Abstract
Centromeres are specified epigenetically, and the histone H3 variant, CENP-A, is assembled into the chromatin of all active centromeres1. Divergence from H3 raises the possibility that CENP-A generates unique chromatin features to physically mark centromere location. The crystal structure of the sub-nucleosomal heterotetramer reported here reveals three distinguishing properties encoded by the residues that comprise the CENP-A Targeting Domain (CATD2): (1) a CENP-A/CENP-A interface that is substantially rotated relative to the H3/H3 interface, (2) a protruding loop L1 of the opposite charge as on H3, and (3) strong hydrophobic contacts that rigidify the CENP-A/H4 interface. Residues involved in the CENP-A/CENP-A rotation are required for efficient incorporation into centromeric chromatin suggesting specificity for an unconventional nucleosome shape. DNA topological analysis indicates that CENP-A-containing nucleosomes are nonetheless octameric with conventional left-handed DNA wrapping, in contrast to other recent proposals3-6. Our results indicate rather that CENP-A marks centromere location by restructuring the nucleosome from within its folded histone core.
Several experiments have identified functionally important portions of CENP-A, while others have supported proposals of its assembly onto DNA in atypical histone arrangements. Early attention was given to the N-terminal tail of CENP-A7,8, but a major contribution of the tail to mammalian CENP-A function has been called into question since an engineered H3 chimera containing the CATD—a portion of the histone fold domain of CENP-A containing 22 amino acid deviations from conventional H32—is sufficient to rescue the lethal mitotic chromosome segregation defect caused by depletion of endogenous CENP-A in mammalian cells9. Recent proposals for atypical centromeric nucleosome arrangements have included, in budding yeast, the replacement of H2A/H2B dimers with a non-histone protein, Scm33, in fission yeast, the removal of H2A/H2B dimers from the nucleosome4, and in fruit flies, a “hemisome” containing only a single copy of each CENP-A, H2A, H2B, and H45. The hemisome proposal was recently extended to include right-handed DNA wrapping of CENP-A-containing histone complexes, opposite of conventional nucleosome wrapping6. In contrast, other studies have suggested that budding yeast, fruit fly, and human versions of CENP-A each exist in their major form in chromatin as nucleosomes containing two copies each of CENP-A, H2A, H2B, and H410-12. Even without deviating from an octameric histone arrangement, CENP-A-containing nucleosomes could potentially mark centromere location by altering the topology of the DNA (e.g. switching handedness of DNA wrapping) and/or the topography of the nucleosome (e.g. structural alterations conferred by CENP-A). Indeed, CENP-A rigidifies the histone complexes into which it assembles, both prior to and after loading onto DNA, a physical property conferred by its CATD2,13. Understanding the physical basis of CENP-A-mediated centromere function—in maintaining centromere location and providing the chromatin foundation for the mitotic kinetochore—has been hampered by a lack of crystal or NMR structures of the CENP-A protein.
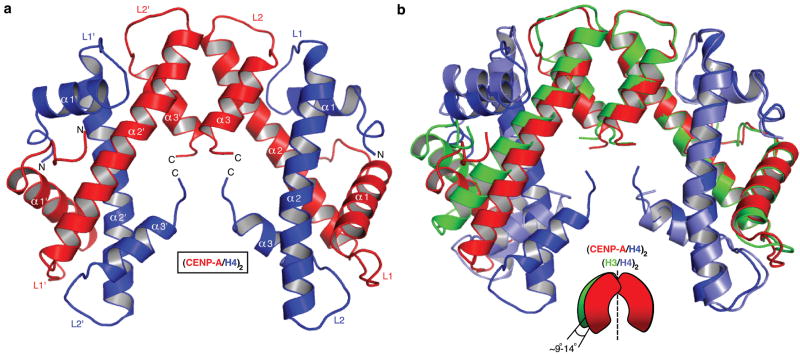
We crystallized the partially proteolyzed CENP-A/H4 complex and collected X-ray diffraction data to 2.5 Å resolution (Fig. S1). We then engineered a recombinant version with N-terminal truncations corresponding to the trypsinized version and obtained crystals that diffracted to 2.1 Å resolution (Fig. 1a, crystallographic R-factor: 0.180; Rfree: 0.241 [Supplementary Table 1]). The CENP-A/H4 dimer halves from each of the two structures overlay nearly perfectly (rmsd=0.5 Å) with a 5° difference in rotation at the CENP-A/CENP-A interface (Fig. S2). The structures show that CENP-A uses the C-terminal portion of its α2 helix and its α3 helix to form a stable tetramer, as does the counterpart (H3/H4)2 heterotetramer from within the canonical nucleosome. We use the H3 and H4 chains from within the canonical nucleosome structure14,15 in our comparisons with the (CENP-A/H4)2 heterotetramer structure, but we could have used conventional structures obtained of octamers lacking DNA or when applicable the trimeric Asf1/H3/H4 structures, since the overall helical fold and structured loops are nearly identical despite crystallization under widely divergent conditions (Fig. S3). As in the H3/H3 interface, the CENP-A/CENP-A interface is comprised of hydrophobic interactions and critical hydrogen bonding network (Fig. S4).
