Abstract
The biosynthesis of the endocannabinoid anandamide and related N-acyl ethanolamine (NAE) lipids is complex and appears to involve multiple pathways, including: 1) direct release of NAEs from N-acyl phosphatidyl ethanolamine (NAPE) precursors by the phosphodiesterase NAPE-PLD, and 2) double O-deacylation of NAPEs followed by phosphodiester bond hydrolysis of the resulting glycero-phospho (GP)-NAEs. We recently identified GDE1 as a GP-NAE phosphodiesterase that may be involved in the second pathway. Here, we report the generation and characterization of GDE1(−/−) mice, which are viable and overtly normal in their cage behavior. Brain homogenates from GDE1(−/−) mice exhibit a near-complete loss of detectable GP-NAE phosphodiesterase activity; however, bulk brain levels of AEA and other NAEs were unaltered in these animals. To address the possibility of compensatory pathways, we generated GDE1(−/−)/NAPE-PLD(−/−) mice. Conversion of NAPE to NAE was virtually undetectable in brain homogenates from these animals as measured under standard assay conditions, but again, bulk changes in brain NAEs were not observed. Interestingly, significant reductions in the accumulation of brain NAEs, including anandamide, were detected in GDE1(−/−)/NAPE-PLD(−/−) mice treated with a FAAH inhibitor that blocks NAE degradation. Finally, we determined that primary neurons from GDE1(−/−)/NAPE-PLD(−/−) mice can convert NAPEs to NAEs by a pathway that is not preserved following cell homogenization. In summary, combined inactivation of GDE1 and NAPE-PLD results in partial disruption of NAE biosynthesis, while also pointing to the existence of an additional enzymatic pathway(s) that converts NAPEs to NAEs. Characterization of this pathway should provide clarity on the multifaceted nature of NAE biosynthesis.
Introduction
The endocannabinoid system plays a key role in both central and peripheral tissues, where it participates in diverse physiological processes including nociception, inflammation, appetite, and metabolism1–4. This system is comprised primarily of the cannabinoid G-protein coupled receptors CB1 and CB2, their two endogenous arachidonate-containing lipids (“endocannabinoids”) anandamide (arachidonoyl ethanolamine, AEA) and 2-arachidonoyl glycerol (2-AG), and the enzymatic pathways responsible for endocannabinoid biosynthesis and degradation5–6.
Classical neurotransmitters, such as amino acids, monoamines, and neuropeptides, are water-soluble and are stored in vesicles prior to release into the synaptic cleft upon neuronal depolarization. In contrast, lipid messengers such as the endocannabinoids are considered to be too hydrophobic for storage in vesicles and are therefore believed to be synthesized “on demand” from phospholipid precursors7. The signaling potential of endocannabinoids is consequently under strict regulation by the enzymes and proteins that catalyze their biosynthesis, transport, and degradation5, 8–10. The identification and characterization of these enzymes is crucial to obtaining a complete understanding of endocannabinoid signaling and its role in mammalian physiology, as well as to ascertain the potentially distinct functions played by AEA and 2-AG.
AEA is a member of a large class of amidated lipids termed the N-acyl ethanolamines (NAEs)11 and is the principal member of this family with CB1 agonist activity12–13. Early studies on changes to the lipidome resulting from ischemic shock in dog heart and brain tissue revealed rapid simultaneous accumulation of NAEs and an unusual phospholipid species, N-acyl phosphatidyl ethanolamines (NAPEs), suggesting a precursor-product relationship between these two classes of lipids14–15. Several alternative hypotheses concerning the biosynthesis of NAEs have been proposed including direct conjugation of ethanolamine with fatty acids or acyl CoAs16–17; however, multiple lines of evidence point to NAPEs as the endogenous precursors to NAEs18. In the early 1980s, several pathways were proposed by which NAEs could be released from NAPEs, including direct hydrolysis by a D-type phospholipase, as well as single- or double-O-deacylation followed by phosphodiesterase activity on the resulting lysophosphatidyl- or glycero-phospho-N-acyl ethanolamines (GP-NAEs)19. Direct release from NAPEs was found to be activated by millimolar concentrations of calcium in vitro, and this D-type phospholipase activity could be detected in multiple rodent tissues, from where it was ultimately purified and molecularly characterized20–22. This enzyme, dubbed NAPE-PLD, was found to be exquisitely selective for NAPEs in vitro and highly expressed in tissues that utilize endocannabinoid signaling, such as nervous tissue and testis.
Consistent with a role for NAPE-PLD in NAE biosynthesis, mice lacking this enzyme [NAPE-PLD(−/−) mice] display profound reductions in very long-chain saturated and mono-unsaturated NAEs in the central nervous system23. Interestingly, however, poly-unsaturated NAEs, including AEA, were unaltered in the nervous system of NAPE-PLD(−/−) mice. This finding engendered the hypothesis that multiple enzymatic pathways are involved in the conversion of NAPEs to NAEs in vivo, with NAPE-PLD controlling the biosynthesis of long-chain saturated NAEs, and additional enzymes participating in the generation of shorter chain and poly-unsaturated NAEs (including AEA). Attempts to detect an additional D-type phospholipase activity in NAPE-PLD(−/−) tissues were unsuccessful, but robust O-deacylation of NAPE was observed, suggesting that lysoNAPEs or GP-NAEs might serve as direct precursors to NAEs24. In support of this deacylation hypothesis, it was observed that inhibitors of A/B-type phospholipases blocked the release of NAE from NAPE and lysoNAPE in brain homogenates from NAPE-PLD(−/−) mice indicating that total O-deacylation must occur prior to phosphodiester bond cleavage under these in vitro conditions.
