Abstract
Incontinentia pigmenti (IP) is a rare X-linked dominant disorder characterized by highly variable abnormalities of the skin, eyes and central nervous system. A mutation of the nuclear factor-κB essential modulator (NEMO) located at Xq28 is believed to play a role in pathogenesis and the mutation occurs mostly in female patients due to fatal consequence of the mutation in males in utero. This study was designed to identify the common NEMO rearrangement in four Korean patients with IP. Deletion of exons 4 to 10 in the NEMO, the most common mutation in IP patients, was detected in all of the patients by the use of long-range PCR analysis. This method enabled us to discriminate between NEMO and pseudogene rearrangements. Furthermore, all of the patients showed skewed XCI patterns, indicating pathogenicity of IP was due to cells carrying the mutant X chromosome. This is the first report of genetically confirmed cases of IP in Korea.
Keywords: Incontinentia Pigmenti, NEMO, IKBKG
INTRODUCTION
Incontinentia pigmenti (IP; MIM 308300) is a rare X-linked dominant disorder characterized by abnormal skin pigmentation, retinal detachment and central nervous system defects (1, 2). This syndrome is typically lethal in male but female patients survive as cells expressing the mutant X-chromosome are selectively inactivated. Such negative selection usually results in IP female patients showing an extremely skewed X-chromosome inactivation (XCI) pattern as compared to approximately 10% in a healthy population (3, 4).
The pathogenesis of IP is associated with mutations in the nuclear factor κB essential modulator (NEMO), also known as IKBKG (IKK-gamma gene). The NEMO encodes a regulatory component of the IkB kinase complex that is required for the activation of the transcription factor NF-κB and is fundamental for cell development, survival and function (5). The gene, located on chromosome Xq28, is composed of 10 exons with three alternative noncoding exons. A highly homologous pseudogene, so-called 'ΔNEMO', also exists within a 35.5 kb duplicated fragment opposite to NEMO, and contain NEMO exons 3 to 10. A large deletion of NEMO exons 4 to 10 is found in approximately 80% of IP patients, whereas other mutations such as small nucleotide substitution, deletion and insertion account for a small proportion of patients (5, 6). The deletion alters sequence after nucleotide 399 (from ATG) in the NEMO mRNA and leads to a truncated protein containing the first 133 N-terminal amino acids (1). Although many IP patients have been described in the literature (7), there are no genetically confirmed patients in Korea. In this study, we performed a genetic analysis for Korean patients clinically diagnosed with IP and found the common genomic rearrangement that involved the deletion of exons 4 to 10 in NEMO.
CASE REPORT
Clinical findings
Four unrelated female patients with clinical features of IP were evaluated. All patients were diagnosed shortly after birth as presenting with progressive erythema, vesicular rash and linear hyperpigmentation on the limbs and trunk. Patient 1 was a 15-month-old girl who presented with a linear vesicular eruption on the extremities since birth. On examination, the patient had vesicles and crusted lesions on the extremities, accompanied by swirled hyperpigmentation on the arm, foot and abdomen (Fig. 1A). A fundus examination revealed retinal hemorrhage and macular hypopigmentation in both eyes (Fig. 1B, C). There was no family history of skin disease. Patient 2 was a 19-month-old girl, with a recurrent linear vesicular eruption on the extremities since birth. The patient had no family history of skin disease. Hematological analysis on admission showed marked eosinophilia (32%) with leukocytosis (23,000/µL). A fundus examination revealed retinal hemorrhage in the left eye and developmental milestones were delayed. The mother of the patient had multiple miscarriages. Patient 3 was a 21-month-old girl who also presented with characteristic features of IP including hyperpigmented skin lesions, hypodontia and a family history of skin disease. Slightly delayed developmental milestones were also observed. The mother was noted to have similar skin manifestations and the maternal grandmother had two episodes of miscarriage. Patient 4 was a 27-yr-old female who had one previous miscarriage at 13 weeks gestation. She was diagnosed with IP based on skin manifestations and a family history of skin disease during early childhood (her mother and older sister had similar skin manifestation). A physical examination was unremarkable other than skin manifestation.
