Abstract
Mismatch repair proteins modulate the cytotoxicity of several chemotherapeutic agents. We have recently proposed a “death conformation” of the MutS homologous proteins that is distinguishable from their “repair conformation.” This conformation can be induced by a small molecule, reserpine, leading to DNA-independent cell death. We investigated the parameters for a small reserpine-like molecule that are required to interact with MSH2/MSH6 to induce MSH2/MSH6-dependent cytotoxic response. A multidisciplinary approach involving structural modeling, chemical synthesis, and cell biology analyzed reserpine analogs and modifications. We demonstrate that the parameters controlling the induction of MSH2/MSH6-dependent cytotoxicity for reserpine-analogous molecules reside in the specific requirements for methoxy groups, the size of the molecule, and the orientation of molecules within the protein-binding pocket. Reserpine analog rescinnamine showed improved MSH2-dependent cytotoxicity. These results have important implications for the identification of compounds that require functional MMR proteins to exhibit their full cytotoxicity, which will avoid resistance in MMR-deficient cells.
1. Introduction
Mismatch repair is one of the key DNA repair systems in the cell that repair incorrectly formed base pairs and insertion/deletion loops during replication, significantly increasing the stability of the genome [1–5]. Additional importance comes from the contribution of defects in MMR proteins to cancer development, progression and therapeutic success. In addition to the recognition of replication errors, key MMR proteins MSH2/MSH6 recognize a number of DNA lesions that are irreparable by these proteins, which, in turn, initiate a cell death pathway. The role of MMR proteins in the cell death response to cytotoxic agents has been the subject of much debate [6, 7]. Whether DNA damage is recognized in form of a mismatch at the site of the damage, or DNA damage itself is recognized remains subject of investigation and may vary with the nature of the damage. Recent data suggest that some types of DNA lesions are recognized without the requirements for a mismatch [8]. As a result of their ability to recognize certain DNA lesions and their participation in the initiation of cell death, MMR proteins modulate the response to certain chemotherapeutic agents. This participation is particularly evident in cells with defects in MMR proteins which reduce the sensitivity to chemotherapeutic agents sufficiently to prove to be a problem for the treatment of tumors [1, 6, 9–12].
The exact mechanism of the cytotoxic response initiated by MSH2/MSH6 interactions with DNA damage remains under investigation and speculation. Recent data suggest that the response pathway depends on the nature of the lesion rather than the unification of different signals into a common pathway [13]. At least two hypotheses have been put forward to address the mechanism behind the MMR protein-dependent cell death: The “futile cycles of repair” hypothesis is based on the formation of a mismatch opposite the DNA damage, followed by attempted repair events that retain the damage and the formation of DNA strand breaks as factual initiators of cell death [1, 4, 14]. The second hypothesis suggests a direct involvement of MMR proteins in the initiation of the cell death event. We and others have identified separation-of-function mutants that demonstrate that repair-defective mutants of MMR proteins retain the ability to induce cell death, suggesting that repair is not required for cytotoxic response to all types of DNA damage [8, 15–17], but also suggesting that specific MMR proteins form recognition complexes for both mismatch repair, and at least some types of DNA damage. Computational modeling predicted the formation of a “death conformation” with distinctly different features to the “repair conformation” of MutS and its homologs (MSH). This model of a MutS “death conformation” was validated in extensive mutational analyses on eukaryotic proteins [8]. Although the mechanistic details of this pathway are largely unknown, targeting the apparent recognition complex for cell death, that is, the “death conformation,” has shown initial successes [18]. Hence, we aim to exploit this pathway via small molecule binding to the “death conformation.”
Recently, we have demonstrated that the MSH2-dependent cell death response can be induced by small molecules that mimic binding to DNA damage and promote the induction of this cell death [18]. The prototype for this induction of MSH2-dependent cell death without genotoxic insult is reserpine, a drug previously in clinical use for hypertension. The predicted binding of the molecule to the protein occurs in the DNA binding pocket and induces specific conformational changes that allow access to the “death conformation” of the protein [18]. At present, the direct binding of reserpine, and its derivatives, to the “death conformation” has been predicted, and the ability of reserpine and rescinnamine to induce MSH2-dependent apoptosis has been verified experimentally in this work as well as in an earlier study [18]. Unfortunately, it does not seem possible, with present technology, to unequivocally demonstrate physical association between MSH2/MSH6 and either reserpine or rescinnamine. Attempts to use SPR and calorimetry (data not shown) have lacked the precision to detect these small alkaloids binding to such a large proteins complex. We expect that the technology used to detect such direct binding of small molecules to large protein complexes will be refined over the next few years, and that presently challenging complexes, such as the proposed MSH2/MSH6/rescinnamine or reserpine complex, will be directly studied. For now, given the circumstantial evidence reported herein and in [18] we consider the hypothesis that reserpine binds to MSH2/MSH6 as predicted to be a reasonable starting point for further studies. Here, we determine the parameters for small molecules to initiate the MMR protein-dependent cell-death response, where the cell-death response is measured via two distinct standard assays, MTS assay and caspase activation, which provide multiple pieces of evidence for the induction of cell-death. Based on our models, we have hypothesized that specific structural parameters are required of a small molecule to mimic the interaction of MSH2/MSH6 with DNA damage, show the correct orientation in the protein-binding pocket and initiate MSH2-dependent cell death. In a combination of computer-based structural modeling, chemical synthesis, and cell biology, we have identified these distinct requirements, which consist of the size of the molecule, the presence and location of methoxy groups, and the orientation of the molecule within the protein-binding pocket. While the computational modeling assumes that the small molecules bind in a specific pocket and in specific orientations, as predicted, the cellular assays do not require such assumptions. These small-molecule structural parameters will provide the foundation for any subsequent searches for new small molecule compounds that initiate the specific MSH2-dependent cell death targeted in this approach.
2. Materials and Methods
2.1. Virtual Screening
AutoDock 3.0 was used to perform 3D docking into structural models generated from the molecular dynamics simulations. Docking grids from vdW and electrostatic interactions were generated using standard AutoDock charges, vdW, and electrostatic parameters on cubic grids, 18.75 Å on a side. The grids were centered on the midpoint of the line connecting the two guanine nitrogens that are crosslinked in the platinated simulation, but the grids were generated in the absence of DNA and the platinum adduct. The mol2 files for the small molecules were generated using MARVIN and assigned charges and rotatable bonds using AutoDock, which was also used to perform the preparatory work for the grid docking. The ligands were docked using default Autodock Lamarckian genetic algorithm parameters except the number of runs was increased to 256. The structure with the lowest estimated inhibition constant, K i, as calculated by AutoDock was used as the docked pose [15].
