Abstract
Background
Epidermal growth factor receptor (EGFR) mutations in non-small cell lung cancer (NSCLC) predict better outcome to EGFR tyrosine kinase inhibitors (TKIs). The most common mutations are exon 19 deletions (most frequently E746-A750) and L858R point mutation in exon 21. Here, we evaluated the accuracy of novel EGFR mutation specific antibodies in a Japanese cohort with NSCLC and compared to direct DNA sequencing and clinical outcome.
Materials and methods
Immunohistochemistry (IHC) using antibodies specific for the E746-A750 and L858R mutations in EGFR was performed on tissue microarrays of tumors from 70 gefitinib treated NSCLC patients. Extracted DNA was sequenced for mutational analysis of EGFR exons 18 to 21.
Results
DNA sequencing showed EGFR mutations in 41 patients (58.6%), and exon 19 deletions in 18 patients (25.7%), 61% (11/18) had a deletion in the range of E746-A750) and 12 (17.1%) had exon 21 mutations (L858R). IHC showed, for the E746-A750 and L858R mutations, sensitivity (81.8% and 75%), specificity (100%, 96.6%), PPV (100%, 81.8%) and NPV (96.7%, 94.9%). Analysis for objective response rates (ORR) and survival were not correlated to IHC staining, although the combined staining showed non-significant trends towards better overall survival for patients with EGFR mutations.
Conclusions
The mutation specific IHC antibodies have high sensitivity and specificity for pre-defined EFGR mutations and may be suitable for screening for these pre-defined mutations. However, negative IHC results require further mutation analyses prior to excluding EGFR-targeted therapy.
Keywords: EGFR, Biomarkers, Lung Cancer, NSCLC, Mutation
Introduction
Epidermal Growth Factor Receptor (EGFR) mutation status has a critical role in the treatment algorithm of advanced Non-Small Cell Lung Cancer (NSCLC) in 1st, 2nd and 3rd line therapy. EGFR tyrosine kinase inhibitors (TKIs) have recently been shown to be superior to chemotherapy in EGFR mutated NSCLC patients1-4 and gefitinib is approved in Europe for patients harboring EGFR mutations. Therefore, evaluating the EGFR mutation status is thought to be highly important before any therapy decision is undertaken in advanced NSCLC.
Activating mutations in exons 18-21 of EGFR were initially identified in NSCLC patients with clinical response to gefitinib5, 6. These somatic mutations in the kinase domain of EGFR exist in approximately 10%-16% of NSCLC specimens in the United States and Europe3 and 30-50% in Asia7 with 28 distinct mutations8, 9. The exon 19 deletions (including E746-A750) account for ∼45% of the total mutations. Eleven different mutations, resulting in deletion of three to seven amino acids, have been detected in exon 19 and all are centered around the uniformly deleted codons for amino acids 747-749. The second major mutation group observed is the missense mutations found in exon 21 (39-45%), followed by mutations in exon 20 and 18 (6-10%). Among the missense mutations in exon 21, the point mutation, L858R, accounts for 39% of the total mutations in exon 21. Patients with EGFR mutations have a greater response rate to EGFR-TKIs (60-80%) than patients with EGFR wild type tumors or unknown mutation status (10-20%)10. Clinically, there seem to be differences in outcome based on the type of mutations. Patients with exon 19 deletions demonstrate a higher response rate and longer survival with EGFR-TKI therapy than patients with point mutations in exon 2110-13. EGFR mutations tend to be associated with adenocarcinoma, East Asian ethnicity and never smokers.
There are many methods to detect mutations (i.e. DxS EGFR Mutation Kit®, high-resolution melting analysis14-16). However the most common is direct sequencing of the PCR-amplified exon sequences. While these methods provide information about numerous genetic mutations, they are not always available. Most recently, immunohistochemisty (IHC) mutation specific antibodies have been developed for EGFR mutations in exon 19 and 21, and encouraging data has been presented17, 18. In this study we tested the performance of IHC based methodology to define EGFR mutation in a retrospective cohort of 70 Japanese patients and validated the data with DNA sequencing.
Materials and Methods
Patients
The study included 70 patients treated with gefitinib as monotherapy (250 mg day/1) for their recurrent diseases after they had undergone surgery between November 1997 to July 2007 at the Tokyo Medical University Hospital. Their clinical characteristics are detailed in Table 1. All patients were Japanese, aged between 27 and 88 years (mean 59.9 years), 36 (51%) male, 41 (48%) smokers and 29 (41%) never smokers. Progression free survival (PFS) and overall survival (OS) were counted from the time of gefitinib therapy to progression or death accordingly. The median survival time was 15.3 months (range 0.1–77.5 months). The median time to progression was 7.5 months (range 0.1-43.3 months). All patients had histologically confirmed NSCLC (57 adenocarcinoma, 7 squamous cell carcinoma, 4 large cell carcinoma, and 2 other NSCLC) with measurable, locally advanced or metastatic disease, progressing or relapsing after the complete resection. On pathological staging at surgery (TNM Classification of malignant tumors seventh edition19), 11 patients were staged as IA, 10 as IB, 8 as IIA, 3 as IIB, 28 as IIIA, 7 as IIIB and 3 as IV.
Table 1.
