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80(4):1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss L, Kekule AS, Jakubowski U, Burgelt E, Hofschneider PH. The HBV-producing cell line HepG2-4A5: a new in vitro system for studying the regulation of HBV replication and for screening anti-hepatitis B virus drugs. Virology. 1996;216(1):214–218. doi: 10.1006/viro.1996.0049. [DOI] [PubMed] [Google Scholar]
- Wong-Staal F, Yang JP, Chang J, Troung K, Lee H, Tang H, Rijnbrand R. 15th International Symposium on Hepatitis C Virus & Related Viruses; San Antonio, TX: 2008. [Google Scholar]
- Wunschmann S, Medh JD, Klinzmann D, Schmidt WN, Stapleton JT. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J Virol. 2000;74(21):10055–10062. doi: 10.1128/jvi.74.21.10055-10062.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Rodrigo A, Villanueva RA, Thomas DL, Wakita T, Lemon SM. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci U S A. 2006;103(7):2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel MB, Koutsoudakis G, Schnober EK, Haberstroh A, Blum HE, Cosset FL, Wakita T, Jaeck D, Doffoel M, Royer C, Soulier E, Schvoerer E, Schuster C, Stoll-Keller F, Bartenschlager R, Pietschmann T, Barth H, Baumert TF. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46(6):1722–1731. doi: 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102(26):9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Gastaminza P, Chung J, Stamataki Z, Isogawa M, Cheng G, McKeating JA, Chisari FV. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J Virol. 2006;80(22):11082–11093. doi: 10.1128/JVI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.