Abstract
A navigator-gated 3D spiral cine displacement encoding with stimulated echoes (DENSE) pulse sequence for imaging 3D myocardial mechanics was developed. In addition, previously-described 2D post-processing algorithms including phase unwrapping, tissue tracking, and strain tensor calculation for the left ventricle (LV) were extended to 3D. These 3D methods were evaluated in 5 healthy volunteers, using 2D cine DENSE and historical 3D myocardial tagging as reference standards. With an average scan time of 20.5 ± 5.7 minutes, 3D data sets with a matrix size of 128 × 128 × 22, voxel size of 2.8 × 2.8 × 5.0 mm3, and temporal resolution of 32 ms were obtained with displacement encoding in three orthogonal directions. Mean values for end-systolic mid-ventricular mid-wall radial, circumferential, and longitudinal strain were 0.33 ± 0.10, −0.17 ± 0.02, and −0.16 ± 0.02, respectively. Transmural strain gradients were detected in the radial and circumferential directions, reflecting high spatial resolution. Good agreement by linear correlation and Bland-Altman analysis was achieved when comparing normal strains measured by 2D and 3D cine DENSE. Also, the 3D strains, twist, and torsion results obtained by 3D cine DENSE were in good agreement with historical values measured by 3D myocardial tagging.
Keywords: Three dimensional, DENSE, myocardial mechanics, cardiac function, stimulated echo, strain
INTRODUCTION
Quantitative imaging of myocardial motion and strain is of growing importance. In addition to conventional applications such as ischemia detection (1,2) and evaluation of myocardial mechanics related to cardiac surgery (3–5), newer applications include quantifying mechanical dyssynchrony in heart failure (6,7) and measuring the functional effects of experimental therapies such as stem cells (8). Potential advantages of quantitative methods are that they may improve diagnostic sensitivity and specificity (9), may reduce subjectivity and intra- and inter-observer variability, and may facilitate statistical comparisons of cardiac function between different experimental groups.
While quantitative two-dimensional (2D) imaging is more common than three-dimensional (3D) imaging, motion of the left ventricle (LV) is, in fact, complex and three-dimensional. The myocardial strain tensor has significant components in the radial, circumferential and longitudinal directions, as well as important off-diagonal components, reflecting the 3D contraction, twist, and torsion that are integral to efficient pump function. A technique that is 3D both with respect to spatial coverage and motion measurement is required for a complete assessment of LV motion. Three-dimensional myocardial tagging MRI has previously been used to noninvasively measure the 3D mechanics of the LV in detail (10–13), and these data provide a reference standard against which new methods may be compared. However, strain analysis of 3D tagged MR images is laborious and time consuming due to the need to detect tag lines, rendering the technique impractical for routine use.
Cine (Displacement Encoding with Stimulated Echoes) DENSE is a newer MRI technique for quantitative imaging of myocardial motion (14,15). This technique encodes tissue displacement (typically relative to an end-diastolic reference time) into the phase of the MR image (14). Since displacement is encoded in the phase, the estimation of tissue motion does not require tag detection, and displacement and strain analysis is less time-consuming for DENSE than for conventional tagging. Previously, 2D cine DENSE has been described and validated using 2D myocardial tagging as a reference standard (15). The purposes of the present study were to develop 3D cine DENSE and evaluate this method in healthy volunteers.
METHODS
All scans in this study were performed on a 1.5T MRI system (Avanto, Siemens Medical Solutions, Erlangen, Germany) with a six-channel phased-array radiofrequency (RF) coil. All human imaging was conducted under protocols approved by the institutional review board at the University of Virginia. Informed consent was obtained from all subjects and all imaging was compliant with the Health Insurance Portability and Accountability Act.
Pulse Sequence and Image Reconstruction
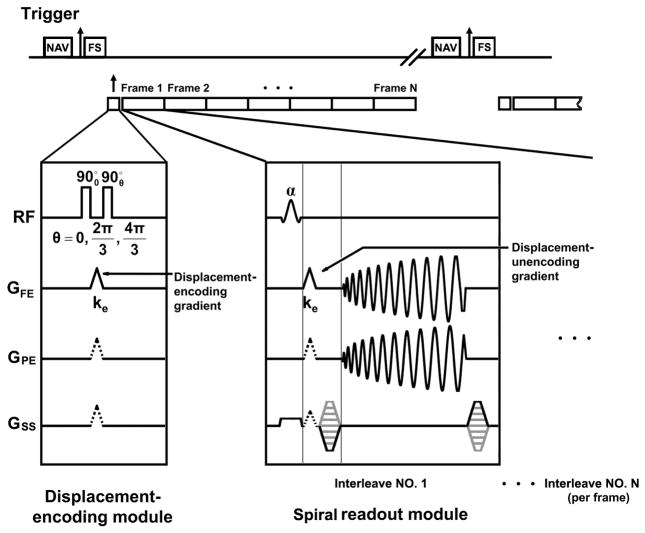
To facilitate volumetric coverage of the entire LV as well as relatively high spatial and temporal resolution compared to previous 3D myocardial tagging studies (10–13), a free-breathing navigator-gated method was employed for 3D cine DENSE data acquisition. Also, to minimize echo time (TE) and increase signal-to-noise ratio (SNR) compared to an echo-planar approach, data were sampled using a spiral k-space trajectory (16). The specific design details of the sequence are shown in Fig. 1. The navigator echo, formed using two orthogonal slice selective 90o and 180o RF pulses (17), was acquired at the end of the cardiac cycle, so as not to interfere with displacement encoding or imaging during the onset of myocardial contraction. The navigator data were used to accept or reject the DENSE data acquired in the subsequent heart beat. Immediately following R-wave detection and just before application of the initial displacement-encoding pulses, a fat suppression RF pulse was applied. The purpose of this pulse was to suppress the contribution of fat to the displacement-encoded magnetization to be stored along the longitudinal axis, resulting in fat suppression for stimulated-echo images throughout the cardiac cycle (18). The displacement-encoding module was applied next, and supported either simple or balanced multi-point displacement encoding (for optimal phase SNR (19)), and two- or three-point phase cycling for suppression of additional artifact-generating echoes (20). The displacement-encoding module was followed by successive (multiphase) application of a readout module, which employed an interleaved stack-of-spirals trajectory to sample the 3D k-space after application of the DENSE unencoding gradients. DENSE displacement-encoding gradients were designed using the shortest possible time, and unencoding gradients were combined with phase-encoding gradients in the slice-select direction to minimize TE. Ramped flip angle was implemented to approximately equalize the SNR at all cardiac phases (21). Two 3D acquisitions with different TEs, where each used a single spiral interleave per 3D partition, were also acquired for field mapping and spiral off-resonance correction (deblurring).