Figure 1. Crystal structure of the (CENP-A/H4)2 heterotetramer.
a, Ribbon diagram of the (CENP-A/H4)2 heterotetramer. The structure has clear electron density for residues 59-134 of CENP-A and 24-92 of H4. b, Overlay of (CENP-A/H4)2 with (H3/H4)2 (PDB ID 1KX515) was done with a secondary structure mapping (SSM) algorithm performed with one CENP-A molecule and one H3 molecule from each complex. CENP-A and H3 molecules on the right are overlaid. Similar results are obtained if one molecule of H4 from each complex is used for SSM (rmsd=~0.9 Å in both cases).
An overlay of one CENP-A molecule and one H3 molecule (Fig. 1b) reveals that the (CENP-A/H4)2 heterotetramer is rotated 9-14° between dimer pairs relative to the (H3/H4)2 heterotetramer in the nucleosome. This results in substantial differences in the relative position of residues within the heterotetramer (Fig. S5), including a predicted compaction of its maximal dimension (Dmax) by ~5 Å (Fig. S6). To test whether the CENP-A/CENP-A rotation is dictated by contacts within the crystal lattice, or rather differences in favored conformations in solution we performed small-angle x-ray scattering studies (SAXS) and found that (CENP-A/H4)2 (Dmax=75 Å) is indeed substantially compacted relative to (H3/H4)2 (Dmax=85 Å)(Fig. S6).
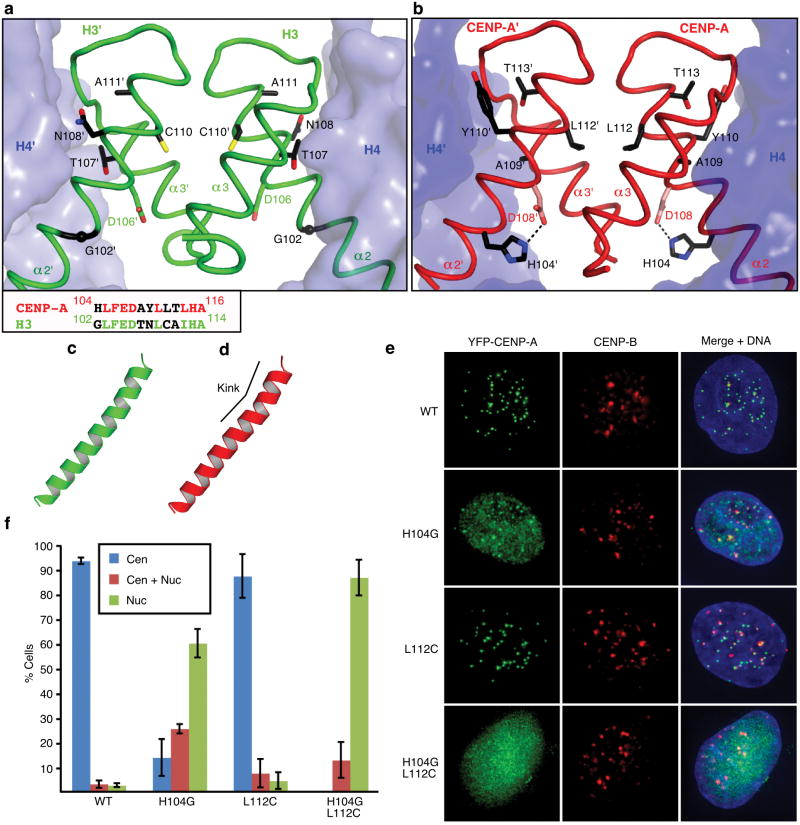
We hypothesized that the rotational divergence requires residues that are specific to CENP-A. There are total of five nonconserved residues between CENP-A and H3 in the portion of the α2 helix within the four-helix bundle that forms the CENP-A/CENP-A interface. We found two of these residues (His104 and Leu112) to introduce clear deviation relative to the H3/H3 interface (Fig. 2a,b). His104 is involved in an intramolecular interaction with Asp108, one turn up the α2 helix, that accommodates a CENP-A-specific kink in the α2 helix (Fig. 2c,d), and Leu112 extends an aliphatic side chain deep into the hydrophobic portion of the four-helix bundle (Figs. 2b and S4). The single H104G mutation causes a defect in efficient centromere targeting, but the individual L112C substitution has no effect (Fig. 2e,f). The combination (H104G, L112C), however, abolishes efficient targeting to centromeres and localizes to bulk chromatin. Thus, these two alterations that are involved in stabilization of the rotated heterotetramer provide essential contributions unique to centromere specifying nucleosomes.
