Abstract
γ-Radiation-induced intrastrand guanine-thymine cross-link, G[8,5-Me]T, hinders replication in vitro and is mutagenic in mammalian cells. Herein we report in vitro translesion synthesis of G[8,5-Me]T by human and yeast DNA polymerase η (hPol η and yPol η). dAMP misincorporation opposite the cross-linked G by yPol η was preferred over correct incorporation of dCMP, but further extension was 100-fold less efficient for G∗:A compared to G∗:C. For hPol η, both incorporation and extension were more efficient with the correct nucleotides. To evaluate translesion synthesis in the presence of all four dNTPs, we have developed a plasmid-based DNA sequencing assay, which showed that yPol η was more error-prone. Mutational frequencies of yPol η and hPol η were 36% and 14%, respectively. Targeted G → T was the dominant mutation by both DNA polymerases. But yPol η induced targeted G → T in 23% frequency relative to 4% by hPol η. For yPol η, targeted G → T and G → C constituted 83% of the mutations. By contrast, with hPol η, semi-targeted mutations (7.2%), that is, mutations at bases near the lesion, occurred at equal frequency as the targeted mutations (6.9%). The kind of mutations detected with hPol η showed significant similarities with the mutational spectrum of G[8,5-Me]T in human embryonic kidney cells.
1. Introduction
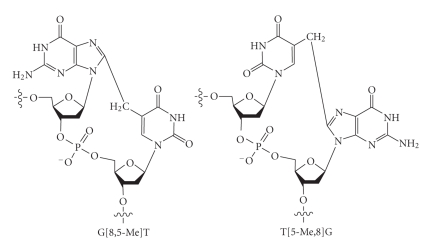
DNA-DNA interstrand and intrastrand cross-links are strong blocks of DNA replication, and understanding the details of polymerase bypass of these complex lesions is of major interest [1–5]. The double base DNA lesions are formed at substantial frequency by ionizing radiation and by metal-catalyzed H2O2 reactions (reviewed in [6]). A major DNA damage, in anoxic conditions, is an intrastrand cross-linked species in which C8 of Gua is linked to the 5-methyl group of an adjacent thymine, but the G[8,5-Me]T cross-link is formed at a much higher rate than the T[5-Me,8]G cross-link (Figure 1) [7]. Additional thymine-purine cross-links have been isolated from γ-irradiated DNA in oxygen-free aqueous solution [8]. Wang and coworkers identified structurally similar guanine-cytosine and guanine-5-methylcytosine cross-links in DNA exposed to γ- or X-rays [9–11]. The G[8,5-Me]T cross-link is formed in a dose-dependent manner in human cells when exposed to γ-rays [12], and the G[8,5]C cross-link is formed at a slightly lower level [13].
Figure 1.
Chemical structures of the two γ-radiation-induced intrastrand cross-links, G[8,5-Me]T and T[5-Me,8]G.
These intrastrand cross-links destabilize the DNA double helix [14], and UvrABC, the excision nuclease proteins from Escherichia coli, can excise them [15, 16]. Using purified DNA polymerases, it was shown that G[8,5-Me]T and G[8,5]C are strong blocks of replication in vitro [12, 17]. For G[8,5-Me]T, primer extension is terminated after incorporation of dAMP opposite the 3′-T by exo-free Klenow fragment and Pol IV (dinB) of Escherichia coli whereas Taq polymerase is completely blocked at the nucleotide preceding the cross-link [17]. However, yeast polymerase η (yPol η), a member of the Y-family DNA polymerase from Saccharomyces cerevisiae, can bypass both G[8,5-Me]T and G[8,5]C cross-links with reduced efficiency [12, 18]. For both these two lesions, nucleotide incorporation opposite the 3′-base of the cross-link is accurate, but the incorporation of dAMP and dGMP is favored opposite the cross-linked G by yPol η over that of the correct nucleotide, dCMP [12, 18].
We have recently compared translesion synthesis of G[8,5-Me]T with T[5-Me,8]G in simian and human embryonic kidney cells and found that both cross-links are strongly mutagenic and that the two lesions show interesting pattern of mutations, which included high frequency of semitargeted mutations that occurred a few bases 5′ or 3′ to the cross-link [19]. One can anticipate a role of one or more Y-family DNA polymerases in bypassing these replication blocking lesions, and we noted that purified human DNA polymerase η (hPol η) incorporates dCMP preferentially opposite the G of G[8,5-Me]T cross-link, in contrast to yPol η which incorporates dAMP and dGMP much more readily [12, 19]. However, the previous preliminary studies did not examine the kinetics of polymerase extension beyond the lesion site; nor were the full-length extension products analyzed. The kinetics of nucleotide incorporations are influenced by DNA damages, not only at the lesion site but at least up to 3 bases 5′ to the lesion [20]. Therefore, incorporation pattern opposite the lesion provides only part of the information on lesion bypass. In the current paper, we have evaluated translesion synthesis of the G[8,5-Me]T cross-link by these two DNA polymerases more critically by determining single nucleotide incorporation kinetics and characterizing the full-length extension products in the presence of all four dNTPs. We report herein that G[8,5-Me]T bypass by yPol η is much more error-prone than hPol η. We also show that the mutational signatures of these two polymerases are different.
2. Materials and Methods
2.1. Materials
[γ-32P] ATP was supplied by Du Pont New England Nuclear (Boston, MA). Recombinant human and yeast DNA polymerases η were purchased from Enzymax, LLC. (Lexington, KY). EcoR V restriction endonuclease, T4 DNA ligase, and T4 polynucleotide kinase were obtained from New England Bioloabs (Beverly, MA). E. coli DL7 (AB1157, l a cΔU169, u v r +) was from J. Essigmann (MIT, Cambridge, MA). The pMS2 phagemid was a gift from Masaaki Moriya (SUNY, Stony Brook, NY).
