Abstract
Diarrhea is a major public health problem that affects the development of children. Anthropometric data were collected from 274 children with (N = 170) and without (N = 104) diarrhea. Stool specimens were analyzed by conventional culture, polymerase chain reaction for enteroaggregative Escherichia coli (EAEC), Shigella, Cryptosporidium, Entamoeba, and Giardia species, and by enzyme-linked immunosorbent assay for fecal lactoferrin levels. About 50% of the study population was mildly to severely malnourished. Fecal lactoferrin levels were higher in children with diarrhea (P = 0.019). Children who had EAEC infection, with or without diarrhea, had high mean lactoferrin levels regardless of nutritional status. The EAEC and Cryptosporidium were associated with diarrhea (P = 0.048 and 0.011, respectively), and malnourished children who had diarrhea were often co-infected with both Cryptosporidium and EAEC. In conclusion, the use of DNA-biomarkers revealed that EAEC and Cryptosporidium were common intestinal pathogens in Accra, and that elevated lactoferrin was associated with diarrhea in this group of children.
Introduction
Diarrhea is a principal cause of morbidity and mortality in children < 5 years of age in developing countries, where acute watery diarrhea accounts for nearly two million diarrhea-related deaths annually in this age group.1,2 In the last decades, however, while mortality caused by diarrhea has been decreasing worldwide mainly because of improved hygiene, morbidity attributable to diarrhea remains high.2–4 A vicious cycle ensues between diarrhea and malnutrition, and studies have shown that malnutrition with frequent diarrheal episodes slows cognitive and physical development of children.5,6 One mechanism for this is that diarrheagenic pathogens damage intestinal epithelium and reduce its absorptive function, leading to nutrient depletion and malnutrition.6
Obstacles to recognition of at-risk children are several. The plight of sub-optimally breast fed and malnourished children is often largely invisible because they are only mildly or moderately undernourished.7 Additionally, anthropometric measurements are not routinely performed to identify malnourished children in most clinics and hospitals in Ghana, thereby missing the opportunity for diagnosis and appropriate management.8 The most established method to identify those with malnutrition is by the use of z-scores, with the reference population defined for the study country or from the standard international reference chart of the National Center for Health and the World Health Organization (WHO).9
The agents capable of causing infectious diarrhea and the mechanisms responsible for disease pathogenesis are generally known, but the true prevalence of these agents in developing countries is poorly understood.10 For example, in most sub-Saharan African countries including Ghana, microbiological methods for clinical investigation of diarrheal diseases are usually restricted to identifying conventional enteric bacteria such as Salmonella and Shigella, Escherichia coli (E. coli) isolates are often not fully characterized because of the lack of resources. Additional pathogens of potential importance include enteroaggregative E. coli (EAEC), which is associated with diarrhea in several contexts: traveler's diarrhea,11,12 pediatric diarrhea,13 foodborne outbreaks,14 human immunodeficiency virus,15 symptomatic and asymptomatic cases,16 acute and persistent diarrhea,5,17 among others; and Cryptosporidium, Entamoeba histolytica, and Giardia, which are parasitic causes of diarrhea.18–20 Unfortunately, investigation of diarrhea caused by these parasites in most developing countries largely depends on expert microscopy, where technical competence is necessary.
The objective of this study was, first, to determine the prevalence of EAEC, Shigella spp., Cryptosporidium spp., E. histolytica, and Giardia lamblia in children < 5 years of age with and without diarrhea in southern Ghana. Second, this study aims to determine whether these enteropathogens were associated with intestinal inflammation in either nourished or malnourished children.
Materials and Methods
Ethical clearance.
The study was reviewed and approved by the Institutional Review Board of the University of Ghana Medical School, Ghana. Participation was voluntary and enrollment was subject to parents/guardians' approval, through signature or by thumb printing their names after the purpose of the study was explained to them.
Study design, population, and settings.
This was a prospective cross-sectional study carried out between August 2007 and May 2008, of children ≤ 5 years of age consulting at the Princess Marie Louise Children's Hospital (PML), Accra, Ghana. Consecutive children from whom consent was given by their caregivers were included in the study. The group of children with diarrhea was recruited from the outpatient clinic and these children were brought to the hospital for acute health care. The control group of children without diarrhea was also from the outpatient clinic, but these children were visiting for routine child welfare care. No follow-up was done after the initial recruitment as a part of this study.
Interviews and diarrhea definition.
A structured questionnaire was used to obtain information on the children from their parents/guardians. Information that was obtained included demographic data, duration of diarrhea, residence/location, breast feeding status, and medication taken before visiting the hospital. Diarrhea was defined as the passage of three or more unformed stools within a 24-h period. Diarrhea duration lasting < 14 days was defined as acute and those lasting ≥ 14 days, persistent. The control (non-diarrhea) group consisted of children who have not passed three or more unformed stools at least within the 24-h period before enrollment.
Anthropometric data and nutritional status assessment.
Height or length measurements in centimeter to the nearest one decimal were performed for children above or below 2 years of age, respectively. Weight measurements in kilogram to the nearest one decimal were performed using a 25 kg Salter hanging scale (CMS Weighing equipment, High Holborn, London, UK). The Z-score, weight-for-age (WAZ), height-for-age (HAZ), and weight-for-height (WHZ) were calculated by use of software designed for nutrition studies (EPINUT, World Health Organization, Geneva; Epi Info version 6.0, Centers for Disease Control and Prevention, Atlanta, GA). These anthropometric Z-scores are a measure of SD above or below the median for the international reference population. Z-score values were used to determine the nutritional status of children on the basis of the following definition: WAZ, well nourished (> −1), mild (−2 to −1), moderate (−3 to −2), and severe (< −3) malnutrition; HAZ, normal height (≥ −2), moderate stunting (−3 < −2), and severe stunting (< −3); WHZ, normal weight (≥ −2), moderate wasting (−3 < −2), and severe wasting (< −3).
Specimen processing and microbiological analysis.
A stool specimen from each participating child was collected into a sterile container and processed within 2 h of collection. Routine enteric bacteria were cultured on MacConkey (MAC), Salmonella-Shigella (SS), and deoxychocolate (DCA) agars (Oxoid, Columbia, MD), using standard techniques. Selenite F broth was used as pre-enrichment for Salmonella before sub-culturing onto MAC, SS, and DCA. Bacterial colonies after an overnight incubation period at 37°C were identified by standard biochemical methods and stored on Muellar Hinton slopes for further analysis. No microscopy was performed for the detection of parasites. One aliquot of a fresh stool specimen from each child was kept frozen at −20°C in cryo-vials (deidentified) until sent to the Center for Global Health, University of Virginia for further analysis.
Fecal DNA extraction.
