Abstract
This study was performed to determine the nationwide antigenic diversity of Orientia tsutsugamushi in South Korea. Sequence analysis was performed around variable domains I and II of a 56-kDa protein-encoding gene. We used eschar to overcome the disadvantages of conventional serotyping. The serological passive hemagglutination assay (PHA) was assessed based on the genotyping results. We analyzed 153 isolates from scrub typhus patients in major endemic areas and found that Boryong was the major strain (68.6%). New strains were also identified: Taguchi (19.6%), Kanda/Kawasaki (9.2%), and UAP7 (1.3%). PHA yielded significantly fewer positive results among Kawasaki strains (P < 0.001), which are not included in the PHA antigen panel. In South Korea, Boryong was still the predominant strain, but the sequence analysis identified new changes in minor strains (30.1%). This antigenic drift had a negative effect on the PHA results. Periodic surveillance of the contemporary strains using sequence analysis is needed.
Introduction
Scrub typhus, or tsutsugamushi disease, is an endemic disease in the Asia-Pacific region and shows various clinical manifestations. The causative organism, Orientia tsutsugamushi, enters the host through chigger bites on the skin, leaving a necrotic eschar lesion. The eschar is a good source of O. tsutsugamushi genetic material and is available for a relatively long time compared with the buffy coat, which is only useful early in the infection.1–3 O. tsutsugamushi has over 20 antigenically distinct, regionally distributed serotypes.4 Knowledge of serotype epidemiology is important for accurate serologic diagnosis and future vaccine development. Conventional serotyping of O. tsutsugamushi is limited to reference laboratories because of the complexity of the procedure and bio-safety considerations. This is one of the reasons that the prevalence data from endemic regions are sporadic4 and that clinicians are not easily engaged in the clinical studies related to the serotypes or genotypes of O. tsutsugamushi. Since the first report of the nucleotide sequence of a 56-kDa protein-encoding gene of O. tsutsugamushi from the Karp strain, molecular methods have been used to determine variants of O. tsutsugamushi strains.5–7 Although the correlation between the genotype and serological antigen type has not been entirely sorted out, gene-sequence analysis has become the major tool for genetic characterization of Orientia strains.4 The genotyping method also has the advantage of detecting unknown serotypes that are not identified by assays that are based on reactions with monoclonal antibodies against known strains.
Scrub typhus is a communicable disease with a heavy disease burden in South Korea. Since the first confirmation of scrub typhus in native Koreans in 1985, the officially reported annual incidence of scrub typhus has increased markedly, with 6,057 cases reported in 2008 (http://stat.cdc.go.kr).8,9
We carried out this study to survey the prevalence of one time point cross-sectional genotypes of O. tsutsugamushi infecting humans throughout South Korea. Eschar was used as the source of O. tsutsugamushi. We analyzed whether genotypic variation had an effect on the performance of the diagnostic serological test widely used in clinical practices in South Korea.
Materials and Methods
Study subjects.
The study was conducted prospectively from September to December 2006, when scrub typhus was at the peak of its seasonal outbreak. Patients were recruited from eight community-based hospitals (Sanggye Paik Hospital, Ilsan Paik Hospital, Pusan Paik Hospital, Dongguk University Ilsan Hospital, Dankook University Hospital, Namwon Medical Center, Chonbuk National University Hospital, and Sunlin Hospital) located at the major endemic areas of the country, with the exception of the northeastern area. We included clinically suspected scrub typhus patients over 16 years of age who had eschars and at least two of the following manifestations: fever, maculopapular skin rash, regional lymphadenopathy, headache, myalgia, cough, or abdominal discomfort. The eschar specimens were collected on either an inpatient or outpatient basis. The area where scrub typhus was acquired was estimated based on the clinical time sequence and the outdoor activity of the patient. The definite diagnosis of scrub typhus was based on the nucleotide sequence of a 56-kDa antigen gene of O. tsutsugamushi from each patient. Informed consent was obtained from the patients, and the study protocol was approved by the institutional review board at each participating hospital.
Specimen collection and DNA extraction.
Most of the eschars were recovered during the dry stage to leave minimal damage to the normal skin tissue. Template DNA was extracted from eschars stored at −70°C using a commercial kit according to the manufacturer's protocol (QIAamp DNA Mini Kit; QIAGEN, Valencia, CA). The template solution was recovered at a final volume of 200 μL.
DNA amplication by polymerase chain reaction.