Recent work from our lab has identified candidate enzymes that can perform each step in this proposed deacylation-phosphodiesterase pathway for NAE biosynthesis. The previously uncharacterized alpha-beta hydrolase enzyme ABH4 acts as a B-type phospholipase selective for (lyso)NAPE and produces GP-NAE from NAPE in vitro. The resulting GP-NAE is a substrate for the metal-dependent enzyme GDE125 (glycerophosphodiester phosphodiesterase 1, also known as MIR16), which was previously characterized in vitro as a glycerophosphodiesterase that acts on multiple glycerophosphodiester substrates26–27. To test the contribution of this pathway to NAE biosynthesis in vivo, we herein report the generation and characterization of mice bearing a targeted disruption of the GDE1 gene.
Results and Discussion
Generation of GDE1(−/−) mice
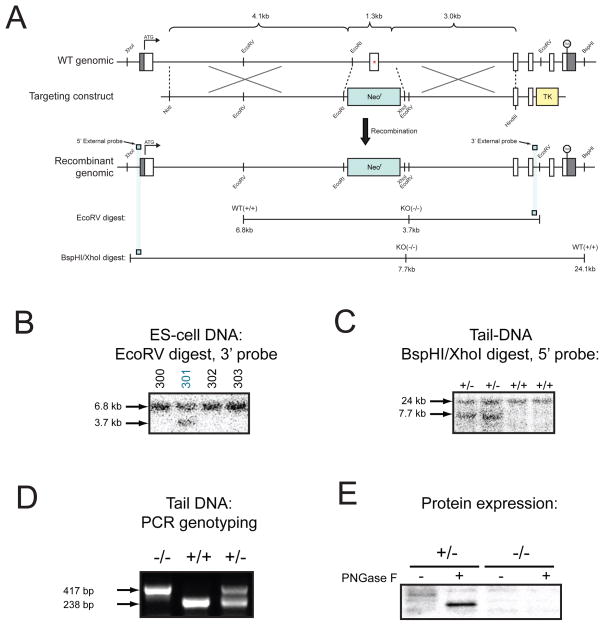
Construction of mice bearing a disruption in the GDE1 gene was achieved using a standard targeting strategy directly in embryonic stem (ES) cells from the C57BL/6 background. A targeting construct was designed to disrupt exon two of the GDE1 gene locus, which contains residues that are required for catalytic activity (Figure 1A). A homologously recombined ES-cell clone was identified by Southern blotting (Figure 1B) and used to generate chimeric mice on an albino C57BL/6 background. These chimeric mice gave germline transfer of the mutated locus (Figure 1C), and the resulting pups served as founders for the GDE1(−/−) strain. Western blotting of tissue extracts from GDE1(−/−) brains confirmed loss of the GDE1 protein (Figure 1E).
Figure 1. Generation of GDE1(−/−) mice.
(A) The genomic structure of the GDE1 gene locus on chromosome 7 is shown, along with the targeting construct used to delete exon 2 (which contains several residues essential for catalysis, Glu97, Asp99 and His112) and the final recombined locus. Only relevant restriction sites are designated. The locations of DNA probes used for Southern blotting 5’ and 3’ to the targeting construct are also shown. (B) Southern blot of EcoRV-digested ES cell DNA showing the heterozygous (targeted) clone #301. (C) Sothern blot of BspHI/XhoI-digested tail-DNA from heterozygous and wild-type mice demonstrating germ-line transmission. (D) PCR strategy in which the wild-type locus is identified by a 238 bp band and the targeted locus is identified by a 417 bp band. (E) Western blot of the particulate fraction from brain tissue confirming loss of GDE1 protein. GDE1 is a PNGaseF-sensitive glycosylated protein.
Characterization of GP-NAE and NAE metabolism in GDE1(−/−) mice
GDE1(−/−) mice were born at the expected Mendelian frequency, were viable and healthy, and showed no overt differences in their cage behavior compared to wild-type littermates. Initial biochemical characterization of the GDE1(−/−) mice focused on measurement of GP-NAE phosphodiesterase activity in brain homogenates (Figure 2A). These experiments revealed that GDE1 is responsible for nearly all (> 95%) of the detectable GP-NAE phosphodiesterase activity in brain tissue. Additionally, we found that conversion of lysoNAPE to NAE was also impaired in tissue extracts (Figure 2B), confirming that conversion of lysoNAPE to NAE proceeds through a GP-NAE intermediate in vitro. Previously, we described an experiment in which brain homogenates accumulated “endogenous” GP-NAEs in an MAFP-sensitive, EDTA-induced manner, and we hypothesized that GDE1, a metal-dependent enzyme, was the EDTA-sensitive phosphodiesterase responsible for this accumulation25. To test this hypothesis, a similar experiment was performed in which brains from GDE1(+/−) and (−/−) mice were incubated with or without EDTA and the accumulation of GP-NAE was measured by LC-MS/MS (Figure 2C). Consistent with previous studies, GP-NAEs accumulated in brain tissue from GDE1(+/−) mice in an EDTA-dependent manner. Interestingly, GP-NAE levels in GDE1(−/−) brains were basally high after 4 hour incubation and equivalent in magnitude to those observed in EDTA-treated GDE1(+/−) brains. No additional EDTA-induced accumulation of GP-NAEs was observed in GDE1(−/−) brains. These results confirm that GDE1 is the principal EDTA-sensitive GP-NAE phosphodiesterase in brain homogenates.
