Abstract
Protein ubiquitination is a post-translational modification (PTM) that regulates various aspects of protein function by different mechanisms. Global profiling of PTMs usually relies on purification and sequencing of PTM-containing peptides from proteolytic digests. Characterization of ubiquitination has lagged behind that of smaller PTMs, such as phosphorylation and acetylation, largely because of the difficulty of isolating and identifying peptides derived from the ubiquitinated portion of proteins. To address this issue, we have generated a monoclonal antibody that can enrich for peptides containing lysine residues modified by diglycine, an adduct left at sites of ubiquitination after trypsin digestion. We use mass spectrometry to identify 374 diglycine-modified lysines on 236 ubiquitinated proteins from HEK293 cells, including 80 proteins containing multiple sites of ubiquitination. Seventy-two percent of these proteins and 92% of the ubiquitination sites do not appear to have been reported previously. Ubiquitin remnant profiling of the multi-ubiquitinated proteins proliferating cell nuclear antigen (PCNA) and tubulin α-1A reveals differential regulation of ubiquitination at specific sites by microtubule inhibitors, demonstrating the effectiveness of our method to characterize the dynamics of lysine ubiquitination.
Protein ubiquitination occurs on a wide variety of eukaryotic proteins and affects processes ranging from protein degradation and subcellular localization to gene expression and DNA repair1. The process of ubiquitination involves the transfer of ubiquitin to a target protein utilizing E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases1. This process typically leads to the formation of an amide linkage comprising the ε-amine of lysine of the target protein and the C terminus of ubiquitin, and can involve ubiquitination at distinct sites within the same protein although the roles of ubiquitination at distinct sites are incompletely understood. The human genome is predicted to encode 16 E1, 53 E2 and 527 E3 proteins2, which underscores the likely importance of ubiquitination in molecular signaling.
In most cases, proteins suspected to be ubiquitinated have been identified based on their susceptibility to proteasome-mediated degradation, as evidenced by their increased levels following application of proteasome inhibitors. These proteins are immunopurified and ubiquitin adducts are confirmed by anti-ubiquitin immunoblotting3. Mutagenesis experiments can identify ubiquitination sites4. Global identification of ubiquitinated proteins has been performed by purifying ubiquitinated proteins, using ubiquitin-binding proteins such as anti-ubiquitin antibodies5, or by purifying hexahistidine (His6)-tagged ubiquitin-protein conjugates6. The enriched set of proteins are then proteolyzed and subjected to tandem mass spectrometry (MS/MS) to identify ubiquitinated proteins. However, as only one or a few lysines are typically modified in any ubiquitinated protein, most peptides do not exhibit any ubiquitin-derived modifications7.
Alternatively, proteolytic digests can be screened for peptides that contain remnants of ubiquitin modification. Digestion of ubiquitin-conjugated proteins results in peptides that contain a ubiquitin remnant derived from the ubiquitin C-terminus. The three C-terminal residues of ubiquitin are Arg-Gly-Gly, with the C-terminal glycine conjugated to the lysine in the target. After trypsinolysis, ubiquitin is cleaved after arginine, resulting in a Gly-Gly dipeptide remnant on the conjugated lysine. Therefore, tryptic digests will include peptides that contain a diglycine-modified lysine, indicating the prior conjugation of ubiquitin to that region of the target protein. The diglycine-modified lysine serves as a signature of ubiquitination and also identifies the specific site of modification. Sequencing of ubiquitin remnant–containing peptides in tryptic digests has been used to identify 110 ubiquitination sites from yeast expressing His6-ubiquitin7. Despite the availability of these approaches for several years, analysis of the Swiss-Prot database indicates that only 255 mammalian proteins have been reported to be ubiquitinated based on experimental evidence. In most cases, the ubiquitination sites have not been identified.
Here we describe a novel approach to identify ubiquitinated proteins and ubiquitination sites using an antibody that selectively binds to the diglycine remnant in peptides generated from tryptic digestion of biological samples. Using this immunoaffinity approach coupled to nanoLC-MS/MS, we have identified 236 ubiquitinated proteins and 374 ubiquitination sites in HEK293 cells. Of these ubiquitinated proteins, 170 have not previously been known to be ubiquitinated. Our experiments demonstrate an immunoaffinity profiling strategy that will have broad utility in characterizing the occurrence and extent of ubiquitination in diverse tissues and disease states.
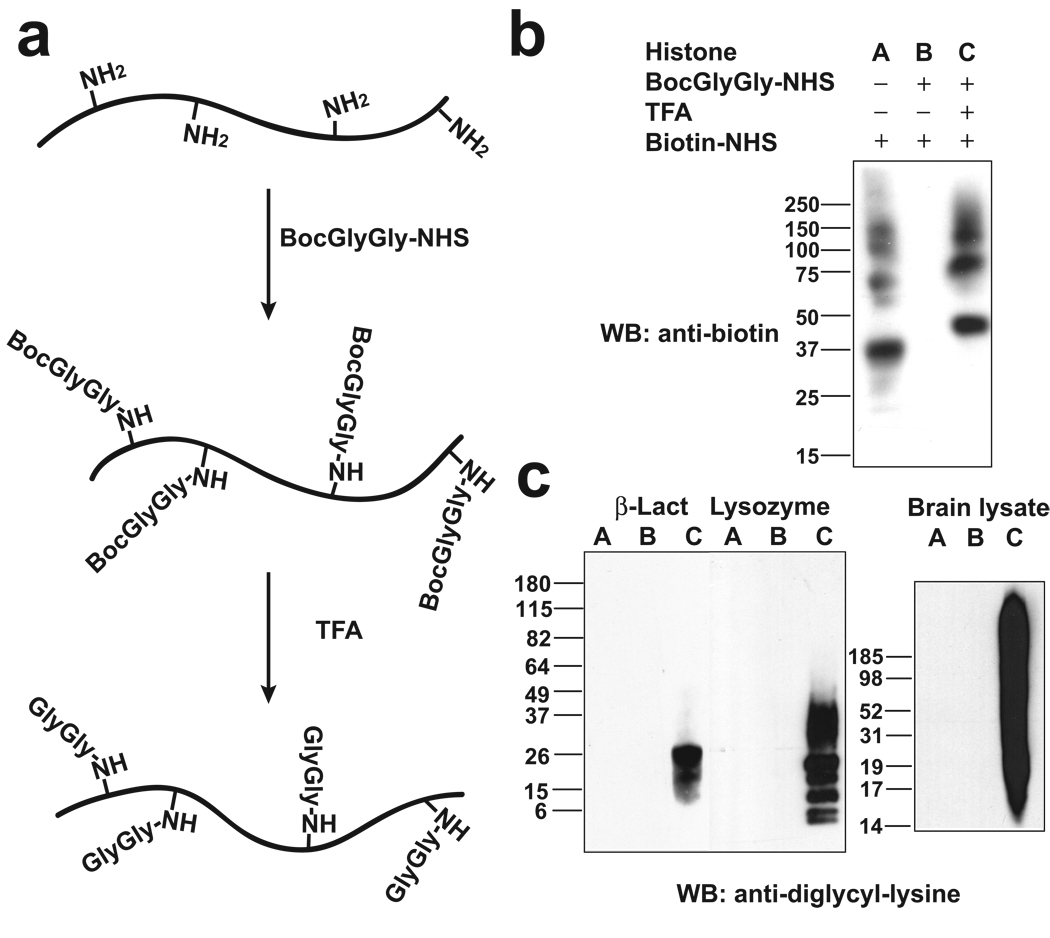
To generate an antibody that recognizes peptides containing the ubiquitin remnant, we prepared a protein antigen containing diglycine-modified lysines. First, the lysine-rich histone III-S was reacted with Boc-Gly-Gly-NHS to form an amide-linked Boc-Gly-Gly adduct on amines (Fig. 1a). Nearly complete modification of the amines was confirmed by the reduction in labeling of the Boc-Gly-Gly-modified protein by the lysine-modifying reagent biotin-NHS, as assessed by anti-biotin immunoblotting (Fig. 1b). The modified protein was treated with TFA to remove the Boc moiety. Quantitative conversion of the Boc-Gly-Gly adduct, which does not contain an amine, to Gly-Gly, which contains an amine, was confirmed by the reactivity of the TFA-treated protein with biotin-NHS (Fig. 1b).