We used the QIAamp stool kit (Qiagen, Valencia, CA) to extract genomic DNA from frozen stool specimens with some minor modifications. The modifications included the addition of dry beads (MO BIO Laboratory Inc., Carlsbad, CA) to weighed stool specimen before the addition of lysis buffer (ASL). The mixture was bead-beated for 2 minutes to make a uniform homogeneous mixture with the lysis buffer. Additionally, we incubated mixtures at 80°C instead of the 70°C recommended by the manufacture to lyse enteric pathogens. For each stool aliquot, between 15 and 20 μg or μL of stool was used depending on stool consistency. DNAs were also extracted from appropriate control organisms. All DNAs were kept frozen at −80°C until needed for analysis.
Quantitative real-time polymerase chain reaction (PCR).
A single-plex quantitative PCR for each gene pair (Table 1) consisted of 5 μL template, 1 μL of each 6.2 μM primer, 12.5 μL of SYBR-Green −490 (Bio-Rad Laboratories, Beltsville, MD), and PCR grade water to a reaction volume of 25 μL. Reactions for each sample were performed using the Bio-Rad iQCycler Real-Time Detection System in Bio-Rad iCycler 96-well plates, where positive and negative controls were included with each reaction set. Table 1 shows the target genes, locations, and annealing temperature for each primer set. The results were analyzed with a user-defined threshold of 200 PCR baseline-subtracted curve-fit relative fluorescence units. Melt curve (ct) data collection and analysis was enabled at cycles 3 and 4, with an increase in set point temperatures after cycle 2 by 0.5°C.
Table 1.
Target genes screened from fecal DNA*
| Strain | Gene target | Location | Primer sequence (5′–3′) | PCR size (bp) | Annealing temperature (°C) | Source/reference |
|---|---|---|---|---|---|---|
| EAEC | aaiC | Chromosome | CTTCTGCTCTTAGCAGGGAGTTTG AAGCGTGAAATGCCTGAGGA | 123 | 47.5 | Nataro's Laboratory |
| aatA | Plasmid | CCTRTGTTGATGCTCGAGAGA CKTTCCTCCTCCTCAAGGACAT | 118 | 55 | Nataro's Laboratory | |
| aap | Plasmid | CTTGGGTATCAGCCTGAATG AACCCATTCGGTTAGAGCAC | 310 | 45 | Cerna and others21 | |
| aggR | Plasmid | CTAATTGTACAATCGATGTA ATGAAGTAATTCTTGAAT | 308 | 45 | Czeczulin and others22 | |
| Shigella/EIEC | ipaH | Plasmid | GTTCCTTGACCGCCTTTCCGATACCGTC GCCGGTCAGCCACCCTCTGAGAGTAC | 619 | 60.5 | Sethabutr and others23 |
| Crypto-sporidium | 18s rRNA | Chromosome | CTCCACCAACTAAGAACGGCC TAGAGATTGGAGGTTGTTCCT | 213 | 60 | Gene ID cgd7_23024 |
| E. histolytica | Eh | Chromosome | AACAGTAATAGTTTCTTTGGTTAGTAAAA CTTAGAATGTCATTTCTCAATTCAT | 134 | 60 | Haque and others25 |
| Giardia lamblia | P241 | Chromosome | CATCCGCGAGGAGGTCAA GCAGCCATGGTGTCGATCT | 74 | 60 | Guy and others26 |
PCR = polymerase chain reaction; EAEC = enteroaggregative Escherichia coli; EIEC = enteroinvasive E. coli.
We sought multiple loci for EAEC (aap, aatA, aggR, and aaiC) and single loci for Shigella, Cryptosporidium, and Giardia species. Standard cultures with known numbers of E. coli 042 and 17-2, Shigella, Cryptosporidium, and Giardia oocysts were used as reference and positive controls. Water and E. coli K-12 were used as negative controls. Melt curve analysis was used to determine positivity of samples using a user defined threshhold.
Intestinal inflammation assessment.
Intestinal inflammations were quantitatively assessed from frozen stool specimens using the IBD SCAN (TechLab, Blacksburg, VA) according to the manufacturer's instructions. Stool specimens were allowed to thaw and were serially diluted 10-fold and analyzed by a polyclonal antibody-based enzyme-linked immunosorbent assay (ELISA) method. The detailed procedure is described elsewhere,27 and absorbance of each assay well was measured spectrophotometrically at 450 and 620 nm (A450/620). Fecal lactoferrin concentrations in μg/mL were determined by comparison with a standard curve using purified human lactoferrin and analyzed by linear regression in Microsoft excel (Redmond, WA). The lowest dilution of a specimen with an absorbance at 450/620 nm within the linear portion of the curve was used to determine the lactoferrin concentration. The final lactoferrin concentration was obtained by multiplying the dilution factor by the concentration. A positive control (purified human lactoferrin) and a negative control (washing buffer) was included in each batch of stools analyzed and linear regression was performed separately for each batch using standard controls. This assessment was performed only on subjects with adequate stool specimens and where necessary, the experiment was repeated. Among EAEC-infected malnourished children, two stool specimens (out of > 200 specimens analyzed) showed lactoferrin values that were 30–50 times more than the mean value for their subject group, were therefore designated as outliers, and not included in the statistical analysis. No satisfactory explanation for results on these two specimens was evident, because no diarrhea was present in one, and breast feeding was unlikely in both.
Statistical analysis.
To avoid any experimental biases, stool specimens were coded before testing and only decoded for purposes of analysis. Statistical analyses were performed using SPSS software (version 17.0, SPSS, Chicago, IL) and Epi-Info. Statistical tests included χ2 for associations of pathogens with age groups, diarrhea, and non-diarrhea and the paired t test for associations with lactoferrin level. Odds ratio (OR) and 95% confidence intervals (CI) are reported for all 2 × 2 comparisons. Two-tailed tests were used and P < 0.05 was considered statistically significant.
Results
Study population and nutritional status.
Within the 9-month study period, 287 children ≤ 5 years of age were recruited with only 13 excluded from analysis because of insufficient data. Of the 274 children included for analysis, 170 (62%) were with and 104 (38%) were without diarrhea; there were more males 156 (56.9%) than females. Acute and persistent diarrhea included 85.3% (145/170) and 7.6% (13/170) of total cases, respectively. Duration of symptoms in the remaining children with diarrhea (7.1%) was not recorded.
At least one anthropometric measurement was taken for 269 out of the 274 children analyzed. Of 269 children from whom weight measurements were recorded, 134 (49.8%) showed mild to severe malnutrition (WAZ < −1) (93/168 [55.4%] in children with and 41/101 [40.6%] without diarrhea, OR = 1.82 [95% CI, 1.102–2.988], P = 0.023). Of 170 children from whom height or length measurements were recorded, 61 (35%) showed moderate to severe stunting (HAZ < −2) (38/86 [44.2%] in children with and 23/84 [27.4%] without diarrhea, OR = 2.10 [95% CI, 1.110–3.972], P = 0.026). Of 161 children from whom both weight and height measurements were recorded, 37 (22.9%) showed moderate to severe wasting (WHZ < −2) (24/82 [29.3%] in children with and 13/79 [16.5%] without diarrhea, OR = 2.101 [95% CI, 0.989–4.454], P = 0.062). The mean age, weight, and height were 15.1/14.6 months, 9.8/9.5 kg, and 88.0/84.0 cm (diarrhea/non-diarrhea, respectively). Table 2 shows the baseline characteristics of the study population.