To genotype the O. tsutsugamushi strains, a gene fragment covering the variable domains (VDs) I and II of the 56-kDa antigen gene was amplified as previously described.10,11 A set of primers, tsu-A (forward, 5′-TTT CGA ACG TGT CTT TAA GC-3′; nucleotide position −285 to −266 from the start codon of the 56-kDa gene based on the Gilliam strain) and tsu-B (reverse, 5′-ACA GAT GCA CTA TTA GGC AA-3′; 847–865), was used for the initial amplification. All polymerase chain reaction (PCR) reagents were used as recommended by the supplier (AccuPower PCR PreMix; Bioneer, Taejon, South Korea). Briefly, 10 μL of template DNA and 250 μM of both primer tsu-A and primer tsu-B were used in each 50-μL reaction mixture. PCR was initiated at 94°C for 2 minutes. Thermocycling was performed for 30 cycles of denaturation at 94°C for 1 minute, annealing at 55°C for 1.5 minutes, and extension at 72°C for 2 minutes, with a final extension at 72°C for 7 minutes (GeneAmp PCR system 2400; Perkin-Elmer, Norwalk, CT). The final PCR products, estimated to be 1,151 bp in size, were confirmed by 2% agarose gel electrophoresis and then purified (QIAquick Gel Extraction Kit; QIAGEN, Valencia, CA).
Sequence determination and phylogenetic analysis.
Direct sequencing of the PCR product was performed using three sequencing primers, tsu-E (forward, 5′-GTT GGA GGA ATG ATT ACT GG-3′; nucleotide position 124–143 from the start codon of the 56-kDa gene of the Gilliam strain), tsu-F (reverse, 5′-AGC GCT AGG TTT ATT AGC AT-3′; 731–749), and tsu-H (forward, 5′-GTT TAG AAT GGT TAC CAC-3′; −53 to −36).11,12 The entire VDs I and II were expected to be included in the final sequence result. Sequencing reactions were performed on an MJ Research PTC-225 Peltier Thermo Cycler using an ABI PRISM BigDye Terminator Cycle Sequencing Kit with AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA) following the protocols supplied by the manufacturer.
Initially, a nucleotide sequence with a maximum pairwise identity score with the 56-kDa antigen gene was identified from GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) for each sequence examined. The complete pool of reference sequences consisted of both the matching strains from GenBank and the general prototype strains. Then, both the study sequences and reference sequences were aligned using CLUSTAL W.13 After the alignment, both the 5′ and 3′ ends of the aligned nucleotide sequences were trimmed to contain the entire VDs I and II of the 56-kDa gene and show even length endings.
Phylogenetic relationships among the study sequences and reference sequences were constructed by the neighbor-joining method and performed with 1,000 bootstrap replications to evaluate the reliability of the constructions.14,15 A similarity matrix was constructed (Lasergene version 7.0, DNAStar, Madison, WI). The reference strains (accession number) from GenBank were Karp (M33004), Kato (M63382), Kawasaki (M63383), Kuroki (M63380), Shimokoshi (M63381), Boryong (AM494475), Yonchon (U19903), Sxh951 (AF050669), TA686 (U80635), TA678 (U19904), TA716 (U19905), TA763 (U80636), Ikeda (AF173033), Oishi (AF173037), Taguchi (AF173038), Kanda (AF173039), Omagari (AF173040), Matsuzawa (AF173043), Mori (AF173044), Okazaki (AF173045), Jecheon (AF430143), UAP7 (AF302995), and Hirahata (AF201835). The Gilliam (M33267) sequence was obtained from the literature.16
Serological assay.
Baseline and follow-up blood samples were taken from patients at 7- to 14-day intervals. Paired sera were used for serological tests by the passive hemagglutination assay (PHA). The PHA was performed with a commercial kit (Genedia Tsutsu PHA II, GreenCross SangA; Yongin city, Kyunggi, South Korea) designed for the quantitative and qualitative detection of antibodies against O. tsutsugamushi in human sera.17 The test panel was comprised of antigens from the strains Karp, Gilliam, and Boryong, which are known to be prevalent in South Korea. A positive result was defined as a four-fold or greater change in the titer of the paired sera or a titer ≥ 1:80 in a single serum sample. The cut-off value of ≥ 1:80 was defined as a positive result by the manufacturer (Genedia Tsutsu PHA II, GreenCross SangA; Yongin city, Kyunggi, South Korea).
Statistical analysis.
Pearson's χ2 test was used for the categorical variables. A P value < 0.05 was considered statistically significant.
Results
Genotypes of O. tsutsugamushi.
