Abstract
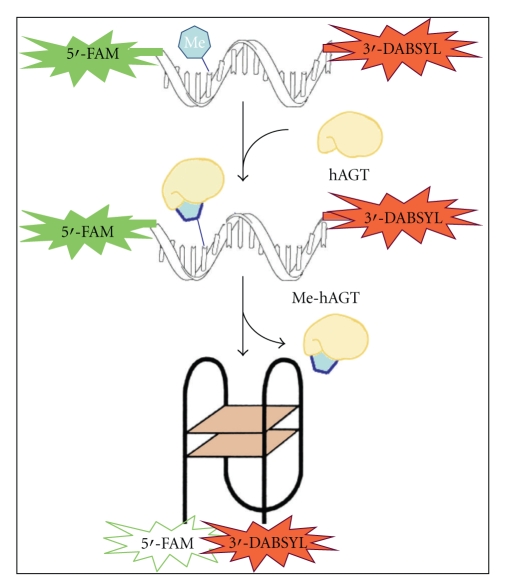
Human O6-alkylguanine-DNA alkyltransferase (hAGT) is a DNA repair protein that reverses the effects of alkylating agents by removing DNA adducts from the O6 position of guanine. Here, we developed a real-time fluorescence hAGT activity assay that is based on the detection of conformational changes of the thrombin-binding aptamer (TBA). The quadruplex structure of TBA is disrupted when a central guanine is replaced by an O6-methyl-guanine. The sequence also contains a fluorophore (fluorescein) and a quencher (dabsyl) attached to the opposite ends. In the unfolded structure, the fluorophore and the quencher are separated. When hAGT removes the methyl group from the central guanine of TBA, it folds back immediately into its quadruplex structure. Consequently, the fluorophore and the quencher come into close proximity, thereby resulting in decreased fluorescence intensity. Here, we developed a new method to quantify the hAGT without using radioactivity. This new fluorescence resonance energy transfer assay has been designed to detect the conformational change of TBA that is induced by the removal of the O6-methyl group.
1. Introduction
Alkylating agents are chemotherapeutic anticancer drugs that produce their cytotoxic effect by generating adducts at multiple sites in DNA [1]. A subset of alkylating agents, which includes nitrosoureas and temozolamide, have a preference for alkylating guanine at the O6 position, which is the most relevant in terms of mutagenesis and carcinogenesis [2–9]. In particular, 1,3-bis-(2-chloroethyl)-1-nitrosourea (BCNU) attacks initially at the O6 guanine position, causing its rearrangement in a cyclic intermediate thus giving rise to N1,O6-ethanoguanine [10]. Finally, a cross-link with the opposite cytosine is formed, and, as a consequence, DNA replication is blocked, producing G2/M arrest [11]. In addition to the well-known side effects and limitations of chemotherapeutic agents, these substances also present problems of acquired tumor resistance. In particular, the DNA-repair human O6-alkylguanine DNA alkyltransferase (hAGT or MGMT) is responsible for removing alkyl adducts from the O6 position of guanines, thereby blocking the cytotoxic effects of the alkylating agents and making a crucial contribution to the resistance mechanism [12–14]. It is well established that tumor cells show greater expression of this protein than healthy cells. Thus, this increased expression appears to be predictive of a poor response to chemotherapeutic drugs. This effect has been observed in a large number of cancers, ranging from colon cancer, lung tumors, breast cancer, pancreatic tumors, non-Hodgkin's lymphoma, myeloma, and glioblastoma multiforme, among others [15–17]. In addition methylation of the hAGT promoter and consequently the complete depletion of hAGT, it has been associated with longer survival in patients with gliomas undergoing combined radiation-chemotherapy treatment [18, 19]. Therefore, pharmacological inhibition of hAGT has the potential to enhance the cytotoxicity of a diverse range of anticancer agents [20].
hAGT is a 22-kDa (207 AA) DNA-binding protein that contains a highly conserved internal cysteine, which acts as the acceptor site for alkyl groups. The S-alkyl-Cys formed is not regenerated and the protein, which behaves as a suicidal enzyme, inactivates itself in the dealkylation process [21, 22]. This single turnover of hAGT renders it vulnerable to inactivation. On the basis of this observation, intense research effort has been devoted to the identification of small molecules capable of inhibiting hAGT activity and significantly enhancing the cytotoxic effect of BCNU in prostate, breast, colon, and lung tumor cells [20].
Several methods are available to characterize the mechanism of action of hAGT and its activity. Moreover, they also have the capacity to evaluate the capacity of small molecules to inhibit hAGT. Most of these methods are based on radioactivity assays while others are based on multiple-step enzymatic reactions [23–26].
G-quadruplexes are a family of four-stranded DNA structures stabilized by the stacking of guanine tetrads, in which four planar guanines form a cyclic array of hydrogen bonds [27]. These G-rich regions are connected by lateral, central, or diagonal loops of diverse sizes and composition that form base-pairing alignments, which in turn stack with the terminal G-tetrads, thus further stabilizing G-quadruplex structures [28]. Another key element in G-quadruplex formation is the presence of monovalent cations, which stabilize the negative electrostatic potential created by the guanine O6 oxygen atoms within the quadruplex core [29, 30]. However, most divalent cations have the capacity to induce the dissociation of G-quadruplex structures [31]. Finally, modifications in the base composition of the tetrads are poorly tolerated by these structures. As an example, inosine [32] and O6-methylguanine [33], both nonnatural bases can form a smaller number of hydrogen bonds and thus destabilize the G-quadruplex.