Table 2.
Baseline characteristics of study population*
| Characteristic | Diarrhea N (%) | Non-diarrhea N (%) |
|---|---|---|
| Age/months | N = 170 | N = 104 |
| 0–6 | 26 (15.3) | 35 (33.7) |
| 7–12 | 54 (31.8) | 30 (28.8) |
| 13–24 | 77 (45.3) | 24 (23.1) |
| 25–60 | 13 (7.6) | 15 (14.4) |
| Sex | N = 170 | N = 104 |
| Male | 97 (57.1) | 73 (42.9) |
| Female | 59 (56.7) | 45 (43.3) |
| Weight/kg | N = 170 | N = 104 |
| 2.5–4.9 | 10 (5.9) | 4 (3.8) |
| 5.0–9.9 | 121 (71.2) | 75 (72.1) |
| 10–19.9 | 34 (20.0) | 22 (21.2) |
| 20–86.0 | 5 (2.9) | 3 (2.9) |
| WAZ | N = 168 | N = 101 |
| Normal (> −1) | 75 (44.6) | 60 (59.4) |
| Mild (−1 to −2) | 33 (19.6) | 19 (18.8) |
| Moderate (−2 to −3) | 32 (19.0) | 12 (11.9) |
| Severe (< −3) | 28 (16.7) | 10 (9.9) |
| HAZ | N = 86 | N = 84 |
| Normal (≥ −2) | 48 (55.8) | 61 (72.6) |
| Moderate (−2 to −3) | 6 (7.0) | 11 (13.1) |
| Severe (< −3) | 32 (37.2) | 12 (14.3) |
| WHZ | N = 82 | N = 79 |
| Normal (≥ −2) | 58 (70.3) | 66 (83.5) |
| Moderate (−2 to −3) | 6 (7.3) | 8 (10.1) |
| Severe (< −3) | 18 (22.0) | 5 (6.3) |
WAZ = weight-for-age; HAZ = height-for-age; WHZ = weight-for-height.
Microbiological studies.
In only 1 of 170 diarrhea stool specimens was Shigella recovered as an enteric bacterial pathogen from culture. This strain was serotyped with Shigella polyvalent anti-sera (Mast Group Ltd., Merseyside, UK) and was Shigella flexneri. In the entire study population, E. coli was the predominant bacterium obtained from culture (79.6%), followed by Klebsiella spp. (5.1%). Other commensals included 9.8% of the total and no bacteria grew in 5.5% of the total stool specimens cultured.
Pathogen detection by real-time PCR.
Table 3 shows bacterial and parasitic agents detected from fecal DNA by real-time PCR in children with and without diarrhea. The EAEC was defined as positivity for any of the four EAEC virulence genes sought (aap, aatA, aggR, and aaiC). Although EAEC was significantly associated with diarrhea (147/170 versus 80/104, OR 1.917 [95% CI = 1.024–3.592], P = 0.048), it was also found in similar frequencies in both nourished and malnourished children.
Table 3.
Organisms detected by real-time PCR from fecal DNA*
| Diarrhea (N = 170) | Non-diarrhea (N = 104) | Odds ratio [95% CI] P value† | |||||
|---|---|---|---|---|---|---|---|
| No. (%) | WN (N = 75) | MN (N = 95) | No. (%) | WN (N = 60) | MN (N = 41) | ||
| Any infection | |||||||
| EAEC | 147 (86.5) | 66 | 79 | 80 (76.9) | 49 | 30 | 1.917 (1.018–3.612) 0.048 |
| Shigella/EIEC | 6 (3.5) | 5 | 1 | 0 (0) | – | – | n/a |
| Cryptosporidium spp. | 14 (8.7) | 4 | 10 | 1 (1.0) | 1 | 0 | 9.244 (1.197–71.371) 0.011 |
| E. histolytica | 5 (3.0) | 2 | 3 | 0 (0) | – | – | n/a |
| Giardia spp. | 0 (0) | – | – | 0 (0) | – | – | – |
PCR = polymerase chain reaction; WN = well nourished (WAZ > 1); MN = malnourished (WAZ < 1); EAEC = enteroaggregative Escherichia coli; EIEC = enteroinvasive E. coli; n/a = not applicable.
P value is between diarrheal and non-diarrheal stool specimen.
In 6 out of 170 diarrheal stool specimens, the ipaH gene, which is expressed by both Shigella and enteroinvasive E. coli (EIEC) was detected. The ipaH gene was not detected in fecal DNA from any of the children without diarrhea. The numbers were, however, too small to assess statistical significance. Five out of 6 of the children in whom the ipaH gene was detected were well-nourished (Table 3).
Cryptosporidium spp. was the most frequently detected protozoan parasite in fecal DNA and was associated with diarrhea (14/170 versus 1/104, OR = 9.244 [95% CI 1.197–71.371], P = 0.011). Cryptosporidiosis was also primarily (10 out of 14) detected in children who were malnourished and had diarrhea (Table 3). Entamoeba histolytica was only detected in children with diarrhea 5 out of 170 (2.9%) and Giardia was not detected in either sub-populations.
We observed children who were co-infected, with two or more pathogens detected in the stool, predominantly in children who had diarrhea. The EAEC-Cryptosporidium was the most prevalent (7.6%, 13/170), followed by EAEC-Shigella/EIEC (2.9%, 5/170), and EAEC-E. histolytica (2.4%, 4/170). Cryptosporidium-E. histolytica and Cryptosporidium-Shigella/EIEC co-infection each formed 0.6% (1/170), and one child who had diarrhea was co-infected with EAEC-Cryptosporidium-Shigella/EIEC (0.06%, 1/170). There was no obvious trend in the distribution of pathogens by age in the two sub-populations, especially for EAEC (Table 4).
Table 4.
Distribution of pathogens by age*
| Any infection | Diarrhea (N = 170) | Non-diarrhea (N = 104) | ||||||
|---|---|---|---|---|---|---|---|---|
| Age category/months | Age category/months | |||||||
| 0–6 (N = 26) | 7–12 (N = 54) | 13–24 (N = 77) | 25–60 (N = 13) | 0–6 (N = 35) | 7–12 (N = 30) | 13–24 (N = 24) | 25–60 (N = 15) | |
| EAEC | 24 (92.3) | 42 (77.7) | 69 (89.6) | 12 (92.3) | 25 (71.4) | 24 (80) | 18 (75) | 13 (86.7) |
| Cryptosporidium | 3 (11.5) | 4 (7.4) | 6 (7.8) | 13 (7.7) | – | – | 1 (4.2) | – |
| E. histolytica | – | 2 (3.7) | 3 (3.9) | – | – | – | – | – |
| Shigella/EIEC | 1 (3.8) | 1 (1.8) | 3 (3.9) | 1 (7.7) | – | – | – | – |
| Giardia spp. | – | – | – | – | – | – | – | – |
EAEC = enteroaggregative Escherichia coli; EIEC = enteroinvasive E. coli.