Figure 1. Development of monoclonal antibodies that selectively recognize diglycine-modified lysines.
(a) Schematic illustration of antigen synthesis. The ε-amine of lysines in histone was modified by Boc-Gly-Gly-NHS and then the Boc group was removed by TFA. The lysines in the final protein contain Gly-Gly adducts on the ε-amine of all lysine residues.
(b) Validation of the synthesis of Gly-Gly-modified histone. To monitor the reaction of histone with Boc-Gly-Gly-NHS, the presence of amines, such as those in unmodified lysine, was detected by reacting the protein with the amine-modifying agent biotin-NHS, and subsequent Western blotting with an anti-biotin antibody. Amines in histone were nearly completely lost after treatment with Boc-Gly-Gly-NHS, indicating near complete modification of all the lysines in histone. Removal of the Boc protecting group with TFA results in the formation of an amine at the N-terminus of the Gly-Gly adduct. This step was essentially complete, as the TFA-treated protein exhibited nearly complete recovery of amine reactivity. The position of the bands in the different samples is slightly shifted due to the different molecular weight and number of positive charges in the modified and unmodified samples. The bands above 50 kDa represent impurities in the histone sample.
(c) The specificity of the GX41 monoclonal antibody was evaluated by Western blotting β-lactoglobulin, lysozyme, or rat brain lysate, in which the lysines were either unmodified (A), or modified with Boc-Gly-Gly (B) or Gly-Gly-(C) adducts, respectively.
The diglycine-modified histone was injected into mice, and hybridoma lines were screened for antibodies that specifically recognize proteins containing diglycine-modified lysines. Hybridoma line GX41 generated monoclonal antibodies that exhibited pronounced specificity for peptides containing the diglycine-modified lysines. The antibodies failed to interact with unmodified lysozyme, lactoglobulin (Fig. 1c), or proteins modified with Boc-Gly-Gly. However, the antibody recognized Gly-Gly-modified proteins obtained after removal of the Boc group. These results indicate that the antibody recognizes Gly-Gly-modified amines, and suggests that the Gly-Gly adduct must contain a free amine. Similarly, the antibody exhibited negligible reactivity with rat brain lysate (Fig. 1c), or brain lysate modified with Boc-Gly-Gly, but exhibited significant reactivity with Gly-Gly-modified proteins from brain lysate. Notably, the brain lysate includes highly abundant proteins containing internal Gly-Gly peptide sequences, such as β-actin, glyceraldehyde-3-phosphate dehydrogenase and α-tubulin, as well as histone H2A which contains an internal Gly-Gly-Lys sequence, indicating that internal Gly-Gly sequences are not recognized by the antibody. Additionally, peptides that contain Gly-Gly as the first two amino acids are not recognized (Supplementary Fig. 1). Together, these data indicate that the antibody recognizes Gly-Gly sequences which contain an unmodified primary amine, and which are present as an adduct on the ε-amine of lysine.
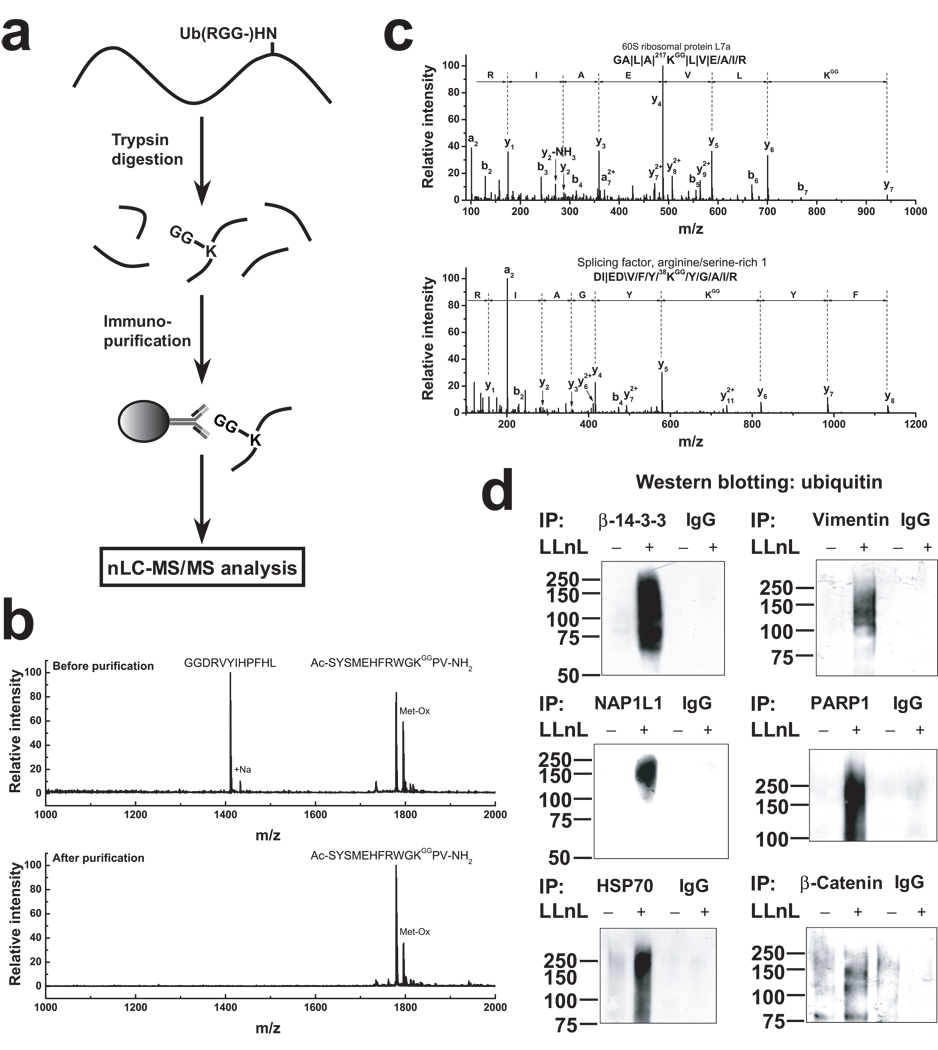
We next investigated whether the anti-diglycyl-lysine antibody was able to immunoprecipitate peptides containing Gly-Gly-modified lysine. A flow chart for sample preparation, immunoprecipitation, and MS/MS analysis is shown in Fig. 2a. We prepared a peptide containing an N-terminal Gly-Gly sequence (GGDRVYIHPFHL), and a peptide containing a diglycyl adduct on lysine (Ac-SYSMEHFRWGK*PV-NH2, K* and Ac represent Gly-Gly-modified lysine and an acetyl group, respectively). An equimolar mixture of the peptides was immunoprecipitated with the anti-diglycyl-lysine antibody, resulting in selective enrichment (≥50×) of the peptide containing the Gly-Gly-modified lysine (Fig. 2b). Additionally, this peptide was quantitatively immunoprecipitated with a nearly 100% yield (Supplementary Fig. 2). These experiments demonstrated that the GX41 antibody is capable of enriching peptides containing diglycine-modified lysines and does not immunoprecipitate peptides containing a Gly-Gly sequence at their N termini.
Figure 2. Profiling immunopurified ubiquitin remnant-containing peptides to identify ubiquitinated proteins.