Given the potential relevance of hAGT as a prognostic marker of cancer and as a therapeutic target, and that its substrate O6-methylguanine disrupts the formation of G-quadruplex structures [33], here, we developed a new fluorescence activity assay for hAGT. For this purpose, we selected the thrombin-binding aptamer (TBA) as our G-quadruplex model. TBA is a well-known 15 mer that adopts a chair-like structure consisting of two G-tetrads connected by two lateral TT loops and a central TGT loop [34]. The modification of its sequence in positions 5 or 6 by replacing a guanine of the tetrad for an O6-methylguanine disrupts folding, leaving it in an extended conformation. Giving that the two conformations bring the two ends of TBA together take them further apart, our working hypothesis was that the incorporation of fluorescence probes results in a measurable change in intensity. The final aim of this fluorescence assay was to measure the DNA repair activity of hAGT and thus facilitate the search for new and more potent inhibitors which enhance chemotherapeutic drugs. Although several methods have been described to quantify hAGT activity [23–26], none of them use the conformational change of G-quadruplex for this purpose. Here, we describe the development of a straightforward, rapid, one-step fluorescence resonance energy transfer (FRET) assay.
2. Materials and Methods
2.1. Chemicals
5′-Fluorescein CE phosphoramidite (6-FAM), 3′-Dabsyl CPG, O6-methylguanine (O6-MeG) and Gdmf phosphoramidites were purchased from Link technologies (UK) and Glen Research (USA). O6-methylguanine was protected with the isobutyryl group [35]. Standard phosphoramidites and ancillary reagents were purchased from Applied Biosystems (Europe).
The matrices for MALDI-TOF experiments were 2′,4′,6′-trihydroxiacetophenone monohydrate (THAP) and ammonium citrate dibasic.
Solvents for chromatographic analysis were prepared using triethylamine, acetic acid, and acetonitrile as mobile phase. All other chemicals were of analytical reagent grade and were used as supplied. Ultrapure water (Millipore, USA) was used in all experiments.
2.2. Instrumentation
Semipreparative reverse phase (RP) HPLC was performed on a Waters chromatography system using Nucleosil semipreparative 120 C18 (250 × 8 mm) columns. Analytical RP-HPLC was performed using an XBridge OST C18 2.5 μm column and a Nucleosil Analytical column 120 C18 (250 × 4 mm). Oligonucleotides were detected by UV absorption at 260 nm. Mass spectra were recorded on a MALDI Voyager DETM RP time-of-flight (TOF) spectrometer (Applied Biosystems, USA) with a nitrogen laser at 337 nm using a 3-ns pulse. Fluorometric measurements were performed on a spectrofluorometer multidetection microplate reader Biotek FL × 800 and a Jasco FP6200. Molecular absorption spectra between 220 and 550 nm were recorded with a Jasco V650 spectrophotometer. The temperature was controlled with a Jasco ETC 272T Peltier device. Hellma quartz cuvettes (0.5 and 1.0 cm path length, 500 or 1000 μL volume) were used.
2.3. Oligonucleotide Synthesis
Oligonucleotide sequences (shown in Table 1) were synthesized on an ABI 3400 DNA Synthesizer (Applied Biosystems, USA) using a 200-nmol scale synthesis and following the standard protocols. We used dimethylformamidino-protected guanine Gdmf phosphoramidite for all the syntheses. 5′-Fluorescein CE phosphoramidite (6-FAM), O6-methylguanine (O6-MeG) and Gdmf phosphoramidites were from commercial sources. The isobutyryl group was used to protect the amino group of O6-MeG [35]. The quencher group was introduced at the 3′ end using the controlled pore glass functionalized with a 3′-Dabsyl derivative CPG. O6-MeG-containing oligonucleotides were deblocked using concentrated aqueous ammonia overnight at room temperature following the manufacturer's instructions. The resulting products were desalted by Sephadex G-25 (NAP-10, GE Healtcare, USA) and purified by reversed-phase HPLC using Nucleosil columns. The yields and purities obtained for the products were around 85% for 5-O6-MeG-TBA and 6-O6-MeG-TBA, and >98% for the rest of the sequences. The length and homogeneity of the oligonucleotides were checked by MALDI-TOF. 5-O6-MeG-TBA [M] = 4731.7 (expected 4737.8), 6-O6-MeG-TBA [M] = 4729.9 (expected 4737.8), MB-5-O6-MeG-TBA [M] = 5828.49 (expected 5826.0), MB-6-O6-MeG-TBA [M] = 5826.80 (expected 5826.0).
Table 1.
