Abstract
Epidermal growth factor receptor (EGFR) signaling plays an important role in non-small cell lung cancer (NSCLC) and therapeutics targeted against EGFR have been effective in treating a subset of patients bearing somatic EFGR mutations. However, the cancer eventually progresses during treatment with EGFR inhibitors, even in the patients who respond to these drugs initially. A large variety of distinct irreversible inhibitors have been developed, which may combat therapeutic resistance. Nonetheless, major challenges in tailoring patient-specific treatment regimens involve predicting the most effective inhibitors and monitoring for acquisition of resistance. A patient-customized, predictive diagnostic that quantifies the effects of specific anti-EGFR therapies may improve outcomes in cancers where EGFR plays a mechanistic role. In this study we used an EGFR-phosphorylatable peptide, AEEEEYFELVAKKK, immobilized within a polyacrylamide hydrogel as a substrate for profiling the activation status of EGFR in the cellular extracts of erlotinib-resistant cancer cells. The hydrogel array was able to detect therapeutic resistance as well as identify inhibitors capable of combating therapeutic resistance. These findings establish the potential of this protein-acrylamide copolymer hydrogel array to not only evaluate EGFR status in cancer cell lysates but also to screen for the most promising therapeutics for individual patients and monitor effects of treatment on acquisition of resistance to EGFR inhibitors.
Keywords: Peptide array, EGFR tyrosine kinase, polyacrylamide, EGFR inhibitors, acquired resistance
1. Introduction
Lung cancer is the leading cause of death globally. The first line of treatment involving platinum-based chemotherapeutics yields a response rate of only 30% in advanced non small cell lung cancer (NSCLC) (Schiller, 2002). Frequent abnormal amplification or activation of epidermal growth factor receptor (EGFR) in NSCLC has been correlated with hallmarks of cancer including increased cell proliferation, angiogenesis, invasion and metastasis (Laskin and Sandler, 2004; Yarden, 2001). EGFR is a member of the ErbB family of receptor tyrosine kinases (RTKs) which is comprised of EGFR (ErbB1), HER2/neu (ErbB2), HER3 (ErbB3), and HER4 (ErbB4). Ligand binding leads to homo or heterodimerization of these receptors which in turn initiates downstream signaling cascades involving the phosphatidylinositol-3-kinase (PI3K), Akt, mitogen activated protein kinase (MAPK), phospholipase cγ and STAT pathways (Laskin and Sandler, 2004; Yarden, 2001).
Identification of the oncogenic potential of EGFR spawned the development of several therapeutics directed against EGFR. Two small molecule EGFR tyrosine kinase inhibitors (EGFR-TKI), gefitinib (Iressa, AstraZeneca International) and erlotinib (Tarveca, OSI Pharmaceuticals) have been evaluated in patients with NSCLC (Inoue et al. 2006; Shepherd et al. 2005). These ATP competitive, reversible EGFR-TKIs have been effective only in a small subset of NSCLC patients (10-15% of Caucasian and 30-40% of Asian) bearing somatic mutations in the kinase domain of EGFR (Sequist et al. 2007). EGFR mutations associated with increased sensitivity to erlotinib and gefitinib are found in exons 18-21, a region that encodes the EGFR tyrosine kinase domain (Lynch et al. 2004; Paez et al. 2004; Pao et al. 2004). Deletions in exon 19 and the L858R mutation constitute around 90% of these mutations. These mutations impart increased affinity for gefitinib or erlotinib as well as decreased affinity for ATP. Nevertheless, patients initially responding to TKI therapy invariably develop resistance to these drugs, thereby limiting the progression-free survival to approximately 9-13 months with a median survival of 2 years (Rossell et al. 2009). Studies have been undertaken to identify the biomolecular properties associated with drug resistance in tumor specimens and also in resistant cell lines. Different molecular techniques involving direct sequencing, subcloning or cycleave PCR, PCR- restriction fragment length polymorphism (PCR-RFLP), Scorpion amplified refractory mutation system (SARMS) as well as immunohistochemical detection using mutation specific antibodies have been employed to identify the mechanisms of de novo or acquired resistance (Balak et al. 2006; Engleman et al. 2006; Kitamura et al. 2010; Kobayashi et al. 2005; Kosaka et al. 2006; Kuang et al. 2009; Pao et al. 2005). Moreover, mutation scanning based on enzymatic digestion of PCR products by SURVEYOR enzymes combined with HPLC chromatography or real time melting curve analysis has also been used for mutational analysis (Kuang et al. 2009; Li J 2007). These studies revealed that 50% of drug resistant tumors are associated with the emergence of a secondary mutation, substitution of methionine for threonine at the position 790 (T790M), in the EGFR kinase domain (Kobayashi et al. 2005; Pao et al. 2005). By increasing ATP affinity, the T790M mutation negates the sensitivity of reversible TKIs and generates a resistance to the achievable clinical doses of the drugs. Studies have also identified the presence of other secondary mutations in the resistant tumors, including D716Y, L747S, E884K, and T854A, although these mutations occur less frequently than T790M (Balak et al. 2006; Choong et al. 2006; Costa et al. 2008). An additional survival mechanism adopted by NSCLC cells in 20% of therapeutic resistance to EGFR-TKIs involves amplification of the MET proto-oncogene (Bean et al. 2007; Engelman et al. 2007a). The molecular mechanism involved in 30-40% of drug resistance cases is yet to be unraveled, illustrating the need to develop assays to directly monitor EGFR activity in cancer cells treated with EGFR-TKIs.