One hundred ninety-two patients with eschar were enrolled from eight hospitals. Eschar specimens from all the patients were subjected to PCR. PCR was repeated several times for the specimens that yielded negative results to obtain the maximum number of positive results, and ultimately, 186 eschar specimens were PCR-positive. From these 186 PCR products, 153 sequences were appropriately determined and yielded nucleotide sequences corresponding to the 56-kDa antigen gene. According to the study design, these 153 patients were confirmed to have scrub typhus, and their data were used for the analysis.
The median age of the patients was 63 years (range = 18–91), and 90 patients (58.8%) were female; 55 patients (35.9%) had one or more underlying chronic diseases. These diseases included hypertension (21.6%), diabetes mellitus (8.5%), and other less frequent diseases such as congestive heart failure, cerebrovascular diseases with residual sequelae, chronic liver disease, and chronic obstructive lung disease requiring maintenance therapy. All patients survived 30 days.
The geographic distribution of the evaluated infections included seven provinces and covered most of the major endemic southern and western plain areas of South Korea (Chonbuk Province, 38.6%; Chungnam Province, 32.7%; Kyungnam Province, 9.8%; Chonnam Province, 2.6%) (Table 1). Northeastern and mideastern regions of the country are mostly mountainous, and cases from these areas were rare.
Table 1.
Genotypic distribution of O. tsutsugamushi strains causing human infections in South Korea (N = 153)
| Location (province) | Genotype | Total (%) | ||||
|---|---|---|---|---|---|---|
| Boryong | Taguchi | Kanda/Kawasaki | Jecheon | UAP7 | ||
| Kyunggi | 4 | 1 | – | 1 | 2 | 8 (5.2) |
| Chungbuk | 1 | 1 | – | – | – | 2 (1.3) |
| Chungnam | 33 | 11 | 6 | – | – | 50 (32.7) |
| Chonbuk | 51 | 5 | 2 | 1 | – | 59 (38.6) |
| Chonnam | 2 | 2 | – | – | – | 4 (2.6) |
| Kyungbuk | 3 | 9 | 3 | – | – | 15 (9.8) |
| Kyungnam | 11 | 1 | 3 | – | – | 15 (9.8) |
| Total (%) | 105 (68.6) | 30 (19.6) | 14 (9.2) | 2 (1.3) | 2 (1.3) | 153 (100) |
Genotypes matching the 153 recovered sequences were retrieved from GenBank. All of the sequences showed 99.6–100% homology with the corresponding reference sequences based on the 56-kDa antigen gene (Table 2). The length of the nucleotide sequences used for the comparison was 722 bp from the start codon of the 56-kDa antigen gene, using the Karp strain (M33004) as the reference for nucleotide position, and this sequence covered the entire VDs I and II. However, the study sequences were approximately 690-bp long, because they contained fewer nucleotide insertions than the reference sequences to which they were aligned. The majority of the isolates belonged to the Boryong strain (68.6%; 105 amplicons) and the Taguchi strain (19.6%; 30 amplicons). In addition, 14 sequences (9.2%) showed 100% homology for both the Kanda and Kawasaki strains. The Jecheon (1.3%; two amplicons) and UAP7 (1.3%; two amplicons) strains were the least common (Table 1). In regard to the reference strains, it was found that the Kuroki (M63380) and Boryong (AM494475) strains and the Kanda (AF173039) and Kawasaki (M63383) strains had identical sequences for the gene fragment analyzed in this study. These strains are antigenically different but could not be differentiated by our method.18,19 Only the Boryong strain, and not the Kuroki strain, is mentioned in this study, because it is a long-term predominant strain and there has been no evidence of divergence. The term Kanda/Kawasaki is used for the 14 cases of Kanda or Kawasaki strains.
Table 2.
