Abstract
Background
Crohn’s disease (CD) and ulcerative colitis (UC) are associated with expression differences in genes involved in immune function, wound healing, and tissue remodeling. Micro-RNAs (miRNAs) are small, noncoding RNAs that act as potent negative regulators of gene expression and are differentially expressed in chronic inflammatory diseases, including UC. We examined the expression of miRNAs in tissues from different intestinal regions and in patients with active ileal and colonic CD.
Methods
Colonoscopic pinch biopsies were obtained from the terminal ileum, cecum, transverse colon, sigmoid colon, and rectum of normal, healthy adults and from the ileum and sigmoid colon of patients with active ileal and colonic CD. miRNA expression was assessed using miRNA microarray and validated by mature miRNA quantitative reverse-transcription polymerase chain reaction (RT-PCR).
Results
Ten intestine region-specific miRNAs were identified. Three miRNAs were increased and one miRNA was decreased in the terminal ileum as compared to the colon. Six other miRNAs expressed varying levels of expression among the colon regions. Five miRNAs were found to be differentially expressed in tissues of patients with active colonic CD, with three increased and two decreased as compared to normal, healthy controls. Similarly, four miRNAs were found to be significantly increased in tissues of patients with active ileal CD.
Conclusions
The expression differences between ileal CD, colonic CD, and previously identified UC-associated miRNAs support the likelihood that miRNAs influence differing inflammation-related gene expression in each inflammatory bowel disease (IBD) subtype and may form the basis for future diagnostic tests and therapeutic targets for IBD.
Keywords: microRNA, Crohn’s disease, inflammatory bowel diseases, gene expression, microarray
Crohn’s disease (CD) and ulcerative colitis (UC) are the two major types of chronic idiopathic inflammatory bowel disease (IBD). While both are thought to arise as a consequence of an aberrant host immune response to gut flora in genetically predisposed individuals, CD and UC differ with respect to clinical presentation, genetic associations, and gene expression patterns.1 UC affects only the colon, while CD can affect any part of the gastrointestinal tract, with the terminal ileum most commonly involved. Since 2005, genome-wide association studies have identified multiple shared as well as distinct genetic risk factors for CD and UC.2 Furthermore, genome-wide mRNA expression studies have demonstrated that CD and UC differ regarding respective mRNA expression profiles.3–5 These differences in gene expression patterns have also been corroborated by proteomics studies comparing CD and UC.6–8
While the regulation of inflammatory gene expression is not fully understood, microRNAs (miRNAs) are increasingly recognized as important posttranscriptional regulators of gene expression.9 Mature miRNAs are short (19–24 nucleotides) noncoding RNAs that are processed from longer pri-miRNAs transcripts. In the cytoplasm, mature miRNA is incorporated into the RNA-induced silencing complex (RISC) where it recognizes and binds to complementary sequences in the 3′ untranslated region (3′ UTR) of the target mRNAs, resulting in suppression of translation and/or degradation of mRNA.10,11
Since the first human miRNA, Let-7, was discovered in 2000,12 ≈700 human miRNAs have been identified. Each miRNA may regulate hundreds of different protein-coding messenger RNA (mRNA), and conversely, a given mRNA sequence may be targeted by several miRNAs.13 Overall, miRNAs are thought to contribute to the regulation of at least one-third of all protein-encoding mRNAs in humans.14
miRNAs have been implicated in many biological processes, including development, determination of cell fate, metabolism, and hematopoiesis.15 While alterations in miRNA expression have been most widely studied in cancer, growing evidence indicates a significant role of miR-NAs in immune function.16 For example, miRNAs have been shown to influence the expression of cytokines,17 proteins involved in Toll-like receptor and cytokine receptor activation,18,19 and T-cell function.20 Furthermore, there is increasing evidence that miRNAs are altered in chronic inflammatory and autoimmune diseases.21
We recently demonstrated that certain miRNAs are differentially expressed in the tissues of patients with active UC, finding that eight miRNAs were significantly increased and three miRNAs were decreased in sigmoid colon tissues with active UC.22 However, altered expression of miRNAs in CD has not been fully investigated. In this study we examined whether there is intestinal region-specific miRNA expression and whether this expression is altered in ileal CD (Crohn’s ileitis) and colonic CD (Crohn’s colitis).
