Abstract
Apolipoprotein A-I Milano (apoA-IMilano) is a naturally occurring human mutation of wild type apoliprotein A-I (apoA-IWT) having cystine substituted for arginine173. Two molecules of apo-IWT form discs with phospholipid having a defined relationship between the apoA-IWT molecules. ApoA-IMilano forms cystine homodimers that would not allow the protein to adopt the conformation reported for apoA-IWT. The conformational constraints for dimeric apoA-IMilano recombinant high density lipoprotein (rHDL) discs made with phospholipid were deduced from a combination of chemical cross-linking and mass spectrometry. Lysine-selective homo-bifunctional cross-linkers were reacted with homogeneous rHDL having diameters of 78Å and 125Å. After reduction, cross-linked apoA-IMilano was separated from monomeric apoprotein by gel electrophoresis then subjected to in-gel trypsin digest. Cross-linked peptides were confirmed by MS/MS sequencing. The cross-links provided distance constraints that were used to refine models of lipid-bound dimeric apoA-IMilano. These studies suggest that a single dimeric apoA-IMilano on 78Å diameter rHDL girdles the edge of a phospholipid disc assuming a “belt” conformation similar to the “belt” region of apoA-IWT on rHDL. However, the C-terminal end of dimeric apoA-IMilano wraps around the periphery of the particle to shield the fatty acid chains from water rather than folding-back onto the “belt” as does apoA-IWT. The two apoA-IMilano dimers on the 125Å diameter rHDL encircle the periphery of a phospholipid disc, but appear to reside on the surface of a laminar micelle.
HDL plays a central role in a process called reverse cholesterol transport (1, 2) where apolipoprotein A-I (apoA-I)1, the principal protein associated with HDL, collects and organizes phospholipid and cholesterol that have been removed from extra-hepatic cells. There is a large body of evidence demonstrating an inverse correlation between plasma HDL and the risk of developing atherosclerosis (3–6). However, all carriers of an apoA-I mutation in which cysteine is substituted for arginine at 173, named apoA-I Milano (apoA-IMilano), have low levels of plasma HDL cholesterol and apoA-I. The low levels of plasma HDL would suggest that carriers of the Milano mutation would be at high risk for atherosclerosis, but the contrary was reported (7, 8). Clinical trials in which repeated intravenous infusions of lipid-complexed apoA-IMilano were administered to patients with coronary disease showed a reduction in the volume of atherosclerotic lesions (9, 10), while several animal studies have demonstrated that intravenous injection of apoA-IMilano regressed atherosclerosis in apoE knockout mice (11–14), rabbits (15–17) and pigs (18). Several animal studies employing targeted replacement of apoA-I with apoA-IMilano have supported the hypothesis that apoA-IMilano was more athero-protective (19) than wild-type apoA-I (apoA-IWT) while other studies have suggested that apoA-IMilano was no more effective than apoA-IWT (20, 21).
It is not known to what extent the apoA-IMilano mutation at residue 173 alters the lipid-bound conformation, but molecular modeling indicates that monomeric apoA-IMilano on rHDL discs assumes a “belt” configuration (22) similar to that reported for apoA-IWT (23). Because apoA-IMilano is present in plasma as a homodimer or as a herterodimer with apoA-II, it has been assumed that the Cys173-Cys173 disulfide linkage between helix 7 of one monomer and helix 7 of the second apoA-IMilano molecule in the homodimer forces the two apoA-I monomers on rHDL to assume a conformation that is different (24) from that suggested for apoA-IWT (23, 25–31). Therefore, there would be no overlap of the helix 5 to helix 5 region that is a major feature of the lipid-bound conformation (32–36).
Considerable agreement exits among the various published models of apoA-IWT on POPC rHDL, suggesting that repeats 2 through 8, called the central region, of lipid-bound apoA-IWT assumes an antiparallel orientation with a helix 5 to helix 5 registry, e.g., central amphipathic helices on each apoA-IWT monomer are next to one another (22, 23, 25–29, 32, 36–42). The results of Wu et al. (29) and Martin et al. (43) suggest that the two apoA-IWT chains do not wrap around the phospholipid disc forming smooth loop, but imply that there is additional protein structure important for LCAT activation and reverse cholesterol transport. Both Wu et al. (26)(29) and Bhat et al. (23, 25) suggest that the C-terminal end is somewhat folded back onto the central region or “belt” as proposed by Bhat et al. (23, 25). However, the principal debate or differences between the models center on ascertaining the conformation assumed by the N- and C-terminal ends.
All models to date suggest that the N- and C-terminal ends are close to one another. However, Wu et al. (29) suggest that the N-terminal end is relatively disordered while Bhat et al. (23, 25) have suggested that both the N- and C-termini are relatively ordered and interact with the central part of the apoA-IWT molecule. Since the results from chemical cross-linking/mass spectrometry (CCL/MS) yield only a time averaged conformation it appears that the N-terminal ends of apoA-IWT spend a significant amount of time close to the “belt,” but does not rule out appreciable mobility for the N-terminus that would allow this region to extend into solution and to possibly overlap with its own C-terminus. Importantly, it should be noted, that CCL is conducted at low cross-linker to protein ratios, thus only 1 or 2 cross-links per apoA-I molecule are formed. This ensures that the formation of cross-links do induce artifactual change to the native conformation of the protein. Therefore, CCL/MS provides a snapshot of the proteins conformation at the time the covalent bonds are formed and provides a powerful method for determining 3-dimensional experimental coordinates of proteins conformation in solution.
In the present report we show our findings regarding the conformation of disulfide coupled apoA-IMilano dimers on rHDL made from POPC. Dimeric apoA-IMilano complexed with POPC yielded rHDL with diameters of 78Å and 125Å that were analyzed using CCL/MS techniques. In answer to the question “could the unique structure of apoA-IMilano affect the biochemical properties of rHDL,” we find that the conformation for apoA-IMilano on rHDL had many similarities to the conformation of apoA-IWT on rHDL, particularly in the “belt” region, but that there were also significant differences in the folding of the N- and C-terminal ends or “buckles” when bound to lipid.
EXPERIMENTAL PROCEDURES
Materials
Bis(sulfosuccinimidyl)suberate (BS3), dithiobis(succinimidylpropionate) (DSP), disuccinimidylglutarate (DSG), bis-N-succinimidyl(pentaethyleneglycol) ester (BS(PEG)5) and ethyleneglycol(bis(succinimidylsuccinate)) (EGS) were from ThermoFisher. POPC and Me2SO were from Sigma-Aldrich. Sequencing grade modified trypsin and restriction enzymes were from Promega. RapiGest SF was obtained from Waters Inc. Formic acid was from Sigma-Aldrich. Sodium desoxycholate, potassium chloride, optima grade methanol, chloroform, acetonitrile, and glacial acetic acid were from Fisher Scientific. Mark 12 molecular weight standards and Simply Blue Safestain were from Invitrogen. Ultrafree-15 centrifugal and Biomax 10K membranes were from Millipore Corp.
Preparation and Purification of rHDL from POPC and Human ApoA-IMilano
Dimeric human apoA-IMilano was purified from the plasma of human carriers in the laboratory of Dr. Laura Calabresi (24). ApoA-IMilano purity was analyzed by both 12% SDS PAGE and mass spectrometry before use. Preparations of all rHDL was carried out as previously described (44) using the cholate dialysis method 45. Briefly, all preparations employed sodium cholate:POPC at a molar ratio of 1:1 with about 5 mg of dimeric apoA-IMilano. 125Å diameter particles containing 2 dimers of apoA-IMilano were prepared from a starting mixture of 120:1, POPC:dimeric apoA-IMilano, while rHDL particles having a diameter of 78Å containing one dimer of apoA-IMilano were prepared from starting molar ratio of 40:1, POPC:dimeric apoA-IMilano. POPC rHDL were prepared after reducing dimeric apoA-IMilano to monomeric apoA-IMilano with 100 mM DTT that was added 30 min before mixing with POPC. The molar ratios of POPC:monomeric apoA-IMilano were 120:1 and 40:1, respectively, like those employed to prepare rHDL particles from wild-type apoA-I (23, 25). After removal of sodium cholate by extensive dialysis the particles were purified by FPLC (44).
4–30% Gradient Gel Electrophoresis
The diameters of POPC containing rHDL were determined using nondenaturing 4–30% gradient PAGE as previously described (42, 46). Gels were run at 2800 V/hr to ensure that the particles had migrated to equilibrium then fixed and stained (47). rHDL particle size was determined by comparison to protein standards of known Stokes’ diameter (46) using a standard mixture from GE Healthcare catalogue No. 17-0445-01.
Cross-linking of rHDL
Cross-linkers were dissolved in dry Me2SO to a final concentration of 5 µg/µL and used within 5 min of preparation. The cross-linkers were added at molar ratios of 2:1 and 10:1 with a final rHDL apoA-I particle concentration of 0.4 µg/µL in 10 mM sodium phosphate pH 7.4 (25). After adding the cross-linker, the reaction was incubated for 5 min at 37°C then quenched by adding 1 M Tris, pH 7.4, giving a final concentration of 50 mM Tris. Samples were dialyzed against 10 mM ammonium bicarbonate, pH 7.4, at 4 °C to remove excess cross-linker. All samples were stored at −20 °C until processed.
SDS PAGE and In-gel Trypsin Digest
Products from CCL apoA-IMilano rHDL were separated on 12% SDS-PAGE under both non-reducing (23) and reducing conditions. Comparison of product migration between reducing and non-reducing conditions allowed the distinction between “true” chemical cross-links and disulfide bonded monomers. Protein bands were excised from the gel, minced and repeatedly dehydrated with acetonitrile. The gel pieces were rehydrated with a cold, freshly prepared solution containing 20 ng/µL trypsin in 10 mM ammonium bicarbonate, pH 7.8, 0.1 % (w/v) RapiGest SF and 1 mM CaCl2, as previously described (23, 25). The final trypsin to apoA-IMilano mass ratio was 1:20. Samples were incubated on ice for 10 min to minimize autolysis while trypsin diffuses into the gel. Then the samples were digested for 18 h at 37 °C.
Peptide Isolation and ES/Q-TOF Mass Spectrometry
Following digestion the aqueous solution was removed and the gel pieces covered with 200 µL of extraction solvent (acetonitrile/formic acid/water, v/v/v, 50:5:45). After incubating the slices for 10 min the solvent was transferred to a fresh tube. The extraction process was repeated and the combined aliquots acidified to an HCl:apoA-I ratio of 1:10 (v/v) using 500 mM HCl. After incubating the acidified solution for 35 min at 37°C precipitate was removed by centrifuging for 10 min at 13,000 rpm. The supernatant was transferred to a fresh tube before processing for mass spectrometry.