Patient characteristics (n = 70)
| Characteristics | n | (%) |
|---|---|---|
| Total | 70 | (100) |
| Median age (range) | 59.9 | (27-88) |
| Median survival time (range) | 15.3 m | (0.1-77.5) |
| Median progressive free survival time (range) | 7.5 m | (0.1-43.3) |
| Gender | ||
| Male: Female | 36 : 34 | |
| Smoking history | ||
| Never | 29 | (41.4) |
| Smoker | 41 | (58.6) |
| Histology | ||
| Adenocarcinoma | 57 | (81.4) |
| Squamous cell carcinoma | 7 | (10.0) |
| Large cell carcinoma | 4 | (5.7) |
| Other NSCLC | 2 | (2.9) |
| pTNM stage* at surgery | ||
| IA | 11 | (15.7) |
| IB | 10 | (14.3) |
| IIA | 8 | (11.4) |
| IIB | 3 | (4.3) |
| IIIA | 28 | (40) |
| IIIB | 7 | (10) |
| IV | 3 | (4.3) |
| Type of resection | ||
| Bi-lobectomy | 3 | (4.3) |
| Lobectomy | 59 | (84.3) |
| Wedge | 7 | (10) |
| Tracheoplasty | 1 | (1.4) |
| EGFR mutation status | ||
| Mutation type | 41 | (58.6) |
| exon18 | 1 | (1.4) |
| exon19 | 18 | (25.7) |
| exon20 | 18 | (25.7) |
| exon21 | 12 | (17.1) |
| Wild type | 29 | (41.4) |
TNM classification 7th edition19
Assessment of tumor regression was conducted according to the response evaluation criteria in solid tumors criteria (RECIST) 20. Tumor response for gefitinib therapy was assessed by CT scan, with a confirmatory evaluation repeated in patients with complete response (CR), partial response (PR), stable disease (SD) and progressive disease (PD) at least 4 weeks after the initial determination of response. During treatment, assessments were performed every 4 weeks for the first 4 months and then every 8 weeks until disease progression. The RECIST20 recommended that duration of SD should specify the minimal time interval required between two measurements for determination of stable disease. Therefore disease control rate (DCR = CR + PR + SD) was evaluated at 12 weeks. Of the studied patients, 6 (9%) received gefitinib as their 1st systemic anticancer therapy after relapse, 33 (47%) received gefitinib as 2nd line therapy and 31 (44%) patients as ≥3rd line. For objective response rate (ORR) calculation, we considered only patients who were treated by gefitinib for at least four weeks (N=62).
Tissue Microarrays
Tissue microarrays (TMA) were constructed from the primary resected tumors according to the procedure described in previous reports 21, 22. Briefly, the most representative tumor areas were carefully selected and marked on the H&E-stained slide to construct the microarrays, and the TMA were assembled using a tissue arraying instrument (J.M and T.N, Department of Anatomic Pathology, Tokyo Medical University Hospital, Tokyo, Japan). Samples of the specimens were routinely obtained by collecting three replicate core samples of the tumor (core size of 1.2 mm) from different areas. Normal liver tissues were used for control and slide orientation purposes.
DNA sequencing for detection of mutations in the EGFR gene
Genomic DNA was extracted from tumor surgical specimens in formalin-fixed, paraffin-embedded blocks. Exons 18-21 of the TK domain in the EGFR gene were sequenced with the primers for exon 18: CCTTGTCTCTGTGTTCTTGT (Forward), CTGCGGCCCAGCCCAGAGGC (Reverse), exon 19: CATGTGGCACCATCTCACA (Forward), CCACACAGCAAAGCAGAAAC (Reverse), exon 20: CTCCCTCCAGGAAGCCTACGTGAT (Forward), TTTGCGATCTGCACACACCA (Reverse), exon 21: CAGGGTCTTCTCTGTTTCAG (Forward), TAAAGCCACCTCCTTACTTT (Reverse). PCR consisted of 30 cycles with an annealing temperature of 72°C in a 4.5 μl reaction mixture containing 0.125 μl TaKaRa Ex Taq HS, 100 ng Template DNA, and 0.5 μl of each primer (SRL, Tachikawa-shi, Tokyo, Japan). PCR products were then sequenced with a 3100 Genetic Analyzer (Applied Biosystems, Chuou-ku, Tokyo, Japan).
EGFR mutation specific IHC staining
Serial 4-μm-thick tissue sections were cut from the TMA for IHC based EGFR exon 19 and exon 21 mutation analysis. Histological classification was determined on HE stained sections according to the World Health Organization criteria23. The slides were baked at 55°C overnight, then deparaffinized in xylene and rehydrated through a graded series of ethanol concentrations. Slides were labeled with antibody and protocol-specific bar-codes and loaded into a Benchmark® XT (Ventana Medical Systems, Inc, Tucson, AZ) automated stainer. The slides were treated with Standard Cell Conditioning 1 for 60 minutes (VMSI). The primary antibodies, EGFR E746-A750del (catalog number 2085, Cell Signaling Technologies (CST), Danvers, MA) and EGFR L858R (catalog number 3197, CST), were both diluted 1:100 with SignalStain® antibody diluent (CST) and manually applied to the TMA slides. The slides were then incubated at 37°C for 1 hour. The UltraviewTM Universal DAB detection kit (VMSI) was used with an extra washing step selected. The slides were counterstained with Hematoxylin and Bluing at 4 min each. The slides were washed with mild soapy water and then dehydrated in Ethyl Alcohol (Surgipath®, Richmond, IL) and Xylene (Surgipath®) before applying coverslips.