Some EGFR the secondary mutations, such as L747S or D761Y, confer substantially less resistance to gefitinib or erlotinib compared with the T790M mutation, and administering alternative EGFR-TKIs can be beneficial (Choong et al. 2006; Costa et al. 2008). One study showed that while switching to erlotinib overcame gefitinib resistance in a NSCLC patient with L858R+L747S mutations, it failed for a gefitinib refractory patient with the T790M mutation (Choong et al. 2006). Similarly another report demonstrated that a switch from erlotinib to gefitinib yielded a positive response in a lung adenocarcinoma patient with L858R+E884K mutations (Costa et al. 2008). However, none of the reversible EGFR-TKIs are effective in patients expressing EGFR with the T790M mutation. Thus it appears that the precise nature of the secondary mutations determines the success of these TKIs. However, the realization that cancer cells with T790M EGFR mutation still depend on EGFR for survival spawned the development of a gamut of irreversible EGFR-TKIs. These second generation irreversible EGFR-TKIs, including CL-387,789, HKI-272, and PF00299804, inhibit EGFR phosphorylation by affecting a Michael addition reaction with the cysteine residue in the ATP binding pocket of the EGFR kinase domain. The covalent attachments ensure a higher occupancy of ATP binding site and thus enable these TKIs to inhibit the activation of T790M EGFR (Engelman et al. 2007b; Zhou et al. 2009). Other second generation irreversible inhibitors which have shown promise at different stages of clinical development include BIBW-2992 (EGFR/HER2 dual inhibitor), CI-1033 (pan-EGFR inhibitor) and EKB-569 (pan-EGFR inhibitor).
However, there are some serious issues which prevent a smooth transition of these TKIs from preclinical studies to clinical therapies. Due to the involvement of different resistance mechanisms, a major challenge involves identifying the mechanism of resistance in individual patients. This is because a general therapeutic strategy to overcome EGFR-TKI resistance will not be effective in treating all resistant patients. For example, patients with amplified MET expression will not respond to EGFR-TKI therapy. Similarly, treating patients bearing secondary EGFR mutations or having some other activated kinase pathway with MET inhibitor will be unsuccessful. Hence, there is a need of a diagnostic tool which can differentiate the patients who may benefit from switching to different EGFR-TKIs from patients who will need different therapeutic strategies to overcome the drug resistance.
Here we report the development of a hydrogel-based peptide array capable of quantitatively assessing EGFR tyrosine kinase activity and the sensitivity of different EGFR inhibitors in extracts of cells that have acquired drug resistance. In this study, we have used the EGFR-phosphorylatable peptide sequence AEEEEYFELVAKKK as the substrate for profiling EGFR kinase activity. As opposed to the conventional method of genotyping cancer specimens and cancer cell lines, the peptide array will not identify mutations but instead will detect alterations in the kinase activity due to acquired drug resistance. Sequencing is regarded as the gold standard for mutational analysis. However, allelic dilution due to the presence of a small fraction of mutated alleles in the background of large excess of normal alleles often renders the sequencing/mutation screening technique inadequate (Engelman et al. 2006; Li et al. 2007). Recent effort directed towards immunohistochemical detection of EGFR mutations with mutation-specific antibodies as an alternative of the mutational analysis revealed limited clinical utility of these antibodies (Kitamura et al. 2010). Since, EGFR-TKI resistant cancer cells with secondary mutations not only express active EGFR but also depend on EGFR for survival, an assay measuring activity of EGFR carrying mutations may provide a more accurate report of the disease state than these conventional techniques. AEEEEYFELVAKKK has been reported to be the optimal peptide for EGFR kinase (Songyang et al. 1995). However, to the best of our knowledge, this peptide has never been used to quantitatively assess the activity of EGFR carrying primary or secondary mutations. Here, we report the ability of the hydrogel based peptide array to profile EGFR kinase activity and assess the sensitivity of different inhibitors in the extracts of cells with wild type EGFR (NCI-H23), cells with activating mutations (NCIH1650, PC9) and also in cells having acquired resistance to erlotinib and gefitinib (H1650-ER and PC9-GR).
2. Materials and Methods
2.1 Preparation and purification of peptide
Amino acids, CLEAR-Amide resin and 2-(6-Chloro-1-H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium haxafluorophosphate (HCTU) were obtained from Peptides International (Louisville, KY, USA). Piperidine, N-methylmorpholine (NMM), trifluoroacetic acid (TFA), 1,2-Ethanedithiol (EDT), Triisopropylsilane (TIS) were obtained from Sigam-Aldrich (St. Louis, MO, USA). All reagents were used as received without any purification. The peptide, AEEEEYFELVAKKK, was synthesized on an automated synthesizer, Prelude™ (Protein Technologies, Inc.), using solid-phase method based on Fmoc-chemistry. Cleavage of the crude peptide was performed with the mixture of TFA/ddH2O/EDT/TIS (94:2.5:2.5:1 v/v) at room temperature. The crude peptide was then precipitated and washed with cold diethyl ether three times. The crude peptides were tested using ABI 4700 MALDI TOF/TOF mass spectrometry (Applied Biosystems) to confirm the correct molecular masses and Agilent 1200 Series LC/MS system for purity. Purification was done through a preparative C18 column in the Agilent 1200 LC/MS system if necessary.
2.2 Cell Culture and Lysate Production
Human lung cancer cell lines NCI-H23 and NCI-H1650 (hence forth referred to as H23 and H1650) were obtained from ATCC (Manassas, VA). PC9 and PC9-GR cells were kind gift from Dr. Pasi A. Janne (Dana-Faeber Cancer Institue, Boston, Massachusetts). The cells were maintained in RPMI-1640 supplemented with 10% FBS and glutamine. During the culture, the media were changed every other day. The cells were passaged every 5-6 days using Trypsin-EDTA (0.25% trypsin, 1mM EDTA). To lyse the cells, the cells were washed 2X with cold phosphate buffered saline (PBS) and then 1 mL of cold lysis buffer containing 1X protease inhibitor cocktail, 1 mM phenylmethylsulfonyl fluoride, and 1 mM activated sodium orthovanadate was added to each cell flask. The cells were incubated on ice for 15 min with occasional swirling then removed from the plate with a cell scraper and transferred to a microcentrifuge tube. The cell lysate was then clarified by centrifuging at 14,000 g at 4°C for 15 min. Total protein concentration was determined using a BCA protein assay kit (Pierce, Rockford, IL) and the lysates were stored at −80°C until further use.