Figure 2. GP-NAE phosphodiesterase activity is lost in GDE1(−/−) tissue.
(A) Near-complete loss of conversion of 14C-GP-NAE to 14C-NAE is evident from a representative thin layer radiochromatogram (left) and quantified data from replicate experiments (right). (B) Loss of conversion of 14C-lysoNAPE to 14C-NAE is evident from a representative thin layer radiochromatogram (left) and quantified data from replicate experiments (right). Note also the accumulation of the GP-NAE intermediate in the assay with GDE1(−/−) tissue. All assays were performed in PBS buffer with tissue homogenates from brains of GDE1(+/+) and (−/−) mice. (C) Ex vivo assay demonstrating that EDTA induces GP-NAE accumulation in brain homogenates due to disruption of GDE1 activity. Brain tissue from GDE1(+/−) and (−/−) mice was homogenized in the presence or absence of 10 mM EDTA (which inhibits GDE1) and incubated at room temperature for 4 hours. The addition of EDTA leads to a rapid accumulation of GP-NAEs in wild-type brain, as previously reported. GP-NAE accumulation was observed in brain tissue from GDE1(−/−) mice with- or without EDTA, demonstrating that GDE1 is the EDTA-sensitive GP-NAE phosphodiesterase in wild-type tissue. n = 4 mice per group. Results are presented as means ± standard error. Asterisks designate p < 0.05, p < 0.01, p < 0.005 (student’s t-test) for *, **, and ***, respectively. Asterisks represent difference from wild-type in panels (A) and (B), and difference from EDTA-treated GDE1(+/−) tissue in panel (C).
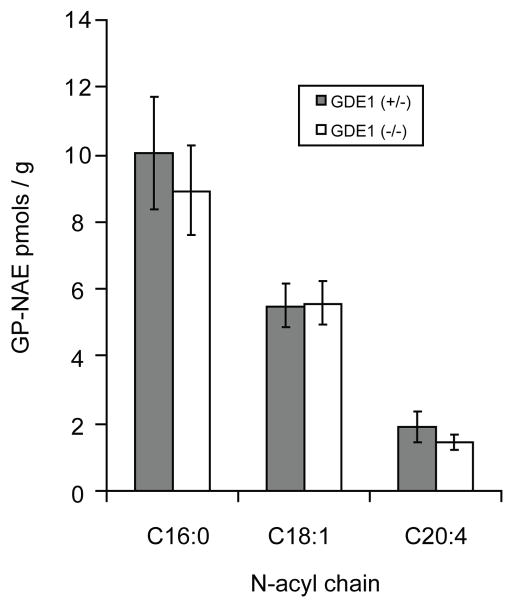
To determine if the deacylation-phosphodiesterase pathway is operant in vivo, we undertook targeted measurements of endogenous metabolites via LC-MS/MS. First, we measured GP-NAEs in the brains of GDE1(+/−) and (−/−) animals but, surprisingly, observed no significant differences between these genotypes (Figure 3). Basal NAE levels were also unchanged in GDE1(−/−) brains (Figure 4).
Figure 3. GP-NAE levels are unchanged in GDE1(−/−) brains.
Endogenous GP-NAEs were measured in brain extracts from GDE1(+/−) and (−/−) mice via LC-MS/MS. No significant differences were observed. n = 8 mice per group. Results are presented as means ± standard error.
Figure 4. Bulk NAE levels in brains from mice lacking GDE1 and/or NAPE-PLD.
Brains from wild-type, GDE1(−/−), NAPE-PLD(−/−) or GDE1(−/−)/NAPE-PLD(−/−) mice were subjected to organic extraction and endogenous NAE levels were measured by LC-MS/MS. No significant differences were observed between wild-type and GDE1(−/−) or between NAPE-PLD(−/−) and GDE1(−/−)/NAPE-PLD(−/−) brains.
Characterization of GP-NAE and NAE metabolism in GDE1(−/−), NAPE-PLD(−/−), and GDE1(−/−)/NAPE-PLD(−/−) mice
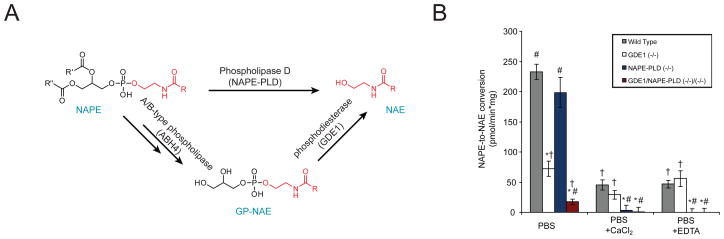
As multiple enzymes are known to utilize NAPE as a substrate (Figure 5A), we next crossed GDE1 and NAPE-PLD lines to generate GDE1(−/−)/NAPE-PLD(−/−) double knockout mice to measure the combined effects of ablation of these enzymes on NAE metabolism. Brain tissue from wild-type, double-knockout, and each single-knockout was assayed for the ability to release 14C-NAE from 14C-NAPE. As has been previously reported23, the choice of assay conditions affected the relative contributions of these two pathways, with the presence or absence of calcium proving particularly dramatic. Furthermore, we found that selection of reaction buffer and inclusion of detergents also influenced the relative contributions made by each pathway (Supplemental Figures 1 and 2). Depending on the assay conditions employed, absence of either NAPE-PLD or GDE1 resulted in major reductions in NAPE-to-NAE conversion, but, under all conditions tested, the double-knockout showed near-total loss of NAPE-to-NAE conversion in brain homogenates (Figure 5B). Similar results were obtained in testis homogenates (not shown).
