Abstract
Commercial HIV-1 RNA viral load assays have been routinely used in developed countries to monitor antiretroviral treatment (ART). However, these assays require expensive equipment and reagents, well-trained operators, and established laboratory infrastructure. These requirements restrict their use in resource-limited settings where people are most afflicted with the HIV-1 epidemic. Inexpensive alternatives such as the Ultrasensitive p24 assay, the Reverse Transcriptase (RT) assay and in-house reverse transcription quantitative polymerase chain reaction (RT-qPCR) have been developed. However, they are still time-consuming, technologically complex and inappropriate for decentralized laboratories as point-of-care (POC) tests. Recent advances in microfluidics and nanotechnology offer new strategies to develop low-cost, rapid, robust and simple HIV-1 viral load monitoring systems. We review state-of-the-art technologies used for HIV-1 viral load monitoring in both developed and developing settings. Emerging approaches based on microfluidics and nanotechnology, which have potential to be integrated into POC HIV-1 viral load assays, are also discussed.
Keywords: HIV-1, viral load, resource-limited settings, point-of-care
1. Introduction
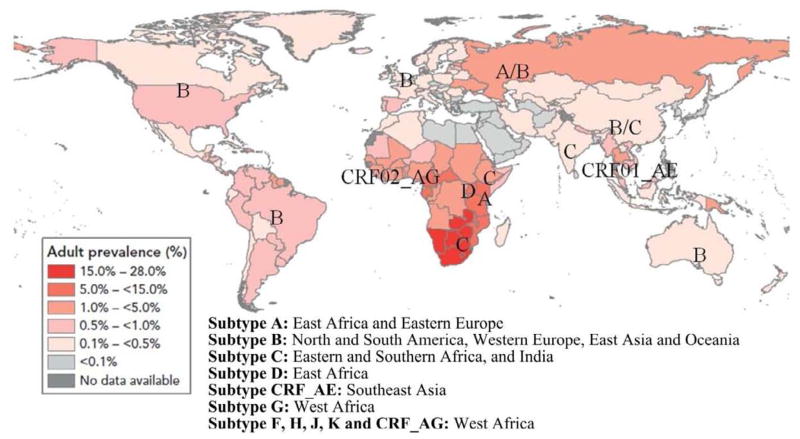
HIV/AIDS has caused 25 million deaths since the first case was reported in 1981, with 2 million HIV-related deaths in 2007 only (UNAIDS, 2008). In addition, there are 33 million people living with HIV-1 worldwide (UNAIDS, 2008). Of them, 94 % of these infected people are from Sub-Saharan Africa (67 %), South and Southeast Asia, Latin America, Eastern Europe and Central Asia (Figure 1), leading to enormous economic burden and humanitarian disaster. Typically, patients in these areas have limited resources and insufficient access to antiretroviral treatment (ART) which can effectively suppress HIV-1 replication and delay disease progression (O’Brien et al., 1996). To save lives, the World Health Organization (WHO) is rapidly expanding access to ART in developing countries, originally aiming at universal access by 2010 (World Health Organization, 2006b). However, this is a challenging goal since only 4 million AIDS patients, approximately 42 % of people who need ART, were actually receiving it by the end of 2008 (World Health Organization, 2009), in part, due to lack of cost-effective ART monitoring.
Figure 1. Global distribution of HIV-1 infection in 2007 (adapated from UNAIDS, 2008).
By the end of 2007, it is estimated that 33 million people were infected with HIV-1. Sub-Saharan Africa accounts for 67 % of infections, with prevalence in adults up to 28 %. Main subtypes are shown as previously reported (Taylor et al., 2008).
Currently, there are two approaches to monitor HIV-1 ART, i.e., HIV-1 viral load in plasma which indicates viral replication in infected individuals and CD4+ T lymphocyte count which reflects the functionality of the host immune system (Mellors et al., 1997). In developed countries, these two parameters are closely monitored (every 3–6 months) to gauge ART efficacy and manage patient adherence (Hammer et al., 2008). HIV-1 viral load is generally monitored using commercial RNA assays, such as Roche COBAS®, Abbott RealTime, Siemens Versant™ and bioMerieux NucliSens®; CD4+ cell count is monitored using flow cytometry. However, implementation of these assays requires high-cost equipment (e.g., thermal cyclers), highly skilled personnel, and expensive reagents ($ 50–100 per test in the US), which is not suitable for resource-limited settings (Fiscus et al., 2006). Due to the lack of viral load monitoring, the WHO recommends guidelines of combining CD4+ cell count with a disease staging system to initiate ART in resource-limited settings. According to these guidelines, ART is initiated when CD4+ cells fall below 200/μL with WHO Stage I or II diseases, or below 350/μL with WHO Stage III diseases, or when WHO Stage IV diseases emerge irrespective of CD4+ cell count (World Health Organization, 2006a). However, the WHO guidelines often lead to late identification of virological ART failure (Mee et al., 2008; van Oosterhout et al., 2009), which may allow for accumulation of drug-resistant strains and reduce the efficacy of second-line drugs (Vekemans et al., 2007). Therefore, there is an urgent need for viral load monitoring to manage AIDS patients on ART (Calmy et al., 2007; Harries et al., 2010; Kuritzkes et al., 2007; Usdin et al., 2010).
Inexpensive alternatives to RNA viral load assays have been developed such as the Ultrasensitive p24 assay (Schupbach et al., 1996), the ExaVir™ RT viral load assay (Malmsten et al., 2003), and real-time reverse transcription quantitative-PCR (RT-qPCR) (Drosten et al., 2006; Rouet et al., 2005). However, these assays still have drawbacks. For example, RT-qPCR, despite reduced cost (approximately $ 20 per test), still requires air conditioning and skilled operators. This method is suitable for centralized laboratories, but not for district clinics in resource-limited settings. Mine et al. has recently demonstrated good acceptability of the ExaVir™ RT viral load assay version 2.0 at a district hospital laboratory in Botswana (Mine et al., 2009). Nevertheless, the throughput of this assay is low, with turnaround time of 2 days and up to 180 samples per week per operator in the improved version 3.0 (Labbett et al., 2009). Thus, recent advances focus on the development of portable detection systems towards the POC testing (Lee et al., 2010a; Tang and Hewlett, 2010; Tang et al., 2010; Tanriverdi et al., 2010). Yet, no such point-of-care (POC) HIV-1 viral load test has become commercially available.
Here, we review emerging technologies for developing HIV-1 viral load assays for resource-limited settings. We first present the markers for HIV-1 viral load monitoring and then report the improvements in HIV-1 viral load assays for developed countries and their inexpensive counterparts for developing countries. We also discuss miniaturized PCR chips and immunoassays, portable amplification systems and upcoming new approaches such as bio-barcode amplification (BCA), microfluidics-based virus detection and quantification, which have been developed for POC viral load testing.
2. Markers for HIV-1 viral load monitoring
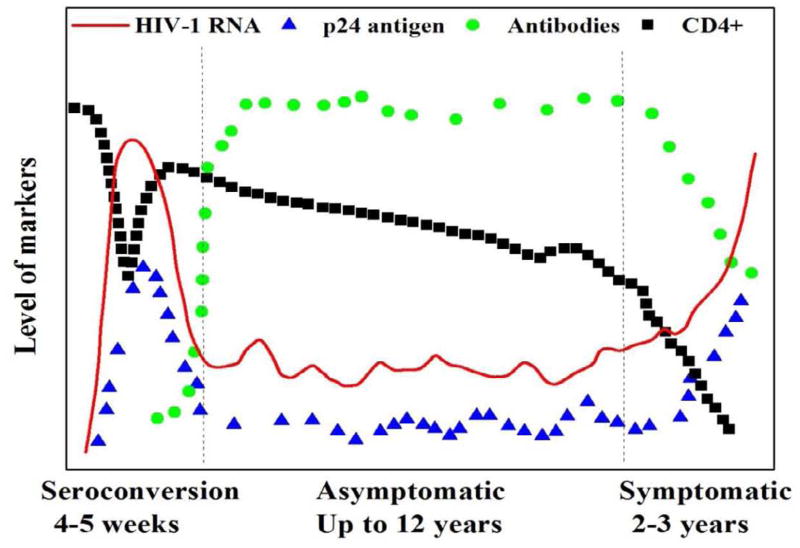
Clinically, the natural course of HIV-1 infection is divided into seroconversion, asymptomatic and symptomatic stages. At each stage, different diagnostic markers, including HIV-1 RNA, DNA, antigen, antibody, reverse transcriptase (RT) and CD4+ cells (Figure 2), can be specifically used to improve the safety of blood products, to screen for acute HIV-1 infection in high-risk populations, to early diagnose infected infants born to HIV-positive mothers, and to gauge ART efficacy. For example, diagnosis of HIV-infected infants cannot be made using serological assays until 18 months after birth due to passively transferred maternity HIV-1 specific antibodies (Chantry et al., 1995).
Figure 2. Diagnostic markers during the natural course of HIV-1 infection (adapted from Chang et al., 2006).
The natural course of HIV-1 infection can be divided into three stages: seroconversion, asymptomatic and symptomatic. HIV-1 RNA (red line) peaks during seroconversion, then decreases and remains at a low level and shows no clinical symptoms. HIV-1 p24 antigen (blue triangles) demonstrates the same trend as HIV-1 RNA. HIV-1 specific antibody (green circles) is produced during seroconversion and reaches a plateau at the asymptomatic stage. The number of CD4+ cells (black squares) drops rapidly and rebounds during seroconversion and it gradually decrease as AIDS develops.
Viral load is defined as the level of HIV-1 RNA in plasma, which indicates HIV-1 replication within infected individuals. Following infection, the level of HIV-1 RNA increases significantly and reaches its peak of approximately 107 copies/mL (Figure 2). Despite high level of viral replication at the seroconversion stage, the host immune system is still intact, which is indicated by a high level of CD4+ cell count, and can effectively suppress HIV-1 replication. Once HIV-1 specific antibodies are produced during seroconversion, HIV-1 viral load is kept at a low level throughout the asymptomatic stage. However, HIV-1 gradually compromises the host immune system, which is shown by a decreasing CD4+ cell count. As the disease progresses, HIV-1 viral load rebounds due to the impaired host immunity. HIV-1 viral load can be suppressed until drug-resistant strains emerge and dominate.