MATERIALS AND METHODS
Human Intestinal Tissues
Normal, healthy individuals undergoing colonoscopy for colorectal cancer screening and patients with CD were recruited for colonoscopic pinch biopsies using a protocol approved by the Johns Hopkins University Institutional Review Board. Pinch biopsies from the terminal ileum, cecum, transverse colon, sigmoid, and rectum were obtained from six normal healthy individuals undergoing screening colonoscopies. Additional sigmoid pinch biopsies were obtained from seven normal healthy individuals. Pinch biopsies from the ileum were obtained from six patients with chronically active CD. Pinch biopsies from the sigmoid colon were obtained from five patients with chronically active Crohn’s colitis. Diagnoses of active CD were confirmed by histopathology conducted on parallel biopsies taken within 10 cm of the research specimens. Clinical characteristics of patients enrolled in the study are summarized in Table 1.
TABLE 1.
Clinical Characteristics of Patients
| Healthy Control | Crohn’s Disease Sigmoid | Crohn’s Disease TI | |
|---|---|---|---|
| Number patients | 13 | 5 | 6 |
| Male/Female | 6/7 | 3/2 | 5/1 |
| Age (years) | 54.6 | 32.6 | 40.3 |
| Mean (range) | (38–68) | (23–51) | (28–64) |
| Duration IBD (years) | n.a. | 10.2 | 9.2 |
| Mean (range) | (1–22) | (1–27) | |
| Medications | |||
| 5-ASA | 0 | 3 (60%) | 3 (50%) |
| Antibiotics | 0 | 1 (20%) | 2 (33.3%) |
| Steroids | 0 | 0 | 1 (16.7%) |
| Immunomodulators | 0 | 0 | 2 (33.3%) |
| Biologics | 0 | 1 (20%) | 1 (16.7%) |
Total RNA and miRNA Enrichment
Pinch biopsies were placed immediately into 1 mL of Trizol reagent (Invitrogen, La Jolla, CA) and total RNA was extracted. The total RNAs were separated into small RNA fraction and large RNA fragments (>200 nucleotides) using the PureLink miRNA Isolation Kit (Invitrogen). The small RNA fraction was measured using RediPlate 96 RiboGreen RNA Quantitation Kit (Invitrogen). The RNA samples were stored at −80°C.
miRNA Microarray
The miRNA expression profile in the small RNA fraction from each patient was established using the NCode Multi-Species miRNA Microarrays, v. 2 (Invitrogen). This array contains three replicate subarrays, each detecting 467 unique human miRNAs and various controls. A total of 48 miRNA microarray assays were performed.
Briefly, 500 ng of small RNAs, mixed with NCode miRNA Microarray Controls, were labeled with Oyster-550 or Oyster-650 using the Flashtag RNA labeling kit (Genisphere, Hatfield, PA). The labeled RNA was hybridized to an NCode miRNA microarray slide at 52°C for 16 hours. Arrays were scanned using a GenePix 4000B scanner (Molecular Devices, Palo Alto, CA) and raw hybridization intensities were obtained. The background subtracted median fluorescence intensity was used for normalization based on dChip software (http://www.dchip.org/). When comparing two groups, findings were considered significant if 1) fold change was ≥2; 2) t-test, P-value was <0.05; and 3) difference in fluorescence intensity between the two group means were >100 arbitrary units.
Quantitative Reverse-transcription Polymerase Chain Reaction (qRT-PCR)
For validation of miRNA expression, the NCode SYBR green miRNA qRT-PCR Kit (Invitrogen) was used. Briefly, 200 ng of small RNA was converted to cDNA. For miRNA qPCR, the reverse primer was the NCode miRNA universal qPCR primer (Invitrogen). Forward primers were obtained (Operon Technologies, Alameda, CA) and are listed in Table 2. The cycles passing threshold (Ct) were recorded. The expression of each target miRNA in tissues was calculated relative to U6B, a ubiquitously expressed small nuclear RNA that has been widely used as an internal control. Data are presented as target miRNA expression = 2ΔCt, with ΔCt = (U6B Ct − target miRNA Ct).
TABLE 2.