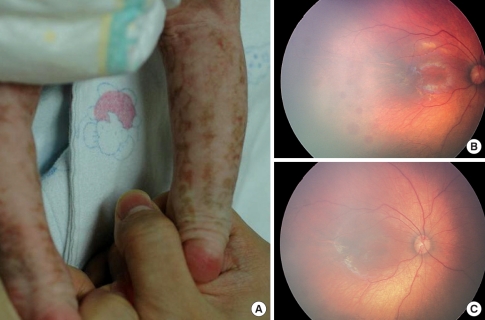
Fig. 1.
Clinical features of Patient 1. (A) Lower leg of Patient 1, showing the hyperpigmented lesion along with Blaschko's lines on the posterior part of the lower limbs. Fundus photographs of the left eye (B) and right eye (C), showing retinal hemorA rhage and macular hypopigmentation.
A recurrent mutation in NEMO
After obtaining written informed consent, PCR-based analyses were performed in the four patients, the parents of Patient 3 and two healthy control subjects in February, 2008. This study was approved by the Samsung Medical Center Institutional Review Board. Multiplex PCR was performed to identify the common deletion as either NEMO or ΔNEMO by using two forward primers (Int3s and Rep3s) and one single reverse primer (L2Rev), described by Steffann et al. (Fig. 2) (8). A 1045-bp sized product was expected for the deletion involving exons 4 to 10 of either the NEMO or ΔNEMO (Int3s and L2Rev), whereas a 733-bp product was expected in all tested participants as an internal amplification control (Rep3s and L2Rev). Confirmatory long-range PCR was also performed to detect the specific genetic rearrangement of the NEMO, described by Bardaro et al. (9). A 2.6-kb product was expected for the pathological NEMO deletion with a forward primer (In2) and a reverse primer (JF3R). In a comparative PCR assay performed with JF3R and a ΔNEMO-specific reverse primer (Rev-2), a 2.5-kb product was expected for the ΔNEMO deletion.
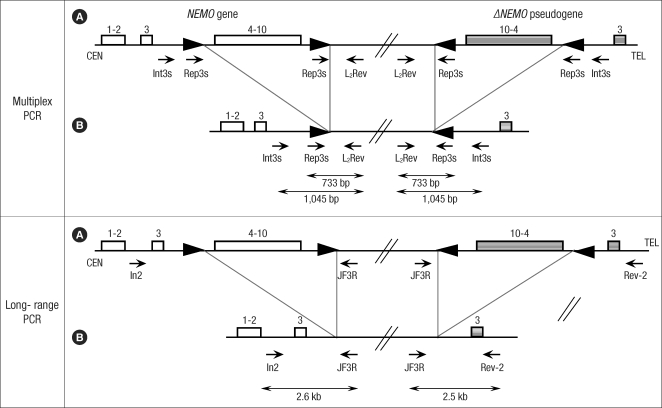
Fig. 2.
Schematic representation of NEMO and ΔNEMO pseudogene location in Xq28. (A) NEMO gene (▭), ΔNEMO pseudogene (▒), (B) rearranged NEMO gene (▭), ΔNEMO pseudogene (▒). In multiplex PCR, forward primers are Int3s (5'-CCA CTC AGG GCT TAG AGC GC-3') and Rep3s (5'-CTC TTT TGA CAA GAA CAC CGG A-3'). Int3s, located within intron 3, matches with the wild-type and rearranged NEMO and pseudogene as well. Rep3s matches with two direct repeats (▸) on both the wild-type NEMO and pseudogene, while it matches with the unique remaining direct repeat on the rearranged NEMO and pseudogene. L2Rev (5'-TCG GAG ACA CAG GAA CCA GCA-3') is the reverse primer. In long-range PCR, In2 (5'-GAG GAC CAA TAC CGA GCA TC-3') and JF3R (5'-CTC GGA GAC ACA GGA ACC AGC A-3') primers amplify a 2.6-kb gene-specific band. JF3R and Rev-2 (5'-GCC ATC TGT TTT TGC GTG TG-3') primers reveal a 2.5-kb pseudogene-specific band.