HIV-1 p24 antigen and RT enzyme have also been used as surrogate markers for viral load monitoring in developing countries. As shown in (Figure 2), the level of p24 antigen is highly correlated with HIV-1 viral load throughout the natural course of HIV-1 infection. In the first four to five weeks of seroconversion, the level of p24 antigen in plasma reaches its peak, in parallel with the highest level of HIV-1 RNA. Once HIV-1 specific antibodies are produced, the level of p24 antigen drops significantly due to the formation of antigen-antibody complex. At the symptomatic stage, (e.g., Mycobacterium tuberculosis or Kaposi’s sarcoma), the level of p24 antigen increases in parallel with HIV-1 RNA. In addition to HIV-1 p24 antigen levels, studies have shown that the level of RT has a close correlation with HIV-1 RNA in patients receiving ART (Jennings et al., 2005; Labbett et al., 2009; Malmsten et al., 2003; Stevens et al., 2005).
Recently, HIV-1 particles have been utilized as a viral load marker in microfluidic systems (Kim et al., 2009). In theory, assays quantifying the HIV-1 particle are more accurate than RNA viral load assays, because they reduce the quantification bias arising from RNA degradation and amplification inhibition in nucleic acid testing. HIV-1 RNA is prone to degradation due to ubiquitous RNases and freeze-thaw cycles. As such, Michael et al. quantified cell-free HIV-1 stocks representing Group A-G by direct viral counting under a transmission electron microscope for a reliable calibration (Michael et al., 1999). The calibrated viral standards showed a good performance in evaluating the Amplicor Monitor™, version 1.5 (Roche).
3. HIV-1 viral load assays for developed countries
Commercial RNA viral load assays have been widely used as ‘gold standards’ to quantify HIV-1 RNA in plasma in developed settings. Four major diagnostic companies, Roche, Abbott, bioMerieux and Siemens, have developed HIV-1 viral load assays (de Mendoza and Soriano, 2009). Compared to previous end-point detection versions, these companies have adapted the real-time technologies based on fluorescence to achieve simultaneous amplification and detection instead of end-point detection (de Mendoza et al., 2005; Ruelle et al., 2009; Schumacher et al., 2007; Tang et al., 2007a). The real-time technologies offer more accurate quantification, a wider dynamic range, and a higher throughput. In addition, these companies have included automated sample preparation to reduce manual time and prevent carry-over contamination. Most importantly, these assays extend the subtype coverage to detect increasing numbers of non-B strains reported in developed countries. In this section, we will review the use of the real-time technologies, which rely on fluorescence to achieve simultaneous amplification and quantification, in these standard viral load assays.
3.1. Reverse transcriptase-quantitative polymerase chain reaction (RT-PCR)
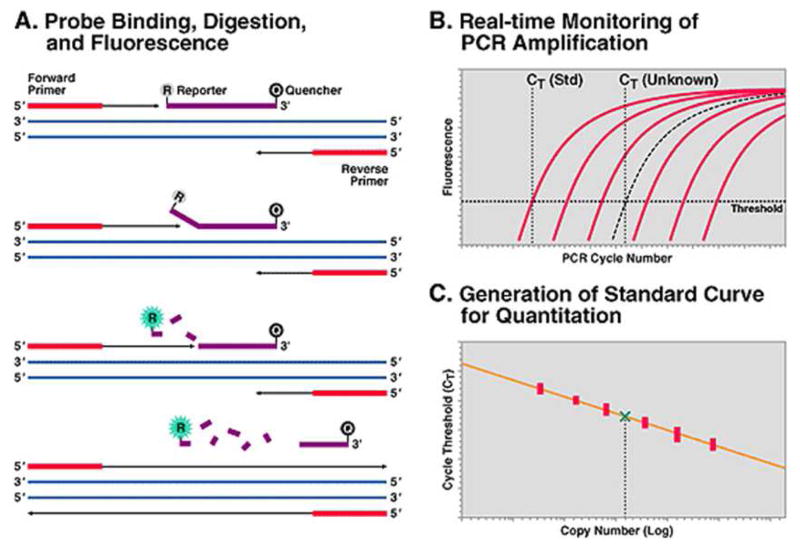
The COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test, the latest product from Roche, obtained FDA approval in 2007(Roche, 2007). In this system, a highly conserved region within the Gag gene is first reverse-transcribed into cDNA, which, in turn, is amplified by PCR. During the PCR amplification, the use of TaqMan probes enables real-time detection (Liegler and Grant, 2006). The TaqMan probe is a dual-labeled DNA sequence of 25–30 nucleotides in length, with a fluorescence reporter at the 5′ end and a quencher at the 3′ end. Due to the proximity between them, the fluorescence emitted by the reporter is negatively suppressed by the quencher. During amplification, Taq DNA polymerase elongates the forward primer and cleaves the TaqMan probe, which hybridizes to the amplified HIV-1 cDNA. The cleaved TaqMan probe allows for dissociation of the reporter from the quencher, resulting in a detectable fluorescence emission (Figure 3A). When the fluorescence exceeds the background signal, the cycle number of PCR is recorded and used for quantification (Figure 3B). The more HIV-1 RNA in plasma, the earlier the fluorescence exceeds the background signal, and the lower CT values are. Compared to the CT values from quantification standards, the viral load of each sample is calculated (Figure 3C). The assay can quantify viral load ranging from 48 to 107 copies/mL and reliably detect HIV-1 subtypes A–D, F–H and CRF01_AE (originally subtype E).
Figure 3. Schematic illustration of the TaqMan technology for quantification (reproduced from Liegler and Grant, 2006).
(A) Principle for PCR amplification and the TaqMan probe. In each cycle of PCR, a new DNA fragment is amplified by DNA polymerase with the aid of forward and reverse primers. During amplification, a TaqMan probe binding to the DNA template is digested by DNA polymerase, releasing fluorescence. (B) Amplification plots of quantification standards and an unknown sample. Six external standards are amplified in parallel with an unknown sample. The increase of fluorescence from each reaction is recorded during the amplification. (C) Quantification of the unknown sample by comparing with the standard curve. The CT values (where fluorescence of each reaction exceeds the background) of quantification standards are plotted against HIV-1 viral load. The viral load of the unknown sample is indicated by the green check.
The RealTime HIV-1 assay (Abbott), another FDA-approved HIV-1 viral load assay, shares the same principles as the Roche COBAS®, in which HIV-1 RNA first undergoes reverse transcription and is then quantified by PCR. However, there are three major differences between them. First, the RealTime assay targets a highly conserved integrase gene within pol, which is different from the amplification region (gag) in Roche assays. Second, the RealTime uses a partially double-stranded probe to achieve real-time detection, rather than a TaqMan probe in the Roche COBAS® (Tang et al., 2007a). Third, the RealTime uses a non-competitive internal control, an armored RNA containing sequence derived from the hydroxypyruvate reductase gene of pumpkin, to indicate potential amplification inhibition. In the Roche COBAS®, a competitive internal control is used. Most importantly, the RealTime assay has wider subtype coverage. In addition to subtypes from group M such as A–D, F–H, CRF01_AE, and CRF02_AG, the RealTime assay can also detect and quantify HIV-1 isolates from groups N and O (Tang et al., 2007a), which are not detected by the Roche COBAS® or other FDA-approved viral load assays.
3.2. Nucleic acid sequence-based amplification (NASBA)
The NucliSens® EasyQ HIV-1 assay has been developed by bioMerieux and CE-marked to replace NucliSens® HIV-1 QT, which is an end-point detection system. Both systems are based on the isothermal amplification of HIV-1 Gag gene by NASBA, which is developed based on HIV-1 replication (Compton, 1991; Kievits et al., 1991). At the early phase of NASBA, a special antisense primer with a tail of T7 RNA polymerase promoter anneals to HIV-1 RNA and initiates the synthesis of antisense cDNA by RT. Concurrently, RNase H specifically digests HIV-1 RNA in the RNA/DNA hybrid. When a sense primer binds to the newly synthesized antisense cDNA, RT starts the production of double-stranded cDNA, forming an intact T7 RNA polymerase promoter at the 5′ end. T7 RNA polymerase recognizes the promoter and transcribes hundreds of copies of antisense HIV-1 RNA. At the late phase of NASBA, anti-sense RNA undergoes a circle of reverse transcription, RNase H digestion, cDNA synthesis and RNA transcription, yielding millions of anti-sense RNA molecules within minutes.
For real-time detection and quantification, the EasyQ assay uses a special DNA probe termed as molecular beacon (Deiman et al., 2002; Yao et al., 2005). It has a stem-loop secondary structure and is labeled with a fluorophore at the 5′ end and a quencher at the 3′ end. In the absence of amplicons, the molecular beacon does not release fluorescence due to proximity between the fluorophore and the quencher. Upon binding to amplicons during amplification, the secondary structure of molecular beacon unfolds, releasing fluorescence proportional to the level of HIV-1 RNA in clinical samples. By comparing the fluorescence emitted from clinical samples with that emitted from quantification standards, the viral load can be quantified.
3.3. Branched DNA (bDNA)
The Versant™ HIV-1 RNA 3.0 assay (Siemens Medical Solutions) is actually probe-based signal amplification rather than sequence amplification (Collins et al., 1997). Through nucleic acid hybridization, HIV-1 RNA is captured in a microtiter plate and detected using an amplified chemiluminescence signal. In brief, HIV-1 RNA is first released from virions and captured by synthetic oligonucleotide probes pre-coated onto a microtiter plate. A set of capture extenders comprising of the Pol gene are used to bind to HIV-1 RNA and the coated capture probes. With the linkage of target probes, which also reside in the Pol gene, HIV-1 RNA, target probes and pre-amplifier probes are sandwiched. Furthermore, addition of amplifier probes and probes labeled with alkaline phosphatase forms a branched DNA complex and results in signal amplification. The amount of HIV-1 RNA is calculated by comparing the chemiluminescence signal emitted from specimens with quantification standards of known concentrations.