2.2. Molecular Dynamics Simulations
The structures used for the docking are the centroids from the final equilibrated clusters of two molecular dynamics simulations. The clusters were obtained by K-means clustering with a 1 Å radius cutoff on the alpha-carbon positions of structures in the molecular dynamics trajectories. The simulation protocol is largely the same as described elsewhere, differing only in the use of a constant pressure algorithm, and differing simulation lengths, and is briefly summarized here. The X-ray structure of Escherichia coli MutS in complex with DNA was the initial starting point; hydrogen atoms were added using the building facility of CHARMM and TIP3P water molecules and counterions were added using the solvate package of the visual molecular dynamics (VMD) package. The CHARMM27 force field was used for the entire complex with additional parameters added based on preexisting cisplatin parameters. The platinum crosslinks two adjacent guanines. The simulation was performed in NAMD using standard parameters: a 2.0 fs timestep using SHAKE on all bonds to hydrogen atoms, a 12 Å cutoff, Particle Mesh Ewald with a 128 grid points on a side, Berendsen's constant pressure algorithm with a target pressure of 1.01325 bar, a compressibility of 45.7 mbar, a relaxation time of 1 ps, and a pressure frequency of 40 fs, and a coordinate save frequency of 200 fs; all as implemented in NAMD. The simulation protocol consisted of 250 ps of thermal equilibration to 300 K, followed by a 10 ns production simulation 15. The partition coefficients were calculated using extended group contribution approach. In this approach, a chemical structure is automatically decomposed into fragments and atom types, and the contributions of different fragments and atom types are summed together, along with correction terms to account for interactions between different fragments [19].
2.3. Chemical Synthesis
Methyl reserpate and benzoyl reserpine were prepared from reserpine as previously described [20]. All other reserpine analogs were generated in a similar fashion by condensation of methyl reserpate with the commercially available acid chloride. All new compounds were purified by flash chromatography and characterized by proton and carbon NMR spectroscopy and mass spectrometry (see Supplementary Material for specific compounds).
2.4. Cell Viability Assays
HEC59 cells (msh2 deficient) and the paired cell line with chromosome 2 transfer HEC59 (2) have been extensively characterized previously [21]. In addition to their msh2 defect, these cells contain dysfunctional p53. The cells were grown in standard growth media of DMEM-F12 +10% FBS. Cells were plated in 96 well plates at an appropriate concentration in 100 μl media and incubated overnight. Media was replaced with fresh media containing drug and allowed to incubate for 24 hours at indicated concentrations. Untreated cells received fresh media with vehicle only. One solution reagent (CellTiter 96(r) AQueous One Solution) was added to existing media (20 μl/well) and allowed to incubate 3-4 hrs. A plate reader was used to record the absorbance at 490 nm.
Dose-dependent response to nineteen increasing concentrations of the respective compound was determined. OD measurements were used to determine cell viability at each of the concentrations as percentage of control and analyzed for IC50 values using GraphPad Prism 4TM. Each compound was analyzed in triplicate. Graphs represent the mean values and standard deviations of three independent experiments.
3. Results
3.1. Functional MSH2- and MSH6 Are Required to Induce Full Cytotoxic Response to Reserpine and Rescinnamine
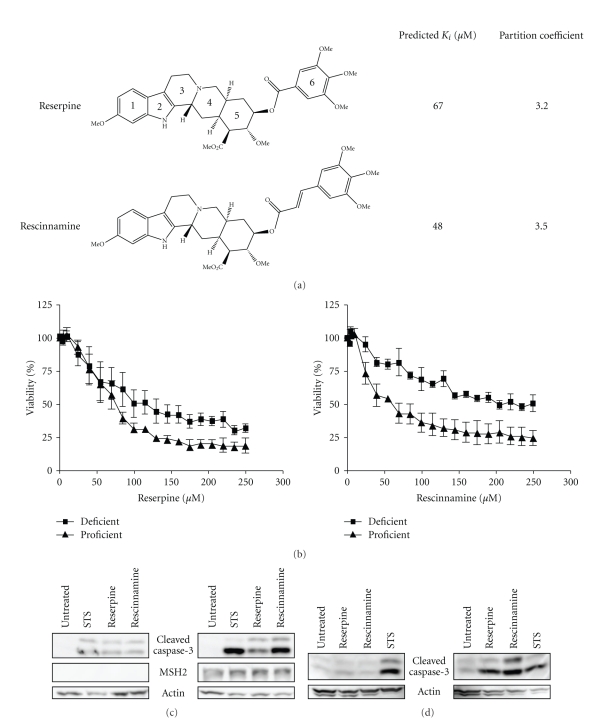
Reserpine (Figure 1(a)), a small molecule identified through computational modeling has been predicted to bind MutS homologs mimicking DNA damage [18]. We have used LC-MS total ion chromatography to provide evidence for the binding of reserpine to yeast Msh2/Msh6 (see Figure 1 in Supplementary Material available online at doi:10.4061/2011/162018). Data analysis was performed for m/z 609, which is characteristic of reserpine alone. The chromatogram of pure reserpine standard showed one prominent peak that eluted at 1.62 minutes with a mass/charge of 609.325 (suppl. Figure 1). This peak was fragmented by the later MS sectors and two fragments—397.08 and 194.82 were reproducibly observed. When the reserpine was mixed in a one-to-one ratio with the protein sample, the peak very reproducibly eluted with a longer retention time (1.88 minutes) relative to pure reserpine. The fact that we detected the peak at 609 could lead to several conclusions: The detected reserpine may represent never bound or excess unbound reserpine. However, no consistent longer migration times would be expected. If the presence of protein simply blocked sites of interaction on the stationary phase, it would be surprising to see this as a highly reproducible effect. An alternate explanation would be that the detected reserpine indeed had protein bound, but that the bound reserpine had been liberated (unbound) during the electrospray stage of the analysis. Since no excess reserpine was present in the mix, we would not expect two peaks (unbound, excess, and bound reserpine). Taken together, our data indicate that reserpine was bound to protein through the LC column, and freed during the electrospray stage. These data indicate a relatively weak binding of reserpine to the protein. The weakness of interaction may not be surprising, given that the compound is much smaller than the normal DNA substrate, and hence provides much fewer opportunities for interactions. This does provide indirect evidence that reserpine does bind, albeit, potentially weakly to the protein complex, which suggests that modification of reserpine may be fruitful to improve targeting of the MSH2-dependent pathway.
