Abstract
Substantial evidence indicates that exposure to bisphenol A (BPA) during early development may increase breast cancer risk later in life. The changes may persist into puberty and adulthood, suggesting an epigenetic process being imposed in differentiated breast epithelial cells. The molecular mechanisms by which early memory of BPA exposure is imprinted in breast progenitor cells and then passed onto their epithelial progeny are not well understood. The aim of this study was to examine epigenetic changes in breast epithelial cells treated with low-dose BPA. We also investigated the effect of BPA on the ERα signaling pathway and global gene expression profiles. Compared to control cells, nuclear internalization of ERα was observed in epithelial cells preexposed to BPA. We identified 170 genes with similar expression changes in response to BPA. Functional analysis confirms that gene suppression was mediated in part through an ERα-dependent pathway. As a result of exposure to BPA or other estrogen-like chemicals, the expression of lysosomal-associated membrane protein 3 (LAMP3) became epigenetically silenced in breast epithelial cells. Furthermore, increased DNA methylation in the LAMP3 CpG island was this repressive mark preferentially occurred in ERα-positive breast tumors. These results suggest that the in vitro system developed in our laboratory is a valuable tool for exposure studies of BPA and other xenoestrogens in human cells. Individual and geographical differences may contribute to altered patterns of gene expression and DNA methylation in susceptible loci. Combination of our exposure model with epigenetic analysis and other biochemical assays can give insight into the heritable effect of low-dose BPA in human cells.
Keywords: Bisphenol A, Estrogen, DNA methylation, Epigenetics, Breast cancer
Introduction
Bisphenol A (BPA), first synthesized by A. P. Dianin in 1891, has been widely used as a cross-linking reagent in the manufacture of epoxy resins since 1950s (Vogel, 2009). It is extensively used in a board range of products, including toys, water pipes, drinking bottles, baby bottles, food containers, tubing, and dental sealants (Welshons et al., 2006). Presently, the worldwide production of BPA exceeds 3 billion kilograms per year (Vandenberg et al., 2009). Studies have shown that BPA can be released from incomplete polymerization upon heating or leached out through normal use (Mountfort et al., 1997; Kang et al., 2003; Goodson et al., 2004). Because of its ubiquity in environment, low levels of BPA can be detected in 92.6% of urine samples (≥ 6 years of age ranging from 0.4 – 149 μg/L) in the National Health and Nutrition Examination Survey (NHANES) 2003–2004 (Calafat et al., 2008; CDC, 2009). Animal studies have shown that these low levels of BPA exposure may alter developmental programs of sensitive end organs, like mammary and prostate gland, during critical stages of early development (Markey et al., 2001; Nikaido et al., 2004; Timms et al., 2005). The changes may persist into puberty and adulthood, suggesting an imprinting process being imposed in differentiated breast epithelial cells (Markey et al., 2001; Munoz-de-Toro et al., 2005).
The molecular mechanisms by which early memory of BPA exposures can be imprinted in breast progenitor cells and then passed onto their epithelial progeny are not well understood. One distinct possibility is through epigenetic remodeling of DNA structure without altering the nucleotide sequence itself. The changes, including DNA methylation, frequently occur in GC-rich promoter CpG islands of transcriptionally repressed genes (Ohm and Baylin, 2007; Widschwendter et al., 2007). Studies have suggested that DNA methylation of a promoter CpG island, or promoter hypermethylation, can be initiated in progenitor genomes and heritably passed onto the differentiated progeny (Jones and Baylin, 2007; Marotta and Polyak, 2009). This epigenetic process is known to cause phenotypic variations among individuals and contributes to the development of pathological conditions, like cancer (Feinberg et al., 2006; Esteller, 2007).
Previous studies of epigenetic effects of BPA preexposure mainly rely on animal models and epidemiological surveys (Ho et al., 2006; Dolinoy et al., 2007; Prins et al., 2008; Yaoi et al., 2008). Whereas these observations strongly implicate that the exposure to low-dose BPA is potentially harmful to human health, the challenge encountered is validation studies of the findings in primary human cells. In this regard, we have recently established a human preexposure model for epigenetic studies (Cheng et al., 2008; Hsu et al., 2009). In the model, breast progenitor cells were first exposed to different environmental chemicals, and then these cells were differentiated into epithelial cells in the absence of these environmental stimulants. We hypothesize that slow-dividing progenitor cells have a longer life span and thus are more susceptible to environmental injuries and can transmit this injured memory to their differentiated progeny through epigenetic mechanisms. In our previous studies, the preexposure to 17β-estradiol (E2) and diethylstilbestrol (DES) may trigger epigenetic repression of protein-coding genes and non-coding microRNAs, some of which exhibit promoter hypermethylation in breast cancer cells (Cheng et al., 2008; Hsu et al., 2009).
Here, we extended this preexposure study to evaluate epigenetic effects of low-dose BPA in human breast epithelial cells. As a result of chronic exposure to BPA, activation of estrogen receptor α (ERα)-mediated signaling and subsequent alterations of responsive gene expression were observed in the differentiated epithelial progeny. This heritable influence on gene expression was similarly observed in ERα-positive breast tumors.
Materials and Methods
Tissue samples and cell culture
Breast tissues, obtained from individuals undergoing mastectomy or reduction mammoplasty, were collected in accordance with the protocols approved by the Institutional Review Boards of the Ohio State University and the National Taiwan University Hospital. For isolation of breast progenitor cells, non-cancerous tissues (age: 17–42 years old) were enzymatically dissociated via collagenase digestion as described previously (Hsu et al., 2009). Single cells were grown into floating spherical colonies (2–10,000 cells per colony), called mammospheres, in ultra-low attachment dishes (Corning, Lowell, MA) in serum-free medium. These mammospheres, enriched in breast progenitor cells (Dontu et al., 2004), were exposed with BPA (4 nM) (Sigma, St. Louis, MO), diethylstilbestrol (DES, 70 nM), daidzein (10 μM), 1,3,5-tris(4-hydroxyphenyl)-4-propyl-1H-pyrazole (PPT, 0.1 nM), 4-nonylphenol (NP, 1 μM), N-butyl-benzyl phthalate (BBP, 10 μM), di(2-ethylhexyl)-phthalate (DEHP, 10 μM), 4,4′-dichloro-biphnyl (PCB, 0.1 nM) or DMSO in phenol red-free medium for 3 weeks (medium changed twice a week). The concentration of each chemical was selected based on the literature review. Cell viability assay indicated that there is no toxicity effect of each compound under the concentration we selected (HarrEus et al., 2002; Rohrdanz et al., 2002; Buteau-Lozano et al., 2008) After the exposure, mammospheres were washed with PBS to remove BPA and then placed on a collagen-coated dish in phenol red-free DMEM/F12 medium containing 5% charcoal-dextran-treated FBS (Hyclone, Waltham, MA) for 2–3 weeks. Under this condition, progenitor cells were differentiated into breast epithelial cells, or called mammosphere-derived epithelial cells (MDECs). A panel of 48 breast cancer cell lines, procured through the Integrative Cancer Biology Program of the National Cancer Institute, were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and routinely propagated in culture dishes for epigenetic analyses.