2.2. Methods
2.2.1. Synthesis and Characterization of Oligonucleotides
The lesions containing oligonucleotides have been synthesized and characterized as reported in [15]. Unmodified oligonucleotides were analyzed by MALDI-TOF MS analysis, which gave a molecular ion with a mass within 0.005% of theoretical whereas adducted oligonucleotides were analyzed by ESI-MS in addition to digestion followed by HPLC analysis.
2.2.2. Construction of 26-mer and 36-mer Containing G[8,5-Me]T Cross-Link
The 26-mer G[8,5-Me]T template, 5′-GTGCGTGTTTGTATCGCTTGCAGGGG-3′, was constructed by ligating a 5′-phosphorylated 14-mer, 5′-ATCGCTTGCAGGGG-3′ (~7.5 nmol), to the G[8,5-Me]T cross-linked 12-mer, 5′-GTGCGTGTTTGT-3′ (~5 nmol), in the presence of an 18-nucleotide complementary oligonucleotide, 5′-GCAAGCGATACAAACACG-3′ (~7.5 nmol), as described [19, 21]. Similarly, a 12-mer, 5′-CCUGGAAGCGAU-3′ (~7.5 nmol), a 5′-phosphorylated G[8,5-Me]T 12-mer (~5 nmol), and a 5′-phosphorylated 12-mer, 5′-AUCGCUGCUACC-3′ (~7.5 nmol), were annealed to a complementary 26-mer, 5′-GCAGCGATACAAACACGCACATCGCT-3′ (~7.5 nmol), and ligated in the presence of T4 DNA ligase to prepare a G[8,5-Me]T cross-linked 36-mer, 5′-CCUGGAAGCGAUGTGCGTGTTTGTAUCGCUGCUACC-3′. The oligonucleotides were separated by electrophoresis on a 16% polyacrylamide-8 M urea gel. The ligated product bands were visualized by UV shadowing and excised. The 26-mers and the 36-mers were desalted on a Sephadex G-25 (Sigma) column and stored at −20°C until further use.
2.2.3. In Vitro Nucleotide Incorporation and Chain Extension
To determine the nucleotide preferentially incorporated opposite G[8,5-Me]T cross-link, the steady-state kinetic analyses were performed by the method of Goodman and coworkers [22, 23]. The primed template was obtained by annealing 5-fold molar excess of the modified or control 26-mer template (~20 ng) to a complementary 5′-32P-labeled primer. Primer extension under standing start conditions was carried out with hPol η or yPol η (6.4 nM) with individual dNTPs or a mixture of all four dNTPs in 25 mM Tris-HCl buffer (pH 7.5), 5 mM MgCl2, and 5 mM dithiothreitol at 37°C for various times. The reactions were terminated by adding an equal volume of 95% (v/v) formamide, 20 mM EDTA, 0.02% (w/v) xylene cyanol, and 0.02% (w/v) bromophenol blue and heating at 90°C for 2 min, and the products were resolved on a 20% polyacrylamide gel containing 8 M urea. The DNA bands were visualized and quantitated using a Phosphorimager. The dNTP concentration and time of incubation were optimized to ensure that primer extension was less than 20%. The K m and k cat were extrapolated from the Michaelis-Menten plot of the kinetic data.
2.2.4. Analysis of the Full-Length Bypass Products Using pMS2 Vector
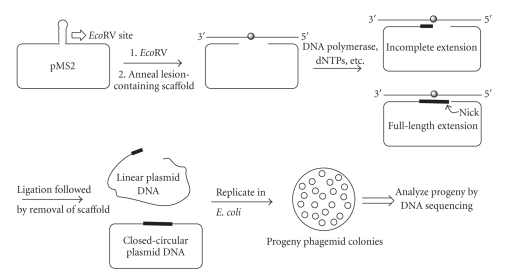
The ss pMS2 shuttle vector DNA (58 pmols, 100 μg) was digested with an excess of EcoR V (300 pmol, 4.84 μg) for 1 h at 37°C followed by room temperature overnight. A 36-mer scaffold oligonucleotide containing the G[8,5-Me]T cross-link (or a control) was annealed overnight at 16°C to form the gapped DNA. The gapped plasmid was incubated with hPol η or yPol η and a mixture of all four dNTPs (25 mM each) in 25 mM Tris-HCl buffer (pH 7.5), 5 mM MgCl2, and 5 mM dithiothreitol at 37°C for various times. DNA ligase (200 units) was added, and the pMS2 mixture containing the DNA polymerase, dNTPs, and so forth, was ligated overnight at 16°C. The scaffold oligonucleotide was digested by treatment with uracil glycosylase and exonuclease III, the proteins were extracted with phenol/chloroform, and the DNA was precipitated with ethanol. The final construct was dissolved in deionized water and used to transform E. coli DL7 cells. The transformants were randomly picked and analyzed by DNA sequencing.