Primers Used for Quantitative Real-time PCR
| Name | Direction | Primer (5′-3′) |
|---|---|---|
| Universal qPCR primer | Reverse | NCode miRNA First-strand cDNA Synthesis Kit (Invitrogen) |
| Let-7d | Forward | AGAGGTAGTAGGTTGCATAGT |
| Let-7i | Forward | TGAGGTAGTAGTTTGTGCTGT |
| miR-22 | Forward | AAGCTGCCAGTTGAAGAACTGT |
| miR-106a | Forward | AAAAGTGCTTACAGTGCAGGTAGC |
| miR-107 | Forward | AGCAGCATTGTACAGGGCTATCA |
| miR-126 | Forward | TCGTACCGTGAGTAATAATGC |
| miR-16 | Forward | TAGCAGCACGTAAATATTGGCG |
| miR-191 | Forward | CAACGGAATCCCAAAAGCAGCT |
| miR-19b | Forward | TGTGCAAATCCATGCAAAACTGA |
| miR-200c | Forward | TAATACTGCCGGGTAATGATGG |
| miR-20a | Forward | TAAAGTGCTTATAGTGCAGGTAG |
| miR-21 | Forward | TAGCTTATCAGACTGATGTTGA |
| miR-215 | Forward | ATGACCTATGAATTGACAGAC |
| miR-223 | Forward | TGTCAGTTTGTCAAATACCCC |
| miR-23a | Forward | ATCACATTGCCAGGGATTTCC |
| miR-23b | Forward | ATCACATTGCCAGGGATTACC |
| miR-26a | Forward | TTCAAGTAATCCAGGATAGGC |
| miR-29a | Forward | TAGCACCATCTGAAATCGGTT |
| miR-31 | Forward | GCAAGATGCTGGCATAGCTG |
| miR-320 | Forward | AAAAGCTGGGTTGAGAGGGCGAA |
| miR-422b | Forward | CTGGACTTGGAGTCAGAAGGCC |
| miR-594 | Forward | ATCTGGGGTGGCCTGTGACTTT |
| miR-629 | Forward | GTTCTCCCAACGTAAGCCCAGC |
| U6B | Forward | CGCAAGGATGACACGCAAATTCG |
Statistical Analysis
Experimental results are expressed as mean values ± standard error. Statistical analyses for qRT-PCR were performed using unpaired, two-tailed Student’s t-tests and one-way analysis of variance (ANOVA) for multiple group comparisons (GraphPad Prism 5, San Diego, CA). P-values < 0.05 were considered significant.
RESULTS
Region-specific miRNA Expression in the Gut
The previous demonstration of intestinal region-specific mRNA expression led us to hypothesize that miRNAs also demonstrate region-specific expression differences.23 To test this hypothesis we performed miRNA microarray analyses on endoscopic pinch biopsies obtained from the terminal ileum, cecum, transverse colon, sigmoid colon, and rectum in six consecutive patients undergoing screening colonoscopy. A total of 30 miRNA microarrays profiles were generated and data analyzed.
Most of the 467 unique miRNAs probed demonstrated undetectable expression or no expression differences. The most highly expressed 5% of miRNAs demonstrated expression levels at least 4-fold greater than the background signal. We identified 13 miRNAs with high expression levels in both the terminal ileum and all four colon regions, i.e., with expression levels at least 4-fold greater background in all regions tested (Table 3). The expression levels of 12 of these 13 miRNAs, let-7a, let-7b, let-7c, miRs-143, -192, -194, -200c, -200b, -24, -26a, -30d, and -375, demonstrated no significant differences among any regions studied. Of these 13 highly expressed miRNAs, only miR-320 demonstrated regional expression differences, with significantly higher levels in the four colon regions as compared to the terminal ileum.
TABLE 3.