Immunofluorescence staining
Approximately 5,000 MDECs were seeded on a collagen I-coated coverslip (BD Biosciences, San Jose, CA) for overnight and then fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 10 min. After blocking with 3% bovine serum albumin (Fisher Scientific, Pittsburgh, PA) for 1 h, the coverslip was incubated with anti-ERα antibody (D-12, 1:50) (Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4 °C. The corresponding secondary FITC-conjugated antibody (Invitrogen, Carlsbad, CA) was applied followed by 4′,6-diamidino-2-phenylindole staining (DAPI) (Invitrogen) to localize cell nuclei. The images were captured by confocal laser microscope (Zeiss LSM510) (Zeiss, Thornwood, NY), and percentages of ERα subcellular localization were calculated in 10 different optical fields (~100 cells) by two independent researchers.
Western blot analysis
MDECs preexposed to BPA or DMSO were collected and protein lysates were made. 30 μg of lysate were immunoblotted with antibody against phospho-p42/44 MAPK (1:1000), phospho-Akt (1:2000) (Cell Signaling Technology). GAPDH (Santa Cruz Biotechnology) was used as loading control. Cy5-conjugated goat anti-rabbit and Cy3-conjugated goat anti-mouse antibody (GE Healthcare, Pittsburgh, PA) were used for multiplex detection. The membranes were scanned by Typhoon 9400 scanner (GE Healthcare).
Gene expression microarray
Total RNAs of ten independent MDECs, including BPA-preexposed and control, were isolated with Trizol (Invitrogen) according to the manufacturer’s instructions. RNA (5 μg/sample) was used for microarray hybridization to the Affymetrix Human Genome U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA) by the Microarray Core Facility at the Ohio State University Comprehensive Cancer Center (Columbus, OH). Gene expression estimates of the 54,675 probe sets on arrays were obtained using robust multi-array analysis (RMA) method with quantile normalization and background correction (Irizarry et al., 2003). Gene expression microarray data of 48 breast cancer cell lines (BCC48, Neve et al., 2006) and breast tumors (GSE2109, International Genomics Consortium, http://www.intgen.org/expo) were available for downloading. Qualtile normalization and background correction were also applied to these individual datasets.
Comparison between BPA-preexposed and control samples was performed using BRB Array Tools software developed by the Biometric Research Branch of the National Cancer Institute (http://linus.nci.nih.gov/BRB-ArrayTools). A paired t-test with random variance model was applied in order to identify differentially expressed genes between BPA-preexposed and control samples. Initial filtering was performed by selecting genes with P < 0.05 and by removing genes with lower expression for all samples (genes with expression values less than or equal to 100 for all samples were removed). An un-paired t-test was applied for the BCC48 and GSE2109 datasets in order to identify differentially expressed genes between ERα-positive and ERα-negative samples. The final 170 loci were obtained by restricting the initial list of candidates to commonly expressed genes in breast cancer based on two microarray datasets, BCC48 and GSE2109. Functional and network analyses of these genes were performed using Ingenuity Systems’ IPA software (Ingenuity Systems Inc., www.ingenuity.com).
Epigenetic treatments and reverse transcription-quantitative PCR (RT-qPCR)
MCF-7 breast cancer cells were treated with 1 μM 5-aza-2′-deoxycytidine (DAC) in phenol red-free MEM containing 10% FBS and 6 ng/ml insulin. During the final 24 h, some cells were additionally treated with 0.5 μM trichostatin A (TSA). RNA (1 μg) was isolated and reversely transcribed into cDNA using the SuperScript III Reverse Transcriptase (Invitrogen). RT-qPCR was performed by using 2x SYBR Green Master Mix (Applied Biosystems, Foster City, CA) on a 7500 Real-Time PCR System apparatus (Applied Biosystems). Levels of the 36B4 mRNA transcript were also measured as internal controls (Akamine et al., 2007). The reactions were performed in triplicate, and the standard deviation was calculated using the Comparative Method (ABI Prism 7700 Sequence Detection System User Bulletin #2). Primer sequences and conditions for amplification are available in Supplemental Table S1.
DNA methylation analysis by Pyrosequencing
To determine methylation levels of candidate genes in samples, the Pyrosequencing system (Qiagen, Valencia, CA) was used to detect methylated CpG sites in sequencing reactions (Tost and Gut, 2007). Genomic DNA (500 ng) was treated with sodium bisulfite using the EZ DNA Methylation kit (Zymo Research, Orange, CA). Bisulfite-treated DNA was amplified with specific primers for each gene of interest. The Pyro Mark Assay Design program and the Pyro Q-CpG software were used for primer designs and data analysis, respectively. Average methylation levels of individual CpG sites for each DNA sample were calculated.
Statistical analysis
All data derived from subcellular localization, RT-qPCR, and Pyrosequencing were presented as mean ± SD of n independent measurements. Statistical comparisons between two groups (DMSO vs. BPA) were made by Student’s t test using GraphPad Prism 6 (GraphPad Software, La Jolla, CA). To avoid any violation of normal distribution assumption for DNA methylation analysis, we applied non-parametric Mann-Whitney rank-sum test (GraphPad Prism 6). A significance was assigned if P < 0.05.
Results
Effect of low-dose BPA on the nuclear localization of ERα in MDECs
Environmental chemicals, such as BPA, are known to act as estrogenic ligands that activate or deactivate gene transcription in breast epithelial cells (Soto et al., 2006; Dairkee et al., 2008). To determine whether BPA causes an estrogen-like effect, we performed immunofluorescence analyses in MDECs (un-exposed) transiently treated with different doses (ranges: 1 – 1,000 nM) of BPA at 0, 5, 30, 60, and 120 min (Figure 1A). BPA, as a weak estrogenic ligand, caused maximized ERα internalization at 30 min in a higher dose (1,000 nM) of exposure.