3. Results
3.1. In Vitro Bypass by DNA Polymerase η
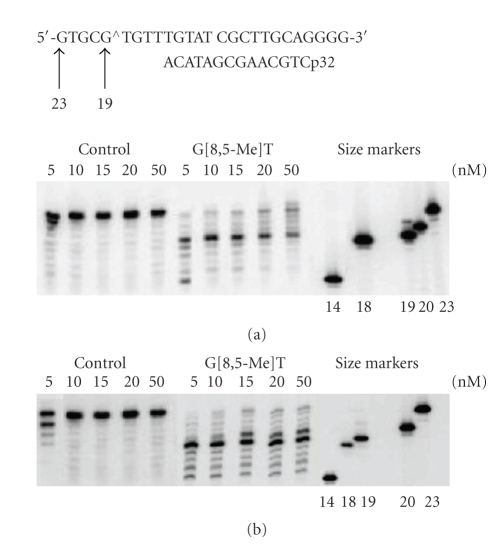
A 26-mer template, 5′-GTGCGTGTTTGTATCGCTTGCAGGGG-3′, which contained the G[8,5-Me]T cross-link (GT) at the 5th and 6th bases from the 5′ end, was constructed. The DNA sequence of the first 12-nucleotides in this template was taken from codon 272–275 of the p53 gene, in which the G[8,5-Me]T cross-link was incorporated at the second and third nucleotide of codon 273, a well-known mutational hotspot for human cancer [24]. We used both running start and standing start conditions to evaluate bypass of the cross-link. Template-primer complex (50 nM) was incubated with increasing concentration of hPol η and yPol η at 37°C for 30 min in the presence of all four dNTPs (100 μM). For the running start experiments, a 5′-32P-radiolabeled 14-mer primer, 5′-CTGCAAGCGATACA-3′, was annealed to the template so that it was 3 bases 3′ to the cross-link. As shown in Figure 2, G[8,5-Me]T was a strong block of both DNA polymerases. With 5 nM hPol η, 80% of the control template extended to a 22-mer and a 23-mer (full-length) products whereas for G[8,5-Me]T less than 1% extended to the full-length product, and a major block was at the cross-linked G (19-mer). With 20 nM hPol η, nearly 75% was blocked after incorporating a base opposite the cross-linked G (19-mer), and the full-length product increased only to ~10%. The full-length product increased to ~18% with 50 nM hPol η. In similar experiment using yPol η, unlike the human enzyme, the major blocks were at 19-mer and 20-mer (i.e., opposite the cross-linked G and its 5′ neighbor). With 50 nM yPol η, 8% of the primer extended to full-length 23-mer product.
Figure 2.
Extension of a 14-mer primer by varying concentration (5, 10, 15, 20, and 50 nM) of hPol η (a) and yPol η (b) on control and G[8,5-Me]T templates in the presence of all four dNTPs. The experiments were carried out at 37°C for 30 min.
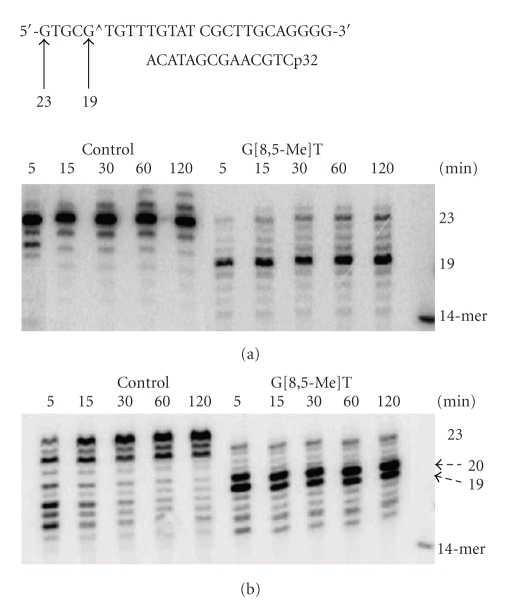
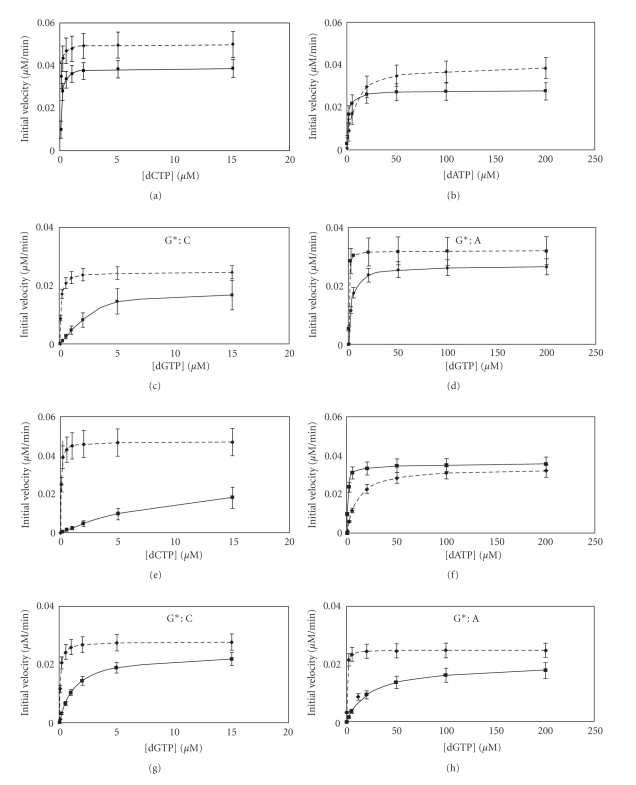
With concentrations of hPol η and yPol η at 50 nM, a substantial fraction (18% and 8%, resp.) of the primer extended to full-length products in 30 min. So we chose to use 50 nM Pol η concentrations for the subsequent experiment. As shown in Figure 3, in the presence of all four dNTPs, extension of a 14-mer primer on the control template rapidly generated a full-length extension product (a 23-mer) as well as a blunt-end addition product (a 24-mer) in 5 min with 50 nM hPol η whereas the extension of the primer stalled after adding a base opposite the cross-linked T and G, generating a 19-mer. It is interesting that hPol η did not stall before either of the cross-linked bases, but it was unable to continue synthesis only after incorporating a dNMP opposite the cross-linked G. Longer incubation allowed further extension, including a small fraction of full-length product, but even after 2 h the 19-mer band was the most pronounced extension product. The result was qualitatively similar with yPol η, except that the extent of full-length product was only marginally increased with time and it stalled both after incorporation of a nucleotide opposite the cross-linked G (19-mer) and after incorporation of a nucleotide opposite its 5′-neighbor (20-mer). Standing start experiments were carried out, and the amount of extension of the primer by one nucleotide was plotted with increasing dNTP concentration to determine the initial velocity of the polymerase-catalyzed reaction, which is shown in Figure 4. From these plots, the steady-state kinetic parameters, K m and k cat, for nucleotide incorporation opposite cross-linked G and the same for the control were determined (Tables 1 and 2). For hPol η, catalytic efficiency (k cat/K m) of dCMP incorporation was 17-fold decreased opposite the cross-linked G whereas extension to the next base was decreased 5-fold relative to control. By contrast, for yPol η dCMP incorporation was decreased 1,000-fold, and extension to the next base was decreased 12-fold relative to control. This suggests that yPol η had more difficulty in bypassing G[8,5-Me]T than hPol η. As was reported before [12, 19], in contrast to hPol η, which incorporates the correct nucleotide preferentially opposite G[8,5-Me]T, yPol η was much more error-prone, and insertion of dAMP opposite the cross-linked G was favored over that of the correct nucleotide, dCMP (Tables 1 and 2). In fact, dAMP misincorporation opposite the cross-linked G was more than 20-times more efficient than dCMP incorporation by yPol η. However, with yPol η the extension was 100-fold slower for G*:A pair compared to G*:C pair whereas the same for hPol η was about 13-fold slower. In each case, the higher catalytic efficiency was due to a much smaller K m. When nucleotide incorporation fidelity opposite the cross-linked G and its 5′ base was considered, dCMP incorporation over dAMP misincorporation was 200-fold more efficient for hPol η whereas the same was only 5-fold more efficient for yPol η. Nevertheless, it seems that dCMP was preferred opposite the cross-linked G for bypass of G[8,5-Me]T by both DNA polymerases although the ability to discriminate against the wrong nucleotide by yPol η was not high.