Figure 5. NAPE conversion to NAE in brain lysates from mice lacking GDE1 and/or NAPE-PLD.
(A) Pathway diagram showing potential routes for NAPE conversion to NAE. Note that both NAPE-PLD and GDE1 can liberate NAE from its esterified precursors (NAPE and GP-NAE, respectively). (B) Brain tissue from wild-type, GDE1(−/−), NAPE-PLD(−/−), or GDE1(−/−)/NAPE-PLD(−/−) mice was homogenized and assayed for the ability to convert NAPE to NAE in PBS buffer in the presence or absence of 10 mM CaCl2 or EDTA via thin layer radiochromatography. In the presence of calcium or EDTA, tissue lacking NAPE-PLD shows complete loss of NAPE-to-NAE conversion. However, assays performed without these additives show no impairment from loss of NAPE-PLD and a large reduction in activity in tissue lacking GDE1. Under all conditions, tissue from the GDE1(−/−)/NAPE-PLD(−/−) displayed negligible activity. The results are presented as means ± standard error. n = 4 mice/group. The symbols above the bars designate p < 0.05 (student’s t-test) with * for different from wild-type, # for different from GDE1(−/−) samples and † for different from NAPE-PLD(−/−) samples.
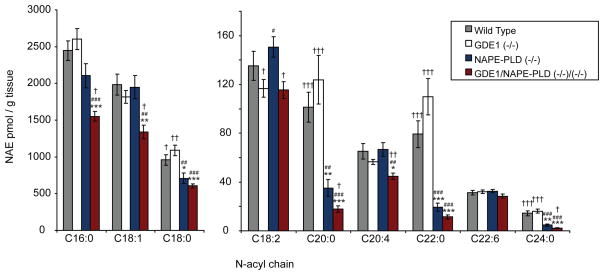
Despite the near-complete loss of NAPE-to-NAE conversion in brain homogenates from GDE1(−/−)/NAPE-PLD(−/−) double knockout mice, no significant differences in bulk brain levels of NAEs were observed between NAPE-PLD(−/−) and the GDE1(−/−)/NAPE-PLD(−/−) mice (Figure 4). We next asked whether the rate of NAE production might be impaired by GDE1 and/or NAPE-PLD disruption by treating mice with the recently described FAAH inhibitor PF-384528. FAAH inhibition has previously been shown to produce a steady and dramatic rise in NAE levels that plateau after about 2–3 hours29–30. We therefore measured brain NAE levels in various mouse models at 3 hours following treatment with PF-3845. Interestingly, at this time point, GDE1(−/−)/NAPE-PLD(−/−) mice displayed significant reductions in several NAEs including the endocannabinoid anandamide (Figure 6). The magnitude of these reductions was rather modest (~25–40%), but clearly suggests that GDE1 and NAPE-PLD, together, contribute to NAE biosynthesis. Similar effects on NAE accumulation were observed in testis from these mice (Supplemental Figure 3). Furthermore, the fact that these reductions were only observed following disruption of both GDE1 and NAPE-PLD indicates that these pathways have the potential to crosstalk with one another to regulate NAE production.
Figure 6. Impaired NAE accumulation is observed following FAAH inhibition in brains from mice lacking GDE1 and NAPE-PLD.
Mice were treated with the selective FAAH inhibitor PF-3845 (10 mg/kg, i.p.) and, after 3 hours, sacrificed and brain NAE levels measured. Significant reductions were observed for several NAEs, including the endocannabinoid anandamide (C20:4), in brains from GDE1(−/−)/NAPE-PLD(−/−) mice. The results are presented as means ± standard error. n = 4 mice/group. The symbols above the bars designate p < 0.05, p < 0.01, p < 0.005 (student’s t-test) with *, **, *** for different from wild-type, #, ##, ### for different from GDE1(−/−) samples and †, ††, ††† for different from NAPE-PLD(−/−) samples, respectively.