Sequences of TBA oligonucleotide derivatives used in the development of the hAGT fluorescence assay. MeG represents O6-methylguanine. MB represents the fluorophore group (FAM), labelled in the 5′ end, and the quencher group (Dabsyl), labelled in the 3′ end of the sequence.
| Abbreviation | Sequence |
|---|---|
| TBA | 5′-GGT TGG TGT GGT TGG-3′ |
| 5-O6-MeG-TBA | 5′-GGT TMe GG TGT GGT TGG-3′ |
| 6-O6-MeG-TBA | 5′-GGT TG Me GTGT GGT TGG-3′ |
| C-TBA | 5′-CCA ACC ACA CCA ACC-3′ |
| MB-TBA | 5′-FAM-GGT TGG TGT GGT TGG-Dabsyl-3′ |
| MB-5-O6-MeG-TBA | 5′-FAM-GGT T Me GG TGT GGT TGG- Dabsyl-3′ |
| MB-6-O6-MeG-TBA | 5′-FAM-GGT TG Me G TGT GGT TGG- Dabsyl-3′ |
| 3-HP-TBA | 5′-A CCT TTT GGT TGG TGT GGT TGG-3′ |
| 6-HP-TBA | 5′-C CAA CCT TTT GGT TGG TGT GGT TGG-3′ |
| 9-HP-TBA | 5′-A CAC CAA CCT TTT GGT TGG TGT GGT TGG-3′ |
The DNA-strand concentration was determined by absorbance measurements (260 nm) by calculating extinction coefficients. Oligonucleotide samples were kept at 4°C until further use. Double-stranded O6-MeG-TBA was formed by annealing equimolar concentrations of complementary oligonucleotide strands at 72°C for 5 min and then allowed to slowly cool to room temperature.
2.4. Melting Temperature Studies
Melting curves were measured by monitoring the absorbance hyperchromicity at 295 and 495 nm in a JASCO V650 spectrophotometer equipped with a Peltier temperature-controlling unit. UV/Vis absorption spectra were recorded at 1°C/min intervals, with a 1-min equilibration time at each temperature; the sample was heated over the range 20–80°C. The buffer solutions used were 10 mM sodium cacodylate pH 7.0 and 100 mM KCl. Sample concentration was around 4 μM. Each sample was allowed to equilibrate at the initial temperature without any external control of temperature for 5 min before the melting experiment began. The melting temperatures (Tm) are the average value of at least one pair of Tm experiments.
2.5. CD Spectra
Samples were prepared as described for the melting experiments by UV spectroscopy. Measurements were conducted in 10 mM sodium cacodylate pH 7.0 and 100 mM KCl. Sample concentration was between 1–4 μM. The CD spectra were recorded on a Jasco J-810 spectropolarimeter attached to a Julabo F/25HD circulating water bath in 1 cm path-length quartz cylindrical cells. Spectra were recorded at room temperature using a 100 nm/min scan rate, a spectral band width of 1 nm and a time constant of 4 s. All the spectra were corrected with the buffer blank and plotted using Origin software.
2.6. Human Recombinant hAGT Protein
Full-length hAGT was overexpressed and purified as previously described [36]. Briefly, hAGT protein cloned in the pet-21a (+) (Novagen) vector was expressed in the E. coli strain Rosetta. Once the culture reached an OD600 value of 0.98, hAGT was induced by adding 1 mM IPTG (Sigma) and left for 4 h at 30°C. The pellet from a 1-L culture was disrupted by sonication and centrifuged. The supernatant was filtered, loaded into a HiTrap FF column (GE Healthcare) with buffer 350 mM NaCl, 20 mM Tris pH 8, 20 mM imidazole, and 1 mM BME, and then eluted with an imidazole (Fluka) gradient up to 500 mM in the same buffer. Finally, the protein was loaded into a Superdex 75 16/60 column (GE Healthcare) with the following buffer: 200 mM NaCl (Merck), 20 mM Tris pH 8.0 (Merck), 10 mM DTT (Sigma) and 0.1 mM EDTA (Sigma). The protein was concentrated to 2 mg/mL in this buffer and kept at −20°C in the presence of 40 % glycerol. The same protocol was used for the purification of the inactive mutant hAGT-C145S, cloned in the pet-28a (+) vector (Novagen), and expressed in the E. coli strain BL21.
2.7. HPLC hAGT Assay
Assays were conducted using double-stranded 5-O6-MeG and 6-O6-MeG TBA paired with TBA complementary sequence or using single-stranded 5-O6-MeG and 6-O6-MeG TBA.
In order to measure the dealkylation of O6-MeG, 50 pmol of the O6-MeG-TBA substrates were incubated with a range of concentration of hAGT (16 nM to 1.6 μM) to a final volume of 120 μL in a reaction buffer (200 mM NaCl, 50 mM Tris pH 8.0, 1 mM DTT, and 5 mM EDTA). Several incubation times were tested (30, 60, 90, 120, 360, and 1440 min) at 37°C and the reaction was stopped by heating the samples at 72°C for 5 min. The reaction products were analyzed by HPLC on a Nucleosil analytical column at 60°C or room temperature, depending on the substrates used in the experiment (double- or single-stranded TBA). The HPLC flow rate was 1 mL/min, and a gradient of 10%–40% acetonitrile for 20 min was used.