Figure 3.
Extension of a 14-mer primer by 50 nM hPol η (a) and yPol η (b) on control and G[8,5-Me]T templates in the presence of all four dNTPs for the indicated time at 37°C.
Figure 4.
Single nucleotide incorporation and extension assay. Template-primer (50 nM) was incubated with 6.4 nM hPol η or yPol η for various times with increasing concentrations of dNTP. Steady-state kinetics for single nucleotide incorporation opposite cross-linked (G) (solid line) or control (G) (dashed line) are shown in (a), (b), (e), and (f). (a) and (b) represent dCTP and dATP incorporations, respectively, for hPol η whereas (e) and (f) represent the same for yPol η. Steady-state kinetics for dGTP incorporation opposite (C) immediately following the cross-linked (G) (solid line) or control (G) (dashed line) are shown in (c), (d), (g), and (h). (c) and (d) represent extension of G*:C and G*:A pairs, respectively, for hPol η whereas (g) and (h) represent the same for yPol η. Error bars show the standard deviation of at least three experiments.
Table 1.
Kinetic parameters for dCTP and dATP incorporation and chain extension by human DNA polymerase η (6.4 nM) on an undamaged and G[8,5-Me]T cross-link containing substrate.
| dNTP | k cat (min−1) | K m (μM) | k cat/K m (μM−1 min−1) | F inc a | X:G | k cat (min−1) | K m (μM) | k cat/K m (μM−1 min−1) | F ext a |
|---|---|---|---|---|---|---|---|---|---|
| 5′-GTGCGTGTTTGTATCGCTTGCAGGGG-3′ | 5′-GTGCGTGTTTGTATCGCTTGCAGGGG-3′ | ||||||||
| ACAAACATAGCGAACp32 | X ACAAACATAGCGAACp32 | ||||||||
| Nucleotide Incorporation | Chain Extensionb | ||||||||
|
| |||||||||
| Undamaged substrate | |||||||||
| dCTP | 7.6 ± 0.1 | 0.02 ± 0.003 | 380 | 1.0 | C:G | 3.9 ± 0.2 | 0.09 ± 0.005 | 43.4 | 1.0 |
| dATP | 6.25 ± 0.01 | 6.5 ± 0.03 | 0.96 | 2.5 × 10−3 | A:G | 4.7 ± 0.1 | 5.1 ± 0.03 | 0.9 | 2.1 × 10−2 |
|
| |||||||||
| G[8,5-Me]T-containing substrate | |||||||||
| dCTP | 2.43 ± 0.02 | 0.11 ± 0.002 | 22.1 | 5.8 × 10−2 | C:G* | 2.0 ± 0.02 | 0.24 ± 0.004 | 8.3 | 0.2 |
| dATP | 1.75 ± 0.1 | 1.07 ± 0.01 | 1.63 | 4.2 × 10−3 | A:G* | 1.7 ± 0.01 | 2.6 ± 0.03 | 0.65 | 1.5 × 10−2 |
aFidelity (F) of incorporation or extension was determined by the following equation: (k cat/K m)incorrect/(k cat/K m)correct.
bSteady-state kinetics for dGTP incorporation opposite C immediately following the X:G or X:G* pair was determined.
Table 2.
Kinetic parameters for dCTP and dATP incorporation and chain extension by yeast DNA polymerase η (6.4 nM) on an undamaged and G[8,5-Me]T cross-link containing substrate.
| dNTP | k cat (min−1) | K m (μM) | k cat/K m (μM−1 min−1) | F inc a | X:G | k cat (min−1) | K m (μM) | k cat/K m (μM−1 min−1) | F ext a |
|---|---|---|---|---|---|---|---|---|---|
| 5′-GTGCGTGTTTGTATCGCTTGCAGGGG-3′ | 5′-GTGCGTGTTTGTATCGCTTGCAGGGG-3′ | ||||||||
| ACAAACATAGCGAACp32 | X ACAAACATAGCGAACp32 | ||||||||
| Nucleotide Incorporation | Chain Extensionb | ||||||||
|
| |||||||||
| Undamaged substrate | |||||||||
| dCTP | 7.3 ± 0.02 | 0.04 ± 0.002 | 182.5 | 1.0 | C:G | 4.4 ± 0.04 | 0.07 ± 0.001 | 62.8 | 1.0 |
| dATP | 5.3 ± 0.03 | 9.5 ± 0.002 | 0.56 | 3.1 × 10−3 | A:G | 3.7 ± 0.04 | 1.3 ± 0.01 | 2.8 | 4.4 × 10−2 |
|
| |||||||||
| G[8,5-Me]T-containing substrate | |||||||||
| dCTP | 1.99 ± 0.001 | 11.2 ± 0.01 | 0.17 | 9.3 × 10−4 | C:G* | 1.6 ± 0.1 | 0.31 ± 0.002 | 5.2 | 8.2 × 10−2 |
| dATP | 2.2 ± 0.01 | 0.59 ± 0.005 | 3.72 | 2.0 × 10−2 | A:G* | 1.2 ± 0.009 | 22.0 ± 1.0 | 0.05 | 7.9 × 10−4 |
aFidelity (F) of incorporation or extension was determined by the following equation: (k cat/K m)incorrect/(k cat/K m)correct.