2.3 Kinase Inhibitors
Gefitinib and erlotinib were procured from LC labs (Woburn, MA), CL-387,785 from EMD Chemicals (Gibbstown, NJ) and BIBW-2992 and CI-1033 from Chemietek (Indianapolis, IN).
2.4 Establishment of erlotinib resistant clones
To establish resistant clones, H1650 cells were cultured continuously in the presence of increasing concentrations of erlotinib. Starting with a concentration of 2.5 μM, exposure dose was doubled every 15 days until the final concentration of 20 μM was achieved. The cells were maintained in continuous culture with the final dose of erlotinib for 30 days. Following which, the resistance phenotype of the pools was characterized by cell proliferation assay. The resistant pool was then used to establish the clones. For the purpose, these cells were seeded in 20 wells of a 96 well plate such that each well had only a single cell. These cells were maintained in media with 20 μM erlotinib until they formed colonies. Cells were then expanded to 6 well plate. The established clones were further maintained in culture with 20 μM erlotinib for another 30 days. The cell viability was then measured by challenging them with varying concentrations of erlotinib. Prior to any experiment, the cells were cultured in media without erlotinib for at least a week.
2.5 Cell viability assay
H1650, H1650-ER1, H1650-ER2 and H1650-ER4 cells were seeded at a density of 5 × 103 cells/well in 96 well plates. After 24 hr, EGFR-TKIs (erlotinib, gefitinib, genistein, PKI-166, CL-387,785, CI-1033 and BIBW-2992) at varying concentrations were added and the cells were incubated further for 48 hr. The cells were then washed with PBS and cell viability was measured using a XTT assay kit (Sigma, St. Louis, MO).
2.6 Clonogenic Assay
H1650, H1650-ER1, H1650-ER2 and H1650-ER4 cells were seeded at a density of 25 cells per well in 6 well plates and incubated overnight. After 24 hr, erlotinib was added so as to achieve the concentration of 10 and 50 μM and the cells were incubated for 15 days. Media was changed twice weekly. After 15 days, the cells were stained with 0.1% crystal violet for 5 min at the room temperature and the number of colonies was counted. Only colonies containing at least 50 cells were counted. The number of colony forming units in the treated cells was expressed as percentages of untreated cells. The experiment was repeated twice.
2.7 PCR and sequencing
RNA from H1650, H1650-ER1, H1650-ER2 and H1650-ER4 cells was extracted using RNeasy Mini kit (Qiagen, Valencia, CA) and cDNA was generated using high capacity cDNA reverse transcription kit (Applied Biosystems). PCR was conducted to amplify the exons 18-21 using the primers designed by Willmore-Payne et al. Each PCR reactions contained 2 μL of cDNA, 2 μL of primers, 1 μL of 10 μM dNTPs, 1 μL of 50 mM MgCl2, 5 μL of Thermo-pol buffer and 1 μL of Taq polymerase in 50 μL reaction volume. PCR cycling parameters were: one cycle of 95°C for 1 min, 50 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 40 s, followed by one cycle of 72°C for 1 min. The PCR products were then purified and followed by sequencing.
2.8 Fabrication of hydrogel based protein arrays
Glass slides were acryl functionalized and peptide sequence AEEEEYFELVAKKK was acryl labeled as described in detail previously (Brueggemeier et al. 2005; Ghosh et al. 2009). Briefly, 1 μL spots were created onto the glass slides by polymerizing the prepolymer (4% (w/v) acrylamide/bisacrylamide, 0.1% (w/v) Irgacure-2959 (Ciba Specialty Chemicals), 15% (w/v) glycerol, and 500 ng of peptide) with UV (365 nm) illumination at 1500 μW/cm2 for 15 min. The glass slide containing the polymerized array was washed in TBST (10 mM Tris-HCl, 100 mM NaCl and 0.15% Tween-20) and then in DI water prior to storing in 1X EGFR kinase buffer (20 mM HEPES pH:7.2, 10 mM MnCl2, 1 mM DTT, 15 mM MgCl2 and 40 μg/mL BSA) at 4°C.
2.9 Array based kinase assay
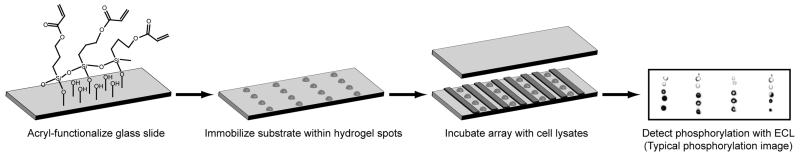
To carry out the kinase assay, the kinase buffer was aspirated from the slides and the slides were dried under a stream of compressed air. As illustrated in Figure 1, reaction chambers were created by placing 3.5 × 0.5 cm spacers, made from Dura Lar sheets of 350μm thickness in between the spots (for 60 μL reactions) and placing a clean glass slide over the spots. The reaction mixture consisting of 50 μg of cell lysates, 6 μL of 1 mM ATP, 6 μL of 100 μg/mL EGF, 20 μL of 3X EGFR kinase assay buffer, 6 μL of 50% glycerol and water (total volume of 60 μL) was then applied to the reaction chamber. Cell lysate protein concentration was 0.6 μg/μL of reaction mixture.
Figure 1.
Quantitative profiling of phosphorylated status of EGFR and sensitivities of EGFR to inhibitors using a hydrogel-immobilized peptide assay. Hydrogel spots were created on the functionalized slides by exposing the prepolymer (with or without peptide) to UV light. Immobilized peptide was then incubated with different reaction mixtures, loaded in parallel in different reaction chambers. Following the reaction, the phosphorylation of the substrates was detected by ECL. After development, average pixel value of each spot and the background signal was measured and the difference between the two signals was reported as the pixel value for the spot.