To identify candidate cross-linked peptides for MS/MS sequencing, survey scans were performed on each peptide mixture using a Waters Q-TOF API-US mass spectrometer equipped with a Waters CapLC. Acquisition was controlled by Mass-Lynx 4.0 software (23, 25). Peptides were loaded onto a PLRP-S trapping column, 0.5 mm diameter × 2.0 mm length, packed with 3 micron diameter particles having a pore diameter of 100 Å. Peptides were loaded onto the column in water/acetonitrile/formic acid (97:3:0.2) at 500 nL/min then separated by gradient elution: solvent A (25 mM formic acid in 97% water and 3% acetonitrile) and solvent B solvent B (25 mM formic acid in 3% water 97% acetonitrile). The gradient profile was as follows: 2% solvent B for 3 min, then a linear increase to 40% B at 90 min and then to 80% B in 5 min. At 95 min the solvent composition was ramped to 2% B over 5 min and then equilibrated with 2% B for 30 min before the next injection. Peptides were eluted at 470 nL/min. Positive ion electrospray survey scans were recorded in the continuum mode using a scan window from 300 to 1500 m/z with an accumulation time of 2 s. The source temperature was 80°C and the cone and capillary voltages were 45 V and 3.5 kV, respectively. The experimental m/z was corrected for the +2 charge state using apoA-I tryptic fragment 7, m/z = 806.8969, and for the +1 charge state using apoA-I tryptic fragment 12, m/z 831.4365. The survey ion scan was deconvoluted to give a list of +1 charge states that eluted from the column. This experimental list was sorted against a list of all possible lysine-to-lysine crosslinks for the particular chemical cross-linker used in an experiment. All experimental ions within ±0.051 m/z of the theoretical ions were sequenced. Product ion MS/MS spectra were acquired in the continuum mode from 50 to 1800 m/z using a data directed charge-state selective collision energy and an accumulation time of 2s. Sequence analysis of the MS/MS spectra was performed with a fragment ion tolerance of ± 0.05 m/z.
Molecular Modeling of Lipid-Bound ApoA-IMilano
The molecular modeling for apoA-IWT has been described in previous publications (23, 25) using the X-ray crystal coordinates for lipid-free Δ43-apoA-I (34) that were joined with the first 43 amino acids of the N-terminal end. Structural studies of unmodified lipid-free apoA-I were used to guide the 1–43 conformation (48–52). Using a similar approach to our elucidation of lipid-bound conformation of apoA-IWT three pieces of information were used to model the spatial relationship of the two molecules of apoA-IMilano on the POPC-containing particles. The first step was to orient the two molecules to accommodate the disulfide cross-link between cysteines at position 173 on each monomer. Then the cross-linked positions on the individual apoA-I molecules were oriented to their correct distances. To do this we used the maximum distance of Cα–Lysine-(cross-linker)-Cα–Lysine calculated for each of the cross-linkers: 26.0 Å, 26.6 Å, 30.7 Å and 36.3Å for BS3, DSP, EGS and BS(PEG)5, respectively. Third, apoA-I was bent only at the proline or glycine-glycine sites between the amphipathic segments of apoA-I. Tools available in Swiss-PdbViewer OSX v4.0.1 (http://www.expasy.org/spdbv/) were used to optimize the conformations while pdb files were manipulated using PyMOL version 1.2r3 (http://www.pymol.org). Swiss-PdbViewer, PyMOL along with Visual Molecular Dynamics (VMD) for Mac OSX version 1.8.7 (53), were used to generate the molecular figures shown in the manuscript.
RESULTS
Characterization and Cross-linking of 78 Å and 125 Å rHDL Containing ApoA-IMilano
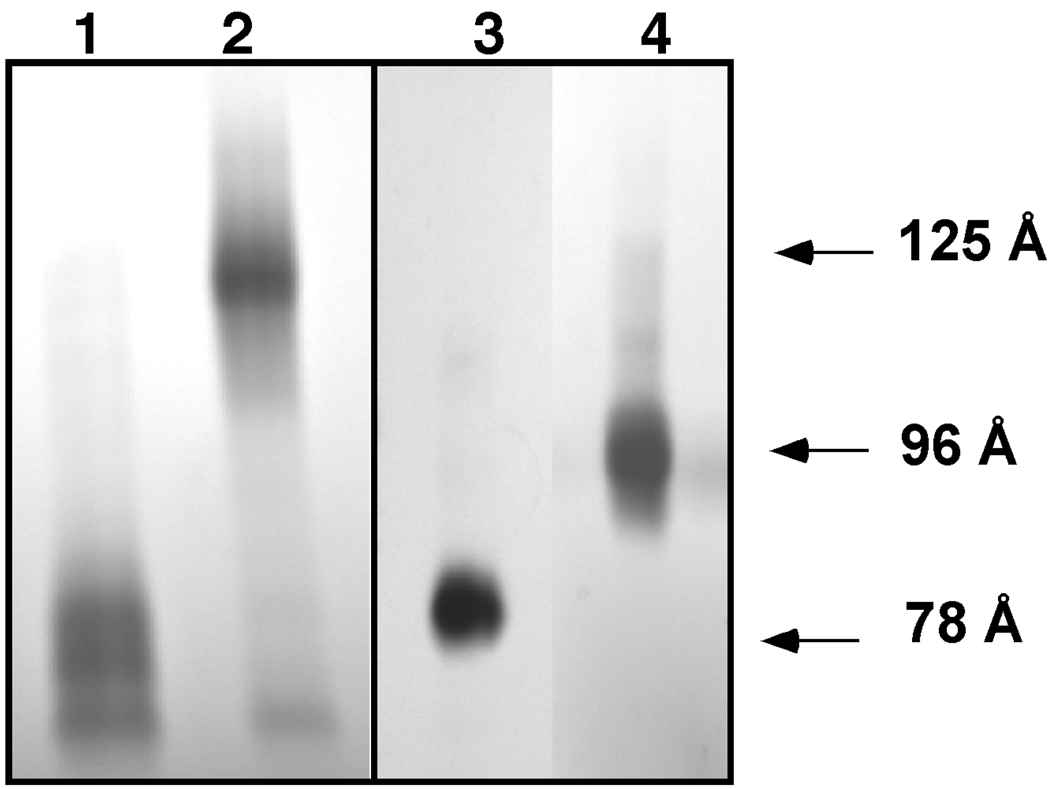
To probe the affect that the R173C mutation would impose on the formation of recombinant HDL, we made POPC containing rHDL with both dimer apoA-IMilano and monomeric apoA-IMilano. Interestingly, when the rHDL was made with increasing amounts of POPC and intact dimeric apoA-IMilano particles with two different diameters were obtained. The first was a 78Å diameter particle containing a total of 76 molecules of POPC and 1 dimer of apoA-IMilano per particle, while the other was a 125Å diameter rHDL containing approximately 209 molecules of POPC and 2-dimers of apoA-IMilano per particle (44), as shown in the non-denaturing 4–30% GGE in Figure 1, Lanes 1 and 2. However, when dimeric apoA-IMilano was reduced before being added to POPC, 80 and 96 Å rHDL were formed, as shown in Figure 1, Lanes 3 and 4. The 80 and 96 Å rHDL particles contain a total of 55 and 150 molecules of POPC per particle, respectively, and both contain 2 monomers of apoA-IMilano per particle. These data strongly suggest a conformational restriction imposed by the helix 7 to helix 7 cystine linkage present in the apoA-IMilano dimer appears to cause a significant change in the ability of the apoprotein to stably accommodate phospholipids.
FIGURE 1. Non-denaturing 4–30% Gradient Gel Electrophoresis of Recombinant HDL (rHDL) Containing POPC and ApoA-IMilano.
Lane 1, purified 78Å diameter rHDL prepared using dimeric apoA-IMilano, as described in “Experimental Procedures” and containing a total of 76 POPC to 1 apoA-IMilano dimer per particle; Lane 2, purified 125Å diameter rHDL containing a total of 209 POPC, with 2 apoA-IMilano dimers per particle; Lane 3, purified 80 Å rHDL prepared with “reduced” apoA-IMilano monomers and having a total of 55 POPC to two apoA-I monomers; Lane 4, purified 96 Å rHDL prepared with “reduced” apoA-IMilano monomers and having a total of 150 POPC to two apoA-I monomers. All rHDL preps were purified by FPLC. Approximately 6 µg of protein was loaded into each lane. High molecular weight markers from GE Healthcare (17-0445-01) were used to calculate each rHDL diameter: 170Å, ferritin; 124Å, catalase; 98Å, lactate dehydrogenase; 82Å, and albumin 71Å.
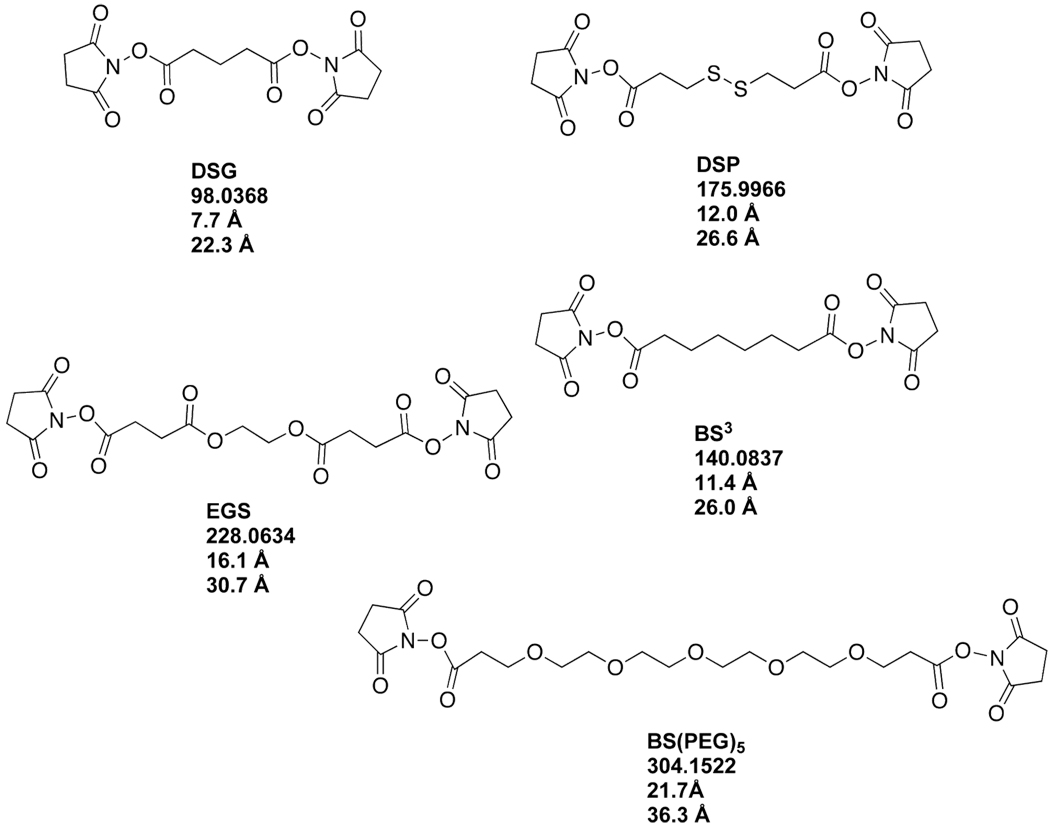
Purified rHDL containing dimeric apoA-IMilano were treated with the homobifunctional, amine-specific cross-linkers BS3, DSG, DSP, EGS or BS(PEG)5. The chemical structures of these cross-linkers are shown in Figure 2 along with their carbonyl to carbonyl mass, and arm length in Å, and the Cα to Cα length in Å that includes the lengths of two lysine side chains. Each of these cross-linkers were used at a mol ratio of 10:1 per rHDL apoA-IMilano. Because the size and composition of rHDL containing monomeric apoA-IMilano was the same as that found for rHDL made with apoA-IWT (25) further studies using these particles were not conducted.
FIGURE 2. Chemical Structure of the Lysine-reactive Homobifunctional Cross-linkers Used in These Studies.
Shown are the chemical structure, spacer arm molecular weight, spacer arm length (Å) and Cα-cross-linker-Cα distance (Å) for each cross-linker used in these studies. Each cross-linker contains an amine-reactive NHS ester that can react with any of the 21 lysines found in a single molecule of apoA-I. The cross-linkers and their abbreviations are: DSG, disuccinimidylglutarate; DSP, dithiobis(succinimidylpropionate); BS3, bis(sulfosuccinimidyl)suberate; EGS, ethyleneglycolbis(succinimidylsuccinate); and BS(PEG)5, Bis-N-succinimidyl(pentaethyleneglycol) ester. Note: only the cross-linker DSP contains a disulfide linkage that can be “reduced” by treatment with dithiothreitol or beta-mercaptoethanol.