2.8. Fluorescence Assay for hAGT Activity
The fluorescence assay was performed in a multidetection microplate reader biotek FLx800 in optical 96-well Optical btm reaction plates (Nunc, USA). Full-length hAGT recombinant protein (207 amino acids) was used for the assay and the hAGT-C145S inactive mutant was used as a negative control. The reaction was performed in a total volume of 50 μL in each well, incubating increasing concentrations of hAGT (5, 10, 20, 40, 60, and 80 nM) enzyme in reaction buffer (200 mM NaCl, 50 mM Tris pH 8.0, 1 mM DTT, 5 mM EDTA, and 20 mM KCl). The assay of hAGT was then initiated by the addition of 5 μL of different concentrations (5, 10, 25, and 50 nM) of fluorescently labelled MB-O6-MeG-TBA substrate and this solution was then placed in a microplate reader system. Fluorescence was measured every minute for 20 min or 40 min at excitation and emission wavelengths of 485 and 535 nm, respectively. Averages over three readings were taken for each condition tested. Each experiment was performed in triplicate.
3. Results and Discussion
The aim of this study was to develop a real-time hAGT activity assay based on the detection of differences between extended and folded conformations of TBA. Our working hypothesis was that the quadruplex structure of TBA is disrupted when a central guanine is replaced by an O6-methylguanine. The TBA sequence also contains a fluorophore and a quencher group attached to 5′ or 3′ end, respectively. In the presence of O6-methylguanine, methylated TBA unfolds and the fluorophore and the quencher become physically separated beyond the Förster distance. When the repair protein hAGT is added to the methylated aptamer, the enzyme removes the methyl group from the mutated guanine, thus allowing TBA to fold back into its chair-like conformation. As a result, the quencher comes closer to the fluorophore and blocks its fluorescence, as illustrated in Figure 1. This loss of fluorescence is quantified as a direct measurement of hAGT activity.
Figure 1.
Scheme of fluorescence-based hAGT assay. The substrate is the thrombin-binding aptamer modified by an O6-methylguanine (Me) in extended conformation, with a fluorophore and quencher in the opposite ends of the sequence. The refold of the G-quadruplex structure of TBA is dependent on the removal of the methyl adduct by alkyl-guanine-DNA-O6-alkyltransferase (hAGT). This repair moves the quencher and the fluorophore molecules in close proximity and blocks fluorescence.
3.1. Synthesis of Modified G-Quadruplex Sequences
The G-quadruplex sequences used in this study are shown in Table 1. Oligonucleotide synthesis was performed by the solid-phase 2′-cyanoethyl-phosphoramidite method [37]. Ammonia treatment was performed at room temperature overnight to minimize decomposition of O6-methylguanine. For the same reason, the dimethylformamidino group was selected for the protection of 2′-deoxyguanosine [38]. The sequences were characterized by HPLC and mass spectrometry, which provided the expected molecular weights. The yields of the isolated molecular beacon oligonucleotides after HPLC purification and desalting were in the range of those obtained for unmodified oligonucleotides.
3.2. Thermal Stability of Methylguanine-Modified TBA
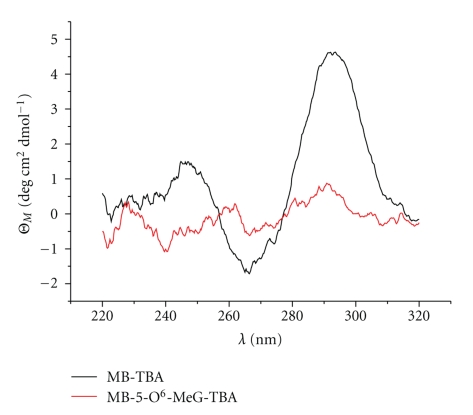
In order to induce the unfolding of the quadruplex structure of TBA, we introduced an O6-methyl-guanine at position 5 or 6 of TBA. Melting curves of the modified sequences were performed by UV spectroscopy and compared with the unmodified sequence. These experiments were carried out at pH 7 in buffer containing 10 mM sodium cacodylate and 100 mM KCl, which is predicted to stabilize G-quadruplexes structures. We did not observe a melting temperature at 295 nm for 5-O6-MeG-TBA or 6-O6-MeG-TBA and the corresponding molecular beacons (see Figures S1–S4 in Supplementary Data Material available online at doi: 10.4061/2010/632041). The absence of this transition is consistent with the disruption of the quadruplex structure. These results contrast with the melting temperature of native TBA (Tm 48°C) and MB-TBA (Tm 46°C). Moreover, circular dicroism of methylated-TBA derivatives did not show the presence of the maximum at 295 nm, which is characteristic of the antiparallel quadruplex of TBA (Figure 2 and Figure S5). These results confirmed that methylation of guanine in either of the two positions of the TBA sequence prevents the formation of the chair structure. This observation confirms our working hypothesis.
Figure 2.
CD spectra of MB-TBA and MB-5-O6-MeG-TBA at 25°C. Buffer conditions: 10 mM sodium cacodylate pH 7.0 and 100 mM KCl, sample concentration 1 μM.
