Abstract
Ctf7/Eco1-dependent acetylation of Smc3 is essential for sister chromatid cohesion. Here, we use epitope tag-induced lethality in cells diminished for Ctf7/Eco1 activity to map cohesin architecture in vivo. Tagging either Smc1 or Mcd1/Scc1, but not Scc3/Irr1, appears to abolish access to Smc3 in ctf7/eco1 mutant cells, suggesting that Smc1 and Smc3 head domains are in direct contact with each other and also with Mcd1/Scc1. Thus, cohesin complexes may be much more compact than commonly portrayed. We further demonstrate that mutation in ELG1 or RFC5 anti-establishment genes suppress tag-induced lethality, consistent with the notion that the replication fork regulates Ctf7/Eco1.
Keywords: Cohesin, sister chromatids, RFC complexes, CTF7/ECO1, MCD1/SCC1, SMC1
INTRODUCTION
Cohesins tether together sister chromatids from early S-phase until anaphase onset and are essential for both high fidelity chromosome transmission and transcription regulation. Cohesin complexes consist of Smc1, Smc3, Mcd1/Scc1 (herein Mcd1) and Scc3/Irr1 and accessory factors Pds5 and Rad61/WAPL [1]. While many of the components that make up the cohesin complex are known, the structure of this complex in vivo remains under intense debate. Based on early electron microscopy (EM) studies and subsequent biochemical assays, a popular model of cohesin is that Smc1 and Smc3 dimerize by hinge-hinge interactions and that their separated head domains are bridged by Mcd1. From this, a huge triangular ring is posited to result with in an inner diameter of 35 nm [2,3]. More recent Atomic Force Microscopy, Fluorescence Resonance Energy Transfer (FRET) and EM studies suggest that Smc1 and Smc3 head domains may remain in contact with one another and that their coiled-coil domains fold back to form shortened structures in which the ATPase heads are positioned closely to the hinges [4–6]. These subunit interactions provide for numerous configurations including one ring, bracelets, snaps, two-ring hand-cuffs and C-clamps [7–12].
A major obstacle in discerning the physiologically relevant cohesin architecture in vivo is that there is evidence supporting both one ring and two ring models. Both models rely heavily on analyses of epitope-tagged subunits. Intriguingly, these studies reveal that alternate tags produce cohesin complexes of different biochemical properties [3,5,13]. A critical issue thus becomes the extent to which these tagged cohesin proteins give rise to complexes that behave like wild type complexes in vivo. In either case, chromatid-bound cohesins must be converted to a pairing competent state by the establishment factor Ctf7/Eco1 (herein Ctf7). Ctf7 is an acetyltransferase that modifies Smc3 to produce sister chromatid pairing – linking establishment and cohesin architecture to sister chromatid pairing [14].
Here, we pursue a novel assay to demonstrate in vivo that epitope-tagged cohesin subunits previously used to support one-ring embrace models are unfit when challenged genetically in vivo. We uncovered cohesin tag-induced lethality in cells diminished for Ctf7 acetyltransferase activity. In turn, we exploited this in vivo cohesin architecture assay to ascertain subunit domains proximal to the site of Smc3 acetylation and thus potential regulators of Ctf7-dependent acetylation reactions. In addition, we demonstrate the molecular basis of anti-establishment factors (Rfc5 and Elg1) in regulating sister chromatid pairing reactions [15–16, Maradeo, Garg and Skibbens - submitted].
MATERIALS AND METHODS
Strain Construction
All strains in this study are W303 background (Table 2). To generate Smc1-13MYC integration product PCR was preformed using primers TTGACCTATATAGATATTATTAGTTATTTGACGGGTTATAGCAGAGGTTGGTT TCATAGAGAATTCGAGCTCGTTTAAAC and AGACAGCAACAAGAAAACTCGTCGAAGATCATAACTTTGGACTTGAGCAATT ACGCAGAACGGATCCCCGGGTTAATTAA with pFA6a-13MYC-KANMx6 Longtine vectors.
Table 2.