Figure 1.
(a) Structures of reserpine and rescinnamine with their binding constants and calculated partition coefficients (see Methods for details). (b) MTS cell survival assay shows MSH2-dependent cell viability after concentration-dependent reserpine (left) and rescinnamine (right) treatments. MSH2-deficient cell response is shown with squares, proficient response with triangles. (c) Western blot shows caspase-3 cleavage in Hec59 (MSH2-deficient; left panel) and Hec59(2) (MSH2-proficient; right panel) cells upon treatment with reserpine (85 μM) and rescinnamine (60 μM). The 17 and 19 kDa cleavage products of caspase-3 are depicted. Staurosporine (STS) was used as a positive control; actin was used as a loading control. (d) Western blot displays caspase-3 cleavage in DLD1 (MSH6-deficient; left panel) and DLD1(2) (MSH6-proficient; right panel) cells treated with reserpine and rescinnamine.
Cell viability assays show that even though reserpine decreases viability of both MSH2-deficient (IC50 93 μM) and MSH2-proficient (IC50 61 μM) cells after 24-hr treatment, it preferentially kills MSH2-proficient cells (Figure 1(b)). The 1.5-fold difference in cell viability between proficient and deficient cells is reminiscent of the activity of cisplatin, though reserpine activity is observed considerably earlier. Rescinnamine (Figure 1(a)), a derivative of reserpine that adds additional length via the substitution of the trimethoxybenzoyl group with a trimethoxycinnamoyl group, likewise requires functional MSH2 for full reduction of cell viability (MSH2-deficient: IC50 115 μM; MSH2-proficient: IC50 38 μM) (Figure 1(b)). The increase in length, without alteration of functional groups, doubled MSH2-dependent cytotoxicity to a 3-fold difference in cell viability between proficient and deficient cells.
We next determined if the decrease in mitochondrial integrity, as determined by the MTS assay for cell viability is indicative of the induction of an apoptotic response pathway. Cell extracts of treated cells were analyzed for the specific activation of caspase-3, a proapoptotic protein commonly used as an indicator for apoptotic cell death. The treatment of MSH2-deficient cells with reserpine (85 μM), rescinnamine (60 μM) or the control compound Staurosporine for 24 hours induces caspase-3 cleavage equally and only weakly above the untreated background control (Figure 1(c)). Drug concentrations were chosen based on the IC50 in these cells, which is reflective of the different toxicity of the compounds. In contrast, in MSH2-proficient cells, caspase-3 activation is significantly stronger. The difference in the efficiency in inducing cell death response between reserpine and rescinnamine is apparent, as rescinnamine induces stronger caspase-3 activation than reserpine (Figure 1(c)).
MSH2 forms a functional heterodimer with MSH6 in its ability to recognize DNA lesions and initiate response pathways. We next determined if MSH6 is also required in the initiation of proper responses to small molecules, such as reserpine and rescinnamine. MSH6-deficient and MSH6-deficient cells were treated with reserpine (85 μM) and rescinnamine (60 μM) for 24 hours. The detection of cleaved caspase-3 in extracts from both cell lines revealed activation primarily in the MSH6-proficient cells, consistent with a functional requirement of the protein for cytotoxic response (Figure 1(d)).
3.2. Combinatorial Treatment of Cisplatin and Rescinnamine Abrogates MSH2-Dependence of Cytotoxic Response and Eliminates Chemotolerance
Cisplatin is a chemotherapeutic that primarily forms (1,2)-intrastrand crosslinks between adjacent guanines in DNA. This compound was previously used as a model agent to characterize the MSH2/MSH6-dependent induction of a protein “death conformation” and cytotoxic response [8]. Since reserpine was predicted to likewise stabilize the MSH2/MSH6 “death conformation” [18], and rescinnamine showed an improved activity over reserpine, we asked whether a combination of rescinnamine with cisplatin would improve the activity of either drug alone.
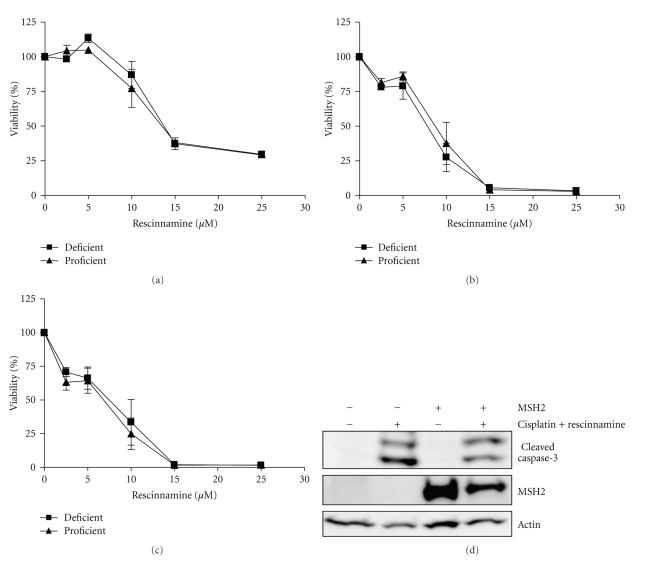
MSH2-deficient and MSH2-proficient cells were treated with sublethal doses of cisplatin (10 μM) and increasing concentrations of rescinnamine (0–25 μM). Cell viability was determined after 24, 48, and 72 hours by MTS assay (Figure 2). Twenty-four hours after treatment (Figure 2(a)), a rapid decrease in cell viability was observed that led to the complete elimination of viable cells after 48 hours at much lower concentration of either cisplatin or rescinnamine alone (Figure 1(b)) [18]. This effect on cell viability was indistinguishable for MSH2-proficient and MSH2-deficient cells. Overall cell viability was reminiscent of the treatment of MSH2-proficient cells with rescinnamine alone, and increased tolerance to the drug was eliminated by the concomitant treatment with cisplatin (Figure 2(a)). Increased exposure to the combination treatment eradicated cell viability entirely (Figures 2(b) and 2(c)), which was previously not observed for much higher concentrations of either drug alone [13, 18]. Full eradication of cells was observed after 48 or 72 hour treatment with 20 μM cisplatin and 15 μM rescinnamine.
