Abstract
Bartonella spp. prevalence in small mammals and their ectoparasites was investigated in Taiwan. Blood samples were obtained from 66 rats, 20 shrews, 276 mites (Laelaps spp.), 74 fleas (Xenopsylla cheopis), 81 lice (Polyplax spp.), and 47 ticks (41 Dermacentor spp. and 6 Ixodes spp.). Bartonellae were isolated or detected in 27 (31.4%) animals. Bartonella DNA was detected in 48 (64.9%) fleas and 11 (64.7%) pooled lice samples, but not in mite and tick samples. Bartonella phoceensis, B. queenslandensis, B. tribocorum, B. elizabethae, and B. rattimassiliensis were isolated or detected in bacteremic mammals. For the first time in Taiwan, B. tribocorum, B. elizabethae, B. queenslandensis, and a B. rochalimae-like strain were detected in fleas, and B. tribocorum, B. phoceensis, and B. rattimassiliensis were detected in lice obtained from small mammals. A broader range of Bartonella species was identified in the ectoparasites than in the small mammals.
Introduction
Numerous Bartonella species have been isolated from rodents, including B. birtlesii,1 B. doshiae,2 B. elizabethae,3 B. grahamii,2 B. phoceensis,4 B. rattimassiliensis,4 B. taylorii,2 B. tribocorum,5 B. washoensis,6 B. vinsonii subsp. arupensis, B. vinsonii subsp. vinsonii,7,8 B. rochalimae,9 B. rattaustraliani, B. queenslandensis, B. cooperplainsensis7 and many more unnamed species. In Thailand, two rodents (Rattus surifer) were infected with a new genotype of Bartonella (Candidatus Bartonella thailandensis).8 Several rodent-associated Bartonella species have been linked to human diseases. Bartonella elizabethae has been detected in a human with endocarditis;10 B. grahamii DNA was detected in ocular fluids of a patient with neuroretinitis11 and antibodies to B. grahamii were detected in a patient with bilateral retinal artery branch occlusions;12 B. vinsonii subsp. arupensis was isolated from a cattle rancher with bacteremia and fever13 and was associated with two patients with endocarditis;14,15 B. washoensis was isolated from a human case of fever and myocarditis;6 and B. rochalimae caused bacteremia, fever, and splenomegaly in a human patient.16
In Asia, many Bartonella species have been isolated from a wide range of small mammals. Bartonella isolates were obtained from seven rodent species of four genera in China: Apodemus (A. chevrieri, A. draco, and A. latronum), Eothenomys (E. miletus), Rattus (R. norvegicus and R. flavipectus), and Mus (M. pahari). Cultures obtained from Rattus rats were genetically related to B. elizabethae, a recognized human pathogen.17,18
In Japan, B. grahamii has been isolated from Apodemus speciosus and A. argenteus; B. tribocorum and B. elizabethae from Apodemus mice and R. rattus; B. rattimassiliensis and B. phoceensis from R. rattus; B. taylorii from A. speciosus and Myodes (formerly Clethrionomys) voles. Two novel Bartonella species groups were isolated from A. speciosus and A. argenteus.19 In Russia, B. grahamii and/or B. taylorii were isolated from six species of small mammals (A. flavicollis, A. uralensis, M. glareolus, M. musculus, Microtus arvalis, and Sorex araneus).20 In Thailand, most of the isolates from rodents Bandicota indica, R. losea, and R. rattus were closely related to B. grahamii and B. elizabethae. Two additional isolates from B. indica clustered together and were nearly identical to an isolate from A. draco collected in southern China.21 In a recent study from Thailand, one isolate from a Berylmys berdmorei was close to B. tribocorum, and several Bartonella species were detected in rodents.22 Bartonella coopersplainensis, B. elizabethae, B. phoceensis, B. rattimassiliensis, B. tribocorum, and an unknown genogroup were detected from Rattus spp., and Bartonella species isolated from Bandicota spp. included B. coopersplainensis, B. rattimassiliensis, and B. tribocorum.22 Bartonella from Mus spp. were clustered with B. coopersplainensis or B. rattimassiliensis.22 In Taiwan, a recent study in small mammals reported isolation of Bartonella species from 41.3% of 310 small mammals. Bartonella rattimassiliensis was isolated from R. norvegicus and Suncus murinus, B. tribocorum from R. norvegicus, S. murinus and R. rattus, B. grahamii, and B. elizabethae from R. norvegicus and R. losea, and B. phoceensis from R. norvegicus.23
In vectors, several Bartonella species were detected from hematophagous arthropods feeding on rodents. Ctenophthalmus nobilis, a rodent flea, has been demonstrated as a competent vector for transmission of B. grahamii and B. taylorii in rodents.24 However, Bartonella DNA has also been detected in lice,25,26 fleas,25 Mesostigmatid mites,27 and ticks27 collected on small mammals; these ectoparasites are considered potential vectors for various Bartonella species. The role of these potential vectors in Bartonella transmission among rodent hosts needs to be confirmed.
The objectives of this study were to investigate the prevalence of Bartonella infection in small mammals in Taiwan and in the ectoparasites infesting these animals, especially in rodent populations close to domestic animal farms; and to determine Bartonella infections with respect to animal sex and age and presence or absence of ectoparasites. This investigation could help in better understanding the epidemiology of Bartonella infection in small mammals in Taiwan, and thus would help provide better strategies for disease control.
Materials and Methods
Collection of blood from small mammals.
Blood samples and ectoparasites were obtained from 88 trapped small mammals, 62 R. norvegicus, 21 S. murinus, and 5 R. rattus, during July 2008–April 2009. The small mammals were anesthetized with zoletil 50 (Virbac Laboratories, Carros, France) by intramuscular injection. Approximately 2 mL of whole blood from each small mammal was obtained by cardiac puncture into plastic tubes containing EDTA and frozen at −80°C before testing. Animals were then euthanized as described.23 Animal species, sex, estimated age, and presence or absence of ectoparasites were recorded for each animal at the time of sampling.
Collection and morphologic identification of ectoparasites.