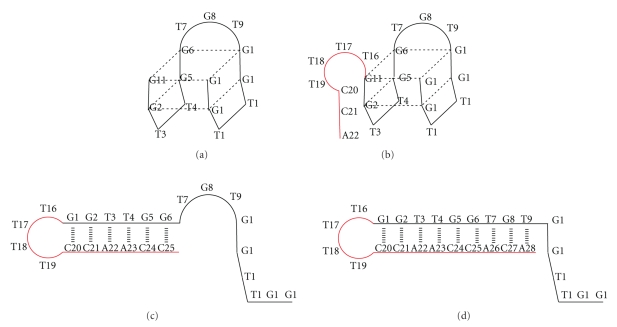
Given that the DNA repair activity exerted by hAGT is higher in double-stranded DNA than single-stranded DNA [39], we studied the stability and the quadruplex formation of elongated self-complementary TBA derivatives (see Table 1). We designed several TBA oligonucleotides elongated in their 3′ end with a subset of self-complementary sequences of diverse sizes. The purpose of these elongations was to check whether the extended sequences have the capacity to form a double strand helix of different lengths and strengths with themselves without disrupting the chair-like structure. For 6-HP-TBA and 9-HP-TBA, we found that the corresponding Tm at 260 nm were 63ºC and 71.8°C, respectively. This observation confirms our hypothesis of a double helix structure that increases in strength as the sequence length increases. However, we did not detect a melting temperature of these two sequences at 295 nm. The absence of a transition at 295 nm is consistent with the absence of an antiparallel quadruplex structure. In contrast, 3-HP-TBA, corresponding to an overhang of only 3 nucleotides, gave a melting temperature of 46°C at 295 nm, which is slightly but similar to that obtained for natural TBA. This result indicates that 3-HP-TBA forms an antiparallel quadruplex instead of a duplex because the overhang is too short to break the chair-like structure. Circular dichroism confirms the quadruplex structure of 3-HP-TBA and the absence of quadruplex structure in 6-HP-TBA and 9-HP-TBA (Figure S6). Models of all these structures are shown in Figure 3.
Figure 3.
Schematic representation of the structure of several derivatives of thrombin-binding aptamers (TBAs) prepared in this study. TBA sequence is shown in black, and T4 loop with different sizes of the complementary sequences are shown in red. (a) Native TBA; (b) TBA-hairpin containing three overhanging complementary nucleotides (3-HP-TBA); (c) TBA-hairpin containing six overhanging complementary nucleotides (6-HP-TBA); (d) TBA-hairpin containing nine overhanging complementary nucleotides (9-HP-TBA). Between 3–6 complementary nucleotides are required to disrupt the intramolecular quadruplex to form an intramolecular duplex.
3.3. HPLC Analysis of DNA Repair Activity of hAGT
In order to observe the efficiency of hAGT to remove alkyl groups from single-stranded oligonucleotides, we performed several assays to determine the optimal conditions of the reaction.
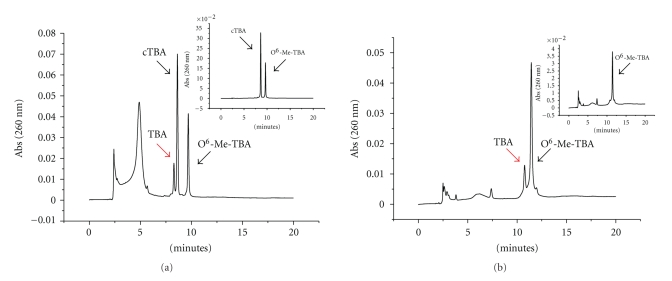
For this purpose, the full-length hAGT was first over-expressed and purified as described previously [36]. Increasing concentrations of the protein were incubated with double-stranded 5-O6-MeG and 6-O6-MeG TBA, annealed with its complementary sequence. Figure 4(a) shows the HPLC profile of the final product of the reaction with double-stranded 5-O6-MeG-TBA. In order to separate the two strands of the TBA substrate (Figure 4(a) top right panel), HPLC analyses were done at 60°C. After incubation with hAGT, we detected a peak with a shorter retention time, which corresponds to the restored TBA sequence caused by the removal of the methyl group. We obtained the same results when we used double-stranded 6-O6-MeG-TBA as a substrate (data not shown). As expected, hAGT activity was not affected by the position of the alkyl group within the sequence. We then tested hAGT activity over single-stranded methylated TBA in the previously optimized conditions and obtained similar results to those shown in Figure 4(b). The top right panel shows the HPLC chromatogram in the absence of hAGT. In this case, the HPLC analyses were performed at room temperature because the substrate was single-stranded TBA. Our results confirmed that hAGT has the capacity to remove methyl groups from single- or double-stranded TBA with the same efficacy. Therefore, we selected single-stranded TBA as substrate for the development of our fluorescence assay.
Figure 4.
HPLC analysis of hAGT activity over double- and single-stranded TBA before and after incubation with hAGT. (a) Repair of double-stranded 5-O6-MeG-TBA by hAGT. The peak labelled as cTBA corresponds to the complementary sequence of 5-O6-MeG-TBA. The top right panel shows HPLC chromatogram in absence of hAGT. The gradient was from 10%–40% acetonitrile for 20 min at 60°C. (b) Repair of single- stranded 5-O6-MeG-TBA by hAGT. The inserted panel shows HPLC chromatogram in the absence of hAGT. Gradient used: 10%–40% acetonitrile, 20 min at room temperature.