Evidence for additional NAPE-to-NAE enzymatic pathways in neurons
Taken together, our results with GDE1(−/−) and NAPE-PLD(−/−) mice are somewhat perplexing. Mice lacking both GDE1 and NAPE-PLD display wild-type levels of NAEs and only modestly compromised rates of NAE production, despite being totally impaired in the ability to release NAE from NAPE in vitro. Two possible explanations for this situation present themselves: (1) NAEs do not, in fact, come from NAPEs in vivo, or (2) NAEs are released from NAPEs in vivo by an enzyme or enzymes whose activities are not maintained in vitro. To explore the latter possibility, we sought to develop a live-cell system in which the conversion of NAPE to NAE could be monitored in situ. To achieve this goal, we cultured primary neurons from neonatal wild-type, GDE1(−/−), NAPE-PLD(−/−), or GDE1(−/−)/NAPE-PLD(−/−) pups and grew them for 6 days in vitro. Neurons were then treated with the FAAH inhibitor PF-3845 (to prevent turnover of NAE) and 14C-NAPE was added to the media. In this way, we observed clear time-dependent release of NAE from NAPE in live neurons (Figure 7A). Previous studies have monitored the formation of NAE in neurons by employing either radiolabeled ethanolamine or arachidonate7, 31, which do not provide information on the direct precursor of NAE biosynthesis. Under our experimental conditions, the presence of a FAAH inhibitor prevented hydrolysis of the amide bond linking 14C-fatty acid to phosphatidylethanolamine thereby ensuring that all NAE formed must be released from NAPE. Once this experimental paradigm was established, we tested the ability of neurons from single- and double knockout animals to release NAPE to NAE. To our surprise, we found that all genotypes including GDE1(−/−)/NAPE-PLD(−/−) released NAE from NAPE at equivalent levels (Figure 7B and C). This result implies the existence of an additional enzyme(s) distinct from NAPE-PLD and GDE1 that can release NAEs from NAPEs. To test whether this in situ-active enzymatic activity requires O-deacylation or whether it operates directly on NAPE, we employed a non-hydrolyzable analogue of NAPE that contains ether-linkages in the sn-1 and sn-2 positions24. This ether-NAPE analogue did not serve as an effective substrate for NAE formation (Figure 7D), suggesting that the in situ (GDE1- and NAPE-PLD-independent) pathway for NAE generation requires O-deacylation prior to phosphodiester bond cleavage.
Figure 7. In situ NAE release from NAPE in primary neuron cultures from mice lacking GDE1 and NAPE-PLD.
(A) Time-dependence of NAE release in the presence or absence of the FAAH inhibitor PF-3845. NAE release in the absence of neurons was negligible on this timescale (not shown). (B) Assays conducted in primary neurons for 6 hours in the presence of PF-3845. Comparison of wild-type neurons to neurons from GDE1(−/−) or NAPE-PLD(−/−) mice reveals no impairment in the ability to release NAE from NAPE in situ. (C) Comparison of NAPE-PLD(−/−) neurons to GDE1(−/−)/NAPE-PLD(−/−) neurons reveals wild-type levels of NAE release in GDE1(−/−)/NAPE-PLD(−/−) neurons. (D) NAE release from 14C-NAPE or an ether-linked (non-hydrolyzable) NAPE analogue in neurons from GDE1(−/−)/NAPE-PLD(−/−) mice reveals the necessity of de-acylation prior to NAE release. These data represent single experiments performed in triplicate, representative of multiple experiments. *, p < 0.05.
Conclusion
In summary, brain tissue from mice lacking GDE1 and NAPE-PLD shows a near-complete loss in NAPE conversion to NAE. Despite this result, however, bulk brain levels of NAEs were unaltered in mice lacking GDE1 and NAPE-PLD. A defect in the rate of NAE production was observed in GDE1(−/−)/NAPE-PLD(−/−) mice, suggesting that both GDE1 and NAPE-PLD make partial contributions to the biosynthesis of anandamide and other NAEs in vivo. Considering further that all of our measurements were made with bulk brain tissue, it is also possible that GDE1 and/or NAPE-PLD play even more substantial roles in anandamide/NAE biosynthesis in specific neuronal circuits or brain regions.
When NAPE metabolism was studied in live neurons (“in situ”), we discovered that neurons lacking both GDE1 and NAPE-PLD are capable of converting NAPEs to NAEs. This result supports the existence of additional NAPE-catabolizing enzymes that are operant in situ, and likely in vivo, but artificially inactivated in brain homogenates. When considering the history of other phosphodiesterase enzymes, one finds that this situation is not exceptional. The characterization of the prototypical mammalian phospholipase D (PLD1) involved years of frustration during which activity in intact cell systems was observed, but lost upon cell-lysis. In fact, several reports similar to this one can be found in the literature in which PLD activity was measured in intact cell preparations, but could not be reconstituted in vitro24, 32–33. The breakthrough discovery came when it was found that PLD1 activity was dramatically enhanced in vitro by performing substrate assays under precise conditions: addition of the non-hydrolyzable GTP analogue GTPγS, addition of the ADP-ribosylation factor (ARF) protein, as well as substrate presentation in mixed liposomes that include phosphatidylinositol-4,5-bisphosphate (PIP2)34. Indeed, omission of any of these factors resulted in almost complete loss of activity. That enzyme, PLD1, is selective for phosphatidylcholine (PC), although to our knowledge it has not been tested with N-acylated substrates. Additionally, there are another six PLD isoforms in mammalian genomes, five of which are virtually unstudied due to inability to show activity in vitro. Furthermore, there are several GDE1 homologues that, to our knowledge, have not yet been shown to display activity in vitro. Finally, other pathways have been proposed for NAPE catabolism involving C-type phospholipase (PLC) activity (resulting in phospho-NAE, which is a substrate for one or more phosphatases)35–36, although direct measurement of NAPE-PLC activity has not yet been described. This does not necessarily argue against a PLC pathway, since mammalian C-type phospholipases have also been shown to have complex requirements for co-factors and protein activators for reconstitution of activity in vitro 37. Looking forward, the ability of various co-factors to stimulate endogenous NAPE catabolic pathway(s) should be explored to more clearly define their composition (e.g., do they involve A/B-, C-, or D-type phospholipases, or some combination thereof?) and possible role in anandamide biosynthesis in vivo. Finally, considering the recent discovery of several classes of endogenous N-acyl amino acids in brain tissue38, it is worth investigating whether alternative pathways for anandamide/NAE production may exist that do not involve NAPEs as obligate precursors.