Ectoparasites attached to the small mammals, such as ticks, mites, fleas, and lice, were carefully pulled out with tweezers or a flea comb and then put in individual vials (all parasites from the same animal were put into the same vial). In the laboratory, each parasite was given an identification number and then stored in separate microtubes containing 70% ethanol. The parasite species, stage, and sex were determined by using an optical microscope (Stemi 2000-C; Zeiss, Oberkochen, Germany) and morphologic keys,28–31 and the results were recorded in a spreadsheet.
Isolation of Bartonella spp. in small mammals.
Two culture methods, direct culture and pre-enrichment culture, were used to isolate Bartonella spp. from each small mammal blood sample. For direct culture, 100 µL of blood were cultured on chocolate agar plates and incubated in an atmosphere of 5% CO2 at 35°C and examined weekly for at least four weeks.32 For pre-enrichment culture, blood samples were incubated in liquid Bartonella alpha-Proteobacteria growth medium and incubated in a water-saturated atmosphere of 5% CO2 at 35°C as described.33 After a seven-day incubation period, 100 µL of the sample from the liquid medium were then sub-cultured on chocolate agar plates as described above and examined weekly for at least four weeks. The rest of the blood was stored at −80°C until DNA extraction.
DNA extraction from isolated colony, small mammals, and ectoparasites.
For each chocolate plate that yielded suspected Bartonella isolates, two colonies were randomly selected for DNA extraction. Each of the two colonies picked from the original plate was heated at 100°C for 10 minutes in 50 µL of sterile water. After centrifugation for 8 minutes at 18,000 × g, supernatants were kept at −20°C until tested by polymerase chain reaction (PCR) as a DNA template. Two hundred microliters of each whole blood sample was used to extract DNA by using the DNeasy Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
When the number of ectoparasites was ≤ 5 for each developmental stage on a given animal, every ectoparasite on this animal was collected and subjected to DNA extraction. If the number of ectoparasites was > 5 on the sampled animal, 5 nymph ticks, 5 adult mites, and 5 adult fleas attached to that animal were randomly selected for molecular detection.
Because larval and nymph ticks, adult and nymph mites, adult fleas, and nymph lice were small, whole ectoparasites or pooled tick larvae (≤ 10), pooled mite nymphs (≤ 10), and pooled louse nymphs (≤ 10) were used for DNA extraction. When > 50 ectoparasites were collected from an animal, only 5 pools of 10 larvae or 10 nymphs were selected for molecular detection. DNA extraction was performed as described above.
Polymerase chain reaction and sequencing for Bartonella detection in isolated colonies and small mammal blood, and DNA of ectoparasites.
A PCR of each DNA sample from isolated colonies, animal blood, and ectoparasites were performed with Bartonella-specific primers of the citrate synthase (gltA) gene as described,34 followed by direct sequencing of purified PCR products. The PCR amplification was performed as follows, after minor modifications: 1 cycle for 10 minutes at 95°C; 45 cycles for 30 seconds at 95°C, 1 minute at 57°C, and 2 minutes at 72°C; and 1 cycle for 5 minutes at 72°C for a final extension. The PCR amplification was carried out in a 25-μL reaction volume that contained 2.5 μL of extracted DNA, 2.5 μL of 10× reaction buffer containing 15 mM MgCl2, 2 μL of dNTP master mix (1.25 mM of each of the four dNTPs), 0.25 μL (10 μM) of each primer, 0.125 μL of Gold Taq DNA polymerase, and 17.375 μL of sterile water. Bartonella henselae Houston-1 (ATCC no. 49882) and double-distilled water were used as positive and negative controls for Bartonella detection. After electrophoresis in 2% agarose gel for 2 hours at 100 volts, the gels were stained with ethidium bromide and PCR products were visualized by ultraviolet fluorescence.
Sequences of positive Bartonella PCR products from isolated colonies, animal blood, and ectoparasites were determined by using an ABI 3730 XL DNA Analyzer (Mission Biotech Co. Ltd., Taipei, Taiwan).
Phylogenetic and statistical analyses.
Phylogenetic and molecular evolutionary analyses were conducted by using MEGA version 4 (http://www.megasoftware.net/). After sequence alignment, DNA samples having the same sequence were assigned a group number to build the phylogenetic tree. Determination of the Bartonella species with the highest DNA similarity value was performed by comparing the sequence of each group with corresponding fragments deposited in GenBank. Phylogenetic analysis was performed on the aligned DNA sequences using neighbor-joining method by Kimura 2-parameter model, and bootstrap support was calculated by using 1,000 bootstraps.35,36
Data were analyzed by using StatXact version 8.0 (Cytel Inc., Cambridge, MA), and exact chi-square tests were used to evaluate the association between 1) the infection status, species, sex, age, and location of an animal; 2) either the presence of Bartonella DNA in ticks, mites, fleas, or lice and the species, sex, and age of an animal, the sex of the flea or mite, and stage of the tick or mite; 3) Bartonella infection in animals and ectoparasite infestation status for these animals; and 4) Bartonella infection in animals and the presence of Bartonella DNA-positive ectoparasites. A two-tailed Fisher's exact test was used when expected numbers of observations were < 5. The Mann-Whitney test was performed to evaluate differences in ectoparasite burden between farm types conditional on rodent species (Rattus spp. versus Suncus spp.) and between rodent species conditional on farm type. A P value < 0.05 was considered to be statistically significant.
Additional PCR and sequencing for Bartonella detection.
After phylogenetic and molecular evolutionary analyses, sequences identified to have a sequence similarity value < 95% with known Bartonella species were tested with other genes for further molecular identification. PCR analyses of the two 16S–23S internal transcribed spacer (ITS) regions, RNA polymerase (rpoB), cell division (ftsZ), and 16S rDNA genes were performed using five pairs of Bartonella-specific primers as described.37–40 For the ITS region, two bands from the ITS pair 1 region and two bands from the ITS pair 2 region after gel expression were excised. DNA was then extracted from the gel and sequenced. The PCR reaction mixtures, gel expression, and sequence performance were the same as described above.
Results
Collection of blood from small mammals.