Percent sequence similarity between study sequences and reference strains
| Study strains | Reference strains | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S004 | S010 | S017 | S048 | S170 | S189 | S275 | S284 | S314 | Boryong | Gilliam | Jecheon | Kanda | Karp | Kawasaki | Taguchi | UAP7 | |
| S004 | – | 91 | 92.1 | 82.2 | 91.3 | 82 | 91.4 | 82.2 | 91.3 | 91.4 | 83.4 | 99.9 | 82 | 96.3 | 82 | 82.2 | 92.1 |
| S010 | – | 91.2 | 81.9 | 99.4 | 81.9 | 99.6 | 82 | 99.4 | 99.6 | 88.3 | 90.9 | 81.9 | 90.1 | 81.9 | 82 | 91.2 | |
| S017 | – | 82.5 | 91.2 | 82.4 | 91.4 | 82.5 | 91.2 | 91.4 | 83.6 | 92 | 82.4 | 90.8 | 82.4 | 82.5 | 100 | ||
| S048 | – | 81.4 | 99.7 | 81.6 | 99.9 | 81.4 | 81.6 | 88.6 | 82 | 99.7 | 81.6 | 99.7 | 99.9 | 82.5 | |||
| S170 | – | 81.3 | 99.9 | 81.4 | 99.7 | 99.9 | 87.7 | 91.1 | 81.3 | 90.1 | 81.3 | 81.4 | 91.2 | ||||
| S189 | – | 81.4 | 99.9 | 81.3 | 81.4 | 88.6 | 81.9 | 100 | 81.5 | 100 | 99.9 | 82.4 | |||||
| S275 | – | 81.6 | 99.9 | 100 | 87.8 | 91.3 | 81.4 | 90.3 | 81.4 | 81.6 | 91.4 | ||||||
| S284 | – | 81.4 | 81.6 | 88.7 | 82 | 99.9 | 81.6 | 99.9 | 100 | 82.5 | |||||||
| S314 | – | 99.9 | 87.7 | 91.1 | 81.3 | 90.1 | 81.3 | 81.4 | 91.2 | ||||||||
Only representative sequences of clustered groups in Figure 1 were used for comparison, and clearly redundant identical sequences were not listed. S170 and S275 were from SNU-I, S314 was from SNU-II, S189 was from SNU-III, and S284 was from SNU-IV. S = SNU.
Phylogenetic relationships based on the alignment of VDs I and II of the 56-kDa antigen gene were determined to analyze the genetic divergence between the study sequences and reference sequences (Figure 1). The study sequences constituted two major clustered groups, the Boryong strain (SNU-I and -II) and Kawasaki strains (SNU-III and -V), with the remaining isolates being scattered. Although the cluster SNU-III was typed as a Kanda/Kawasaki strain and the cluster SNU-IV was typed as a Taguchi strain, both SNU-III and SNU-IV were called Kawasaki strains because of their close similarity (99.7–99.9%). The classic Kato strain was not found. The nucleotide sequences established in this study were deposited in GenBank under accession numbers EU523383–EU523531.
Figure 1.
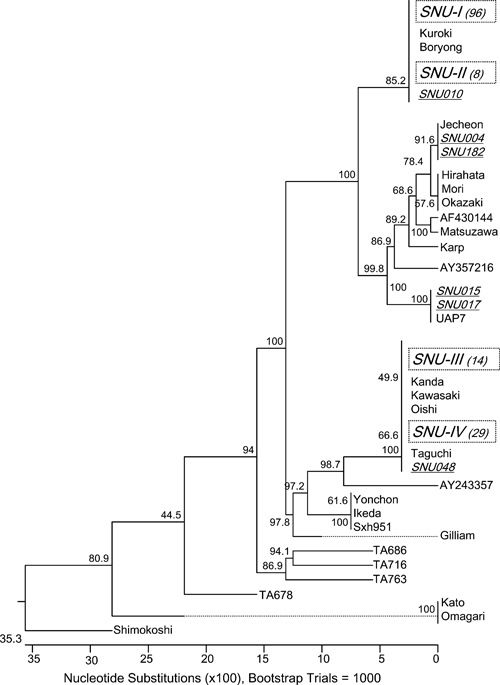
The phylogenetic tree was constructed by the neighbor-joining method based on VD I and II of the 56-kDa antigen gene of O. tsutsugamushi isolated from humans in South Korea. Large clusters of similar nucleotide sequences are grouped by dotted boxes and indicated as SNU-I to -IV. A basic local alignment search tool (BLAST) search confirmed the identification of SNU-I and SNU-II as Boryong strains, SNU-III as a Kanda/Kawasaki strain, and SNU-IV as a Taguchi strain. The numbers within parentheses indicate the number of isolates identified. Scattered isolates are indicated by individual underlined names such as SNU***. Reference strains were retrieved from GenBank. The numbers at the nodes indicate the percent reliability of each branch of the tree.
Serological assay.
PHA results were available for 136 of the total 153 patients. Among the 95 patients infected with Boryong strains, 75 had a positive PHA result and 20 had a negative result compared with 9 positive and 31 negative PHA results among the 40 patients infected with Kawasaki strains (Kanda/Kawasaki + Taguchi). The PHA, thus, yielded significantly fewer positive results among Kawasaki strains, which are not included in the current PHA test panel (P < 0.001) (Table 3).
Table 3.