Strains used in this study are W303 background.
| YMM629 | MATα eco1-1:ADE rfc5-1:LEU ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | Sugimoto et al 1998 |
| YMM630 | MATa eco1-1:ADE rfc5-1:LEU ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | Sugimoto et al 1998 |
| YMM902 | Matα SMC1-13MYC:KAN ade2 his3 leu2 trp1 ura3 | This study |
| YMM708 | MATα SCC3-18MYC:TRP ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Ciosk et al 2000* |
| YMM833 | MATa MCD1-18MYC:TRP elg1::KAN ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | Michaelis et al 1997* |
| YMM864 | MATα MCD1-18MYC:TRP eco1-1:ADE rfc5-1:LEU ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM926 | MATa eco1-1:ADE ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM927 | MATa eco1-1ADE rfc5-1:LEU ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM928 | MATa eco1-1:ADE SMC1-13MYC:KAN ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM929 | MATa eco1-1:ADE SMC1-13MYC:KAN ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM930 | MATα eco1-1:ADE SMC1-13MYC:KAN rfc5-1:LEU ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM931 | MATα eco1-1:ADE SMC1-13MYC:KAN rfc5-1:LEU ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM932 | MATa eco1-1:ADE SMC1-13MYC:KAN rfc5-1:LEU ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM935 | MATa eco1-1ADE ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM936 | MATα eco1-1:ADE rfc5-1:ADE ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM937 | MATα eco1-1:ADE SCC3-18MYC:TRP ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
| YMM940 | MATa eco1-1:ADE SCC3-18MYC:TRP rfc5-1:LEU ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | This study |
Backcrossed to W303
Genetic Analysis
Diploid strains were sporulated in 0.3% potassium acetate and tetrads dissected on YPD agar media. The genotypes of the resultant spores were analyzed for wild type, single, double, triple and quadruple mutants. Isolated spores were grown at log phase in YPD followed by 10-fold serial dilutions and then maintained on YPD agar plates at a range of temperatures. For Table 1A, first MCD1-18MYC elg1 mutant cells were crossed to eco1-1 rfc5-1 mutant cells, sporulated, dissected and genotypes analyzed. No MCD1-18MYC eco1-1 double mutant strains were recovered but both MCD1-18MYC eco1-1 rfc5-1 and MCD1-18MYC eco1-1 elg1 triple mutants were obtained. To bias the recovery of a MCD1-18MYC eco1-1 double mutant, MCD1-18MYC elg1 was crossed to MCD1-18MYC eco1-1 rfc5-1, sporulated, dissected and genotypes analyzed.
Table 1A. mcd1:myc rfc5-1 ctf7eco1-1 X mcd1:myc elg1.
MCD1-18MYC are synthetically lethal in combination with ctf7eco1-1 mutation. The lethality of ctf7eco1-1 MCD1-18MYC can be bypassed with mutations in either elg1 or rfc5-1.
| GENOTYPE | OBSERVED | EXPECTED |
|---|---|---|
| mcd1:myc | 11 | 15 |
| mcd1:myc elg1 | 10 | 15 |
| mcd1:myc rfc5-1 | 11 | 15 |
| mcd1:myc ctf7eco1-1 | 0 | 15 |
| mcd1:myc elg1 rfc5-1 | 17 | 15 |
| mcd1:myc elg1 ctf7eco1-1 | 13 | 15 |
| mcd1:myc rfc5-1 ctf7eco1-1 | 9 | 15 |
| mcd1:myc elg1 rfc5-1 ctf7eco1-1 | 13 | 15 |
| DEAD | 36 | 15 |
| TOTAL | 120 |
RESULTS AND DISCUSSION
Sister chromatid cohesion, via conversion of cohesins to a pairing competent state, is established during S phase by Ctf7-dependent Smc3 acetylation [17–18]. Numerous studies employ cohesin-tagged subunits to characterize cohesin structure and acetylation. However, studies that challenge the physiological relevance of these constructs in vivo are severely limited. In the process of generating new strains to further characterize cohesion and acetylation regulation, we discovered that C terminally MYC-tagged Mcd1 is lethal when combined with ctf7 alleles. Strains containing MCD1-MYC were crossed to cells containing ctf7eco1-1, sporulated, dissected and the resulting tetrads analyzed. Of an expected 24 MCD1-MYC ctf7eco1-1 spores, no viable isolates were recovered (Table 1A, data not shown). In contrast, MCD1-MYC expression is not lethal in combination with other cohesion mutant strains (scc2, scc4 or pds5), obviating arguments that the lethality reported here is simply based on compounded cohesion defects [19,20]. Nor is MCD1-HA expression lethal in ctf7 mutant strains, augmenting the specificity of the synthetic lethality reported here [21]. These results reveal that mcd1-myc is a severe but cryptic mutant allele of MCD1 and that the MYC-based C-terminal extension specifically diminishes the ability of Ctf7/Eco1 to acetylate Smc3 – the only known essential substrate for Ctf7 acetylation.