Figure 2.
Combination treatment of cisplatin and rescinnamine drastically reduces cell viability within 24 hours (a) and completely eliminates all cells at 48 and 72 hours (b and c, resp.) when cells are treated with cisplatin (10 μM) and increasing concentrations of rescinnamine (0–25 μM). Cell viability is accessed via MTS assays; MSH2-deficient cells (■) and MSH2-proficient (▴) cell response is shown. Mean values and standard deviations of three independent experiments are shown. (d) Immunoblotting demonstrates caspase-3 cleavage in Hec59 (MSH2-deficient; lanes 1 and 2) and Hec59(2) (MSH2-proficient; lanes 3 and 4) cells upon treatment with 24 hours of cisplatin (10 μM) and rescinnamine (60 μM). The 17 and 19 kDa cleavage products of caspase-3 are depicted. Staurosporine (STS) was used as a positive control; actin was used as a loading control.
The IC50 values of the combination treatment confirm this result. The addition of cisplatin resulted in a 7.3-fold decrease in IC50 value for rescinnamine after 24 hours, and a 9.7-fold decrease after 48 hours in MSH2-deficient cells. The same levels of rescinnamine reduced cell viability in MSH2-proficient cells (Table 1). An additional 24 hours of treatment for a total of 72 hours did not significantly alter the IC50 any further. The combination treatment results in a rapid, MSH2-independent demise of cell viability. These findings are confirmed by immunoblotting; upon treatment with 24 hours of rescinnamine (60 μM) and cisplatin (10 μM) comparable levels of caspase-3 cleavage in MSH2-deficient and MSH2-proficient cells are observed (Figure 2(d)). At 48 and 72 hours, all cells treated with the combination treatment were eliminated and no lysates could be collected and assessed for caspase-3 cleavage.
Table 1.
IC50 values for response of MSH2-deficient and MSH2-proficient cells to Reserpine analogs.
| Compound | IC50 [μM] | Fold difference | |||
|---|---|---|---|---|---|
| MSH2-deficient | 95% CL | MSH2-proficient | 95% CL | ||
| cisplatin | 75 | 46–130 | 47 | 35–64 | 1.6 |
| reserpine | 93 | 74–120 | 61 | 54–70 | 1.5 |
| rescinnamine | 87 | 53–150 | 39 | 32–47 | 2.6 |
|
| |||||
| rescinnamine + 10 μM cisplatin (24 h) | 12 | 11–13 | 11 | 10–13 | 1.1 |
| rescinnamine + 10 μM cisplatin (48 h) | 9.1 | 7–12 | 9.7 | 9–11 | 0.94 |
| rescinnamine + 10 μM cisplatin (72 h) | 9.9 | 9–11 | 9.8 | 5−13–214 | 1.01 |
|
| |||||
| benzoyl | 110 | 100–120 | 132 | 107–160 | 0.83 |
| 3-methoxy benzoyl | 83 | 74–92 | 88 | 75–104 | 0.94 |
| 4-methoxy benzoyl | 68 | 58–81 | 67 | 59–75 | 1.01 |
| 3,4-dimethoxy benzoyl | 85 | 78–93 | 66 | 59–74 | 1.3 |
| 3,5-dimethoxy benzoyl | 36 | 28–48 | 38 | 30–49 | 0.95 |
| cinnamoyl | 150 | 92–250 | 130 | 107–160 | 1.2 |
| methylenedioxybenzoyl | 36 | 32–41 | 51 | 45–58 | 0.71 |
| deserpidine | 59 | 51–69 | 48 | 46–51 | 1.2 |
3.3. Structural Predictions and Validation of Parameters Required for MSH2-Dependent Cell Death
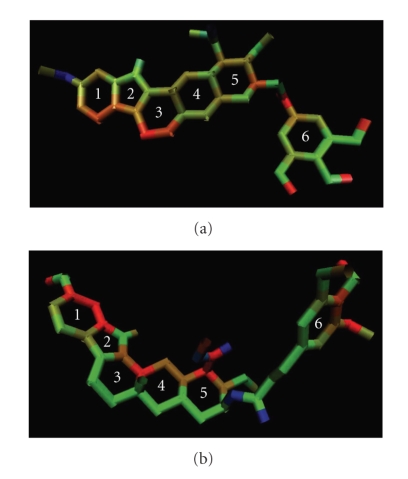
Next, we wanted to identify the specific parameters that are required for the MSH2-dependent induction of cell death, and when altered, reduce or abolish this function. Modeling of reserpine and rescinnamine into the MutS structure (Figure 3) predicted functional groups and parts of the molecules with favorable (red) or unfavorable (blue) interactions with the protein. Green depicts intermediate importance in the interaction.
Figure 3.
Structures of reserpine (a) and rescinnamine (b) analyzed by computational modeling depicting favorable (red) and unfavorable (blue) interactions with amino acids within MutS-binding pocket. Green indicates interactions of intermediate binding relevance.
Differences in the predicted favorable and unfavorable interactions of reserpine and rescinnamine, respectively, represent the difference in their activity. The methoxy group on ring 1 of reserpine is predicted to be nonessential for interactions, while, in rescinnamine, it is proposed to be important for interactions. As for ring 6, all three methoxy groups in reserpine are predicted to have favorable interactions, but only two of them are highly favorable for interactions between rescinnamine and MutS/MSH proteins. In reserpine, the carbon of ring 5 that connects to carbon chain leading to ring 6 is pertinent for interactions. Nonetheless, in rescinnamine, that system is unfavorable.