The results of passive hemagglutination assay for 136 genotypes of O. tsutsugamushi
| PHA results | Genotypes | |||
|---|---|---|---|---|
| Boryong | Taguchi | Kanda/Kawasaki | Jecheon | |
| PHA-positive | 75* | 7 | 2 | 0 |
| Single titer ≥ 1:80 | 44 | 3 | 1 | 0 |
| Follow-up titer rise ≥ four times | 31 | 4 | 1 | 0 |
| PHA-negative | 20 | 22 | 9 | 1 |
| Negative single titer | 11 | 9 | 2 | 0 |
| Negative follow-up titer rise | 9 | 13 | 7 | 1 |
| Total | 95 | 29 | 11 | 1 |
P value < 0.001 for Boryong strain vs. Kawasaki strains (Taguchi + Kanda/Kawasaki).
PHA = passive hemagglutination assay.
Discussion
Our study determined the nationwide O. tsutsugamushi genotypes causing human infections in South Korea. The antigenic diversity of O. tsutsugamushi has several implications. The serotypes of O. tsutsugamushi are related to vaccine development, clinical virulence, and appropriateness of the antigen panel used in the serological assay. Therefore, periodic surveillance of the circulating contemporary serotypes in actual human infections is important. However, the conventional surveillance approach has complex problems, including laborious procedures and biosafety, which need to be overcome to be readily accessible for investigators outside of the reference laboratory. To manage this problem, we chose genotyping based on the nucleotide sequence of the 56-kDa antigen gene and used clinical eschar as a source of O. tsutsugamushi. Furthermore, we expected that genotyping might further differentiate serotypes that cannot be further subclassified by the monoclonal antibody method.
The antigenic diversity of O. tsutsugamushi in South Korea has been reported in several studies. In a nationwide prevalence study in the early 1990s using the monoclonal antibody method, Chang20 reported that 82.8% of 137 human cases were caused by the Boryong serotype and found less frequent cases caused by the Karp (13 cases; 9.7%) and Gilliam (10 cases; 7.5%) serotypes.20 Several other published and unpublished data with small sample sizes show that the Boryong serotype is consistently predominant, but the other identified serotypes were rare and inconsistent.21 Ree and others22 characterized isolates (N = 41) from mite pools from 1997 to 1999 using serotype-specific nested PCR.22 The Boryong strain (82.9%) was predominant throughout the country, Karp strains (12.2%) were found in central South Korea, and the other strains were a mixture of both Boryong and Karp. No other strain was isolated. Shim and others23 surveyed the genotypes isolated from wild rodents (60 isolates) and mite pools (129 isolates) in central and southern South Korea from 2005 to 2007 using the PCR-restriction fragment-length polymorphism method and sequence analysis of the 56-kDa antigen gene.23 Boryong was the most common strain, with rates of 76.7% in rodents and 82.9% in mites. The other identified stains were Gilliam (10%), Pajoo (8.3%), and Karp (5%) in rodents and Karp (11.6%), Pajoo (3.1%), Yoncheon (1.6%), and Kawasaki (0.8%) in mites.
Our study is mostly an update for human infections and covers most of the endemic regions in South Korea. Our sequences contained the complete VDs I and II of the 56-kDa antigen gene, which shows the most variation in amino acid sequence. The Boryong strain was still dominant (68.6%), but previously unknown strains (Taguchi, Kanda/Kawasaki, and UAP7) were identified in 30.1% of the cases. Altogether, Boryong is undoubtedly the predominant strain in South Korea. However, there have been discrepancies regarding the minor strains, and the typing methods are not comparable. Taguchi and Kanda/Kawasaki strains, which are clustered as Kawasaki strains, have not been previously reported in South Korea. These strains made up a relatively large portion of the human infections in our study, but they are not included in the antigen panel for the serological assays used in South Korea. We do not have a baseline for previous O. tsutsugamushi genotype data that is comparable with our results. This could be an indication of the changes in human cases that have taken place over the last two decades. Taguchi and UAP7 have never been isolated from rodents or mites in South Korea. Only one Kawasaki strain was isolated by another investigator during the same period as our study,23 which may provide supporting evidence for the Kawasaki strains identified in our study.
Human infection by UAP7 has not been previously reported in the literature. This study is the first demonstration of UAP7 in a human infection since its initial isolation from rodents in Japan.12 The two UAP7 cases were from northern South Korea and had severe clinical manifestations, including pneumonia. In a previous study, UAP7 reacted with an anti–Karp-specific monoclonal antibody and produced PCR products with a Karp type-specific primer pair.12 Although we cannot extrapolate its frequency because of the limited number of UAP7 cases in our study, immunological methods would not distinguish between this strain and the Karp strain frequently isolated in northern South Korea.