(a) Schematic of the strategy for identifying ubiquitinated proteins by immunoprecipitation of peptides containing diglycyl-lysine and MS analysis.
(b) Confirmation of antibody specificity using two peptides, GGDRVYIHPFHL and Ac-SYSMEHFRWGK*PV-NH2 (K* representing Gly-Gly-modified lysine). An equimolar amount (0.3 nmol) of the two peptides were mixed and immunoprecipitated with immobilized anti-diglycyl-lysine monoclonal antibody. MALDI-TOF-MS analysis was carried out for the starting material and the antibody-purified material. An enrichment factor of at least 50 is estimated from the relative MS signal of the two peptides before and after immunoprecipitation.
(c) Representative annotated MS/MS spectra of a ubiquitin remnant-containing peptide obtained by immunoprecipitation from a HEK293 cell lysate. The sequence of the ubiquitinated peptide is indicated and the fragment ions are labeled. The diglycine-modified lysine and its position are indicated in red. The symbols, \, / and |, represent b-ions, y-ions, and both b-ions and y-ions, respectively.
(d) Biochemical verification of ubiquitinated proteins. Proteins were immunoprecipitated using target-specific antibodies and the immunoprecipitate was Western blotted with an anti-ubiquitin antibody. IgG was used as a control for nonspecific immunoprecipitation. The proteasome inhibitor LLnL was added to allow accumulation of the ubiquitinated protein.
We next sought to assess the diversity of lysine ubiquitination in cultured cells. To distinguish diglycine remnants derived from ubiquitin from those originating from less common ubiquitin-like proteins (such as ISG15 and NEDD8, which also leave a diglycine remnant on lysines after trypsinization8), we used HEK293 cells expressing His6-tagged ubiquitin. Ubiquitinated proteins were purified by immobilized metal-affinity chromatography, prior to proteolysis and anti-diglycyl-lysine immunopurification. Ubiquitin remnant-containing peptides were subjected to LC-MS/MS followed by database searching and spectral validation. To minimize alterations in ubiquitination levels after cell lysis, 5 mM chloroacetamide was included in lysis buffer to inhibit deubiquitinase and ubiquitin ligase activity9. To measure post-lysis ubiquitination, we spiked a lysate with excess glutathione S-transferase (GST). This protein showed no detectable level of ubiquitination (Supplementary Fig. 3) suggesting that negligible ubiquitination occurred after cell lysis.
MS/MS spectra of ubiquitin remnant-containing peptides exhibited normal y- and b-ion series, typically with a pair of ions separated by a mass of 242.14 Da, consistent with the masses of a lysine residue (128.09) and a Gly-Gly adduct (114.04 Da) on the ε-amine of lysine. Most peptides contained a single diglycine-modified lysine (Fig. 2c), while 17 peptides contained two diglycine-modified lysines. The majority (> 92%) of ubiquitin remnant-containing peptides have a +3 or +4 charge (Supplementary Fig. 4), which reflects the additional charge from the N-terminal amine on the Gly-Gly adduct. Gly-Gly-modified lysines at the C-terminus of peptides were also detected (~2% of total) (Supplementary Fig. 5), and reflect utilization of the Gly-Gly-modified lysine as a substrate for trypsin, as has been described previously10.
In total, 374 diglycine-modified lysines on 236 ubiquitinated mammalian proteins were identified, of which 72% were not previously reported to be ubiquitinated based on the PTM annotation in the Swiss-Prot database. Similarly, 92% of the ubiquitination sites that were identified were not previously known. Among the identified proteins, 156 proteins have one ubiquitination site and 80 have two or more ubiquitination sites (Supplementary Table 1 and Supplementary Fig. 6). To validate the ubiquitination detected using the ubiquitin remnant profiling approach, we selected a subset of six proteins identified by MS and assessed whether they were ubiquitinated in cells. Lysates from HEK293 cells were immunoprecipitated with antibodies specific for the protein under investigation and immunoblotted using an anti-ubiquitin antibody (Fig. 2d). In these experiments, the HEK293 cells were not transfected with plasmids expressing His6-tagged ubiquitin. In each case, the immunopurified protein exhibits anti-ubiquitin immunoreactivity consistent with the endogenous ubiquitination of these proteins.
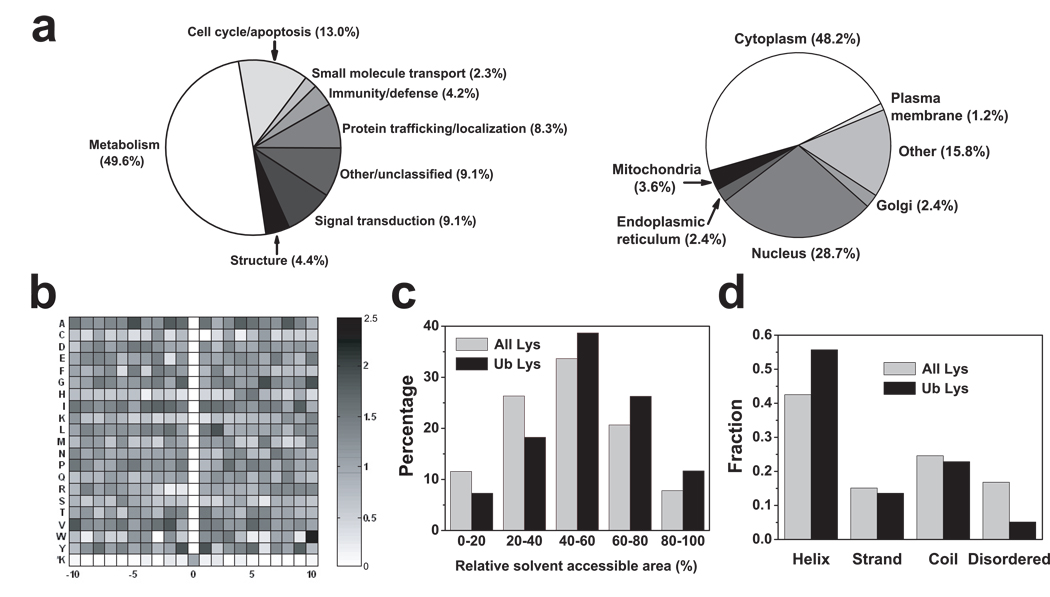
The ubiquitination targets include disease-related proteins, such as 14-3-3ε, ataxin, β-catenin, BRCA1-associated protein, and TTRAP (TRAF and TNF receptor-associated protein). The proteins identified by ubiquitin remnant profiling have roles in numerous biological processes, of which the largest number involve metabolism, cell cycle/apoptosis, and signal transduction (Fig. 3a). Additionally, proteins were found that have roles in protein trafficking/localization, protein structure, and the immune system, consistent with previously reported roles for ubiquitination11–14. Many ubiquitin-conjugating enzymes, ubiquitin ligases, and 26S proteasome regulatory subunits are ubiquitinated, consistent with previous studies that many proteins involved in proteasome degradation pathways are ubiquitinated15, 16. Some proteins found to be ubiquitinated extend findings regarding the role of ubiquitination in certain cellular processes. For example, while histone H2 ubiquitination has been described3, we find that histone H1, H3, and H4 isoforms are also ubiquitinated, as well as histone acetyltransferases and histone deacetylase subunits, supporting the idea that ubiquitin contributes to epigenetic gene regulation through multiple pathways. Many heat shock proteins, such as HSP70, HSP105, and HSC71, are ubiquitinated, linking ubiquitination to stress responses. Several heterogeneous nuclear ribonucleoproteins are ubiquitinated, showing that ubiquitination influences mRNA processing, metabolism, transport, and splicing. Our studies also identify numerous transcription factors, splicing factors, DNA repair proteins, and kinases, supporting the well-characterized role for ubiquitination in regulating cellular signal transduction.