Figure 2. The residues involved in the rotated CENP-A/CENP-A interface are essential for centromere targeting.
Detail of H3/H3, a, and CENP-A/CENP-A, b, interfaces with side chains of 5 non-conserved residues in α2 helix at the interface. An alignment of residues in the C-terminal portion of the α2 helix are shown in the box at lower left in panel a. c and d, the α2 helices of H3, c, and CENP-A, d. e, Centromere targeting of WT and mutant versions of CENP-A. f, Quantitation of targeting experiment. For each version of CENP-A, 3 or more experiments were conducted in which a total of >250 cells were analyzed for localization (Cen = exclusively centromeric; Cen + Nuc = both centromere and nucleoplasm staining; Nuc = nucleoplasm only [i.e. no enrichment at centromeres]). Values are plotted +/- s.d.
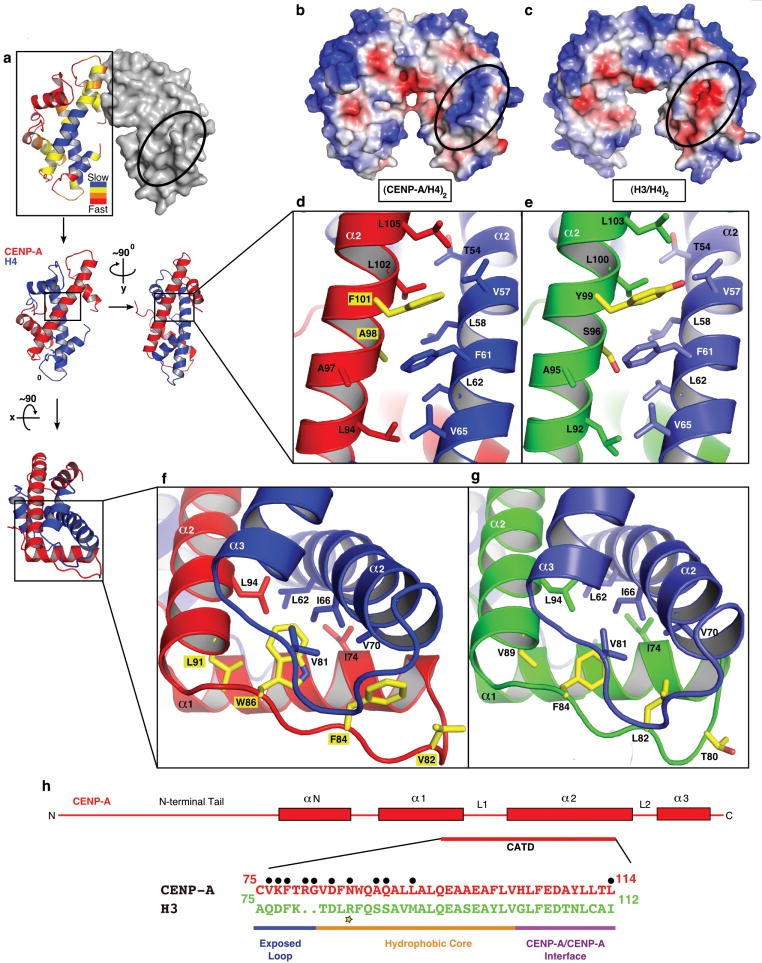
The (CENP-A/H4)2 crystal structure also reveals a bulge at the N-terminal portion of loop L1 of CENP-A that extends positions 80 (Arg) and 81 (Gly) beyond the corresponding surface in H3/H4 (Fig. S7). This part of the heterotetramer remains exposed on the surface following canonical nucleosome assembly and is the site of a prominent posttranslational modification (H3-Lys79 methylation) that regulates Sir3 binding16,17. CENP-A dramatically alters its corresponding surface with the replacement of aspartate with lysine at position 77 and the addition of Arg80 within its extended L1 bulge (Fig. S7). These changes render the CENP-A surface with charge and shape features that distinguish the complexes into which it assembles from its canonical counterparts containing H3 (Fig. 3a-c).