Fig. 1.
Timing diagram for the 3D cine DENSE pulse sequence. A navigator echo is played out before the ECG trigger so as not to interfere with displacement encoding prior to or imaging during the onset of myocardial contraction (marked as the NAV block in the diagram). Immediately after detection of the R wave, a spectrally-selective fat suppression pulse is applied (marked as the FS block in the diagram), which is followed by the displacement-encoding module. A segmented data acquisition, which includes displacement-unencoding gradients and a 3D stack-of-spirals k-space trajectory, is used to sample the displacement-encoded longitudinal magnetization at multiple cardiac phases. In this diagram, the displacement-encoding gradients are applied in the frequency-encoding direction (the gradient waveform shown in the solid line on the GFE axis). However, more generally, displacement encoding can be applied in any direction by also applying the gradients shown in dashed lines on the GPE and/or GSS axes, respectively. Applying gradients in multiple directions is employed for balanced multi-point displacement encoding. In practice, to minimize the echo time, the displacement-unencoding gradients are combined with spatial encoding gradients, but are shown separately in this diagram for clarity.
Displacement-encoded phase images of the stimulated echo were reconstructed online. First, deblurring of the 3D stack-of-spirals data set was performed (22). Specifically, a Fourier transform (FT) was performed in the z direction, and then 2D off-resonance correction was performed partition-by-partition using linear fit coefficients calculated using the field maps (22). Two-dimensional in-plane gridding and FT were applied to the deblurred k-space data to obtain complex 3D image volumes. Cancellation of interference from artifact-generating echoes and isolation of the stimulated echo were achieved by combination of the phase cycling data sets (20). Decoding of the multi-point displacement-encoded data (19), integrated with phase-difference multi-coil reconstruction (23), was performed to obtain the displacement-encoded phase images in the x, y and z directions at each cardiac phase. The corresponding overall magnitude image at each cardiac phase was also calculated using the square root of the sum of the squares of stimulated echo data for all encoding directions and all coils.
Computation of Myocardial Mechanics from 3D Cine DENSE Images
Analysis of myocardial mechanics was performed offline using algorithms developed in MATLAB (Mathworks Inc., Natick, MA, United States). The postprocessing of cine DENSE data typically includes the following steps: segmentation of the LV, phase unwrapping, and tissue tracking (24). Subsequently, the calculation of strain, twist, torsion, and other metrics of motion and function are performed. For the 3D data acquired in this study, previous 2D algorithms (24) were generalized and extended to 3D.
In the present study, 3D segmentation of the LV myocardium was performed manually, using the magnitude-reconstructed images. For phase unwrapping, the spatiotemporal guided-floodfill algorithm developed for 2D cine DENSE (24) was extended to three spatial dimensions plus time, and was applied to the voxels of the segmented myocardium. The unwrapped phase data were then converted to Eulerian displacement by dividing by the displacement encoding frequency, and the 3D Eulerian displacement of each voxel was computed by means of vector addition of the three orthogonal 1D displacement data.
Next, to compute Lagrangian displacement trajectories, the tissue tracking algorithms previously described for 2D data (24) were extended to 3D. Scattered data interpolation using radial basis functions of linear splines was performed to calculate the 3D motion trajectories of LV voxel centers identified at the initial acquired cardiac phase (25). These trajectories were then slightly smoothed using 10th order polynomial functions.
A 3D strain analysis algorithm was also implemented. Briefly, for each voxel of interest (VOI) at the first cardiac phase, N nearest available neighbor voxel positions (N ≤ 12) were identified in the closest three partitions within the myocardial contours. The distance vector matrix V0 at the initial time was formed by assembling the distance vectors pointing from the VOI to its N neighbors. Assuming uniform and isotropic strain within the space specified by the VOI and its neighbors, the deformed distance vector matrix Vn at any cardiac phase n is expressed by Vn = FV0, where F is the 3D deformation gradient tensor. F was solved using singular value decomposition, which is insensitive to ill-conditioned cases. The 3D Lagrangian finite strain tensor E was then calculated as E = (FTF − I)/2, where the superscript T denotes the matrix transpose operation, and I is the identity matrix. For each voxel within the myocardium, this algorithm uses all available neighboring voxels in 3D to calculate the strain tensor. Using this approach, a strain tensor is computed for all myocardial voxels. Since there are typically 3 – 4 voxels across the wall at the first cardiac phase, strain can be estimated at 3 layers.
E was then decomposed into the local radial, circumferential and longitudinal (RCL) coordinate system, defined with respect to the LV contours at the first cardiac phase (10), to obtain normal strains Err, Ecc, and Ell, respectively. The shear strains Erc, Erl, and Ecl were also calculated. The directions of the first, second and third principal strains and the corresponding eigenvalues (E1, E2 and E3) were found after diagonalization of E.
The twist angle of a VOI was defined as the angle between the radial lines connecting the center of the slice to the center of the VOI at end diastole and at each cardiac phase. A positive angle was defined as counterclockwise rotation when viewed from the base of the LV.
Torsion, defined as the normalized gradient of twist in the longitudinal direction, was calculated as the difference between the twist angles of the apex and the base of the LV, normalized by the ratio of the long-axis length to the average short-axis radius of the LV.
For each partition, the myocardium was divided into subendocardial, mid-wall and subepicardial layers, and strain-time curves of each layer were examined separately. In addition, standard LV segments were identified based on the conventions of the American Heart Association (AHA) (26) and were used to characterize the strains and twist angles in different regions at end systole. Paired student t-test and analysis of variance (ANOVA) were performed to evaluate the statistical significance when necessary. In general, one-way ANOVA was performed. However, if the data failed the normality test, then ANOVA on ranks was used.