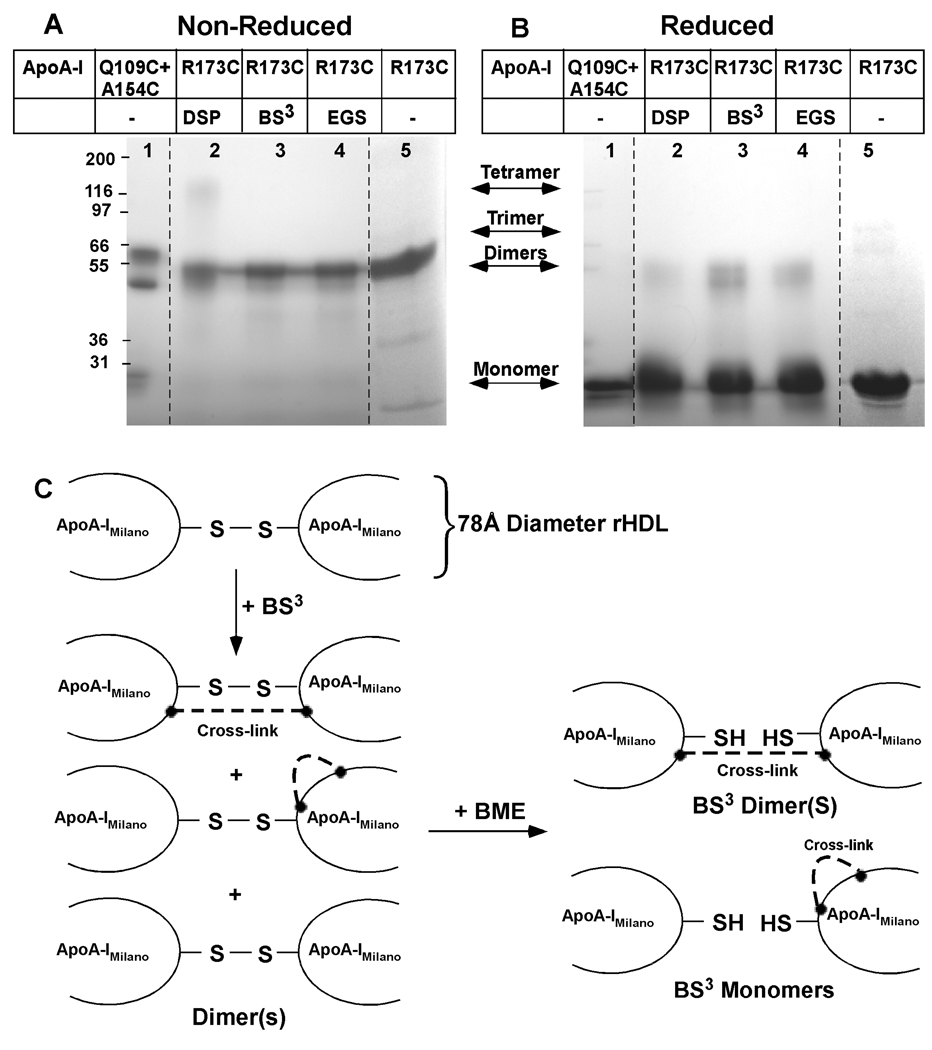
Cross-linked protein from 78Å diameter POPC rHDL particles carrying dimeric apoA-IMilano were separated by 4–30% non-reducing gradient SDS PAGE as shown in Figure 3A or under reducing conditions as shown in Figure 3B. Each gel shows the product distribution following treatment with DSP, Lanes 2; BS3, Lanes 3; EGS, Lanes 4 and rHDL containing unmodified apoA-IMilano, Lanes 5. A mixture of the homodimers, Q109C apoA-I and A154C apoA-I, are shown in Lanes 1 to indicate the extremes of aberrant molecular size migration induced by the alternative conformations of apoA-I dimers, as described previously (25).
FIGURE 3. 4–30% Gradient SDS PAGE Analysis of Cross-linked 78Å Diameter POPC:ApoA-IMilano rHDL.
Panel A shows migration under non-reducing conditions. Panel B shows migration under reducing conditions after treatment with βME. Lane 1 shows the positional specific migration of a mixture of Q109C and A154C apoA-I homodimers (25). Lanes 2–4 show 78Å diameter POPC:apoA-IMilano treated with DSP (Lane 2), BS3 (Lane 3), and EGS (Lane 4). Lane 5, contains uncross-linked apoA-IMilano. Mark 12 (Invitrogen) molecular weight markers were used. All samples were treated with molar ratio of 10:1 cross-linker to apoA-I for 5 min at 37 °C, as described under “Experimental Procedures.” The dashed lines indicate that lanes have been added or removed. A lane between lanes 1 and 2 was removed. Lane 5 from another SDS PAGE analysis was added to the figure to show the migration of dimeric apoA-IMilano. Panel C, illustrates the pattern of cross-linking for 78Å diameter rHDL particles that carry a single disulfide-linked dimer of apoA-IMilano. Red lines depict chemical cross-links which can be intramolecular or intermolecular. The labels monomer, dimer trimer, tetramer indicate the where apoA-IMilano or multimers having 2, 3 or 4 apoA-IMilano would migrate on the gel.
As mentioned before, apoA-IMilano exists as a cysteine disulfide bridged dimer in rHDL which was evident from the product band mobility, as shown in Figure 3A. Therefore, to distinguish between CCL formed between lysines and disulfide cross-links from bridged cysteines present in the starting apoA-IMilano, each cross-linked sample was reduced with beta-mercaptoethanol (βME) before SDS PAGE, as shown in Figure 3B. Since DSP is the only “reducible” cross-linker used, >90% of dimeric bands in Lane 2 were lost. However, Lanes 3 and 4 showed that after reduction with βME cross-linked products from treatment with BS3 and EGS still remained. A pictorial representation of these data are shown in Figure 3C.
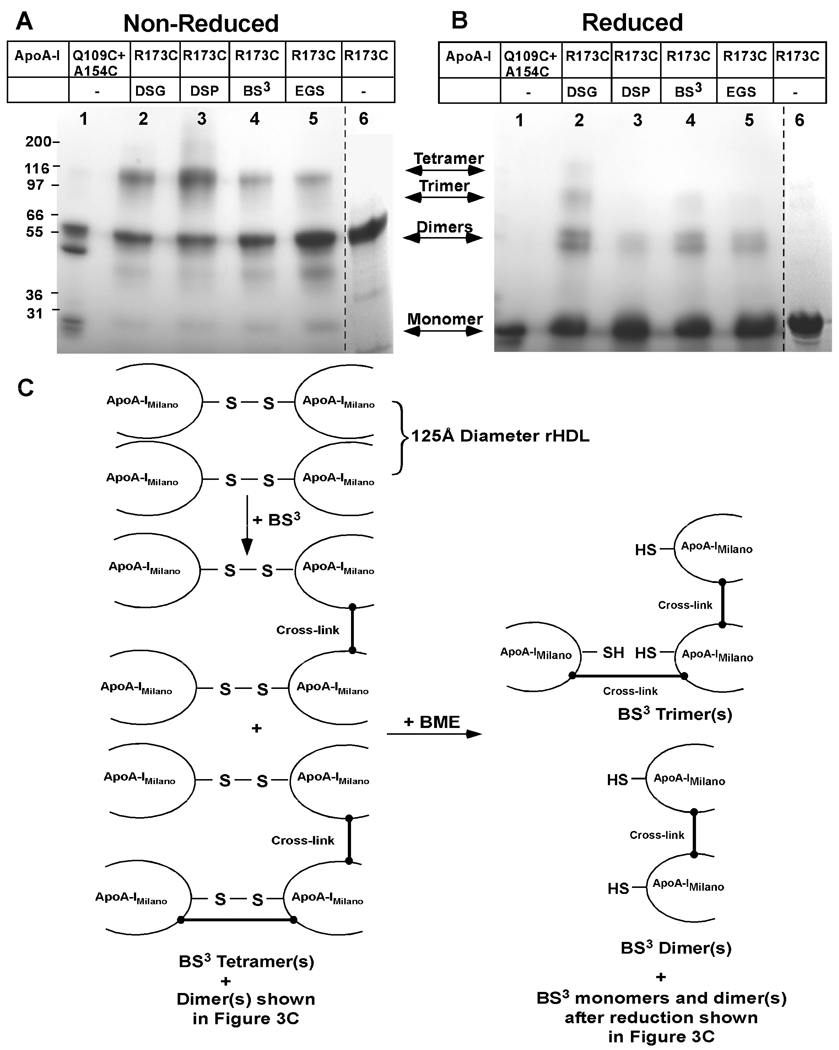
Cross-linked protein from 125Å diameter POPC rHDL particles carrying dimeric apoA-IMilano were separated by 4–30% non-reducing gradient SDS PAGE as shown in Figure 4A or under reducing conditions as shown in Figure 4B. Each gel shows the product distribution following treatment with DSG, Lanes 2; DSP, Lanes 3; BS3, Lanes 4; EGS, Lanes 5, and rHDL containing apoA-IMilano, Lanes 6. Figure 4A shows that cross-linking with DSG, DSP, BS3 and EGS gives protein migrating at a molecular weight consistent with a tetramer composed of 4 molecules of apoA-IMilano. Analysis of 125Å diameter rHDL using reductive SDS PAGE yielded mostly monomer, however, Figure 4B shows that non-reducible dimers were present in preparations cross-linked by BS3 and EGS and both dimers and trimers present after treatment with DSG. Figure 4C illustrates how chemical cross-linking could yield a tetramer and a dimer and how reduction of these species yields trimeric and dimeric apoA-IMilano. Overall, Figures 3C and 4C suggest that some dimers from 125Å diameter rHDL might be formed from different lysine-lysine intermolecular cross-links compared to those formed on 78Å diameter rHDL.
FIGURE 4. 4–30% Gradient SDS PAGE Analysis of Cross-linked 125Å Diameter POPC:ApoA-IMilano rHDL.
Panel A shows migration under non-reducing conditions. Panel B shows migration under reducing conditions after treatment with βME. Lane 1 shows the positional specific migration of a mixture of Q109C and A154C apoA-I homodimers (25). Lanes 2–5 show 125Å diameter POPC:apoA-IMilano treated with DSG (Lane 2), DSP (Lane 3) BS3 (Lane 4), and EGS (Lane 5). Lane 6 contains uncrosslinked apoA-IMilano. All samples were treated with 10:1 cross-linker to protein for 5 min at 37 °C, as described under “Experimental Procedures.” The dashed lines indicate that lanes have been added or removed. Lane 6 from another SDS PAGE analysis was added to the figure to show the migration of dimeric apoA-IMilano. Panel C, illustrates the pattern of cross-linking for 125Å diameter rHDL particles containing 2 disulfide-linked dimers of apoA-IMilano. Note, that in the absence of inter-particle cross-linking only the 125Å diameter rHDL particle can generate a product that is composed of 4 cross-linked apoA-IMilano, a tetramer. Red and blue line colors suggest that some lysine-lysine cross-links may be unique to the 125Å diameter rHDL.
Identification of Lysine-Lysine Cross-links from ApoA-IMilano
Cross-linked samples were separated under reducing conditions using 4–30% SDS PAGE. The protein bands were excised and digested with trypsin (23, 25). The extracted peptides were analyzed by ES-MS/MS. After mass correction a peptide list was generated from the survey scan and compared to a theoretical list of all possible peptide intermolecular and intramolecular cross-links for each CCL. After mass correction candidate peptides were assessed by assuming the masses of the experimental and theoretical cross-inked were within ±20 ppm. Candidate peptides identified from the survey scans were sequenced by MS/MS. In these experiments the difference between theoretical and experimental values for each cross-linked peptide was ≤8 ppm.