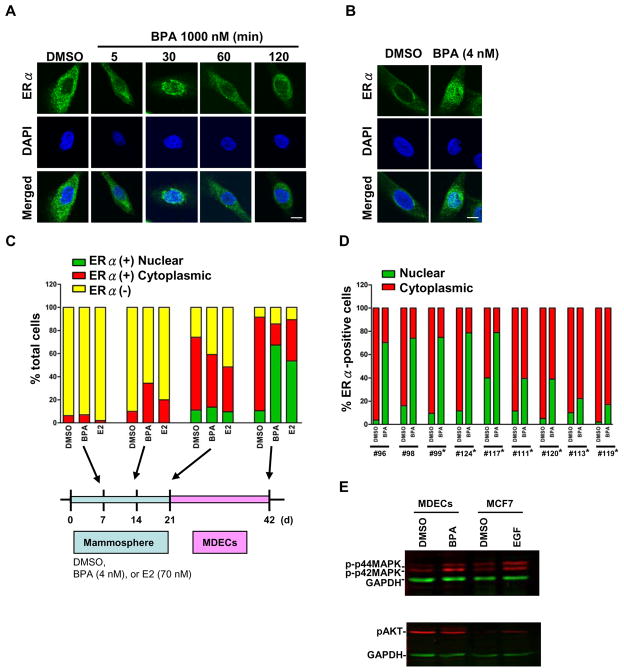
Fig. 1.
Preexposure of MDECs to bisphenol A (BPA) and immunofluorescence analysis of nuclear ERα. (A) Subcellular localization of ERα in MDECs on acute BPA treatment. MDECs were exposed to BPA (1000 nM) for the indicated time periods. The observed translocation of ERα protein (green) from the cytoplasm to nucleus is indicative of functional estrogen signaling. Nuclei were stained with DAPI (blue). Bar = 10 μm. (B) Mammospheres were treated with BPA 4 nM or DMSO for 3 weeks. After the exposure, mammospheres were washed with PBS to remove BPA and then placed on a collagen coated dishes for differentiation. Immunofluorescence staining showed that translocation of ERα protein (green) from the cytoplasm to nucleus was observed. Nuclei were stained with DAPI (blue). Bar = 10 μm. (C) Increased internalization of ERα. in BPA-preexposed MDECs (#124). Mammospheres were treated with DMSO, BPA (4 nM) or E2 (70 nM) for 3 weeks. The distribution of ERα in mammospheres was monitored each week as indicated in the bottom graphic (7, 14, and 21 d). After the exposure, BPA was removed, and progenitor cells underwent epithelial differentiation in the collagen-coated dishes for 3 weeks. ERα localization was also monitored as indicated in figure (42 d). Yellow bars indicate the percentage of ERα-negative cells within the total population. Green bars (nuclear ERα) and red bars (cytoplasmic ERα) represent ERα-positive cells within total population. (D) Increased nuclear localization of ERα in BPA-preexposed MDECs. After the preexposure to BPA (4 nM) or DMSO, MDECs were subjected to immunofluorescence analysis. (C–D) The percentage of subcellular localization of ERα-positive cells, independently scored by two researchers, is shown. These results were collected from 9 independent sets of MDECs samples. *, indicates samples were also subjected to gene expression analysis. (E) BPA induced p42/44 phosphorylation. Phosphorylated levels of p42/44 MAPK and Akt were analyzed in MDECs preexposed to BPA (4 nM) or DMSO by western blotting. GAPDH was used as loading control.
Because prolonged exposure of breast progenitor cells to xenoestrogens also causes ERα internalization in their differentiated progeny (Hsu et al., 2009), we determined whether BPA has this effect. Progenitor-containing mammospheres from an individual (#124) were continuously exposed to 4 nM BPA for 3 weeks. After the exposure, BPA was removed, and progenitor cells underwent epithelial differentiation in the collagen-coated dishes for 2–3 weeks. Immunofluorescence analysis showed an increase of ERα-positive population during the mammosphere and MDEC stages (Figure 1C). After 7-d preexposure, the majority (>90%) of mammospheres were ERα-negative (yellow bar in Figure 1C). However, cell lineages were greatly shifted from ERα-negative to ERα-positive (green-plus-red bar, > 80%) after 42-d incubation. Among ERα-positive cells at 42-d, nuclear expression of ERα (green bar/green-plus-red bar, 78%) in BPA-preexoposed MDECs was increased compared to that of control (DMSO) cells (11.6%), suggesting that BPA preexposure contributes to ERα internalization in MDECs (Figure 1B). As a control, we also observed similar effect in E2 (70 nM)-preexposed MDECs.
When the analysis was extended to different primary MDECs (n = 9), we noticed individual variations in response to this low-dose BPA preexposure. As shown in Figure 1D, five (#96, 124, 98, 99, and 117) of these MDEC sets exhibited greater effects (up to 80%) of ERα internalization compared to the other four sets (#111, 120, 113, and 119) showing lesser effects (18 – 40%). This initial result suggests that as a weak estrogenic ligand, high-dose BPA (at least 1000 nM) is needed to acutely activate ERα-mediated signaling while chronic exposure of a lower dose (4 nM) can similarly bring about this signal transduction in breast epithelial cells. Furthermore, our observations indicate that the genetic background of individuals may influence differential responses to the exposure of low-dose BPA.
It is known that other exogenous stimulants, such as growth factors, may act through mitogen-activated protein kinase (MAPK) or Akt pathways to promote the nuclear internalization of ERα for transcriptional regulation in proliferating cells (Lannigan, 2003; Murphy et al., 2009). In this regard, we observed an increased level of phospho-p42/44 MAPK, likely attributed to this internalization in BPA-preexposed MDECs without further ligand stimulation (Figure 1E).