Evaluation of 3D Cine DENSE in Normal Volunteers
The 3D cine DENSE pulse sequence, reconstruction methods, and mechanics analysis were evaluated in 5 healthy volunteers (all male, age ranged from 21 to 45 years with a mean of 30.0 ± 8.5 years). Imaging parameters included field of view (FOV) = 360 × 360 × 140 mm3, voxel size = 2.8 × 2.8 × 5.0 mm3, ramped flip angles with the last angle = 20°, repetition time (TR) = 16 ms, TE = 1.3 ms, temporal resolution = 32 ms, and cardiac phases = 22. The imaging volume was oriented such that the 3D partition direction was aligned with the long axis of the LV. Fourteen 3D partitions were acquired, and zero-padding to 28 partitions was performed during reconstruction. After FT in the partition direction, 3 partitions at each end of the volume were discarded to avoid aliasing. The resulting 3D image matrix size was 128 × 128 × 22. The balanced four-point displacement encoding strategy was used with a displacement-encoding frequency ke = 0.06 cycles/mm (19). Three-point phase cycling was used to suppress artifact-generating echoes (20). For a single 3D partition corresponding to a single displacement encoding direction and a single phase cycling point, a total of 6 spiral interleaves were acquired. Two interleaves were acquired per heartbeat, so 3 heartbeats were needed to acquire a complete set of spirals. In addition, for each 3D partition, 2 heartbeats were used to acquire data for two low spatial resolution 3D field maps for deblurring. Therefore, the number of navigator-accepted heartbeats to complete the entire 3D acquisition is (3 × 4 × 3 + 2) × 14 = 532. During the scan, the volunteers breathed freely. The navigator echo acceptance window center was placed at end expiration, with a ± 3-mm acceptance window width.
In addition to 3D imaging, 2D breath-hold spiral cine DENSE images (16) with closely-matched parameters were also acquired for comparison. Specifically, 2D short-axis views at basal, mid-ventricular, and apical levels, and long-axis two-chamber and four-chamber views were acquired. The slice thickness was 8 mm. For the short-axis images, the distances between the basal and mid-ventricular levels and between the mid-ventricular and apical levels were 15 mm. Simple three-point displacement encoding was used with an encoding frequency ke = 0.1 cycles/mm (19). For artifact suppression, two- point phase cycling with through-plane dephasing was utilized (27).
Comparison of Strain Results From 2D and 3D Cine DENSE Data
A comparison of the normal strains measured using 2D and 3D cine DENSE was performed for 16 segments of the standard AHA 17-segment model (all segments except the apex cap) (26). For Err and Ecc comparisons, the short-axis 2D cine DENSE images were used, and for the Ell comparison the long-axis 2D cine DENSE data were used. Because just two long-axis views were acquired, the Ell comparisons were made only for 12 segments. The segments were selected from locations in 3D cine DENSE data sets that matched the locations of the 2D cine DENSE slices. Mean values of Err, Ecc, and Ell of these segments were compared using all cardiac phases by means of linear regression. Bland-Altman analysis was also performed.
RESULTS
Pulse Sequence, Image Reconstruction, and Strain Analysis
The total scan time of the navigator-gated 3D cine DENSE acquisition depended on each individual’s heart rate and the navigator efficiency. For the 5 healthy volunteers, the navigator acceptance percentage was 48.0 ± 15.7%, and the mean scan time was 20.5 ± 5.7 minutes. Using navigator gating, signal loss and other artifacts due to respiratory motion were greatly reduced compared to data acquired without any kind of compensation for respiratory motion (data not shown).
Example magnitude-reconstructed and phase-reconstructed 3D cine DENSE images at end-systole from one volunteer are shown in Fig. 2. The upper row contains short-axis images that were reconstructed online, including a magnitude image (Fig. 2a) and phase images with displacement encoded in the horizontal (Fig. 2b), vertical (Fig. 2c), and through-plane directions (Fig. 2d). The lower row contains corresponding images reformatted offline in a long-axis four-chamber view. The average SNR for magnitude-reconstructed images of the mid-ventricular partition at the earliest and the latest cardiac phases for the 5 healthy volunteers were 81.4 ± 12.5 and 34.1 ± 7.7, respectively.
Fig. 2.
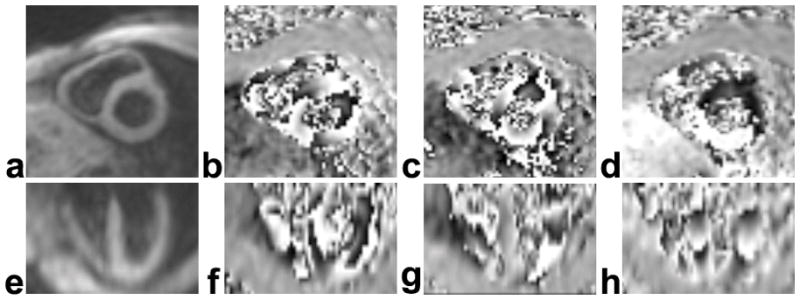
Example magnitude- (a,e) and phase-reconstructed 3D spiral cine DENSE images (b–d, f–h) of the heart at end-systole. The 3D volume was oriented along the principal axes of the LV. Online image reconstruction depicted short-axis planes of the LV, as shown in the magnitude- (a) and phase-reconstructed (b–d) images in the upper row. The lower row contains corresponding data reformatted offline in a long-axis four-chamber view. The images in (b) and (f) were encoded for displacement in the horizontal direction of the short-axis plane, in (c) and (g) were encoded for displacement in the vertical direction of the short-axis plane, and in (d) and (h) were encoded for displacement in the longitudinal direction.