2.9.1 Comparing the effects on inhibitors on EGFR kinase activity
EGFR-TKIs at varying concentrations were incubated with cell lysates for 30 min at room temperature prior to performing the kinase assay.
2.10 Detection and quantification of phosphorylated substrates
After the reaction, the slides were washed with TBST for 30 min. Then the slides were blocked in TBST containing 1% BSA for 1 hr at room temperature. This was followed by incubation of the slides with 4G10 anti-phosphotyrosine (Upstate Biotech) antibody for 1 hr at room temperature. After incubation with primary antibody, the slides were washed 3X with TBST then incubated with horseradish peroxidase (HRP) conjugated goat anti-mouse secondary antibody (Invitrogen). Enhanced chemiluminescence (ECL) technique was used to detect HRP activity. After development, average pixel value of each spot and the background signal in the near vicinity was measured using Image J software (NIH, Bethesda, MD) and the difference between the two signals was reported as the pixel value for the spot.
2.11 Western Blot Analysis
Following incubation with or without EGFR-TKIs for 5 hr, H1650, H1650-ER1, H1650-ER2 and H1650-ER4 cells were harvested and cellular protein concentration was quantified using BCA protein assay (Pierce, Rockford, IL). 30 μg of proteins were subjected to 10% SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. After blocking the membranes with 0.1% BSA in TBST for 1 hr at room temperature, the membranes were probed with primary anti-EGFR antibody (Santa Cruz Biotechnologies), anti-erbB2 antibody (Cell Signaling Technology), anti-Akt antibody (Cell Signaling Technology), anti-p-Akt antibody (Cell Signaling Technology), anti-MAPK antibody (Cell Signaling Technology), anti-p-MAPK antibody (Cell Signaling Technology) and horseradish peroxidase-conjugated secondary antibody respectively for 1 hr at room temperature. Protein levels were detected by ECL (Pierce).
3. Results and Discussion
To develop an assay to quantify EGFR activity in cancer cell lysates, acryl-labeled peptide (AEEEEYFELVAKKK) was attached to the functionalized glass slides by copolymerization with acrylamide in a free-radical polymerization. Previous studies from our lab have shown that immobilization of peptides or fusion proteins within hydrogel networks helps maintain the native form of the substrate due to the presence of hydrated environment within the hydrogel (Brueggemeier et al. 2005; Ghosh et al. 2009). Hydrophilic hydrogel passivates the glass surface and thus reduce the hydrophobic interactions between the glass and the protein. Moreover, the characteristic three dimensional network of the hydrogel increases the substrate capacity to a great extent. These characteristics provide a probe with high binding capacity and sensitivity, ultimately improving signal-to-noise ratio (Brueggemeier et al. 2004; Brueggemeier et al. 2005; Ghosh et al. 2009).
3.1 Detection of EGFR kinase activity in cell lysates
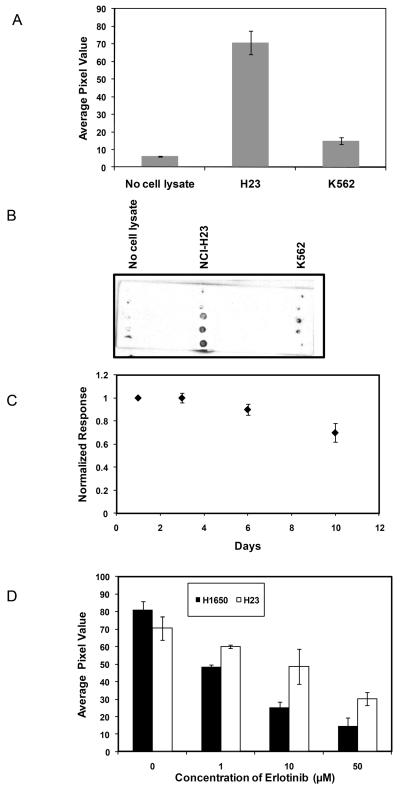
To investigate the ability of the peptide to act as a specific substrate for EGFR kinase, we assessed the phosphorylation of the immobilized peptide following incubation with H23 cell lysate, K562 cell lysate, and a no lysate buffer control. H23 is a NSCLC cell line that expresses wild type EGFR while K562 is a human chronic myeloid leukemia cell line that does not express EGFR but expresses very high levels of the constitutively active tyrosine kinase Bcr-Abl (Naruse et al. 2002). Figure 2 illustrates the phosphorylation of the substrate by the cell lysates, using an anti-phosphotyrosine antibody followed by ECL detection as readout. Substrate phosphorylation was significantly higher for H23 cells (ECL value: 71 ± 7) as compared to K562 cells (ECL value: 15 ± 2, p < 0.005). This observation suggests that the immobilized peptide substrate was phosphorylated primarly by EGFR, and that the other tyrosine kinases present in the cell lysates do not substantially contribute toward substrate phosphorylation. In the absence of cell lysate, very little ECL signal was detected (ECL value: 6 ± 0.2).
Figure 2.
Specificity of immobilized peptide phosphorylation by EGFR. (A) H23 lysate, K562 lysate, and no lysate control were assayed for EGFR activity. ECL values represent the extent of substrate phosphorylation. (B) Typical image of phosphorylation of immobilized peptide after incubation with H23, K562 cell lysates and no lysate control for 1 hr. (C) Comparison of substrate phosphorylation following incubation in PBS buffer for 1-10 days. (D) Comparison of the effects of erlotinib on the phosphorylation of immobilized peptide by H23 (cell with wild type EGFR) and H1650 (mutant EGFR) cell lysate. Each data point represents the mean of three independent experiments in which each experiment contained three replicates. The error bars represent s.e.m. (n=3).