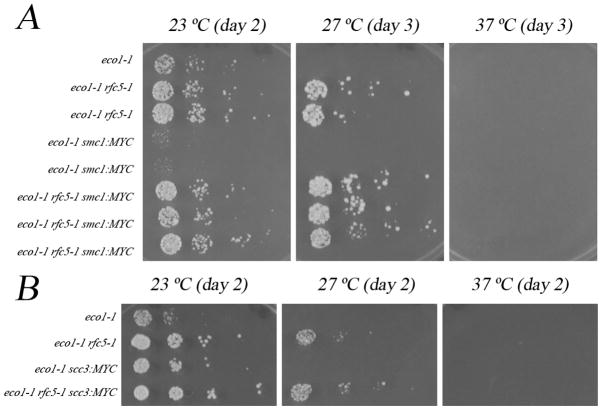
The head domain of Smc1, composed of both N- and C-termini, interacts closely with the C terminus of Mcd1 [3]. If the bulky MYC adduct on Mcd1 significantly diminishes Ctf7 access to Smc3, then MYC-tagging Smc1 C terminus within ctf7 mutant cells should phenocopy this lethality. To test this prediction, we crossed strains containing SMC1-13MYC allele into ctf7eco1-1 mutant cells, sporulated, dissected and analyzed the resulting tetrads. Of the expected 9 SMC1-13MYC ctf7eco1-1 spores, 7 spores were inviable. After prolonged incubation, we were able to recover 2 viable spores, but these are dramatically growth-compromised such that both are near-inviable at temperatures that support robust growth of ctf7eco1-1 single mutant cells (Figure 1A, Table 1B). These findings reveal that smc1-myc is also a severe but cryptic allele of SMC1 and suggests that the bulky 13MYC moiety extending from the C-terminus of Smc1 similarly blocks efficient Ctf7-dependent Smc3 acetylation. In combination, these results support a compacted cohesin structure such that Smc1 and Smc3 head domains are in direct contact with each other and also with Mcd1. These results appear to undermine a popular model in which Smc1 and Smc3 are separate but bridged by Mcd1 to form a huge open ring-like structure capable of entrapping two sister chromatids (Figure 2).
Figure 1.
A) SMC1-13MYC exhibits conditional growth defects in combination with ctf7eco1-1 mutant cells that can be rescued with mutation in rfc5. 10 fold serial dilutions of single, double and triple mutant cells. Colony growth shown for cells on rich medium plates grown at 23 °C, 27 °C, and 37 °C for number of days indicated. B) SCC3-18MYC ctf7eco1-1 mutants exhibit no conditional growth phenotypes. 10 fold serial dilutions of single, double and triple mutant cells. Colony growth shown for cells on rich medium plates grown at 23 °C, 27 °C, and 37 °C for number of days indicated.
Table 1B. smc1:myc X ctf7eco1-1 rfc5-1.
SMC1-13MYC exhibit reduced viability in combination with ctf7eco1-1 mutation.
| GENOTYPE | OBSERVED | EXPECTED |
|---|---|---|
| Wild Type | 7 | 9 |
| smc1:myc | 10 | 9 |
| rfc5-1 | 8 | 9 |
| ctf7eco1-1 | 11 | 9 |
| ctf7eco1-1 rfc5-1 | 8 | 9 |
| smc1:myc ctf7eco1-1 | 2 | 9 |
| smc1:myc rfc5-1 | 10 | 9 |
| smc1:myc ctf7eco1-1 rfc5-1 | 3 | 9 |
| DEAD | 13 | 9 |
| TOTAL | 72 |
Figure 2.
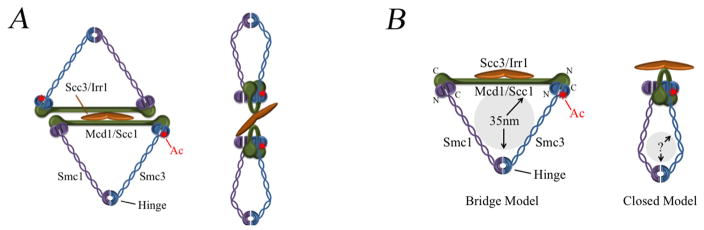
2A) Left: Dimerization cohesins in ‘Bridge’ conformation. Right: Dimerization of cohesins in ‘Closed’ conformation. Both models reflect evidence of Mcd1-Mcd1 dimerization but that a single copy of Scc3 is present in a cohesin complex [4]. Note that several other pairing-competent cohesin structures (one ring, two ring, C-clamps, bracelet-type spirals) are equally feasible and posited in the literature. 2B) Left: Separated head domains of Smc1 (purple) and Smc3 (blue) are bridged by Mcd1 (green) to create a huge triangular ring structure of up to 35 nm. Right: Smc1 and Smc3 head domains are in intimate contact with each other and also with Mcd1 to produce a greatly diminished ring lumen. Numerous studies suggest that folding of Smc1 and Smc3 arms positions the hinge region near to the head domains and thus eliminates an embrace-type ring lumen (1). In either model, Scc3/(orange) binds only through Mcd1/Scc1 and is distal from Smc1/3 head domains. Red sunburst – acetylation site on Smc3; N = N-terminus; C = C-terminus.