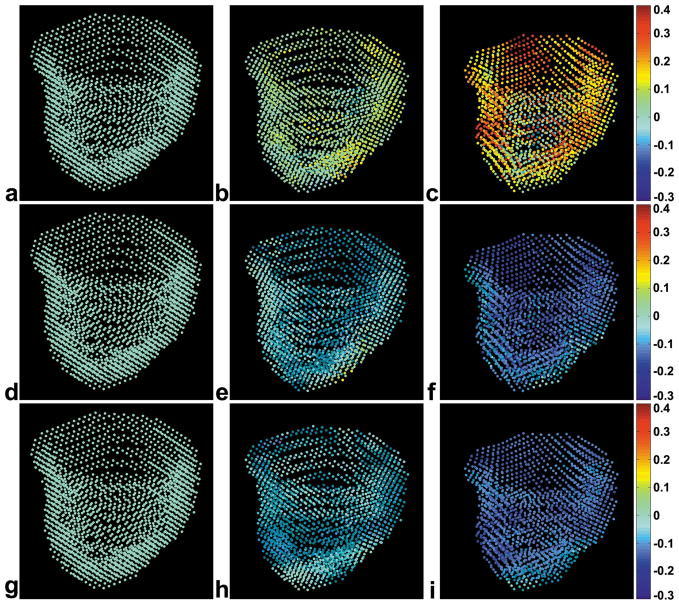
Three-dimensional strain at each voxel within the myocardium was computed for both the local RCL coordinate system and the principal coordinate system. Example high-resolution 3D displacement renderings for one volunteer at end-diastole, mid-systole, and end-systole, with color encoded for Err, Ecc and Ell, are shown in Fig. 3a–3c, Fig. 3d–3f and Fig. 3g–3i, respectively. The spatiotemporal evolution of strain through different cardiac phases can be appreciated as the heart contracts during systole. Multiphase data are also displayed as corresponding 3D displacement movies in online supplemental data.
Fig. 3.
Example 3D voxel-wise displacement and strain displays of the LV at end-diastole (a,d,g), mid-systole (b,e,h) and end-systole (c,f,i) are shown. The location of each dot represents the 3D position of an element of myocardium, and the color of each dot represents the strain value. Radial strain (Err) is shown in the top row (a–c), circumferential strain (Ecc) is shown in the middle row (d–f), and longitudinal strain (Ell) is shown in the bottom row (g–i). The Err values are mostly positive, which represents local tissue stretching in the radial direction. The Ecc and Ell values are mostly negative, representing local tissue shortening in the circumferential and longitudinal directions, respectively. Multiphase data are displayed as corresponding 3D displacement rendering movies in online supplemental data.
3D Cine DENSE-derived Myocardial Mechanics for Normal Volunteers
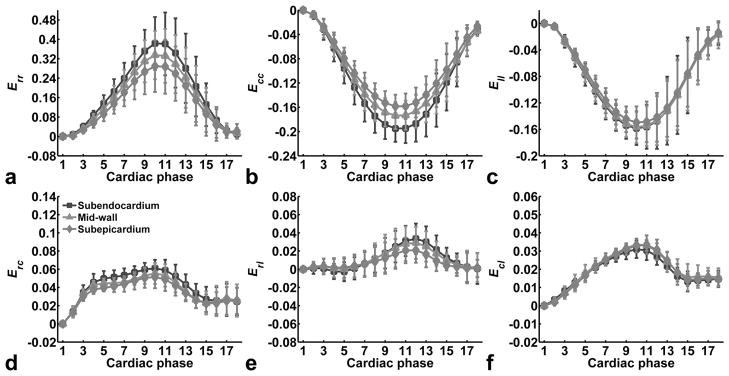
Mean strain-time curves for the normal strains Err, Ecc, and Ell, as well as for the shear strains Erc, Erl and Ecl from the mid-ventricular partitions of the 3D cine DENSE data for 5 volunteers are shown in Fig. 4a–4f, respectively. Some phases near the end of the cardiac cycle were dropped because not all the data sets have 22 cardiac phases, depending on the heart rate of subjects. The high spatial resolution of these images enables the detection of differences in strain between the subendocardial, mid-wall and subepicardial layers. Transmural gradients of strain are evident for Err and Ecc (Fig. 4a and 4b), while no strain gradient is observed for Ell (Fig. 4c). Specifically, ANOVA on ranks indicated significant differences of Err for different layers, with greater Err in the subendocardial layer compared to the subepicardial layers (p < 0.01). Similarly, one-way ANOVA indicated significant differences of Ecc for different layers, with greater shortening in the subendocardium compared to the subepicardium (p < 0.05). In contrast, no differences of Ell were detected for different layers by ANOVA.
Fig. 4.
Mean strain-time curves for the normal strains Err, Ecc, and Ell, and for the shear strains Erc, Erl and Ecl for the mid-ventricular slice for all 5 normal volunteers. Data are shown separately for subendocardial, mid-wall and subepicardial layers, demonstrating the measurement of transmural strain gradients for some strains. Data are shown as mean ± standard deviation.
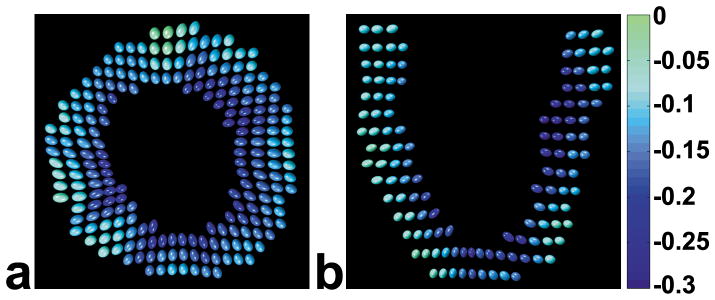
In Fig. 5, 3D ellipsoids are used to visualize the 3D strain tensor as well as tissue displacement. Specifically, the position of each ellipsoid is determined by its displacement from the end diastolic to the end systolic position (as in Fig. 3), and the lengths and orientations of the principal axes of the ellipsoids are determined by the lengths and orientations of the principal strains. In addition, the ellipsoids are color coded according to Ecc. End-systolic ellipsoid strain tensor maps are shown for one volunteer in a mid-ventricular short-axis plane in Fig. 5a, and in a long-axis plane in Fig. 5b. In both views, the direction of the first principal strain generally points toward the center of the LV, and tissue shortening is observed perpendicular to that direction. Transmural variations of strain are evident between the subepicardial and subendocardial layers, with greater radial lengthening and circumferential shortening occurring in the subendocardial compared to the subepicardial layer. Multiphase data are displayed as corresponding ellipsoid movies in online supplemental data.
Fig. 5.