Scoring methodology
IHC staining was scored according to the University of Colorado IHC H-score criteria with assessment of staining intensity (0-4) multiplied by the percentage of positive cells (0-100%) for each intensity for a final IHC score of 0-400. Tumors with H score ≥ 20 (i.e. 5%) were interpreted as positive and tumors with an H score < 20 (5%) were interpreted as negative. IHC staining overview was performed by a pathologist (M.P) and a trained reader (Y.K) at the University of Colorado Cancer Center. The final score per patient was calculated by the two readers using the core with the maximum value for each patient.
Statistic analysis
Standard descriptive statistics and Kaplan-Meier survival curves were used. Differences in survival were determined by the log-rank test. Proportions were compared by means of χ2 analysis. The differences were considered to be statistically significant when the p value < 0.05. SPSS for Windows Version 12.0 (SPSS Inc., Chicago, IL) was also used to calculate the sensitivity, specificity and survival data.
Results
For 70 NSCLC patients, the median interval between surgery and gefitinib treatment for the recurrence disease was 14.2 months (range 3 to 82.9 months). The majority of patients (81.4%) had adenocarcinoma histology. DNA sequencing mutation analysis showed EGFR mutations in 41 patients (58.6%): 18 patients (25.7%) had an exon 19 deletion, 18 patients (25.7%) had an exon 20 mutation, 12 patients (17.1%) had an exon 21 mutation (6 had 2573 T>K and 6 had 2573 T>G), and one patient (1.4%) had an exon 18 mutation. (Table 1) Among the 18 patients with exon 19 deletion, 7 had a deletion other than E746-A750. (Table 2-1) Overlapping mutations existed between exon 19 and exon 20 (4 cases), exon 20 and exon 21 (3 cases) and exon 18 and exon 20 (1 case).
Table 2.
| Table 2-1. IHC and direct sequencing details (N=70) | ||||||
|---|---|---|---|---|---|---|
| Sample No. | IHC | H score max | Direct sequencing | Nucleotide number and sequence | Amino-acid change | |
| Exon 19 deletions | 56 | Del | 400 | Del | 2236-2250del15 | del E746–A750 |
| 49 | Del | 280 | Del | 2235-2249del15 | del E746–A750 | |
| 41 | Del | 230 | Del | 2235-2249del15 | del E746–A750 | |
| 46 | Del | 205 | Del | 2235-2249del15 | del E746–A750 | |
| 69 | Del | 190 | Del | 2235-2249del15 | del E746–A750 | |
| 16 | Del | 110 | Del | 2235-2249del15 | del E746–A750 | |
| 3 | Del | 100 | Del | 2235-2249del15 | del E746–A750 | |
| 43 | Del | 100 | Del | 2235-2249del15 | del E746–A750 | |
| 61 | Del | 100 | Del | 2235-2249del15 | del E746–A750 | |
| 27 | Wt | 5 | Del | 2235-2249del15 | del E746–A750 | |
| 4 | Wt | 0 | Del | 2236-2250del15 | del E746–A750 | |
| 42 | Wt | 0 | Del | 2239-2253del15* | delL747-T751 | |
| 50 | Wt | 0 | Del | 2239-2253del15* | delL747-T751 | |
| 67 | Wt | 0 | Del | 2239-2253del15* | delL747-T751 | |
| 24 | Wt | 0 | Del | 2240-2257del18* | del L747-P753insS | |
| 40 | Wt | 0 | Del | 2240-2257del18* | del L747-P753insS | |
| 55 | Wt | 0 | Del | 2240-2257del18* | del L747-P753insS | |
| 18 | Wt | 0 | Del | 2253-2276del24* | S752-I759 | |
| Exon 21 mutations | 32 | L858R | 370 | L858R or Wt | 2573 T>K | L858R or Wt |
| 68 | L858R | 360 | L858R | 2573 T>G | L858R | |
| 1 | L858R | 240 | L858R | 2573 T>G | L858R | |
| 66 | L858R | 240 | L858R | 2573 T>G | L858R | |
| 51 | L858R | 230 | L858R or Wt | 2573 T>K | L858R or Wt | |