Experimental
Generation of GDE1(−/−) mice
The GDE1 locus was obtained as part of a BAC clone from the RPCI-23 library of genomic DNA from a female C57BL/6J mouse. The targeting construct was generated using PCR-amplified 5’ and 3’ homologous recombination fragments surrounding exon 2 of the GDE1 gene, which were subcloned into NotI/EcoRI and XhoI/HindIII sites in the pKO-NTKV vector. Homologous recombinant Bruce-4 stem cell clones (C57BL/6 strain of origin) were identified by Southern blot analysis, and one such clone was used to generate chimeric mice on an albino C57BL/6 background, several of which yielded germline transmission of the mutated gene.
In vitro enzyme assays
All assays were performed in tissue homogenates. Mice were anesthetized with isoflurane and killed by decapitation. Tissues were immediately removed and snap-frozen in liquid N2. Frozen tissue was homogenized in phosphate buffered saline (PBS, except where noted) using a dounce-homogenizer and centrifuged at 1000 × g to remove tissue debris. The supernatant from this centrifugation step was adjusted to 2mg/ml with PBS and stored in single-use aliquots at −80°C. Except where noted, assays were performed in glass vials in PBS at 37 °C for 30 minutes in 100 μL total volume. Final protein concentration for enzyme assays was 1mg/mL for NAPE and lysoNAPE substrates, and 0.1mg/mL for GP-NAE substrates. Substrates (1,2-dioleoyl-sn-glycero-3-phospho(N-[1’-14C]-palmitoyl)ethanolamine, 1-oleoyl-2-hydroxy-sn-glycero-3-phospho(N-[1’-14C]-palmitoyl)ethanolamine, and 1,2-dihydroxy-sn-glycero-3-phospho(N-[1’-14C]-palmitoyl)ethanolamine) were prepared as previously described 24–25. Substrates were stored as 2mM stock in EtOH at −80°C and 5uL was added to each enzyme assay (for 100μM final concentration). After 30 minutes, 1.5 mL of 2:1 CHCl3:MeOH was added to quench the reactions and an additional 0.4 mL of 1% formic acid was added. Vials were mixed and briefly centrifuged to separate phases and the organic (bottom) phase was removed to fresh vials and evaporated to dryness under N2. Lipids thus extracted were resuspended in 20 μL 2:1 CHCl3:MeOH and spotted on thin layer silica plates and developed in 40:10:1 CHCl3:MeOH:28% NH4OH. Distribution of radioactivity on the plates was quantified by a phosphorimaging device (Packard) and products were identified by comparison to synthetic standards. Heat-denatured or no-protein controls were included with all assays and these values were subtracted as background. As noted in the text, some assays included one of the following additives: 10 mM EDTA, 10 mM CaCl2 or 0.1% Triton X-100.
Primary neuron culture and in situ assays
Culturing of primary neurons was performed essentially as previously described 39. Briefly, P0 pups were taken from their mothers and kept at ~37 °C prior to dissection. Pups were tailed and PCR-genotyped within 2 hours of separation from their mothers. Pups with the desired genotypes were sacrificed and forebrains were dissected and dissociated by incubation for 7 minutes at 37 °C in digestion solution containing 6 mg/mL trypsin, 0.5 mg/mL DNAse, 137 mM NaCl, 5 mM KCl, 7 mM Na2HPO4, and 25 mM HEPES, pH 7.2. The dissociated cells were washed once with Hank’s balanced salt solution (HBS) containing 20% fetal bovine serum (FBS) and once in serum-free HBS, and further dissociated by gentle trituration in HBS containing 12 mM MgSO4 and 0.5 mg/mL DNAse. The cell suspension was centrifuged for 3 minutes at 1000 × g and plated in Matrigel-coated 6-well dishes in MEM supplemented with glutamine, insulin, transferrin, glucose, and 10% FBS. Cells from the forebrain of a single pup were used to plate 6 wells in a 6-well dish. After ~12 hours, the media was removed and replaced with MEM supplemented with glucose, transferrin, B27, glutamine, and 5% FBS (“growth media”). After 3 days of culture (when glial cells reached ~40–50%), 50% of the culture-medium was replaced with fresh growth medium containing 10μM Ara-C (for 5μM final concentration) to prevent glial proliferation. The cultures were maintained in medium containing 5μM Ara-C. After 6 days in vitro, 2 μL PF-3845 in DMSO or DMSO was added to a final concentration of 10 μM and cells were returned to the incubator. After 1 hour, media was removed, and replaced with 1 mL of fresh growth media containing 10 μM PF-3845 as well as 14C-labeled substrate at 20 μM (final concentration), and neurons were returned to the incubator for various time points, typically 6 hours. After 6 hours, each well was scraped and neurons and media were transferred to a glass vial containing 6 mL of 2:1 CHCl3:MeOH and 1 mL of 1% formic acid for 8 mL total. Vials were vortexed to mix, centrifuged to separate phases, and the organic (bottom) phase was removed to a fresh glass vial and evaporated to dryness under a stream of N2. Dried reactions were then analyzed by thin layer radiochromatography as described above.