Table 1 shows all the cross-links identified from 78Å diameter POPC rHDL using DSP and BS3 while Table 2 shows all of the cross-links identified from 125Å diameter POPC rHDL using BS3, DSG, EGS, and BS(PEG)5. As an internal control, unreduced cross-linked samples were also analyzed. The disulfide cross-link between the two Cys173 was detected and sequenced and is listed as T26–27/T26–27 where T indicates that the peptide derived from tryptic cleavage and the subscript refers to the individual tryptic peptides that make up the connected parts. To help ascertain whether the cross-links were intramolecular or intermolecular the single scan intensity at the m/z of the sequenced monoisotopic ion were extracted from each total ion chromatogram and divided by the intensity of the ubiquitous intramolecular cross-link, T20–22, (lysines 133–140) that has been identified using most CCLs. This calculation was performed for each type of CCL use in these studies. Because DSP has a disulfide bridge, Figure 2, that would be reduced under the reducing conditions of the analysis, it was not used for this analysis. The results for both 78Å and 125Å diameter rHDL are summarized in Table 3. After disulfide reduction 2 protein species were obtained from 78Å diameter rHDL and 3 from 125Å diameter rHDL: M-monomeric apoA-IMilano, D-dimeric apoA-IMilano, and T-tetrameric apoA-IMilano. As discussed in our previous publications (23, 25) CCL yielded 2 dimers that were combined as a single entity to simplify analysis in Table 3.
Table 1.
Cross-links Identified from 78Å Diameter rHDL
| Sequenced m/z |
Peptide m/z Experimenta l MH+1 |
Peptide m/z Theoretical MH+1 |
m/z error |
Cross- linker |
ID | Cross- linked positions |
Peptides1 | |
|---|---|---|---|---|---|---|---|---|
| 684.68 | +32 | 2052.0244 | 2052.0146 | 0.0098 | DSP | T2–4 | 12–23 | 11–27 |
| 854.863 | +2 | 1708.7122 | 1708.7596 | −0.0474 | DSP/BS3 | T11–13 | 88–94 | 84–96 |
| 954.793 | +3 | 2862.3544 | 2862.3647 | −0.0103 | DSP/BS3 | T1α/T2–3 | 1–12 | 1–10, 11–23 |
| 1170.063 | +2 | 2339.1122 | 2339.1198 | −0.0076 | DSP/BS3 | T20–22 | 133–140 | 132–149 |
| 913.14 | +3 | 2737.4373 | 2737.4365 | 0.0008 | BS3 | T5–6/T35–36 | 40–239 | 28–45, 239–243 |
| 872.80 | +3 | 2616.4028 | 2616.4037 | −0.0009 | BS3 | T6–8 | 45–59 | 41–61 |
| 783.08 | +3 | 2347.2388 | 2347.2509 | −0.0121 | BS3 | T31–32 | 206–208 | 196–215 |
| 820.10 | +3 | 2458.3232 | 2458.3303 | −0.0071 | BS3 | T13–14/T17–18 | 96–118 | 95–106, 117–123 |
| 760.89 | +4 | 3040.5620 | 3040.5547 | 0.0073 | BS3 | T13–14/T13–14 | 96–96 | 95–106, 95–106 |
| 793.66 | +4 | 3171.6382 | 3171.6636 | −0.0254 | BS3 | T7–8/T20–21 | 59–133 | 46–61, 132–140 |
| 777.76 | +5 | 3884.9016 | 3884.9288 | −0.0272 | S-S | T26–27/T26–27 | 173–173 | 161–177, 161–177 |
Starting and ending amino acids for each peptide component are given.
The charge on the sequenced ion is shown in the second column.
Ions from both cross-linkers were sequenced, but only the m/z for the DSP cross-linked ions are shown.
Table 2.
Cross-links Identified from 125Å Diameter rHDL.
| Sequenced m/z |
Peptide m/z Experimental MH+1 |
Peptide m/z Theoretical MH+1 |
m/z Error |
Cross- linker |
ID | Cross- linked positions |
Peptides1 | |
|---|---|---|---|---|---|---|---|---|
| 836.912 | +23 | 1672.8584 | 1672.8468 | 0.0116 | BS3/EGS | T11–13 | 88–94 | 84–96 |
| 731.39 | +3 | 2192.1384 | 2192.1565 | −0.0181 | BS3 | T13–14/T35–36 | 96–239 | 95–106, 239–243 |
| 718.02 | +3 | 2152.0525 | 2152.0713 | −0.0188 | BS3 | T15–16/T35–36 | 107–239 | 107–116, 239–243 |
| 768.32 | +3 | 2303.1929 | 2303.2069 | −0.0140 | BS3/EGS/ BS(PEG)5 |
T20–22 | 133–140 | 132–149 |
| 820.10 | +3 | 2458.3291 | 2458.3303 | −0.0012 | BS3 | T13–14/T17–18 | 96–118 | 95–106, 117–123 |
| 913.142 | +3 | 2737.4216 | 2737.4365 | −0.0149 | BS3/DSG | T5–6/T35–36 | 40–239 | 28–45, 239–243 |
| 812.41 | +3 | 2435.2029 | 2435.2305 | −0.0276 | EGS | T31–32 | 206–208 | 196–215 |
| 1052.56 | +2 | 2104.0852 | 2104.0813 | 0.0039 | EGS | T2–4 | 12–23 | 11–27 |
| 1066.04 | +2 | 2131.0386 | 2131.0196 | 0.0190 | BS(PEG)5 | T1α/T35–36 | α-239 | 1–10, 239–243 |
| 799.72 | +3 | 2397.1863 | 2397.1939 | −0.0076 | BS(PEG)5 | T1α/T17–18 | α-118 | 1–10, 117–123 |
| 970.14 | +3 | 2908.4302 | 2908.3887 | 0.0415 | BS(PEG)5 | T1α/T14–15 | α-106 | 1–10, 97–107 |
| 972.01 | +4 | 3884.8782 | 3884.9288 | −0.0506 | S-S | T26–27/T26–27 | 173–173 | 161–177, 161–177 |
Starting and ending amino acids for each peptide component are given.
Ions from both cross-linkers were sequenced, but only the m/z for BS3 cross-linked ions are shown.
The charge on the sequenced ion is shown in the second column.
Table 3.
Relative Intensities of Cross-linked Peptides.
| Sequenced m/z |
Cross- linker |
ID | Lysine #1 | M2 | D2,3 | T2 |
|---|---|---|---|---|---|---|
| 78Å Diameter rHDL | ||||||
| 836.96 | BS3 | T11–13 | 88–94 | 0.2 | 0.1 | |
| 942.84 | BS3 | T1α/T2–3 | 1–12 | 0.1 | 0.3 | |
| 913.14 | BS3 | T5–6/T35–36 | 40–239 | 0.1 | 0.7 | |
| 872.80 | BS3 | T6–8 | 45–59 | 0.1 | 0.2 | |
| 783.08 | BS3 | T31–32 | 206–208 | 0.8 | 0.8 | |
| 820.10 | BS3 | T13–14/T17–18 | 96–118 | 0.0 | 0.2 | |
| 760.89 | BS3 | T13–14/T13–14 | 96–96 | 0.0 | 0.4 | |
| 793.66 | BS3 | T7–8/T20–21 | 59–133 | 0.0 | 0.5 | |
| 768.42 | BS3 | T20–22 | 133–140 | 1.0 | 1.0 | |
| 777.76 | S-S | T26–27/T26–27 | 173–173 | P | P | |
| 125Å Diameter rHDL | ||||||
| 836.91 | BS3 | T11–13 | 88–94 | 1.3 | 1.0 | 0.9 |
| 731.39 | BS3 | T13–14/T35–36 | 96–239 | 0.0 | 0.5 | 1.3 |
| 718.02 | BS3 | T15–16/T35–36 | 107–239 | 0.0 | 0.6 | 3.3 |
| 1152.04 | BS3 | T20–22 | 133–140 | 1.0 | 1.0 | 1.0 |
| 820.10 | BS3 | T13–14/T17–18 | 96–118 | 0.0 | 0.0 | 0.4 |
| 913.14 | BS3 | T5–6/T35–36 | 40–239 | 0.3 | 2.4 | 1.3 |
| 1066.04 | BS(PEG)5 | T1α/T35–36 | 1α-239 | 0.0 | 0.0 | 0.4 |
| 799.72 | BS(PEG)5 | T1α/T17–18 | 1α-118 | 0.1 | 0.4 | 1.6 |
| 970.14 | BS(PEG)5 | T1α/T14–15 | 1α-106 | 0.1 | 0.3 | 0.3 |
| 822.77 | BS(PEG)5 | T20–22 | 133–140 | 1.0 | 1.0 | 1.0 |
| 812.41 | EGS | T31–32 | 206–208 | 1.5 | 1.7 | 1.0 |
| 1052.56 | EGS | T2–4 | 12–23 | 2.1 | 2.8 | 1.4 |
| 797.74 | EGS | T20–22 | 133–140 | 1.0 | 1.0 | 1.0 |
| 777.76 | S-S | T26–27/T26–27 | 173–173 | P | P | P |
The numbers indicate the lysines that were bridged by the cross-linker.
Within a single analysis for monomer-M, dimer-D or tetramer-T for each cross-linker, the ion current for each m/z was divided by the ion current for the ubiquitous T20–22 intramolecular cross-linked peptide.
D represents the sum of the intensity for the two dimer bands D1 and D2 that has been previously discussed (25).
Sometimes the sites of cross-linking in peptides, like T11–13 where lysines 88 and 94 are connected, suggest that they are intramolecular cross-links in regions that would be suspected of having α-helical character with the lysines located on the same face of the peptide. These peptides have been characterized as intrapeptide cross-links (54). Others of this group are T2–4, T6–8 and T20–22. The cross-linked peptide T31–32 has lysines adjacent one another at positions 206 and 208 that do not span a region of α-helix. Other cross-links may arise from intramolecular cross-links between lysines that are close together in space, but near one another in the primary sequence or from intermolecular cross-links between the two protein strands. As an example compare Tα/T2–3 and T7–8/T20–21 from 78Å diameter rHDL. In support, Table 3 shows that T1α/T2–3 was present in all protein bands, i.e., it is distributed in both monomer and dimer bands like the intrapeptide cross-link T20–22. T1α/T2–3 was detected only in dimer bands, D (Table 3), suggesting that it is an intermolecular CCL. An unusual case was T5–6/T35–36 which has a small presence in the monomer, M, but much larger contribution in D from 78Å diameter rHDL and D and tetramer, T, from 125Å diameter rHDL. These observations suggest that the conformation of apoA-IMilano puts lysines 40 and 239 close together giving both intra- and intermolecular association, one product associated with apoA-IMilano monomer and the other with apoA-IMilano dimer.