| 31 | L858R | 200 | L858R or Wt | 2573 T>K | L858R or Wt | |
| 12 | L858R | 100 | L858R or Wt | 2573 T>K | L858R or Wt | |
| 21 | L858R | 80 | L858R | 2573 T>G | L858R | |
| 60 | L858R | 20 | L858R | 2573 T>G | L858R | |
| 65 | L858R | 215 | Wt | |||
| 58 | L858R | 180 | Wt | |||
| 62 | Wt | 0 | L858R | 2573 T>G | L858R | |
| 5 | Wt | 0 | L858R or Wt | 2573 T>K | L858R or Wt | |
| 33 | Wt | 0 | L858R or Wt | 2573 T>K | L858R or Wt | |
| Table 2-2. Immunoreactivity of E746-A750 specific and L858R specific antibodies | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample No. | IHC | Amino acid change | H score | Staining area (%) per intensity | ||||||
| max | mean | 0 | 1+ | 2+ | 3+ | 4+ | ||||
| E746-A750 specific antibody | 56 | Del | del E746–A750 | 400 | 400 | 0 | 0 | 0 | 0 | 100 |
| 49 | Del | del E746–A750 | 280 | 225 | 0 | 4 | 68 | 28 | 0 | |
| 41 | Del | del E746–A750 | 230 | 116.7 | 50 | 0 | 33 | 17 | 0 | |
| 46 | Del | del E746–A750 | 205 | 198.3 | 2 | 0 | 96 | 2 | 0 | |
| 69 | Del | del E746–A750 | 190 | 163.3 | 0 | 67 | 13 | 10 | 10 | |
| 16 | Del | del E746–A750 | 110 | 105 | 0 | 95 | 5 | 0 | 0 | |
| 3 | Del | del E746–A750 | 100 | 36.7 | 65 | 33 | 2 | 0 | 0 | |
| 43 | Del | del E746–A750 | 100 | 33.3 | 67 | 33 | 0 | 0 | 0 | |
| 61 | Del | del E746–A750 | 100 | 63.3 | 37 | 63 | 0 | 0 | 0 | |
| 27 | Wt (FN) | del E746–A750 | 5 | 3.33 | 98 | 2 | 0 | 0 | 0 | |
| L858R specific antibody | 32 | L858R | L858R or Wt | 370 | 350 | 0 | 0 | 0 | 50 | 50 |
| 68 | L858R | L858R | 360 | 256.7 | 0 | 0 | 63 | 17 | 20 | |
| 1 | L858R | L858R | 240 | 138.3 | 10 | 62 | 8 | 20 | 0 | |
| 66 | L858R | L858R | 240 | 230 | 0 | 0 | 70 | 30 | 0 | |
| 51 | L858R | L858R or Wt | 230 | 173.3 | 0 | 37 | 53 | 10 | 0 | |
| 31 | L858R | L858R or Wt | 200 | 133.3 | 0 | 67 | 33 | 0 | 0 | |
| 12 | L858R | L858R or Wt | 100 | 66.7 | 33 | 67 | 0 | 0 | 0 | |
| 21 | L858R | L858R | 80 | 36.7 | 63 | 37 | 0 | 0 | 0 | |
| 60 | L858R | L858R | 20 | 16.7 | 83 | 17 | 0 | 0 | 0 | |
| 65 | L858R (FP) | Wt | 215 | 138.3 | 0 | 0 | 94 | 3 | 3 | |
| 58 | L585R (FP) | Wt | 180 | 146.7 | 0 | 55 | 43 | 2 | 0 | |
other deletions; L858R = 2573 T>G, and 2573 T>K (K = G or T)
EGFR mutation specific antibody IHC staining
Expression of EGFR, both E746-A750 deleted and L858R point mutated, was evaluated in all 70 patients by immunohistochemisty. The mutation specific antibodies have distinct immunoreactivity for the plasma membrane of the tumor cells, with some weak cytoplasmic staining, as presented in Figure 1. Table 2-1 summarizes all the cases with either positive direct sequencing or positive IHC staining. In addition, as tumors tend to be heterogeneous in their mutational status, table 2-2 explore staining intensity over the examined core.
Figure 1.
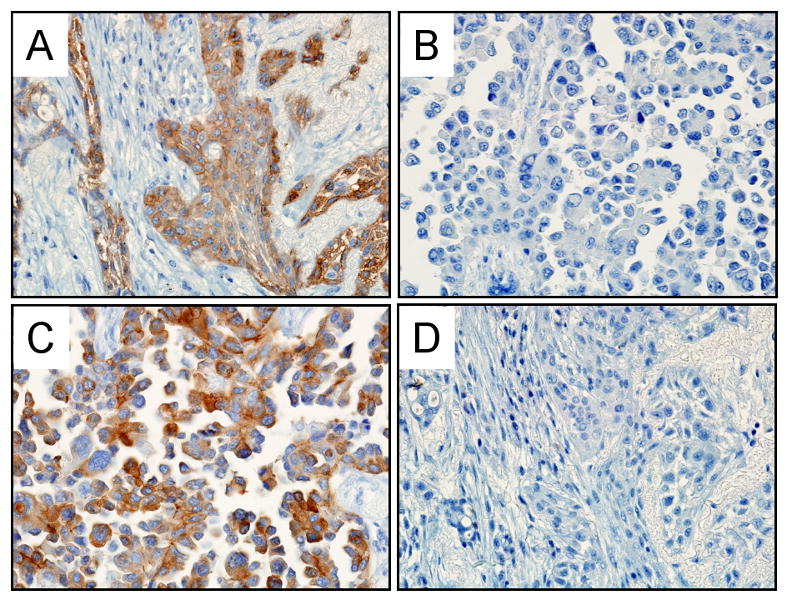
Representative pictures of EGFR expression by immunohistochemical staining (40× magnification). A. DNA sequence E746-A750 deletion specimen stained with the E746-A750 specific antibody. Immunoreactivity is positive in the tumor cells but absent in non-tumor cells. B. DNA sequence L858R point mutation case. The tumor cells show no immunoreactivity with the E746-A750 specific antibody. C. L858R point mutation tumor showing tumor cells positive for L858R specific antibody. D. E746-A750 mutation case displaying no immunoreactivity in the tumor or non-tumor cells with the L858R specific antibody.