Effect of low-dose BPA on differential gene expression in MDECs
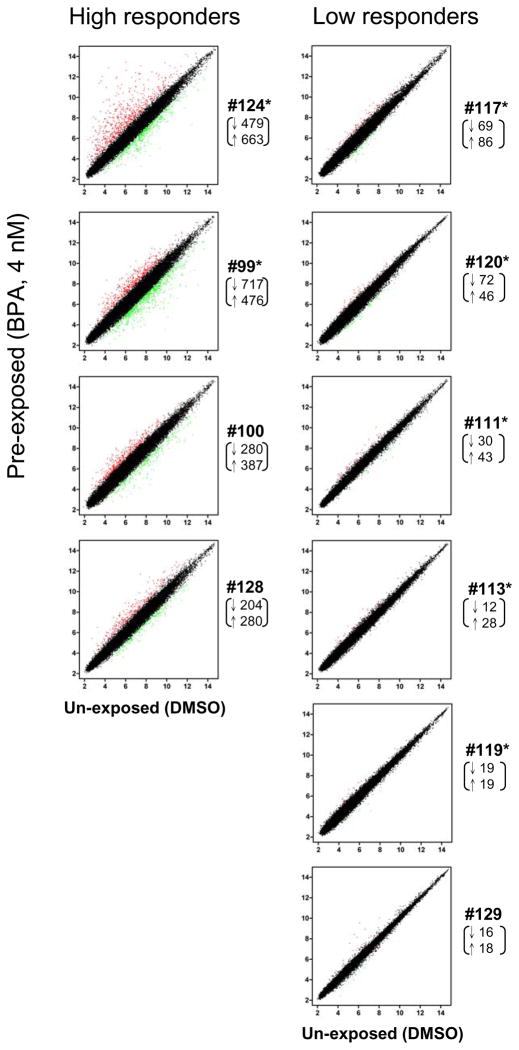
To investigate whether this effect altered gene expression, we conducted microarray analysis in ten sets of preexposed (BPA, 4 nM) and control MDECs using the Affymetrix Human Genome U133 Plus 2.0 Array. One set of the MDECs (#119) was removed from gene expression analysis because of the low nuclear localization of ERα. Differential expression of genes at P < 0.05 within 9 set samples was scored, yielding a total of 2,234 candidate loci (1,162 down-regulated and 1,072 up-regulated) likely influenced by this BPA preexposure. Scatter plots and the number of differentially expressed genes for individual MDECs are presented in Figure 2. Consistent with the observation of ERα internalization, we observed individual variations of gene expression in these primary MDECs preexposed to low-dose BPA. In this regard, greater numbers of differentially expressed genes were seen in #124, 99, 100, and 128 (i.e., the high-responder group) while the rest of six primary MDECs had fewer changes of expression (i.e., the low- responder group). We additionally compared these expression profiles with the status of ERα internalization available for seven MDEC sets (#124, 99, 117, 120, 111, 113 and 119). Though not statistically significant, we observed a general trend that greater degrees of ERα internalization seemed to be associated with increased numbers of differentially expressed genes in MDECs.
Fig. 2.
Gene expression profiling of ten sets of the MDECs. A total of genes in control (DMSO) and BPA-preexposed cells are shown in the scatter plot. The number of significant down-regulated (↓, green dots) and up-regulated (↑, red dots) genes (2 fold differences between DMSO vs. BPA) are shown below the ID#. *, samples were also analyzed for ERα nuclear localization.
Effect of BPA-influenced gene signatures in ERα-positive breast cancer
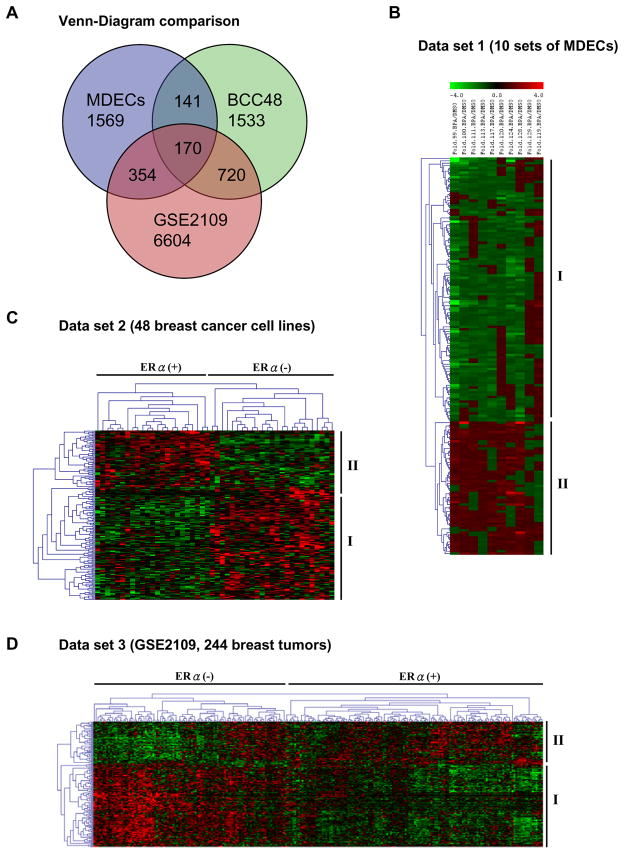
In silico analysis was conducted to determine whether specific expression profiles of BPA-influenced genes are associated with the development of breast cancer. When the 2,234 candidate loci were compared with those of two microarray datasets, BCC48 (Neve et al., 2006) and GSE2109 (International Genomics Consortium, http://www.intgen.org/expo), we found a total of 170 BPA-influenced genes (57 up-regulated and 113 down-regulated), the aberrant expression of which may contribute to breast tumorigenesis (Figure 3A and B; see also Supplemental Table S2). Hierarchical clustering of 48 breast cancer cell lines (i.e., BCC48) and 244 breast tumors (i.e., GSE2109) revealed that specific up- and down-regulated patterns of these 170 genes are distinctly related to ERα-positive cell lines (Figure 3C and D). This observation further indicates that 1) BPA may aberrantly regulate gene expression through an ERα-dependent pathway and 2) this regulatory mechanism may be epigenetically imprinted in ERα-positive breast cancer.
Fig. 3.
Comparison of gene expression profiles in BPA-preexposed MDECs, 48 breast cancer cell lines (Neve et al. 2006) and breast tumor samples (GSE2109). (A) Venn diagram showing 170 common genes identified in separate analyses of the three different data sets, i.e., MDECs (2234 genes altered by BPA), breast cancer cell lines (2564 genes preferential difference between ERα-positive and ERα-negative), and primary tumor samples (7848 genes preferential difference between ERα-positive and ERα-negative). Gene tree cluster analysis was performed on the 170 genes altered by BPA in MDECs (B), ERα-stratified cell line (C) and patient tumor data sets (D). This analysis identified 113 repressed (panel B, group I) and 57 activated (panel B, group II) genes in BPA-preexposed MDECs. Group I genes were also identified in ERα-positive breast cancer cell lines (panel C) and primary tumor samples (panel D); group II genes were identified in ERα-negative breast cancer cell lines (panel C) and primary tumor samples (panel D). Color bar, magnitude of gene expression; green, repression; red, stimulation.