Molecular Models for ApoA-IMilano Folding
The apoA-I primary and secondary structure was constructed from coordinates for lipid-free Δ43-apoA-I (34) joined with 1–43 amino acids, initially as an α-helical segment, and arginine at position 173 was replaced with a cysteine. The two molecules were oriented to accommodate the disulfide cross-link between cysteines at position 173 on each protein. Then the chemical cross-links were oriented to the correct distances based on whether these cross-links were intramolecular or intermolecular. We used the maximum distance of Cα–Lysine-(cross-linker)-Cα-Lysine calculated for each of the cross-linkers to establish the appropriate separation. To accommodate the cross-links apoA-I was bent only at the proline or glycine-glycine sites between the amphipathic segments of apoA-I. Of particular interest are the sequenced cross-links T5–6/T35–36 (lysines 40 and 239), shown in Figure 5 Panel A, that span one end of the molecule to the other and suggest N- and C-termini are proximate. Other cross-links like, T13–14/T17–18 (lysines 96 and 118), shown in Figure 5 Panel B, along with the intermolecular cross-link between amino acids 173 define the antiparallel orientation of the two apoA-IMilano chains. One assumption that we made was that the cross-links represented only a single conformation.
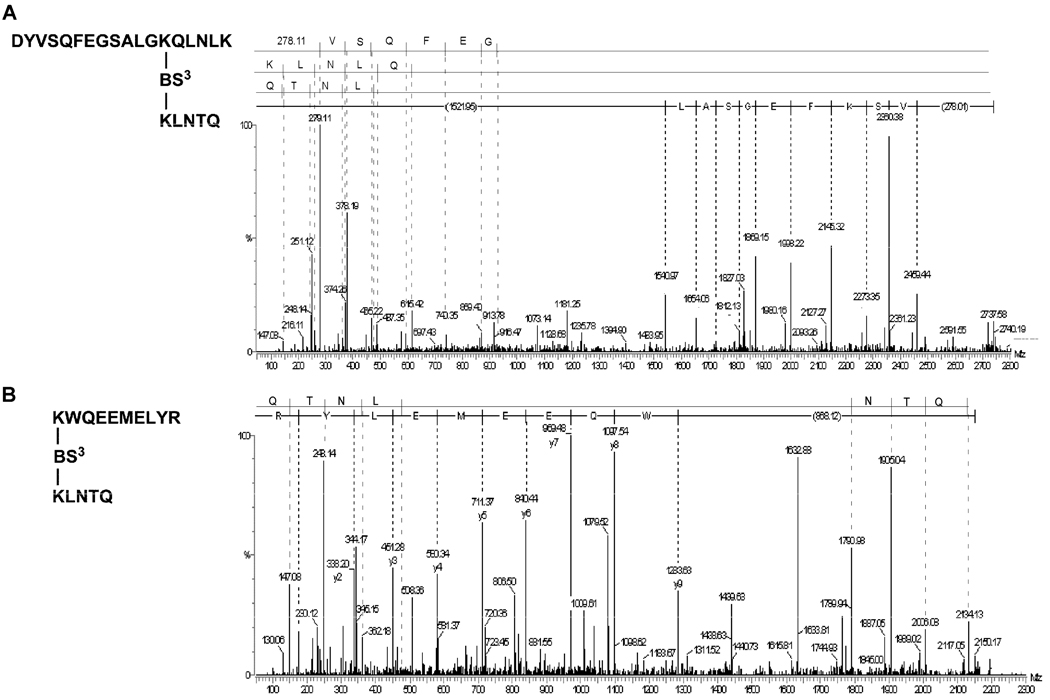
FIGURE 5. Verification of Q-TOF-MS/MS Spectrum.
Panel A shows the Q-TOF-MS/MS spectrum of the T5–6/T35–36 (lysines 40–239) cross-linked peptide from 78Å diameter rHDL treated with BS3. The MS/MS spectrum was obtained from the triply charged ion having m/z = 913.14 that gives a singly protonated mass of 2737.4348 Da. Panel B shows the Q-TOF-MS/MS spectrum of T15–16/T35–36 (lysines 107–239) peptide cross-link from 125Å diameter rHDL treated with BS3. The MS/MS spectrum was obtained from the triply charged ion having m/z = 718.02 and a singly protonated mass of 2152.0525 Da. The sequences are shown using single letter abbreviations for the amino acids. y-Series start with either K or R. Details are reported in the section on “Experimental Procedures.”
DISCUSSION
To construct a 3-dimensional model of apoA-IMilano we assumed that the conformation of the central region of the dimer would be similar to apoA-IWT monomers on 80Å and 96Å diameter rHDL. Using these assumptions and the chemical cross-links as physical constraints we obtained a particle with an outer diameter of about 78Å and an internal diameter of about 57Å. Using the POPC bilayer area of 68.3Å2 (55) this internal region would hold about 37 POPC per particle face for a total of 74 POPC per particle. Experimental measurement show there are 76 POPC per particle. However, if the “belt buckle” conformation proposed for apoA-IWT (25) is used as a preliminary model, the frame-shift induced by the cysteine-173 to cysteine-73 disulfide cross-link leaves part of the hydrophobic region on the edge of the particle exposed to aqueous solvent. We speculate that to compensate, the C-terminal end covers the hydrophobic edge of the particle putting the C-terminal and N-terminal ends close to one another as shown by the lysine-40 to lysine-239 cross-link. This change in the C-terminal conformation may explain why the 78Å apoA-IWT rHDL is apparently a less stable particle as suggested by the large signature on separation by non-reducing gradient gel electrophoresis (Figure 1). We propose that the N-terminal ends of dimeric of apoA-IMilano fold back similar to that reported for 80Å apoA-IWT rHDL (25).
The BS3 intermolecular cross-links between lysines-96 and between lysine-59 and lysine-133 on each apoA-IMilano strongly suggests an antiparallel alignment for the two strands. If the conformation is an antiparallel “belt-like loop,” similar to that suggested for 80Å diameter apoA-IWT (25), then lysines at position 96 on each strand would be close to one another. Furthermore, an intramolecular cross-link between lysines-40 and lysine-239 suggest that the N-terminal regions are close to their respective C-terminus. After connecting the cross-links listed above lysines 40 and 239 on the opposite strands were next to one another. In Table 3 the reduced, digested peptides from CCL apoA-I dimers, that run at about 55 kDa, show a large amount of the lysine 40-lysine-239 cross-link compared to the digested peptides from apoA-IMilano that runs as a monomer at 28 kDa, consistent with the hypothesis that the N-terminal end of one apoA-I molecule is close to the C-terminal end of the second molecule and that they can be coupled to give an intermolecular cross-link.
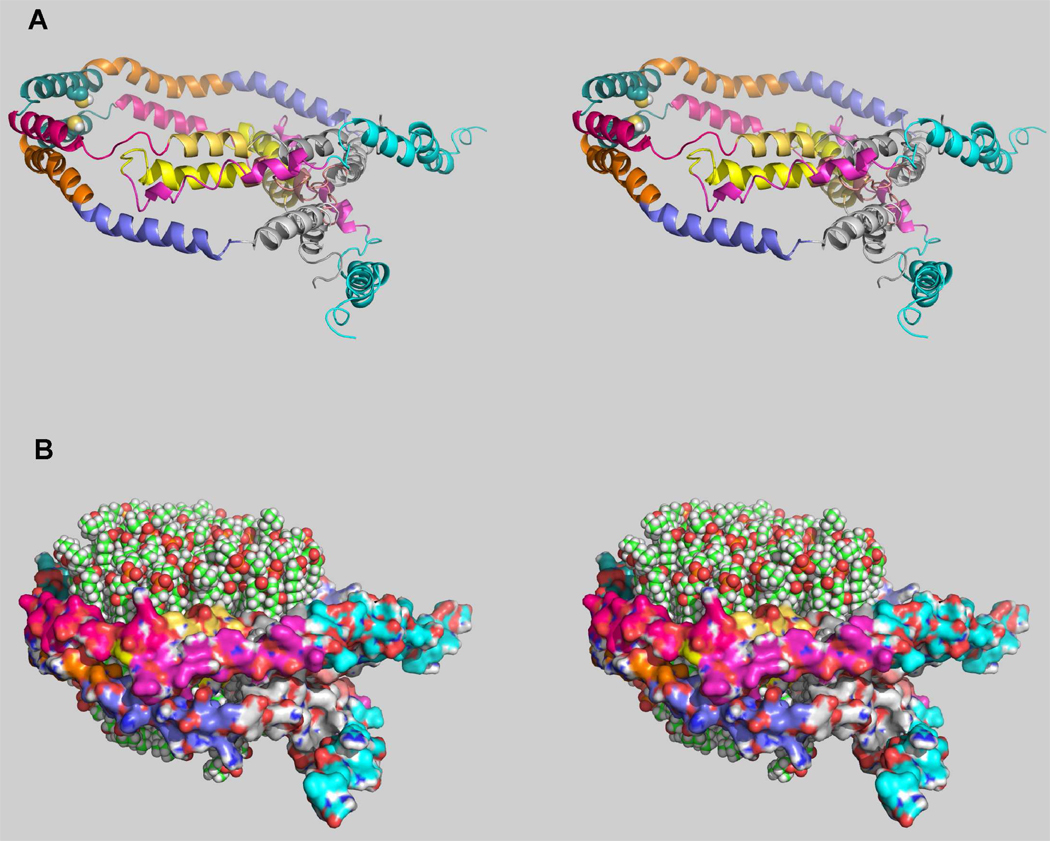
Figure 6 shows the conformation we propose for the orientation of a single apoA-IMilano dimer bound to a 78Å diameter rHDL. The “belt” conformation of apoA-IMilano on the rHDL model is saddle-shaped like the starting coordinates used for the model and similar to the conformation that was reported after modeling phospholipid and modified forms of apoA-IWT that was missing either the first 40 amino acids (56) or the first 11 to 22 amino acids (57). The affect of the disulfide crosslink is apparent by comparing these results to those obtained from particles prepared with monomeric apoA-IMilano. Our preliminary results using CCL/MS of monomeric apoA-IMilano rHDL support a “belt” model (data not shown) identical to that reported for apoA-IWT (23, 25) and calculated for apoA-IMilano (22).
FIGURE 6. Conformation for Dimeric ApoA-IMilano on a 78Å Diameter POPC rHDL Particle.
Panels A and B show three the 3-dimensional conformation for 1 dimer of apoA-IMilano bound to 78Å diameter POPC rHDL particle. MS/MS sequenced cross-links (Table 1) were used as molecular constraints for modeling the lipid-bound conformation of dimeric apoA-IMilano on 78 Å particles. Panel A, shows dimeric apoA-IMilano is locked by a single cysteine-cysteine disulfide bond at position 173 on each monomer. Although the structure resembles the antiparallel “belt-like” conformation as has been reported for 80Å apoA-IWT on POPC rHDL (25), the C-terminal end of each monomer does not fold back on itself as it does for apoA-IWT, but instead the C-terminal end wraps around the periphery. Each helical region is shown in a different color. Panel B, shows the bilayer contains 76 molecules of POPC and the model was generated using the membrane plugin, version 1.1, by I. Balabin available in VMD version 1.8.7 (53). Protein is shown as a surface for simplicity.
In contrast to a “belt” of protein wrapped around a bilayer of lipid, Wu et al. (28) suggested a “double super helix” model for the conformation of apoA-IWT on 96Å diameter rHDL composed of POPC, cholesterol and apoA-IWT in a ratio of 89:10:1. The relationship between the two monomer strands is antiparallel with the helix 5 to helix 5 regions, amino acids 121 to 142, adjacent one to another. The protein conformation can be described as having an open helical shape reminiscent of the shapes derived from computational methods with low phospholipid to apoA-I ratios (56). For apoA-IMilano the intramolecular cross-link between lysine-40 and lysine-239 show that the N- and C-terminal regions of a single apoA-I strand are close together and, therefore, apoA-IMilano may not preferentially adopt a “double super helix” conformation. Silva et al. (26) have proposed that there are two possible registries for the apoA-I monomers: the first is the helix 5 to helix 5 discussed above and the second is a helix 5 to helix 2 registry. The disulfide bond fixes a helix 7 to helix 7 registry that does not allow overlap between helix 5 and helix 2. These data strongly suggest a conformational restriction imposed by the helix 7 to helix 7 cysteine linkage present in the apoA-IMilano dimer, which may lead to a significant change in the ability of the apoprotein to stably accommodate phospholipids particularly evident by the diffuse banding seen for the 78Å rHDL containing apoA-IMilano dimer (Figure 1, Lane 1).