Overall, 88 small mammals were trapped in 3 counties in central Taiwan (Table 1). Of these 88 mammals, blood samples could not be obtained from 2 animals (one R. norvegicus at one swine farm and one S. murinus at one cattle farm), but ectoparasites were obtained when present from all 88 animals. Among these samples, 42 (31 Rattus and 11 Suncus) were collected from 7 cattle farms (median = 6 small mammals/farm, range = 1–18 small mammals/farm) and 29 animals (24 Rattus sp. and 5 Suncus) from 6 swine farms (median = 5 animals/farm, range = 1–9 animals/farm). Overall, a higher ectoparasite burden for the small mammals, mainly rats, captured on cattle farms (rats: median = 7 ectoparasites/animal, range = 0–107 ectoparasites/animal; shrews: median = 0 ectoparasite/animal, range = 0–5 ectoparasites/animal) was observed compared with the animals captured on swine farms (rats: median = 0 ectoparasite/animal; shrews: median = 0 ectoparasite/animal, range = 0–8 ectoparasites/animal). Interestingly, among Rattus spp., there was a significant difference between farm types (P = 0.004) but not among Suncus spp. (P = 0.64) and between small mammals species on the cattle farms (P = 0.0002) but not on the swine farms (P = 0.97).
Table 1.
Bartonella spp. detection in small mammals, Taiwan*
| Animal species | No. animals (no. PCR/culture+, %) | Sex | Age | Location | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F | M | A | J | CF | SF | S | AS | H | GF | DF | ||
| Rattus norvegicus | 61 (22, 36) | 35 (11)† | 26 (11) | 40 (18) | 21 (4) | 28 (19) | 22 (1) | 2 (1) | 5 | 2 | 1 (1) | 1 |
| Suncus murinus | 20 (4, 20) | 13 (2) | 7 (2) | 3 (1) | 17 (3) | 10 (3) | 5 (1) | 4 | 1 | 0 | 0 | 0 |
| Rattus rattus | 5 (1, 20) | 0 | 5 (1) | 3 (1) | 2 | 3 (1) | 1 | 1 | 0 | 0 | 0 | 0 |
| Total | 86 (27, 31.4) | 48 (13) | 38 (14) | 46 (20) | 40 (7) | 41 (23) | 28 (2) | 7 (1) | 6 (0) | 2 (0) | 1 (1) | 1 (0) |
Values in parenthesis are number of positive animals; last number in second column is a percentage. A = adult; J = juvenile; CF = cattle farm; SF = swine farm; S = university campus; AS = animal shelter; H = housing unit; GF = goose farm; DF = duck farm.
Two small mammals, 1 R. norvegicus, and 1 Suncus murinus could not be bled but were examined for ectoparasites.
Collection and morphologic identification of ectoparasites.
Ectoparasites were obtained from 54 (61.4%) of the 88 small mammals (Table 2). Of the 54 animals, 43 (79.6%) were R. norvegicus, 9 (16.7%) were S. murinus, and 2 (3.7%) were R. rattus. Seventeen (31.5%) animals were adult females, 12 (22.2%) were adult males, 15 (27.8%) were juvenile females, and 10 (18.5%) were juvenile males. From these ectoparasites, 276 mites, 74 adult fleas, 81 nymph lice, and 47 ticks were selected for identification and DNA extraction (Table 2). All mites belonged to the genus Laelaps. All fleas and lice were identified as Xenopsylla cheopis and Polyplax spp., respectively.Most of the ticks belonged to the genus Dermacentor, and only six nymphs were identified as Ixodes spp.
Table 2.
Bartonella spp. detection in ectoparasites, Taiwan
| Ectoparasite | Species | Stage | No. ectoparasites (PCR+)* | Rattus norvegicus (PCR+) | Suncus murinus (PCR+) | Rattus rattus (PCR+) |
|---|---|---|---|---|---|---|
| Flea | Xenopsylla cheopis | Adult | 74 (48; 64.8%) | 60 (46) [F: 37 (31) and M: 23 (15)] | 14 (2) [F: 7 (1) and M: 7(1)] | 0 |
| Louse | Polyplax spinulosa | Nymph | 81 in 17 pools (11; 64.7%) | 16 (11) | 1 | 0 |
| Mite | Laelaps echianinus | Adult | 38 (0) | 0 | 33 [F 32 and M 1] | 5 [F 5] |
| Laelaps spp. | Nymph | 238 in 41 pools (0) | 39 | 1 | 1 | |
| Tick | Ixodes spp. | Nymph | 6 (0) | 0 | 6 | 0 |
| Dermacentor spp. | Nymph | 6 (0) | 5 | 1 | 0 | |
| Dermacentor spp. | Larvae | 35 in 5 pools (0) | 4 | 1 | 0 |
PCR = polymerase chain reaction.
Isolation of Bartonella spp. in small mammals and DNA extraction from isolated colonies, small mammal blood, and ectoparasites.
The blood of 23 (26.7%) animals yielded suspected Bartonella colonies on chocolate agar plates, including 5 (5.8%) animals that also yielded suspected Bartonella colonies when using the pre-enrichment culture technique.
DNA was extracted from 44 colonies obtained from 23 animal blood samples (2 colonies for 21 animals and 1 colony for 2 animals) by direct culture and from 10 colonies obtained from 5 animals (2 colonies for each animal) by pre-enrichment culture. DNA was extracted from all 86 animal blood samples, 38 selected adult mites, 74 adult fleas, and 12 nymph ticks. The 238 nymph mites, 81 nymph lice, and 35 tick larvae were combined in 41, 17, and 5 pools, respectively (range = 1–10 nymphs or larvae/pool), and DNA was extracted from these pools.
Polymerase chain reaction for Bartonella detection from DNA of isolated colonies, small mammal blood, and ectoparasites.
The 54 DNA samples (44 from direct culture and 10 from pre-enrichment culture) from the 23 animal blood culture–positive samples were confirmed to be positive for Bartonella by using the gltA primers. Of the 86 DNA samples from the small mammals, Bartonella DNA was detected in 18 (21%) animals by using the same primers. Bartonella spp. were detected in 23 (56.1%) of the 41 animals for which blood was obtained on the cattle farms (19 R. norvegicus, 3 S. murinus, and 1 R. rattus (Table 1). One R. norvegicus and one S. murinus (7.1%) for the 28 animals for which blood was obtained on the swine farms were bacteremic for Bartonella spp. One (14.3%) of the seven small mammals from the university campus and the R. norvegicus from a farm raising geese were also detected as positive for Bartonella spp. by PCR (Table 1). No Bartonella species were detected in animals from the animal shelter, housing units, and the duck farm.