3.4. Fluorescence hAGT Activity Assay
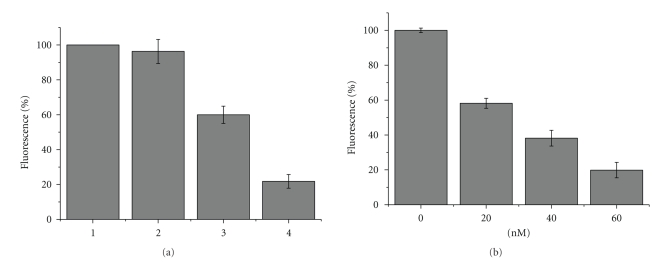
On the basis of the results obtained in the melting temperature and the HPLC experiments, we tested the effectiveness of our proposed model to the DNA repair activity of hAGT by means of fluorescence. First of all, we determined the minimum amount of TBA required to achieve a detectable and reliable difference in intensity compared to the background. As expected, the negative control natural TBA quadruplex gave low background fluorescence, because of the proximity of the fluorophore and the quencher groups in the chair-like structure (Figure 5(a)). Although the fluorescence of MB-TBA was low, this fluorescence was subtracted from the fluorescence value in each experiment. The concentration of 5 nM of fluorescently labelled MB-O6-MeG-TBA was considered the optimal concentration as the fluorescence signal was intense and only a small amount of hAGT protein was required to achieve a substantial decrease in fluorescence in 20–40 min. In the optimal conditions, the presence of hAGT produced a remarkable decrease in fluorescence intensity, caused by the demethylation of the O6 position of guanines, thereby allowing the TBA to form its typical quadruplex structure, which brings together the fluorophore and the quencher groups, as occurred in the negative control. The rate of decrease in fluorescence intensity correlated directly with the amount of hAGT in the reaction mixture (Figure 5(b)). Moreover, the inactive mutant hAGT-C145S did not exhibit any decrease in fluorescence (Figure 5(a)). This result was expected because of the inability of this mutated protein to repair the modified TBA. Figure 5(b) shows the fluorescence intensities for the real time hAGT assay. All these observations are consistent with the hypothesis and design of our FRET method.
Figure 5.
Fluorescence intensity of the real-time hAGT assay, measured at excitation and emission wavelengths of 485 nm and 535, respectively. (a) Illustration of the different controls with 5 nM MB-5-O6-MeG-TBA: (1) Positive control of MB-5-O6-MeG-TBA in the absence of hAGT; (2) Intensity in presence of the inactive mutant hAGT-C145S; (3) Decrease in fluorescence when adding 40 nM active hAGT; and (4) basal fluorescence of 5 nM of MB-TBA. (b) Decrease in intensity caused by the activity of hAGT at a range of concentrations (0, 20, 40, and 60 nM), in all of them the basal fluorescence of MB-TBA was subtracted.
4. Conclusion
Although radioactivity has been widely used in the search of potential inhibitors of hAGT [23, 24], this technique does not allow real-time data acquisition and, in addition, requires radioactive materials. Here, we developed a sensitive fluorescence method that allows the quantification of hAGT activity in a single step and in a straightforward manner. Although another fluorescence method has already been developed for this purpose [25], it requires the addition of a restriction enzyme, followed by the addition of an exonuclease after the hAGT activity reaction. Consequently, although it is a real-time assay, a three-step reaction is required before observing an increase in fluorescence. In contrast, in our assay, a change in fluorescence is detected in a single step (homogeneous), and this method does not depend on the activity of two additional enzymes.
Our assay is based on the detection of conformational changes of TBA in the presence or absence of O6-methylguanines in its structure. The quadruplex structure of TBA is disrupted when a central guanine is replaced by an O6-methylguanine. Fluorophore groups can be added to the modified sequence in order to detect the conformational changes by fluorescence. The variation in fluorescence can be quantified as a direct measurement of hAGT activity. In addition, this technique requires lower amounts of substrate and does not call for the use of radioactive materials. Furthermore, this method can be easily transferred to a high throughput experiment for the evaluation of small molecules as potential hAGT inhibitors [36]. Research in this direction is currently underway in the laboratory.
Supplementary Material
Supplementary data contains the UV, CD spectra and melting curves of TBA derivatives.
Acknowledgments
This work was supported by the Fondo de Investigaciones Sanitarias (Grant PI06/1250), by the Dirección General de Investigación Científica y Técnica (Grant BFU2007-63287, CTQ2010-20541), the Generalitat de Catalunya (2009/SGR/208), the COST project (G4-net, MP0802), the Instituto de Salud Carlos III (CIBER-BNN, CB06_01_0019), and a SNS Miguel Servet contract from the Instituto de Salud Carlos III. The authors thank Tanya Yates for editing the paper and the reviewer for the detailed revision of the paper. This work is dedicated to the memory of Ángel Ramírez Ortiz who was a great mentor, colleague, and friend.
Abbreviations
- BME:
2-mercaptoethanol
- Dabsyl:
3-(N-4′-sulfonyl-4-(dimethylamino)-azobenzene)-3-aminopropyl
- dmf:
Dimethylformamidino group
- DTT:
Dithiothreitol
- EDTA:
Ethylenediaminetetraacetic acid
- FAM:
Fluoresceine
- AGT:
O 6-alkylguanine-DNA alkyltransferase
- hAGT:
Human AGT
- HPLC:
High performance liquid chromatography
- IPTG:
Isopropyl β-D-1-thiogalactopyranoside
- MB:
Molecular Beacon
- O6-MeG:
O6-methylguanine
- TBA:
Thrombin-binding aptamer
- cTBA:
Complementary TBA
- TEAA:
Triethylammonium acetate
- TFA:
Trifluoroacetic acid
- Tris:
Tris(hydroxymethyl)aminomethane
- Tm:
Melting temperature
- UV:
Ultraviolet.