There are two differences in the cross-link pattern for large diameter particles compared to the pattern for the 78Å diameter particles. Two BS3 cross-links present in the 78Å particles, lysine-96 to lysine-96 and lysine-59 to lysine-133 were not present in the 125Å diameter rHDL, while two new BS3 cross-links appear in the larger particles, connections between lysine-96 and lysine-239 and between lysine-107 and lysine-239. Three cross-links from BS(PEG)5 were identified for the 125Å diameter particles. Of these cross-links, Table 3 suggests that 1α-239 was exclusively an intermolecular cross-link, 1α-106 was exclusively an intramolecular cross-link, while 1α-118 has both intermolecular and intramolecular components. That the 1α position has yielded so many cross-links suggests that it may have conformation freedom not available to the other positions in the molecule.
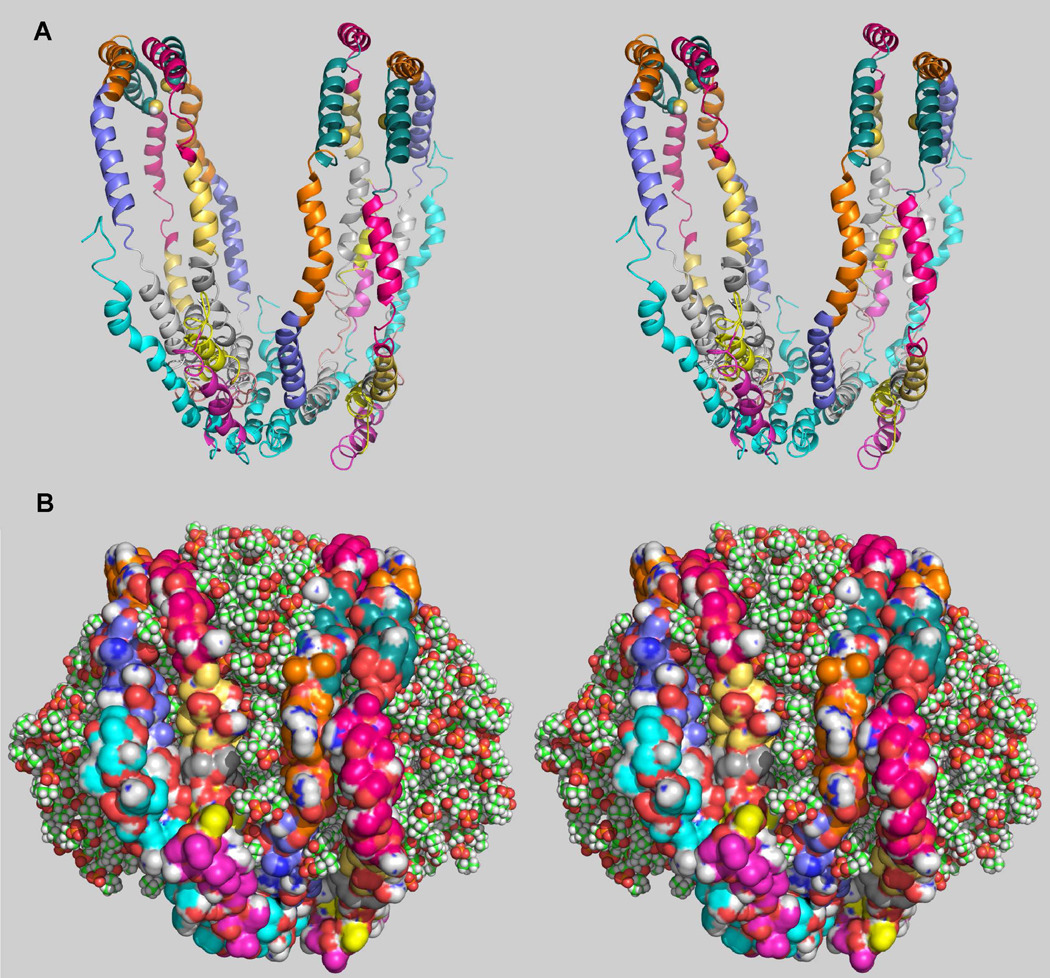
Our first hypothesis was that the protein strands encircling 125Å diameter rHDL had a greater diameter compared to 78Å diameter rHDL. Three sets of cross-links, the intermolecular disulfide link 173 to 173, the intramolecular and intermolecular lysine-40 to lysine-239, the intermolecular lysine-96 to lysine-239 and lysine-107 to lysine-239 suggested that the diameter of dimeric apoA-IMilano in 125Å diameter rHDL was closer to the diameter of the 96Å-diameter rHDL. From Tables 2 and 3 we assume that cross-links lysine-96 to lysine-118 and aspartic acid-1α to lysine-239 were involved in tying together the two dimers. We speculate that two dimeric apoA-IMilano molecules were stacked adjacent one another oriented such that lysine-96 on one of the apoA-IMilano dimer strands was close to lysine-118 of the second dimer. Figure 7 shows our proposed conformation for 2 dimers of apoA-IMilano bound to a 125Å diameter rHDL. For the model shown in Figure 7 the maximum atom to atom diagonal distance from the top strand to the bottom strand was about 96Å.
FIGURE 7. Conformation for 125Å Diameter POPC rHDL Particle Containing 2 Dimers of ApoA-IMilano.
Panels A and B show three the 3-dimensional conformation for 2 molecules of dimeric apoA-IMilano bound to 125Å diameter POPC rHDL particle. MS/MS sequenced cross-links (Table 2) were used as molecular constraints for modeling the lipid-bound conformation of dimeric apoA-IMilano. Panel A, each of the 2 dimeric apoA-IMilano is locked by a single cysteine-cysteine disulfide bond at position 173. Each helical region is shown in a different color. Panel B, shows the bilayer contains 209 molecules of POPC and the model was generated using the membrane plugin, version 1.1, by I. Balabin available in VMD version 1.8.7 (53). Each apoA-IMilano monomer within a dimer is shown as a surface for simplicity.
The orientation of the lipid poses a curious conundrum for the 125Å diameter rHDL. In the smaller particles apoA-IMilano is envisioned to enclose the bilayer associating with the non-polar edge of the POPC bilayer. We address the question of lipid packing by first noting the depth of a POPC membrane bilayer is roughly 36Å. For a single pair of apoA-I molecules computational studies suggest that POPC hydrophobic regions associate with the hydrophobic region of the amphipathic helix (22, 28, 39, 41, 56). Previous studies of the binding of amphipathic α-helical peptides 2F and 4F to DOPC multilayers (58) indicated that these peptides bury themselves so that the center of the α-helix is located about 17.1Å from the hydrophobic face, about the level of the glycerol backbone, 17.6Å (59). High resolution NMR analyses of the DMPC/2F and DMPC/4F complexes suggested that the α-helical peptide was aligned perpendicular to the acyl chains (60, 61). If the apoA-I protein strands align themselves perpendicular to the fatty acyl chains of POPC our “belt” model for the 78Å diameter POPC/apoA-IMilano complex is consistent with our cross-linking results. However, a flat bilayer structure does not seem to be well suited for representing the larger diameter particle. To obtain a structure having the hydrophobic region of the amphipathic α-helices interacting with the fatty acyl chains suggests that POPC behaves like a laminar micelle (62) and that the POPC/apoA-IMilano complex assumes an ellipsoidal shape.
Several studies have suggested less effective LCAT-catalyzed remodeling of small apoA-IMilano-containing particles compared to the remodeling of apoA-IWT-containing rHDL (63–65). The regions of apoA-I that have been associated with LCAT binding (47, 66–73), most of helix 6 (amino acids 143–164), were located closer-together in the models for both apoA-IMilano-containing particles compared to the same regions in apoA-IWT-containing discs (25, 73). This conformational change may well affect the binding of LCAT to rHDL (64, 65, 74).
In conclusion, we report low resolution 3-dimensional conformations for dimeric apoA-IMilano bound to POPC rHDLs. The results suggest that the overall protein conformation is a “belt,” similar to the conformation proposed for apoA-IWT. In addition, both apoA-IMilano and apoA-IWT appear to have an N-terminal region that spends a finite amount of time folded back over the “belt” or central region of the protein. The feature unique to apoA-IMilano compared to apoA-IWT is that for apoA-IMilano the C-termini appears to wrap around the periphery of the particle. Functionally, these models suggest that the regions associated with LCAT activity (helices 5–7) have been pushed together and thus possibly reducing exposure of the region which is usually accessible to LCAT and thus vital for activation of the enzyme.
ACKNOWLEDGEMENT
The Waters Q-TOF mass spectrometer was purchased with funds from NIH Shared Instrumentation Grant 1S10RR17846 (MJT). The MS analyses were performed in the Mass Spectrometer Facility of the Comprehensive Cancer Center of Wake Forest University School of Medicine supported in part by NCI center grant 5P30CA12197.
These studies were supported by grants from the National Institutes of Health (NIH) HL-49373 and HL-64163 (MST) and from the American Heart Association (MJT).
Footnotes
Abbreviations: 2F, peptide Ac-DWLKAFYDKVAEKLKEAF-NH2; 4F, peptide Ac-DWFKAFYDKVAEKFKEAF-NH2; apoA-I, apolipoprotein A-I; apoA-IMilano, mutant apolipoprotein A-I having R173C; βME, beta-mercaptoethanol; BS3, bis(sulfosuccinimidyl)suberate; BS(PEG)5, bis-N-succinimidyl(pentaethylene glycol) ester; CCL, lysine-selective chemical cross-linker; CCL/MS, chemical cross-linking combined with mass spectrometry; DMPC, 1,2-dimyristoylphosphotidylcholine; DOPC, dioleoylphosphotidylcholine; DSG, disuccinimidylglutarate; DSP, dithiobis(succinimidylpropionate); DTT, dithiothreitol; EGS, ethylene glycol bis(succinimidylsuccinate); HDL, high density lipoprotein; LC, liquid chromatography; MS/MS, tandem mass spectrometry; nHDL, nascent HDL; POPC, 1-palmitoyl-2-oleoylphosphotidylcholine; QTOF-MS, quadrupole time of flight mass spectrometer; rHDL, recombinant HDL; SANS, small-angle neutron scattering; SAXS, small-angle X-ray scattering; wild-type apoA-I, apoA-IWT.