For the multiplex PCR analysis, all tested individuals showed the 733-bp product, whereas only the four female patients and the mother of Patient 3 showed the 1045-bp product (Fig. 3A). For the long-range PCR analysis, the expected 2.6-kb product was seen in only the four patients and the mother of Patient 3 (Fig. 3B). Also, in a comparative PCR assay performed with JF3R and a ΔNEMO-specific reverse primer (Rev-2), the expected 2.5-kb product was not detected in all tested individuals. These results indicate that the deletion was in the NEMO and not in the ΔNEMO.
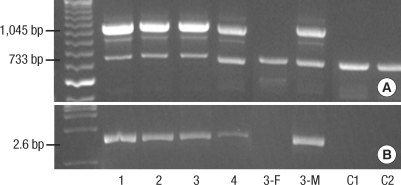
Fig. 3.
Representative results of multiplex-PCR and long-PCR analysis. (A) Multiplex PCR products in all participants and controls (C1, C2). A 1045-bp band corresponding to DNA amplification between the Int3s and L2Rev primers indicates the presence of the common rearrangement found only in IP affected individuals. A 733-bp product serves as an internal control of DNA amplification (Rep3s and L2Rev primers). (B) Result of long-range PCR analysis. The specific NEMO deletion corresponds to the 2.6-kb band (In2 and JF3R primers) detected in the patients as well as in the mother of Patient 3 (3-M). 3-F, the father of Patient 3.
X-inactivation analysis
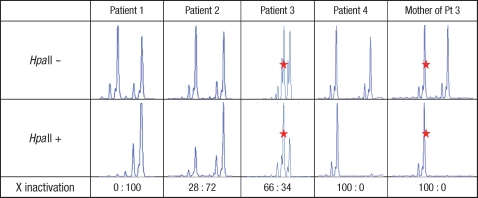
The XCI pattern was determined by PCR analysis of a polymorphic CAG repeat in the HUMARA (10). After digestion with the methylation sensitive enzyme HpaII, a PCR product is obtained only from the inactive X-chromosome. The PCR products were analyzed by GeneScan Software. XCI was calculated as the ratio between the intensities of the PCR products of the two alleles with the smallest allele given first. The presence of skewed XCI was considered if the ratio was ≥65:35, and considered extremely skewed if the ratio was ≥80:20 (11). The heterozygous alleles of the X-chromosomes in each non-digested sample were shown with a CAG repeat polymorphism (Fig. 4). After DNA was digested with HpaII, Patient 1, Patient 4 and the mother of Patient 3 showed only one of the heterozygous alleles, indicating that these had a highly skewed XCI. Patient 2 and Patient 3 showed one distinct peak and one faint peak. These results indicate that all affected individuals had a skewed XCI, although the degree of skewing was different.
Fig. 4.
XCI analyses in affected female individuals. After digestion with HpaII, a PCR product is obtained from the inactive X chromosome only. The red star (⋆) in Patient 3 indicates the maternal allele (273-bp).
DISCUSSION
Bardaro et al. have reported the use of a long-range PCR method that discriminates between rearrangement in the NEMO and ΔNEMO, thus enabling molecular diagnosis for IP (9). However, the absence of an internal control for PCR precludes discrimination between PCR failure in rearranged individuals and the absence of NEMO or ΔNEMO rearrangement, giving false-negative results. Subsequently, Steffann et al. proposed the use of a multiplex PCR method to overcome these limitations (8).