Of the 74 adult fleas and 17 nymph lice pooled DNA samples, the Bartonella gltA gene was detected in 48 (64.9%) adult fleas and 11 (64.7%) lice pooled DNA samples, but was not detected in mite and tick DNA samples (Table 2). Of the 48 Bartonella PCR-positive adult fleas, 46 (95.8%) were obtained from R. norvegicus and 2 (4.2%) were obtained from S. murinus. The 11 Bartonella PCR-positive louse pooled DNA samples were obtained from R. norvegicus.
Sequencing and phylogenetic analyses.
For the gltA gene, complete sequences could be interpreted for all 54 animal blood isolate DNA samples, but only for 13 (72%) of the 18 PCR-positive samples from animal blood DNA extracts. For ectoparasites, complete sequences could be interpreted for 47 (97.9%) of the 48 PCR-positive samples for the flea DNA samples and for all 11 PCR-positive samples for the lice pooled DNA samples. Strains used as references for comparison of the gltA gene are shown in Figure 1. Most of the sequences were identical or nearly identical to B. rattimassiliensis (99–100%), B. queenslandensis (99%), B. tribocorum (96–100%), B. elizabethae (95–96%), B. phoceensis (94–100%) and B. rochalimae-like [strain Bart. Sp 1-1C from Taiwan] (96–100%) (Table 3).
Figure 1.
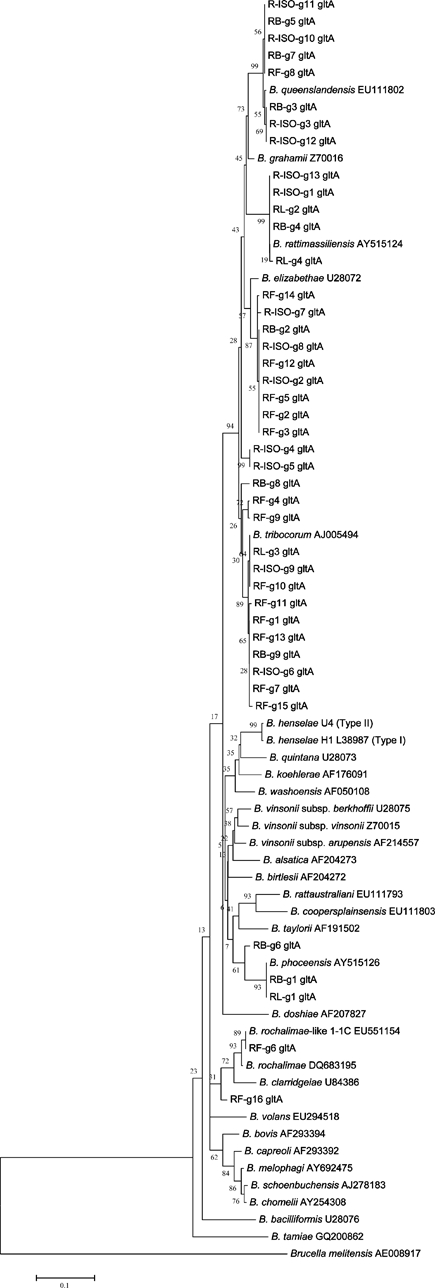
Phylogenetic tree based on 224 basepairs of the citrate synthase (gltA) of Bartonella strains, Taiwan, and constructed by using MEGA version 4 software (http://www.megasoftware.net/). The tree was obtained by using the neighbor-joining method and the Kimura-2 parameter model. Bootstrap support was calculated by using 1,000 bootstrap values.35,36
Table 3.
Sequence homology of Bartonella spp., Taiwan*
| Specimen | B. rattimassiliensis, No. (%) | B. queenslandensis, No. (%) | B. tribocorum, No. (%) | B. elizabethae, No. (%) | B. phoceensis, No. (%) | B. rochalimae-like,† No. (%) |
|---|---|---|---|---|---|---|
| Blood culture | ||||||
| Direct plating | 8 (100) | 8 (99) | 21 (96–99) | 7 (95–96) | 0 | 0 |
| Pre-enrichment | 2 (100) | 8 (99) | 0 | 0 | 0 | 0 |
| Blood DNA | 1 (100) | 4 (99) | 3 (97–99) | 1 (96) | 4 (94–100) | 0 |
| Fleas | 0 | 2 (99) | 24 (97–99) | 18 (96) | 0 | 3 (96–100) |
| Lice | 2 (99–100) | 0 | 1 (100) | 0 | 8 (100) | 0 |
Values are number of sequences and % similarity by source of samples
Strain 1-1C closely related to B. rochalimae detected in Taiwan (9).
By direct culture, two Bartonella species (B. elizabethae and B. tribocorum) were isolated from the blood of the same rodent (R85). However, different Bartonella species were isolated from the same rodent when using the direct culture and the pre-enrichment methods. For instance, rodent R30 and rodent R35 were respectively co-infected with B. elizabethae or B. tribocorum and B. queenslandensis.
When combining the sequencing results from the isolates obtained by the two culture methods and the PCR segments of Bartonella-positive animal blood, there were 27 (31.4%) bacteremic small mammals. Six R. norvegicus captured on three cattle farms were co-infected by two or three Bartonella species. Rodents R4, R6, and R72 were infected with B. rattimassiliensis and B. phoceensis, rodent R30 was infected with B. queenslandensis and B. elizabethae, and rodent R85 was infected with B. elizabethae and B. tribocorum. Rodent R35 was infected with B. queenslandensis, B. tribocorum, and B. phoceensis. Overall, B. phoceensis, B. queenslandensis, B. tribocorum, B. elizabethae, and B. rattimassiliensis were detected in 22 R. norvegicus, B. tribocorum from 4 S. murinus, and B. queenslandensis from 1 R. rattus (Supplementary Table 1, available at www.ajtmh.org).