Figure 3. Surface and internal structural features unique to CENP-A-containing complexes.
a, The rates of H/DX remapped from the original data2 are plotted onto the crystal structure of the (CENP-A/H4)2 heterotetramer (left dimer half is shown as ribbon and right dimer half is in surface representation). b and c, Calculated electrostatic potential on the surface of (CENP-A/H4)2, b, and (H3/H4)2, c. The region corresponding to the positively charged bulge in the L1 of CENP-A is circled in each structure. d and e, Hydrophobic interactions between the α2 helices of CENP-A (red) and H4 (blue), d, and α2 helices of H3 (green) and H4 (light blue) e. f and g, Hydrophobic interactions at the helical bundle surrounding the interface of L1 of CENP-A and L2 of H4, f, and L1 of H3 and L2 of H4, g. h, Diagram of the structural differences between CENP-A and H3. Black circles indicate surface exposed residues. The yellow star highlights Arg83 from H3 that inserts into the minor groove of nucleosomal DNA14.
CENP-A-containing complexes are rigidified compared to their canonical counterparts. This was first observed in (CENP-A/H4)2 heterotetramers2 using hydrogen/deuterium exchange-mass spectrometry (H/DX-MS), where protection from exchange is consistent with local conformational inflexibility. Upon mapping the original H/DX-MS data2 onto the structure of the (CENP-A/H4)2 heterotetramer, we confirmed that the region that is ~10-fold slower to exchange (the regions in blue in Fig. 3a) than any region of conventional (H3/H4)2 heterotetramers consists of the interface of the α2 helix from CENP-A and the α2 and α3 helices of H4.
Slowing of exchange at this rigid interface could occur through tighter packing of local residues (or other tertiary structure changes) that restrict solvent accessibility. Alternatively, it could occur by inter-chain contacts in CENP-A/H4 pairs that restrict, relative to the H3/H4 pair, local unwinding of the affected helices (note that transient unfolding of secondary structure is required for exchange of amide protons18). The crystal structure of the (CENP-A/H4)2 heterotetramer strongly argues that the latter mechanism explains the rigidity of centromeric nucleosomes and sub-nucleosomal CENP-A complexes. The organization, packing, and inter-backbone distances at the corresponding interfaces of CENP-A/H4 and H3/H4 pairs are nearly identical (Fig. 3d-g). In both pairs, the interface is primarily comprised of hydrophobic interactions. On either end of the CENP-A/H4 interface, there are key substitutions in CENP-A relative to H3 that increase local hydrophobicity (Fig. 3d-g). At the contact point between the α2 helices from CENP-A and H4, two CENP-A-specific changes (Ala98 and Phe101) remove polarity (Fig. 3d,e). Likewise, on the other end of its major binding surface with H4, four CENP-A-specific changes (Val82, Phe84, Trp86, and Leu91) include several residues that add hydrophobicity and/or extend deeper into the hydrophobic bundle at the junction of L1 and the α2 helix of CENP-A and the α2 and α3 helices of H4 (Fig. 3f,g). Either end of the CENP-A/H4 interface is thus stitched together with increased hydrophobic contacts that substantially prevent the flexibility that allows regional conformational sampling of partially unfolded states in conventional H3-containing complexes. Immediately past each end of the hydrophobic stitches are the two other major distinguishing structural features conferred by CENP-A: the positively charged bulge of the extended loop L1 that is adjacent to the N-terminal end stitch and the residues responsible for the rotation of the CENP-A/CENP-A interface that is adjacent to the C-terminal end stitch (Fig. 3h).