Exon 19 deletion
IHC staining with the EGFR exon 19 E746-A750 deletion specific antibody was seen in 9 out of 11 patients (Table 2 & 3). Since the exon 19 specific antibody was designed specifically for the E746-A750 deletion we considered only those 11 patients that had DNA sequence verified deletions, 2236-2250 or 2235-2249, that resulted in E746-A750 amino acid deletions as sequence positive. We observed a discrepancy relative to DNA sequencing in 2 cases (#4 and #27) (Table 2 & 3). Case #27 had IHC score of 5, but was scored as negative due to limited staining and the predetermined cut-off of 20. Case #4 was completely negative by IHC. The sensitivity, specificity, positive predictive values (PPV) and the negative predictive values (NPV) to detect exon 19 E746-A750 deletions were 81.8%, 100%, 100%, and 96.7% respectively (Table 4). If also the other 7 deletions beyond the pre-defined antibody target were considered the sensitivity, specificity, PPV and NPV were 50%, 100%, 100%, and 85.2% respectively (Table 4). The heterogeneity of staining with the mutation specific antibodies was evaluated. With regard to the E746-A750 specific antibody the areas of poorly differentiated cells were stained more strongly than well differentiated areas.
Table 3.
IHC for E746-A750 and L858R (N=70)
| DNA sequencing for exon 19 | |||||
|---|---|---|---|---|---|
| E746-A750 | Other deletions | Wt | |||
| IHC | Positive | 9 | 0 | 0 | 9 |
| exon 19 | Negative | 2 | 7 | 52 | 61 |
| 11 | 7 | 52 | 70 | ||
| DNA sequencing for exon 21 | ||||
|---|---|---|---|---|
| L858R* | Wt | |||
| IHC | Positive | 9 | 2 | 11 |
| exon 21 | Negative | 3 | 56 | 59 |
| 12 | 58 | 70 | ||
L858R = 2573 T>G, and 2573 T>K (K = G or T)
Table 4.
Detection capabilities for the EGFR mutation-specific antibodies.
| Sensitivity | Specificity | Accuracy | PPV | NPV | |
|---|---|---|---|---|---|
| E746-A750 | 81.8% | 100% | 97.1% | 100% | 96.7% |
| All exon 19 mt | 50% | 100% | 87.1% | 100% | 85.2% |
| L858R* | 75% | 96.6% | 92.9% | 81.8% | 94.9% |
| E746-A750 + L858R | 78.3% | 95.7% | 90% | 90% | 90% |
| Any mutation of All Ex19 mt + L858R | 43.9% | 99.9% | 64.3% | 90% | 54% |
PPV=Positive predictive values, NPV=Negative predictive values
Including 2573 T>K (K=G or T)
Exon 21 L858R
Out of the 12 EGFR L858R mutated cases detected by DNA sequencing, 11 were seen by the EGFR L858R mutation specific antibody (Table 2 & 3). DNA sequencing of exon 21 showed that nucleotide 2573 was mutated from T to G in 6 patients but in 6 other patients it was equivocal if there was a mutation because both a T and G were seen at this position, thus labeled K. However, all the 12 cases were considered as L858R mutation based on DNA sequencing since it was not possible the discriminate the 2573K cases. Thus, for exon 21 specific L858R mutation, 9 out of 12 patients with EGFR L858R mutation were stained positive by IHC. However, two other cases showed positive staining discordant with the DNA sequencing (case #58, 65, Table 2 & 3) and 3 cases that showed no IHC staining were discordant with DNA sequencing (case #5, 33, 62). Therefore the sensitivity, specificity, PPV, and NPV of the L858R antibody are 75%, 96.6%, 81.8%, and 94.9% respectively (Table 4). However, the 2 cases of the discordant negative cases (#5, #33, Table 2) had a direct sequencing of 2573 T>K. Therefore, based on the negative IHC staining, if one retrospectively classified those patients as negative for EGFR mutation, the sensitivity, specificity, PPV, and NPV of the L858R antibody are 90%, 96.7%, 81.8%, and 98.3% respectively. The heterogeneity of staining with the L858R specific antibody, poorly differentiated areas, such as the solid central area, was more strongly than well differentiated areas, such as those showing lepidic growth.
Focusing on the clinical need to detect any of the common EGFR mutations, we calculated the yield of double staining (E746-A750 and L858R) and found a 78.3% sensitivity and 95.7% specificity for detecting the pre-defined EGFR mutations and 43.9% and 99.9% to detect any exon19 deletion and L858R mutation (Table 4).
Clinical outcome
Sixty two patients were treated by gefitinib for at least 4 weeks and were considered for outcome analysis (DCR) at 12 weeks. Among them, one (1.6%) had CR, 14 (22.6%) had PR, 38 (61.3%) had SD as the best response and 9 (14.5%) had PD. Overall Objective Response rate (ORR) was 24.2% at 8 weeks. Analyzing overall and progression-free survival (Figure 2) by positive or negative IHC staining, for either of the antibodies, produced a non-significant trend toward a favorable outcome with positive staining. Overall 2-year and 5-year survivals with expression of dual staining were 61.7% and 36.2% versus 22.9% and 15.7% in the negative IHC group (NS) respectively. Two years PFS rate was 47.0% versus 24.4% respectively (NS).
Figure 2.