bSteady-state kinetics for dGTP incorporation opposite C immediately following the X:G or X:G* pair was determined.
3.2. Analysis of the Full-Length Bypass Products
Although steady-state kinetics provides useful information on the ability to incorporate a nucleotide opposite a lesion and further extension, it is important to determine the sequences of full-length bypass products in the presence of all four dNTPs. In mammalian cells, replication of G[8,5-Me]T-containing DNA also generates significant level of semitargeted mutations [19], and it would be of interest to determine if pol η causes errors not only opposite the cross-link but also near the lesion. Guengerich and colleagues have developed an elegant LC-ESI/MS/MS-based method to analyze the polymerase extension products [25–30]. In the current paper, we report a plasmid-based approach to accomplish the same goal. The principle of this approach is shown in Scheme 1. The pMS2 plasmid was linearized by digestion with EcoR V. A scaffold 36-mer, containing two 12-nucleotide regions complementary to the two ends of the digested plasmid, was annealed to generate a gapped circular DNA, in which the G[8,5-Me]T cross-link was located in the middle of a 12-nucleotide gap. The scaffold G[8,5-Me]T-36-mer contained the same local DNA sequence near the G[8,5-Me]T cross-link as the 26-mer used in the steady-state kinetic assay. It also contained several uracils replacing thymines at the two ends where it annealed with the plasmid. The circular scaffold plasmid DNA was incubated with 50 nM hPol η or yPol η and a mixture of all four dNTPs (25 mM each) in 25 mM Tris-HCl buffer (pH 7.5), 5 mM MgCl2, and 5 mM dithiothreitol at 37°C for various times. We expected a large fraction of the control construct to extend to full-length circular product whereas a much smaller fraction of the cross-linked construct was able to do the same. The full-length extension product extended up to the 3′ end of the circular DNA, and the nick between the two ends was sealed by ligation overnight at 16°C in the presence of an excess of T4 DNA ligase to generate covalently closed circular ss plasmid. Although the DNA polymerase was not inactivated, both hPol η and yPol η were inefficient in continuing further extension at 16°C (data not shown). The scaffold 36-mer was digested by treatment with uracil DNA glycosylase and exonuclease III. The removal of the lesion-containing scaffold was considered critical to avoid any potential in vivo replication of the lesion. Therefore, we analyzed the products by agarose gel electrophoresis after uracil DNA glycosylase followed by exonuclease III treatment and confirmed that the plasmid was quantitatively linearized when either Pol η or DNA ligase was absent (data not shown). The proteins were extracted with phenol and chloroform, and the DNA was precipitated with ethanol. The DNA was used to transform repair-competent E. coli DL7, and the transformants were analyzed by DNA sequencing.
Scheme 1.
General protocol for analyzing the full-length extension products.
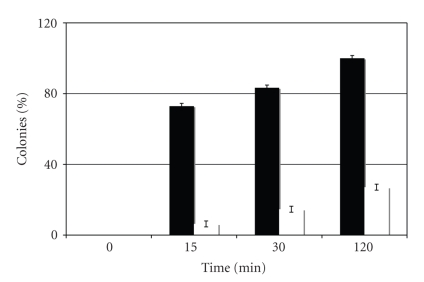
The number of colonies recovered upon transformation in E. coli of the plasmid incubated with hPol η for different times is shown in Figure 5. Since linear ss DNA is inefficient in transfecting E. coli, no colonies were recovered from the zero time point from both the control and the G[8,5-Me]T scaffold whereas increasing numbers of colonies were recovered as incubation times with the DNA polymerase were increased. The number of colonies reflected the extent of full-length product that was ligated, and relative to the control 36-mer scaffold, the G[8,5-Me]T scaffold generated only 9% progeny at 15 min, which increased to 18% at 30 min and to 27% after 2 h (Figure 4). (For this calculation, the number of colonies obtained from the 120 min extension of the control 36-mer was considered 100%.) This suggests that with increased time of incubation, more DNA polymerase can bypass the G[8,5-Me]T cross-link, as we have also noted in the primer extension experiment with the G[8,5-Me]T 26-mer.
Figure 5.
The number of colonies obtained from extension by hPol η of a control scaffold (black) was compared with the G[8,5-Me]T scaffold (white) at different time points in the bar graph. The number of colonies obtained from the 120 min extension of the control 36-mer was arbitrarily considered 100%. The zero time point showing no colonies ensures that colonies only originated from the extension products.