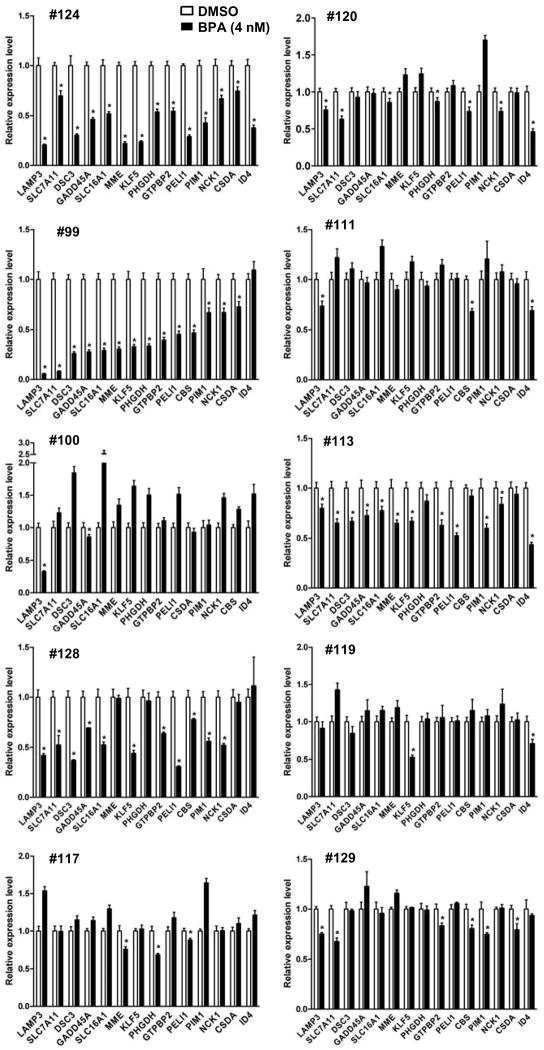
To validate this potential imprinting effect, we choose 15 down-regulated genes for expression analysis (Figure 4). The reason to focus on these loci was that the BPA-influenced repression might be associated with hypermethylation of their CpG islands, which are located in the transcription start sites of these selected genes. First, RT-qPCR was used to confirm the expression status of these loci in the aforementioned six MDECs preexposed to BPA (4 nM). The expression of these loci was consistently down-regulated in three high-responders, #124, 99, and 128 (P < 0.05). Down-regulation of these loci, however, could not be confirmed in one high-responder (#100), likely attributed to a small sampling of down-regulated loci. Though this down-regulation was also seen in the low-responder group by the sensitive RT-qPCR assay, the repressive effect was usually less apparent (e.g., #120 and 113). In the rest of low-responders (#111, 129, 117, and 119), significant changes of expression between pre-exposed and control MDECs were not noted.
Fig. 4.
Validation of differentially expressed loci by RT-qPCR. Gene-specific RT-qPCR on 10 independent sets of DMSO and BPA-preexposed MDECs was conducted to validate 15 down-regulated loci. Data were analyzed by ΔΔCt method using 36B4 as the internal control and are presented relative to DMSO treatment for each MDEC sample. Mean ± SD (n = 3). *, P < 0.05 (Student’s t test) for down-regulated genes compared with DMSO treated control.
Epigenetic repression of a BPA-influenced locus, LAMP3, in ERα-positive breast cancer cells
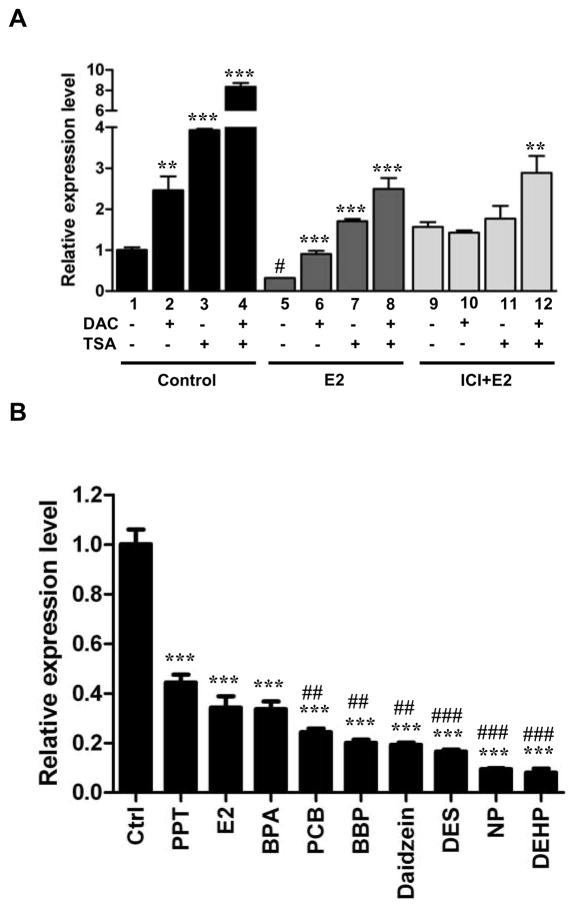
To further investigate a potential role of epigenetic repression, we focused the expression analysis on a candidate gene, lysosomal-associated membrane protein 3 (LAMP3), in the well-characterized ERα-positive MCF-7 cell line. (see functional analysis of this gene in MCF-7 cells in supplemental Figure S1). A low level of LAMP3 expression was detected in MCF-7 cells. To determine whether this reduced expression is mediated by epigenetic mechanisms, we treated these cells with the demethylating agent DAC (1 μM) and/or the histone deacetylase inhibitor TSA (0.5 μM), known to reactivate epigenetically repressed genes (Dworkin et al., 2009; Huang et al., 2009). As shown in Figure 5A (lanes 1–4), the expression of LAMP3 was significantly reactivated by single treatments (i.e., DAC or TSA, P < 0.01). Furthermore, synergistic re-expression of this gene was observed in cells with the combined treatment (DAC plus TSA, P < 0.001). Additional results of 7 other repressed genes are presented in supplemental Figure S2.
Fig. 5.
Epigenetic reactivation of ERα-mediated LAMP3 repression in MCF-7. (A) MCF-7 were treated with DAC (1 μM), TSA (1 μM) and/or ERα antagonist, ICI182780 (ICI, 1 μM) 6 hr before E2 stimulation. Total RNA was subjected to RT-qPCR analysis. 36B4 was used as internal control. Mean ± SD; **, P < 0.01, ***, P < 0.001 compared with DMSO treated control (lane 1–4), E2 treated alone (lane 5–8), and ICI+E2 alone (lane 9–12). #, P < 0.001 compared lane 5 (E2 treatment) with lane 1 (DMSO). (B) Mammospheres were exposed to E2 (70 nM), diethylstilbestrol (DES, 70 nM), daidzein (10 μM), 1,3,5-tris(4-hydroxyphenyl)-4-propyl-1H-pyrazole (PPT, 0.1 nM), 4-nonylphenol (NP, 1 μM), N-butyl-benzyl phthalate (BBP, 10 μM), di(2-ethylhexyl)-phthalate (DEHP, 10 μM), and 4,4′-dichloro-biphnyl (PCB, 0.1 nM) for 3 weeks. After the exposure, the MDECs were subjected to RT-qPCR analysis for LAMP3 expression. Mean ± SD; ***, P < 0.001 compared with DMSO treated control. ##, P < 0.01, ###, P < 0.001 compared with BPA treated sample.