To investigate the efficacy of the peptide to act as substrate of EGFR, phosphorylation of peptide was compared to that of GST-Eps15 in the presence of cell lysates. The peptide was found to be more consistently phosphorylated when incorporated in polyacrylamide gels (data not shown). All the experiments in this study were carried out using the peptide as substrate. Moreover, to characterize the stability of polyacrylamide copolymerization attachment, the slides with the polymerized spots were incubated in 1X EGFR buffer at 4° C over a time ranging from 1 to 10 days. To investigate the stability and diffusion of the peptide within the hydrogel spots, the phosphorylation of the peptide upon incubation with H23 cell lysate at different time points was compared to that observed on day 1. Approximately 70% of the phosphorylation was retained after 10 days (Figure 2C).
To investigate the ability of the peptide to profile activation status of mutant EGFR as well as to quantify the sensitivity of mutants to the EGFR inhibitor erlotinib, the substrate was incubated with H23 or H1650 cell lysates for 1 hr in the presence of 0-50 μM erlotinib. H1650 is a NSCLC cell line that expresses EGFR containing a deletion mutation (ΔE746-A750) in the kinase domain. As shown in Figure 2D, substrate phosphorylation was significantly attenuated when H1650 was incubated with erlotinib. This indicates that erlotinib had effectively reduced EGFR kinase activity in H1650 (IC50: ~1.5 μM). However, erlotinib was much less effective on H23. This observation agrees with reports that erlotinib is much more active against EGFR containing activating mutations as compared to wild type EGFR (Sequist et al. 2007; Rossell et al. 2009; Wang et al. 2008).
3.2 Establishment and characterization of erlotinib resistant clones
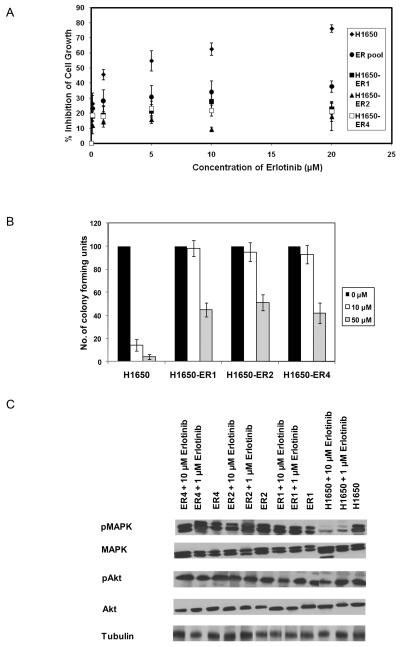
NSCLC cell line H1650 was used to generate resistance to the EGFR inhibitor erlotinib. As described in the Materials and Methods section, cells were exposed to erlotinib at 2 μM and the dose was progressively doubled every 15 days until the final concentration of 20 μM was reached. The cells were cultured at this concentration of erlotinib for 1 month. We then determined the resistance of this pool of H1650 cells by challenging it to different concentrations of erlotinib. As shown in Figure 3A, higher viability at all erlotinib concentrations was observed in the pool of erlotinib-resistant cells. Individual H1650 clones were then isolated and characterized from this resistant pool. We characterized the resistant phenotype of the clones by quantifying cell proliferation at different concentrations of erlotinib. A much higher proliferative rate and significantly higher IC50 were observed for these cells (IC50 > 20 μM) as compared with the sensitive parental H1650 cells (IC50 ~2.4 μM) (Fig. 3A). The resistant phenotype was also confirmed by clonogenic assay (Fig. 3B). Under continuous exposure of 10 μM and 50 μM of erlotinib, parental cells formed very few colonies while significantly greater numbers of colonies were observed in case of the resistant clones.
Figure 3.
Establishment and characterization of erlotinib-resistant H1650 cells. (A) Dose-response curves of parental H1650 cells, a resistant pool, and clones isolated from the resistant pool following incubation with varying concentration of erlotinib for 48 hr. (B) Colony formation assay: H1650, H1650-ER1, H1650-ER2 and H1650-ER4 cells were seeded at a density of 25 cells per well in 6 well plates and cultured in the presence of varying concentration of erlotinib. After 15 days, the number of colonies was counted. Only colonies containing at least 50 cells were counted. Numbers of colony forming units in the treated cells were expressed as percentages of untreated cells. (C) Extracts of the cells treated with 0, 1 and 10 μM erlotinib for 5 hr and were analyzed for phospho and total MAPK and Akt by Western blot. Each data point represents the mean of three (A) or two (B) independent experiments. The error bars represent s.e.m.
To examine the effect of erlotinib upon downstream targets of EGFR which mediate the proliferative and survival pathways, the expression and activation state of MAPK and Akt was studied. Treatment with erlotinib resulted in attenuation in MAPK phosphorylation in the parental cell line (Fig. 3C). In resistant clones (H1650-ER1, H1650-ER2 and H1650-ER4) the phosphorylation of MAPK was maintained in the presence of erlotinib. However, in both the parental cell line as well as the resistant clones, erlotinib dosage did not affect the phosphorylation of Akt. Maintenance of Akt phosphorylation in H1650 in presence of EGFR inhibitor may be due to the loss of phosphatase and tensin homolog (PTEN) (Sos et al. 2009).
Sequencing revealed the persistence of the deletion mutation ΔE746-A750 within the kinase domain of EGFR in both H1650 and the resistant clones; however, no additional mutation was observed in the EGFR open reading frame in resistant cells.