REFERENCES
- 1.Yancey PG, Bortnick AE, Kellner-Weibel G, De La Llera-Moya M, Phillips MC, Rothblat GH. Importance of Different Pathways of Cellular Cholesterol Efflux. Arterioscler Thromb Vasc Biol. 2003;23:712–719. doi: 10.1161/01.ATV.0000057572.97137.DD. [DOI] [PubMed] [Google Scholar]
- 2.Libby P. Managing the risk of atherosclerosis: The role of high-density lipoprotein. Am. J. Card. 2001;88:3N–8N. doi: 10.1016/s0002-9149(01)02145-2. [DOI] [PubMed] [Google Scholar]
- 3.Kwiterovich PO. The antiatherogenic role of high-density lipoprotein cholesterol. Am. J. Cardiol. 1998;82:13Q–21Q. doi: 10.1016/s0002-9149(98)00808-x. [DOI] [PubMed] [Google Scholar]
- 4.Boden WE. High-density lipoprotein cholesterol as an independent risk factor in cardiovascular disease: Assessing the data from Framingham to the Veterans Affairs high-density lipoprotein intervention trial. Am. J. Card. 2000;86:19L–22L. doi: 10.1016/s0002-9149(00)01464-8. [DOI] [PubMed] [Google Scholar]
- 5.Harper CR, Jacobson TA. New perspectives on the management of low levels of high-density lipoprotein cholesterol. Arch.Intern.Med. 1999;159:1049–1057. doi: 10.1001/archinte.159.10.1049. [DOI] [PubMed] [Google Scholar]
- 6.Libby P. What have we learned about the biology of atherosclerosis? The role of inflammation. Am J Cardiol. 2001;88:3J–6J. doi: 10.1016/s0002-9149(01)01879-3. [DOI] [PubMed] [Google Scholar]
- 7.Gualandri V, Orsini GB, Cerrone A, Franceschini G, Sirtori CR. Familial Associations of Lipids and Lipoproteins in a Highly Consanguineous Population: The Limone Sul Garda Study. Metabolism. 1985;34:212–221. doi: 10.1016/0026-0495(85)90003-4. [DOI] [PubMed] [Google Scholar]
- 8.Sirtori CR, Calabresi L, Franceschini G, Baldassarre D, Amato M, Johansson J, Salvetti M, Monteduro C, Zulli R, Muiesan ML, Agabiti-Rosei E. Cardiovascular status of carriers of the apolipoprotein A-I(Milano) mutant: the Limone sul Garda study. Circulation. 2001;103:1949–1954. doi: 10.1161/01.cir.103.15.1949. [DOI] [PubMed] [Google Scholar]
- 9.Nissen SE, Tsunoda T, Tuzcu EM, Schoenhagen P, Cooper CJ, Yasin M, Eaton GM, Lauer MA, Sheldon WS, Grines CL, Halpern S, Crowe T, Blankenship JC, Kerensky R. Effect of recombinant ApoA-I Milano on coronary atherosclerosis in patients with acute coronary syndromes: a randomized controlled trial. Jama. 2003;290:2292–2300. doi: 10.1001/jama.290.17.2292. [DOI] [PubMed] [Google Scholar]
- 10.Tardif JC, Gregoire J, L'Allier PL, Ibrahim R, Lesperance J, Heinonen TM, Kouz S, Berry C, Basser R, Lavoie MA, Guertin MC, Rodes-Cabau J. Effects of reconstituted high-density lipoprotein infusions on coronary atherosclerosis: a randomized controlled trial. JAMA. 2007;297:1675–1682. doi: 10.1001/jama.297.15.jpc70004. [DOI] [PubMed] [Google Scholar]
- 11.Shah PK, Nilsson J, Kaul S, Fishbein MC, Ageland H, Hamsten A, Johansson J, Karpe F, Cercek B. Effects of recombinant apolipoprotein A-IMilano on aortic atherosclerosis in apolipoprotein E-deficient mice. Circulation. 1998;97:780–785. doi: 10.1161/01.cir.97.8.780. [DOI] [PubMed] [Google Scholar]
- 12.Shah PK, Kaul S, Nilsson J, Cercek B. Exploiting the vascular protective effects of high-density lipoprotein and its apolipoproteins: an idea whose time for testing is coming, part II. Circulation. 2001;104:2498–2502. doi: 10.1161/hc4501.098468. [DOI] [PubMed] [Google Scholar]
- 13.Shah PK, Kaul S, Nilsson J, Cercek B. Exploiting the vascular protective effects of high-density lipoprotein and its apolipoproteins: an idea whose time for testing is coming, part I. Circulation. 2001;104:2376–2383. doi: 10.1161/hc4401.098467. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Sharifi BG, Pan T, Song L, Yukht A, Shah PK. Bone marrow transplantation shows superior atheroprotective effects of gene therapy with apolipoprotein A-I Milano compared with wild-type apolipoprotein A-I in hyperlipidemic mice. J Am Coll Cardiol. 2006;48:1459–1468. doi: 10.1016/j.jacc.2006.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiesa G, Monteggia E, Marchesi M, Lorenzon P, Laucello M, Lorusso V, Di Mario C, Karvouni E, Newton RS, Bisgaier CL, Franceschini G, Sirtori CR. Recombinant apolipoprotein A-I(Milano) infusion into rabbit carotid artery rapidly removes lipid from fatty streaks. Circ Res. 2002;90:974–980. doi: 10.1161/01.res.0000018422.31717.ee. [DOI] [PubMed] [Google Scholar]
- 16.Marchesi M, Booth EA, Davis T, Bisgaier CL, Lucchesi BR. Apolipoprotein A-IMilano and 1-palmitoyl-2-oleoyl phosphatidylcholine complex (ETC-216) protects the in vivo rabbit heart from regional ischemia-reperfusion injury. J Pharmacol Exp Ther. 2004;311:1023–1031. doi: 10.1124/jpet.104.070789. [DOI] [PubMed] [Google Scholar]
- 17.Marchesi M, Booth EA, Rossoni G, Garcia RA, Hill KR, Sirtori CR, Bisgaier CL, Lucchesi BR. Apolipoprotein A-IMilano/POPC complex attenuates post-ischemic ventricular dysfunction in the isolated rabbit heart. Atherosclerosis. 2008;197:572–578. doi: 10.1016/j.atherosclerosis.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Kaul S, Rukshin V, Santos R, Azarbal B, Bisgaier CL, Johansson J, Tsang VT, Chyu KY, Cercek B, Mirocha J, Shah PK. Intramural delivery of recombinant apolipoprotein A-IMilano/phospholipid complex (ETC-216) inhibits in-stent stenosis in porcine coronary arteries. 2003 doi: 10.1161/01.CIR.0000074042.19447.B1. [DOI] [PubMed] [Google Scholar]
- 19.Parolini C, Chiesa G, Zhu Y, Forte T, Caligari S, Gianazza E, Sacco MG, Sirtori CR, Rubin EM. Targeted replacement of mouse apolipoprotein A-I with human ApoA-I or the mutant ApoA-IMilano. Evidence of APOA-IM impaired hepatic secretion. J Biol Chem. 2003;278:4740–4746. doi: 10.1074/jbc.M207335200. [DOI] [PubMed] [Google Scholar]
- 20.Parolini C, Chiesa G, Gong E, Caligari S, Cortese MM, Koga T, Forte TM, Rubin EM. Apolipoprotein A-I and the molecular variant apoA-I(Milano): evaluation of the antiatherogenic effects in knock-in mouse model. Atherosclerosis. 2005;183:222–229. doi: 10.1016/j.atherosclerosis.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 21.Lebherz C, Sanmiguel J, Wilson JM, Rader DJ. Gene transfer of wild-type apoA-I and apoA-I Milano reduce atherosclerosis to a similar extent. Cardiovasc Diabetol. 2007;6:6–15. doi: 10.1186/1475-2840-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klon AE, Jones MK, Segrest JP, Harvey SC. Molecular belt models for the apolipoprotein A-I Paris and Milano mutations. Biophys J. 2000;79:1679–1685. doi: 10.1016/S0006-3495(00)76417-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhat S, Sorci-Thomas MG, Alexander ET, Samuel MP, Thomas MJ. Intermolecular contact between globular N-terminal fold and C-terminal domain of ApoA-I stabilizes its lipid-bound conformation: studies employing chemical cross-linking and mass spectrometry. J Biol Chem. 2005;280:33015–33025. doi: 10.1074/jbc.M505081200. [DOI] [PubMed] [Google Scholar]
- 24.Calabresi L, Vecchio G, Longhi R, Gianazza E, Palm G, Wadensten H, Hammarstrom A, Olsson A, Karlstrom A, Sejlitz T, et al. Molecular characterization of native and recombinant apolipoprotein A-IMilano dimer. The introduction of an interchain disulfide bridge remarkably alters the physicochemical properties of apolipoprotein A-I. J Biol Chem. 1994;269:32168–32174. [PubMed] [Google Scholar]
- 25.Bhat S, Sorci-Thomas MG, Tuladhar R, Samuel MP, Thomas MJ. Conformational adaptation of apolipoprotein A-I to discretely sized phospholipid complexes. Biochemistry. 2007;46:7811–7821. doi: 10.1021/bi700384t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva RA, Hilliard GM, Li L, Segrest JP, Davidson WS. A mass spectrometric determination of the conformation of dimeric apolipoprotein A-I in discoidal high density lipoproteins. Biochemistry. 2005;44:8600–8607. doi: 10.1021/bi050421z. [DOI] [PubMed] [Google Scholar]
- 27.Davidson WS, Hilliard GM. The spatial organization of apolipoprotein A-I on the edge of discoidal high density lipoprotein particles - A mass spectrometry study. J. Biol. Chem. 2003;278:27199–27207. doi: 10.1074/jbc.M302764200. [DOI] [PubMed] [Google Scholar]
- 28.Wu Z, Gogonea V, Lee X, Wagner MA, Li XM, Huang Y, Undurti A, May RP, Haertlein M, Moulin M, Gutsche I, Zaccai G, Didonato JA, Hazen SL. The Double Super Helix model of high density lipoprotein. J Biol Chem. 2009 doi: 10.1074/jbc.M109.039537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Z, Wagner MA, Zheng L, Parks JS, Shy JM, 3rd, Smith JD, Gogonea V, Hazen SL. The refined structure of nascent HDL reveals a key functional domain for particle maturation and dysfunction. Nat Struct Mol Biol. 2007;14:861–868. doi: 10.1038/nsmb1284. Erratum: Nat Struct Mol Biol. 2008 2015: 2330. [DOI] [PubMed] [Google Scholar]
- 30.Panagotopulos SE, Horace EM, Maiorano JN, Davidson WS. Apolipoprotein A-I adopts a belt-like orientation in reconstituted high density lipoproteins. J Biol Chem. 2001;276:42965–42970. doi: 10.1074/jbc.M106462200. [DOI] [PubMed] [Google Scholar]
- 31.Maiorano JN, Jandacek RJ, Horace EM, Davidson WS. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 2004;43:11717–11726. doi: 10.1021/bi0496642. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Lyles DS, Thomas MJ, Pan W, Sorci-Thomas MG. Structural determination of lipid-bound ApoA-I using fluorescence resonance energy transfer. J Biol Chem. 2000;275:37048–37054. doi: 10.1074/jbc.M005336200. [DOI] [PubMed] [Google Scholar]
- 33.Li HH, Thomas MJ, Pan W, Alexander E, Samuel M, Sorci-Thomas MG. Preparation and incorporation of probe-labeled apoA-I for fluorescence resonance energy transfer studies of rHDL. J Lipid Res. 2001;42:2084–2091. [PubMed] [Google Scholar]
- 34.Borhani DW, Rogers DP, Engler JA, Brouillette CG. Crystal structure of truncated human apolipoprotein A-I suggests a lipid-bound conformation. Proc Natl Acad Sci U S A. 1997;94:12291–12296. doi: 10.1073/pnas.94.23.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tricerri MA, Behling Agree AK, Sanchez SA, Bronski J, Jonas A. Arrangement of apolipoprotein A-I in reconstituted high-density lipoprotein disks: an alternative model based on fluorescence resonance energy transfer experiments. Biochemistry. 2001;40:5065–5074. doi: 10.1021/bi002815q. [DOI] [PubMed] [Google Scholar]