In contrast to signal amplification, Siemens has recently developed a nucleic acid amplification assay, namely Versant™ HIV-1 RNA 1.0 kPCR (Ruelle et al., 2009). In this system, a dual-labeled DNA probe forms a folded secondary structure, keeping fluorescence from being released due to the proximity between the fluorophore at the 5′ end and the quencher at the 3′ end. During amplification, fluorescence increases, due to destruction of the secondary structure, in proportion to HIV-1 viral load in clinical samples.
3.4. Comparison of FDA-approved viral load assays
Commercial RNA assays have similar sensitivity, specificity, dynamic range and throughput (details see Table 1), as have been extensively evaluated (Elbeik et al., 2002; Galli et al., 2005; Ginocchio et al., 2003; Murphy et al., 2000; Swanson et al., 2007). Murphy et al. found that Roche COBAS® Amplicor HIV-1 Monitor™ 1.5 gave consistently higher viral load values than bDNA 3.0 and that bDNA was less specific than NucliSens® QT and Amplicor Monitor™ 1.5 (Murphy et al., 2000). However, in another large scale study (n=1000), no aberrant difference was observed between the Versant™ HIV-1 RNA 3.0 and the COBAS® Amplicor HIV-1 Monitor™ 1.5 when clinical samples were grouped according to viral load ranging from < 50 copies/mL, 50–250 copies/mL and 250–500,000 copies/mL (Galli et al., 2005). This indicates that the discordance in viral load as observed by Murphy et al. may be caused by sampling variation. In addition, the subtype coverage is essential to viral load quantification, especially in developing countries where non-B subtypes dominate. Recent studies have shown that the Abbott RealTime HIV-1 assay have better quantification on HIV-1 group O and N, which may be underestimated or missed by the Versant™ HIV-1 RNA 3.0 or the Amplicor HIV-1 Monitor™ 1.5 (Swanson et al., 2007; Tang et al., 2007a). This advantage may be attributable to the use of pol, which is more conservative than gag, and the use of a non-competitive internal control in the Abbott RealTime assay.
Table 1.
Comparison of commercial assays for monitoring HIV-1 viral load
| Assay | Company | Technology | Diagnostic markers | Sample Volume (mL) | Linear Range (copies/mL) | Subtype/group | Cost ($) | Throughput (hours/test) | Applicable settings | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| COBAS AmpliPrep/COBAS TaqMan assay a | Roche | RT-qPCR | RNA (gag) | 0.85 | 48 to 10,000,000 | Group M: A-H | 50–100 | 6 | Developed | (Fiscus et al., 2006; Roche, 2007) |
| RealTime HIV-1 Assay a | Abbott | RT-qPCR | RNA (pol) | 1.0 | 40 to 10,000,000 | Group M: A-H Groups N & O | 50–100 | 6 | Developed | (Abbott, 2007; Fiscus et al., 2006; Tang et al., 2007a) |
| Versant™ HIV-1 RNA 3.0 a b | Siemens | bDNA | RNA (gag) | 1.0 | 75 to 500,000 | Group M: A-G | 50–100 | 22 | Developed | (Fiscus et al., 2006; Gleaves et al., 2002; Rouet et al., 2001) |
| NucliSens® HIV-1 QT a | BioMerieux | NASBA | RNA (gag) | 1.0 | 176 to 3,470,000 | Group M: A-D | 50–100 | 8 | Developed | (Fiscus et al., 2006) (Bonard et al., 2003; Fiscus et al., 2006; |
| Ultrasensitive p24 assay b | PerkinElmer | p24 EIA | P24 antigen | 0.05 | 5,000 upwards | All | 5–19 | 6 | Developing | Fiscus et al., 2007; Jennings et al., 2005) |
| ExaVir™ RT assay | Cavidi | Reverse transcriptase assay | RT | 1.0 | 200 upwards | All | 14–23 | 48–72 | Developing | (Labbett et al., 2009; Mine et al., 2009) |
| LTR-based HIV-1 RNA RT-PCR | Biocentric | RT-qPCR | RNA (LTR) | 0.2 | 300 to 10,000,000 | All | 10–20 | 4 | Developing | (Rouet et al., 2007; Rouet et al., 2008) |
Note.
These assays have FDA approval. NASBA: nucleic acid sequence-based amplification; bDNA: branched DNA; EIA: enzyme immunoassay; RT-PCR: reverse transcriptase and polymerase chain reaction; COBAS®: comprehensive bioanalytical systems.
The Versant™ and the Ultrasensitive p24 assay have a specificity of 97.2% and 98.5, respectively, when used for early diagnosis of HIV-1 infection in infants (Fiscus et al., 2007; Rouet et al., 2001).
4. Alternative HIV-1 viral load assays for resource-limited settings
Monitoring HIV-1 viral load in resource-limited settings faces multiple challenges (Fiscus et al., 2006; Rouet and Rouzioux, 2007). First, viral load monitoring is only available in reference laboratories, where various clinical trials are financially supported by national and international efforts. Second, provincial hospitals and local clinics lack basic laboratory infrastructure such as reliable power and water supply, refrigeration and air conditioning. Third, technicians from decentralized laboratories and clinical staff are rarely trained to perform technologically complex molecular assays. Fourth, variations of environmental temperature and humidity cannot be controlled during transportation and storage. Fifth, robust methods and equipment are needed to withstand harsh conditions outside of main hospitals due to lack of maintenance and technical support.
To provide viral load monitoring in resource-limited settings, inexpensive alternatives (Table 1) have been developed, such as the ExaVir™ RT assay (Cavidi) (Jennings et al., 2005; Labbett et al., 2009; Mine et al., 2009; Stevens et al., 2005), the Ultrasensitive p24 assay (PerkinElmer) (Bonard et al., 2003; Boni et al., 1997; Jennings et al., 2005; Pascual et al., 2002; Prado et al., 2004; Stevens et al., 2005) and in-house RT-qPCR (Drosten et al., 2006; Rouet et al., 2005). The ExaVir™ assay and the Ultrasensitive p24 assay are based on enzyme-linked immunosorbent assay (ELISA), measuring the activity of RT and the level of p24 antigen, respectively. In-house RT-qPCR methods utilize the same TaqMan technology as Roche, but target another highly conserved region within the long terminal repeat (LTR). These alternative methods have significantly reduced cost ($ 13–20), compared with commercial viral load assays ($ 50–100). In addition, the ExaVir™ assay and the Ultrasensitive p24 assay are technologically simpler than commercial RNA viral load assays, and thus do not require highly skilled operators. On the downside, the ExaVir™ assay and the Ultrasensitive p24 assay are time-consuming and labor-intensive. Although in-house RT-qPCR methods are highly correlated with commercial viral load assays, they still need expensive thermal cyclers and highly trained operators, which restricts their application in de-centralized laboratories.
4.1. Reverse transcriptase assay
The Cavidi ExaVir™ RT assay is an ELISA-based assay to measure the RT activity, which in turn correlates with the level of HIV-1 RNA (Malmsten et al., 2003). In brief, HIV-1 virions are first purified from plasma specimens using a solid phase extraction manifold to remove contaminants and inhibitors. Purified HIV-1 virions are then lysed to release RT. The released RT is added to a microtiter plate, which is coated with poly-A oligonucleotides. RT incorporates BrdUTP into cDNA, using the poly-A oligonucleotides as templates. After incubation at 33 °C for 40 hours, an alkaline phosphatase-conjugated anti-BrdU antibody and a substrate are sequentially added to achieve colorimetric detection. The color intensity measured at a wavelength of 405 nm correlates with the RT activity. The RT activity of unknown samples is compared to those obtained from quantification standards, reporting the viral load as fg/RT/mL or as HIV-1 RNA equivalents/mL.
Recently, ExaVir™ version 2.0 has demonstrated acceptability regarding sensitivity, complexity, and cost at district-based hospital laboratories in Botswana (Mine et al., 2009). Meanwhile, this study also pinpoints the encountered challenges such as requirement for 1 mL of plasma, the lengthy testing procedure and the limited number of samples per operator per day. With improved performance, the ExaVir™ version 3.0, has a detection limit of as low as 200 RNA copies/mL, shortened turnaround time (48–72 h) and increased throughput (120 to 180 samples per week per operator) (Labbett et al., 2009). Most importantly, the cost of ExaVir™ RT assay can be reduced to $ 13.66 per test given a large ordering volume. The ExaVir™ RT assay quantifies samples with viral load more than 400 RNA copies/mL measured by the Roche COBAS® and the Abbott RealTime, demonstrating its utility of monitoring viral load in resource-limited settings. In another study, the RT assay was strongly correlated with the Roche Amplicor (r = 0.8554), leading to implementation at 4 district hospital laboratories in Botswana (Mine et al., 2009). However, it should be noted that the ExaVir™ RT assay is a functional assay; heavily mutated RT or co-infection with other retroviruses (e.g., human T-lymphotropic virus-I) may interfere with its performance.
4.2. Ultrasensitive p24 assay
The PerkinElmer Ultrasensitive p24 assay is also based on ELISA to measure the level of HIV-1 p24 antigen, which has been shown to be strongly correlated with HIV-1 RNA (Schupbach et al., 1996). Prior to the capture of p24 by a specific antibody-coated 96-well plate, specimens are lysed by a special kit buffer and incubated at 100 °C for 5 minutes to release p24 molecules to a maximum degree from HIV-1 virions and from the p24/antibody complex in plasma (Boni et al., 1997). The captured p24 antigen is recognized by a biotinylated anti-p24 antibody, followed by the colorimetric detection step using a streptavidin-horseradish peroxidase conjugate. The emitted color intensity of each sample is compared to intensities emitted from external standards of known concentrations, determining the level of p24.
Although the Ultrasensitive p24 assay has shown a good correlation with commercial RNA assays in some settings, discordant results have also been observed in other studies. Pascual et al. observed that the Ultrasensitive p24 assay detected 66.7 % of the specimens with viral load less than 10,000 copies/mL, and 87 % of the specimens with viral load of between 10,000 and 100,000 copies/mL (Pascual et al., 2002). In another study, only 27 % of the specimens were detected with viral load less than 1,000 copies/mL (Lombart et al., 2005). In addition, studies have shown that the Ultrasensitive p24 assay can not reflect the response to ART as well as nucleic acid viral load assays using samples collected from patients undergoing structured treatment interruption (Prado et al., 2004) or using CRF02_AG strains dominant samples (Bonard et al., 2003). The discordant results may be due to inefficient dissociation between p24 and antibody, binding of the immune complex to erythrocytes or HIV-1 genetic diversity (Bonard et al., 2003; Prado et al., 2004). Therefore, further improvement and evaluation are needed so that the Ultrasensitive p24 assay can be reliably used for viral load monitoring in resource-limited settings.