References
- 1.Middleton MR, Margison GP. Improvement of chemotherapy efficacy by inactivation of a DNA-repair pathway. Lancet Oncology. 2003;4(1):37–44. doi: 10.1016/s1470-2045(03)00959-8. [DOI] [PubMed] [Google Scholar]
- 2.Sabharwal A, Middleton MR. Exploiting the role of O6-methylguanine-DNA-methyltransferase (MGMT) in cancer therapy. Current Opinion in Pharmacology. 2006;6(4):355–363. doi: 10.1016/j.coph.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Pegg AE, Singer B. Is O6-alkylguanine necessary for initiation of carcinogenesis by alkylating agents? Cancer Investigation. 1984;2:p. 18. [Google Scholar]
- 4.Saffhill R, Margison GP, O’Connor PJ. Mechanisms of carcinogenesis induced by alkylating agents. Biochimica et Biophysica Acta. 1985;823(2):111–145. doi: 10.1016/0304-419x(85)90009-5. [DOI] [PubMed] [Google Scholar]
- 5.Singer B. O-Alkyl pyrimidines in mutagenesis and carcinogenesis: occurrence and significance. Cancer Research. 1986;46(10):4879–4885. [PubMed] [Google Scholar]
- 6.Jaeckle KA, Eyre HJ, Townsend JJ, et al. Correlation of tumor O6 methylguanine-DNA methyltransferase levels with survival of malignant astrocytoma patients treated with bis-chloroethylnitrosourea: a Southwest Oncology Group study. Journal of Clinical Oncology. 1998;16(10):3310–3315. doi: 10.1200/JCO.1998.16.10.3310. [DOI] [PubMed] [Google Scholar]
- 7.Belanich M, Pastor M, Randall T, et al. Retrospective study of the correlation between the DNA repair protein alkyltransferase and survival of brain tumor patients treated with carmustine. Cancer Research. 1996;56(4):783–788. [PubMed] [Google Scholar]
- 8.Foote RS, Mitra S, Pal BC. Demethylation of O6-methylguanine synthetic DNA polymer by an inducible activity in Escherichia coli. Biochemical and Biophysical Research Communications. 1980;97(2):654–659. doi: 10.1016/0006-291x(80)90314-9. [DOI] [PubMed] [Google Scholar]
- 9.Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Research. 1990;50(19):6119–6129. [PubMed] [Google Scholar]
- 10.Tong WP, Kirk MC, Ludlum DB. Formation of the cross-link 1-[N3-deoxycytidyl],2-[N1-deoxyguanosinyl]ethane in DNA treated with N,N’-bis(2-chloroethyl)-N-nitrosourea. Cancer Research. 1982;42(8):3102–3105. [PubMed] [Google Scholar]
- 11.Yan L, Donze JR, Liu L. Inactivated MGMT by O6-benzylguanine is associated with prolonged G2/M arrest in cancer cells treated with BCNU. Oncogene. 2005;24(13):2175–2183. doi: 10.1038/sj.onc.1208250. [DOI] [PubMed] [Google Scholar]
- 12.Brent TP, Houghton PJ, Houghton JA. O6-alkylguanine-DNA alkyltransferase activity correlates with the therapeutic response of human rhabdomyosarcoma xenografts to 1-(2-chloroethyl)-3-(trans-4-methylcyclohexyl)-1-nitrosourea. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(9):2985–2989. doi: 10.1073/pnas.82.9.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tagliabue G, Citti L, Massazza G, Damia G, Giavazzi R, D’Incalci M. Tumour levels of O6-alkylguanine-DNA-alkyltransferase and sensitivity to BCNU of human xenografts. Anticancer Research. 1992;12(6B):2123–2125. [PubMed] [Google Scholar]
- 14.Pepponi R, Marra G, Fuggetta MP, et al. The effect of O6-alkylguanine-DNA alkyltransferase and mismatch repair activities on the sensitivity of human melanoma cells to temozolomide, 1,3-bis(2-chloroethyl)1-nitrosourea, and cisplatin. Journal of Pharmacology and Experimental Therapeutics. 2003;304(2):661–668. doi: 10.1124/jpet.102.043950. [DOI] [PubMed] [Google Scholar]
- 15.Gerson SL. Clinical relevance of MGMT in the treatment of cancer. Journal of Clinical Oncology. 2002;20(9):2388–2399. doi: 10.1200/JCO.2002.06.110. [DOI] [PubMed] [Google Scholar]
- 16.Margison GP, Povey AC, Kaina B, Santibáñez Koref MF. Variability and regulation of O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2003;24(4):625–635. doi: 10.1093/carcin/bgg005. [DOI] [PubMed] [Google Scholar]
- 17.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nature Reviews Cancer. 2004;4(4):296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 18.Hegi ME, Diserens A-C, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. The New England Journal of Medicine. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 19.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. The New England Journal of Medicine. 2000;343(19):1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 20.Pegg AE, Swenn K, Chae M-Y, Dolan ME, Moschel RC. Increased killing of prostate, breast, colon, and lung tumor cells by the combination of inactivators of O6-alkylguanine-DNA alkyltransferase and N,N’-bis(2-chloroethyl)-N-nitrosourea. Biochemical Pharmacology. 1995;50(8):1141–1148. doi: 10.1016/0006-2952(95)00249-y. [DOI] [PubMed] [Google Scholar]