EAEC virulence genes distribution.
Of the four genes associated with EAEC, aatA was the most frequently detected (67.2%) of all fecal DNA, followed by aap (59.9%), aggR (42.7%), and aaiC (33.6%) (Table 5). The EAEC's plasmid gene aap was significantly associated with diarrhea (OR = 2.506 [95% CI, 1.516–4.144], P < 0.001) and the chromosomal gene aaiC was not (OR = 1.639 [95% CI, 0.0962–2.792], P = 0.086) (Table 5). Multiple gene combinations were also observed in EAEC infections in our study population, and the presence of any three genes was associated with diarrhea (OR = 2.101 [95% CI, 1.261–3.502], P = 0.006). We did not find any of the EAEC virulence genes associated with malnutrition (WAZ < −1) (P > 0.05).
Table 5.
EnteroaggregativeEscherichia coli (EAEC) virulence factor-positive in stool samples*
| Characteristic | Diarrhea (N = 170) N (%) | Non-diarrhea (N = 104) N (%) | Total (N = 274) N (%) | OR [95% CI] | χ2 | P value |
|---|---|---|---|---|---|---|
| EAEC virulence-related gene | ||||||
| aaiC | 64 (37.6) | 28 (26.9) | 92 (33.6) | 1.639 [0.962–2.792] | 3.327 | 0.086 |
| aggR | 79 (46.5) | 38 (36.5) | 117 (42.7) | 1.508 [0.914–2.486] | 2.602 | 0.131 |
| aatA | 118 (69.4) | 66 (63.5) | 184 (67.2) | 1.307 [0.780–2.188] | 1.036 | 0.354 |
| aap | 116 (68.2) | 48 (46.2) | 164 (59.9) | 2.506 [1.516–4.144] | 13.093 | < 0.001 |
| EAEC gene combination | ||||||
| Any 1 gene | 147 (86.5) | 80 (76.9) | 227 (82.8) | 1.917 [1.018–3.612] | 4.139 | 0.048 |
| Any 2 genes | 130 (76.5) | 73 (70.2) | 203 (74.1) | 1.380 [0.797–2.391] | 1.325 | 0.259 |
| Any 3 genes | 84 (49.4) | 33 (31.7) | 117 (42.7) | 2.101 [1.261–3.502] | 8.224 | 0.006 |
| All 4 genes | 34 (20.0) | 13 (12.5) | 47 (17.2) | 1.750 [0.876–3.496] | 2.554 | 0.110 |
OR = odds ratio; 95% CI = confidence interval; P < 0.05 is significant.
Fecal lactoferrin levels.
Figure 1 shows enteric pathogens detected in children with/without diarrhea and the distribution of fecal lactoferrin levels. Generally, the mean lactoferrin levels were lower in children who had diarrhea and were also malnourished compared with those with diarrhea but were nourished. Regardless of the enteric pathogen detected, fecal lactoferrin levels were relatively high (manufacturer's cut-off value = 7.24 μg/mL). Especially for EAEC infection, both controls and patients had a wide range of lactoferrin levels, regardless of whether they were nourished or malnourished (Figure 1).
Figure 1.
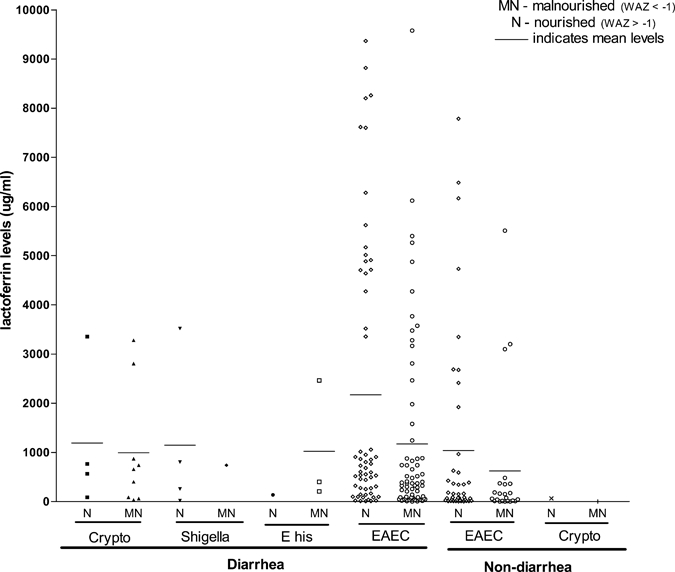
Enteric pathogens detected and fecal lactoferrin (LF) levels. Crypto = Cryptosporidium spp.; E his = E. hiostolytica; EAEC = enteroaggregative Escherichia coli. *Breast-feeding may cause moderately (15 ≤ 120 μg/mL) increased LF (Lima and others, unpublished observation).
Children with diarrhea had significantly higher fecal lactoferrin levels (N = 143; 1658.9 ± 204.2 μg/mL) compared with those without diarrhea (N = 84; 935.5 ± 194.4 μg/mL) (P = 0.019). The aatA gene and the presence of any one or two genes of EAEC were also significantly (P < 0.05) associated with elevated fecal lactoferrin levels (Table 6). In comparing diarrhea with non-diarrhea stool specimens, EAEC's chromosomal gene aaiC showed the highest fold-increase in fecal lactoferrin level (2.7) followed by the aap gene (2.5). Additionally, detection of a multiple virulence gene of EAEC in a stool was associated with an increased fold-rise in the mean fecal lactoferrin level between children with and without diarrhea (Table 6).
Table 6.
EAEC genes detected and fecal lactoferrin levels*
| Characteristic | Diarrheal stool n (mean ± SE) μg/mL | Non-diarrheal stool n (mean ± SE) μg/mL | Fold-rise† | P value |
|---|---|---|---|---|
| EAEC gene | ||||
| aaiC | 56 (1021.0 ± 251.6) | 25 (821.2 ± 402.1) | 1.2 | 0.667 |
| aggR | 66 (1713.9 ± 292.5) | 33 (934.1 ± 329.2) | 1.8 | 0.104 |
| aatA | 98 (1706.1 ± 244.1) | 54 (927.1 ± 259.1) | 1.8 | 0.044 |
| aap | 101 (1656.7 ± 245.9) | 39 (901.2 ± 261.6) | 1.8 | 0.080 |
| EAEC gene combinations | ||||
| Any 1 gene | 123 (1633.5 ± 215.4) | 66 (1175.4 ± 356.8) | 1.4 | 0.029 |
| Any 2 gene | 107 (1679.8 ± 231.1) | 60 (892.6 ± 234.3) | 1.9 | 0.028 |
| Any 3 gene | 73 (1504.9 ± 269.7) | 30 (1132.8 ± 394.8) | 1.3 | 0.451 |
| Any 4 gene | 30 (1319.5 ± 400.4) | 11 (370.5 ± 285.5) | 3.6 | 0.175 |
| All specimens | 143 (1658.9 ± 204.2) | 84 (935.5 ± 194.4) | 1.8 | 0.019 |
EAEC = enteroaggregative Escherichia coli.