In this study, all affected individuals showed an identical genomic rearrangement involving a deletion of exons 4 to 10 of the NEMO. It has been reported that in Western countries, an identical genomic deletion accounts for 80% of the identified mutations in the NEMO (5). As the same mutation was found in Korean and Japanese IP patients (12, 13), the deletion may be the most common mutational hot spot irrespective of the ethnic background, although the number of the patients was limited.
In this study, the method proposed by Bardaro et al. has been demonstrated to be reproducible; however, we experienced difficulty to identify optimal PCR conditions for DNA amplification. In addition, long-range PCR is a time-intensive and labor-intensive method as compared with multiplex PCR. Therefore, we suggest that the multiplex PCR proposed by Steffann et al. may be useful for screening the existence of the NEMO rearrangement for clinically suspicious IP patients, and the use of long-range PCR proposed by Bardaro et al. should be considered as a confirmatory diagnostic tool for patients with positive findings with screening multiplex PCR testing.
All the four patients showed the classical cutaneous signs of IP and the observed clinical manifestation spectrum was similar to that previous Korean IP study report by Kim et al. (14). For the XCI analysis, all affected individuals showed a skewed XCI pattern. Two patients (Patients 1 and 4) and the mother of Patient 3 showed an extremely skewed XCI and two patients (Patients 2 and 3) showed mild skewing of XCI. The XCI phenotype might be related to the disease phenotype, since Patients 2 and 3 showed relatively severe clinical manifestations such as developmental delay, hypodontia and retinopathy, although all affected individuals had the same mutation. Martinez-Pomar et al. (15) described an IP patient with severe clinical manifestations in the first 30 months of life. At that time, the patient showed a random XCI pattern. At the age of 42 months, however, all clinical signs disappeared, and a completely skewed XCI pattern was observed. Therefore, XCI could modify the clinical phenotype of IP, but has limitations to explain all of the phenotypic manifestations of IP.
This is the first report of genetically confirmed IP patients in Korea. The patients showed a common genomic rearrangement involving the deletion of exons 4 to 10 in NEMO and the mutation spectrum was similar to that previously reported. Although only a small number of patients was investigated, this study indicates that the genetic analysis of the NEMO may be helpful for rapid confirmation of IP diagnosis, prenatal diagnosis and carrier detection.
Footnotes
This work was supported by the Samsung Biomedical Research Institute grant (# SBRI C-A8-205-1) and by a grant of the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare, and Family Affairs, Republic of Korea (A080588).
References
- 1.Berlin AL, Paller AS, Chan LS. Incontinentia pigmenti: a review and update on the molecular basis of pathophysiology. J Am Acad Dermatol. 2002;47:169–187. doi: 10.1067/mjd.2002.125949. [DOI] [PubMed] [Google Scholar]
- 2.Landy SJ, Donnai D. Incontinentia pigmenti (Bloch-Sulzberger syndrome) J Med Genet. 1993;30:53–59. doi: 10.1136/jmg.30.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parrish JE, Scheuerle AE, Lewis RA, Levy ML, Nelson DL. Selection against mutant alleles in blood leukocytes is a consistent feature in Incontinentia Pigmenti type 2. Hum Mol Genet. 1996;5:1777–1783. doi: 10.1093/hmg/5.11.1777. [DOI] [PubMed] [Google Scholar]