Ellipsoid visualization of 3D end-systolic LV function for one volunteer. In short-axis (a) and long-axis (b) views reconstructed from full volumetric 3D data sets, displaced (relative to their end-diastolic positions) ellipsoids represent both motion and 3D strain. For 3D strain, the lengths and orientations of the principal axes of the ellipsoids are determined by the lengths and orientations of the principal strains. Also, the ellipsoids are color coded according to Ecc. The direction of the first principal strain generally points toward the center of the LV. Transmural gradients of strain are evident, with greater radial lengthening and circumferential shortening occurring in the subendocardium vs the subepicardium. Multiphase data are displayed as corresponding ellipsoid movies in online supplemental data.
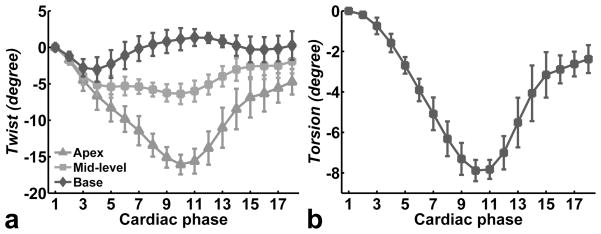
Twist angles for basal, mid-ventricular, and apical levels are plotted vs. time in Fig. 6a. The most apical slice rotates clockwise as viewed from the base (negative angle) during systole. The mid-ventricular slice also rotates clockwise as viewed from the base, but the amount of rotation is less than at the apex. The most basal slice rotates slightly clockwise in early systole and then turns counterclockwise in later systole. All the slices rotate back to their original position at the end of the cardiac cycle. The evolution of LV torsion as a function of time in the cardiac cycle is shown in Fig. 6b.
Fig. 6.
(a) Twist angle as a function of time at the apex, mid-level and base of the LV. Systolic rotation of the apex is clockwise when viewed from the base, whereas the base rotates first clockwise and then counterclockwise. LV torsion as a function of time is shown in (b). Data are from 5 normal volunteers. Data are shown as mean ± standard deviation.
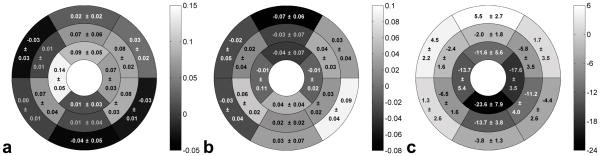
Regional variation in end systolic values of Erc, Erl and twist angle were observed in the normal volunteers. Bull’s eye plots of these parameters employing the AHA 16-segment model display these differences in Fig. 7a–7c, respectively. Specifically, 3D cine DENSE showed that Erc generally increased from base to apex (Fig. 7a), with statistical significance indicated by one-way ANOVA (p < 0.01). Also, Erl was found to be significantly greater in the postero-lateral wall compared to the antero-septum (Fig. 7b), (p < 0.05) when using a t-test to compare the mean of all postero-lateral segments to the mean of all antero-septal segments. For all other strains, no significant differences were detected for different LV regions. For twist angle, one-way ANOVA showed significant differences among basal, mid-ventricular and apical levels (p < 0.01), with greater clockwise twist at the apex compared to the base (Fig. 7c). In addition, regional variations in twist angle can be observed within a slice, especially at the base (Fig. 7c).
Fig. 7.
Bull’s eye plots at end systole for (a) Erc, (b) Erl, and (c) twist angle (in degrees). Data are from 5 normal volunteers. Sectors are gray-scale coded according to the mean values of strains or twist angle, and are marked with mean ± standard deviation. Regional variation was observed for these parameters. Specifically, Erc increased from base to apex, Erl was greater in the postero-lateral wall compared to the antero-septum, and twist angles were higher at the apex compared to the base.
Comparison of Strain Results using 2D and 3D Cine DENSE
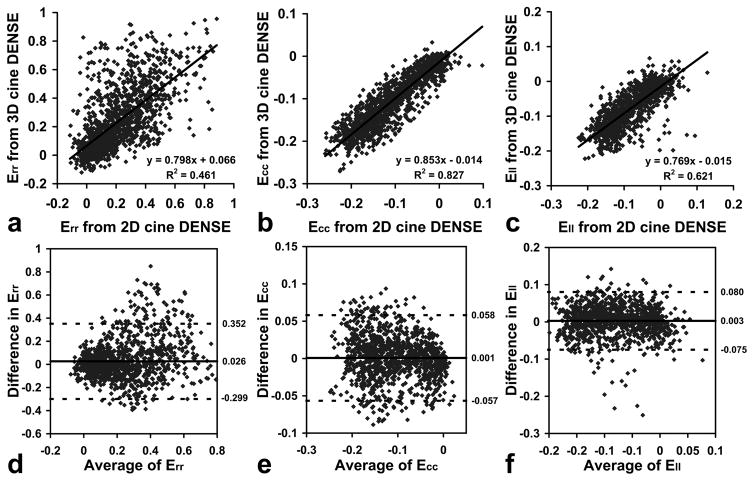
Good linear correlations between normal strains obtained using 3D and 2D cine DENSE data at multiple cardiac phases and in different LV segments were generally found, including a slope of 0.798, an intercept of 0.066 and R2 = 0.461 for Err (Fig. 8a), a slope of 0.853, an intercept of −0.014 and R2 = 0.827 for Ecc (Fig. 8b), and a slope of 0.769, an intercept of −0.015 and R2 = 0.621 for Ell (Fig. 8c). Bland-Altman plots also demonstrate good agreement between 3D and 2D cine DENSE data, where the mean differences between 3D and 2D cine DENSE data were 0.026 for Err (Fig. 8d), 0.001 for Ecc (Fig. 8e), and 0.003 for Ell (Fig. 8f).
Fig. 8.
Scatter plots (a–c) and Bland-Altman plots (d–f) show correlations and agreement for normal strains measured with both 3D and 2D cine DENSE at multiple cardiac phases and in different LV segments (16 segments for Err and Ecc, and 12 segments for Ell). (a–c): Scatter plots and linear regressions for Err, Ecc and Ell, respectively. (d–f): Bland-Altman plots for Err, Ecc and Ell, respectively. Data are from 5 normal volunteers.