4.3. In-house RT-qPCR assays
Since RT-qPCR is commonly used in commercial viral load assays, some researchers have developed low-cost in-house RT-qPCR for HIV-1 RNA quantification (Drosten et al., 2006; Rouet et al., 2005). These assays target a highly sequence-conserved region within the LTR to maximize the subtype coverage, which are more diverse in resource-limited settings than in developed countries (Figure 1). Rouet et al. has shown that in-house RT-qPCR correlates well with the Versant™ kit (r = 0.901; P < 0.001) and the Monitor™ test (r = 0.856; P < 0.001), with samples dominant in a mosaic subtype of CRF02_AG (Rouet et al., 2005). In the second generation of this assay, a shorter minor groove binding probe and a shorter forward primer were utilized. For 898 specimens with diverse subtypes, viral load values obtained by this modified assay are highly correlated with those obtained by the Roche Amplicor Monitor™ and the Versant™ 3.0 (Rouet et al., 2007). Another group in Germany demonstrates comparable detection as the Roche Amplicor Monitor™, BioMerieux NucliSens® QT, Bayer Versant™ 3.0, when evaluating 1487 specimens from Brazil, India, South Africa and Germany (Drosten et al., 2006). This study also shows a better coverage of Group N and O, which are not detected by the Roche Amplicor Monitor™. The major benefit of these two methods is cost reduction, lowered to $ 20 per test (Drosten et al., 2006; Rouet et al., 2005). Although the test cost is significantly reduced compared to commercial viral load assays, in-house RT-PCR assays still need high-cost equipment, solid laboratory infrastructure and well-trained operators, restricting their use to district laboratories in rural areas. Recently, the RT-qPCR method developed by Rouet et al. has become commercially available (Biocentric, Bandol, France) (Rouet et al., 2008).
5. Advances in developing HIV-1 POC viral load assays for resource-limited settings
To facilitate decision-making on initiation of ART or identification of virological ART failure, an HIV-1 POC viral load assay is urgently needed in rural areas (Lee et al., 2010b; Usdin et al., 2010; Yager et al., 2008). Although inexpensive viral load assays can be performed at centralized laboratories, they are very difficult to implement at rural areas due to the need for basic laboratory instrument such as a 96-well plate washer and reader for the RT assay. To address this challenge, one of the options is to store blood samples using filter paper forming dry blood spots (DBS), and transporting them to centralized laboratories (Cassol et al., 1997; Johannessen et al., 2009).
However, the turnaround time from sample-to-answer may take up to several weeks (Calmy et al., 2007). So ideally, instrument-free or portable systems that can be performed used by health care workers with minimal training and that give a readout in less than 30 minutes will be preferred (Fiscus et al., 2006; Rouet and Rouzioux, 2007; Usdin et al., 2010). These systems will facilitate clinical decisions while patient are on site. In addition, HIV-1 POC viral load assays need to be performed using a low-cost disposable device (ideally less than $ 10). These assays should not be affected by non-B HIV-1 subtypes which are dominant in developing countries. This goal is feasible since the consumption of enzymes or gold particles is very small in a miniaturized system. Because of low cost, the self-contained device with pre-loaded reagents can be discarded after use to prevent cross contamination.
To address these challenges, new approaches based on nanotechnology and microfluidics have been developed towards viral detection in a POC testing format. These new approaches include bio-barcode amplification (BCA)-based assays (Kim et al., 2008; Tang et al., 2007b), microfluidics-based viral detection and quantification (Kim et al., 2009; Liu et al., 2005; Wang et al., 2010b; Ymeti et al., 2007), miniaturized PCR chips (Easley et al., 2006; Lee et al., 2008), miniaturized immunoassays (Lee et al., 2009; Lee et al., 2004; Sia et al., 2004) and POC HIV-1 amplification systems (Lee et al., 2010a; Tang and Hewlett, 2010; Tang et al., 2010; Tanriverdi et al., 2010). These approaches bear similar characteristics, such as low cost, short turnaround time, simple procedure and high sensitivity, to detect analytes of interest. In addition, the miniaturized systems, such as functionalized cartridges, can be easily run by less-trained health care workers at the POC. These studies demonstrate the feasibility of implementing such assays for viral load monitoring or diagnosis of HIV-1 infection in infants born to HIV-infected mothers.
5.1. BCA-based assays
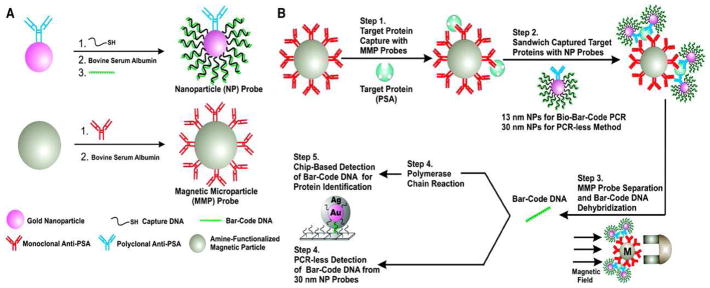
BCA was initially developed by Mirkin’s group based on immuno-PCR to detect trace amount of prostate-specific antigen (PSA) with the aid of gold nanoparticles (NPs) and magnetic microparticles (MMPs) (Nam et al., 2003). In this system (Figure 4), MMPs, heavily functionalized with PSA monoclonal antibody, are used to capture and concentrate PSA. NPs, dual-labeled with polyclonal antibody against PSA and barcode DNA, form a PSA-antibody immune complex on the surface of MMPs. After separation by a magnetic field, unwanted reaction components are removed and the PSA immune complex is concentrated. Barcode DNA is then eluted from NPs and subject to scanometric detection with/without PCR. This method detected PSA at 30 attomolar; if combined with PCR, this method detected PSA at 3 attomolar, which was 100-fold more sensitive than conventional immuno-PCR (Nam et al., 2003). Thus, the use of NPs enables BCA to detect trace amount of protein without nucleic acid amplification. Most importantly, BCA can also be adapted for DNA detection with a PCR-like sensitivity without nucleic acid amplification (Nam et al., 2004).
Figure 4. Schematic illustration of the BCA technology for DNA detection (reproduced from Nam et al., 2003).
(A) Functionalization of gold nanoparticles (NPs) and magnetic microparticles (MMPs). NPs (in pink) are dual-labeled with polyclonal antibody against prostate-specific antigen (PSA, in blue) and many copies of barcode DNA (in green). MMPs (in grey) are heavily conjugated to PSA monoclonal antibody (in red). (B) Barcode DNA mediated protein separation and scanometric detection with/without nucleic acid amplification by PCR.
Recently, researchers have developed a BCA-based p24 assay to early detect HIV-1 during seroconversion (Kim et al., 2008; Tang and Hewlett, 2010; Tang et al., 2007b). In brief, the p24 antigen is captured by an anti-p24 antibody-coated microtiter plate instead of antibody-coated magnetic particles. The captured p24 antigen is recognized by a biotinylated antibody, forming a sandwich format. Streptavidin-labeled gold nanoparticles are bound to the secondary antibody via biotin-streptavidin interaction, which also mediates the binding of biotin-labeled barcode DNA to the gold nanoparticles. In the end, barcode DNA is detected by a modified scanometric method, in which silver ions increase the sensitivity 100-fold. The detection limit of this method was 0.1pg/mL, which was approximately 150-fold more sensitive than the conventional p24 ELISA (10–15pg/mL), and this assay detected HIV-1 infection 3 days earlier than conventional serological assays (Tang et al., 2007b). Alternatively, gold nanoparticles can be replaced with europium nanoparticles to simplify this method and reduce the testing time (Tang and Hewlett, 2010). In another study, Kim et al. detected 111 HIV-1 infections out of 112 samples using a BCA-basedp24 assay, and only detected 23 HIV-1 infections using the conventional p24 ELISA (Kim et al., 2008). For samples with viral load above 50 copies/mL, the BCA results were well correlated (n = 92, r2 = 0.8882) with the Roche Amplicor Monitor™. However, there was no association between these two methods when the viral load was below 50 copies/mL.
5.2. Microfluidics-based intact virus detection and quantification
Microfluidics-based detection has been utilized to address global health issues (Lee et al., 2010b). From a viral load point of view, several methods have the potential to be integrated into a POC device for resource-limited settings because of high sensitivity and short turnaround time. Liu et al. built a microbead-based microfluidic system for virus capture and detection (Liu et al., 2005). In this system, primary antibody-coated microbeads significantly increase the surface-to-volume ratio and thus improve the capturing efficiency. The primary antibody-virus complex interacts with a secondary antibody, which is conjugated with quantum dots. The captured viruses are detected under a microscopy-based imaging system, and the quantification is measured by the fluorescence intensity. Using this system, the detection limit can be improved to 22 ng/mL (giving results within 30 minutes) in comparison with 360 ng/mL in conventional ELISA (2–3 hours per test) (Liu et al., 2005).
Ymeti et al. developed a Young interferometer-based sensor for direct virus detection and quantification (Ymeti et al., 2007). This sensor is based on the use of a monochromatic light, which is wave-guided into 4 parallel channels (i.e., 3 measuring channels and 1 reference channel) by means of Y-junctions. These 4 channels were labeled with 3 different target-specific antibodies, and a reference antibody, allowing for multiplex detection. When a viral target is captured in a channel, the monochromatic light interferes, generating a target-specific phase pattern on a screen. The change in the phase pattern is associated with the amount of target captured in the channel. In the study, only one channel was used to detect human simplex virus-1 (HSV-1) using an anti- HSV-1 glycoprotein G monoclonal antibody. This sensor showed dynamic range between 8.5 × 102 to 8.5 × 106 particles/mL, with a correlation coefficient of 0.98. These two assays achieve sensitivity comparable to conventional ELISA, which shows the potential to replace conventional ELISA such as the Ultrasensitive p24 antigen assay at the POC.