Highly Expressed miRNAs in Ileum and Colona
| Terminal Ileum | Cecum | Trans.Colon | Sigmoid | Rectum | |
|---|---|---|---|---|---|
| Let-7a | 1469 ± 576 | 997 ± 530 | 1134 ± 912 | 939 ± 167 | 1246 ± 638 |
| Let-7b | 1869 ± 516 | 1684 ± 555 | 1855 ± 596 | 1711 ± 649 | 2170 ± 254 |
| Let-7c | 1313 ± 224 | 1407 ± 340 | 1632 ± 752 | 1310 ± 301 | 1581 ± 243 |
| miR-143 | 496 ± 139 | 409 ± 129 | 436 ± 140 | 510 ± 163 | 551 ± 165 |
| miR-192 | 1906 ± 905 | 1489 ± 569 | 1381 ± 176 | 1598 ± 642 | 1536 ± 611 |
| miR-194 | 3060 ± 1070 | 1799 ± 555 | 1992 ± 779 | 2075 ± 691 | 1872 ± 605 |
| miR-200b | 542 ± 205 | 832 ± 64 | 745 ± 153 | 717 ± 129 | 695 ± 181 |
| miR-200c | 1817 ± 568 | 2065 ± 428 | 2478 ± 1301 | 2128 ± 552 | 2579 ± 867 |
| miR-24 | 565 ± 160 | 504 ± 65 | 377 ± 160 | 450 ± 110 | 577 ± 177 |
| miR-26a | 599 ± 255 | 437 ± 93 | 381 ± 187 | 486 ± 53 | 704 ± 262 |
| miR-30d | 391 ± 35 | 456 ± 251 | 397 ± 99 | 385 ± 80 | 403 ± 100 |
| miR-375 | 749 ± 360 | 631 ± 257 | 729 ± 188 | 857 ± 272 | 812 ± 250 |
| miR-320 | 491 ± 105b | 1264 ± 630 | 1164 ± 589 | 972 ± 631 | 962 ± 424 |
Background-subtracted intensity (arbitrary unit) was obtained by microarray and presented as mean ± SD.
Significant difference as compared to four locations of colon as 1) fold change was ≥2; 2) t-test, P-value was <0.05; and 3) difference in fluorescence intensity between the two group means were >100 arbitrary units.
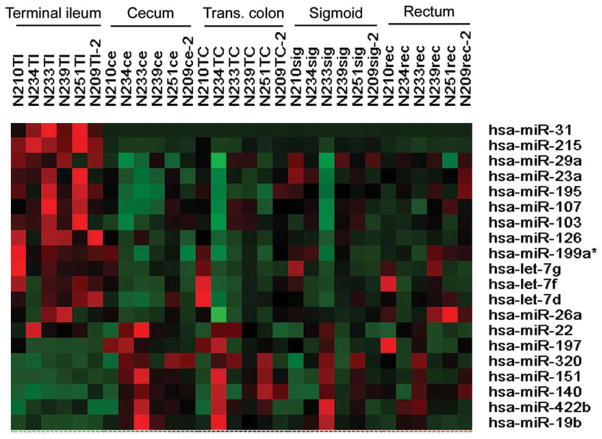
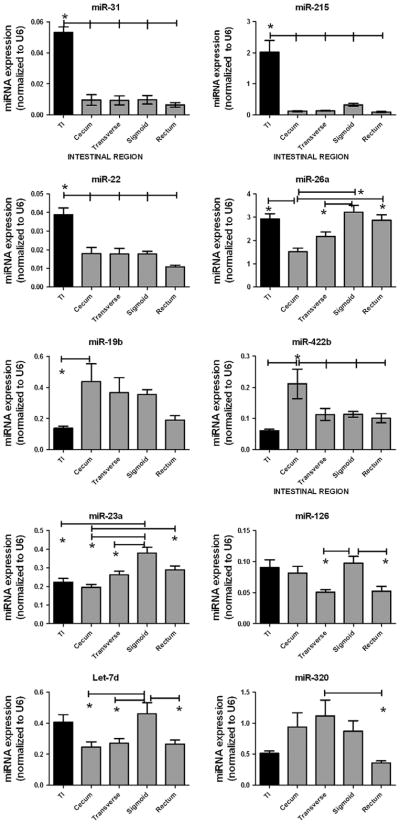
These miRNA microarray data also demonstrated 20 other miRNAs exhibiting region-specific expression (Fig. 1). By conducting mature miRNA qPCRs we validated regional expression differences in 10 of these 20 miRNAs (Fig. 2). Specifically, miR-22, miR-31, and miR-215 were significantly increased in the terminal ileum as compared to all four colonic regions. Additionally, miR-19b showed a 3.2-fold decrease in expression in the terminal ileum relative to the neighboring cecum.
FIGURE 1.
Heatmap of differential miRNA expression in the normal gut. Twenty miRNAs were identified in the miRNA microarray profiling of intestinal regions from six consecutive patients. Red indicates higher than mean intensity (black) across all samples, green represents lower than mean intensity (black).
FIGURE 2.
Mature miRNA validation in the normal gut. Ten miRNAs were validated by mature miRNA qRT-PCR. Data are presented as mean ± SEM (*P < 0.05).