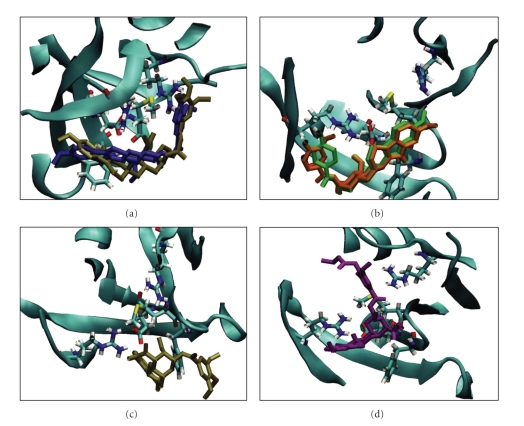
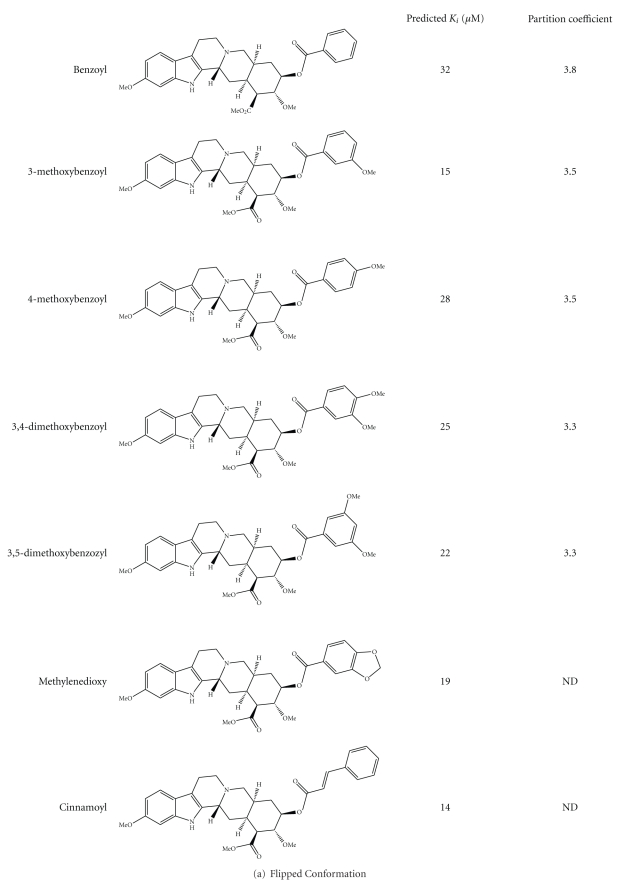
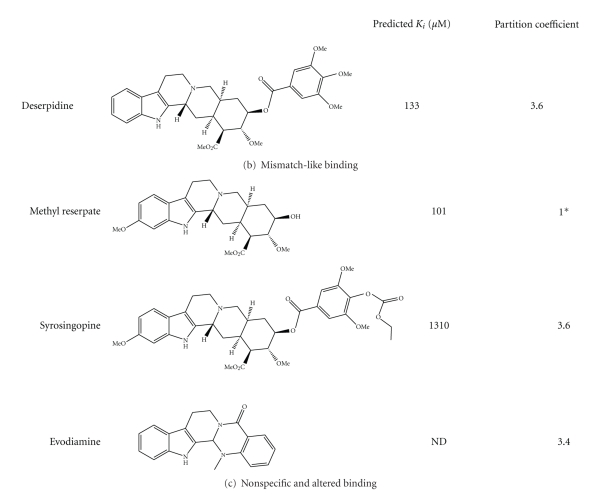
These computational predictions were subsequently experimentally analyzed to determine the validity of the prediction. Derivatives of reserpine and rescinnamine were purchased or synthesized (if not commercially available) (see Table 1). Through structural predictions, the compounds were subdivided into four distinct modes of binding to MutS/MSH proteins: reserpine-like conformation, flipped conformation, mismatch-like conformation, and nonspecific conformation (see Figure 5).
Figure 5.
Predicted modes of binding of reserpine and rescinnamine derivatives include: (a) reserpine-like; (b) flipped; (c) mismatch-like; (d) nonspecific or altered.
3.3.1. Reserpine-Like Conformation
Reserpine was identified utilizing the cisplatin-induced “death conformation” of MSH2/MSH6 [8, 18], predicting a similar interaction with the DNA-binding pocket of MutS (Figure 3(a)). Likewise, rescinnamine is predicted to show similar interactions (Figure 3(b)). The space occupied by either molecule largely overlaps with that of DNA. The molecules are stabilized by hydrogen bonds between their methoxy groups and at least three amino acids (G38, R58, R108 of MutS). These residues were previously identified as being important for the interaction with cisplatinated DNA [8]. The phenylalanine (F36) shown to be indispensable for mismatch repair is far removed from the small molecules and shows no significant binding activity [8, 22]. This lack of significance is reminiscent of its failure to exhibit a significant effect on the binding to cisplatinated DNA or, when mutated, on the cytotoxic response to cisplatin [8]. Both molecules show a characteristic bend. Previously, it was suggested that an acquired or preexisting bend in the ligand for MutS homologous proteins is required for robust binding to their substrates [23]. The existence of such a bend may be a prerequisite for small molecules to specifically bind to MutS homologous proteins and induce a specific response. However, the predicted orientation of the bend in the small molecules appears to be inverted compared to DNA, which may suggest that the actual presence of a bend is not a requirement for cytotoxic response. Among all the compounds tested, reserpine and rescinnamine are the only molecules predicted to preferentially bind in this specific conformation and exhibit these specific interactions with the protein binding pocket.
3.3.2. Flipped Conformation
To determine the functional requirements of a small molecule to induce MSH2-dependent cell death, we constructed a number of analogs based on the reserpine structure. We hypothesized, based on the structural predictions, that the methoxy groups on ring 6 will have significant impact. We constructed several compounds that either lack all or some of the methoxy groups, resulting in seven different molecules: benzoyl, 3-methoxybenzoyl, 4-methoxybenzoyl, 3,4-dimethoxybenozyl, 3,5-dimethoxybenzoyl, methylendioxybenzoyl, and cinnamoyl resperine (Figure 4(a)). Interestingly, the structural predictions suggest a preferred altered binding orientation of these molecules, even for those lacking only one methoxy group (3,4-dimethoxy benzoyl; 3,5-dimethoxy benzoyl). This altered binding mode would flip the molecule and have it oriented the opposite way to the reserpine/rescinnamine. The binding of each one of the five molecules in this orientation is predicted to be strong, with predicted K i values of 15–32 μM. Experimental determination of the cell death demonstrates the ability of these compounds to induce cell death, however in an MSH2-independent manner (Figure 6(a), Table 1). The 3,4-dimethoxy benzoyl compound is an exception and the only molecule that initiates a significant MSH2-dependent cell death response. The IC50 values for this compound result in 85 μM in MSH2-deficient cells versus 66 μM in MSH2-proficient cells, with a 1.3-fold difference, reminiscent of the response observed with reserpine (Table 1). When forced by computational modeling into the “correct” orientation, the predicted binding constant is 93 μM, which would substantiate the cell biology data and suggest a poorer binding to MutS than reserpine. The limitations of the resolution of structural modeling make it impossible to distinguish the binding affinity of this compound from the one of other methoxy compounds in the “correct” orientation.