5.3. Microfluidics-based virus count
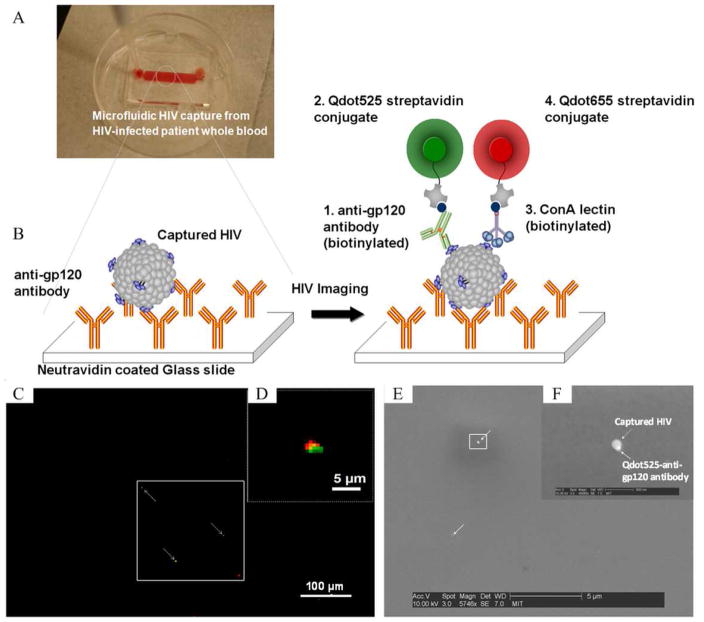
With rapid progress in the areas of microfluidics and nanotechnology, direct virus imaging/counting has become feasible (Alyassin et al., 2009; Kim et al., 2009; Wang et al., 2010a). As shown in Figure 5, 10 μL of unprocessed whole blood is pumped into a Poly-(methyl methacrylate) (PMMA) based microfluidic chamber which is previously coated with an anti-gp120 antibody. If present, HIV-1 particles are captured by the anti-gp120 antibody. The captured HIV-1 particles are then sandwiched by a biotinylated anti-gp120 antibody, which binds to a streptavidin-conjugated green quantum dot. Meanwhile, the captured HIV-1 particles are co-recognized by a biotinylated ConA lectin, which interacts with a streptavidin-conjugated red quantum dot. Under a fluorescence microscope, HIV-1 particles are indicated by co-location of a green and red florescence signal, which are emitted by the green and red quantum dots, respectively (Kim et al., 2009; Wang et al., 2010b).
Figure 5. A microfluidics-based viral load device to capture and image HIV-1 particles (adapted from Kim et al., 2009).
(A) A top-down image of a microfluidic device containing whole blood. (B) Schematic illustration of the HIV-1 capturing/imaging strategy. On a glass slide, anti-gp120 antibody is immobilized. HIV-1 particles, which have gp120 antigen protruding on the surface, are captured via specific antigen-antibody interaction. The captured particles are then co-recognized by biotinylated anti-gp120 and ConA lectin, which are conjugated to quantum dots 525 (solid green circle) and 655 (solid red circle), respectively. (C–D) Under a fluorescence microscope, HIV-1 particles are identified by co-located green and red light, and quantified using an imaging system. (E–F) SEM images of the captured HIV particle with Qdot525/anti-gp120 antibody.
This method starts with a small volume of (10–100 μL) whole blood samples and detects HIV-1 particles directly, eliminating the process of nucleic acid isolation required in nucleic acid testing. As a qualitative test, this is particularly useful to identify HIV-1 infected infants born to high-risk mothers, because small volumes of whole blood can be collected from a fingerprick. Most importantly, the combination with a quantitative imaging system enables this method to count HIV-1 particles (Wang et al., 2010a). As a quantitative test, this method eliminates the need for complex nucleic acid amplification, potentially cutting the cost of one device down to $ 5. These features are encouraging to develop a microfluidics-based POC viral load test. However, the issue of imaging fluorescent quantum dots under a fluorescence microscope remains to be solved since it is not practical at POC settings. The ultimate goal would be to develop a viral load microchip using a portable lensless imaging system as we previously reported for CD4+ T cell counts, eliminating the need for a fluorescence microscope (Moon et al., 2009; Ozcan and Demirci, 2008).
Recently, paper-based analytical devices (μPADs) have been proposed to create inexpensive diagnostic tools for developing world (Martinez et al., 2010; Martinez et al., 2008; Zhao and van der Berg, 2008). In comparison to other technologies, this platform can significantly reduce the fabrication cost to less than $ 0.01 per μPAD. These paper based approaches for microfluidics may achieve impact at the POC including viral load assays.
5.4. Miniaturized PCR chips
Miniaturized PCR chips are designed to carry out PCR in a microfluidic device (Zhang and Xing, 2007). Due to a large surface-to-volume ratio and fast mixing in the device, PCR can be completed within 30 minutes, with reduced reagent consumption. Recently, an integrated PCR chip is developed, which contained 3 functional chambers dedicated to sample preparation, amplification and detection (Easley et al., 2006). A microchannel was used to direct liquid flow through 5 valves to these 3 chambers preloaded with reagents. When whole blood was introduced into the device, nucleic acid of Bacillus anthracis was rapidly extracted within 9 minutes. A 211-bp DNA fragment was successfully amplified by PCR and detected by subsequent electrophoresis. In another study, a polymer-based microfluidic device and a portable analyzer were developed for the detection of HIV-1 RNA by RT-PCR (Lee et al., 2008). The miniaturized chip contained two modules; one for reverse transcription and PCR, and another for chemiluminescence detection. Micro pinch valves were used to help maintain high inner pressure and control the liquid flow. The portable analyzer included infrared-based temperature control and on-chip optical detection. Results showed satisfactory yields comparable to conventional RT-PCR, demonstrating the feasibility to achieve rapid viral load results at the POC.
Although strides have been made in miniaturized PCR chips to reduce cost and achieve rapid detection, several critical technical issues remain to achieve viral load quantification. First, sample preparation is extremely difficult to be integrated into such a sample-in-answer-out platform, because sample preparation involves cell and/or virus lysis, release of target nucleic acids, and removal of unwanted protein contaminants. These manipulation steps necessitate appropriate use of micropumps, plugs and mixers, which are difficult to integrate into microfluidic-based assays. Second, although large surface-to-volume ratio and efficient diffusion facilitate the sample mixing, this may also cause sub-optimized PCR reaction or even PCR inhibition (Gonzalez et al., 2007). This phenomenon may also be caused by incompatible materials that are used for chip assembly, such as silicon and silicon nitride (Kolari et al., 2008). Third, carryover contamination remains to be overcome for high throughput operation in the design of continuous-flow PCR and droplet PCR.
5.5. Miniaturized immunoassays
Miniaturized immunoassays have been developed for the detection of analyte-specific antigens or antibodies (Bhattacharyya and Klapperich, 2007; Lee et al., 2004; Sia et al., 2004). To overcome the technical difficulties in microfluidics-based ELISA, antibody-conjugated gold colloids and a solution containing silver nitrate and hydroquinone were used (Sia et al., 2004). The gold colloids catalyzed and cascaded the reduction of silver ions to silver atoms, forming a silver film, the opacity of which was associated with the concentration of analytes. Using this immunoassay, the level of HIV-1 specific antibody in the samples can be differentiated. In another study, Lee et al. used nanoarrays to detect HIV-1 p24 antigen (Lee et al., 2004). In this system, dip-pen nanolithography was used to conjugate anti-p24 antibody onto a gold thin film. HIV-1 p24 antigen, if present in plasma samples, would be captured by a monoclonal anti-p24 antibody on the gold film. To amplify the signal, p24 antigen was further sandwiched by gold nanoparticles which were heavily functionalized with a polyclonal anti-p24 antibody. This nanoarray-based immunoassay showed a much lower detection limit (0.025 pg/mL) than conventional ELISA (5 pg/mL) and it even detected p24 antigen when HIV-1 viral load was below 50 RNA copies/mL.
More recently, Lee and colleagues reported a fully automated immunoassay system using whole blood (Lee et al., 2009). This system consists of a lab-on-a-disc (LOD) and a portable analyzer. The disc was preloaded with reagents and buffers necessary for performing ELISA; the analyzer included essential subunits such as a rotation controller, a laser position controller, a temperature controller and an optical detection unit. The automation was made via sequential centrifugation to separate plasma from whole blood and transfer it through chambers, where mixing, incubating, washing or colorimetric reading was performed. Each chamber was localized by using a laser irradiated ferrowax microvalve. Compared to conventional ELISA detecting HBsAg or Anti-HBs antibody, the LOD showed a superior performance regarding the handling time, dynamic range and detection limit. Despite reduced reagent consumption, these two miniaturized ELISA methods have improved sensitivity in the detection of targets of interest. These systems may be explored as POC viral load monitoring tools.
5.6. Portable HIV-1 RNA amplification systems
At the POC settings, automated, portable and self-contained HIV-1 viral load systems would be more suitable. Tanriverdi et al. reported a Liat platform, which can automatically process whole blood, extract HIV-1 RNA, amplify HIV-1 RNA by RT-PCR and report viral load based on fluorescence reading within 88 minutes (Tanriverdi et al., 2010). This assay had sensitivity (50 copies/mL) comparable to commercial RNA assays and the correlation efficiency was 0.92 and 0.88 when compared to the Versant bDNA and the COBAS assay, respectively. Dipstick-based nucleic acid detection has also been integrated into portable cartridges which contain reagents for HIV-1 RNA amplification either by reverse-transcription helicase-dependent amplification (RT-HDA) (Tang et al., 2010) or NASBA (Lee et al., 2010a). Both systems explore isothermal RNA amplification to eliminate the need for thermal cyclers. Notably, the former assay is termed as IsoAmp, which achieves DNA separation by helicase other than by heating and subsequent amplification by DNA polymerase. Both IsoAmp and SAMBA have shown satisfactory detection of HIV-1 RNA at 50 and 200 copies/mL, respectively. However, these two systems have shown saturated signals on dipsticks (end-point detection), which may restrict them from accurately reporting viral load over a wide dynamic range (e.g., 200–1,000,000 copies/mL). Nevertheless, the concept of using a closed system either in a tube or in a cartridge is representative of the current trend in developing POC viral load assays.