DISCUSSION
A 3D cine DENSE sequence was developed that provides volumetric anatomical coverage and encodes for 3D tissue displacement during a navigator-gated free-breathing scan. In addition, post-processing algorithms were developed for tissue tracking and strain analysis of 3D cine DENSE data. These methods were evaluated for the quantitative assessment of LV mechanics in 5 healthy volunteers. Navigator gating was effective for reducing respiratory artifact in the healthy volunteers that participated in this study. The navigator pulses were timed to occur near the end of the cardiac cycle, prior to R-wave detection, so as not to interfere with displacement encoding and data acquisition during the onset of contraction, which may be important to detect in the setting of electrical conduction delays and mechanical dyssynchrony. A stack-of-spirals k-space trajectory provided high SNR and efficient data sampling, while field mapping and off-resonance correction reduced blurring. Balanced displacement encoding also served to increase the SNR of DENSE phase images (19). Fat suppression and three-point phase cycling were other important pulse sequence design features that served to reduce artifacts from unwanted signals. While segmentation of the LV was performed manually in this study and was time consuming, the subsequent calculation of the Lagrangian displacement field (as well as all derivative measurements of mechanics based on displacement such as strain, twist, and torsion) was automatic, without the need for any manual intervention such as tag detection and without applying filters to the raw data such as in harmonic phase (HARP) processing (28). The latter practice leads to a loss of spatial resolution, which, in practice, is often substantial (29). Completely eliminating the need for tag detection without filtering the raw data represents an important advance in quantitative imaging of myocardial mechanics.
The 3D cine DENSE measurements of the mechanics of the normal human LV from this study are in good agreement with detailed measurements previously reported using 3D myocardial tagging. For example, our mean end-systolic mid-ventricular mid-wall Err, Ecc, and Ell values were 0.33 ± 0.10, −0.17 ± 0.02 and −0.16 ± 0.02, respectively. These are similar to the corresponding values of 0.42 ± 0.11, −0.19 ± 0.02 and −0.15 ± 0.02 inferred from the data of Moore et al (10), and 0.18 ± 0.04, −0.20 ± 0.01 and −0.16 ± 0.01 inferred from the data of Young et al (11). The 3D cine DENSE method, using this particular set of parameters, also provided high spatial resolution compared to previous techniques (10–13), albeit using a relatively long scan time. For example, in the present study, Err, Ecc, Ell values at end systole were measured as 0.38 ± 0.10, −0.20 ± 0.02 and − 0.16 ± 0.03, respectively, for the mid-ventricular subendocardial layer, and 0.29 ± 0.11, −0.16 ± 0.02 and −0.15 ± 0.02, respectively, for the mid-ventricular subepicardial layer. This transmural gradient of Ecc across the wall of the heart measured by 3D cine DENSE is in agreement with Moore et al (10). In addition, while Moore et al lacked the spatial resolution to measure a similar transmural gradient of Err (10), our technique was able to detect this variation.
High spatial resolution was also helpful in measuring the shear strains. For example, we measured mean end-systolic mid-ventricular mid-wall Erc, Erl and Ecl values of 0.06 ± 0.01, 0.02 ± 0.02 and 0.03 ± 0.00, respectively. These measurements are similar to the corresponding values of 0.05 ± 0.03, 0.05 ± 0.03 and 0.04 ± 0.02 inferred from the data of Moore et al (10), and 0.03 ± 0.01, 0.01 ± 0.02 and 0.04 ± 0.01 inferred from the data of Young et al (11). Also, our Erc strain-time curve has similar range and shape compared to those reported by Ubbink et al (30) and Bovendeerd et al (31). In contrast to the curves for Erc and Ecl, we observed that Erl develops later and peaks later. This pattern of Erl evolution is consistent with the data reported by Cheng et al (32,33). In addition, our twist value of −6.4 ± 1.4 degrees for the mid-ventricular slice at end systole is similar to the corresponding value of 5.8 ± 1.3 degrees (where the different sign is due to different definitions for a positive rotation angle) inferred from the data of Young et al (11), and 3.8 ± 1.6 degrees reported by Russel et al (34). Also our torsion value at end systole was −7.9 ± 0.5 degree, which is similar to the value of 7.7 ± 1.4 degrees reported by Russel et al (34), where again the different signs are due to different definitions of positive rotation angle. The range and shape of our curves for twist and torsion also match closely with those reported by Russel et al (34). For visualizing the strain tensor maps at high spatial resolution, we used an ellipsoid map. Superquadric glyphs may be even better for strain tensor visualization (35).
To validate the 3D cine DENSE data, we also scanned the same volunteers using short- and long-axis 2D cine DENSE MRI. These comparisons showed excellent correlations for Ecc and Ell, but only a moderate correlation for Err. The moderate correlation for Err occurred because 2D estimates of Err do not account for the curvature of the LV as a function of longitudinal position, whereas 3D estimates do account for this factor. For this reason, it is likely that the 3D estimates of Err are more accurate than the 2D estimates. In addition, the standard deviation for Err is greater than for Ecc and Ell. This may also have contributed to the poorer correlation for Err.
Two limitations of the study were the relatively long scan time and the use of manual segmentation of the LV. However, since the SNR of the images was high (between 34 and 81), methods such as outer volume suppression (18), parallel imaging and sparse sampling may be applicable in the future and may reduce the total scan time by at least a factor of 2. To address the issue of segmentation, we plan to extend a previously described motion-guided segmentation algorithm for 2D cine DENSE (36) to the 3D case. Using these algorithms, analysis of 3D cine DENSE images may be considerably more rapid.
In conclusion, 3D cine DENSE is a promising method for quantitatively measuring 3D myocardial mechanics. A navigator-gated free breathing method was effective for reducing respiratory artifact. Since displacement is encoded into the phase of the isolated stimulated echo, neither tag detection nor filtering the raw data are required, and image analysis is less time consuming while maintaining spatial resolution. Myocardial mechanics measured by 3D cine DENSE are similar to historical 3D myocardial tagging data, the reference standard, and the high spatial resolution may reveal even more detail than tagging such as a transmural gradient of Err. Direct comparison of strain with 2D cine DENSE also demonstrates the accuracy of the 3D cine DENSE data. With future developments in automatic segmentation and faster pulse sequences, 3D cine DENSE may be suitable for routine use.