To investigate whether this epigenetic repression could be attributed to an estrogen-mediated pathway, MCF-7 cells were additionally treated with E2 and/or an ERα antagonist, ICI182780. The subsequent E2 treatment led to re-silencing of LAMP3, suggesting a role of estrogen signaling in mediating this epigenetic repression (Figure 5A, lanes 5–8). Treatment of ICI182780 abolished the down-regulation, additionally indicating that this regulation is partly mediated through an ERα-dependent pathway (Figure 5A, lane 9). The repression was partially attenuated in the presence of additional epigenetic treatments (i.e., DAC and TSA, lanes 10–12).
Based on the results of these pharmacological experiments, our observations suggest that 1) estrogen signaling initiates the repression of the BPA-influenced loci in breast epithelial cells; 2) this repression is partly mediated trough an ERα-dependent pathway; and 3) persistent repression of the BPA-influenced loci in cancer cells may be further maintained by DNA methylation and histone modifications.
LAMP3 repression in MDECs preexposed to other estrogen-like chemicals
To determine whether long-term exposure of other estrogen-like chemicals can additionally initiate this epigenetic repression, mammospheres were exposed to diethylstilbestrol (DES, 70 nM), daidzein (10 μM), 1,3,5-tris(4-hydroxyphenyl)-4-propyl-1H-pyrazole (PPT, 0.1 nM), 4-nonylphenol (NP, 1 μM), N-butyl-benzyl phthalate (BBP, 10 μM), di(2-ethylhexyl)-phthalate (DEHP, 10 μM), and 4,4′-dichloro-biphnyl (PCB, 0.1 nM) for 3 weeks. After the exposure, MDECs were subjected to RT-qPCR analysis for LAMP3 expression. As shown in Figure 5B, downregulation of LAMP3 was confirmed in MDECs preexposed to these estrogen-like chemicals (2.3 to 12.5-fold decrease). Suppressive effects varied for the different environmental exposures, indicating differential sensitivity of progenitors to these chemicals.
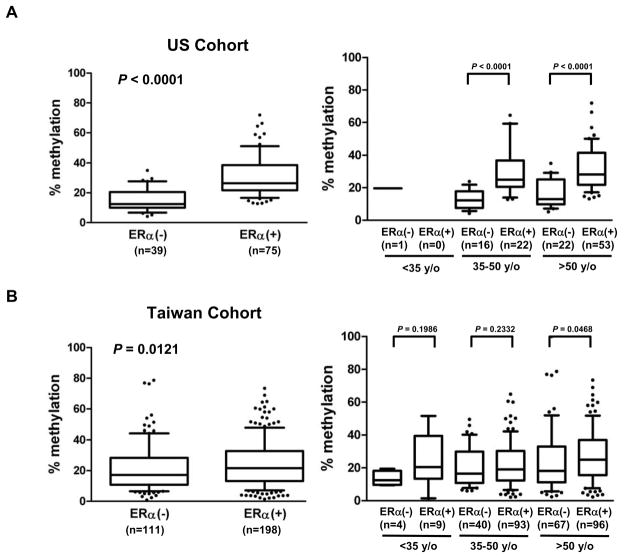
Promoter hypermethylation of LAMP3 in ERα-positive breast cancer
To confirm the in vitro epigenetic findings, we conducted DNA methylation analysis in the promoter CpG island regions of LAMP3 loci, in 48 breast cancer cell lines, 484 primary breast tumors (Taiwan cohort, n = 336; US cohort, n = 148), and 10 noncancerous breast tissues as normal controls. Pyrosequencing analysis of LAMP3 (9 CpG sites) revealed that DNA methylation levels were significantly increased in breast cancer cell lines relative to those of normal controls (Supplemental Figure S3B). Moreover, promoter hypermethylation of LAMP3 (P = 0.008) was significantly associated with the ERα-positive status. In close agreement with these results, hypermethylation of LAMP3 was observed in ERα-positive tumors in the US cohort (P < 0.0001) (Figure 6A, and Supplemental Figure S3A) and in the Taiwan cohort (Figures 6B, and Supplemental S3A). The cut-off points of age groups used in the further analysis were based on menopausal status - premenopausal (age <50 years) and postmenopausal (age >50 years) groups. The young age group defined by age <35 years appears to have distinct biological characteristics and display poor prognosis compared to those ≥35 years. Interestingly, while the hypermethylation event occurred in both age groups (35–50 and >50 years old) in the US cohort, this trend was only seen in the old age group (>50 years old) of the Taiwan cohort. Association of this hypermethylation with other clinicopathological features of patients was not apparent.
Fig. 6.
DNA methylation analysis of LAMP3. Quantitative methylation profiles of tumor samples from the US and Taiwan cohorts are shown in supplemental figure S3A. (A) Box plots indicate that the level of LAMP3 promoter methylation is positively correlated with ERα status in primary tumors from the US cohort (left panel). A positive correlation between LAMP3 methylation and ERα status is also observed in patient age 35–50 years, and >50 years (right panel). (B) Box plots indicate that the level of LAMP3 promoter methylation is positively correlated with ERα status in primary tumors from the Taiwan cohort (left panel). Further analysis shows a positive correlation between LAMP3 methylation and ERα status is observed in the age >50 years (right panel).
Discussion
When acutely exposed to estrogenic ligands, signal transduction is mediated in part through nuclear hormone receptors, such as ERα (Bjornstrom and Sjoberg, 2005). We have previously shown that the hallmark of this signal transduction is the translocation of cytoplasmic ERα into the nucleus of a normal breast epithelial cell (Hsu et al., 2009). Unlike E2 and DES, BPA is considered to be a weak estrogenic ligand based on the present immunofluorescence analysis and previous receptor binding assays (Okada et al., 2008). In our case, up to 1000 nM BPA is needed to initiate this ligand-dependent function, which mobilizes ERα into the nucleus for transcriptional activation and deactivation (Bjornstrom and Sjoberg, 2005; Okada et al., 2008).