Moreover, six miRNAs, miRs-26a, -422b, -23a, -126, -320, and let-7d, demonstrated colon region-specific miRNA expression differences (Fig. 2). For example, miR-422b was expressed at equal levels in the terminal ileum, transverse colon, sigmoid colon, and rectum but was significantly increased in the cecum. Meanwhile, miR-26a was significantly decreased in the cecum as compared to the terminal ileum, sigmoid colon, and the rectum and significantly decreased in the transverse colon as compared to the sigmoid colon. Overall, these data support the hypothesis that specific miRNAs exhibit unique expression levels in different intestinal regions, with the most dramatic differences seen in the terminal ileum compared to the colon.
Identification of miRNAs Differentially Expressed in Crohn’s Colitis
We previously demonstrated the miRNA expression in sigmoid colon pinch biopsies of patients with active UC differed from that in normal adults as well as from that in patients with microscopic colitis, CD, infectious colitis, and irritable bowel syndrome.22 However, a direct comparison of the miRNA expression from colonic biopsy tissues of CD patients with healthy subjects was not performed in that previous study. In the current study we focused on further comparing miRNA expression in sigmoid colon pinch biopsies of five patients with chronically active CD and 13 normal healthy adults to determine whether miRNA expression patterns in patients with active Crohn’s colitis differ from healthy adults.
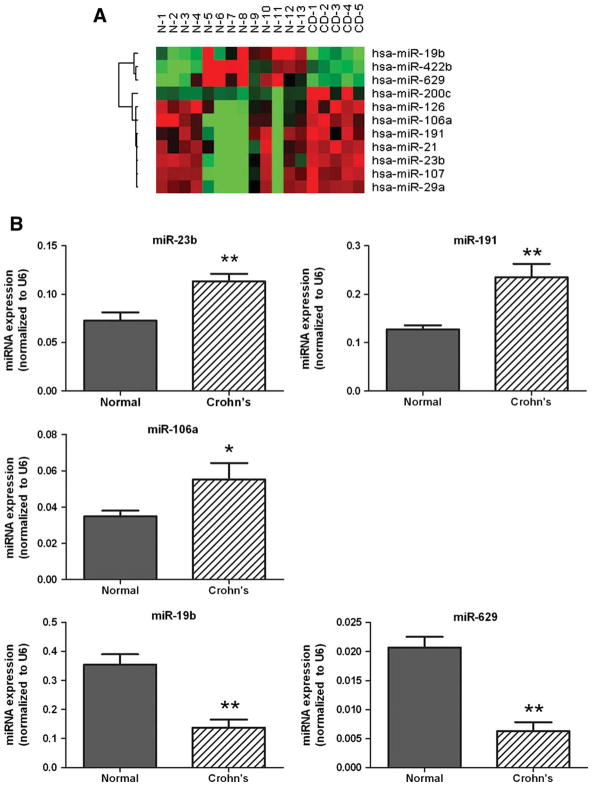
Our initial miRNA microarray profiling identified eight miRNAs, miR-21, miR-23b, miR-29a, miR-106a, miR-107, miR-126, miR-191, and miR-200c, with increased expression in tissues from Crohn’s colitis patients compared to normal healthy adults (Fig. 3A). Validation by mature miRNA qPCR confirmed that miR-23b, miR-106, and miR-191 were increased in active Crohn’s colitis tissues (Fig. 3B). These miRNAs were not previously identified as UC-associated miRNAs and their lack of altered expression in active UC was confirmed (data not shown).
FIGURE 3.
Identification of active colonic CD-associated miRNAs. (A) Heatmap of 11 miRNAs identified by miRNA microarray as differentially expressed in colonic CD. Red indicates higher than mean intensity (black) across all samples, green represents lower than mean intensity (black). (B) Mature miRNA qRT-PCR validation of five miRNAs differentially expressed in colonic CD. Data are presented as mean ± SEM (*P < 0.05).
The initial miRNA microarray profiling also identified three miRNAs, miR-19b, miR-422b, and miR-629, showing diminished expression in tissues from Crohn’s colitis patients as compared to normal healthy adults (Fig. 3A). Validation by mature miRNA qPCR confirmed that miR-19b and miR-629, but not miR-422b, were underexpressed in active Crohn’s colitis tissues (Fig. 3B). MiRs-19b and -629 were 2.6- and 3.5-fold decreased, respectively, in Crohn’s colitis tissues as compared to normal healthy control tissues. Analogous to the three upregulated miRNAs, these two downregulated miRNAs had not been previously identified as UC-associated miRNAs and their lack of altered expression in active UC was confirmed (data not shown).