5.7. POC CD4+ cell count
The number of CD4+ T lymphocytes is essential to determine the immune state of HIV-infected individuals. To realize CD4+ cell counting at the POC, functionalized microfluidic devices have been developed to capture CD4+ T lymphocytes from 10 microliters of whole blood (Jokerst et al., 2008; Moon et al., 2009; Rodriguez et al., 2005). In brief, anti-CD4+ antibody was immobilized via biotin-streptavidin interaction on the surface of a microchannel. The CD4+ T lymphocytes were captured from whole blood and then tagged with fluorescence. Then, they were counted under fluorescence microscopy (Jokerst et al., 2008; Rodriguez et al., 2005). Since a fluorescence microscope is not affordable and not practical at the POC, a light microscope can be used, which however involves tedious manual counting (Cheng et al., 2007). Recently, Cheng et al. demonstrated the feasibility of measuring the number of CD4+ cells based on impedance sensing (Cheng et al., 2009). Although this method significantly reduces the testing time, it may suffer from patient-to-patient variation or the requirement for stringent storage conditions. Alternatively, CD4+ cell counts can be obtained using a lensless, ultra wide-field cell array based on shadow imaging (LUCAS) (Ozcan and Demirci, 2008), which enables rapid CD4+ cell counting within 10 minutes using a portable battery-powered charge-coupled device (CCD) camera (Moon et al., 2009). The development of POC CD4+ quantification devices shows the potential of pairing with POC viral load testing to facilitate clinical decision-making on site.
To facilitate ART expansion in developing countries, POC viral load assays as well as POC CD4 assays are urgently needed. Despite tremendous progress on these areas, no commercial POC viral load assays have become readily available yet. The main challenges for developing such assays are to minimize the cost, sample-to-answer time, instrument dependence and required human skills while maximizing the robustness of these assays to be more tolerant to various storage and testing conditions. In addition, these assays should not be interfered by emerging HIV-1 subtypes, which are dominant in developing countries. To realize POC viral load testing, miniaturized amplification/detection systems in disposable devices (e.g., cartridges) potentially have advantages over traditional viral load assays in further decreasing the cost (reagents and/or nanoparticles) and improving portability. However, clinical trials of these disposable devices at the POC are urgently needed to further characterize their performances including sensitivity, dynamic range, accuracy and throughput.
6. Summary and Prospective
In developed countries, commercial RNA-based assays have been widely used to monitor HIV-1 viral load in plasma since it can gauge the efficacy of ART and monitor the progression of AIDS. Recent advances in commercial viral load assays include: 1) use of the fluorescence-based real-time technology, 2) improved quantification of HIV-1 subtypes within group M, and detection of groups N and O (Abbott RealTime), and 3) automation of sample preparation. Notably, the fluorescence-based real-time technologies allow for a higher throughput and a wider dynamic range. However, the implementation of such assays requires skilled workers, solid infrastructure, and sufficient capital, which prevents them from being practical for resource-limited settings.
To implement cost-effective viral load monitoring in resource-limited settings, inexpensive alternatives such as the Ultrasensitive p24 assay, the ExaVir™ RT assay and in-house RT-qPCR have been developed. The Ultrasensitive p24 assay needs to be evaluated because of variability, which may arise from incomplete disassociation of the p24-antibody complex in plasma. Both the ExaVir™ RT assay and in-house RT-qPCR have demonstrated a good correlation with commercial viral load assays. Further, ExaVir™ RT version 3.0 improves the detection limit and throughput in comparison with previous versions and in-house LTR-based RT-qPCR method has become commercially available. Nevertheless, these two methods still require long turnaround time (e.g., up to 48 hours for ExaVir™ RT 3.0) or well-trained operators (e.g., RT-qPCR).
To provide HIV-1 viral load monitoring at POC settings, new approaches based on microfluidics and nanotechnology have been developed to further reduce cost, complexity and turnaround time. Generally, two strategies have been used, either miniaturization (e.g., cartridge with pre-loaded reagents) of the current quantification methods such as RT-PCR, NASBA and ELISA, or development of new quantification methods such as BCA or microfluidics-based viral counting. However, detailed aspects should be considered carefully at the early stage, including the quantification markers (RNA, p24 or virus), the types of clinical samples (whole blood or plasma), and the detection strategy (nucleic acid amplification or signal amplification). In addition, production cost, stability of reagents and throughput should be taken into account during the design. Of these emerging approaches, a cartridge with preloaded reagents, in which nucleic acid amplification can be performed with the aid of an automated portable system, may deliver POC viral load testing in the future. Alternatively, microfluidics-based viral counting, once integrated into a portable lensless imaging system, may achieve POC viral load testing without nucleic acid amplification.
Acknowledgments
We acknowledge support from MIT Deshpande Center, W.H. Coulter Foundation Young Investigator Award,. We also acknowledge support from NIH (R01AI081534) and NIH (R21AI087107). This work was supported by Center for Integration of Medicine and Innovative Technology (CIMIT) under U.S. Army Medical Research Acquisition Activity Cooperative Agreement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott. Abbott RealTime HIV-1 (package insert) 2007. [Google Scholar]
- Alyassin MA, Moon S, Keles HO, Manzur F, Lin RL, Haeggstrom E, et al. Rapid automated cell quantification on HIV microfluidic devices. Lab Chip. 2009;9:3364–9. doi: 10.1039/b911882a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A, Klapperich CM. Design and testing of a disposable microfluidic chemiluminescent immunoassay for disease biomarkers in human serum samples. Biomed Microdevices. 2007;9:245–51. doi: 10.1007/s10544-006-9026-2. [DOI] [PubMed] [Google Scholar]
- Bonard D, Rouet F, Toni TA, Minga A, Huet C, Ekouevi DK, et al. Field evaluation of an improved assay using a heat-dissociated p24 antigen for adults mainly infected with HIV-1 CRF02_AG strains in Cote d’Ivoire, West Africa. J Acquir Immune Defic Syndr. 2003;34:267–73. doi: 10.1097/00126334-200311010-00002. [DOI] [PubMed] [Google Scholar]
- Boni J, Opravil M, Tomasik Z, Rothen M, Bisset L, Grob PJ, et al. Simple monitoring of antiretroviral therapy with a signal-amplification-boosted HIV-1 p24 antigen assay with heat-denatured plasma. Aids. 1997;11:F47–52. doi: 10.1097/00002030-199706000-00001. [DOI] [PubMed] [Google Scholar]
- Calmy A, Ford N, Hirschel B, Reynolds SJ, Lynen L, Goemaere E, et al. HIV viral load monitoring in resource-limited regions: optional or necessary? Clin Infect Dis. 2007;44:128–34. doi: 10.1086/510073. [DOI] [PubMed] [Google Scholar]
- Cassol S, Gill MJ, Pilon R, Cormier M, Voigt RF, Willoughby B, et al. Quantification of human immunodeficiency virus type 1 RNA from dried plasma spots collected on filter paper. J Clin Microbiol. 1997;35:2795–801. doi: 10.1128/jcm.35.11.2795-2801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Learmonth K, Dax EM. HIV testing in 2006: issues and methods. Expert Rev Anti Infect Ther. 2006;4:565–82. doi: 10.1586/14787210.4.4.565. [DOI] [PubMed] [Google Scholar]
- Chantry CJ, Cooper ER, Pelton SI, Zorilla C, Hillyer GV, Diaz C. Seroreversion in human immunodeficiency virus-exposed but uninfected infants. Pediatr Infect Dis J. 1995;14:382–7. doi: 10.1097/00006454-199505000-00009. [DOI] [PubMed] [Google Scholar]