3.3 Quantification of acquired resistance by hydrogel based assay
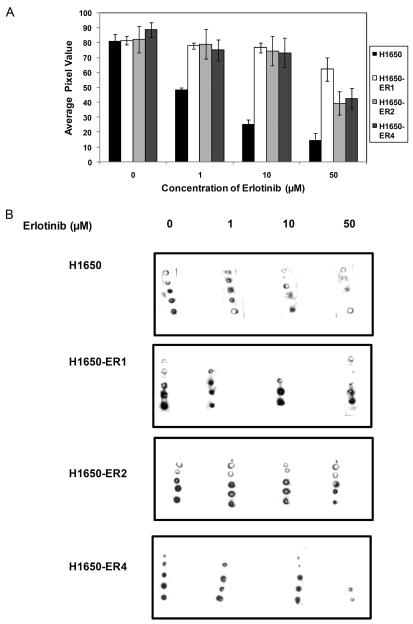
To investigate the ability of the hydrogel-based peptide array to quantitatively assay acquired erlotinib resistance in the clones, the immobilized peptide was incubated with the cellular extracts of clones in the presence of erlotinib. As illustrated in Figure 4A and 4B, substrate phosphorylation was significantly attenuated by erlotinib in the parental cells, indicating a reduction of EGFR kinase activity in the presence of erlotinib (IC50: ~1.5 μM). However, such a profound effect of the inhibitor was not observed in the resistant clones (IC50: > 50 μM) indicating erlotinib was not able to induce a reduction in kinase activity in the clones. More importantly, this study also demonstrates the ability of the immobilized peptide based array to detect acquired resistance in the cells.
Figure 4.
Quantification of acquired resistance to erlotinib by hydrogel based peptide array. (A) Comparison of the effects erlotinib on the phosphorylation of immobilized peptide by H1650, H1650-ER1, H1650-ER2, and H1650-ER4 cell lysates. Each data point represents the mean of three independent experiments in which each experiment contained three replicates. The error bars represent s.e.m. (n=3). (B) Typical images of phosphorylation of immobilized peptide after incubation with H1650, H1650-ER1, H1650-ER2 and H1650-ER4 cell lysates for 1 hr.
3.4 Quantification of sensitivities of different inhibitors in resistant clone and comparison with parental cell
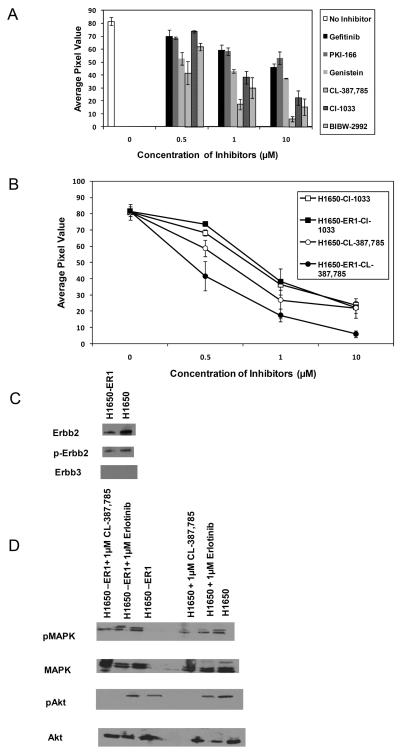
Since many inhibitors that were designed in part to overcome therapeutic resistance to existing RTKs are now in the process of development, any tool capable of evaluating the sensitivities of these inhibitors will aid in drug screening and development. With this goal in mind, we explored the possibility of profiling the sensitivities of different inhibitors on the resistant clones using the hydrogel-based peptide array. Toward this end, we investigated the effect of the reversible ATP competitive inhibitors including gefitinib (EGFR inhibitor), PKI-166 (dual EGFR-ErbB2 inhibitor), and genistein (kinase inhibitor), as well as the irreversible inhibitors CL-387,785 (EGFR inhibitor), CI-1033 (pan-EGFR inhibitor), and BIBW-2992 (dual EGFR-ErbB2 inhibitor). The concentrations of the inhibitors were varied from 0 to 10 μM. Since the three clones have demonstrated a similar resistant phenotype, we assessed the effects of these inhibitors on substrate phosphorylation on only H1650-ER1. Figure 5A demonstrates a dose dependent effect of the inhibitors on substrate phosphorylation. Incubation of H1650-ER1 cell lysate with gefitinib and PKI-166 had little significant effect on substrate phosphorylation (IC50: > 10 μM). Genistein had moderate effect with IC50: ~4 μM. However, the irreversible inhibitors had much more profound effect on substrate phosphorylation. When H1650-ER1 cell lysate was incubated with CL-387,785, CI-1033 and BIBW-2992, the substrate phosphorylation was attenuated significantly (ECL value: 6 ± 2, 22 ± 6 and 15 ± 6 for 10 μM of CL-387,785, CI-1033 and BIBW-2992 respectively) as compared to that in absence of inhibitors (ECL value: 82 ± 3). These data show that the irreversible inhibitors were much more efficient in reducing EGFR kinase activity in cells with therapeutic resistance as compared to the reversible counterparts.
Figure 5.
(A) Comparison of the effects different EGFR inhibitors on the phosphorylation of immobilized peptide by H1650-ER1 cell lysates. Each data point represents the mean of three independent experiments in which each experiment contained three replicates. (B) Quantification of substrate phosphorylation by H1650 and H1650-ER1 cell lysates in the presence of the inhibitors. (C) Expression levels of phopho and total ErbB2 and ErbB3 were determined in the cellular extracts of H1650 and H1650-ER1 by Western blot analysis. (D) Western blot analysis carried out to analyze the effect of erlotinib and CL-387,785 on phosphorylated and total MAPK and Akt levels in H1650 and H1650-ER1 cells. The error bars represent s.e.m. (n=3).
Experiments were also performed to determine the effect of these inhibitors on proliferation of H1650-ER1 cells. Cell death induced by irreversible inhibitors suggests that acquisition of erlotinib resistance did not trigger EGFR independent survival mechanism in these cells. When the effects of these inhibitors on cell proliferation and substrate phosphorylation were compared, comparable IC50 values were obtained (data not shown).