We also observed that long-term exposure of breast progenitor cells to low-dose BPA (4 nM) is capable of triggering ERα internalization later observed in the differentiated progeny. In this case, exogenous stimulants (e.g., growth factors) may elicit ligand-independent activation by promoting the nuclear internalization of phosphorylated ERα for transcriptional regulation in proliferating cells (Lannigan, 2003; Murphy et al., 2009). This ligand-independent genomic function likely co-regulates a subset of target genes (e.g., LAMP3) governed through the ligand-dependent pathway. We speculate that persistent exposure of progenitor cells to low-dose PBA likely renders a permanent alteration of their differentiated transcriptomes that are maintained by epigenetic mechanisms. Deacetylated modifications of histone and promoter hypermethylation are heritably established in an inactive gene while acetylated histone and promoter hypomethylation may be present to mark an active locus (Jones and Baylin, 2007; Vaissiere et al., 2008). However, individuals may have different susceptibility to these epigenetic modifications. Based on our expression profile analysis of primary MDECs, the high-responder group is more sensitive than the low-responder group to the BPA preexposure. Whereas genetic variations in response to xenoestrogens are well documented in different strains of mice or rats (Richter et al., 2007; Tyl, 2009), this study provides the first evidence that differential susceptibility to low-dose BPA exposure may also be present in human populations.
It has been observed that exposure to low-dose BPA during early stages of mammary gland development may increase the risk of developing breast neoplasm in adult animals (Durando et al., 2007; Murray et al., 2007). Supporting this finding, we have identified 170 human candidate genes that may play a critical role in tumorigenesis. Ingenuity pathway analysis has uncovered their putative functions primarily related to aminoacyl-tRNA biosynthesis, circadian rhythm signaling, GM-CSF signaling, and HER-2 signaling, the deregulation of which can promote the development of breast cancer (see Supplemental Figure S4). The expression of some of these BPA-influenced genes may be epigenetically imprinted in breast cancer cells. We were able to validate one candidate gene, LAMP3, the promoter hypermethylation of which is preferentially linked to transcriptional silencing in ERα breast cancer cells. This gene is known to encode proteins associated with cell mobility and adhesion, and its overexpression is usually linked to invasiveness in cancer (Kanao et al., 2005). Since LAMP3 may not be epigenetically silenced in ERα-negative tumors, its aberrant expression could contribute to more aggressive phenotypes in this type of breast cancer. The hypermethylation finding of LAMP3 independently observed in MCF-7 cells and primary breast tumors suggests that this epigenetic event can be initiated in normal breast epithelial cells and then heritably passed on to cancer cells during the course of malignant progression. We further speculate that DNA methylation of LAMP3 is potentially acquired as a result of long-term exposure of progenitor cells to BPA and other xenoestrogens.
Interestingly, promoter hypermethylation of this locus was found to be associated with older ERα-positive breast patients in the US cohort and Taiwan cohort. However, the levels of DNA methylation distribution showed significantly differences between age groups. This epigenetic disparity could be attributed in part to the geographical differences of breast cancer incidence in these cohorts. Compared to the US patient population, there has been an increased trend of ERα-positive young breast cancers (<50 years old) in Taiwan (Lin et al., 2009). Future population study is needed to additionally determine whether different exposure history of BPA and other related chemicals contribute to this epigenetic disparity in the two patient populations.
Conclusions
In the present study, we have shown that the mammospheres exposure system is a valuable tool for validation studies of BPA findings based on animal models. We observed heritable effects of low-dose BPA on the nuclear localization of ERα and differential gene expression in primary MDECs. Long-term exposure of breast progenitor cells to BPA may promote ligand-independent ERα actions in differentiated progeny. Furthermore, genetic variations of individuals may contribute to differential susceptibility of breast epithelial cells to the environmental exposure. We have also identified 170 BPA-influenced genes that likely play a role in the development of ERα-positive breast cancer. These loci are potential biomarkers for assessing the risk of developing breast cancer from exposure to other environmental chemicals.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health [U01 ES015986, R01 CA069065, and R01 ES017594 to T.H.-M.H]. We thank Jeff Apostolos for his technical assistance.
Footnotes
Conflict of interest statement
All authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akamine R, Yamamoto T, Watanabe M, Yamazaki N, Kataoka M, Ishikawa M, Ooie T, Baba Y, Shinohara Y. Usefulness of the 5′ region of the cDNA encoding acidic ribosomal phosphoprotein P0 conserved among rats, mice, and humans as a standard probe for gene expression analysis in different tissues and animal species. J Biochem Biophys Methods. 2007;70:481–486. doi: 10.1016/j.jbbm.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Bjornstrom L, Sjoberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol. 2005;19:833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- Buteau-Lozano H, Velasco G, Cristofari M, Balaguer P, Perrot-Applanat M. Xenoestrogens modulate vascular endothelial growth factor secretion in breast cancer cells through an estrogen receptor-dependent mechanism. J Endocrinol. 2008;196:399–412. doi: 10.1677/JOE-07-0198. [DOI] [PubMed] [Google Scholar]
- Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003–2004. Environ Health Perspect. 2008;116:39–44. doi: 10.1289/ehp.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Fourth national report on human exposure to environmental chemicals. Atlanta, GA: Centers for Disease Control and Prevention; National Center for Environmental Health; Division of Laboratory Sciences; 2009. [Google Scholar]
- Cheng AS, Culhane AC, Chan MW, Venkataramu CR, Ehrich M, Nasir A, Rodriguez BA, Liu J, Yan PS, Quackenbush J, Nephew KP, Yeatman TJ, Huang TH. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68:1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dairkee SH, Seok J, Champion S, Sayeed A, Mindrinos M, Xiao W, Davis RW, Goodson WH. Bisphenol A induces a profile of tumor aggressiveness in high-risk cells from breast cancer patients. Cancer Res. 2008;68:2076–2080. doi: 10.1158/0008-5472.CAN-07-6526. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, El-Ashry D, Wicha MS. Breast cancer, stem/progenitor cells and the estrogen receptor. Trends Endocrinol Metab. 2004;15:193–197. doi: 10.1016/j.tem.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Durando M, Kass L, Piva J, Sonnenschein C, Soto AM, Luque EH, Munoz-de-Toro M. Prenatal bisphenol A exposure induces preneoplastic lesions in the mammary gland in Wistar rats. Environ Health Perspect. 2007;115:80–86. doi: 10.1289/ehp.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin AM, Huang TH, Toland AE. Epigenetic alterations in the breast: Implications for breast cancer detection, prognosis and treatment. Semin Cancer Biol. 2009;19:165–171. doi: 10.1016/j.semcancer.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Goodson A, Robin H, Summerfield W, Cooper I. Migration of bisphenol A from can coatings--effects of damage, storage conditions and heating. Food Addit Contam. 2004;21:1015–1026. doi: 10.1080/02652030400011387. [DOI] [PubMed] [Google Scholar]