Ratio of diarrhea and non-diarrhea mean lactoferrin levels.
Discussion
Nutritional shortfalls among our study population.
Mild to severe malnutrition (WAZ < −1, which reflects both acute and chronic types of malnutrition) was identified in 49.8% of our study population. Severe growth shortfalls occurred in both children with and without diarrhea in the entire study population. Growth faltering in infants from developing countries has been reported to occur as early as 2–3 months of age28,29 in contrast to developed countries,28 a fact attributable to timing of complementary feeding. For example, in the DARLING (Davis Area Research on Lactation, Infant Nutrition and Growth) study, although breast milk intakes were similar, the amount and nutrient density of food consumed after 6 months were lower in Peru than in the United States.28 In 2005, Antwi8 noted a 21.2% incidence of wasting (acute malnutrition) among children seen in Kumasi, Ghana, similar to an overall incidence of 22.9% (37/161) in the current study—both consistent with the 22.1% estimate for Ghana by the World Food Program.30
Approximately 14% (38/269) of the children we studied were severely malnourished (WAZ < −3), and, out of this number, 73.7% had diarrhea. A self-perpetuating vicious cycle in which malnutrition and diarrhea are synergistic is suggested, and may explain their effect on cognitive development of children, especially their semantic fluency.6 Our data supports this link between malnutrition and diarrhea, placing these groups of children at a higher risk of morbidity over time.
Bacterial culture versus real-time PCR for detection of pathogens.
The PCR methodology is more sensitive in screening for pathogens. In sub-Saharan Africa as in many settings, however, the cost involved cannot be passed on to patients and therefore this tool is used mainly in research facilities. In this study, routine bacterial culture detected only one stool positive for S. flexneri and none for Salmonella spp. However, PCR detected six ipaH genes in diarrheal stool specimens. Although, the ipaH gene is expressed by both Shigella and enteroinvasive (EIEC), EIEC is not often detected in Ghana,31 and we speculate that the ipaH gene detected most likely reflects the presence of Shigella spp. that was missed on the culture. The sero-group identified by culture and serology is predominant in Ghana32 as it is in many developing countries.
Cryptosporidium spp. was not only the most prevalent parasite detected, but it is also significantly associated with diarrhea (8.7% versus 1.0%, P = 0.011). Children who were malnourished and had diarrhea often had cryptosporidiosis. Prevalence rates of Cryptosporidium spp. using presumably less sensitive methods like microscopy and ELISA in Ghana,33,34 Liberia,35 Mexico,36 and Guinea Bissau37 reported ranges between 7.7% and 29% in symptomatic children. The current study recorded 8.2% in symptomatic and 1.0% in non-symptomatic patients.
Entamoeba histolytica causes amebic colitis, amebic dysentery, and liver abscess. Modern diagnostic tools like PCR are able to discriminate E. histolytica from the non-pathogenic E. dispar.25 This study detected E. histolytica in 2.9% (5/170) of the children who had diarrhea. No visible blood was observed in any of the stools from these children. All the E. histolytica detected in the current study were from children who had diarrhea. In Bangladesh and elsewhere, relatively few numbers of people infected with E. histolytica develop symptomatic disease.18,38,39
This study did not detect any Giardia in the stool specimens screened for the p241 gene of Giardia spp. The p241 gene target we used is well validated26 and our positive control, which was included in each PCR run was amplified while the negative control was not. Addy and others40 in 2004, found a 3.7% prevalence rate of Giardia in Kumasi, Ghana. In a 7-year study of diarrhea caused by parasites in Guinea Bissau, the most prevalent parasite was Giardia lamblia (14.8%) followed by Cryptosporidium (7.7%).37 Although seasonality, study duration, or geographical location may influence parasite prevalence, we cannot pinpoint additional reasons for the zero prevalence of Giardia in the current study.
Children who had diarrhea were often co-infected, with two or more pathogens detected in the stool, and EAEC-Cryptosporidium was the most prevalent (13/170) in the current study. Among human immunodeficiency virus (HIV)-infected children in South Africa, Samie and others15 identified children who were co-infected with as many as six different species of pathogens. This study detected a child who had diarrhea with EAEC-Cryptosporidium-Shigella/EIEC co-infection. The HIV status of this child is however not known. Relatively fewer children who are > 2 years of age were sampled in this study. However, cryptosporidiosis seemed to be more common in children < 2 years of age, and this is in agreement with an earlier study in Ghana.34
Prevalence of EAEC virulence-associated genes.
Among the EAEC plasmid genes (aap, aatA, and aggR) we tested, only aap was significantly associated with diarrhea (P = 0.0003). A recent publication, which compared molecular probes to the “gold standard” (aggregation of cultured epithelial cells), however, suggested that the aap gene is not restricted to EAEC, but is also detected in diffusely adherent E. coli (DAEC) and in non-pathogenic E. coli.41 We did not observe any significant statistical association between any individual EAEC gene (whether plasmid or chromosome borne) and malnutrition (WAZ < −1).
Information on the presence of EAEC virulence-associated genes in body fluids of persons in Africa is limited, as is the prevalence and distribution of this organism in Ghana. The aggR is known to regulate its own expression and that of several plasmid genes and chromosomal genes of EAEC,42 including the aggregative adherence fimbriae, a dispersin (aap), a dispersin translocator apparatus called aat, and several chromosomal loci including the aaiC.43 In a different population, Huang and others failed to show any association between four EAEC virulence-associated genes (aggA, aspU, aafA and aggR) and clinical illness in travelers from the United States to Mexico.43 Furthermore, in that study, aspU (now designated aatA) was the least prevalent gene among the four EAEC virulence genes studied,43 whereas this same gene (aatA) was the most prevalent (67.2%) in the current study. In South Africa, Samie and others15 found the aap gene to be the predominant among others and also associated with diarrhea. These disparate findings on the relative distribution and importance of EAEC genes in diarrheal illnesses suggest that geographical location, type of exposure, and/or host factors may dictate the nature of EAEC infection. For example, among the risks of contracting traveler's diarrhea, the country of destination was the most important determining factor as reported by Cabada and White.12
Although aatA was the most prevalent gene observed in this study, it was not associated with diarrhea (P > 0.05) (Table 5). Some studies have shown that the novel protein aatA, which is encoded on EAEC virulence plasmid pAA2, localizes to the outer membrane of this bacterium and facilitates export of the dispersin aap across the outer membrane.45,46 Our results may support this notion because aatA and aap were detected in greater than 68% of patients with diarrhea. Further study is required for a firm conclusion. Of interest, the chromosomal gene aaiC did not show any significant differential association between the diarrhea and the non-diarrhea group in the current study (P = 0.068). About 12% (13/104) of children without diarrhea in the current study were positive for all four EAEC genes tested. The presence of 2, 3, or 4 genes in stool specimens of study children suggests a strong association. For example, the presence of three of these genes is associated with a higher OR than one gene alone (Table 5).