Our specimens covered most of the endemic regions of South Korea. Although the number of specimens from the northern and northeastern regions was relatively small, the reported case incidence in those areas was also low. Kim HY (49th ICAAC poster presentation #B-031, unpublished data) characterized isolates (N = 40) from 2006 to 2008 from the northeastern mountainous region of South Korea, which was not covered in our study, using nested PCR and sequence analysis of the final 487 bp of the 56-kDa antigen gene. That study identified 5 cases of the Boryong strain (12.5%), 2 cases of the Taguchi strain (5%), 22 cases of Karp-related strains (55%), 9 cases of Saitama strains (22.5%), and 2 cases of Japanese Karp (JP)-related strains (5%). In this region, the non-Boryong strains Gilliam and Karp were considered dominant.
Eschar is a necrotic tissue containing a mixture of O. tsutsugamushi and host inflammatory cells. Although the prevalence of eschars in scrub-typhus patients varies according to geographic region, they are identifiable in about 90% of patients from East Asian countries, including South Korea.24,25 Genetic material from O. tsutsugamushi can be obtained from the tissue for more than 2 weeks, despite antibiotic treatment.1,26 However, it is difficult to collect eschar during the wet, exudative early phase, and one sometimes obtains only thick exfoliative skin without any bacterial components. Positive results were obtained by PCR for 186 of the initial 192 eschar specimens, but for 33 of these instances, there was not enough PCR product to sequence. This was our first attempt to collect clinical eschars from multiple centers. With more experience and uniform techniques in specimen collection, the yield should be higher. In endemic regions with low eschar prevalence, the buffy coat would be an alternative source of O. tsutsugamushi genetic material, but this would only be successful during the early phase of infection or within 1 week of antibiotic therapy.1,26,27
The PHA method is widely used in South Korea because of its availability.28 The indirect fluorescent antibody assay (IFA) has greater diagnostic sensitivity than PHA, but the overall diagnostic performance is also unsatisfactory.26 Three antigens of Karp, Gilliam, and Boryong strains have been used in the test panels of both serological assays in South Korea. The present study showed that positive results from PHA were significantly lower with Kawasaki strains than with the Boryong strain; an antigen for the latter is included in the PHA test panel. This may explain why PHA has recently displayed lower sensitivity in clinical practice.26 Considering our results, the best antigen panel for a serological assay to be used in South Korea would be comprised of Boryong, Karp, and Kawasaki antigens.
Our study was performed during a single outbreak season, and we only analyzed VDs I and II of the 56-kDa antigen gene. Previous data from the literature that we evaluated used different serotyping or genotyping methods from our study. These are limiting factors in the interpretation of our results. Therefore, periodic surveillance with uniform methods, preferably sequencing of the whole 56-kDa antigen gene, is needed to gain an accurate picture of the contemporary strains in the community.
In summary, we determined the circulating genotypes of O. tsutsugamushi strains causing human infections in South Korea by targeting the 56-kDa antigen gene and using clinical eschar specimens. This enabled us to identify previously unknown strains and determine the interval of change for the current circulating strains of O. tsutsugamushi. The serological assay lacked antigens that are relevant to the circulating strains and had significantly fewer positive results.
Acknowledgments
The results of this study were presented in part at the 48th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy (ICAAC)/Infectious Diseases Society of America (IDSA) 46th Annual Joint Meeting in Washington, DC in 2008.
Disclaimer: The authors have no conflicts of interest in relation to this article.
Footnotes
Financial support: This study was supported by Grant 04-2008-068 from the Seoul National University Hospital (SNUH) Research Fund and funding from the Chonbuk National University Hospital Research Institute of Clinical Medicine.
Authors' addresses: Sang-Won Park, Department of Internal Medicine, Boramae Medical Center, Seoul 156-707, South Korea, E-mail: hswon1@snu.ac.kr. Chi Kug Lee, Department of Internal Medicine, Namwon Medical Center, Gojuk-dong, Namwon, Chonbuk 590-702, South Korea, E-mail: na25pro@hotmail.com. Yee Gyung Kwak, Chisook Moon, and Baek-Nam Kim, Department of Internal Medicine, Inje University College of Medicine, Gaeeum-dong, Busan 614-735, South Korea, E-mails: philmed202@hanmail.net, duomon@hanmail.net, and griuni@chol.com. Eu Suk Kim, Department of Internal Medicine, Dongguk University Ilsan Hospital, Siksa-dong, Goyang, Gyeonggi 410-773, South Korea, E-mail: yonathan@hanafos.com. Jae Myung Kang, Department of Internal Medicine, Handong Global University, Sunlin Hospital, Daeshin-dog, Pohang, Kyungbuk 791-704, South Korea, E-mail: joshuakang@hanmail.net. Chang-Seop Lee, Department of Internal Medicine, Chonbuk National University Medical School, Jeonju, Chonbuk 561-712, South Korea, E-mail: lcsmd@chonbuk.ac.kr.