Fleas obtained from each animal could harbor several Bartonella species. The Bartonella species detected in fleas or lice could be identical or different from the Bartonella species isolated or detected from animals from which fleas or lice were obtained. Bartonella phoceensis was detected in eight Polyplax spp. lice obtained from five Bartonella-positive R. norvegicus, and two of these five rats were also bacteremic for B. phoceensis. Isolates from a Norway rat (R. norvegicus) and a shrew (S. murinus) captured on the swine farms and the two Norway rats captured on the university campus and the goose farm, respectively, were identified as B. tribocorum (Supplementary Table 2, available at www.ajtmh.org).
The phylogenetic tree derived from gltA data set also showed the sequences to be closely related to B. tribocorum, B. rattimassiliensis, B. queenslandensis, B. elizabethae, B. phoceensis, and B. rochalimae (Figure 1).
Risk factors associated with Bartonella spp. infection.
Animal blood.
Among the animal risk factors analyzed (species, sex, age and location) and possible association with bacteremia, only age and location were significant risk factors. The percentage of Bartonella-positive small mammals was significantly higher on cattle farms than on swine farms (cattle farm = 56.1% [23 of 41] versus swine farm = 7.1% [2 of 28]; P < 0.0001) and in adult animals than in juvenile animals (adults = 43.5% [20 of 46] versus juveniles = 17.5% [7 of 40]; P = 0.011).
Fleas.
Bartonella prevalence of fleas obtained from R. norvegicus (76.7% [46 of 60]) was significantly higher than in fleas obtained from S. murinus (14.3% [2 of 14]; P < 0.0001). Fleas obtained from male animals (81.6% [31 of 38]) were more likely to harbor Bartonella spp. than those obtained from female animals (47.2% [17 of 36]; P = 0.0032). Similarly, fleas obtained from adult animals (77.1% [37 of 48]) were more likely to harbor Bartonella spp. than fleas obtained from juveniles (42.3% [11 of 26]; P = 0.0046). Sex of the flea was not identified as a risk factor.
Lice.
Percentages of Bartonella PCR-positive lice obtained from male or female mammals and from juvenile or adult small mammals were not statistically different.
Small mammals and fleas.
Bacteremic small mammals were significantly more likely to be infested by fleas than non-bacteremic mammals (bacteremic animals = 40.7% [11 of 27] versus non-bacteremic animals = 18.6% [11 of 59]; prevalence odds ratio [POR] = 3.0, 95% confidence interval [CI] = 0.96–9.2, P = 0.036). In addition, fleas collected from Bartonella-positive animals were more likely to be Bartonella PCR positive than fleas obtained from Bartonella-negative animals (positive animals = 87.2% [34 of 39] versus negative animals = 37.9% [11 of 29]; POR = 11.1, 95% CI = 3.0–45.9, P < 0.0001).
Small mammals and lice.
Bacteremic animals were more likely than non-bacteremic animals to be infested with lice (bacteremic animals = 33.3% [9 of 27] versus non-bacteremic animals = 6.8% [4 of 59]; POR = 6.9, 95% CI = 1.6–33.5, P = 0.0028). Similarly, lice obtained from Bartonella PCR-positive animals were more likely to be Bartonella PCR positive than lice obtained from Bartonella PCR-negative animals (positive animals = 83.3% [10 of 12] versus negative animals = 20% [1 of 5]) (P = 0.028, by Fisher exact test).
Additional PCR and sequencing for Bartonella spp. detection.
One animal blood DNA sample (R35) and one flea DNA sample (RF85-3) were further analyzed with four other Bartonella genes. Sample RB35 and RF85-3 were PCR positive by using primers specific for the ITS pair 1 and pair 2 regions and ftsZ gene, but not with the other two pairs of primers. For sample RB35, two sequences of the ITS pair 1 region and two sequences of the ITS pair 2 region were obtained. One sequence of ITS pair 1 region and 1 sequence of ITS pair 2 region had 100% similarity with B. tribocorum. The other sequence of the ITS pair 1 region and the other sequence of the ITS pair 2 region had 96% similarity with B. phoceensis. Sequence of the ftsZ gene had 99% similarity with B. phoceensis. When tested by using primers for the gltA gene, rodent R35 was found to be co-infected with three Bartonella species: B. queenslandensis (99% homology), B. tribocorum (99% homology), and B. phoceensis (94% homology). For sample RF85-3, sequences of ITS pair 1 and pair 2 regions and the ftsZ gene had 100% similarity with the B. rochalimae-like strain detected in Taiwan; it had 96% homology with the same strain for the gltA gene.
Discussion
In the present study, several Bartonella species were isolated or detected by PCR in rodents and shrews from Taiwan. Norway rats (R. norvegicus) harbored the largest number of Bartonella species (B. phoceensis, B. queenslandensis, B. tribocorum, B. elizabethae, and B. rattimassiliensis). However, B. tribocorum and B. queenslandensis were also detected in S. murinus and R. rattus, respectively. Our study reports the first isolation of B. queenslandensis from R. norvegicus and B. queenslandensis from R. rattus in Taiwan. These data also confirm the previous report of isolation of strains closely related to B. tribocorum, B. grahamii, B. elizabethae, B. phoceensis, and B. rattimassiliensis in rodents and shrews from central Taiwan.23
An important finding of this study is the wide diversity of Bartonella species identified in the ectoparasites compared with the species isolated from the blood of the small mammals from which these ectoparasites were obtained. One hypothesis could be a selective pressure or competition between Bartonella species, with some species more adapted to their mammalian reservoir in comparison to some species, which are more adapted to the ectoparasite vectors that are possible reservoirs for these Bartonella species. Such an observation was recently reported on a limited number of rodents and their ectoparasites tested.41
A surprising result was the limited value of the pre-enrichment method in detecting Bartonella species from small mammal blood samples. Such results may be caused by our limited experience with this new technique or a lower sensibility of this technique for isolation of rodent Bartonella species compared with a better yield with larger mammals, such as dogs, horses, or humans and/or for different Bartonella species such as B. henselae and B. vinsonii subsp. berkhoffii.33,42–44 Detection of Bartonella species by using PCR to amplify DNA directly from animal blood is often not successful possibly because of the inhibitory effect of hemin.15 Based on the present results, blood culture on chocolate agar seems to be the most sensitive technique for isolation of Bartonella species from small mammals.