Overall survival (A) and progression free survival (B) based on dual mutation specific IHC staining for E746-A750 and L858R.
In order to evaluate the contribution of the technology chosen, direct sequencing or mutation specific IHC, for making clinical decisions, we calculated the clinical outcome for each of the methodologies relative to each of the relevant mutations (Table 5). When guided by DNA sequencing, any of the mutations (all exon 19 deletions, E746-A750 deletions, and L858R mutations) had significantly better ORR and DCR compared to wild type cases (Table 5). On the other hand, if the decision would have been made by the mutation specific IHC technique alone, no significant differences between the positive and negative cases would be found. The same holds true when considering the combination of the two antibodies. This is because of high ORR among cases with negative staining, which is most likely related to other types of EGFR mutations such as in exon 18, exon 20 or in exon 19 but outside the E746-A750 range. Importantly enough, in this cohort, patients harboring EGFR mutations in exon 20 showed a favorable outcome like patients harboring exon 19 mutation or L858R (Table 5).
Table 5.
Outcome (OS, PFS, RR, DCR) to gefitinib therapy per mutation status and technique (N=62)
| Methods | Mutation status | Total no. pts* | OS (month) | PFS (month) | ORR** (n, %) | P-value | DCR*** (n, %) | P-value |
|---|---|---|---|---|---|---|---|---|
| DNA sequencing | Exon 19 all deletions | Mt (n=18) | 38.6 | 12.1 | 8 (44) | 16 (89) | ||
| E746-A750 | Mt (n=11) | 40.8 | 39.8 | 4 (36) | 10 (91) | |||
| L858R (including 2573 T>K) | Mt (n=11) | 34.4 | 19.9 | 4 (36) | 10 (91) | |||
| Exon 18 | Mt (n=1) | 78 | 1 (100) | 1 (100) | ||||
| Exon 20 | Mt (n=15) | 25 | 23.5 | 2 (13) | 14 (78) | |||
| EGFR WT | Wt (n=25) | 12.9 | 7.3 | 1 (4) | 0.009 | 16 (64) | 0.057 | |
| Specific mutation IHC | E746-A750 | Mt (n=9) | 22.6 | 11.6 | 2 (22) | 8 (89) | ||
| Wt (n=53) | 22.1 | 11.6 | 13 (25) | 0.882 | 41 (78) | 0.691 | ||
| L858R | Mt (n=9) | 37.8 | 19.9 | 3 (33) | 8 (89) | |||
| Wt (n=53) | 16.8 | 9.2 | 12 (23) | 0.488 | 42 (79) | 0.499 | ||
| E746-A750 & L858R | Mt (n=18) | 34.4 | 19.9 | 5 (28) | 16 (89) | |||
| Wt (n=44) | 15.7 | 8.9 | 10 (23) | 0.673 | 33 (75) | 0.227 |
Patients with mutation
ORR: Responder= CR+PR
Disease control rate; CR+PR+SD ≥ 12 weeks
P values are between EGFR WT and each of the mutated subtype per section.
Discussion
In this study we evaluated by IHC the sensitivity, the specificity and the association to clinical outcome of novel mutation specific antibodies for exon 19 (E746-A750) and exon 21 (L858R) and compared it to DNA sequencing in a Japanese cohort with advanced NSCLC. The two EGFR mutations represent the majority of EGFR mutations in NSCLC24. The study demonstrated a high specificity (>96%) and sensitivity (>75%; Table 4) to the predetermined mutations, which are targeted by these antibodies.
Recently, Yamamoto, et al25 reported 569 mutations in 2880 lung cancer patients and distribution of EGFR mutations was as follows: 48.2% of exon 19, 42.7% of exon 21, 3.7% of exon 20, and 3.2% of exon 18. Together, exon 19 deletion and exon 21 mutations accounted for about 90% of all EGFR mutations in NSCLC. In our cohort, 27% of the patients had other mutations than these two mutation types, mainly (26%) exon 20 mutation. However, while we know that the E746-A750 and L858R EGFR mutations in exon 19 and 21 are activating mutations sensitive to EGFR TKIs, it is not known at this time whether the other – non-canonical- mutations are sensitive to EGFR TKIs. In this study, however, we showed that patients harboring EGFR mutations in exon 20, also had a favorable outcome like patients harboring exon 19 mutation or L858R (Table 5).
The mutation specific antibodies we used in this IHC study were designed to detect specific mutations, i.e. E746-A750 deletions and L858R point mutations. Indeed, the sensitivity and the specificity of IHC to detect E746-A750 deletion were 81.8% and 100% and 75% and 96.6% for L858R respectively (or 90% and 96.7% for L858R if considering the 2 discussed cases as true negative). However, the IHC detection of exon 19 mutation covered only 61 % of all the exon 19 cases in our cohort. If screening was performed by IHC for both L858R and E746-A750 in our cohort, 18/70 (26%) of the patients would have been diagnosed as having EGFR mutation, which represents only 44% of the current EGFR mutated cases. Therefore, for clinical decisions, since any existence of EGFR mutation might be important, negative IHC will require a validation by other technique before a clinical decision can be made. Previous studies that used these antibodies17, 18 came to similar conclusions. Yu J et al17, who reported overall sensitivity to detect any EGFR mutation in 340 tumors of 92% and specificity of 99%, however the rate of other EGFR mutations was relatively low in their cohort. Brevet et al18 just recently reported sensitivity of 85% to detect exon 19 deletion and of 95% to detect L858R point mutations in exon 21. Our data supports these studies, and was the first to associates this method with the clinical outcome, where positive antibody specific IHC staining, for either of the antibodies, was associated with a non-significant trend toward a favorable outcome with positive staining (figure 2).