Reprint requests: Chang-Seop Lee, Department of Internal Medicine, Chonbuk National University Medical School, 634-18, Keumam-dong, Jeonju, 561-712, Republic of Korea, E-mail: lcsmd@chonbuk.ac.kr.
References
- 1.Kim DM, Kim HL, Park CY, Yang TY, Lee JH, Yang JT, Shim SK, Lee SH. Clinical usefulness of eschar polymerase chain reaction for the diagnosis of scrub typhus: a prospective study. Clin Infect Dis. 2006;43:1296–1300. doi: 10.1086/508464. [DOI] [PubMed] [Google Scholar]
- 2.Kim DM, Yun NR, Yang TY, Lee JH, Yang JT, Choi EN, Park MY, Lee SH. Usefulness of nested PCR for the diagnosis of scrub typhus in clinical practice: a prospective study. Am J Trop Med Hyg. 2006;75:542–545. [PubMed] [Google Scholar]
- 3.Murai K, Okayama A, Horinouchi H, Oshikawa T, Tachibana N, Tsubouchi H. Eradication of Rickettsia tsutsugamushi from patients' blood by chemotherapy as assessed by the polymerase chain reaction. Am J Trop Med Hyg. 1995;52:325–327. doi: 10.4269/ajtmh.1995.52.325. [DOI] [PubMed] [Google Scholar]
- 4.Kelly DJ, Fuerst PA, Ching WM, Richards AL. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis. 2009;48((Suppl 3)):S203–S230. doi: 10.1086/596576. [DOI] [PubMed] [Google Scholar]
- 5.Stover CK, Marana DP, Carter JM, Roe BA, Mardis E, Oaks EV. The 56-kilodalton major protein antigen of Rickettsia tsutsugamushi: molecular cloning and sequence analysis of the sta56 gene and precise identification of a strain-specific epitope. Infect Immun. 1990;58:2076–2084. doi: 10.1128/iai.58.7.2076-2084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuya Y, Yoshida Y, Katayama T, Kawamori F, Yamamoto S, Ohashi N, Tamura A, Kawamura A., Jr Specific amplification of Rickettsia tsutsugamushi DNA from clinical specimens by polymerase chain reaction. J Clin Microbiol. 1991;29:2628–2630. doi: 10.1128/jcm.29.11.2628-2630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuya Y, Yoshida Y, Katayama Y, Yamamoto S, Kawamura A. Serotype-specific amplification of Rickettsia tsutsugamushi DNA by nested polymerase chain reaction. J Clin Microbiol. 1993;31:1637–1640. doi: 10.1128/jcm.31.6.1637-1640.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JS, Ahn C, Kim YK, Lee MH. Thirteen cases of rickettsial infection including 9 cases of tsutsugamushi disease first confirmed in Korea [Korean] J Korean Med Assoc. 1986;29:430–438. [Google Scholar]
- 9.Kweon SS, Choi JS, Lim HS, Kim JR, Kim KY, Ryu SY, Yoo HS, Park O. Rapid increase of scrub typhus, South Korea, 2001–2006. Emerg Infect Dis. 2009;15:1127–1129. doi: 10.3201/eid1507.080399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohashi N, Nashimoto H, Ikeda H, Tamura A. Diversity of immunodominant 56-kDa type-specific antigen (TSA) of Rickettsia tsutsugamushi: sequence and comparative analyses of the genes encoding TSA homologues from four antigenic variants. J Biol Chem. 1992;267:12728–12735. [PubMed] [Google Scholar]
- 11.Enatsu T, Urakami H, Tamura A. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol Lett. 1999;180:163–169. doi: 10.1111/j.1574-6968.1999.tb08791.x. [DOI] [PubMed] [Google Scholar]
- 12.Tamura A, Yamamoto N, Koyama S, Makisaka Y, Takahashi M, Urabe K, Takaoka M, Nakazawa K, Urakami H, Fukuhara M. Epidemiological survey of Orientia tsutsugamushi distribution in field rodents in Saitama prefecture, Japan, and discovery of a new type. Microbiol Immunol. 2001;45:439–446. doi: 10.1111/j.1348-0421.2001.tb02643.x. [DOI] [PubMed] [Google Scholar]