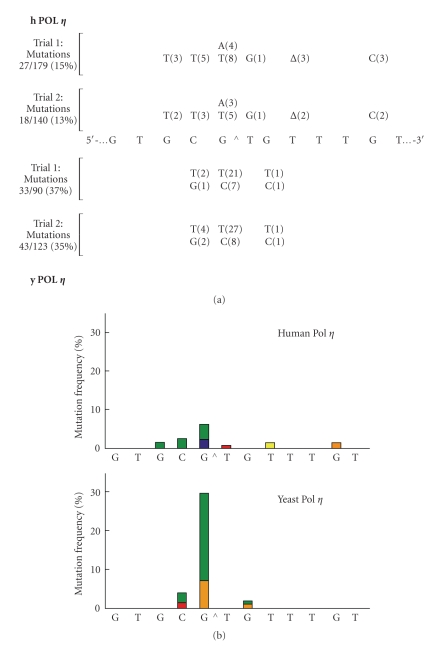
DNA sequencing results of the 2 h incubation products from two independent experiments with hPol η and yPol η are shown in Figure 6. The types and numbers of mutants from two different experiments are shown in Figure 6(a) whereas Figure 6(b) shows the combined result in a bar graph. As noted in the kinetic studies, yPol η was found to be more error-prone than hPol η. Mutational frequencies of yPol η and hPol η were 36% and 14%, respectively, for the G[8,5-Me]T cross-link whereas no mutants were recovered from the control after sequencing in excess of one hundred colonies following extension with each DNA polymerase. The pattern of mutagenesis from the G[8,5-Me]T cross-link was significantly different for these two polymerases. yPol η induced targeted G → T as the major mutagenic event, followed by targeted G → C; these two base substitutions, taken together, constituted 83% of the mutations. By contrast, in the case of hPol η, semitargeted mutations (7.2%) occurred at equal frequency as the targeted mutations (6.9%). With hPol η, though most frequent mutation was G → T (4%), approximately half as many G → A (2.2%) was also detected. It is interesting that even a single targeted G → A could not be detected in the extension by yPol η. Similarly, targeted G → C was completely absent with hPol η. For the cross-linked T, yPol η bypass was completely error-free whereas low (0.6%) level of T → G transversions was detected with hPol η. With yPol η, semitargeted mutations were restricted to the immediate 5′-C and 3′-G of the cross-link, but with hPol η, errors were noted as far as two bases 5′ and five bases 3′ to the cross-link. In sum, despite the similarity of targeted G → T transversions, the mutational profile of the two Y-family DNA polymerases exhibited distinct patterns.
Figure 6.
(a) Types and frequencies of mutations induced by G[8,5-Me]T as determined from the full-length extension products generated by hPol η (top) and yPol η (bottom). It is noteworthy that no mutants were isolated from the control batches after sequencing in excess of one hundred colonies. (b) The combined data in (a) is represented in bar graph showing the percentages of each type of single-base substitution or deletion induced by G[8,5-Me]T by hPol η (top) and yPol η (bottom). The colors represent T (green), A (blue), G (red), C (orange), and one-base deletion (yellow). The T deletion in a run of three thymines by hPol η was arbitrarily shown here at the T closest to the lesion.
4. Discussion
In earlier studies it was shown that hPol η preferentially incorporates the correct nucleotide opposite each of the cross-linked bases whereas yPol η, though accurately incorporates dAMP opposite the cross-linked T, is highly error-prone in nucleotide incorporation opposite the cross-linked G [12, 19]. However, neither the kinetics of further extension of the primer nor the sequences of the full-length extension products were determined. Miller and Grollman [20] have shown that DNA polymerase functions can be affected by replication-blocking lesions remote from the lesion site. In the current investigation, using steady-state kinetics, we determined that though dAMP incorporation opposite the G of G[8,5-Me]T by yPol η was more than 20-fold preferred over dCMP incorporation, further extension of the G*:A pair was 100-fold less efficient than extension of the G*:C pair. As a result, dCMP incorporation followed by further extension was 5-times as efficient as dAMP incorporation by yPol η. For hPol η, on the other hand, it was nearly 200-times as efficient.
In order to characterize the full-length extension products in the presence of all four dNTPs, we developed a novel method to sequence them. In this approach, as shown in Scheme 1, a single-stranded plasmid (e.g., pMS2) containing a restriction endonuclease site in a hairpin region is digested and linearized by the enzyme. A DNA adduct containing scaffold is annealed to the linear DNA to create a gapped plasmid, in which the lesion is situated in the middle of this gap. A DNA polymerase is allowed to extend the 3′-end of the plasmid to fill in the gap, which is then enzymatically ligated to create a closed-circular plasmid or viral genome. The ss circular DNA is replicated in E. coli, and the progeny is subjected to DNA sequencing. The scaffold is quantitatively removed prior to transformation in E. coli to avoid biological processing of the lesion in vivo. The DNA sequencing result of the area which originally contained the gap provides the nature of extension products. It is worth mentioning that other plasmid-based sequencing techniques using PCR amplification have been developed and successfully used in recent years [31, 32]. However, we believe that the hallmark of our current approach is its simplicity. It neither requires expensive instrumentation nor is technically demanding. While the sensitivity of the mass spectral analysis is limited by the signal to noise ratio, which varies from experiment to experiment, the plasmid-based sequencing approach enables determination of misincorporations occurring at a level of less than 1% frequency. However, the sequence determination is dependent on the efficiency of ligation, which is only proficient with full-length extension products. As a result, a limitation of the current plasmid-based approach is that it does not offer any information on the incomplete extension products, which may be readily available by the MS approach. Using this method of sequencing, we showed that yPol η was much more error-prone in bypassing G[8,5-Me]T than hPol η. The targeted G → T was the major type of mutation by both DNA polymerases, but yPol η induced it nearly 6-times more efficiently than hPol η. With hPol η, semitargeted mutations, that is, mutations near the lesion, occurred at approximately equal frequency as the targeted mutations whereas more than 80% of the mutations were targeted mutations with yPol η.
Several studies have established differences between the yeast and the human enzyme. For translesion synthesis of γ-hydroxypropanodeoxyguanosine, yPol η synthesizes past the adduct relatively accurately whereas hPol η discriminates poorly between incorporation of correct and wrong nucleotides opposite the adduct [33]. The mechanistic basis of these two enzymes has been examined, which showed that they differ in several important respects [34]. hPol η has a 50-fold-faster rate of nucleotide incorporation than yPol η but binds the nucleotide with an approximately 50-fold-lower level of affinity. It is unclear how these differences influence the nucleotide incorporation opposite the G[8,5-Me]T cross-link.