Figure 4.
Structures of reserpine and rescinnamine derivatives are shown on the left and their corresponding predicted (calculated, as described in Section 2) binding constants and partition coefficients are displayed on the right. Derivatives are subdivided into their predicted binding modes: (a) flipped; (b) mismatch-like; and (c) nonspecific or altered (see also Figure 5).
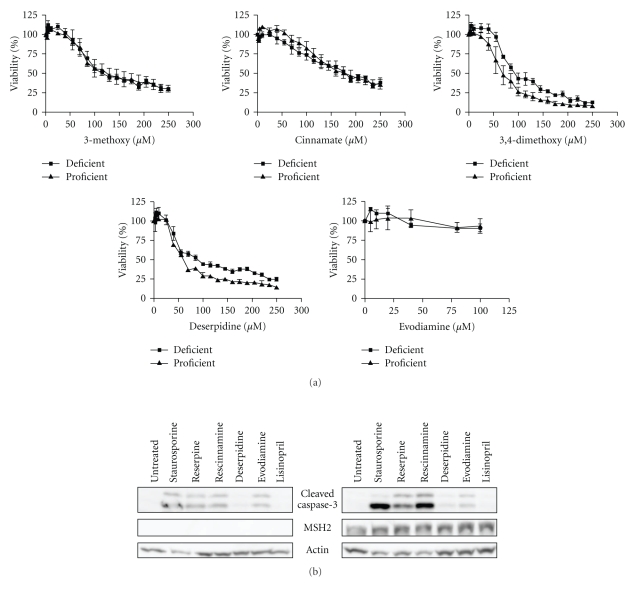
Figure 6.
(a) Derivatives of reserpine and rescinnamine diminish cell viability in an MSH2-independent manner as shown by MTS assays with an exception of 3,4-dimethoxy reserpine, which demonstrates a slight dependence on MSH2. Evodiamine has an insignificant effect on cell viability. One representative from each of the predicted binding mode groups is shown. MSH2-deficient cells (■) and MSH2-proficient (▴) cell response is shown. (b) Caspase-3 cleavage was assessed in Hec59 (MSH2-deficient; left panel) and Hec59(2) (MSH2-proficient; right panel) cells when treated with various compounds. The 17 and 19 kDa cleavage products of caspase-3 are depicted. Staurosporine (STS) was used as a positive control; actin was used as a loading control. Lisinopril is an ACE inhibitor, like rescinnamine, and demonstrates rescinnamine's action as being independent of its antihypertensive properties.
Since ring 6 is attached to the remainder of the protein by a rotatable bond, the 3,4-dimethoxy benzoyl compound is indistinguishable from the 4,5-dimethoxy molecule. Whether the presence of two methoxy groups on this ring is sufficient for the induction of MSH2-dependent cell death, or this rotation and the resulting presence of methoxy groups in all three positions enable the appropriate response is unknown.
Methylenedioxybenzoyl has an additional ring system on ring 6. This compound is predicted to bind to MutS with a K i of 19 μM; however, the predicted binding is also found to be in a flipped conformation. The cell death response is opposite to the damage response generally observed in dependence of mismatch repair proteins. MSH2-deficient cells show a higher sensitivity to the compound than proficient ones (IC50 of 36 μM in MSH2-deficient ones, versus 51 μM in MSH2-proficient cells, Table 1), reversing the resistance phenotype seen with cisplatin and reserpine/rescinnamine.
Cinnamoyl reserpine is a rescinnamine derivative that lacks the methoxy groups on ring 6, similar to the reserpine analog benzoyl. Again, similar to the corresponding reserpine analog, the predicted binding of cinnamoyl reserpine to MutS is predicted to occur in the flipped conformation with a binding constant of 14 μM. The compound induces cell death that is independent of functional MSH2 (IC50 value of 150 μM in MSH2-deficient cells versus 130 μM in MSH2-proficient cells; Figure 6(a), Table 1). This result confirms the structural prediction.
Together, these results suggest that the absence of essential methoxy groups on ring 6 that appear to position the molecules in the correct conformation triggers a different binding mode (Figure 5(b)) that does not result in MSH2-dependent cell death. Removal of these suggested, essential functional groups eliminates the MSH2-dependent cell death response.
3.3.3. Mismatch-Like Binding
The methoxy group on ring 1 was predicted to show unfavorable interactions with the MutS protein for reserpine, but not rescinnamine (Figure 3). A clinically relevant compound exists that has the general structure of reserpine, but lacks this methoxy group (deserpidine, Figure 4(b)). Though eliminating a suggestively unfavorable interaction, the predicted binding constant is roughly 2-fold worse than that for reserpine (130 μM), which suggests that even an “unfavorable” interaction may be required to correctly orient the molecule in the binding pocket.
The predicted binding mode for deserpidine is reminiscent of the binding of mismatched DNA to the protein. The molecule shows a stronger bending than other compounds and altered orientation that results in the molecule wrapping around the phenylalanine involved in coordinating mismatch binding (F36) (Figure 5(c)). The predicted specificity for the “death conformation” is reduced for this molecule, which would further confirm the predicted “mismatched DNA”-like binding mode.
The cell biology of this compound revealed cell death induction with no significant specificity for the presence of MSH2 (IC50 values of 56 μM in MSH2-deficient versus 49 μM in MSH2-proficient cells; Figure 6(b), Table 1), confirming the structural predictions. This result suggests that binding in the “mismatched DNA mode” eliminates MSH2-dependent cytotoxic response.
3.3.4. Nonspecific and Altered Binding
Some compounds are predicted to bind MutS in an entirely altered conformation. Though binding is still observed in or close to the DNA-binding pocket, the general orientation of these compounds is altered when compared to the binding of reserpine/rescinnamine (Figure 5(d)).
Methyl Reserpate lacks the entire ring 6. This compound is predicted to bind in a distance to the actual binding pocket with a K i of 101 μM. The molecule only weakly induces cell death in either MSH2-proficient and MSH2-deficient cells with no preference for either cell line. The cell death response was not sufficient to reliably determine IC50 values.