To date, little evidence supports a model in which Scc3/Irr1 (herein Scc3) is in direct contact with Smc1 or Smc3 head domains other than it’s binding to Mcd1 [3, 5]. To augment the specificity of the mapping results described above, we tested whether expression of Scc3-18MYC is lethal in ctf7 mutant cells. We crossed strains containing SCC3-18MYC to ctf7eco1-1 mutant cells, sporulated, dissected and analyzed the resulting tetrads. In contrast to either mcd1-myc or smc1-myc, expression of SCC3-MYC in ctf7 mutant cells provided for robust growth at permissive temperature identical to that of single mutant ctf7 cells (Figure 1B). Thus, MYC addition to Scc3 is transparent to Ctf7 access to Smc3, positioning Scc3 away from the Smc3 acetylation site.
Anti-establishment factors are in part defined by the discovery that their deletion rescue ctf7 mutant cell phenotypes [15,16,18,22,23]. Likely anti-establishment mechanisms include altering cohesin complex structure to prohibit pairing or sequestering Ctf7 protein to block cohesin acetylation. The tag-induced lethality reported above provides a robust and unique assay to further characterize the recently identified anti-establishment activities of Elg1-RFC and Rfc5 [15,16, Maradeo, Garg and Skibbens - submitted]. If either Elg1 or Rfc5 block Ctf7 access to Smc3, then deletion of ELG1 or mutation in RFC5 (rfc5-1) should bypass the lethality resulting from co-expression of ctf7eco1-1 and mcd1-myc. To test this hypothesis, mcd1-myc elg1double mutant cells were crossed to ctf7eco1-1 rfc5-1 double mutant cells, sporulated, dissected and the resulting tetrads analyzed. While no ctf7eco1-1 mcd1-myc double mutant isolates were recovered, these crosses produced expected frequencies of both ctf7eco1-1 mcd1-myc elg1 and ctf7eco1-1 mcd1-myc rfc5-1 triple mutant strains (Materials and Methods). A similar level of bypass was obtained when we tested for rfc5-1 bypass of ctf7/eco1-1 smc1-myc phenotypes, although we recovered slightly less then the expected number of spores (Table 1B). In contrast to the severe growth defects exhibited by ctf7eco1-1 smc1-myc even at 23°, ctf7eco1-1 smc1-myc rfc5-1 triple mutant cells grew robustly at both 23°C and 27°C. Moreover, ctf7eco1-1 smc1-myc rfc5-1 triple mutant cells significantly out-performed ctf7eco1-1 single mutants at all temperatures tested (Figure 1A). Thus, both ELG1 deletion and rfc5-1 mutation are individually sufficient to rescue cohesin tag-induced defects in Ctf7-dependent Smc3 acetylation and sister chromatid pairing reactions. In combination, these findings suggest a direct mechanism through which Elg1 and Rfc5 oppose establishment at the DNA replication fork by hindering Ctf7/Eco1 accessibility to Smc3
The inability to biochemically recover cohesin-cohesin interactions was previously used to argue against two-cohesin ring models [3]. In this study, we used a genetic assay to reveal that these epitope tagged cohesins do not function as wild type proteins in vivo but instead are in fact severe but cryptic alleles. Tag-induced lethality appears specific to Ctf7 function and thus Smc3 acetylation - given that similar tagged subunits support viability in other cohesin mutants. These findings undermine more speculative but plausible models that tag-induced lethality is based on adversely affecting some other cohesin mechanism (for instance, tag-based alteration of SMC ATPase cycles). Moreover, the tag-induced lethality reported here supports a compact cohesin architecture in vivo in which Smc1 and Smc3 head domains are in intimate proximity to one another and also to Mcd1 - in contrast to a huge ring model (predicated on separate but bridged Smc1,3 heads) conjectured to entrap two chromatin fibers (Figure 2).