One explanation for the high frequency of EAEC-associated virulence genes in symptomatic and asymptomatic patients and the heterogeneity of the different gene assortments found is that EAEC is endemic in our study population. In support of this notion, almost all prior studies recover EAEC from controls47–49 and from individuals with diarrhea.17,50 Pathogenic factors may influence the initiation of a symptomatic phase, determined by and include distinct mechanisms as reviewed by Kaper and others.51 For example, Huang and others suggested that a first exposure to EAEC infection “primes” the immune system to prevent a second infection.44 Furthermore, in their study of traveler's diarrhea, after the initial EAEC infection, only 4 (11%) of the subjects had a subsequent symptomatic EAEC infection.44
Lactoferrin levels.
Lactoferrin is bactericidal to enteric pathogens, modulates the intestinal immune response, and is released by neutrophils into stool in response to infection.52 Of interest in the current study, the mean lactoferrin levels were lower in children who were malnourished and had diarrhea compared with those without malnutrition who had diarrhea. We speculate that the lactoferrin assay may be marginally less sensitive in the setting of malnutrition. Perhaps, the enterocytes are less able to synthesize lactoferrin in children who are malnourished and have diarrhea. This observation needs further investigation for a firm conclusion. In earlier studies, the mean lactoferrin levels for healthy controls were less than 12.8 μg/mL27,52 compared with healthy controls (298.8 μg/mL) in the current study. We do not have immediate explanation for this observation. However, unpublished observations by Lima and others suggest that breast milk may contribute to moderately (15 to 120 μg/mL) increased fecal lactoferrin levels. Some studies have demonstrated an association of EAEC infection with inflammatory cytokines53,54 and several others have associated EAEC with elevated lactoferrin levels.15,55 In children with diarrhea, we found a significant statistical association of elevated fecal lactoferrin with the aap gene, and with the detection of any one or two of the EAEC genes tested (P < 0.05). Between diarrhea and non-diarrhea stool specimens, EAEC's chromosomal gene aaiC was associated with the highest fold-rise in fecal lactoferrin level (2.7) followed by the aap gene (2.5). Additionally, more virulence EAEC genes detected corresponded to a rise in the fecal lactoferrin level (Table 6). The protective function of lactoferrin on infections with enteropathogens have been acknowledged,56 and colonization/infection, particularly by EAEC in the current study probably contributed to the high lactoferrin levels (Table 6).
The current study had some limitations. Accurate height measurements were the most difficult to obtain on an outpatient basis, and this limited two of the three z-scores of malnutrition (WHZ and HAZ). Knowledge of history of antimicrobial exposure is important, however, this information was obtainable in only a few of the patients. Only selected pathogens were assayed using PCR and thus, we may have missed other less common pathogens. That is, no stool analyses for intestinal parasites other than Cryptosporidium, Giardia, and E. histolytica were performed, therefore, missing helmintic infections.
In conclusion, through the use of specific DNA-biomarkers, we were able to determine that EAEC and Cryptosporidium were common intestinal pathogens and that elevated fecal lactoferrin levels were associated with diarrhea in this group of children from southern Ghana.
Acknowledgments
We thank David Kwarteng and the nurses at the Princess Marie Louise Hospital for data collection and anthropometric measurements. Relanna Pinkerton at UVA reviewed all statistical analyses.
Footnotes
Financial support: This study was supported in part by the University of Ghana Medical School, College of Health Sciences, Accra, Ghana and Pfizer-supported funds to the Center for Global Health, University of Virginia, Charlottesville, VA.
Authors' addresses: Japheth A. Opintan, Mercy J. Newman, and Patrick F. Ayeh-Kumi, Department of Microbiology, University of Ghana Medical School, Korle-Bu, Accra, Ghana, E-mails: jyo4g@virginia.edu, newmerci@yahoo.co.uk, and payehkumi@yahoo.com. Raymond Affrim and Rosina Gepi-Attee, Princess Marie Louise Children's Hospital, Accra, Ghana, E-mails: raffrim@yahoo.com and rgepi_attee@yahoo.com. Jesus E. A. D. Sevilleja, James K. Roche, Cirle A. Warren, and Richard L. Guerrant, Center for Global Health, Division of Infectious Diseases and International Health, University of Virginia, Charlottesville, VA, E-mails: emsevilleja@yahoo.com, jkr7m@virginia.edu, ca6t@virginia.edu, and rlg9a@virginia.edu. James P. Nataro, Center for Vaccine Development, Department of Pediatrics and Medicine, University of Maryland, Baltimore, MD, E-mail: jpn2r@virginia.edu.