Figure 3. Bioinformatic analysis of ubiquitinated proteins and ubiquitin-modified lysines.
(a) Pie charts of biological processes and subcellular localization of ubiquitinated proteins analyzed by PANTHER and PENCE Proteome Analyst database, respectively. Some proteins were designated “other” if their localizations or functions were not annotated in the database.
(b) Backbone amino acid sequence analysis of ubiquitinated peptides. A density map of the ratios of the frequencies of each of the 20 amino acids adjacent to the ubiquitinated lysines and adjacent to lysines in general was plotted using MATLAB. Several amino acids are slightly enriched at certain positions, such as Leu at +2, Val at −2, Ala at −5, Gly at +6, and Tyr at −1 and +1, determined by Rosner’s test with a 95% confidence.
(c) Increased solvent accessible area (SAA) of ubiquitinated lysines relative to lysines in general. The distribution of SAA of both populations of lysines indicates an increase in SAA among ubiquitinated lysines. The two distributions are significantly different (student’s t test, p < 0.001). The results were obtained from an analysis of 89 PDB structures (140 ubiquitinated lysines, 3970 total lysines).
(d) Distribution of secondary structures of all lysines and ubiquitinated lysines obtained from an analysis of 89 PDB structures. The disordered region was predicted by DisEMBL for all ubiquitinated proteins identified by our MS experiments. χ2 test: p-value < 0.001.
The subcellular distribution of the detected proteins is likely to reflect, in part, the subcellular fractions that were used for MS/MS analysis. Subcellular localization analysis of the identified proteins indicates that essentially all the ubiquitinated proteins are cytosolic (Fig. 3a, right panel), which is consistent with the general observation that ubiquitination occurs primary in the cytosolic compartment of the cell12. Many of the identified proteins are localized to the nucleus, and several proteins are mitochondrially-localized, suggesting a role for ubiquitination in regulating aspects of mitochondrial function.
We next wanted to gain insight into how lysine ubiquitination might be regulated at the level of primary and secondary structure. Interestingly, ubiquitin remnant-modified lysines have a slight tendency to be localized in regions enriched in small hydrophobic residues, such as Ala, Leu, Ile, Gly, Pro and Val (Supplementary Fig. 7a). Examination of a six amino acid window adjacent to modified lysines revealed that Cys, His, and Lys are found at a ~40% lower frequency than when they are adjacent to lysines in general in the human proteome (Supplementary Fig. 7a). A consensus ubiquitination site analysis using Motif-x17 identified K*XL as a potential motif, and was found 1.8× more commonly among ubiquitinated lysines than lysines in general (Supplementary Fig. 7b). To compare all 20 amino acids for their propensity to be found at specific residues adjacent to ubiquitinated lysines, we prepared a density map that indicates the frequency of each amino acid at any of the ten proximal positions on either side of the ubiquitinated lysines, compared to the frequency of that amino acid next to lysines in general, as assessed by surveying the human proteome (Fig. 3b). This analysis shows that there is only a subtle enrichment for specific residues at some positions, such as leucine at the +2 position, valine at the −2 position, alanine at the −5 position, glycine at the +6 position and tyrosine at the −1 and +1 positions. In contrast, an analysis of ubiquitinated proteins in yeast7 indicates a significant enrichment of Asp, Glu, His and Pro at some positions (Supplementary Fig. 7c). To determine if the sequence of the immunogen affected the specificity of the immunoprecipitated peptides, we generated a similar density map to present the frequency of each amino acid adjacent to the Gly-Gly-modified lysines in immunogen. Although there are marked amino acid preferences adjacent to lysine in immunogen (Supplementary Fig. 7d), these preferences are not seen in the peptides that pulled down by the anti-diglycyl- lysine antibody (Supplementary Fig. 7d), supporting the idea that immunogen sequence does not significantly bias the sequences of the immunoprecipitated peptides the antibody recovers.
We found that ubiquitinated lysines have a slight tendency to appear on protein surfaces in preferred structural contexts. For 89 of the proteins identified in this study, structural information was available in Protein Data Bank (PDB). Measurements of the solvent-accessible area of lysines in these proteins indicate that ubiquitinated lysines tend to be slightly more solvent exposed than other lysines (Fig. 3c, student’s t test, p < 0.001). If lysines with more than 50% surface exposure are considered solvent exposed18, 60% of the ubiquitinated lysines are exposed, which is higher than lysines in general (45%). Overall, ubiquitinated lysines are about 6.5% more exposed than all the lysines. This is in agreement with a ubiquitination site survey for yeast19. Interestingly, in some cases, the ubiquitinated lysine is fully buried (for example, see Supplementary Fig. 8). In these proteins, ubiquitination may be regulated by stimuli that induce the exposure of the lysine to the surface. Secondary structure analysis for all lysines and ubiquitinated lysines indicates that ubiquitinated lysines prefer helical structures compared to all lysines, although ubiquitination sites can also be found in other structural contexts (Fig. 3d). Additional crystal structures of proteins that are susceptible to ubiquitination are needed to fully assess the solvent exposure and structural contexts of ubiquitinated lysines.
Recently, a large number of lysine acetylation sites have been discovered by proteomic approaches20–23. Although only 0.6% of lysines are predicted to be acetylated based on yeast studies24, more than 20% of the lysines that we found to be ubiquitinated are also sites of acetylation. For example, all the ubiquitinated lysines detected in H2B, H3.1 and H4 were reported as acetylated. In the case of tubulin α-1A, four of the six ubiquitinated lysines were reported to be acetylated. The surprisingly high degree of concordance of lysine ubiquitination and acetylation sites suggests that acetylation of a specific lysine residue could serve as a means to prevent lysine ubiquitination25, or vice versa. A BLAST analysis of ubiquitination sites in human proteins against mouse, rat and yeast revealed that modified lysines are statistically more conserved between these species than lysines in general (Supplementary Fig. 9). This suggests that the pathways leading to the ubiquitination of these sites may be evolutionarily conserved.