Supplementary Material
Acknowledgments
This study was supported in part by NIH grant RO1 EB 001763 and Siemens Medical Solutions. We gratefully acknowledge Weitian Chen, PhD for helpful discussions regarding off-resonance correction for the 3D spiral data, and Drew Gilliam, PhD for implementing the spatiotemporal phase unwrapping algorithm for the 3D cine DENSE data. We also appreciate Li Pan, PhD, Steven Shea, PhD and Sven Zuehlsdorff, PhD for their help during the implementation of navigator gating. Lastly, we thank all the volunteers that participated in this study.
Footnotes
Address for reprint requests: Radiology Research, University of Virginia, Room 155 Snyder Building, 480 Ray C Hunt Drive, Charlottesville, VA 22908
References
- 1.Korosoglou G, Futterer S, Humpert PM, Riedle N, Lossnitzer D, Hoerig B, Steen H, Giannitsis E, Osman NF, Katus HA, Korosoglou G, Futterer S, Humpert PM, Riedle N, Lossnitzer D, Hoerig B, Steen H, Giannitsis E, Osman NF, Katus HA. Strain-encoded cardiac MR during high-dose dobutamine stress testing: comparison to cine imaging and to myocardial tagging. J Magn Reson Imaging. 2009;29(5):1053–1061. doi: 10.1002/jmri.21759. [DOI] [PubMed] [Google Scholar]
- 2.Bansal M, Cho GY, Chan J, Leano R, Haluska BA, Marwick TH. Feasibility and accuracy of different techniques of two-dimensional speckle based strain and validation with harmonic phase magnetic resonance imaging. J Am Soc Echocardiogr. 2008;21(12):1318–1325. doi: 10.1016/j.echo.2008.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Setser RM, Smedira NG, Lieber ML, Sabo ED, White RD. Left ventricular torsional mechanics after left ventricular reconstruction surgery for ischemic cardiomyopathy. J Thorac Cardiovasc Surg. 2007;134(4):888–896. doi: 10.1016/j.jtcvs.2007.05.060. [DOI] [PubMed] [Google Scholar]
- 4.Setser RM, White RD, Sturm B, McCarthy PM, Starling RC, Young JB, Kasper J, Buda T, Obuchowski N, Lieber ML. Noninvasive assessment of cardiac mechanics and clinical outcome after partial left ventriculectomy. Ann Thorac Surg. 2003;76(5):1576–1585. doi: 10.1016/s0003-4975(03)00976-7. [DOI] [PubMed] [Google Scholar]
- 5.Kramer CM, Magovern JA, Rogers WJ, Vido D, Savage EB. Reverse remodeling and improved regional function after left ventricular aneurysm repair. J Thorac Cardiovasc Surg. 2002;123(4):700–706. doi: 10.1067/mtc.2002.120710. [DOI] [PubMed] [Google Scholar]
- 6.Bilchick KC, Dimaano V, Wu KC, Helm RH, Weiss RG, Lima JA, Berger RD, Tomaselli GF, Bluemke DA, Halperin HR, Abraham T, Kass DA, Lardo AC. Cardiac magnetic resonance assessment of dyssynchrony and myocardial scar predicts function class improvement following cardiac resynchronization therapy. J Am Coll Cardiol. 2008;1(5):561–568. doi: 10.1016/j.jcmg.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curry CW, Nelson GS, Wyman BT, Declerck J, Talbot M, Berger RD, McVeigh ER, Kass DA. Mechanical dyssynchrony in dilated cardiomyopathy with intraventricular conduction delay as depicted by 3D tagged magnetic resonance imaging. Circulation. 2000;101(1):E2. doi: 10.1161/01.cir.101.1.e2. [DOI] [PubMed] [Google Scholar]
- 8.Amado LC, Schuleri KH, Saliaris AP, Boyle AJ, Helm R, Oskouei B, Centola M, Eneboe V, Young R, Lima JA, Lardo AC, Heldman AW, Hare JM. Multimodality noninvasive imaging demonstrates in vivo cardiac regeneration after mesenchymal stem cell therapy. J Am Coll Cardiol. 2006;48(10):2116–2124. doi: 10.1016/j.jacc.2006.06.073. [DOI] [PubMed] [Google Scholar]
- 9.Gotte MJ, Van Rossum AC, Twisk JWR, Kuijer JPA, Marcus JT, Visser CA. Quantification of regional contractile function after infarction: strain analysis superior to wall thickening analysis in discriminating infarct from remote myocardium. J Am Coll Cardiol. 2001;37(3):808–817. doi: 10.1016/s0735-1097(00)01186-4. [DOI] [PubMed] [Google Scholar]
- 10.Moore CC, Lugo-Olivieri CH, McVeigh ER, Zerhouni EA. Three-dimensional systolic strain patterns in the normal human left ventricle: characterization with tagged MR imaging. Radiology. 2000;214(2):453–466. doi: 10.1148/radiology.214.2.r00fe17453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young AA, Kramer CM, Ferrari VA, Axel L, Reicheck N. Three-dimensional left ventricular deformation in hypertrophic cardiomyophathy. Circulation. 1994;90(2):854–867. doi: 10.1161/01.cir.90.2.854. [DOI] [PubMed] [Google Scholar]
- 12.Ryf S, Spiegel MA, Gerber M, Boesiger P. Myocardial tagging with 3D-CSPAMM. J Magn Reson Imaging. 2002;16(3):320–325. doi: 10.1002/jmri.10145. [DOI] [PubMed] [Google Scholar]
- 13.Rutz AK, Ryf S, Plein S, Boesiger P, Kozerke S. Accelerated whole-heart 3D CSPAMM for myocardial motion quantification. Magn Reson Med. 2008;59(4):755–763. doi: 10.1002/mrm.21363. [DOI] [PubMed] [Google Scholar]
- 14.Aletras AH, Ding S, Balaban RS, Wen H. DENSE: displacement encoding with stimulated echoes in cardiac functional MRI. J Magn Reson. 1999;137(1):247–252. doi: 10.1006/jmre.1998.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D, Gilson WD, Kramer CM, Epstein FH. Myocardial tissue tracking with two-dimensional cine displacement-encoded MRI: development and initial evaluation. Radiology. 2004;230(3):862–871. doi: 10.1148/radiol.2303021213. [DOI] [PubMed] [Google Scholar]