Identification of miRNAs Differentially Expressed in Crohn’s Ileitis
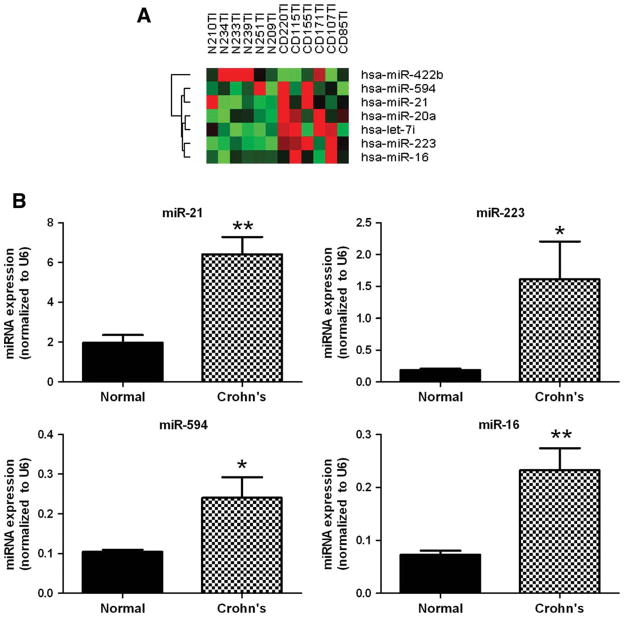
We conducted a similar assessment comparing the miRNA expression in terminal ileal biopsies from six patients with chronically active terminal ileal CD and six normal healthy adults. Initial miRNA microarray profiling identified one microRNA, miR-422b, which exhibited diminished expression in Crohn’s ileitis, and six miRNAs, let-7i, miR-16, miR-20a, miR-21, miR-223, and miR-594, with increased expression in Crohn’s ileitis as compared to normal healthy control tissues (Fig. 4A). Validation by mature miRNA qPCR confirmed that miR-16, miR-21, miR-223, and miR-594 were overexpressed in chronically active terminal ileal CD tissues (Fig. 4B). Specifically, miRs-16, -21, -223, and -594 were 3.2-, 3.3-, 8.6-, and 2.3-fold increased, respectively, in Crohn’s ileitis tissues as compared to normal healthy control tissues. The remaining two miRNAs, miR-422b and let-7i, were not found to be altered significantly by mature miRNA qPCR validation (data not shown).
FIGURE 4.
Identification of active ileal CD-associated miRNAs. (A) Heatmap of seven miRNAs identified by miRNA microarray as differentially expressed in ileal CD. Red indicates higher than mean intensity (black) across all samples, green represents lower than mean intensity (black). (B) Mature miRNA qRT-PCR validation of four miRNAs differentially expressed in ileal CD. Data are presented as mean ± SEM (*P < 0.05).
DISCUSSION
Previous systematic studies have demonstrated intestinal region-specific differences in gene expression.23 The molecular basis underlying region-specific expression of these genes has not yet been established. In this study we confirmed that miRNAs, key negative regulators of post-transcriptional gene expression, are expressed in the gastrointestinal tract and demonstrate intestinal region-specific expression. Overall, these miRNAs expressed in the intestine comprise a small fraction of the total number of known miRNAs encoded in the human genome. These results are analogous to observations of mRNA expression in the gastrointestinal tract. However, our new findings suggest that the influence of region-specific miRNAs on region-specific mRNAs and proteins now merits further investigation.
We previously demonstrated that miRNAs are differentially expressed in the sigmoid colon of patients with active UC.22 These active UC-associated miRNAs demonstrated distinct expression levels relative to tissues from patients with inactive UC, infectious colitis, microscopic colitis, irritable bowel syndrome, Crohn’s colitis, and normal healthy patients. In the current study we determined that tissues from the ileum and the sigmoid colon of CD patients also express distinct miRNAs. Specifically, we showed that five miRNAs were differentially expressed in active Crohn’s colitis and that four miRNAs were differentially expressed in Crohn’s ileitis. These results are consistent with our previous findings regarding differential expression of miRNAs in UC.