Sex of the animals was not identified as a specific risk factor for Bartonella bacteremia in R. norvegicus in a study conducted in southern France.4 Similarly, in our study, sex of the animals was not identified as a risk factor although Bartonella spp. prevalence in males was higher than that in females. Adults were also more likely to be Bartonella PCR positive than juveniles, as reported by Gundi and others,4 in which Bartonella spp. were isolated more often from rats with a weight of 200 grams (considered adults). In contrast, a study of Bartonella infection in squirrels (Spermophilus richardsonii) reported that juvenile squirrels were significantly more likely to be infected with Bartonella spp. than adults.45 In cotton rats (Sigmodon hispidus), there were no differences among the proportion of cotton rats in different age classes infected by different Bartonella genogroups.46 Such differences may be related to the vector infestation rate among the different rodent populations and vector transmission efficiency. Future studies on large-scale rodent and shrew populations and their ectoparasites will be necessary to demonstrate a correlation between Bartonella infection and animal age.
The percentage of Bartonella-positive small mammals was significantly higher on cattle farms than on swine farms, with more than half of the animals trapped on cattle farms being bacteremic for Bartonella spp. Such a difference could result either from the small number of farms tested and the limited number of small mammals trapped on swine and cattle farms or from the level of infestation by ectoparasites. There was no significant difference in the level of ectoparasite infestation of shrews on both types of farms. However, the infestation rate was significantly higher in rats from the cattle farms compared with pig farms. Zoonotic B. elizabethae was only detected from 5 R. norvegicus on cattle farms, which may have contact with domestic animals and humans because the isolates from all other locations were B. tribocorum.
Bartonella tribocorum, B. elizabethae, B. queenslandensis, and Bartonella sp. 1-1C were detected in X. cheopis fleas collected from small mammals. Moreover, B. tribocorum, B. phoceensis, and B. rattimassiliensis were detected in Polyplax spp. lice. Our study is the first report of detection of Bartonella spp. in fleas and lice obtained from small mammals in Taiwan. None of the 274 mites obtained from 37 small mammals and the 47 ticks obtained from 4 small mammals were positive by PCR for Bartonella spp. Interestingly, several rodents or shrews infested with various ectoparasites, as well as the fleas or lice obtained from them, were positive for Bartonella spp. However, the mites or ticks obtained were not positive for Bartonella spp. Such findings raise the possibility that in rodents and shrews in Taiwan, X. cheopis fleas and Polyplax spp. lice could have a greater potential as vectors for Bartonella spp. than Laelaps spp. mites and Dermacentor spp. and Ixodes spp. ticks. However, different tick species, including Haemaphysalis longicornis, H. flava, I. nipponensis, I. turdus, I. persulcatus, and Ixodes spp., and Mesostigmatid mites, have been found to be positive for Bartonella spp. by PCR in South Korea.27 Future studies of larger small mammal populations and ectoparasites infesting them would be useful to better understand the role of the different species of ectoparasites in transmission of Bartonella spp. between animals and possibly to humans.
Small mammals bacteremic for Bartonella spp. were more likely to be infested with fleas and/or lice than non-bacteremic mammals, and fleas or lice obtained from bacteremic animals were more likely to be Bartonella PCR positive than those obtained from non-bacteremic animals. In this study, B. phoceensis, B. queenslandensis, B. tribocorum, B. elizabethae, and B. rattimassiliensis were detected in small mammals and fleas or lice. Bartonella sp. 1-1C, which was previously isolated from R. norvegicus in Taiwan,9 was detected in fleas in the present study. In Taiwan, fleas and lice may be efficient vectors for infection of small mammal hosts with several Bartonella species. However, vector competency of these various flea and lice species has not been determined.
Reeves and others26 hypothesized that B. phoceensis might be associated with or transmitted by Hoplopleura pacifica lice but not P. spinulosa because B. phoceensis was detected only in H. pacifica collected from a B. rattimassiliensis- infected rat. In the present study, B. phoceensis was detected in 8 Polyplax spp. lice obtained from five Bartonella-positive R. norvegicus, and two of these five animals were bacteremic for B. phoceensis. Our findings support the possibility that Polyplax spp. may also be associated with transmission of B. phoceensis.
Small mammals are usually hosts of ectoparasites, such as ticks, in their early stage of life. To decrease the risk of spreading zoonotic Bartonella spp. to humans, it will be important to determine the role of ectoparasites in transmission of these pathogens between animals and possibly to humans and to control the numbers of small mammals surrounding human residential areas and ectoparasites that parasitize them.
Supplementary Material
Note: Supplemental tables are available at www.ajtmh.org.
Footnotes
Financial support: This study was supported by grant 95AS-13.2.1-BQ-B1 from Council of Agriculture, Executive Yuan, Taiwan and grant NSC95-2313-B-005-028-MY2 from the National Science Council, Executive Yuan, Taiwan.
Authors' addresses: Yi-Lun Tsai, Philip H. Kass, and Bruno B. Chomel, Department of Population Health and Reproduction, School of Veterinary Medicine, University of California, Davis, CA 95616, E-mails: yltsai@ucdavis.edu, phkass@ucdavis.edu, and bbchomel@ucdavis.edu. Shih-Te Chuang, Department of Veterinary Medicine, School of Veterinary Medicine, National Chung Hsing University, Taichung 402, Taiwan, E-mail: stchuang@dragon.nchu.edu.tw. Chao-Chin Chang, Graduate Institute of Microbiology and Public Health, School of Veterinary Medicine, National Chung Hsing University, Taichung 402, Taiwan, E-mail: changcc@dragon.nchu.edu.tw.