Next, we compared the effect of CL-387,785 and CI-1033 on kinase activity of H1650-ER1 to that of parental H1650 cell. These two irreversible inhibitors were chosen due to the fact that CL-387,785 had been most effective in reducing EGFR kinase activity in cells with therapeutic resistance and CI-1033 is a pan-EGFR inhibitor. Upon incubation of the immobilized substrate with H1650 in the presence of these inhibitors, a significant reduction in substrate phosphorylation was observed. As illustrated in Figure 5B, these irreversible inhibitors had comparable effects on EGFR kinase activities of H1650 and H1650-ER1 cells. Again, CL-387,785 was found to be more effective than CI-1033. Cell proliferation assays also exhibited similar trends. To gain insight into the mechanism underlying the varying efficacies of different inhibitors on parental H1650 and erlotinib resistant H1650-ER1 cells, we examined the expression levels of ErbB family members in these cells (Fig. 5C). Both cells lines express comparable levels of p-ErbB2. ErbB3 was not expressed by any of these cells. Therefore, we speculate that both H1650 and H1650-ER1 primarily depend on EGFR for survival with a possibility of EGFR-ErbB2 heterodimer formation.
We compared the ability of erlotinib and CL-387,785 to suppress the phosphorylation of downstream effectors Akt and MAPK. While erlotinib exhibited reduced efficacy in suppressing p-AKT and p-MAPK levels in resistant cells compared to the parent H1650 cells, CL-387,785 showed persistent activity (Fig. 5D).
3.5 Detecting resistance in PC9-GR cells
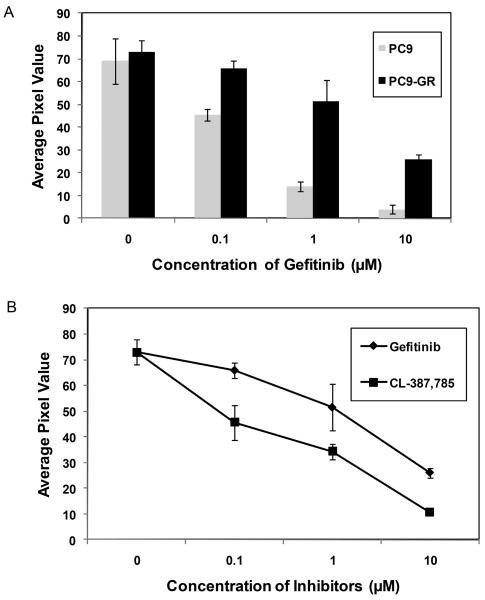
Human lung adenocarcinoma cell line PC9, which contains a 15 bp deletion in exon 19 of EGFR, is hypersensitive to gefitinib. We incubated the immobilized peptide with extracts of PC9 cells and gefitinib-acquired resistant subline PC9-GR cells, bearing a secondary T790M mutation (Zhou et al. 2009) in the presence of gefitinib to further confirm the ability of the hydrogel-based array to detect drug sensitivity and acquired resistance. As illustrated in Figure 6A, gefitinib significantly attenuated substrate phosphorylation at a concentration of 0.1 μM (p < 0.05) indicating reduced EGFR kinase activity (IC50: 0.2 μM). However, such a dramatic effect of gefitinib on PC9-GR cells was not observed (IC50: 4.2 μM).
Figure 6.
Effect of PC9 and PC9-GR cells on the hydrogel based peptide array. (A) Detection of acquired resistance by the peptide array upon incubation of the immobilized substrate with PC9 and PC9-GR cells in the presence of gefitinib. (B) Comparison of the effect of gefitinib and CL-387,785 on substrate phosphorylation when incubated with PC9-GR cells. Each data point represents the mean of three independent experiments in which each experiment contained three replicates. The error bars represent s.e.m. (n=3).
Next we incubated the immobilized substrate with PC9-GR cells in the presence of the irreversible EGFR inhibitor, CL-387,785. This irreversible inhibitor was much more effective in reducing the substrate phosphorylation as compared to gefitinib (IC50: 0.5 vs.4.2 μM) (Fig. 6B). IC50 values thus obtained correspond well with those obtained in cell proliferation assay (Zhou et al. 2009). These experiments demonstrate the ability of the peptide array to predict effectiveness of EGFR TKIs in a hypersensitive cell (PC9) as well as in the resistant subline (PC9-GR). Moreover, these experiments further verified the observations with H1650 and H1650-ER1 cells and substantiated the potential of this hydrogel based peptide array to evaluate the efficacies of different inhibitors to overcome therapeutic resistances.
4. Conclusion
In this study we have developed a robust and efficient hydrogel based bioassay to achieve high sensitivity, high throughput, and quantitative measurement of EGFR kinase activity in NSCLC cells with acquired drug resistance using ECL detection as read out.
The present study initially demonstrated the ability of a peptide, AEEEEYFELVAKKK, immobilized in a hydrogel array to quantify the phosphorylation status of wild-type and mutant EGFR in the presence of EGFR-TKIs. We then established erlotinib resistant H1650 clones and characterized the resistance phenotype by carrying out clonogenic assays as well as measuring the cell viability upon challenging the cells with erlotinib. Subsequently, we challenged the peptide array to detect acquisition of therapeutic resistance. A resistant clone, H1650-ER1, was used to explore the potential of hydrogel based peptide array to quantify the sensitivities of different inhibitors. Of all the inhibitors examined, the irreversible EGFR-TKI CL-387,785 was found to be most effective in inhibiting EGFR kinase activity.