Syrosingopine is a commercially available reserpine analog that has found application as an antihypertensive drug. Its predicted binding to the mismatch repair protein is the worst among the tested compounds, with a predicted K i of 1310 μM. This is largely due to the fact that its substitution on position 4 of ring 6 increases its length considerably, which interferes with the binding to the protein (Figures 4(c) and 5(d)). The binding of the molecule is hence highly “distorted” when compared to the interaction of reserpine/rescinnamine with the protein (compare Figures 5(a) and 5(d)).
Evodiamine, a compound described as aiding in diet-induced obesity [24], only contains ring systems 1-5 and lacks all methoxy groups seen in reserpine/rescinnamine. This compound shows no specific binding in the computational docking experiments, and hence does not result in a predictable binding constant. In the cell system, this molecule shows no significant cytotoxic effect, in either the presence or absence of MSH2 (Figure 6(a)). This is presumably due to the fact that evodiamine cannot establish any of the required interactions with the MMR protein.
Cells were treated with selected reserpine derivatives to determine their effect on cell viability and caspase-3 cleavage. MSH2-proficient and MSH2-deficient cells were treated with increasing concentrations (0–250 μM) of 3-methoxy reserpine, 3,4-dimethoxybenzoyl reserpine, 3,5-dimethoxybenzoyl reserpine, evodiamine, and deserpidine for 24 hours after which time their viability was assessed via MTS assay (Figure 6(a) and data not shown). Upon treatment with 3,5-dimethoxybenzoyl, no MSH2-dependent cell death was observed (data not shown). Upon treatment with 3,4-dimethoxybenzoyl reserpine, however, a 1.3-fold difference between MSH2-proficient and MSH2-deficient cells was observed, reminiscent of the increase in resistance to cisplatin in MSH2-deficient cells. This result indicates the importance of two methoxy groups on ring 6 of the small molecule. This cellular result provides further indirect evidence that the computational model, which is one of direct binding to a specific protein conformation, is valid.
Since rescinnamine exhibits the activity of an ACE inhibitor, we determined if the observed effect on cell viability is due to this activity. In comparison, another, structurally different ACE inhibitor, lisinopril [25], was added and cell viability determined. No effect on cell viability and caspase-3 activation was observed in either MSH2-proficient and MSH2-deficient cells (Figure 6(b)), suggesting a different function for rescinnamine in MSH2-dependent cell death.
4. Discussion
MMR is one of the primary DNA repair pathways that maintain genome stability within every cell. Its significance is displayed by its contribution to carcinogenesis in cells deficient for a key MMR protein. Based on the increasing number of additional protein interactions MMR proteins undergo, they may participate in numerous other cellular functions that remain to be understood. Among these, their participation in the response to cytotoxic agents and the initiation of cell death is of importance, particularly since defects in MMR proteins increase resistance to certain chemotherapeutic agents and lead to the clonal selection of MMR-deficient cells. We have previously described the response of MMR proteins to the chemotherapeutic agent cisplatin and predicted a “death conformation” that is selectively targeted. This “death conformation” can be accessed by a small molecule, reserpine that induces DNA damage-independent cell death. Our multiple cellular assays have provided indirect evidence that this model, which required direct binding of reserpine, is indeed correct. However, we do look forward to the day when direct binding can be observed with either refinements of current technologies, or with brand new technologies. We have here determined the particular parameters that allow reserpine and its analogs to access the predicted “death conformation” of MSH2/MSH6 and induce cell death. These parameters, although predicted based on our model, were verified by cellular assays, which provided indirect evidence for the appropriateness of our model. Our experimental data indicates that the length of the molecule and specific functional groups are required for the induction of cell death. In our model, these parameters are critical to maintain the correct orientation of the small molecules within the MSH2/MSH6-binding pocket. Small changes in this structure of reserpine abrogate the ability to induce MSH2-dependent cell death; which would not be surprising if our model of direct binding were correct. These results suggest that the development of an improved molecule for the induction of cell death that is based on reserpine will need to be closely designed upon the original structure of the molecule. Since rescinnamine proved to be the best compound, future designs would do well to be based on this structure. Since cinnamate, a derivative of rescinnamine without the trimethoxy groups on ring system 6, abrogates the MSH2-dependent cell death response, these groups are required for the observed activity.
5. Conclusions
Our data demonstrate the combining virtual screening with conformational modeling is a novel, valid approach to identify compounds that target highly specific structures and pathways. We show that, in at least the case, molecular dynamics simulations provide molecular conformations that are sufficiently accurate as to allow for targeting of specific conformations associated with specific molecular functions. In particular, we have shown that cell death can be induced by small molecules that have been screened against models for a specific “death” conformation of the MSH2/MSH6 complexes. We have demonstrated the effects of these small molecules, reserpine-analogs, via two distinct cell assays; MTS assay and caspase-activation, which are standard and commonly used tests for cell viability and apoptotic cell death. Furthermore, the combinatorial treatment of reserpine and cisplatin resulted in rapid, MSH2-independent cell death, suggesting that the combination treatment “overloads” the MSH2-dependent system, and the cell induces death via a different pathway. This observation suggests that future mechanistic studies, well beyond the scope of this paper as few of the actors involved are known, would be fruitful in determining the precise signaling pathways induced in the MSH2-dependent and MSH2-independent cell-death pathways. A more detailed mechanistic study of the MSH2-dependent pathway would be useful in potentially providing another protein target. Understanding the mechanism of the MSH2-dependent pathway may provide treatment options for MMR-deficient tumors, which are found in many patients with colorectal cancers.
Taken together, our data determine the parameters required of reserpine analogs to induce MMR-dependent cell death that can be utilized to further develop chemicals targeting this particular cell death pathway.
Supplementary Material
Representative total ion chromatograms (TIC) of (a) reserpine standard (33 nM) and (b) a 1:1 mixture of reserpine (33 nM) and protein. The multiple reaction monitoring (MRM) spectra obtained from each chromatographic peak in (a) and (b) are shown as insets. The average retention time for (a) was 1.68 +/− 0.01 min and for (b) was 1.88 +/− 0.01 min ( replicate injections, alternating between (a) and (b)). The reproducible increase in retention time and decrease in relative abundance (nearly 3-fold) of the peak in (b) suggest the formation of a complex between reserpine and the protein. This complex is thought to partially dissociate during the electrospray process as demonstrated by the decrease in relative abundance of the released reserpine peak compared to (a).