References
- 1.Bryce J, Boschi-Pinto C, Shibuya K, Black RE. WHO estimates of the causes of death in children. Lancet. 2005;365:1147–1152. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- 2.Kosek M, Bern C, Guerrant RL. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull World Health Organ. 2003;81:197–204. [PMC free article] [PubMed] [Google Scholar]
- 3.Bern C, Martines J, de Zoysa I, Glass RI. The magnitude of the global problem of diarrhoeal disease: a ten-year update. Bull World Health Organ. 1992;70:705–714. [PMC free article] [PubMed] [Google Scholar]
- 4.Durley A, Shenoy A, Faruque AS, Suskind R, Ahmed T. Impact of a standardized management protocol on mortality of children with diarrhoea: an update of risk factors for childhood death. J Trop Pediatr. 2004;50:271–275. doi: 10.1093/tropej/50.5.271. [DOI] [PubMed] [Google Scholar]
- 5.Lima AA, Moore SR, Barboza MS, Jr, Soares AM, Schleupner MA, Newman RD, Sears CL, Nataro JP, Fedorko DP, Wuhib T, Schorling JB, Guerrant RL. Persistent diarrhea signals a critical period of increased diarrhea burdens and nutritional shortfalls: a prospective cohort study among children in northeastern Brazil. J Infect Dis. 2000;181:1643–1651. doi: 10.1086/315423. [DOI] [PubMed] [Google Scholar]
- 6.Guerrant RL, Oria RB, Moore SR, Oria MO, Lima AA. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr Rev. 2008;66:487–505. doi: 10.1111/j.1753-4887.2008.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.UNICEF, Ghana . ChildInfo: Monitoring the Situation of Children and Women. Child Nutrition. Statistics by Area/Child Nutrition; 2009. [Google Scholar]
- 8.Antwi S. Malnutrition: missed opportunities for diagnosis. Ghana Med J. 2008;42:101–104. doi: 10.4314/gmj.v42i3.43627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joosten KF, Hulst JM. Prevalence of malnutrition in pediatric hospital patients. Curr Opin Pediatr. 2008;20:590–596. doi: 10.1097/MOP.0b013e32830c6ede. [DOI] [PubMed] [Google Scholar]
- 10.Gomez-Duarte OG, Bai J, Newell E. Detection of Escherichia coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae, and Campylobacter spp. enteropathogens by 3-reaction multiplex polymerase chain reaction. Diagn Microbiol Infect Dis. 2009;63:1–9. doi: 10.1016/j.diagmicrobio.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adachi JA, Jiang ZD, Mathewson JJ, Verenkar MP, Thompson S, Martinez-Sandoval F, Steffen R, Ericsson CD, DuPont HL. Enteroaggregative Escherichia coli as a major etiologic agent in traveler's diarrhea in 3 regions of the world. Clin Infect Dis. 2001;32:1706–1709. doi: 10.1086/320756. [DOI] [PubMed] [Google Scholar]
- 12.Cabada MM, White AC., Jr Travelers' diarrhea: an update on susceptibility, prevention, and treatment. Curr Gastroenterol Rep. 2008;10:473–479. doi: 10.1007/s11894-008-0087-7. [DOI] [PubMed] [Google Scholar]
- 13.Paul M, Tsukamoto T, Ghosh AR, Bhattacharya SK, Manna B, Chakrabarti S, Nair GB, Sack DA, Sen D, Takeda Y. The significance of enteroaggregative Escherichia coli in the etiology of hospitalized diarrhoea in Calcutta, India and the demonstration of a new honey-combed pattern of aggregative adherence. FEMS Microbiol Lett. 1994;117:319–325. doi: 10.1111/j.1574-6968.1994.tb06786.x. [DOI] [PubMed] [Google Scholar]
- 14.Scavia G, Staffolani M, Fisichella S, Striano G, Colletta S, Ferri G, Escher M, Minelli F, Caprioli A. Enteroaggregative Escherichia coli associated with a foodborne outbreak of gastroenteritis. J Med Microbiol. 2008;57:1141–1146. doi: 10.1099/jmm.0.2008/001362-0. [DOI] [PubMed] [Google Scholar]
- 15.Samie A, Obi CL, Dillingham R, Pinkerton RC, Guerrant RL. Enteroaggregative Escherichia coli in Venda, South Africa: distribution of virulence-related genes by multiplex polymerase chain reaction in stool samples of human immunodeficiency virus (HIV)-positive and HIV-negative individuals and primary school children. Am J Trop Med Hyg. 2007;77:142–150. [PubMed] [Google Scholar]
- 16.Nataro JP, Kaper JB, Robins-Browne R, Prado V, Vial P, Levine MM. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr Infect Dis J. 1987;6:829–831. doi: 10.1097/00006454-198709000-00008. [DOI] [PubMed] [Google Scholar]
- 17.Okeke IN, Ojo O, Lamikanra A, Kaper JB. Etiology of acute diarrhea in adults in southwestern Nigeria. J Clin Microbiol. 2003;41:4525–4530. doi: 10.1128/JCM.41.10.4525-4530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haque R, Huston CD, Hughes M, Houpt E, Petri WA., Jr Amebiasis. N Engl J Med. 2003;348:1565–1573. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- 19.Kosek M, Alcantara C, Lima AA, Guerrant RL. Cryptosporidiosis: an update. Lancet Infect Dis. 2001;1:262–269. doi: 10.1016/S1473-3099(01)00121-9. [DOI] [PubMed] [Google Scholar]
- 20.Ortega YR, Adam RD. Giardia: overview and update. Clin Infect Dis. 1997;25:545–549. doi: 10.1086/513745. quiz 550. [DOI] [PubMed] [Google Scholar]
- 21.Cerna JF, Nataro JP, Estrada-Garcia T. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. J Clin Microbiol. 2003;41:2138–2140. doi: 10.1128/JCM.41.5.2138-2140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czeczulin JR, Whittam TS, Henderson IR, Navarro-Garcia F, Nataro JP. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect Immun. 1999;67:2692–2699. doi: 10.1128/iai.67.6.2692-2699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sethabutr O, Venkatesan M, Murphy GS, Eampokalap B, Hoge CW, Echeverria P. Detection of Shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J Infect Dis. 1993;167:458–461. doi: 10.1093/infdis/167.2.458. [DOI] [PubMed] [Google Scholar]
- 24.Cryptosporidium. 2009. DB (version 4.2) Cryptosporidium Genomic Resources: 18s ribosomal RNA.
- 25.Haque R, Roy S, Siddique A, Mondal U, Rahman SM, Mondal D, Houpt E, Petri WA., Jr Multiplex real-time PCR assay for detection of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium spp. Am J Trop Med Hyg. 2007;76:713–717. [PubMed] [Google Scholar]
- 26.Guy RA, Payment P, Krull UJ, Horgen PA. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl Environ Microbiol. 2003;69:5178–5185. doi: 10.1128/AEM.69.9.5178-5185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kane SV, Sandborn WJ, Rufo PA, Zholudev A, Boone J, Lyerly D, Camilleri M, Hanauer SB. Fecal lactoferrin is a sensitive and specific marker in identifying intestinal inflammation. Am J Gastroenterol. 2003;98:1309–1314. doi: 10.1111/j.1572-0241.2003.07458.x. [DOI] [PubMed] [Google Scholar]
- 28.Dewey KG, Peerson JM, Heinig MJ, Nommsen LA, Lonnerdal B, Lopez de Romana G, de Kanashiro HC, Black RE, Brown KH. Growth patterns of breast-fed infants in affluent (United States) and poor (Peru) communities: implications for timing of complementary feeding. Am J Clin Nutr. 1992;56:1012–1018. doi: 10.1093/ajcn/56.6.1012. [DOI] [PubMed] [Google Scholar]
- 29.Sathian U, Joseph A, Waterlow JC. Exclusive breast feeding versus supplementation: a prospective study in a rural south Indian community. Ann Trop Paediatr. 1983;3:157–161. doi: 10.1080/02724936.1983.11748288. [DOI] [PubMed] [Google Scholar]
- 30.World Food Programme WFP World hunger—Ghana. 2006. http://www.wfp.org/country_brief/indexcountry.asp?country=288 Available at.