- 4.Aradhya S, Courtois G, Rajkovic A, Lewis RA, Levy M, Israel A, Nelson DL. Atypical forms of incontinentia pigmenti in male individuals result from mutations of a cytosine tract in exon 10 of NEMO (IKK-gamma) Am J Hum Genet. 2001;68:765–771. doi: 10.1086/318806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smahi A, Courtois G, Vabres P, Yamaoka S, Heuertz S, Munnich A, Israel A, Heiss NS, Klauck SM, Kioschis P, Wiemann S, Poustka A, Esposito T, Bardaro T, Gianfrancesco F, Ciccodicola A, D'Urso M, Woffendin H, Jakins T, Donnai D, Stewart H, Kenwrick SJ, Aradhya S, Yamagata T, Levy M, Lewis RA, Nelson DL. Genomic rearrangement in NEMO impairs NF-kappaB activation and is a cause of incontinentia pigmenti. The International Incontinentia Pigmenti (IP) Consortium. Nature. 2000;405:466–472. doi: 10.1038/35013114. [DOI] [PubMed] [Google Scholar]
- 6.Fusco F, Bardaro T, Fimiani G, Mercadante V, Miano MG, Falco G, Israel A, Courtois G, D'Urso M, Ursini MV. Molecular analysis of the genetic defect in a large cohort of IP patients and identification of novel NEMO mutations interfering with NF-kappaB activation. Hum Mol Genet. 2004;13:1763–1773. doi: 10.1093/hmg/ddh192. [DOI] [PubMed] [Google Scholar]
- 7.Aradhya S, Woffendin H, Jakins T, Bardaro T, Esposito T, Smahi A, Shaw C, Levy M, Munnich A, D'Urso M, Lewis RA, Kenwrick S, Nelson DL. A recurrent deletion in the ubiquitously expressed NEMO (IKK-gamma) gene accounts for the vast majority of incontinentia pigmenti mutations. Hum Mol Genet. 2001;10:2171–2179. doi: 10.1093/hmg/10.19.2171. [DOI] [PubMed] [Google Scholar]
- 8.Steffann J, Raclin V, Smahi A, Woffendin H, Munnich A, Kenwrick SJ, Grebille AG, Benachi A, Dumez Y, Bonnefont JP, Hadj-Rabia S. A novel PCR approach for prenatal detection of the common NEMO rearrangement in incontinentia pigmenti. Prenat Diagn. 2004;24:384–388. doi: 10.1002/pd.889. [DOI] [PubMed] [Google Scholar]
- 9.Bardaro T, Falco G, Sparago A, Mercadante V, Gean Molins E, Tarantino E, Ursini MV, D'Urso M. Two cases of misinterpretation of molecular results in incontinentia pigmenti, and a PCR-based method to discriminate NEMO/IKKgamma dene deletion. Hum Mutat. 2003;21:8–11. doi: 10.1002/humu.10150. [DOI] [PubMed] [Google Scholar]
- 10.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 11.Xinhua B, Shengling J, Fuying S, Hong P, Meirong L, Wu XR. X chromosome inactivation in Rett Syndrome and its correlations with MECP2 mutations and phenotype. J Child Neurol. 2008;23:22–25. doi: 10.1177/0883073807307077. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Kondo H, Uchio E. A case of incontinentia pigmenti in Japan and its genetic examination. Jpn J Ophthalmol. 2007;51:142–145. doi: 10.1007/s10384-006-0412-3. [DOI] [PubMed] [Google Scholar]
- 13.Tada H, Yoshida S, Yamaji Y, Fujisawa K, Ishibashi T. NEMO mutational analysis in a Japanese family with incontinentia pigmenti. Eye. 2007;21:888–890. doi: 10.1038/sj.eye.6702770. [DOI] [PubMed] [Google Scholar]
- 14.Kim BJ, Shin HS, Won CH, Lee JH, Kim KH, Kim MN, Ro BI, Kwon OS. Incontinentia pigmenti: clinical observation of 40 Korean cases. J Korean Med Sci. 2006;21:474–477. doi: 10.3346/jkms.2006.21.3.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Pomar N, Munoz-Saa I, Heine-Suner D, Martin A, Smahi A, Matamoros N. A new mutation in exon 7 of NEMO gene: late skewed X-chromosome inactivation in an incontinentia pigmenti female patient with immunodeficiency. Hum Genet. 2005;118:458–465. doi: 10.1007/s00439-005-0068-y. [DOI] [PubMed] [Google Scholar]