Brain-lipid extraction and tandem MS analysis of NAE and GP-NAE
Endogenous lipid measurements were performed essentially as previously described. Briefly, male mice between 2 and 6 months of age were anesthetized with isoflurane and killed by decapitation. Brains were rapidly removed, sectioned into left and right hemispheres, and immediately frozen in liquid N2. Frozen brains were placed in a Dounce homogenizer with 8 mL of 2:1:1 CHCl3:MeOH:50 mM Tris, pH 8.0, and 200 pmol of 1,2-dihydroxy-sn-glycero-3-phospho(N-pentadecenoyl)ethanolamine (C15:1 GP-NAE) or 2 pmol of arachidonoyl-(d4)-ethanolamine for GP-NAE or NAE measurements, respectively. Lipids were extracted via homogenization with 8–10 strokes of the Dounce homogenizer at which point homogenates were transferred to 8mL glass vials and centrifuged for 10 minutes at 1,400 × g. The organic (bottom) phase was transferred to a clean vial and the remaining aqueous phase was re-extracted with an additional 4 mL of CHCl3. After centrifugation, the organic phases were pooled and evaporated to dryness under a stream of N2. Lipid extracts from each half-brain were then resuspended in 120 μL 2:1 CHCl3:MeOH and transferred to LC-MS vials for immediate analysis or stored at −80°C. 15 μL of resuspended lipid was injected into an Agilent 6410 triple quadrupole mass spectrometer with an Agilent 1200 autosampler. Samples were chromatographed on a Gemini 5 μm C18 column (50 × 4.6 mm, Phenomenex, Torrance, CA) in the negative-ion mode for GP-NAE measurements and the positive-ion mode for NAE measurements at a flow-rate of 0.4 mL/min. GP-NAEs and NAEs were monitored by multiple reaction monitoring (MRM). For GP-NAEs the transition from [M-H]− to [M-H-74]− with collision energy 35V was monitored and compared to the internal C15:1 GP-NAE standard and measurements of endogenous GP-NAEs were corrected for extraction efficiency as previously described25. For NAEs, the MRM transition from [M+H]+ to m/z 62+ (ethanolamine fragment) with collision energy 11V was used, except for the d4-AEA standard which monitored the transition from [M+H]+ to m/z 66+ (d4 ethanolamine fragment). NAEs were quantified by comparison to the d4-AEA standard. The dwell-time was set to 100ms for all lipids. For ex vivo analysis of GP-NAEs, half-brains were homogenized in a Dounce homogenizer in 2 mL of 50 mM Tris pH 8.0 with or without 10 mM EDTA. Samples were incubated at room temperature for 4hr with gentle rotation prior to extraction and GP-NAE quantification as described above.
Supplementary Material
References
- 1.Pacher P, Bátkai S, Kunos G. Pharmacol Rev. 2006;58:389–462. doi: 10.1124/pr.58.3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howlett AC, Breivogel CS, Childers SR, Deadwyler SA, Hampson RE, Porrino LJ. Neuropharmacology. 2004;47:345–358. doi: 10.1016/j.neuropharm.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 3.Walter L, Stella N. Br J Pharmacol. 2004;141:775–785. doi: 10.1038/sj.bjp.0705667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fowler CJ, Holt S, Nilsson O, Jonsson KO, Tiger G, Jacobsson SO. Pharmacol Biochem Behav. 2005;81:248–262. doi: 10.1016/j.pbb.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 5.Ahn K, McKinney MK, Cravatt BF. Chem Rev. 2008;108:1687–1707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Marzo V. Rev Physiol Biochem Pharmacol. 2008;160:1–24. doi: 10.1007/112_0505. [DOI] [PubMed] [Google Scholar]
- 7.Cadas H, Gaillet S, Beltramo M, Venance L, Piomelli D. J Neurosci. 1996;16:3934–3942. doi: 10.1523/JNEUROSCI.16-12-03934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fowler CJ. Fundam Clin Pharmacol. 2006;20:549–562. doi: 10.1111/j.1472-8206.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- 9.Glaser ST, Kaczocha M, Deutsch DG. Life Sci. 2005;77:1584–1604. doi: 10.1016/j.lfs.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Deutsch DG, Ueda N, Yamamoto S. Prostaglandins Leukot Essent Fatty Acids. 2002;66:201–210. doi: 10.1054/plef.2001.0358. [DOI] [PubMed] [Google Scholar]
- 11.Schmid HH. Chem Phys Lipids. 2000;108:71–87. doi: 10.1016/s0009-3084(00)00188-2. [DOI] [PubMed] [Google Scholar]