Acknowlegments
The authors thank Dr. Christa Colyer for valuable discussions and providing LC-MS experiments. The project described was supported by Grants nos. CA101829 and CA129373 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. Further support was provided from the Comprehensive Cancer Center and the Translational Science Institute at Wake Forest University Health Sciences. The computations were performed on the WFU DEAC cluster (http://www.deac.wfu.edu/), which is supported in part by WFU IS and WFU's Associate Provost for Research. Additional storage was provided by IBM through a SUR grant. The authors thank WFU and IBM for their support. S. B. King, F. Salsbury, and K. D. Scarpinato contributed equally to the paper.
References
- 1.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair. 2004;3(8-9):1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 2.Fishel R, Wilson T. MutS homologs in mammalian cells. Current Opinion in Genetics and Development. 1997;7(1):105–113. doi: 10.1016/s0959-437x(97)80117-7. [DOI] [PubMed] [Google Scholar]
- 3.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Current Opinion in Genetics and Development. 1999;9(1):89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 4.Bellacosa A. Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death and Differentiation. 2001;8(11):1076–1092. doi: 10.1038/sj.cdd.4400948. [DOI] [PubMed] [Google Scholar]
- 5.Kunkel TA, Erie DA. DNA mismatch repair. Annual Review of Biochemistry. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 6.Irving JAE, Hall AG. Mismatch repair defects as a cause of resistance to cytotoxic drugs. Expert Review of Anticancer Therapy. 2001;1(1):149–158. doi: 10.1586/14737140.1.1.149. [DOI] [PubMed] [Google Scholar]
- 7.Jiricny J. The multifaceted mismatch-repair system. Nature Reviews Molecular Cell Biology. 2006;7(5):335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 8.Salsbury FR, Jr., Clodfelter JE, Gentry MB, Hollis T, Scarpinato KD. The molecular mechanism of DNA damage recognition by MutS homologs and its consequences for cell death response. Nucleic Acids Research. 2006;34(8):2173–2185. doi: 10.1093/nar/gkl238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aebi S, Kurdi-Haidar B, Gordon R, et al. Loss of DNA mismatch repair in acquired resistance to cisplatin. Cancer Research. 1996;56(13):3087–3090. [PubMed] [Google Scholar]
- 10.Modrich P. Strand-specific mismatch repair in mammalian cells. The Journal of Biological Chemistry. 1997;272(40):24727–24730. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- 11.Aebi S, Fink D, Gordon R, et al. Resistance to cytotoxic drugs in DNA mismatch repair-deficient cells. Clinical Cancer Research. 1997;3(10):1763–1767. [PubMed] [Google Scholar]
- 12.Fink D, Nebel S, Aebi S, et al. The role of DNA mismatch repair in platinum drug resistance. Cancer Research. 1996;56(21):4881–4886. [PubMed] [Google Scholar]
- 13.Topping RP, Wilkinson JC, Scarpinato D. Mismatch repair protein deficiency compromises cisplatin-induced apoptotic signaling. The Journal of Biological Chemistry. 2009;284(21):14029–14039. doi: 10.1074/jbc.M809303200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22(12):1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- 15.Lin DP, Wang Y, Scherer SJ, et al. An MSH2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Research. 2004;64(2):517–522. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- 16.Drotschmann K, Topping RP, Clodfelter JE, Salsbury FR. Mutations in the nucleotide-binding domain of MutS homologs uncouple cell death from cell survival. DNA Repair. 2004;3(7):729–742. doi: 10.1016/j.dnarep.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 17.Yang G, Scherer SJ, Shell SS, et al. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6(2):139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 18.Vasilyeva A, Clodfelter JE, Rector B, Hollis T, Scarpinato KD, Salsbury FR., Jr. Small molecule induction of MSH2-dependent cell death suggests a vital role of mismatch repair proteins in cell death. DNA Repair. 2009;8(1):103–113. doi: 10.1016/j.dnarep.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klopman G, Li J, Wang S, Dimayuga M. Computer automated logP calculations based on an extended group contribution approach. Journal of Chemical Information and Computer Sciences. 1994;34(4):752–781. [Google Scholar]
- 20.Pearce HL, Safa AR, Bach NJ, Winter MA, Cirtain MC, Beck WT. Essential features of the P-glycoprotein pharmacophore as defined by a series of reserpine analogs that modulate multidrug resistance. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(13):5128–5132. doi: 10.1073/pnas.86.13.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umar A, Koi M, Risinger JI, et al. Correction of hypermutability, N-Methyl-N’-nitro-N-nitrosoguanidine resistance, and defective dna mismatch repair by introducing chromosome 2 into human tumor cells with mutations in MSH2 and MSH6 . Cancer Research. 1997;57(18):3949–3955. [PubMed] [Google Scholar]
- 22.Drotschmann K, Yang W, Brownewell FE, Kool ET, Kunkel TA. Asymmetric recognition of DNA local distortion. The Journal of Biological Chemistry. 2001;276(49):46225–46229. doi: 10.1074/jbc.C100450200. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Yang Y, Schofield MJ, et al. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi Y, Nakano Y, Kizaki M, Hoshikuma K, Yokoo Y, Kamiya T. Capsaicin-like anti-obese activities of evodiamine from fruits of Evodia rutaecarpa, a vanilloid receptor agonist. Planta Medica. 2001;67(7):628–633. doi: 10.1055/s-2001-17353. [DOI] [PubMed] [Google Scholar]
- 25.Messerli FH, Kaesser UR. Lisinopril in the treatment of hypertension. Journal of Human Hypertension. 1989;3(1):17–21. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative total ion chromatograms (TIC) of (a) reserpine standard (33 nM) and (b) a 1:1 mixture of reserpine (33 nM) and protein. The multiple reaction monitoring (MRM) spectra obtained from each chromatographic peak in (a) and (b) are shown as insets. The average retention time for (a) was 1.68 +/− 0.01 min and for (b) was 1.88 +/− 0.01 min ( replicate injections, alternating between (a) and (b)). The reproducible increase in retention time and decrease in relative abundance (nearly 3-fold) of the peak in (b) suggest the formation of a complex between reserpine and the protein. This complex is thought to partially dissociate during the electrospray process as demonstrated by the decrease in relative abundance of the released reserpine peak compared to (a).