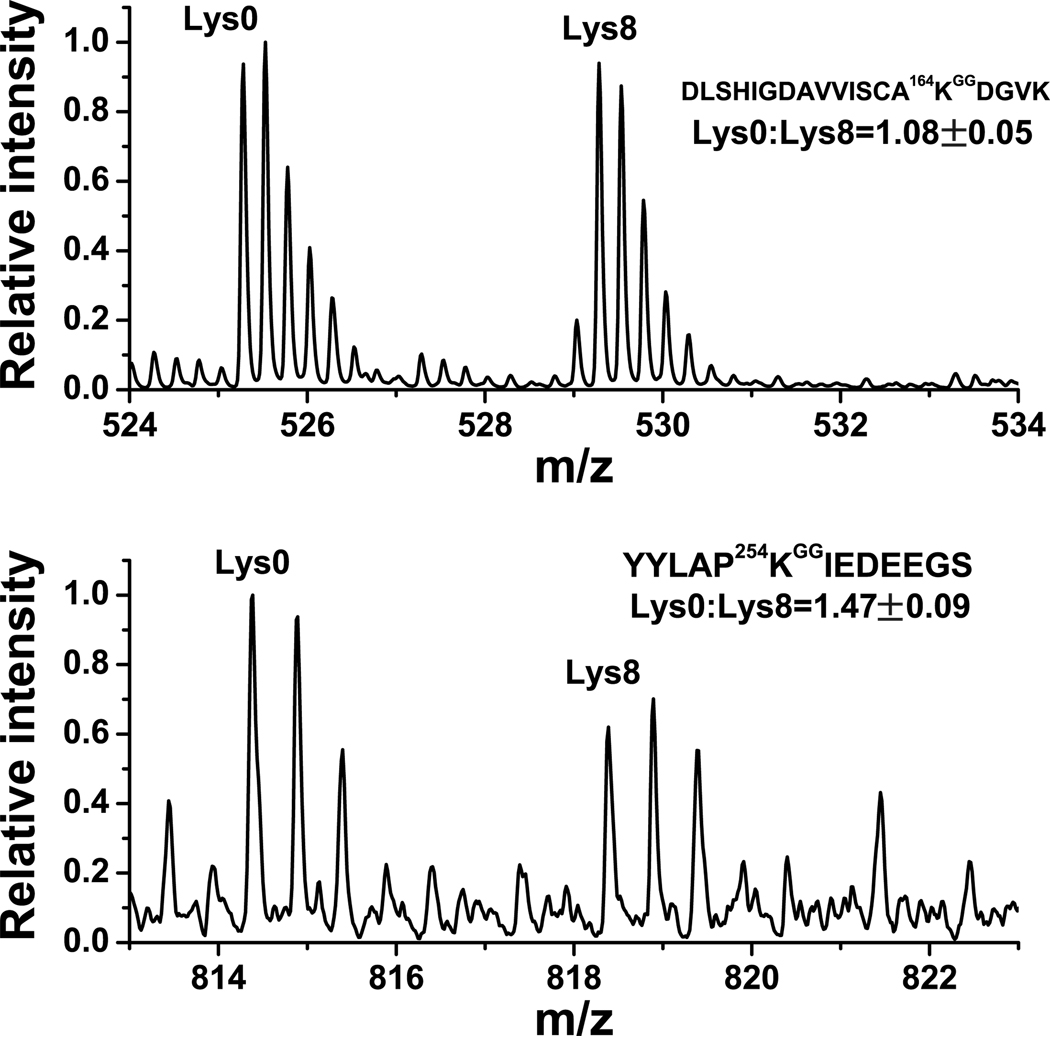
In cases where a protein is ubiquitinated at more than one site, it is particularly challenging to monitor how the ubiquitination at the individual sites is independently regulated. We therefore examined two proteins exhibiting multiubiquitination, tubulin α-1A and PCNA (proliferating cell nuclear antigen), a protein that regulates cell cycle progression26 and has been linked to tumorigenesis27. His6-ubiquitin-expressing HEK293T cells were metabolically labeled with either light (Lys0) or heavy (Lys8) lysine to quantitate ubiquitination using the SILAC (stable isotope labeling by amino acids in cell culture) approach28 (Supplementary Fig. 10). Cells were treated for 16 h with vehicle (Lys8) or 10 µM colchicine (Lys0), an inhibitor of microtubule polymerization that affects cell cycle29. Cells were mixed, lysed and processed as before, and the samples were analyzed by nanoLC-MS to quantify ubiquitination at the PCNA ubiquitination sites that were previously identified in tandem MS based on their retention time, mass-to-charge ratio (m/z), and charge states. Quantification of relative ubiquitination at each modification site was measured by normalization to protein abundance, as measured by the averaged light-to-heavy ratio of unmodified peptides detected from initial mixed cell lysate prior to any affinity purification as described previously30 (Supplementary Fig. 11). Interestingly, following colchicine treatment, the ubiquitination of K164 was unaffected, while the ubiquitination of K254 was increased by 47% (Fig. 4).
Figure 4. Colchicine differentially regulates the ubiquitination of two lysines in PCNA.
HEK293 cells were grown in SILAC media containing either light (Lys0) or heavy (Lys8) lysine, and transfected with a plasmid expressing His6-ubiquitin. Lys0-labeled cells were treated with 10 µM colchicine while Lys8-labeled cells were treated with vehicle for 16 h. An identical amount of cells from both treatments were mixed and processed for MS analysis of ubiquitin remnant-containing peptides. The relative ratio of MS signals between Lys0- and Lys8-labeled peptides was used for relative quantification of the change in ubiquitination at K164 and K254. The observed ratio was normalized to the change in PCNA protein abundance in the two samples by measuring two unmodified PCNA peptides in the initial mixed cell lysate (Supplementary Fig. 11). We found that the ion intensity of the novel ubiquitination site (K254) is about 20% of that of K164, which suggests that its ubiquitination may be less common or more transient than K164, and which may explain why it was not previously detected in mutagenesis studies33. All experiments are the average of experiments repeated three times. Note that the peptide ubiquitinated at K254 is the C-terminal tryptic peptide of the protein so that the last amino acid is not K or R, and the charge state is +2.
We also examined the multiubiquitination of tubulin α-1A. Treatment with colchicine resulted in a similar ~80% decrease in the ubiquitination of K326, K336, and K370. Surprisingly, treatment with vinblastine, which also disrupts microtubules, although through a distinct mechanism31, 32, resulted in an opposite effect on ubiquitination, with a ~40% increase in ubiquitination at each of these sites (Supplementary Figs. 12 and 13). These results highlight how some ubiquitination sites may be ubiquitinated in a dynamic manner, for example in response to specific signals, while other ubiquitination sites may be “constitutive.” In the case of both PCNA and tubulin α-1A, ubiquitin remnant profiling provided insights into how distinct ubiquitination sites respond to different experimental treatments in a manner not readily available using currently available approaches.
The ubiquitin remnant profiling approach described here provides a simple and robust strategy to identify and quantify sites of ubiquitination in cells. It could be used to identify ubiquitination patterns in cells and tissues with altered expression of ubiquitin ligases, deubiquitinating enzymes, as well as to profile changes in ubiquitination elicited by various signaling molecules, drugs, and in disease states. Although the present data utilized cells expressing His6-tagged ubiquitin to reduce the likelihood of obtaining diglycine-modified peptides from ISG15- and NEDD8-modified proteins, ubiquitin-modified proteins could readily be enriched using immobilized ubiquitin-binding proteins, such as S5a, or ubiquitin antibodies5 in cells and tissues not amenable to transfection.
Methods
Antigen synthesis and antibody production
Lysine-rich histone from calf thymus (type III-S, Sigma) was dissolved in 100 mM NaHCO3 buffer (10 ml) at pH 10. 500 µl t-butyloxycarbonyl-Gly-Gly-N-hydroxysuccinimide (50 mM, Boc-Gly-Gly-NHS, ref. 34) in DMSO was added to histone solution and the reaction was carried out at room temperature for 1 h by constant shaking on a plate rotator. This step was repeated three additional times and sample B was obtained. For deprotection of the Boc group, neat trifluoroacetic acid (6 ml, TFA, Sigma) was added and the solution was shaken for 2 h at room temperature. The reaction was stopped by neutralizing with 10 M NaOH dropwise on ice (sample C). Sample A, B and C were dialyzed four times against 20 mM acetic acid followed by lyophilization. The degree of the reaction was accessed by anti-biotin (Sigma) Western blotting after samples A, B, and C were reacted with 5 mM biotin-NHS (Sigma) for 10 min. The same protocol was used to prepare Boc-Gly-Gly- and Gly-Gly-modified β-lactoglobulin, hen egg white lysozyme, rat brain lysate, and peptides (DRVYIHPFHL and Ac-SYSMEHFRWGKPV-NH2) for antibody evaluation.
The antigen was used for antibody production in mouse and hybridoma clones were made by Promab. Cells of monoclonal clones were grown in MDMEM (MegaCell Dulbecco’s Modified Eagles’s Medium, pH 7.2, Sigma) supplemented with 10% fetal bovine serum (FBS), 50 µg/ml of kanamycin, 1 mM glutamine, and cells were split and cell culture supernatant was collected every week.