- Cheng X, Irimia D, Dixon M, Ziperstein JC, Demirci U, Zamir L, et al. A microchip approach for practical label-free CD4+ T-cell counting of HIV-infected subjects in resource-poor settings. J Acquir Immune Defic Syndr. 2007;45:257–61. doi: 10.1097/QAI.0b013e3180500303. [DOI] [PubMed] [Google Scholar]
- Cheng XH, Gupta A, Chen CC, Tompkins RG, Rodriguez W, Toner M. Enhancing the performance of a point-of-care CD4+T-cell counting microchip through monocyte depletion for HIV/AIDS diagnostics. Lab on a Chip. 2009;9:1357–64. doi: 10.1039/b818813k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins ML, Irvine B, Tyner D, Fine E, Zayati C, Chang C, et al. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–84. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–2. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- de Mendoza C, Koppelman M, Montes B, Ferre V, Soriano V, Cuypers H, et al. Multicenter evaluation of the NucliSens EasyQ HIV-1 v1.1 assay for the quantitative detection of HIV-1 RNA in plasma. J Virol Methods. 2005;127:54–9. doi: 10.1016/j.jviromet.2005.03.013. [DOI] [PubMed] [Google Scholar]
- de Mendoza C, Soriano V. Update on HIV viral-load assays: new technologies and testing in resource-limited settings. Future Virol. 2009;4:423–30. [Google Scholar]
- Deiman B, van Aarle P, Sillekens P. Characteristics and applications of nucleic acid sequence-based amplification (NASBA) Mol Biotechnol. 2002;20:163–79. doi: 10.1385/MB:20:2:163. [DOI] [PubMed] [Google Scholar]
- Drosten C, Panning M, Drexler JF, Hansel F, Pedroso C, Yeats J, et al. Ultrasensitive monitoring of HIV-1 viral load by a low-cost real-time reverse transcription-PCR assay with internal control for the 5′ long terminal repeat domain. Clin Chem. 2006;52:1258–66. doi: 10.1373/clinchem.2006.066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, et al. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc Natl Acad Sci U S A. 2006;103:19272–7. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbeik T, Alvord WG, Trichavaroj R, de Souza M, Dewar R, Brown A, et al. Comparative analysis of HIV-1 viral load assays on subtype quantification: Bayer Versant HIV-1 RNA 3.0 versus Roche Amplicor HIV-1 Monitor version 1.5. J Acquir Immune Defic Syndr. 2002;29:330–9. doi: 10.1097/00126334-200204010-00002. [DOI] [PubMed] [Google Scholar]
- Fiscus SA, Cheng B, Crowe SM, Demeter L, Jennings C, Miller V, et al. HIV-1 viral load assays for resource-limited settings. PLoS Med. 2006;3:e417. doi: 10.1371/journal.pmed.0030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiscus SA, Wiener J, Abrams EJ, Bulterys M, Cachafeiro A, Respess RA. Ultrasensitive p24 antigen assay for diagnosis of perinatal human immunodeficiency virus type 1 infection. J Clin Microbiol. 2007;45:2274–7. doi: 10.1128/JCM.00813-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli R, Merrick L, Friesenhahn M, Ziermann R. Comprehensive comparison of the VERSANT HIV-1 RNA 3.0 (bDNA) and COBAS AMPLICOR HIV-1 MONITOR 1.5 assays on 1,000 clinical specimens. J Clin Virol. 2005;34:245–52. doi: 10.1016/j.jcv.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Ginocchio CC, Kemper M, Stellrecht KA, Witt DJ. Multicenter evaluation of the performance characteristics of the NucliSens HIV-1 QT assay used for quantitation of human immunodeficiency virus type 1 RNA. J Clin Microbiol. 2003;41:164–73. doi: 10.1128/JCM.41.1.164-173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleaves CA, Welle J, Campbell M, Elbeik T, Ng V, Taylor PE, et al. Multicenter evaluation of the Bayer VERSANT HIV-1 RNA 3.0 assay: analytical and clinical performance. J Clin Virol. 2002;25:205–16. doi: 10.1016/s1386-6532(02)00011-2. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Grimes R, Walsh EJ, Dalton T, Davies M. Interaction of quantitative PCR components with polymeric surfaces. Biomedical Microdevices. 2007;9:261–6. doi: 10.1007/s10544-006-9030-6. [DOI] [PubMed] [Google Scholar]
- Hammer SM, Eron JJ, Jr, Reiss P, Schooley RT, Thompson MA, Walmsley S, et al. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society-USA panel. Jama. 2008;300:555–70. doi: 10.1001/jama.300.5.555. [DOI] [PubMed] [Google Scholar]
- Harries AD, Zachariah R, van Oosterhout JJ, Reid SD, Hosseinipour MC, Arendt V, et al. Diagnosis and management of antiretroviral-therapy failure in resource-limited settings in sub-Saharan Africa: challenges and perspectives. Lancet Infect Dis. 2010;10:60–5. doi: 10.1016/S1473-3099(09)70321-4. [DOI] [PubMed] [Google Scholar]
- Jennings C, Fiscus SA, Crowe SM, Danilovic AD, Morack RJ, Scianna S, et al. Comparison of two human immunodeficiency virus (HIV) RNA surrogate assays to the standard HIV RNA assay. J Clin Microbiol. 2005;43:5950–6. doi: 10.1128/JCM.43.12.5950-5956.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen A, Garrido C, Zahonero N, Sandvik L, Naman E, Kivuyo SL, et al. Dried blood spots perform well in viral load monitoring of patients who receive antiretroviral treatment in rural Tanzania. Clin Infect Dis. 2009;49:976–81. doi: 10.1086/605502. [DOI] [PubMed] [Google Scholar]
- Jokerst JV, Floriano PN, Christodoulides N, Simmons GW, McDevitt JT. Integration of semiconductor quantum dots into nano-bio-chip systems for enumeration of CD4+T cell counts at the point-of-need. Lab on a Chip. 2008;8:2079–90. doi: 10.1039/b817116e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kievits T, van Gemen B, van Strijp D, Schukkink R, Dircks M, Adriaanse H, et al. NASBA isothermal enzymatic in vitro nucleic acid amplification optimized for the diagnosis of HIV-1 infection. J Virol Methods. 1991;35:273–86. doi: 10.1016/0166-0934(91)90069-c. [DOI] [PubMed] [Google Scholar]
- Kim EY, Stanton J, Korber BT, Krebs K, Bogdan D, Kunstman K, et al. Detection of HIV-1 p24 Gag in plasma by a nanoparticle-based bio-barcode-amplification method. Nanomed. 2008;3:293–303. doi: 10.2217/17435889.3.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Moon S, Kuritzkes DR, Demirci U. Quantum dot-based HIV capture and imaging in a microfluidic channel. Biosens Bioelectron. 2009;25:253–8. doi: 10.1016/j.bios.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolari K, Satokari R, Kataja K, Stenman J, Hokkanen A. Real-time analysis of PCR inhibition on microfluidic materials. Sensors and Actuators B-Chemical. 2008;128:442–9. [Google Scholar]
- Kuritzkes DR, Ribaudo HJ, Squires KE, Koletar SL, Santana J, Riddler SA, et al. Relationship of plasma HIV-1 RNA dynamics to treatment regimen and sex in antiretroviral-native subjects receiving either triple-nucleoside- or efavirenz-containing regimens: AIDS Clinical Trials Group protocol A5166s. J Infect Dis. 2007;195:1169–76. doi: 10.1086/512619. [DOI] [PubMed] [Google Scholar]
- Labbett W, Garcia-Diaz A, Fox Z, Clewley GS, Fernandez T, Johnson M, et al. Comparative evaluation of the ExaVir Load version 3 reverse transcriptase assay for measurement of human immunodeficiency virus type 1 plasma load. J Clin Microbiol. 2009;47:3266–70. doi: 10.1128/JCM.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BS, Lee JN, Park JM, Lee JG, Kim S, Cho YK, et al. A fully automated immunoassay from whole blood on a disc. Lab Chip. 2009;9:1548–55. doi: 10.1039/b820321k. [DOI] [PubMed] [Google Scholar]
- Lee HH, Dineva MA, Chua YL, Ritchie AV, Ushiro-Lumb I, Wisniewski CA. Simple amplification-based assay: a nucleic acid-based point-of-care platform for HIV-1 testing. J Infect Dis. 2010a;201 (Suppl 1):S65–72. doi: 10.1086/650385. [DOI] [PubMed] [Google Scholar]
- Lee K-B, Kim E-Y, Mirkin CA, Wolinsky SM. The Use of Nanoarrays for Highly Sensitive and Selective Detection of Human Immunodeficiency Virus Type 1 in Plasma. Nano Letters. 2004;4:1869–72. [Google Scholar]
- Lee SH, Kim SW, Kang JY, Ahn CH. A polymer lab-on-a-chip for reverse transcription (RT)-PCR based point-of-care clinical diagnostics. Lab Chip. 2008;8:2121–7. doi: 10.1039/b811131f. [DOI] [PubMed] [Google Scholar]
- Lee WG, Kim YG, Chung BG, Demirci U, Khademhosseini A. Nano/Microfluidics for diagnosis of infectious diseases in developing countries. Adv Drug Deliv Rev. 2010b;62:449–57. doi: 10.1016/j.addr.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liegler TJ, Grant RM. [Accessed on September 2, 2009];Nucleic Acid-Based HIV-1 Viral Load Assays. http://hivinsiteucsfedu/InSite?page=kb-02-02-02-01#.