- 12.Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 13.Felder CC, Briley EM, Axelrod J, Simpson JT, Mackie K, Devane WA. Proc Natl Acad Sci U S A. 1993;90:7656–7660. doi: 10.1073/pnas.90.16.7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epps DE, Schmid PC, Natarajan V, Schmid HHO. Biochem Biophys Res Commun. 1979;90:628–633. doi: 10.1016/0006-291x(79)91281-6. [DOI] [PubMed] [Google Scholar]
- 15.Natarajan V, Schmid PC, Reddy PV, Zuzarte-Augustin ML, Schmid HHO. J Neurochem. 1983;41:1303–1312. doi: 10.1111/j.1471-4159.1983.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 16.Colodzin M, Bachur NR, Weissbach H, Udenfriend S. Biochem Biophys Res Commun. 1963;10:165–170. doi: 10.1016/0006-291x(63)90044-5. [DOI] [PubMed] [Google Scholar]
- 17.Deutsch DG, Chin SA. Biochem Pharmacol. 1993;46:791–796. doi: 10.1016/0006-2952(93)90486-g. [DOI] [PubMed] [Google Scholar]
- 18.Sugiura T, Kondo S, Sukagawa A, Tonegawa T, Nakane S, Yamshita A, Waku K. Eur J Biochem. 1996;240:53–62. doi: 10.1111/j.1432-1033.1996.0053h.x. [DOI] [PubMed] [Google Scholar]
- 19.Natarajan V, Schmid PC, Reddy PV, Schmid HH. J Neurochem. 1984;42:1613–1619. doi: 10.1111/j.1471-4159.1984.tb12750.x. [DOI] [PubMed] [Google Scholar]
- 20.Okamoto Y, Morishita J, Tsuboi K, Tonai T, Ueda N. J Biol Chem. 2004;279:5298–5305. doi: 10.1074/jbc.M306642200. [DOI] [PubMed] [Google Scholar]
- 21.Ueda N, Liu Q, Yamanaka K. Biochim Biophys Acta. 2001;1532:121–127. doi: 10.1016/s1388-1981(01)00120-2. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Okamoto Y, Morishita J, Tsuboi K, Miyatake A, Ueda N. J Biol Chem. 2006;281:12325–12335. doi: 10.1074/jbc.M512359200. [DOI] [PubMed] [Google Scholar]
- 23.Leung D, Saghatelian A, Simon GM, Cravatt BF. Biochemistry. 2006;45:4720–4726. doi: 10.1021/bi060163l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon GM, Cravatt BF. J Biol Chem. 2006;281:26465–26472. doi: 10.1074/jbc.M604660200. [DOI] [PubMed] [Google Scholar]
- 25.Simon GM, Cravatt BF. J Biol Chem. 2008;283:9341–9349. doi: 10.1074/jbc.M707807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng B, Chen D, Farquhar MG. Proc Natl Acad Sci U S A. 2000;97:3999–4004. doi: 10.1073/pnas.97.8.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng B, Berrie CP, Corda D, Farquhar MG. Proc Natl Acad Sci U S A. 2003;100:1745–1750. doi: 10.1073/pnas.0337605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson DS, Ahn K, Kesten S, Lazerwith SE, Song Y, Morris M, Fay L, Gregory T, Stiff C, Dunbar JB, Jr, Liimatta M, Beidler D, Smith S, Nomanbhoy TK, Cravatt BF. Bioorg Med Chem Lett. 2009;19:2865–2869. doi: 10.1016/j.bmcl.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, Lazerwith S, Stiff C, Kamtekar S, Bhattacharya K, Zhang Y, Swaney S, Van Becelaere K, Stevens RC, Cravatt BF. Chem Biol. 2009;16:411–420. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fegley D, Gaetani S, Duranti A, Tontini A, Mor M, Tarzia G, Piomelli D. J Pharmacol Exp Ther. 2005;313:352–358. doi: 10.1124/jpet.104.078980. [DOI] [PubMed] [Google Scholar]
- 31.Di Marzo V, Fontana A, Cadas H, Schinelli S, Cimino G, Schwartz JC, Piomelli D. Nature. 1994;372:686–691. doi: 10.1038/372686a0. [DOI] [PubMed] [Google Scholar]
- 32.Cockcroft S. Biochim Biophys Acta. 1992;1113:135–160. [PubMed] [Google Scholar]
- 33.Martin TW, Michaelis K. J Biol Chem. 1989;264:8847–8856. [PubMed] [Google Scholar]
- 34.Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternwels PC. Cell. 1993;75:1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Wang L, Harvey-White J, Huang BX, Kim HY, Luquet S, Palmiter RD, Krystal G, Rai R, Mahadevan A, Razdan RK, Kunos G. Neuropharmacology. 2008;54:1–7. doi: 10.1016/j.neuropharm.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Wang L, Harvey-White J, Osei-Hyiaman D, Razdan R, Gong Q, Chan AC, Zhou Z, Huang BX, Kim HY, Kunos G. Proc Natl Acad Sci USA. 2006;103:13345–13350. doi: 10.1073/pnas.0601832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smrcka AV, Hepler JR, Brown KO, Sternweis PC. Science. 1991;251:804–807. doi: 10.1126/science.1846707. [DOI] [PubMed] [Google Scholar]
- 38.Tan B, O'Dell DK, Yu YW, Monn MF, Hughes HV, Burstein S, Walker JM. J Lipid Res. 2009:M900198–JLR900200. doi: 10.1194/jlr.M900198-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maximov A, Pang ZP, Tervo DGR, Südhof TC. J Neurosci Methods. 2007;161:75–87. doi: 10.1016/j.jneumeth.2006.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.