- HarrEus UA, Wallner BC, Kastenbauer ER, Kleinsasser NH. Genotoxicity and Cytotoxicity of 4-Nonylphenol Ethoxylate on Lymphocytes as Assessed by the Comet Assay. International Journal of Environmental Analytical Chemistry. 2002;82:395–401. [Google Scholar]
- Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Deatherage DE, Rodriguez BA, Liyanarachchi S, Weng YI, Zuo T, Liu J, Cheng AS, Huang TH. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res. 2009;69:5936–5945. doi: 10.1158/0008-5472.CAN-08-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, Miller DS, Huang TH. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanao H, Enomoto T, Kimura T, Fujita M, Nakashima R, Ueda Y, Ueno Y, Miyatake T, Yoshizaki T, Buzard GS, Tanigami A, Yoshino K, Murata Y. Overexpression of LAMP3/TSC403/DC-LAMP promotes metastasis in uterine cervical cancer. Cancer Res. 2005;65:8640–8645. doi: 10.1158/0008-5472.CAN-04-4112. [DOI] [PubMed] [Google Scholar]
- Kang JH, Kito K, Kondo F. Factors influencing the migration of bisphenol A from cans. J Food Prot. 2003;66:1444–1447. doi: 10.4315/0362-028x-66.8.1444. [DOI] [PubMed] [Google Scholar]
- Lannigan DA. Estrogen receptor phosphorylation. Steroids. 2003;68:1–9. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- Lin CH, Liau JY, Lu YS, Huang CS, Lee WC, Kuo KT, Shen YC, Kuo SH, Lan C, Liu JM, Kuo WH, Chang KJ, Cheng AL. Molecular subtypes of breast cancer emerging in young women in Taiwan: evidence for more than just westernization as a reason for the disease in Asia. Cancer Epidemiol Biomarkers Prev. 2009;18:1807–1814. doi: 10.1158/1055-9965.EPI-09-0096. [DOI] [PubMed] [Google Scholar]
- Markey CM, Luque EH, Munoz De Toro M, Sonnenschein C, Soto AM. In utero exposure to bisphenol A alters the development and tissue organization of the mouse mammary gland. Biol Reprod. 2001;65:1215–1223. doi: 10.1093/biolreprod/65.4.1215. [DOI] [PubMed] [Google Scholar]
- Marotta LL, Polyak K. Cancer stem cells: a model in the making. Curr Opin Genet Dev. 2009;19:44–50. doi: 10.1016/j.gde.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Mountfort KA, Kelly J, Jickells SM, Castle L. Investigations into the potential degradation of polycarbonate baby bottles during sterilization with consequent release of bisphenol A. Food Addit Contam. 1997;14:737–740. doi: 10.1080/02652039709374584. [DOI] [PubMed] [Google Scholar]
- Munoz-de-Toro M, Markey CM, Wadia PR, Luque EH, Rubin BS, Sonnenschein C, Soto AM. Perinatal exposure to bisphenol-A alters peripubertal mammary gland development in mice. Endocrinology. 2005;146:4138–4147. doi: 10.1210/en.2005-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LC, Skliris GP, Rowan BG, Al-Dhaheri M, Williams C, Penner C, Troup S, Begic S, Parisien M, Watson PH. The relevance of phosphorylated forms of estrogen receptor in human breast cancer in vivo. J Steroid Biochem Mol Biol. 2009;114:90–95. doi: 10.1016/j.jsbmb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Murray TJ, Maffini MV, Ucci AA, Sonnenschein C, Soto AM. Induction of mammary gland ductal hyperplasias and carcinoma in situ following fetal bisphenol A exposure. Reprod Toxicol. 2007;23:383–390. doi: 10.1016/j.reprotox.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido Y, Yoshizawa K, Danbara N, Tsujita-Kyutoku M, Yuri T, Uehara N, Tsubura A. Effects of maternal xenoestrogen exposure on development of the reproductive tract and mammary gland in female CD-1 mouse offspring. Reprod Toxicol. 2004;18:803–811. doi: 10.1016/j.reprotox.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Ohm JE, Baylin SB. Stem cell chromatin patterns: an instructive mechanism for DNA hypermethylation? Cell Cycle. 2007;6:1040–1043. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Tokunaga T, Liu X, Takayanagi S, Matsushima A, Shimohigashi Y. Direct evidence revealing structural elements essential for the high binding ability of bisphenol A to human estrogen-related receptor-gamma. Environ Health Perspect. 2008;116:32–38. doi: 10.1289/ehp.10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins GS, Tang WY, Belmonte J, Ho SM. Developmental exposure to bisphenol A increases prostate cancer susceptibility in adult rats: epigenetic mode of action is implicated. Fertil Steril. 2008;89:e41. doi: 10.1016/j.fertnstert.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter CA, Birnbaum LS, Farabollini F, Newbold RR, Rubin BS, Talsness CE, Vandenbergh JG, Walser-Kuntz DR, vom Saal FS. In vivo effects of bisphenol A in laboratory rodent studies. Reprod Toxicol. 2007;24:199–224. doi: 10.1016/j.reprotox.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrdanz E, Ohler S, Tran-Thi QH, Kahl R. The phytoestrogen daidzein affects the antioxidant enzyme system of rat hepatoma H4IIE cells. J Nutr. 2002;132:370–375. doi: 10.1093/jn/132.3.370. [DOI] [PubMed] [Google Scholar]
- Soto AM, Maffini MV, Schaeberle CM, Sonnenschein C. Strengths and weaknesses of in vitro assays for estrogenic and androgenic activity. Best Pract Res Clin Endocrinol Metab. 2006;20:15–33. doi: 10.1016/j.beem.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Timms BG, Howdeshell KL, Barton L, Bradley S, Richter CA, vom Saal FS. Estrogenic chemicals in plastic and oral contraceptives disrupt development of the fetal mouse prostate and urethra. Proc Natl Acad Sci U S A. 2005;102:7014–7019. doi: 10.1073/pnas.0502544102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- Tyl RW. Basic exploratory research versus guideline-compliant studies used for hazard evaluation and risk assessment: bisphenol A as a case study. Environ Health Perspect. 2009;117:1644–1651. doi: 10.1289/ehp.0900893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Maffini MV, Sonnenschein C, Rubin BS, Soto AM. Bisphenol-A and the great divide: a review of controversies in the field of endocrine disruption. Endocr Rev. 2009;30:75–95. doi: 10.1210/er.2008-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel SA. The politics of plastics: the making and unmaking of bisphenol a “safety”. Am J Public Health. 2009;99(Suppl 3):S559–566. doi: 10.2105/AJPH.2008.159228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welshons WV, Nagel SC, vom Saal FS. Large effects from small exposures. III. Endocrine mechanisms mediating effects of bisphenol A at levels of human exposure. Endocrinology. 2006;147:S56–69. doi: 10.1210/en.2005-1159. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, Fushiki S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem Biophys Res Commun. 2008;376:563–567. doi: 10.1016/j.bbrc.2008.09.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.