Table 1C. SCC3:MYC X ctf7eco1-1 rfc5-1.
SCC3-18MYC ctf7eco1-1 double allele strains are obtained at the expected frequencies.
| GENOTYPE | OBSERVED | EXPECTED |
|---|---|---|
| Wild Type | 8 | 9 |
| SCC3:MYC | 8 | 9 |
| rfc5-1 | 10 | 9 |
| ctf7eco1-1 | 7 | 9 |
| ctf7eco1-1 rfc5-1 | 10 | 9 |
| SCC3:MYC ctf7eco1-1 | 12 | 9 |
| SCC3:MYC rfc5-1 | 10 | 9 |
| SCC3:MYC ctf7eco1-1 rfc5-1 | 6 | 9 |
| DEAD | 1 | 9 |
| TOTAL | 72 |
Acknowledgments
The authors thank both Skibbens and Cassimeris lab members for comments throughout the course of this study. RVS is supported by awards from the National Institutes of General Medicine (1R15GM083269) and by the Susan G. Komen for the Cure Foundation (BCTR0707708). Any opinions, findings, and conclusions expressed in this study are those of the author(s) and do not necessarily reflect the views of the National Institutes of General Medicine nor those of the Susan G. Komen Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Skibbens RV. Mechanisms of sister chromatid pairing. Int Rev Cell Mol Biol. 2008;269:283–339. doi: 10.1016/S1937-6448(08)01005-8. [DOI] [PubMed] [Google Scholar]
- 2.Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–24. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–88. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 4.Mc Intyre J, Muller EGD, Weitzer S, Snydsman BE, Davis TN, Uhlmann F. In vivo analysis of cohesin architecture using FRET in the budding yeast Saccharomyces cerevisiae. EMBO J. 2007;26:3783–3793. doi: 10.1038/sj.emboj.7601793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang N, Kuznetsov SG, Sharan SK, Li K, Rao PH, Pati D. A handcuff model for the cohesin complex. J Cell Biol. 2008;183:1019–31. doi: 10.1083/jcb.200801157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Surcel A, Koshland D, Ma H, Simpson RT. Cohesin interaction with centromeric minichromosomes shows a multi-complex rod-shaped structure. PLoS One. 2008;3:e2453. doi: 10.1371/journal.pone.0002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skibbens RV. Unzipped and loaded: the role of DNA helicases and RFC clamp-loading complexes in sister chromatid cohesion. J Cell Biol. 2005;169:841–846. doi: 10.1083/jcb.200503129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasmyth K. How might cohesin hold sister chromatids together? Philos Trans R Soc Lond B Biol Sci. 2005;360:483–96. doi: 10.1098/rstb.2004.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philos Trans R Soc Lond B Biol Sci. 2005;360:537–42. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guacci V. Sister chromatid cohesion: the cohesin cleavage model does not ring true. Genes Cells. 2007;12:693–708. doi: 10.1111/j.1365-2443.2007.01093.x. [DOI] [PubMed] [Google Scholar]
- 11.Skibbens RV. Buck the establishment: re-inventing sister chromatid cohesion. Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2010.06.003. (in press) [DOI] [PubMed] [Google Scholar]
- 12.Zhang N, Pati D. Handcuff for sisters: a new model for sister chromatid cohesion. Cell Cycle. 2009;8:399–402. doi: 10.4161/cc.8.3.7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–77. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 14.Skibbens RV. Establishment of Sister Chromatid Cohesion Minireview. Curr Biol. 2009;19:R1126–R1132. doi: 10.1016/j.cub.2009.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maradeo ME, Skibbens RV. The Elg1-RFC Clamp-Loading Complex Performs a Role in Sister Chromatid Cohesion. PLoS One. 2009:4. doi: 10.1371/journal.pone.0004707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parnas O, Zipin-Roitman A, Mazor Y, Liefshitz B, Ben-Aroya S, Kupiec M. The Elg1 Clamp Loader Plays a Role in Sister Chromatid Cohesion. PLoS One. 2009:4. doi: 10.1371/journal.pone.0005497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 18.Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 19.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 20.Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- 21.Milutinovich M, Unal E, Ward C, Skibbens RV, Koshland D. A multi-step pathway for the establishment of sister chromatid cohesion. PLoS Genet. 2007;3:e12. doi: 10.1371/journal.pgen.0030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowland BD, et al. Building Sister Chromatid Cohesion: Smc3 Acetylation Counteracts an Antiestablishment Activity. Mol Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 23.Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol. 2009;19:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]