References
- 1.Bermond D, Heller R, Barrat F, Delacour G, Dehio C, Alliot A, Monteil H, Chomel B, Boulouis HJ, Piemont Y. Bartonella birtlesii sp. nov., isolated from small mammals (Apodemus spp.) Int J Syst Evol Microbiol. 2000;50:1973–1979. doi: 10.1099/00207713-50-6-1973. [DOI] [PubMed] [Google Scholar]
- 2.Birtles RJ, Harrison TG, Saunders NA, Molyneux DH. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol. 1995;45:1–8. doi: 10.1099/00207713-45-1-1. [DOI] [PubMed] [Google Scholar]
- 3.Ellis BA, Regnery RL, Beati L, Bacellar F, Rood M, Glass GG, Marston E, Ksiazek TG, Jones D, Childs JE. Rats of the genus Rattus are reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J Infect Dis. 1999;180:220–224. doi: 10.1086/314824. [DOI] [PubMed] [Google Scholar]
- 4.Gundi VA, Davoust B, Khamis A, Boni M, Raoult D, La Scola B. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J Clin Microbiol. 2004;42:3816–3818. doi: 10.1128/JCM.42.8.3816-3818.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, Lamarque F, Monteil H, Chomel B, Piémont Y. Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. 1998;48:1333–1339. doi: 10.1099/00207713-48-4-1333. [DOI] [PubMed] [Google Scholar]
- 6.Kosoy M, Murray M, Gilmore RD, Jr, Bai Y, Gage KL. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J Clin Microbiol. 2003;41:645–650. doi: 10.1128/JCM.41.2.645-650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gundi VA, Taylor C, Raoult D, La Scola B. Bartonella rattaustraliani sp. nov., Bartonella queenslandensis sp. nov. and Bartonella coopersplainsensis sp. nov., identified in Australian rats. Int J Syst Evol Microbiol. 2009;59:2956–2961. doi: 10.1099/ijs.0.002865-0. [DOI] [PubMed] [Google Scholar]
- 8.Saisongkorh W, Wootta W, Sawanpanyalert P, Raoult D, Rolain JM. “Candidatus Bartonella thailandensis”: a new genotype of Bartonella identified from rodents. Vet Microbiol. 2009;139:197–201. doi: 10.1016/j.vetmic.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Lin JW, Chen CY, Chen WC, Chomel BB, Chang CC. Isolation of Bartonella species from rodents in Taiwan including a strain closely related to ‘Bartonella rochalimae’ from Rattus norvegicus. J Med Microbiol. 2008;57:1496–1501. doi: 10.1099/jmm.0.2008/004671-0. [DOI] [PubMed] [Google Scholar]
- 10.Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, Steigerwalt AG, Weaver RE, Daneshvar MI, O'Connor SP. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerkhoff FT, Bergmans AM, van Der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol. 1999;37:4034–4038. doi: 10.1128/jcm.37.12.4034-4038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serratrice J, Rolain JM, Granel B, Ene N, Conrath J, Avierinos JF, Disdier P, Raoult D, Weiller PJ. Bilateral retinal artery branch occlusions revealing Bartonella grahamii infection [in French] Rev Med Interne. 2003;24:629–630. doi: 10.1016/s0248-8663(03)00224-8. [DOI] [PubMed] [Google Scholar]
- 13.Welch DF, Carroll KC, Hofmeister EK, Persing DH, Robison DA, Steigerwalt AG, Brenner DJ. Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J Clin Microbiol. 1999;37:2598–2601. doi: 10.1128/jcm.37.8.2598-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenollar F, Sire S, Raoult D. Bartonella vinsonii subsp. arupensis as an agent of blood culture-negative endocarditis in a human. J Clin Microbiol. 2005;43:945–947. doi: 10.1128/JCM.43.2.945-947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirillov MIu, Markov AP, Lopyrev IV, Pankratova VN, Levitskiĭ SA, Bashkirov VN, Smirnov GB, Kruglov AN, Osadchaia VA, Frolova GP, Barmina GV, Morozova OA, Kosoy MY. Molecular genetic methods of typing of the bartonellae [in Russian] Mol Gen Mikrobiol Virusol. 2007:8–15. [PubMed] [Google Scholar]
- 16.Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, Mathew SS, Ferraro MJ, Holden JM, Nicholson WL, Dasch GA, Koehler JE. Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med. 2007;356:2381–2387. doi: 10.1056/NEJMoa065987. [DOI] [PubMed] [Google Scholar]
- 17.Ying B, Kosoy MY, Maupin GO, Tsuchiya KR, Gage KL. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am J Trop Med Hyg. 2002;66:622–627. doi: 10.4269/ajtmh.2002.66.622. [DOI] [PubMed] [Google Scholar]
- 18.Li DM, Yu DZ, Liu QY, Gong ZD. Study on the prevalence of Bartonella species in rodent hosts from different environmental areas in Yunnan [in Chinese] Zhonghua Liu Xing Bing Xue Za Zhi. 2004;25:934–937. [PubMed] [Google Scholar]
- 19.Inoue K, Maruyama S, Kabeya H, Yamada N, Ohashi N, Sato Y, Yukawa M, Masuzawa T, Kawamori F, Kadosaka T, Takada N, Fujita H, Kawabata H. Prevalence and genetic diversity of Bartonella species isolated from wild rodents in Japan. Appl Environ Microbiol. 2008;74:5086–5092. doi: 10.1128/AEM.00071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Markov AP, Lopyrev IV, Irkhin AI, Khliap LA, Levitskii SA, Kirillov M, Manuvera VA, Il'ina TS, Pokrovskaia MS, Aleshkin GI, Bashkirov VN, Smirnov GB, Kosoi M. Wild small mammals are the reservoir hosts of the Bartonella genus bacteria in the south of Moscow region [in Russian] Mol Gen Mikrobiol Virusol. 2006;4:8–13. [PubMed] [Google Scholar]
- 21.Castle KT, Kosoy M, Lerdthusnee K, Phelan L, Bai Y, Gage KL, Leepitakrat W, Monkanna T, Khlaimanee N, Chandranoi K, Jones JW, Coleman RE. Prevalence and diversity of Bartonella in rodents of northern Thailand: a comparison with Bartonella in rodents from southern China. Am J Trop Med Hyg. 2004;70:429–433. [PubMed] [Google Scholar]