Hybridoma clone GX41 was obtained after screening a panel of hybridomas to assess their utility in detecting diglycine-modified lysines. Antibodies from each hybridoma clone were first evaluated by Western blotting using Gly-Gly-modified β-lactoglobulin, lysozyme, and rat brain lysate. Clones were selected based on the absence of reactivity with unmodified protein and lysates, absence of reactivity with proteins and lysate modified with Boc-Gly-Gly, and reactivity with Gly-Gly-modified proteins and lysate. The top five clones that were further characterized were based on their ability to recognize the largest number of bands in the Gly-Gly-modified rat brain lysate. Antibodies from these clones were purified and used for immunoprecipitation of ubiquitin remnant-containing peptides from His6-ubiquitin expressing HEK 293 cells, and tandem MS identification of tryptic ubiquitinated peptides to assess the degeneracy of antibodies. Only clone GX41 pulled down peptides that contained each of the 20 amino acids N-terminal to the modified lysine, and each of the 20 amino acids C-terminal to the modified lysine, suggesting that the antibody can bind peptides which contain the diglycyl-lysine in a wide range of sequence contexts, which was supported by subsequent characterization of the amino acid context of the diglycyl-lysine obtained from a larger data set of ubiquitin remnant-containing peptides (Fig. 3b and Supplementary Fig. 7a). The GX41 anti-diglycyl-lysine monoclonal antibody was found to be IgG1κ isotype. This antibody was used for all the experiments in this study.
Antibody purification and coupling
Gly-Gly-modified β-lactoglobulin was coupled to Affi-gel 10 resin (Bio-Rad) in a concentration of 5 mg protein/mL resin in a pH 8 HEPES buffer overnight in 4°C. The resin was quenched by 1 M Tris-HCl (pH 8), washed with three volumes of 10 mM citric acid (pH 3), and phosphate buffered saline (PBS). Cell culture supernatant (50 mL) from monoclonal cell lines was loaded six times into an 8-cm column with 1 mL Affi-gel 10 resin coupled with Gly-Gly-modified β-lactoglobulin in 4°C using a peristaltic pump. The resin was washed three times with 6 ml of 2XPBS and three times with 6 mL of PBS. The antibody was eluted four times with 0.5 mL 10 mM citric acid (pH 3) and immediately neutralized by 50 µL of 1 M HEPES (pH 8). The pH was adjusted to 8.5 and the antibody was concentrated by a 15 mL filter device (30 kD molecular weight cutoff, Millipore). The antibody concentration was measured by Bradford protein assay (Bio-Rad). Typically, 0.1~0.2 mg of antibody was coupled to 20 µL Affi-gel 10 resin according to the method described above. The antibody resin was stored in PBS buffer with 0.1% sodium azide at 4°C.
Cell culture and sample preparation
Human embryo kidney (HEK) 293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 4.5 g/L glucose, 10% FBS, 100 units/mL penicillin G, and 100 µg/mL streptomycin. When the confluence reached ~ 50%, cells were transfected with 10 µg of a His6-tagged ubiquitin plasmid per 10-cm Petri dish using the calcium phosphate transfection method. Cells were used one day after transfection and treated with vehicle or proteasome inhibitor 25 µM N-acetyl-Leu-Leu-norleucinal (LLnL, Calbiochem) in DMSO and incubated for 16 h prior to harvest. The His6-ubiquitin is expressed at a fraction of the level of endogenous ubiquitin (Supplementary Fig. 14) suggesting that it is unlikely to perturb endogenous ubiquitin pathways. The expression of tagged ubiquitin has been widely used in proteomics studies of protein ubiquitination7, 35, 36.
Twenty 10-cm Petri dishes were cultured and cells were washed twice with ice-cold PBS. The cells were detached, collected and centrifuged at 1000 g for 5 min at 4°C. To increase coverage of ubiquitinated proteins, crude lysates, as well as subcellular fractions, including nuclear, membrane, and cytosolic fractions, were prepared for analysis. For the crude lysate, the cell pellet was lysed and His6-tagged proteins were purified by Ni-NTA resin (Qiagen) in native and denaturing conditions according to the manufacturer’s protocol. The lysis buffer contained 5 mM chloroacetamide to alkylate cysteines and to inhibit ubiquitin ligases and deubiquitinases9. The membrane fraction was obtained by centrifuging at 100,000 g for 60 min after removing the nuclear pellet in the presence of 250 mM sucrose. The pellet from nuclear and membrane fraction was dissolved in 8 M urea with 1% triton X-100 and 0.1% SDS and the proteins were purified by Ni-NTA resin in the presence of 10 mM β-mercaptoethanol. After immobilized metal affinity purification, ubiquitinated proteins are significantly enriched (Supplementary Fig. 15).
All the samples after Ni-NTA purification were concentrated on an Amicon YM10 filter device (Millipore) and separated by SDS-PAGE. Gel pieces were treated with 10 mM dithiothreitol at 50°C for 30 min, followed by 55 mM chloroacetamide at room temperature for 45 min, using methods described previously37, except that chloroacetamide was used in place of iodoacetamide. In-gel digestion and peptide extraction were performed as described37.
The lyophilized peptide mixture was dissolved in 300 µL of buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH7.4) and 2 mM EDTA. The sample was boiled for 10 min to deactivate residual trypsin activity. The peptide mixture was incubated with 20 µL antibody resin for 4 h in 4°C, loaded on a micro-spin column (Pierce) six times, washed three times with 2XPBS and three times with PBS, and eluted six times with 20 µL 10 mM citric acid (pH 3). The eluted peptide mixture was concentrated to 20 µL for tandem MS analysis.
For the MALDI-TOF-MS experiment, a sample containing about 0.3 nmol of each peptide, GGDRVYIHPFHL and Ac-SYSMEHFRWGK*PV-NH2 (K* and Ac represent Gly-Gly-modified lysine and an acetyl group, respectively), was prepared and subjected to immunoprecipitation using the agarose-immobilized antibody described above.
For SILAC quantification, five 10-cm dishes of HEK293T cells were grown in the media containing either light lysine (Lys0: 12C6 14N2-Lys) or heavy lysine (Lys8: 13C6 15N2-Lys) (Cambridge Isotope Labs) using previously described procedures for SILAC experiments38. The cells were transfected with His6-ubiquitin plasmid as described above, and treated with vehicle or drugs (10 µM colchicine or 1 µM vinblastine, Sigma) in the presence of LLnL (PCNA: 25 µM for 16 h; tubulin α-1A: 50 µM for 30 min). The cells were mixed and purified under denaturing condition as described above without fractionation. In order to normalize the ubiquitinated peptides by unmodified peptides in the cell lysate, a small amount of initial mixed cell lysate was digested by trypsin followed by tandem MS analysis30.
Mass spectrometric analysis
For MALDI-TOF-MS, samples were desalted by Millipore C18 ZipTip according to manufacturer’s protocol and eluted in a 2 µL solvent with 50% acetonitrile and 0.1% TFA in the presence of 10 mg/mL α-cyano-4-hydroxycinnamic acid (Sigma). The masses of the samples were analyzed in the reflector mode by Voyager-DE PRO MALDI-TOF-MS (Applied Biosystems).
The samples purified from cell lysate were analyzed by nanoLC Q-TOF MS/MS (Agilent 6520) to obtain peptide sequence information using settings as described previously39. Briefly, 8 µL of peptide mixtures were loaded onto an enrichment column with 97% solvent A and 3% solvent B with a flow rate of 3 µL/min. Solvent A consists of 0.1% formic acid (Fluka) and solvent B of 90% acetonitrile (Fisher) and 0.1% formic acid. Peptides were eluted with a gradient from 3% to 40% solvent B in 20 min, followed by a steep gradient to 90% solvent B in 5 min at a flow rate of 0.3 µL/min. Mass spectra were acquired in the positive-ion mode with automated data-dependent MS/MS on the five most intense ions from precursor MS scans and every selected precursor peak was analyzed twice within three minutes. In some runs, a list of previous identified peptides was excluded for MS/MS fragmentation.