When the hPol η mutational spectrum was compared with the mutations detected in human embryonic kidney cells [19], significant similarities in the two results are apparent. Notably, the high frequency of G → T followed by G → A and the semitargeted mutations 5′ to the cross-link such as 5′-C → T and 5′-G → T reflect a similar pattern in the in vitro studies using purified DNA hPol η and the cellular studies. These similarities notwithstanding, certain variations in the mutation profiles are also noteworthy. Targeted T → A and substitutions at adjacent 3′-G and the thymines noted in the mammalian cells were absent in the hPol η extensions. It was suggested that in a cell the binding to proliferating cell nuclear antigen (PCNA) via its PCNA-interacting protein domain is a prerequisite for hPol η's ability to function in translesion synthesis in human cells [35]. Therefore, certain differences between bypass of a DNA damage by a purified hPol η in vitro and that in a cell should be anticipated. Although there is insufficient evidence to conclude that hPol η is responsible for the observed mutations of G[8,5-Me]T in human cells, it seems reasonable to postulate that this Y-family DNA polymerase is one of the DNA polymerases involved in the cellular bypass of this cross-link.
Acknowledgment
This paper was supported by the National Institute of Environmental Health Sciences, NIH (Grant ES013324).
Abbreviations
- G[8,5-Me]T or GT:
Guanine-thymine intrastrand cross-link where C8 of guanine is covalently bonded to the methyl carbon of the 3′-thymine
- T[5-Me,8]G or TG:
Thymine-guanine intrastrand cross-link where C8 of guanine is covalently bonded to the methyl carbon of the 5'-thymine
- hPol η and yPol η:
Represent human and yeast DNA polymerase η, respectively.
References
- 1.Brown JA, Newmister SA, Fiala KA, Suo Z. Mechanism of double-base lesion bypass catalyzed by a Y-family DNA polymerase. Nucleic Acids Research. 2008;36(12):3867–3878. doi: 10.1093/nar/gkn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaisman A, Chaney SG. The efficiency and fidelity of translesion synthesis past cisplatin and oxaliplatin GpG adducts by human DNA polymerase β . Journal of Biological Chemistry. 2000;275(17):13017–13025. doi: 10.1074/jbc.275.17.13017. [DOI] [PubMed] [Google Scholar]
- 3.Vaisman A, Masutani C, Hanaoka F, Chaney SG. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η . Biochemistry. 2000;39(16):4575–4580. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- 4.Kanuri M, Nechev LV, Kiehna SE, et al. Evidence for Escherichia coli polymerase II mutagenic bypass of intrastrand DNA crosslinks. DNA Repair. 2005;4(12):1374–1380. doi: 10.1016/j.dnarep.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Kumari A, Minko IG, Harbut MB, Finkel SE, Goodman MF, Lloyd RS. Replication bypass of interstrand cross-link intermediates by Escherichia coli DNA polymerase IV. Journal of Biological Chemistry. 2008;283(41):27433–27437. doi: 10.1074/jbc.M801237200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Box HC, Dawidzik JB, Budzinski EE. Free radical-induced double lesions in DNA. Free Radical Biology and Medicine. 2001;31(7):856–868. doi: 10.1016/s0891-5849(01)00653-0. [DOI] [PubMed] [Google Scholar]
- 7.Box HC, Patrzyc HB, Dawidzik JB, et al. Double base lesions in DNA X-irradiated in the presence or absence of oxygen. Radiation Research. 2000;153(4):442–446. doi: 10.1667/0033-7587(2000)153[0442:dblidx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 8.Bellon S, Ravanat J-L, Gasparutto D, Cadet J. Cross-linked thymine-purine base tandem lesions: synthesis, characterization, and measurement in γ-irradiated isolated DNA. Chemical Research in Toxicology. 2002;15(4):598–606. doi: 10.1021/tx015594d. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Q, Wang Y. Independent generation of the 5-hydroxy-5,6-dihydrothymidin-6-yl radical and its reactivity in dinucleoside monophosphates. Journal of the American Chemical Society. 2004;126(41):13287–13297. doi: 10.1021/ja048492t. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Q, Wang Y. The reactivity of the 5-hydroxy-5,6-dihydrothymidin-6-yl radical in oligodeoxyribonucleotides. Chemical Research in Toxicology. 2005;18(12):1897–1906. doi: 10.1021/tx050195u. [DOI] [PubMed] [Google Scholar]
- 11.Zhang QQ, Wang Y. Generation of 5-(2′-deoxycytidyl)methyl radical and the formation of intrastrand cross-link lesions in oligodeoxyribonucleotides. Nucleic Acids Research. 2005;33(5):1593–1603. doi: 10.1093/nar/gki301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y, Hong H, Cao H, Wang Y. In vivo formation and in vitro replication of a guanine-thymine intrastrand cross-link lesion. Biochemistry. 2007;46(44):12757–12763. doi: 10.1021/bi7012195. [DOI] [PubMed] [Google Scholar]
- 13.Hong H, Cao H, Wang Y. Formation and genotoxicity of a guanine-cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Research. 2007;35(21):7118–7127. doi: 10.1093/nar/gkm851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu C, Wang Y. Thermodynamic and in vitro replication studies of an intrastrand G[8-5]C cross-link lesion. Biochemistry. 2005;44(24):8883–8889. doi: 10.1021/bi050036+. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z, Colis LC, Basu AK, Zou Y. Recognition and incision of γ-radiation-induced cross-linked guanine-thymine tandem lesion G[8,5-Me]T by UvrABC nuclease. Chemical Research in Toxicology. 2005;18(9):1339–1346. doi: 10.1021/tx050147+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu C, Zhang Q, Yang Z, Wang Y, Zou Y, Wang Y. Recognition and incision of oxidative intrastrand cross-link lesions by UvrABC nuclease. Biochemistry. 2006;45(35):10739–10746. doi: 10.1021/bi060423z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellon S, Gasparutto D, Saint-Pierre C, Cadet J. Guanine-thymine intrastrand cross-linked lesion containing oligonucleotides: from chemical synthesis to in vitro enzymatic replication. Organic and Biomolecular Chemistry. 2006;4(20):3831–3837. doi: 10.1039/b609460k. [DOI] [PubMed] [Google Scholar]