- 13.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 16.Ohashi N, Nashimoto H, Ikeda H, Tamura A. Cloning and sequencing of the gene (tsg56) encoding a type-specific antigen from Rickettsia tsutsugamushi. Gene. 1990;91:119–122. doi: 10.1016/0378-1119(90)90171-m. [DOI] [PubMed] [Google Scholar]
- 17.Kim IS, Seong SY, Woo SG, Choi MS, Chang WH. High-level expression of a 56-kilodalton protein gene (bor56) of Rickettsia tsutsugamushi Boryong and its application to enzyme-linked immunosorbent assays. J Clin Microbiol. 1993;31:598–605. doi: 10.1128/jcm.31.3.598-605.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang JS, Chang WH. Antigenic relationship among the eight prototype and new serotype strains of Orientia tsutsugamushi revealed by monoclonal antibodies. Microbiol Immunol. 1999;43:229–234. doi: 10.1111/j.1348-0421.1999.tb02397.x. [DOI] [PubMed] [Google Scholar]
- 19.Ohashi N, Koyama Y, Urakami H, Fukuhara M, Tamura A, Kawamori F, Yamamoto S, Kasuya S, Yoshimura K. Demonstration of antigenic and genotypic variation of Orientia tsutsugamushi which were isolated in Japan, and their classification into type and subtype. Microbiol Immunol. 1996;40:627–638. doi: 10.1111/j.1348-0421.1996.tb01120.x. [DOI] [PubMed] [Google Scholar]
- 20.Chang WH. Tsutsugamushi Disease in Korea. Seoul, Korea: Seohung Press; 1994. pp. 25–30. [Google Scholar]
- 21.Korea Centers for Disease Control and Prevention Current trends of scrub typhus (Korean) Commun Dis Monthly Rep. 2004;15:245–252. [Google Scholar]
- 22.Ree HI, Kim TE, Lee IY, Jeon SH, Hwang UW, Chang WH. Determination and geographical distribution of Orientia tsutsugamushi serotypes in Korea by nested polymerase chain reaction. Am J Trop Med Hyg. 2001;65:528–534. doi: 10.4269/ajtmh.2001.65.528. [DOI] [PubMed] [Google Scholar]
- 23.Shim SK, Choi EN, Yu KO, Park HJ, Kim CM, Lee KH, Park JK, Park PH, Yoon MH, Park SH, Choi YS, Hwang KJ, Park MY. Characterisation of Orientia tsutsugamushi genotypes from wild rodents and chigger mites in Korea. Clin Microbiol Infect. 2009;15:311–312. doi: 10.1111/j.1469-0691.2008.02254.x. [DOI] [PubMed] [Google Scholar]
- 24.Liu YX, Feng D, Suo JJ, Xing YB, Liu G, Liu LH, Xiao HJ, Jia N, Gao Y, Yang H, Zuo SQ, Zhang PH, Zhao ZT, Min JS, Feng PT, Ma SB, Liang S, Cao WC. Clinical characteristics of the autumn-winter type scrub typhus cases in south of Shandong province, northern China. BMC Infect Dis. 2009;9:82. doi: 10.1186/1471-2334-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogawa M, Hagiwara T, Kishimoto T, Shiga S, Yoshida Y, Furuya Y, Kaiho I, Ito T, Nemoto H, Yamamoto N, Masukawa K. Scrub typhus in Japan: epidemiology and clinical features of cases reported in 1998. Am J Trop Med Hyg. 2002;67:162–165. doi: 10.4269/ajtmh.2002.67.162. [DOI] [PubMed] [Google Scholar]
- 26.Kim DM, Yun NR, Yang TY, Lee JH, Yang JT, Shim SK, Choi EN, Park MY, Lee SH. Usefulness of nested PCR for the diagnosis of scrub typhus in clinical practice: a prospective study. Am J Trop Med Hyg. 2006;75:542–545. [PubMed] [Google Scholar]
- 27.Fournier PE, Siritantikorn S, Rolain JM, Suputtamongkol Y, Hoontrakul S, Charoenwat S, Losuwanaluk K, Parola P, Raoult D. Detection of new genotypes of Orientia tsutsugamushi infecting humans in Thailand. Clin Microbiol Infect. 2008;14:168–173. doi: 10.1111/j.1469-0691.2007.01889.x. [DOI] [PubMed] [Google Scholar]
- 28.Blacksell SD, Bryant NJ, Paris DH, Doust JA, Sakoda Y, Day NP. Scrub typhus serologic testing with the indirect immunofluorescence method as a diagnostic gold standard: a lack of consensus leads to a lot of confusion. Clin Infect Dis. 2007;44:391–401. doi: 10.1086/510585. [DOI] [PubMed] [Google Scholar]