Taken together, our findings of IBD-associated miRNAs and intestinal tissue region-specific miRNAs suggest that miRNAs are involved in the maintenance of intestinal homeostasis and in differences in the pathogenesis of IBD subtypes. In particular, it is striking that there was very little overlap between the expression levels of specific miRNAs in active UC, Crohn’s colitis, and Crohn’s ileitis. We previously demonstrated that none of the active UC-related miRNAs, including miRs-16, -21, -23a, 24, -29a, -126, -192, -195, -375, -422b, and let-7f, are altered in Crohn’s colitis tissues. Similarly, in the current study, none of the Crohn’s colitis-associated miRNAs were previously found to be altered in UC tissues. Of the miRNAs altered in Crohn’s ileitis, we identified only miR-16 and miR-21 as altered in UC but not in Crohn’s colitis.
There is accumulating evidence that miRNAs play a significant role in modulating inflammatory gene expression.24 For example, miR-146 downregulates the expression of tumor necrosis factor receptor-associated factor 6 and interleukin-1 receptor-associated kinase 1, key molecules in cytokine and Toll-like receptor signaling.19 Similarly, our laboratory demonstrated that miR-192, an miRNA significantly downregulated in active UC tissues, is expressed in colonic epithelial cells and regulates inflammatory cytokine-induced macrophage inflammatory peptide 2α expression.22 Further studies are now indicated to determine the cellular localization of the Crohn’s colitis- and Crohn’s ileitis-associated miRNAs, as well as whether these miRNAs regulate genes associated with inflammation or fibrosis.
Our data contribute to the growing evidence that miRNAs are differentially expressed in inflammatory and autoimmune diseases. Specifically, miRNAs have been found to be differentially expressed in psoriasis and atopic eczema,25 rheumatoid arthritis,26–28 asthma,29 systemic lupus erythematosus, and idiopathic thrombocytopenic purpura.30
Of all the CD-associated miRNAs identified in the current study, miR-21 has been particularly widely implicated in the regulation of inflammatory disorders. For example, we previously demonstrated increased miR-21 expression in active UC. Similarly, miR-21 was also found to be increased in the lungs of mice exposed to aerosolized lipopolysaccharide.31 In addition, Sonkoly et al25 reported that miR-21 was among several miRNAs upregulated in psoriasis and atopic eczema. The expression of miR-21 was also increased in peripheral blood cells from patients with systemic lupus erythematosus (SLE) and idiopathic thrombocytopenic purpura (ITP).30
Other CD-associated miRNAs have also been identified in inflammatory disorders. Like miR-21, miR-106 was found to be upregulated in psoriasis,25 while miR-223 was upregulated in chorioamnionitis membranes,32 endometriosis,33 and T-cells from patients with rheumatoid arthritis.34 Finally, miR-16 was found to be upregulated in peripheral blood mononuclear cells of patients with rheumatoid arthritis.35 It can be hypothesized that the overlap of these miR-NAs among multiple inflammatory and autoimmune processes indicates that specific miRNAs function as key regulators of innate and adaptive immune mechanisms by regulating families of immune-related target mRNAs. These new findings now merit further studies to understand the precise role of immune-associated miRNAs.
Taken together, the identification of miRNAs associated with active Crohn’s ileitis, Crohn’s colitis, and UC not only implies distinct pathogenic mechanisms underlying IBD subtypes but also raises the possibility that distinct miRNA expression patterns in IBD subtypes can be used to distinguish these IBD subtypes or to assess disease activity and therapeutic efficacy. Indeed, the use of miRNAs as diagnostic tools and therapeutic targets for disease has been proposed36 and shown to distinguish numerous cancers.37 While we have identified 11 UC-associated miRNAs, five Crohn’s colitis miRNAs and four Crohn’s ileitis miRNAs, further studies on tissue and blood-based miRNAs on larger patient populations are now indicated to determine whether miRNA expression profiles will lead to useful IBD diagnostic tools and therapeutic targets.
Acknowledgments
We thank Dr. Yuriko Mori for discussions.
Supported by Broad Medical Research Program grants IBD-0212 (to F.W. and J.H.K.); National Institutes of Health grant K08DK078046 (to J.H.K.). J.H.K. This research was also supported by the Sherlock Hibbs IBD Research Fund, the M. Alan Guerrieri Family Fund, and the Meyerhoff IBD Center at Johns Hopkins University.