IHC is a well-established method routinely applied in lung cancer diagnosis in clinical laboratories. IHC also leads for the simultaneous analysis of expression level of other proteins or protein modifications. IHC also allows for the analysis of small tissue samples or cytological samples (body fluids, bronchial washings, and fine needle aspirates samples) as well as circulating tumor cells 26. Thus, the detection of E746-A750 and L858R EGFR protein specific mutations by IHC would be a valuable addition to the current protocols used in the diagnosis and treatment of lung cancer and particularly useful for mass screening of NSCLC patients for EGFR mutations.
Recently many studies have reported simple and highly sensitive non-sequencing methods for detecting EGFR mutations in small tumor tissue samples 26-29 in addition to cytological specimens (pleural effusion, aspiration cytological specimens) 30-34. These methods are reported to be quick and convenient. However these methods have limitations, for example, many, if not all, biopsy specimens will also contain non-cancerous lesions (e.g. scar lesion, inflammation, peripheral pre cancerous lesions, etc). The aforementioned methods can not distinguish between the cancerous and non-cancerous material unless the tumor cells are microdissected, while the IHC can be viewed by a pathologist, who discriminates the tissues and assess the cancer cells per se. The discrepant results between IHC and DNA sequencing found in the current study may result from differences in sample size of the TMA cores versus the large specimens used for DNA sequencing. In our study we used TMAs, while previous studies17, 18 specimens from the whole paraffin embedded blocks were used. Thus, a much smaller quantity of tumor specimen was used in our study. EGFR mutations may not be homogeneously dispersed throughout the tumor so that even though a core contains tumor cells it may randomly only include wild type EGFR and miss mutation positive tumor cells compared to whole section assessment.
The limitation of our study pertaining to the clinical outcome analysis, may be due to the heterogeneity of the studied clinical cohort, with regard to type of disease recurrence and line gefitinib treatment. Therefore, even though we have analyzed the outcome per line of therapy (not shown), still, this is not an optimal cohort for analyzing the predictive performance of IHC to the clinical outcome. Likewise, we had a high rate of exon 20 (18/70; 26%) mutations, which decreased the power of the test to predict the clinical outcome, as patients harboring exon 20 EGFR mutation showed a better response to gefitinib in our cohort.
In summary, the accuracy of the used mutation specific IHC for E746-A750 and L858R is high for pre-defined EGFR mutations. We believe that IHC is suitable for screening NSCLC patients for existence of theses pre-defined EGFR mutations, but negative results should be validated further before excluding them from EGFR related therapy. Such IHC method might provide faster and wider test availability, requires less tumor material, allow histologic evaluation and is more cost differential for a positive test
Acknowledgments
We are grateful to members of the Hirsch laboratory for helpful suggestions. We thank Cindy Tran for expert technical assistance with IHC staining.
Antibodies were generously provided by Cell Signaling Technologies, Danvers, MA.
N.P and C.M: Supported by IASLC young investigator award (NP, CM) and by the Fulbright-Schneider Yehuda Danon United State – Israel Education foundation (NP). Supported by NIH/ Lung SPORE grant P50-CA 058187.
Footnotes
Disclosure: F.H: Consultant/Advisory Boards: Astra Zeneca, Roche, Lilly, Pfizer, Boehringer-Ingelheim, Merck Serono, Ventana-Roche, Glasxo Smith Kline, BMS/Imclone, Syndax; Research Funding: OSI, Genentech, AstraZeneca, Merck (USA), Syndax, Ventana-Roche; Patent: EGFR FISH as a predictive marker for EGFR Inhibitors.