Database search of MS/MS spectra for peptide and protein identification
Analysis of MS/MS spectra for peptide and protein identification was performed by protein database searching with Spectrum Mill software (Rev A.03.02, Agilent) against the Swiss-Prot database (v57.2, May 5, 2009) containing a concatenated reverse database with the same entries and the same length for each protein as described by Elias and Gygi40. The use of a decoy database to evaluate the false positive rate for modified peptides may underestimate the false identifications since protein modifications can greatly expand the search space. Raw spectra were first extracted to MS/MS spectra that could be assigned to at least four y- or b-series ions. Scans with the same precursor within a mass window of ±0.4 m/z were merged within a time frame of ±15 s, charges up to a maximum of 7 were assigned to the precursor ion, and the 12C peak was determined by the Data Extractor. Key search parameters were a minimum matched peak intensity of 50%, a precursor mass tolerance of ±20 ppm, and a product mass tolerance of ±40 ppm. A fixed modification was carbamidomethylation (same modification as chloroacetamide) for cysteines and variable modifications were Gly-Gly-modification for lysines and oxidation for methionines. It should be noted that there are potentially a large number of naturally occurring sequence variants in mammals, but very limited data in the databases on these sequences. These variants may be missed or misidentified if the sequence variation lies in the same peptide that contains the diglycine modified-lysine. The maximal number of diglycine modifications was set as two. Trypsin was selected as enzyme for sample digestion and four missed cleavages were allowed during the database search. The threshold used for peptide identification was a Spectrum Mill score of ≥ 9, an SPI% (the percentage of the scored peak intensity) of ≥ 50% and the difference between forward and reverse scores of ≥2. Under these criteria, the false positive rate is less than 1%. In the peptide list (Supplementary Table 1), only the highest scoring member of each peptide group is shown and only peptides with a charge state of 2, 3, 4, and 5 are reported. Finally, all MS/MS spectra were manually validated and the spectra with low quality of fragmentations were discarded.
For SILAC experiments that quantified the relative ubiquitination at specific sites in tubulin α-1A and PCNA, quantification was performed using peak intensities from the MS data on peptides that were previously identified by MS/MS (included within Supplementary Fig. 6 and the manually annotated MS/MS spectra were shown in Supplementary Fig. 16). Peptides were identified based on LC-retention time, m/z and charge state as described41,42. Quantification of relative ubiquitination at each modification site was obtained by normalization to protein abundance, as measured by the averaged light-to-heavy ratio of unmodified peptides detected from initial mixed cell lysate prior to any affinity purification as described previously30.
The two modifications isobaric to the Gly-Gly seen on the Delta Mass PTM website using a 0.02 Da mass window margin of error, which is appropriate for the Q-TOF instrument used in this study, are asparagyl- and hydroxyprolyl-modifications. The ornithyl modification is 0.04 Da larger than the diglycine modification. These rare modifications could be mistaken for the diglycine remnant on lysine. Alternatively, if these residues are adjacent to a lysine, the MS/MS ion series could be misinterpreted to contain a Gly-Gly-modified lysine. However, none of the diglycine-modified lysine residues reported in this study were found in a peptide context that could have matched an alternate tryptic peptide with a lysine adjacent to an asparagine or hydroxyproline.
This paper describes the identification of several hundred diglycine-modified peptides. Because of the large number of diglycine-modified peptides identified by Spectrum Mill, manual verification of every spectra proved to be impractical. Other than the spectra obtained for the tubulin α-1A and PCNA experiments (Supplementary Fig. 16), we have not manually verified all the diglycine-modified lysine residues listed in Supplementary Table 1. To provide an alternate estimate of the accuracy of diglycyl-lysine modification identification in peptides, we used a combination of decoy database searching (described above), as well as the use of a consensus searching approach to report peptides that were identified by Spectrum Mill and at least one other search program. To identify Spectrum Mill-identified peptides that were also matched by other search algorithms, the pkl files for ubiquitinated peptides identified by Spectrum Mill were extracted, converted and merged to a single file in Mascot Generic Format (mgf). The file was searched with other online search programs: Mascot, X!Tandem, and Phenyx, against the Swiss-Prot human database. The Mascot score and expectation value, X!Tandem log (e), and Phenyx p-value are reported in Supplementary Table 1. The cutoff values for positive identifications are: 29 for Mascot score or 0.05 for Mascot expectation value; −1.0 for X!Tandem log (e), and 10−4 for Phenyx p-value. A Venn diagram depicting the number of common and unique ubiquitinated peptides obtained from the three search programs is presented in Supplementary Figure 17. In order to increase the confidence of our identification, only the peptides which were found by at least two search programs were considered as likely ubiquitinated peptides and considered in bioinformatic studies. It should be noted that peptide identification from MS/MS spectra using computational approaches has the potential of producing false positives. Any lysine residue predicted to be modified should be verified experimentally in order to conclusively demonstrate that this site is indeed ubiquitinated.
Biochemical validation of ubiquitination
Some identified ubiquitinated proteins were verified by the conventional immunoprecipitation and immunoblot approach. The target proteins were precipitated using specific antibodies: 14-3-3 (K-19, Santa Cruz), β-catenin (5H10, Millipore), HSP70 (W27, Labvision), NAP1L1 (RB8368, Abgent), PCNA (PC10, Labvision), vimentin (AMF-17b–c, Developmental Studies Hybridoma Bank). Western blotting analysis was carried out against anti-ubiquitin antibody (P4D1, Santa Cruz and SPA-200D, Assay Designs). Full blots for Fig. 2d are presented in Supplementary Figure 18.
Bioinformatic analysis
Protein biological processes were analyzed and clustered by PANTHER43 after converting the Swiss-Prot accession numbers of identified ubiquitinated proteins into RefSeq protein accession numbers by the Database for Annotation, Visualization and Integrated Discovery (DAVID)44 online gene ID conversion function. In total, 385 biological processes were found in the database and the category was further grouped into eight classes.
Protein subcellular localization of the ubiquitinated proteins was extracted from the database provides by PENCE Proteome Analyst45. Note that some proteins were predicted to have multiple subcellular localizations while some proteins have no subcellular information available in the database.
The density map for the diglycine-modified lysines was calculated as follows: A subset of protein sequence, 10 amino acids on either side of modified lysines, was extracted from the whole protein sequence. The frequency of each of the 20 individual amino acids at each position from −10 to +10 was calculated for diglycine-modified lysines and this value was normalized to the frequency of the same amino acid at the same position using all lysines in the human Swiss-Prot database to obtain a relative ratio. If the ratio in one position (say −1) for a specific amino acid (say Pro) is larger than 1, there is a commensurately higher likelihood for Pro at the −1 position to be adjacent to a ubiquitinated lysine. The highest relative ratio detected was 2.1 and the range of the colormap was set from 0 to 2.5. The density map was prepared by MATLAB. The enriched amino acids were obtained by determining the outliers with a 95% confidence using the Rosner’s test46.
To access the structural features of ubiquitinated lysine residues for human proteins, we searched crystal structures for all the ubiquitinated proteins in protein database bank (PDB). In total, 89 PDB structures (Supplementary Table 2) contained lysines that we found are susceptible to ubiquitination (140 modified lysines and 3970 total lysines). In cases when multiple PDB structures for a ubiquitinated protein were reported, the structure with best quality was used. The secondary structure types for lysines were determined using the program DSSP47. H and G were considered to be helix, E and B to be strand, S, T and others for coil. The fraction of each secondary structure type of modified lysines was compared to that of all the lysine residues in 89 PDB structures. The disordered region was predicted by DisEMBL48 for all identified ubiquitinated proteins and the information for modified lysines and all lysines was extracted. The relative solvent-accessible area (SAA) for the modified and all lysines in 89 crystal structures was calculated using NACCESS49 with a probe of 1.4 Å, which corresponds to the size of a water molecule.