- 18.Gu C, Wang Y. LC-MS/MS Identification and yeast polymerase η bypass of a novel γ-irradiation-induced intrastrand cross-link lesion G[8-5]C. Biochemistry. 2004;43(21):6745–6750. doi: 10.1021/bi0497749. [DOI] [PubMed] [Google Scholar]
- 19.Colis LC, Raychaudhury P, Basu AK. Mutational specificity of γ-radiation-induced guanine-thymine and thymine-guanine intrastrand cross-links in mammalian cells and translesion synthesis past the guanine-thymine lesion by human DNA polymerase η . Biochemistry. 2008;47(31):8070–8079. doi: 10.1021/bi800529f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller H, Grollman AP. Kinetics of DNA polymerase I (Klenow fragment exo-) activity on damaged dna templates: effect of proximal and distal template damage on DNA synthesis. Biochemistry. 1997;36(49):15336–15342. doi: 10.1021/bi971927n. [DOI] [PubMed] [Google Scholar]
- 21.McNulty JM, Jerkovic B, Bolton PH, Basu AK. Replication inhibition and miscoding properties of DNA templates containing a site-specific cis-thymine glycol or urea residue. Chemical Research in Toxicology. 1998;11(6):666–673. doi: 10.1021/tx970225w. [DOI] [PubMed] [Google Scholar]
- 22.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. Journal of Biological Chemistry. 1987;262(30):14689–14696. [PubMed] [Google Scholar]
- 23.Mendelman LV, Petruska J, Goodman MF. Base mispair extension kinetics. Comparison of DNA polymerase α and reverse transcriptase. Journal of Biological Chemistry. 1990;265(4):2338–2346. [PubMed] [Google Scholar]
- 24.Hollstein M, Shomer B, Greenblatt M, et al. Somatic point mutations in the p53 gene of human tumors and cell lines: updated compilation. Nucleic Acids Research. 1996;24(1):141–146. doi: 10.1093/nar/24.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stover JS, Chowdhury G, Zang H, Guengerich FP, Rizzo CJ. Translesion synthesis past the C8- and N2-deoxyguanosine adducts of the dietary mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the NarI recognition sequence by prokaryotic DNA polymerases. Chemical Research in Toxicology. 2006;19(11):1506–1517. doi: 10.1021/tx0601455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zang H, Goodenough AK, Choi J-Y, et al. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. Journal of Biological Chemistry. 2005;280(33):29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 27.Eoff RL, Angel KC, Egli M, Guengerich FP. Molecular basis of selectivity of nucleoside triphosphate incorporation opposite O6-benzylguanine by Sulfolobus solfataricus DNA polymerase Dpo4: steady-state and pre-steady-state kinetics and X-ray crystallography of correct and incorrect pairing. Journal of Biological Chemistry. 2007;282(18):13573–13584. doi: 10.1074/jbc.M700656200. [DOI] [PubMed] [Google Scholar]
- 28.Eoff RL, Irimia A, Angel KC, Egli M, Guengerich FP. Hydrogen bonding of 7,8-dihydro-8-oxodeoxyguanosine with a charged residue in the little finger domain determines miscoding events in sulfolobus solfataricus DNA polymerase Dpo4. Journal of Biological Chemistry. 2007;282(27):19831–19843. doi: 10.1074/jbc.M702290200. [DOI] [PubMed] [Google Scholar]
- 29.Eoff RL, Irimia A, Egli M, Guengerich FP. Sulfolobus solfataricus DNA polymerase Dpo4 is partially inhibited by “Wobble” pairing between O6-methylguanine and cytosine, but accurate bypass is preferred. Journal of Biological Chemistry. 2007;282(2):1456–1467. doi: 10.1074/jbc.M609661200. [DOI] [PubMed] [Google Scholar]
- 30.Christov PP, Angel KC, Guengerich FP, Rizzo CJ. Replication past the N5-methyl-formamidopyrimidine lesion of deoxyguanosine by DNA polymerases and an improved procedure for sequence analysis of in vitro bypass products by mass spectrometry. Chemical Research in Toxicology. 2009;22(6):1086–1095. doi: 10.1021/tx900047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiala KA, Suo Z. Sloppy bypass of an abasic lesion catalyzed by a Y-family DNA polymerase. Journal of Biological Chemistry. 2007;282(11):8199–8206. doi: 10.1074/jbc.M610719200. [DOI] [PubMed] [Google Scholar]
- 32.Fang H, Taylor J-S. Serial analysis of mutation spectra (SAMS): a new approach for the determination of mutation spectra of site-specific DNA damage and their sequence dependence. Nucleic Acids Research. 2008;36(18):6004–6012. doi: 10.1093/nar/gkn595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minko IG, Washington MT, Kanuri M, Prakash L, Prakash S, Lloyd RS. Translesion synthesis past acrolein-derived DNA adduct, γ-hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase η . Journal of Biological Chemistry. 2003;278(2):784–790. doi: 10.1074/jbc.M207774200. [DOI] [PubMed] [Google Scholar]
- 34.Washington MT, Johnson RE, Prakash L, Prakash S. The mechanism of nucleotide incorporation by human DNA polymerase η differs from that of the yeast enzyme. Molecular and Cellular Biology. 2003;23(22):8316–8322. doi: 10.1128/MCB.23.22.8316-8322.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Acharya N, Yoon J-H, Gali H, et al. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase η in translesion DNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(46):17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]