- 22.Bai Y, Kosoy MY, Lerdthusnee K, Peruski LF, Richardson JH. Prevalence and genetic heterogeneity of Bartonella strains cultured from rodents from 17 provinces in Thailand. Am J Trop Med Hyg. 2009;81:811–816. doi: 10.4269/ajtmh.2009.09-0294. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh JW, Tung KC, Chen WC, Lin JW, Chien LJ, Hsu YM, Wang HC, Chomel BB, Chang CC. Epidemiology of Bartonella infection in rodents and shrews in Taiwan. Zoonoses Public Health. 2009 doi: 10.1111/j.1863-2378.2009.01234.x. Jun 17. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 24.Bown KJ, Bennet M, Begon M. Flea-borne Bartonella grahamii and Bartonella taylorii in bank voles. Emerg Infect Dis. 2004;10:684–687. doi: 10.3201/eid1004.030455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durden LA, Ellis BA, Banks CW, Crowe JD, Oliver JH., Jr Ectoparasites of gray squirrels in two different habitats and screening of selected ectoparasites for bartonellae. J Parasitol. 2004;90:485–489. doi: 10.1645/GE-3299. [DOI] [PubMed] [Google Scholar]
- 26.Reeves WK, Szumlas DE, Moriarity JR, Loftis AD, Abbassy MM, Helmy IM, Dasch GA. Louse-borne bacterial pathogens in lice (Phthiraptera) of rodents and cattle from Egypt. J Parasitol. 2006;92:313–318. doi: 10.1645/GE-717R.1. [DOI] [PubMed] [Google Scholar]
- 27.Kim CM, Kim JY, Yi YH, Lee MJ, Cho MR, Shah DH, Klein TA, Kim HC, Song JW, Chong ST, O'Guinn ML, Lee JS, Lee IY, Park JH, Chae JS. Detection of Bartonella species from ticks, mites and small mammals in Korea. J Vet Sci. 2005;6:327–334. [PubMed] [Google Scholar]
- 28.Teng K. Economic Insect Fauna of China [in Chinese] Beijing: Science Press; 1993. (Fasc 40: Acari: Dermanyssoideae). [Google Scholar]
- 29.Teng K, Jiang Z. Economic Insect Fauna of China [in Chinese] Beijing: Science Press; 1991. (Fasc 39: Acari: Ixodidae). [Google Scholar]
- 30.Walker A. In: The Arthropods of Humans and Domestic Animals. Walker A, editor. London: Chapman and Hall, London; 1994. pp. 25–48. (Ticks-Ixodidae). [Google Scholar]
- 31.Wall R, Shearer D. Veterinary Ectoparasites: Biology, Pathology and Control. Oxford: United Kingdom: Blackwell Science Ltd; 2001. pp. 162–178. 227. [Google Scholar]
- 32.Chomel BB, Abbott RC, Kasten RW, Floyd-Hawkins KA, Kass PH, Glaser CA, Pedersen NC, Koehler JE. Bartonella henselae prevalence in domestic cats in California: risk factors and association between bacteremia and antibody titers. J Clin Microbiol. 1995;33:2445–2450. doi: 10.1128/jcm.33.9.2445-2450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maggi RG, Duncan AW, Breitschwerdt EB. Novel chemically modified liquid medium that will support the growth of seven Bartonella species. J Clin Microbiol. 2005;43:2651–2655. doi: 10.1128/JCM.43.6.2651-2655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 37.Maillard R, Vayssier-Taussat M, Bouillin C, Gandoin C, Halos L, Chomel B, Piemont Y, Boulouis HJ. Identification of Bartonella strains isolated from wild and domestic ruminants by a single-step PCR analysis of the 16S-23S intergenic spacer region. Vet Microbiol. 2004;98:63–69. doi: 10.1016/j.vetmic.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 38.Renesto P, Gouvernet J, Drancourt M, Roux V, Raoult D. Use of rpoB gene analysis for detection and identification of Bartonella species. J Clin Microbiol. 2001;39:430–437. doi: 10.1128/JCM.39.2.430-437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeaiter Z, Liang Z, Raoult D. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsZ gene sequences. J Clin Microbiol. 2002;40:3641–3647. doi: 10.1128/JCM.40.10.3641-3647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heller R, Artois M, Xemar V, De Briel D, Gehin H, Jaulhac B, Monteil H, Piemont Y. Prevalence of Bartonella henselae and Bartonella clarridgeiae in stray cats. J Clin Microbiol. 1997;35:1327–1331. doi: 10.1128/jcm.35.6.1327-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brinkerhoff RJ, Kabeya H, Inoue K, Bai Y, Maruyama S. Detection of multiple Bartonella species in digestive and reproductive tissues of fleas collected from sympatric mammals. ISME J. 2010;4:955–958. doi: 10.1038/ismej.2010.22. [DOI] [PubMed] [Google Scholar]
- 42.Breitschwerdt EB, Maggi RG, Chomel BB, Lappin MR. Bartonellosis: an emerging infectious disease of zoonotic importance to animals and human beings. J Vet Emerg Crit Care (San Antonio) 2010;20:8–30. doi: 10.1111/j.1476-4431.2009.00496.x. [DOI] [PubMed] [Google Scholar]
- 43.Breitschwerdt EB, Maggi RG, Lantos PM, Woods CW, Hegarty BC, Bradley JM. Bartonella vinsonii subsp. berkhoffii and Bartonella henselae bacteremia in a father and daughter with neurological disease. Parasit Vectors. 2010;3:29. doi: 10.1186/1756-3305-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones SL, Maggi R, Shuler J, Alward A, Breitschwerdt EB. Detection of Bartonella henselae in the blood of 2 adult horses. J Vet Intern Med. 2008;22:495–498. doi: 10.1111/j.1939-1676.2008.0043.x. [DOI] [PubMed] [Google Scholar]
- 45.Jardine C, Waldner C, Wobeser G, Leighton FA. Demographic features of Bartonella infections in Richardson's ground squirrels (Spermophilus richardsonii) J Wildl Dis. 2006;42:739–749. doi: 10.7589/0090-3558-42.4.739. [DOI] [PubMed] [Google Scholar]
- 46.Kosoy M, Mandel E, Green D, Marston E, Jones D, Childs J. Prospective studies of Bartonella of rodents. Part II. Diverse infections in a single rodent community. Vector Borne Zoonotic Dis. 2004;4:296–305. doi: 10.1089/vbz.2004.4.296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