Supplementary Material
Acknowledgements
We thank T. Neubert and G. Zhang (NYU) for useful suggestions, P. Zhou (WCMC) for the His6-ubiquitin plasmid, U. Hengst, A. Deglincerti, R. Almeida, and B. Derakhshan for the assistance during initial cell culturing, S. Gross and Y. Ma (WCMC Mass Spectrometry Core Facility) for helpful discussion in MS/MS analysis, F. Campagne, L. Skrabanek, J. Sun (WCMC Institute for Computational Biomedicine) for instructions and assistance in bioinformatic analysis. The mass spectrometry work was performed at the WCMC Mass Spectrometry Core Facility using instrumentation supported by NIH RR19355 and RR22615. This work was supported by grants from Weill Cornell, NIH-NIMH (MH086128) (S.R.J.), and a pharmacology cancer training grant from the NCI (T32CA062948) (G.X. and J.S.P.).
Footnotes
Accession Information
MS/MS data and the identifications are deposited in the open access public repository PRIDE (http://www.ebi.ac.uk/pride/) with the accession code of 12018.
Contributions. S.R.J. and G.X. conceived and designed the study. G.X. and J.S.P conducted the experiments, and G.X. and S.R.J. analyzed the data. S.R.J. and G.X. wrote the manuscript.
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Note: Supplementary information is available on the Nature Biotechnology website.
REFERENCES
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Xu P, Peng J. Dissecting the ubiquitin pathway by mass spectrometry. Biochim Biophys Acta. 2006;1764:1940–1947. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ericsson C, Goldknopf IL, Daneholt B. Inhibition of transcription does not affect the total amount of ubiquitinated histone 2A in chromatin. Exp Cell Res. 1986;167:127–134. doi: 10.1016/0014-4827(86)90210-7. [DOI] [PubMed] [Google Scholar]
- 4.Galluzzi L, Paiardini M, Lecomte MC, Magnani M. Identification of the main ubiquitination site in human erythroid alpha-spectrin. FEBS Lett. 2001;489:254–258. doi: 10.1016/s0014-5793(00)02333-4. [DOI] [PubMed] [Google Scholar]
- 5.Tomlinson E, Palaniyappan N, Tooth D, Layfield R. Methods for the purification of ubiquitinated proteins. Proteomics. 2007;7:1016–1022. doi: 10.1002/pmic.200601008. [DOI] [PubMed] [Google Scholar]
- 6.Beers EP, Callis J. Utility of polyhistidine-tagged ubiquitin in the purification of ubiquitin-protein conjugates and as an affinity ligand for the purification of ubiquitin-specific hydrolases. J Biol Chem. 1993;268:21645–21649. [PubMed] [Google Scholar]
- 7.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 8.Srikumar T, Jeram SM, Lam H, Raught B. A ubiquitin and ubiquitin-like protein spectral library. Proteomics. 10:337–342. doi: 10.1002/pmic.200900627. [DOI] [PubMed] [Google Scholar]
- 9.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 10.Denis NJ, Vasilescu J, Lambert JP, Smith JC, Figeys D. Tryptic digestion of ubiquitin standards reveals an improved strategy for identifying ubiquitinated proteins by mass spectrometry. Proteomics. 2007;7:868–874. doi: 10.1002/pmic.200600410. [DOI] [PubMed] [Google Scholar]
- 11.Rechsteiner M. Ubiquitin-mediated pathways for intracellular proteolysis. Annu Rev Cell Biol. 1987;3:1–30. doi: 10.1146/annurev.cb.03.110187.000245. [DOI] [PubMed] [Google Scholar]
- 12.Bonifacino JS, Weissman AM. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16:119–126. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Etlinger JD, Li SX, Guo GG, Li N. Phosphorylation and ubiquitination of the 26S proteasome complex. Enzyme Protein. 1993;47:325–329. doi: 10.1159/000468690. [DOI] [PubMed] [Google Scholar]
- 16.Peters JM. Subunits and substrates of the anaphase-promoting complex. Exp Cell Res. 1999;248:339–349. doi: 10.1006/excr.1999.4443. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad S, Gromiha MM. NETASA: neural network based prediction of solvent accessibility. Bioinformatics. 2002;18:819–824. doi: 10.1093/bioinformatics/18.6.819. [DOI] [PubMed] [Google Scholar]
- 19.Catic A, Collins C, Church GM, Ploegh HL. Preferred in vivo ubiquitination sites. Bioinformatics. 2004;20:3302–3307. doi: 10.1093/bioinformatics/bth407. [DOI] [PubMed] [Google Scholar]
- 20.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 21.Gnad F, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SC, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basu A, et al. Proteome-wide prediction of acetylation substrates. Proc Natl Acad Sci U S A. 2009;106:13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prosperi E. Multiple roles of the proliferating cell nuclear antigen: DNA replication, repair and cell cycle control. Prog Cell Cycle Res. 1997;3:193–210. doi: 10.1007/978-1-4615-5371-7_15. [DOI] [PubMed] [Google Scholar]
- 27.Mayer A, et al. The prognostic significance of proliferating cell nuclear antigen, epidermal growth factor receptor, and mdr gene expression in colorectal cancer. Cancer. 1993;71:2454–2460. doi: 10.1002/1097-0142(19930415)71:8<2454::aid-cncr2820710805>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 28.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 29.Jordan MA. Mechanism of action of antitumor drugs that interact with microtubules and tubulin. Curr Med Chem Anticancer Agents. 2002;2:1–17. doi: 10.2174/1568011023354290. [DOI] [PubMed] [Google Scholar]
- 30.Wisniewski JR, et al. Constitutive and dynamic phosphorylation and acetylation sites on NUCKS, a hypermodified nuclear protein, studied by quantitative proteomics. Proteins. 2008;73:710–718. doi: 10.1002/prot.22104. [DOI] [PubMed] [Google Scholar]
- 31.Gigant B, et al. Structural basis for the regulation of tubulin by vinblastine. Nature. 2005;435:519–522. doi: 10.1038/nature03566. [DOI] [PubMed] [Google Scholar]
- 32.Ravelli RB, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 33.Unk I, et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2006;103:18107–18112. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derrien D, et al. Muramyl dipeptide bound to poly-L-lysine substituted with mannose and gluconoyl residues as macrophage activators. Glycoconj J. 1989;6:241–255. doi: 10.1007/BF01050652. [DOI] [PubMed] [Google Scholar]
- 35.Kirkpatrick DS, Weldon SF, Tsaprailis G, Liebler DC, Gandolfi AJ. Proteomic identification of ubiquitinated proteins from human cells expressing His-tagged ubiquitin. Proteomics. 2005;5:2104–2111. doi: 10.1002/pmic.200401089. [DOI] [PubMed] [Google Scholar]
- 36.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 38.de Godoy LM, et al. Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol. 2006;7:R50. doi: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu G, Shin SB, Jaffrey SR. Global profiling of protease cleavage sites by chemoselective labeling of protein N-termini. Proc Natl Acad Sci U S A. 2009;106:19310–19315. doi: 10.1073/pnas.0908958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 41.Silva JC, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 42.Mortensen P, et al. MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J Proteome Res. 9:393–403. doi: 10.1021/pr900721e. [DOI] [PubMed] [Google Scholar]
- 43.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 45.Lu Z, et al. Predicting subcellular localization of proteins using machine-learned classifiers. Bioinformatics. 2004;20:547–556. doi: 10.1093/bioinformatics/btg447. [DOI] [PubMed] [Google Scholar]
- 46.Rosner J. Test of Auditory Analysis Skills (TAAS) in Helping children overcome learning difficulties: A step-by-step guide for parents and teachers. New York: Academic Therapy; 1979. [Google Scholar]
- 47.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 48.Linding R, et al. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 49.Hubbard SJ, Campbell SF, Thornton JM. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J Mol Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.