- 36.Segrest JP, Jones MK, Klon AE, Sheldahl CJ, Hellinger M, De Loof H, Harvey SC. A detailed molecular belt model for apolipoprotein A-I in discoidal high density lipoprotein. J Biol Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 37.Klon AE, Segrest JP, Harvey SC. Comparative models for human apolipoprotein A-I bound to lipid in discoidal high-density lipoprotein particles. Biochemistry. 2002;41:10895–10905. doi: 10.1021/bi020315m. [DOI] [PubMed] [Google Scholar]
- 38.Klon AE, Segrest JP, Harvey SC. Molecular dynamics simulations on discoidal HDL particles suggest a mechanism for rotation in the apo A-I belt model. J Mol Biol. 2002;324:703–721. doi: 10.1016/s0022-2836(02)01143-9. [DOI] [PubMed] [Google Scholar]
- 39.Rocco AG, Gianazza E, Calabresi L, Sensi C, Franceschini G, Sirtori CR, Eberini I. Structural features and dynamics properties of human apolipoprotein A-I in a model of synthetic HDL. J Mol Graph Model. 2009;28:305–312. doi: 10.1016/j.jmgm.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 40.Shih AY, Sligar SG, Schulten K. Molecular models need to be tested: the case of a solar flares discoidal HDL model. Biophys J. 2008;94:L87–L89. doi: 10.1529/biophysj.108.131581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shih AY, Arkhipov A, Freddolino PL, Sligar SG, Schulten K. Assembly of lipids and proteins into lipoprotein particles. J Phys Chem B. 2007;111:11095–11104. doi: 10.1021/jp072320b. [DOI] [PubMed] [Google Scholar]
- 42.Li HH, Lyles DS, Pan W, Alexander E, Thomas MJ, Sorci-Thomas MG. ApoA-I structure on discs and spheres. Variable helix registry and conformational states. J Biol Chem. 2002;277:39093–39101. doi: 10.1074/jbc.M206770200. [DOI] [PubMed] [Google Scholar]
- 43.Martin DD, Budamagunta MS, Ryan RO, Voss JC, Oda MN. Apolipoprotein A-I assumes a "looped belt" conformation on reconstituted high density lipoprotein. J Biol Chem. 2006;281:20418–20426. doi: 10.1074/jbc.M602077200. [DOI] [PubMed] [Google Scholar]
- 44.Calabresi L, Vecchio G, Frigerio F, Vavassori L, Sirtori CR, Franceschini G. Reconstituted high-density lipoproteins with a disulfide-linked apolipoprotein A-I dimer: evidence for restricted particle size heterogeneity. Biochemistry. 1997;36:12428–12433. doi: 10.1021/bi970505a. [DOI] [PubMed] [Google Scholar]
- 45.Jonas A. Lecithin cholesterol acyltransferase. In: Gotto AM Jr., editor. Plasma Lipoproteins. Amsterdam: Elsevier; 1987. pp. 299–333. [Google Scholar]
- 46.Sorci-Thomas MG, Parks JS, Kearns MW, Pate GN, Zhang C, Thomas MJ. High level secretion of wild-type and mutant forms of human proapoA-I using baculovirus-mediated Sf-9 cell expression. J Lipid Res. 1996;37:673–683. [PubMed] [Google Scholar]
- 47.Alexander ET, Bhat S, Thomas MJ, Weinberg RB, Cook VR, Bharadwaj MS, Sorci-Thomas M. Apolipoprotein A-I helix 6 negatively charged residues attenuate lecithin-cholesterol acyltransferase (LCAT) reactivity. Biochemistry. 2005;44:5409–5419. doi: 10.1021/bi047412v. [DOI] [PubMed] [Google Scholar]
- 48.Lagerstedt JO, Budamagunta MS, Oda MN, Voss JC. EPR spectroscopy of site-directed spin labels reveals the structural heterogeneity in the N-terminal domain of apo-AI in solution. J Biol Chem. 2007;282:9143–9149. doi: 10.1074/jbc.M608717200. [DOI] [PubMed] [Google Scholar]
- 49.Zhu HL, Atkinson D. Conformation and lipid binding of the N-terminal (1–44) domain of human apolipoprotein A-I. Biochemistry. 2004;43:13156–13164. doi: 10.1021/bi0487894. [DOI] [PubMed] [Google Scholar]
- 50.Okon M, Frank PG, Marcel YL, Cushley RJ. Heteronuclear NMR studies of human serum apolipoprotein A-I. Part I. Secondary structure in lipid-mimetic solution. FEBS Letters. 2002;517:139–143. doi: 10.1016/s0014-5793(02)02600-5. [DOI] [PubMed] [Google Scholar]
- 51.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 52.Silva RA, Hilliard GM, Fang J, Macha S, Davidson WS. A three-dimensional molecular model of lipid-free apolipoprotein A-I determined by cross-linking/mass spectrometry and sequence threading. Biochemistry. 2005;44:2759–2769. doi: 10.1021/bi047717+. [DOI] [PubMed] [Google Scholar]
- 53.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 54.Bennett KL, Kussmann M, Björk P, Godzwon M, Mikkelsen M, Sørensen P, Roepstorff P. Chemical cross-linking with thiol-cleavable reagents combined with differential mass spectrometric peptide mapping - a novel approach to assess intermolecular protein contacts. Protein Sci. 2000;9:1503–1518. doi: 10.1110/ps.9.8.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z, Bhide SY, Berkowitz ML. Molecular dynamics simulations of bilayers containing mixtures of sphingomyelin with cholesterol and phosphatidylcholine with cholesterol. J Phys Chem B. 2007;111:12888–12897. doi: 10.1021/jp074037i. [DOI] [PubMed] [Google Scholar]
- 56.Catte A, Patterson JC, Jones MK, Jerome WG, Bashtovyy D, Su Z, Gu F, Chen J, Aliste MP, Harvey SC, Li L, Weinstein G, Segrest JP. Novel changes in discoidal high density lipoprotein morphology: a molecular dynamics study. Biophys J. 2006;90:4345–4360. doi: 10.1529/biophysj.105.071456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shih AY, Denisov IG, Phillips JC, Sligar SG, Schulten K. Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophys J. 2005;88:548–556. doi: 10.1529/biophysj.104.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mishra VK, Palgunachari MN, Segrest JP, Anantharamaiah GM. Interactions of synthetic peptide analogs of the class A amphipathic helix with lipids. Evidence for the snorkel hypothesis. J Biol Chem. 1994;269:7185–7191. [PubMed] [Google Scholar]
- 59.Hristova K, Wimley WC, Mishra VK, Anantharamiah GM, Segrest JP, White SH. An amphipathic alpha-helix at a membrane interface: a structural study using a novel X-ray diffraction method. J Mol Biol. 1999;290:99–117. doi: 10.1006/jmbi.1999.2840. [DOI] [PubMed] [Google Scholar]
- 60.Mishra VK, Anantharamaiah GM, Segrest JP, Palgunachari MN, Chaddha M, Sham SW, Krishna NR. Association of a model class A (apolipoprotein) amphipathic alpha helical peptide with lipid: high resolution NMR studies of peptide.lipid discoidal complexes. J Biol Chem. 2006;281:6511–6519. doi: 10.1074/jbc.M511475200. [DOI] [PubMed] [Google Scholar]
- 61.Mishra VK, Palgunachari MN, Krishna R, Glushka J, Segrest JP, Anantharamaiah GM. Effect of leucine to phenylalanine substitution on the nonpolar face of a class A amphipathic helical peptide on its interaction with lipid: high resolution solution NMR studies of 4F-dimyristoylphosphatidylcholine discoidal complex. J Biol Chem. 2008;283:34393–34402. doi: 10.1074/jbc.M806384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peters-Libeu CA, Newhouse Y, Hall SC, Witkowska HE, Weisgraber KH. Apolipoprotein E*dipalmitoylphosphatidylcholine particles are ellipsoidal in solution. J Lipid Res. 2007;48:1035–1044. doi: 10.1194/jlr.M600545-JLR200. [DOI] [PubMed] [Google Scholar]
- 63.Calabresi L, Franceschini G, Burkybile A, Jonas A. Activation of lecithin cholesterol acyltransferase by a disulfide-linked apolipoprotein A-I dimer. Biochem Biophys Res Commun. 1997;232:345–349. doi: 10.1006/bbrc.1997.6286. [DOI] [PubMed] [Google Scholar]
- 64.Bielicki JK, Forte TM, McCall MR, Stoltzfus LJ, Chiesa G, Sirtori CR, Franceschini G, Rubin EM. High density lipoprotein particle size restriction in apolipoprotein A-I(Milano) transgenic mice. J Lipid Res. 1997;38:2314–2321. [PubMed] [Google Scholar]
- 65.Bielicki JK, McCall MR, Stoltzfus LJ, Ravandi A, Kuksis A, Rubin EM, Forte TM. Evidence that apolipoprotein A-IMilano has reduced capacity, compared with wild-type apolipoprotein A-I, to recruit membrane cholesterol. Arterioscler Thromb Vasc Biol. 1997;17:1637–1643. doi: 10.1161/01.atv.17.9.1637. [DOI] [PubMed] [Google Scholar]
- 66.Sorci-Thomas MG, Thomas MJ. The effects of altered apolipoprotein A-I structure on plasma HDL concentration. Trends Cardiovasc Med. 2002;12:121–128. doi: 10.1016/s1050-1738(01)00163-3. [DOI] [PubMed] [Google Scholar]
- 67.Sorci-Thomas M, Kearns MW, Lee JP. Apolipoprotein A-I domains involved in lecithin-cholesterol acyltransferase activation. Structure:function relationships. J Biol Chem. 1993;268:21403–21409. [PubMed] [Google Scholar]
- 68.Sorci-Thomas MG, Thomas M, Curtiss L, Landrum M. Single repeat deletion in apoA-I blocks cholesterol esterification and results in rapid catabolism of D6 and wild-type apoA-I in transgenic mice. J Biol Chem. 2000;275:12156–12163. doi: 10.1074/jbc.275.16.12156. [DOI] [PubMed] [Google Scholar]
- 69.McManus DC, Scott BR, Franklin V, Sparks DL, Marcel YL. Proteolytic degradation and impaired secretion of an apolipoprotein A-I mutant associated with dominantly inherited hypoalphalipoproteinemia. J Biol. Chem. 2001;276:21292–21302. doi: 10.1074/jbc.M100463200. [DOI] [PubMed] [Google Scholar]
- 70.Roosbeek S, Vanloo B, Duverger N, Caster H, Breyne J, De Beun I, Patel H, Vandekerckhove J, Choulders C, Rosseneu M, Peelman F. Three arginine residues in apolipoprotein A-I are critical for activation of lecithin:cholesterol acyltransferase. J. Lipid Res. 2001;42:31–40. [PubMed] [Google Scholar]
- 71.Maiorano JN, Jandacek RJ, Horace EM, Davidson WS. Identification and structural ramifications of a hinge domain in apolipoprotein A-I discoidal high-density lipoproteins of different size. Biochemistry. 2004;43:11717–11726. doi: 10.1021/bi0496642. [DOI] [PubMed] [Google Scholar]
- 72.Sorci-Thomas MG, Curtiss L, Parks JS, Thomas MJ, Kearns MW, Landrum M. The hydrophobic face orientation of apolipoprotein A-I amphipathic helix domain 143–164 regulates lecithin:cholesterol acyltransferase activation. J Biol Chem. 1998;273:11776–11782. doi: 10.1074/jbc.273.19.11776. [DOI] [PubMed] [Google Scholar]
- 73.Sorci-Thomas MG, Bhat S, Thomas MJ. Lecithin:cholesterol acyltransferase. Clin Lipidology/Future Med. 2009;4:113–124. doi: 10.2217/17584299.4.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alexander ET, Tanaka M, Kono M, Saito H, Rader DJ, Phillips MC. Structural and functional consequences of the milano mutation (R173C) in human apolipoprotein A-I. J Lipid Res. 2009;50:1409–1419. doi: 10.1194/jlr.M800578-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]