References
- 1.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Tamura K, Okamoto I, Kashii T, et al. Multicentre prospective phase II trial of gefitinib for advanced non-small cell lung cancer with epidermal growth factor receptor mutations: results of the West Japan Thoracic Oncology Group trial (WJTOG0403) Br J Cancer. 2008;98:907–14. doi: 10.1038/sj.bjc.6604249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–67. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch FR, Dziadziuszko R, Camidge DR, et al. Biomarker Status Correlates with Clinical Benefit: Phase 2 Study of Single-agent Erlotinib (E) or E Intercalated with Carboplatin and Paclitaxel (ECP) in an EGFR Biomarkerselected NSCLC Population. J Thorac Oncol. 2008;3:S267. [Google Scholar]
- 5.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science (New York, NY. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25:587–95. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 8.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–46. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 9.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 10.Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:839–44. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- 11.Jackman DM, Yeap BY, Sequist LV, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12:3908–14. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- 12.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–20. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch FR, Varella-Garcia M, Cappuzzo F, et al. Combination of EGFR gene copy number and protein expression predicts outcome for advanced non-small-cell lung cancer patients treated with gefitinib. Ann Oncol. 2007;18:752–60. doi: 10.1093/annonc/mdm003. [DOI] [PubMed] [Google Scholar]
- 14.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49:853–60. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 15.Fukui T, Ohe Y, Tsuta K, et al. Prospective study of the accuracy of EGFR mutational analysis by high-resolution melting analysis in small samples obtained from patients with non-small cell lung cancer. Clin Cancer Res. 2008;14:4751–7. doi: 10.1158/1078-0432.CCR-07-5207. [DOI] [PubMed] [Google Scholar]
- 16.Takano T, Ohe Y, Tsuta K, et al. Epidermal growth factor receptor mutation detection using high-resolution melting analysis predicts outcomes in patients with advanced non small cell lung cancer treated with gefitinib. Clin Cancer Res. 2007;13:5385–90. doi: 10.1158/1078-0432.CCR-07-0627. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Kane S, Wu J, et al. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clin Cancer Res. 2009;15:3023–8. doi: 10.1158/1078-0432.CCR-08-2739. [DOI] [PubMed] [Google Scholar]
- 18.Brevet M, Arcila M, Ladanyi M. Assessment of EGFR Mutation Status in Lung Adenocarcinoma by Immunohistochemistry Using Antibodies Specific to the Two Major Forms of Mutant EGFR. J Mol Diagn. 2010;12:169–76. doi: 10.2353/jmoldx.2010.090140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobin L, Gospodarowicz M, Wittekind C. TNM classification of malignant tumors. seventh. Wiley-Blackwell; 2009. pp. 136–46. [Google Scholar]
- 20.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 21.Kojika M, Ishii G, Yoshida J, et al. Immunohistochemical differential diagnosis between thymic carcinoma and type B3 thymoma: diagnostic utility of hypoxic marker, GLUT-1, in thymic epithelial neoplasms. Mod Pathol. 2009;22:1341–50. doi: 10.1038/modpathol.2009.105. [DOI] [PubMed] [Google Scholar]
- 22.Nitadori J, Ishii G, Tsuta K, et al. Immunohistochemical differential diagnosis between large cell neuroendocrine carcinoma and small cell carcinoma by tissue microarray analysis with a large antibody panel. Am J Clin Pathol. 2006;125:682–92. doi: 10.1309/DT6B-J698-LDX2-NGGX. [DOI] [PubMed] [Google Scholar]
- 23.Travis W, Brambilla E, HK MH, Harris C. Pthology & Genetice Tumors of the Lung, Pleura, Thymus and Heart. IARC; 2004. [Google Scholar]
- 24.Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98:1817–24. doi: 10.1111/j.1349-7006.2007.00607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto H, Toyooka S, Mitsudomi T. Impact of EGFR mutation analysis in non-small cell lung cancer. Lung Cancer. 2009;63:315–21. doi: 10.1016/j.lungcan.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Yatabe Y, Hida T, Horio Y, Kosaka T, Takahashi T, Mitsudomi T. A rapid, sensitive assay to detect EGFR mutation in small biopsy specimens from lung cancer. J Mol Diagn. 2006;8:335–41. doi: 10.2353/jmoldx.2006.050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasaki H, Endo K, Konishi A, et al. EGFR Mutation status in Japanese lung cancer patients: genotyping analysis using LightCycler. Clin Cancer Res. 2005;11:2924–9. doi: 10.1158/1078-0432.CCR-04-1904. [DOI] [PubMed] [Google Scholar]
- 28.Pan Q, Pao W, Ladanyi M. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn. 2005;7:396–403. doi: 10.1016/S1525-1578(10)60569-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cohen V, Agulnik JS, Jarry J, et al. Evaluation of denaturing high-performance liquid chromatography as a rapid detection method for identification of epidermal growth factor receptor mutations in nonsmall-cell lung cancer. Cancer. 2006;107:2858–65. doi: 10.1002/cncr.22331. [DOI] [PubMed] [Google Scholar]
- 30.Kimura H, Fujiwara Y, Sone T, et al. High sensitivity detection of epidermal growth factor receptor mutations in the pleural effusion of non-small cell lung cancer patients. Cancer Sci. 2006;97:642–8. doi: 10.1111/j.1349-7006.2006.00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimura H, Nakajima T, Itakura M, Shingyoji M, Iizasa T, Kimura H. Successful treatment of lung cancer with gefitinib and EGFR mutation status determination using EBUS-TBNA samples in an extremely old patient. Intern Med. 2009;48:1905–7. doi: 10.2169/internalmedicine.48.2494. [DOI] [PubMed] [Google Scholar]
- 32.Oshita F, Matsukuma S, Yoshihara M, et al. Novel heteroduplex method using small cytology specimens with a remarkably high success rate for analysing EGFR gene mutations with a significant correlation to gefitinib efficacy in non-small-cell lung cancer. Br J Cancer. 2006;95:1070–5. doi: 10.1038/sj.bjc.6603396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Olive I, Monso E, Andreo F, et al. Endobronchial ultrasound-guided transbronchial needle aspiration for identifying epidermal growth factor receptor mutations. Eur Respir J. 2009 doi: 10.1183/09031936.00028109. [DOI] [PubMed] [Google Scholar]
- 34.Nakajima T, Yasufuku K, Suzuki M, et al. Assessment of epidermal growth factor receptor mutation by endobronchial ultrasound-guided transbronchial needle aspiration. Chest. 2007;132:597–602. doi: 10.1378/chest.07-0095. [DOI] [PubMed] [Google Scholar]