As opposed to the conventional methods which focus on genotyping cancer specimens for identifying mutations responsible for enhanced kinase activity or acquired resistance, the hydrogel based peptide assay cannot identify mutations that lead to changes in kinase activity. However, it can detect the changes in kinase activity resulting from unknown mutations. Moreover, it can evaluate the efficacy of different inhibitors and also screen for the most promising therapeutics for individual patients to combat the resistance and monitor treatment progression. Furthermore, with a slew of inhibitors presently at different stages of development, the hydrogel-based peptide array may also act as a drug screening tool.
Acknowledgements
The authors thank National Institutes of Health for providing financial support to this research (grant R01GM074691).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, Chiang A, Yang G, Ouerfelli O, Kris MG, Ladanyi M, Miller VA, Pao W. Clin Cancer Res. 2006;12:6494–501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, Balak M, Chang W-C, Yu C-J, Gazdar A, Pass H, Rusch V, Gerald W, Huang S-F, Yang P-C, Miller V, Ladanyi M, Yang C-H, Pao W. Proc. Natl. Acad. Sci. USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggemeier SB, Kron SJ, Palecek SP. Anal Biochem. 2004;329:180–189. doi: 10.1016/j.ab.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Brueggemeier SB, Stephen DW, Kron SJ, Palecek SP. Biomacromolecules. 2005;6:2765–2775. doi: 10.1021/bm050257v. [DOI] [PubMed] [Google Scholar]
- Choong NW, Dietrich S, Seiwert TY, Tretiakova MS, Nallasura V, Davies GC, Lipkowitz S, Husain AN, Salgia R, Ma PC. Nat Clin Pract Oncol. 2006;3:50–57. doi: 10.1038/ncponc0400. [DOI] [PubMed] [Google Scholar]
- Costa DB, Schumer ST, Tenen DG, Kobayashi S. J Clin Oncol. 2008;26:1182–1184. doi: 10.1200/JCO.2007.14.9039. [DOI] [PubMed] [Google Scholar]
- Engleman JA, Mukohara T, Zejnullahu K, Lifshits E, Borras AM, Gale C-M, Naumov GN, Yeap BY, Jarrell E, Sun J, Tracy S, Zhao X, Heymach JV, Johnson BE, Cantley LC, Janne PA. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. Science. 2007a;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Gale CM, Lifshits E, Gonzales AJ, Shimamura T, Zhoa F, Vincent PW, Naumov GN, Bradner JE, Althaus IW, Gandhi L, Shapiro GI, Nelson JM, Heymach JV, Meyerson M, Wong K-K, Janne PA. Cancer Res. 2007b;67:11924–11932. doi: 10.1158/0008-5472.CAN-07-1885. [DOI] [PubMed] [Google Scholar]
- Ghosh G, Lee AG, Palecek SP. Anal Biochem. 2009;393:205–214. doi: 10.1016/j.ab.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Suzuki T, Fukuhara T, Maemondo M, Kimura Y, Morikawa N, Watanabe H, Saijo Y, Nukiwa T. J Clin Oncol. 2006;24:3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- Kitamura A, Hasoda W, Sasaki E, Mitsudomi T, Yatabe Y. Clin Cancer Res. 2010 doi: 10.1158/1078-0432.CCR-10-0129. doi: 10.1158/1078-0432.CCR-10-0129. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, Kuwano H, Mitsudomi T. Clin Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- Kuang Y, Rogers A, Yeap BY, Wang L, Makrigiorgos M, Vetrand K, Thiede S, Distel RJ, Janne PA. Clin Can Res. 2009;15:2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskin JJ, Sandler AB. Cancer Treat Rev. 2004;30:1–17. doi: 10.1016/j.ctrv.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Li J, Berbeco R, Distel RJ, Janne PA, Wang L, Makrigiorgos MG. Nucleic Acid Research. 2007;35:e84. doi: 10.1093/nar/gkm403. doi:10.1093/nar/gkm403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. N Engl J Med. 2004;50:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Naruse I, Ohmori T, Ao Y, Fukumoto H, Kuroki T, Mori M, Saijo N, Nishio K. Int J. Cancer. 2002;98:310–315. doi: 10.1002/ijc.10173. [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossell R, Moran T, Queralt C, Porta R, Cardenal F, Majem M, Lopez-Vivanco G, Isla D, Provencio M, Insa A, Massuti B, Gonzalez-Larriba JL, Paz-Ares L, Bover I, Garcia-Campelo R, Moreno MA, Catot S, Rolfo C, Reguart N, Palmero R, Sanchez JM, Bastus R, Mayo C, Bertran-Alamillo J, Molina MA, Sanchez JJ, Taron M. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. N Engl J Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Bell DW, Lynch TJ, Haber DA. J. Clin. Oncol. 2007;25:587–595. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- Shepherd FA, Pereira JR, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, Kooten M, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabarbara P. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- Songyang Z, Carraway KL, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Ponder BAJ, Mayer BJ, Cantlet LC. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- Sos ML, Koker M, Weir BA, Hetneck S, Rabinovsky R, Zander T, Seeger JM, Weiss J, Fischer F, Frommolt P, Michel K, Peifer M, Mermel C, Girard L, Peyton M, Gazdar AF, Minna JD, Garraway LA, Kashkar H, Pao W, Meyerson M, Thomas RK. Cancer Research. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-C, Kulp SK, Wang D, Yang C-C, Sargeant AM, Hung J-H, Kashida Y, Yamagichi M, Chang G-D, Chen C-H. Cancer Research. 2008;68:2820–2830. doi: 10.1158/0008-5472.CAN-07-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmore-Payne C, Holden JA, Layfield LJ. Mod. Pathol. 2006;19:634–640. doi: 10.1038/modpathol.3800552. [DOI] [PubMed] [Google Scholar]
- Yarden Y. Eur J Can. 2001;37:S3–S8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- Zhou W, Ercan D, Chen L, Yun C-H, Li D, Capalletti M, Cortot AB, Chiriea L, Iacob RE, Padera R, Engen JR, wong K-K, Eck MJ, Gray NS, Janne PA. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]