References
- 1.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 2.Budarf ML, Labbe C, David G, et al. GWA studies: rewriting the story of IBD. Trends Genet. 2009;25:137–146. doi: 10.1016/j.tig.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Costello CM, Mah N, Hasler R, et al. Dissection of the inflammatory bowel disease transcriptome using genome-wide cDNA microarrays. PLoS Med. 2005;2:e199. doi: 10.1371/journal.pmed.0020199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn’s disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Gen. 2001;10:445–456. doi: 10.1093/hmg/10.5.445. [DOI] [PubMed] [Google Scholar]
- 5.Wu F, Dassopoulos T, Cope L, et al. Genome-wide expression differences between Crohn’s and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 6.Meuwis MA, Fillet M, Geurts P, et al. Biomarker discovery for inflammatory bowel disease, using proteomic serum profiling. Biochem Pharmacol. 2007;73:1422–1433. doi: 10.1016/j.bcp.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 7.Meuwis MA, Fillet M, Lutteri L, et al. Proteomics for prediction and characterization of response to infliximab in Crohn’s disease: a pilot study. Clin Biochem. 2008;41:960–967. doi: 10.1016/j.clinbiochem.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 8.Shkoda A, Werner T, Daniel H, et al. Differential protein expression profile in the intestinal epithelium from patients with inflammatory bowel disease. J Proteome Res. 2007;6:1114–1125. doi: 10.1021/pr060433m. [DOI] [PubMed] [Google Scholar]
- 9.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Chua JH, Armugam A, Jeyaseelan K. MicroRNAs: biogenesis, function and applications. Curr Opin Mol Ther. 2009;11:189–199. [PubMed] [Google Scholar]
- 11.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 12.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S, Saini HK, van Dongen S, et al. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 16.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Asirvatham AJ, Magner WJ, Tomasi TB. miRNA regulation of cytokine genes. Cytokine. 2009;45:58–69. doi: 10.1016/j.cyto.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connell RM, Taganov KD, Boldin MP, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/micro-RNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pauley KM, Cha S, Chan EK. MicroRNA in autoimmunity and autoimmune diseases. J Autoimmun. 2009;32:189–194. doi: 10.1016/j.jaut.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu F, Zikusoka M, Trindade A, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635. e1624. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 23.Bates MD, Erwin CR, Sanford LP, et al. Novel genes and functional relationships in the adult mouse gastrointestinal tract identified by microarray analysis. Gastroenterology. 2002;122:1467–1482. doi: 10.1053/gast.2002.32975. [DOI] [PubMed] [Google Scholar]
- 24.Lodish HF, Zhou B, Liu G, et al. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 25.Sonkoly E, Wei T, Janson PC, et al. MicroRNAs: novel regulators involved in the pathogenesis of Psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakasa T, Miyaki S, Okubo A, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum. 2008;58:1284–1292. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanczyk J, Pedrioli DM, Brentano F, et al. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 28.Tili E, Michaille JJ, Costinean S, et al. MicroRNAs, the immune system and rheumatic disease. Nat Clin Pract Rheumatol. 2008;4:534–541. doi: 10.1038/ncprheum0885. [DOI] [PubMed] [Google Scholar]
- 29.Tan Z, Randall G, Fan J, et al. Allele-specific targeting of micro-RNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai Y, Huang YS, Tang M, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–946. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 31.Moschos SA, Williams AE, Perry MM, et al. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montenegro D, Romero R, Pineles BL, et al. Differential expression of microRNAs with progression of gestation and inflammation in the human chorioamniotic membranes. Am J Obstet Gynecol. 2007;197:289, e281–286. doi: 10.1016/j.ajog.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohlsson Teague EM, Van der Hoek KH, Van der Hoek MB, et al. MicroRNA-regulated pathways associated with endometriosis. Mol Endocrinol. 2009;23:265–275. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fulci V, Scappucci G, Sebastiani GD, et al. miR-223 is overexpressed in T-lymphocytes of patients affected by rheumatoid arthritis. Hum Immunol. 2010;71:206–211. doi: 10.1016/j.humimm.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Pauley KM, Satoh M, Chan AL, et al. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perron MP, Boissonneault V, Gobeil LA, et al. Regulatory RNAs: future perspectives in diagnosis, prognosis, and individualized therapy. Methods Mol Biol. 2006;361:311–326. doi: 10.1385/1-59745-208-4:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]