- 21.Srivenugopal KS, Yuan X-H, Friedman HS, Ali-Osman F. Ubiquitination-dependent proteolysis of O6-methylguanine-DNA methyltransferase in human and murine tumor cells following inactivation with O6-benzylguanine or 1,3-bis(2-chloroethyl)-1-nitrosourea. Biochemistry. 1996;35(4):1328–1334. doi: 10.1021/bi9518205. [DOI] [PubMed] [Google Scholar]
- 22.Xu-Welliver M, Pegg AE. Degradation of the alkylated form of the DNA repair protein, O6-alkylguanine-DNA alkyltransferase. Carcinogenesis. 2002;23(5):823–830. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- 23.Wilson BD, Strauss M, Stickells BJ, Hoal-van Helden EG, Van Helden PD. An assay for O6-alkylguanine-DNA alkyltransferase based on restriction endonuclease inhibition and magnetic bead separation of products. Carcinogenesis. 1994;15(10):2143–2148. doi: 10.1093/carcin/15.10.2143. [DOI] [PubMed] [Google Scholar]
- 24.Dolan ME, Schicchitiano D, Pegg AE. Use of oligodeoxynucleotides containing O6-alkylguanine for the assay of O6-alkylguanine-DNA-alkyltransferase activity. Cancer Research. 1988;48(5):1184–1188. [PubMed] [Google Scholar]
- 25.Moser AM, Patel M, Yoo H, Balis FM, Hawkins ME. Real-time fluorescence assay for O6-alkylguanine-DNA alkyltransferase. Analytical Biochemistry. 2000;281(2):216–222. doi: 10.1006/abio.2000.4554. [DOI] [PubMed] [Google Scholar]
- 26.Wu RS, Hurst-Calderone S, Kohn KW. Measurement of O6-alkylguanine-DNA alkyltransferase activity in human cells and tumor tissues by restriction endonuclease inhibition. Cancer Research. 1987;47(23):6229–6235. [PubMed] [Google Scholar]
- 27.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chemical Society Reviews. 2008;37(7):1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 28.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Current Opinion in Structural Biology. 2009;19(3):239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Hud NV, Smith FW, Anet FAL, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1H NMR. Biochemistry. 1996;35(48):15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 30.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Research. 2006;34(19):5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blume SW, Guarcello V, Zacharias W, Miller DM. Divalent transition metal cations counteract potassium-induced quadruplex assembly of oligo(dG) sequences. Nucleic Acids Research. 1997;25(3):617–625. doi: 10.1093/nar/25.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrovic AG, Polavarapu PL. Quadruplex structure of polyriboinosinic acid: dependence on alkali metal ion concentration, pH and temperature. Journal of Physical Chemistry B. 2008;112(7):2255–2260. doi: 10.1021/jp075873v. [DOI] [PubMed] [Google Scholar]
- 33.Mekmaysy CS, Petraccone L, Garbett NC, et al. Effect of O6-methylguanine on the stability of G-quadruplex DNA. Journal of the American Chemical Society. 2008;130(21):6710–6711. doi: 10.1021/ja801976h. [DOI] [PubMed] [Google Scholar]
- 34.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(8):3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhanot OS, Ray A. The in vivo mutagenic frequency and specificity of O6-methylguanine in φX174 replicative form DNA. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(19):7348–7352. doi: 10.1073/pnas.83.19.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruiz FM, Gil-Redondo R, Morreale A, Ortiz ÁR, Fábrega C, Bravo J. Structure-based discovery of novel non-nucleosidic DNA alkyltransferase inhibitors: virtual screening and in vitro and in vivo activities. Journal of Chemical Information and Modeling. 2008;48(4):844–854. doi: 10.1021/ci700447r. [DOI] [PubMed] [Google Scholar]
- 37.Caruthers MH, Barone AD, Beaucage SL, et al. Chemical synthesis of deoxyoligonucleotides by the phosphoramidite method. Methods in Enzymology. 1987;154:287–313. doi: 10.1016/0076-6879(87)54081-2. [DOI] [PubMed] [Google Scholar]
- 38.Vu H, McCollum C, Lotys C, Andrus A. New reagents and solid support for automated oligonucleotide synthesis. Nucleic Acids Symposium Series. 1990;(22):63–64. [PubMed] [Google Scholar]
- 39.Rasimas JJ, Pegg AE, Fried MG. DNA-binding mechanism of O6-alkylguanine-DNA alkyltransferase: effects of protein and DNA alkylation on complex stability. Journal of Biological Chemistry. 2003;278(10):7973–7980. doi: 10.1074/jbc.M211854200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data contains the UV, CD spectra and melting curves of TBA derivatives.