- 31.Opintan JA, Rima AB, Newman MJ, Okeke IN. Carriage of diarrhoeagenic Escherichia coli by older children and adults in Accra, Ghana. Trans R Soc Trop Med Hyg. 2010;104:504–506. doi: 10.1016/j.trstmh.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 32.Opintan J, Newman MJ. Distribution of serogroups and serotypes of multiple drug resistant Shigella isolates. Ghana Med J. 2007;41:4–8. [PMC free article] [PubMed] [Google Scholar]
- 33.Addy PA, Aikins-Bekoe P. Cryptosporidiosis in diarrhoeal children in Kumasi, Ghana. Lancet. 1986;1:735. doi: 10.1016/s0140-6736(86)91119-0. [DOI] [PubMed] [Google Scholar]
- 34.Adjei AA, Armah H, Rodrigues O, Renner L, Borketey P, Ayeh-Kumi P, Adiku T, Sifah E, Lartey M. Cryptosporidium spp., a frequent cause of diarrhea among children at the Korle-Bu Teaching Hospital, Accra, Ghana. Jpn J Infect Dis. 2004;57:216–219. [PubMed] [Google Scholar]
- 35.Hojlyng N, Molbak K, Jepsen S. Cryptosporidium spp., a frequent cause of diarrhea in Liberian children. J Clin Microbiol. 1986;23:1109–1113. doi: 10.1128/jcm.23.6.1109-1113.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller K, Duran-Pinales C, Cruz-Lopez A, Morales-Lechuga L, Taren D, Enriquez FJ. Cryptosporidium parvum in children with diarrhea in Mexico. Am J Trop Med Hyg. 1994;51:322–325. doi: 10.4269/ajtmh.1994.51.322. [DOI] [PubMed] [Google Scholar]
- 37.Perch M, Sodemann M, Jakobsen MS, Valentiner-Branth P, Steinsland H, Fischer TK, Lopes DD, Aaby P, Molbak K. Seven years' experience with Cryptosporidium parvum in Guinea-Bissau, West Africa. Ann Trop Paediatr. 2001;21:313–318. doi: 10.1080/07430170120093490. [DOI] [PubMed] [Google Scholar]
- 38.Ayeh-Kumi PF, Ali IM, Lockhart LA, Gilchrist CA, Petri WA, Jr, Haque R. Entamoeba histolytica: genetic diversity of clinical isolates from Bangladesh as demonstrated by polymorphisms in the serine-rich gene. Exp Parasitol. 2001;99:80–88. doi: 10.1006/expr.2001.4652. [DOI] [PubMed] [Google Scholar]
- 39.Verweij JJ, Oostvogel F, Brienen EA, Nang-Beifubah A, Ziem J, Polderman AM. Short communication: prevalence of Entamoeba histolytica and Entamoeba dispar in northern Ghana. Trop Med Int Health. 2003;8:1153–1156. doi: 10.1046/j.1360-2276.2003.01145.x. [DOI] [PubMed] [Google Scholar]
- 40.Addy P, Antipim G, Frimpong E. Prevalence of pathogenic Escherichia coli and parasites in infants with diarrhoea in Kumasi, Ghana. East Afr Med J. 2004;81:353–357. doi: 10.4314/eamj.v81i7.9190. [DOI] [PubMed] [Google Scholar]
- 41.Monteiro BT, Campos LC, Sircili MP, Franzolin MR, Bevilacqua LF, Nataro JP, Elias WP. The dispersin-encoding gene (aap) is not restricted to enteroaggregative Escherichia coli. Diagn Microbiol Infect Dis. 2009;65:81–84. doi: 10.1016/j.diagmicrobio.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 43.Huang DB, Jiang ZD, Dupont HL. Association of virulence factor-positive and -negative enteroaggregative Escherichia coli and occurrence of clinical illness in travelers from the United States to Mexico. Am J Trop Med Hyg. 2003;69:506–508. [PubMed] [Google Scholar]
- 44.Nataro JP. Enteroaggregative Escherichia coli pathogenesis. Curr Opin Gastroenterol. 2005;21:4–8. [PubMed] [Google Scholar]
- 45.Imuta N, Nishi J, Tokuda K, Fujiyama R, Manago K, Iwashita M, Sarantuya J, Kawano Y. The Escherichia coli efflux pump TolC promotes aggregation of enteroaggregative E. coli 042. Infect Immun. 2008;76:1247–1256. doi: 10.1128/IAI.00758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheikh J, Czeczulin JR, Harrington S, Hicks S, Henderson IR, Le Bouguenec C, Gounon P, Phillips A, Nataro JP. A novel dispersin protein in enteroaggregative Escherichia coli. J Clin Invest. 2002;110:1329–1337. doi: 10.1172/JCI16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gascon J, Vargas M, Schellenberg D, Urassa H, Casals C, Kahigwa E, Aponte JJ, Mshinda H, Vila J. Diarrhea in children under 5 years of age from Ifakara, Tanzania: a case-control study. J Clin Microbiol. 2000;38:4459–4462. doi: 10.1128/jcm.38.12.4459-4462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valentiner-Branth P, Steinsland H, Fischer TK, Perch M, Scheutz F, Dias F, Aaby P, Molbak K, Sommerfelt H. Cohort study of Guinean children: incidence, pathogenicity, conferred protection, and attributable risk for enteropathogens during the first 2 years of life. J Clin Microbiol. 2003;41:4238–4245. doi: 10.1128/JCM.41.9.4238-4245.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Albert MJ, Faruque AS, Faruque SM, Sack RB, Mahalanabis D. Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. J Clin Microbiol. 1999;37:3458–3464. doi: 10.1128/jcm.37.11.3458-3464.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rappelli P, Folgosa E, Solinas ML, Dacosta JL, Pisanu C, Sidat M, Melo J, Cappuccinelli P, Colombo MM. Pathogenic enteric Escherichia coli in children with and without diarrhea in Maputo, Mozambique. FEMS Immunol Med Microbiol. 2005;43:67–72. doi: 10.1016/j.femsim.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 52.Walker TR, Land ML, Kartashov A, Saslowsky TM, Lyerly DM, Boone JH, Rufo PA. Fecal lactoferrin is a sensitive and specific marker of disease activity in children and young adults with inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2007;44:414–422. doi: 10.1097/MPG.0b013e3180308d8e. [DOI] [PubMed] [Google Scholar]
- 53.Cennimo D, Abbas A, Huang DB, Chiang T. The prevalence and virulence characteristics of enteroaggregative Escherichia coli at an urgent-care clinic in the USA: a case-control study. J Med Microbiol. 2009;58:403–407. doi: 10.1099/jmm.0.005793-0. [DOI] [PubMed] [Google Scholar]
- 54.Greenberg DE, Jiang ZD, Steffen R, Verenker MP, DuPont HL. Markers of inflammation in bacterial diarrhea among travelers, with a focus on enteroaggregative Escherichia coli pathogenicity. J Infect Dis. 2002;185:944–949. doi: 10.1086/339617. [DOI] [PubMed] [Google Scholar]
- 55.Bouckenooghe AR, Dupont HL, Jiang ZD, Adachi J, Mathewson JJ, Verenkar MP, Rodrigues S, Steffen R. Markers of enteric inflammation in enteroaggregative Escherichia coli diarrhea in travelers. Am J Trop Med Hyg. 2000;62:711–713. doi: 10.4269/ajtmh.2000.62.711. [DOI] [PubMed] [Google Scholar]
- 56.Ochoa TJ, Cleary TG. Effect of lactoferrin on enteric pathogens. Biochimie. 2009;91:30–34. doi: 10.1016/j.biochi.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