- 16.Zhong X, Spottiswoode BS, Meyer CH, Epstein FH. Two-dimensional spiral cine DENSE; Proceedings of the 15th Annual Meeting of ISMRM; Berlin, Germany. 2007. p. 756. [Google Scholar]
- 17.Taylor AM, Jhooti P, Wiesmann F, Keegan J, Firmin DN, Pennell DJ. MR navigator-echo monitoring of temporal changes in diaphragm position: implications for MR coronary angiography. J Magn Reson Imaging. 1997;7(4):629–636. doi: 10.1002/jmri.1880070404. [DOI] [PubMed] [Google Scholar]
- 18.Ibrahim E, Stuber M, Fahmy AS, bd-Elmoniem KZ, Sasano T, Abraham MR, Osman NF. Real-time MR imaging of myocardial regional function using strain-encoding (SENC) with tissue through-plane motion tracking. J Magn Reson Imaging. 2007;26(6):1461–1470. doi: 10.1002/jmri.21125. [DOI] [PubMed] [Google Scholar]
- 19.Zhong X, Helm PA, Epstein FH. Balanced multi-point displacement encoding for DENSE MRI. Magn Reson Med. 2009;61(4):981–988. doi: 10.1002/mrm.21851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsao J, Laurent D. N-SPAMM for efficient displacement-encoded acquisition in myocardial tagging. Proceedings of the 13th Annual Meeting of ISMRM; Miami, FL, USA. 2005. p. 273. [Google Scholar]
- 21.Stuber M, Spiegel MA, Fischer SE, Scheidegger MB, Danias PG, Pedersen EM, Boesiger P. Single breath-hold slice-following CSPAMM myocardial tagging. Magn Reson Mater Physics Biol Med. 1999;9(1/2):85–91. doi: 10.1007/BF02634597. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Meyer CH. Off-resonance correction for 3D imaging using a stack of spirals trajectory. Proceedings of the 14th Annual Meeting of ISMRM; Seattle, WA, USA. 2006. p. 2966. [Google Scholar]
- 23.Bernstein MA. Phase difference reconstruction. In: Bernstein MA, King KF, Zhou XJ, editors. Handbook of MRI pulse sequences. Burlington, San Diego, London: Elsevier; 2004. pp. 558–567. [Google Scholar]
- 24.Spottiswoode BS, Zhong X, Hess AT, Kramer CM, Meintjes EM, Mayosi BM, Epstein FH. Tracking myocardial motion from cine DENSE images using spatiotemporal phase unwrapping and temporal fitting. IEEE Trans Med Imaging. 2007;26(1):15–30. doi: 10.1109/TMI.2006.884215. [DOI] [PubMed] [Google Scholar]
- 25.Hardy RL. Theory and applications of the multiquadric-biharmonic method: 20 years of discovery. Comput Math Applic. 1990;19(8/9):163–208. [Google Scholar]
- 26.Cerqueira MD, Weissman NJ, Dilsizian V, Jacobs AK, Kaul S, Laskey WK, Pennell DJ, Rumberger JA, Ryan T, Verani MS. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart: a statement for healthcare professionals from the cardiac imaging committee of the council on clinical cardiology of the American Heart Association. Circulation. 2002;105:539–542. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 27.Zhong X, Spottiswoode BS, Cowart EA, Gilson WD, Epstein FH. Selective suppression of artifact-generating echoes in cine DENSE using through-plane dephasing. Magn Reson Med. 2006;56(5):1126–1131. doi: 10.1002/mrm.21058. [DOI] [PubMed] [Google Scholar]
- 28.Osman NF, Kerwin WS, McVeigh ER, Prince JL. Cardiac motion tracking using CINE harmonic phase (HARP) magnetic resonance imaging. Magn Reson Med. 1999;42(6):1048–1060. doi: 10.1002/(sici)1522-2594(199912)42:6<1048::aid-mrm9>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garot J, Bluemke DA, Osman NF, Rochitte CE, McVeigh ER, Zerhouni EA, Prince JL, Lima JA. Fast determination of regional myocardial strain fields from tagged cardiac images using harmonic phase MRI. Circulation. 2000;101(9):981–988. doi: 10.1161/01.cir.101.9.981. [DOI] [PubMed] [Google Scholar]
- 30.Ubbink SWJ, Bovendeerd PHM, Delhaas T, Arts T, van de Vosse FN. Towards model-based analysis of cardiac MR tagging data: relation between left ventricular shear strain and myofiber orientation. Med Image Anal. 2006;10:632–641. doi: 10.1016/j.media.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Bovendeerd PHM, Kroon W, Delhaas T. Determinants of left ventricular shear strain. Am J Physiol Heart Circ Physiol. 2009;297:H1058–H1068. doi: 10.1152/ajpheart.01334.2008. [DOI] [PubMed] [Google Scholar]
- 32.Cheng A, Nguyen TC, Malinowski M, Langer F, Liang D, Daughters GT, Ingels NB, Miller DC. Passive ventricular constraint prevents transmural shear strain progression in left ventricle remodeling. Circulation. 2006;114(suppl I):I79–I86. doi: 10.1161/CIRCULATIONAHA.105.001578. [DOI] [PubMed] [Google Scholar]
- 33.Cheng A, Langer F, Nguyen TC, Malinowski M, Ennis DB, Daughters GT, Ingels NB, Miller DC. Transmural left ventricular shear strain alternations adjacent to and remote from infracted myocardium. J Heart Valve Dis. 2006;15(2):209–218. [PubMed] [Google Scholar]
- 34.Russel IK, Gotte MJ, Kuijer JP, Marcus JT. Regional assessment of left ventricular torsion by CMR tagging. J Cardiovasc Magn Reson. 2008;10:26. doi: 10.1186/1532-429X-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ennis DB, Kindlman G, Rodriguez I, Helm PA, McVeigh ER. Visualization of tensor fields using superquadric glyphs. Magn Reson Med. 2005;53(1):169–176. doi: 10.1002/mrm.20318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spottiswoode BS, Zhong X, Lorenz CH, Mayosi B, Meintjes EM, Epstein FH. Motion guided segmentation for cine DENSE MRI. Med Imaging Ana. 2009;13(1):105–115. doi: 10.1016/j.media.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.