- Liu WT, Zhu L, Qin QW, Zhang Q, Feng H, Ang S. Microfluidic device as a new platform for immunofluorescent detection of viruses. Lab Chip. 2005;5:1327–30. doi: 10.1039/b509086e. [DOI] [PubMed] [Google Scholar]
- Lombart JP, Vray M, Kafando A, Lemee V, Ouedraogo-Traore R, Corrigan GE, et al. Plasma virion reverse transcriptase activity and heat dissociation-boosted p24 assay for HIV load in Burkina Faso, West Africa. AIDS. 2005;19:1273–7. doi: 10.1097/01.aids.0000180098.58017.48. [DOI] [PubMed] [Google Scholar]
- Malmsten A, Shao XW, Aperia K, Corrigan GE, Sandstrom E, Kallander CF, et al. HIV-1 viral load determination based on reverse transcriptase activity recovered from human plasma. J Med Virol. 2003;71:347–59. doi: 10.1002/jmv.10492. [DOI] [PubMed] [Google Scholar]
- Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Diagnostics for the developing world: microfluidic paper-based analytical devices. Anal Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- Martinez AW, Phillips ST, Wiley BJ, Gupta M, Whitesides GM. FLASH: a rapid method for prototyping paper-based microfluidic devices. Lab Chip. 2008;8:2146–50. doi: 10.1039/b811135a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee P, Fielding KL, Charalambous S, Churchyard GJ, Grant AD. Evaluation of the WHO criteria for antiretroviral treatment failure among adults in South Africa. AIDS. 2008;22:1971–7. doi: 10.1097/QAD.0b013e32830e4cd8. [DOI] [PubMed] [Google Scholar]
- Mellors JW, Munoz A, Giorgi JV, Margolick JB, Tassoni CJ, Gupta P, et al. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–54. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- Michael NL, Herman SA, Kwok S, Dreyer K, Wang J, Christopherson C, et al. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J Clin Microbiol. 1999;37:2557–63. doi: 10.1128/jcm.37.8.2557-2563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine M, Bedi K, Maruta T, Madziva D, Tau M, Zana T, et al. Quantitation of human immunodeficiency virus type 1 viral load in plasma using reverse transcriptase activity assay at a district hospital laboratory in Botswana: a decentralization pilot study. J Virol Methods. 2009;159:93–7. doi: 10.1016/j.jviromet.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Moon S, Keles HO, Ozcan A, Khademhosseini A, Haeggstrom E, Kuritzkes D, et al. Integrating microfluidics and lensless imaging for point-of-care testing. Biosens Bioelectron. 2009;24:3208–14. doi: 10.1016/j.bios.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DG, Cote L, Fauvel M, Rene P, Vincelette J. Multicenter comparison of Roche COBAS AMPLICOR MONITOR version 1.5, Organon Teknika NucliSens QT with Extractor, and Bayer Quantiplex version 3.0 for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2000;38:4034–41. doi: 10.1128/jcm.38.11.4034-4041.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JM, Stoeva SI, Mirkin CA. Bio-bar-code-based DNA detection with PCR-like sensitivity. J Am Chem Soc. 2004;126:5932–3. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–6. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- O’Brien WA, Hartigan PM, Martin D, Esinhart J, Hill A, Benoit S, et al. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. Veterans Affairs Cooperative Study Group on AIDS. N Engl J Med. 1996;334:426–31. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- Ozcan A, Demirci U. Ultra wide-field lens-free monitoring of cells on-chip. Lab Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- Pascual A, Cachafeiro A, Funk ML, Fiscus SA. Comparison of an assay using signal amplification of the heat-dissociated p24 antigen with the Roche Monitor human immunodeficiency virus RNA assay. J Clin Microbiol. 2002;40:2472–5. doi: 10.1128/JCM.40.7.2472-2475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado JG, Shintani A, Bofill M, Clotet B, Ruiz L, Martinez-Picado J. Lack of longitudinal intrapatient correlation between p24 antigenemia and levels of human immunodeficiency virus (HIV) type 1 RNA in patients with chronic hiv infection during structured treatment interruptions. J Clin Microbiol. 2004;42:1620–5. doi: 10.1128/JCM.42.4.1620-1625.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche . COBAS AmpliPrep/COBAS TaqMan HIV-1 Test. 2007. [Google Scholar]
- Rodriguez WR, Christodoulides N, Floriano PN, Graham S, Mohanty S, Dixon M, et al. A microchip CD4 counting method for HIV monitoring in resource-poor settings. PLoS Med. 2005;2:e182. doi: 10.1371/journal.pmed.0020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet F, Chaix ML, Nerrienet E, Ngo-Giang-Huong N, Plantier JC, Burgard M, et al. Impact of HIV-1 genetic diversity on plasma HIV-1 RNA Quantification: usefulness of the Agence Nationale de Recherches sur le SIDA second-generation long terminal repeat-based real-time reverse transcriptase polymerase chain reaction test. J Acquir Immune Defic Syndr. 2007;45:380–8. doi: 10.1097/QAI.0b013e3180640cf5. [DOI] [PubMed] [Google Scholar]
- Rouet F, Ekouevi DK, Chaix ML, Burgard M, Inwoley A, Tony TD, et al. Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus type 1 infection in a West African resource-limited setting. J Clin Microbiol. 2005;43:2709–17. doi: 10.1128/JCM.43.6.2709-2717.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet F, Menan H, Viljoen J, Ngo-Giang-Huong N, Mandaliya K, Valea D, et al. In-house HIV-1 RNA real-time RT-PCR assays: principle, available tests and usefulness in developing countries. Expert Rev Mol Diagn. 2008;8:635–50. doi: 10.1586/14737159.8.5.635. [DOI] [PubMed] [Google Scholar]
- Rouet F, Montcho C, Rouzioux C, Leroy V, Msellati P, Kottan JB, et al. Early diagnosis of paediatric HIV-1 infection among African breast-fed children using a quantitative plasma HIV RNA assay. Aids. 2001;15:1849–56. doi: 10.1097/00002030-200109280-00015. [DOI] [PubMed] [Google Scholar]
- Rouet F, Rouzioux C. HIV-1 viral load testing cost in developing countries: what’s new? Expert Rev Mol Diagn. 2007;7:703–7. doi: 10.1586/14737159.7.6.703. [DOI] [PubMed] [Google Scholar]
- Ruelle J, Jnaoui K, Lefevre I, Lamarti N, Goubau P. Comparative evaluation of the VERSANT HIV-1 RNA 1.0 kinetic PCR molecular system (kPCR) for the quantification of HIV-1 plasma viral load. J Clin Virol. 2009;44:297–301. doi: 10.1016/j.jcv.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Schumacher W, Frick E, Kauselmann M, Maier-Hoyle V, van der Vliet R, Babiel R. Fully automated quantification of human immunodeficiency virus (HIV) type 1 RNA in human plasma by the COBAS AmpliPrep/COBAS TaqMan system. J Clin Virol. 2007;38:304–12. doi: 10.1016/j.jcv.2006.12.022. [DOI] [PubMed] [Google Scholar]
- Schupbach J, Flepp M, Pontelli D, Tomasik Z, Luthy R, Boni J. Heat-mediated immune complex dissociation and enzyme-linked immunosorbent assay signal amplification render p24 antigen detection in plasma as sensitive as HIV-1 RNA detection by polymerase chain reaction. AIDS. 1996;10:1085–90. [PubMed] [Google Scholar]
- Sia SK, Linder V, Parviz BA, Siegel A, Whitesides GM. An integrated approach to a portable and low-cost immunoassay for resource-poor settings. Angew Chem Int Ed Engl. 2004;43:498–502. doi: 10.1002/anie.200353016. [DOI] [PubMed] [Google Scholar]
- Stevens G, Rekhviashvili N, Scott LE, Gonin R, Stevens W. Evaluation of two commercially available, inexpensive alternative assays used for assessing viral load in a cohort of human immunodeficiency virus type 1 subtype C-infected patients from South Africa. J Clin Microbiol. 2005;43:857–61. doi: 10.1128/JCM.43.2.857-861.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson P, Huang S, Abravaya K, de Mendoza C, Soriano V, Devare SG, et al. Evaluation of performance across the dynamic range of the Abbott RealTime HIV-1 assay as compared to VERSANT HIV-1 RNA 3.0 and AMPLICOR HIV-1 MONITOR v1.5 using serial dilutions of 39 group M and O viruses. J Virol Methods. 2007;141:49–57. doi: 10.1016/j.jviromet.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Tang N, Huang S, Salituro J, Mak WB, Cloherty G, Johanson J, et al. A RealTime HIV-1 viral load assay for automated quantitation of HIV-1 RNA in genetically diverse group M subtypes A-H, group O and group N samples. J Virol Methods. 2007a doi: 10.1016/j.jviromet.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Tang S, Hewlett I. Nanoparticle-based immunoassays for sensitive and early detection of HIV-1 capsid (p24) antigen. J Infect Dis. 2010;201 (Suppl 1):S59–64. doi: 10.1086/650386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Zhao J, Storhoff JJ, Norris PJ, Little RF, Yarchoan R, et al. Nanoparticle-Based biobarcode amplification assay (BCA) for sensitive and early detection of human immunodeficiency type 1 capsid (p24) antigen. J Acquir Immune Defic Syndr. 2007b;46:231–7. doi: 10.1097/QAI.0b013e31814a554b. [DOI] [PubMed] [Google Scholar]
- Tang W, Chow WH, Li Y, Kong H, Tang YW, Lemieux B. Nucleic acid assay system for tier II laboratories and moderately complex clinics to detect HIV in low-resource settings. J Infect Dis. 2010;201 (Suppl 1):S46–51. doi: 10.1086/650388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanriverdi S, Chen L, Chen S. A rapid and automated sample-to-result HIV load test for near-patient application. J Infect Dis. 2010;201 (Suppl 1):S52–8. doi: 10.1086/650387. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS. [Accessed November 20, 2009];Report on the global AIDS epidemic. 2008 Available at: http://data.unaids.org/pub/GlobalReport/2008/JC1510_2008GlobalReport_en.zip.
- Usdin M, Guillerm M, Calmy A. Patient needs and point-of-care requirements for HIV load testing in resource-limited settings. J Infect Dis. 2010;201 (Suppl 1):S73–7. doi: 10.1086/650384. [DOI] [PubMed] [Google Scholar]
- van Oosterhout JJG, Brown L, Weigel R, Kumwenda JJ, Mzinganjira D, Saukila N, et al. Diagnosis of antiretroviral therapy failure in Malawi: poor performance of clinical and immunological WHO criteria. Trop Med Int Health. 2009;14:856–61. doi: 10.1111/j.1365-3156.2009.02309.x. [DOI] [PubMed] [Google Scholar]
- Vekemans M, John L, Colebunders R. When to switch for antiretroviral treatment failure in resource-limited settings? AIDS. 2007;21:1205–6. doi: 10.1097/QAD.0b013e3281c617e8. [DOI] [PubMed] [Google Scholar]
- Wang S, Moon S, Demirci U. Development of a microfluidic system for measuring HIV-1 viral load. Proc SPIE - Int Soc Opt Eng. 2010a doi: 10.1117/12.853132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, SangJun M, Demirci U. Development of a microfluidic system for measuring HIV-1 viral load. Proc SPIE - Int Soc Opt Eng. 2010b doi: 10.1117/12.853132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. [Accessed on November 25, 2009];Antiretroviral therapy for HIV infection in adults and adolescents. 2006a Available at: http://www.who.int/hiv/pub/guidelines/artadultguidelines.pdf. [PubMed]
- World Health Organization. [Accessed on November 15, 2009];Towards universal access by 2010. 2006b Available at: http://www.who.int/hiv/toronto2006/towardsuniversalaccess.pdf. [PubMed]
- World Health Organization. Scaling up priority HIV/AIDS interventions in the health sector-Progress report. [Accessed on November 15, 2009];2009 Available at: http://www.who.int/hiv/pub/tuapr_2009_en.pdf.
- Yager P, Domingo GJ, Gerdes J. Point-of-care diagnostics for global health. Annu Rev Biomed Eng. 2008;10:107–44. doi: 10.1146/annurev.bioeng.10.061807.160524. [DOI] [PubMed] [Google Scholar]
- Yao J, Liu Z, Ko LS, Pan G, Jiang Y. Quantitative detection of HIV-1 RNA using NucliSens EasyQ HIV-1 assay. J Virol Methods. 2005;129:40–6. doi: 10.1016/j.jviromet.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Ymeti A, Greve J, Lambeck PV, Wink T, van Hovell SW, Beumer TA, et al. Fast, ultrasensitive virus detection using a Young interferometer sensor. Nano Lett. 2007;7:394–7. doi: 10.1021/nl062595n. [DOI] [PubMed] [Google Scholar]
- Zhang C, Xing D. Miniaturized PCR chips for nucleic acid amplification and analysis: latest advances and future trends. Nucleic Acids Res. 2007;35:4223–37. doi: 10.1093/nar/gkm389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, van der Berg A. Lab on paper. Lab Chip. 2008;8:1988–91. doi: 10.1039/b814043j. [DOI] [PubMed] [Google Scholar]