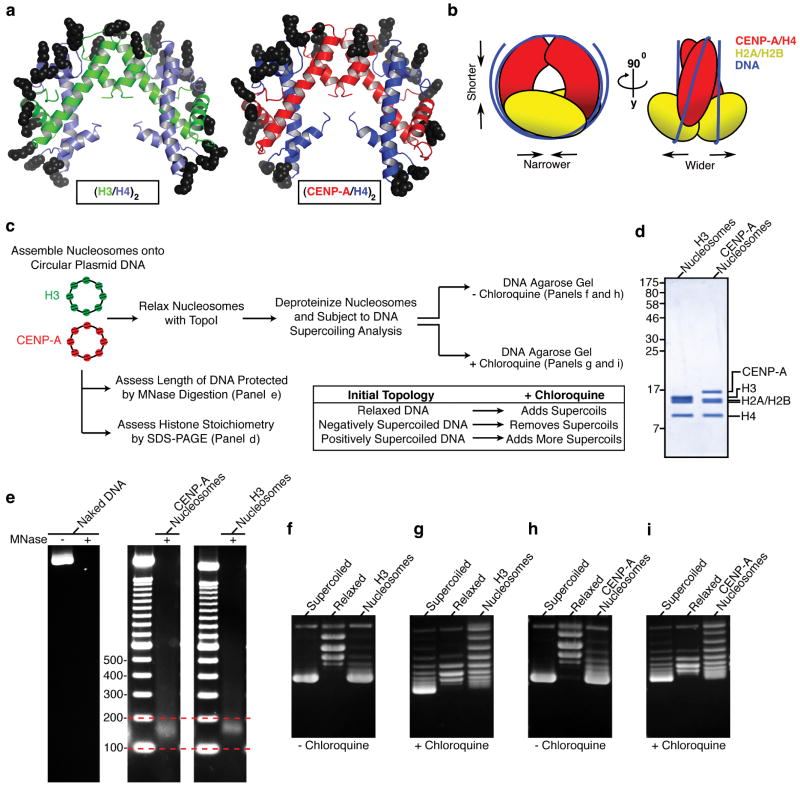
The majority of the basic residues on the DNA binding surface of the (H3/H4)2 heterotetramer are preserved in (CENP-A/H4)2 (Fig. 4a) and oriented in a manner favoring conventional left handed DNA wrapping. The compact nature of the (CENP-A/H4)2 heterotetramer (Fig. 1), if preserved upon nucleosome formation, will cause the radius of curvature of nucleosomal DNA to decrease slightly. More pronounced is the movement of the two H2A/H2B dimer-binding sites relative to each other. In conventional nucleosomes, each H2A/H2B dimer docks onto the (H3/H4)2 heterotetramer by forming a four-helix bundle between H2B and H414. Since the side-chain substitutions involved in the CENP-A/CENP-A interface are also critical for incorporation into centromeric chromatin, we favor a working model where the CENP-A/CENP-A interface maintains the rotation that is present in free heterotetramers (Fig. 4b). In this model, H2A/H2B dimers and the gyres of DNA would be positioned further from the dyad axis of the nucleosome in order to avoid steric clashes between the two H2A/H2B dimers. The requisite H2A/H2B repositioning is likely to be accomplished by a minor rotation at the H2B/H4 four-helix bundle. Thus, our working model predicts that the CENP-A nucleosome wraps DNA in a conventional left-handed manner but with alterations to its overall shape relative to canonical counterparts.
Figure 4. The (CENP-A/H4)2 heterotetramer assembles with H2A/H2B dimers into an octameric nucleosome with conventional handedness of DNA wrapping.
a, Basic residues that form contacts with nucleosomal DNA are highlighted on (H3/H4)2 (left) and the putative DNA binding surface of (CENP-A/H4)2 (right). b, Model highlighting alterations in nucleosome structure based on the structure of (CENP-A/H4)2. c, Scheme for nucleosome assembly and analysis. d, Histone content of assembled H3- and CENP-A-containing nucleosomes. e, Digestion of nucleosome arrays with micrococcal nuclease reveals that both H3- and CENP-A-containing nucleosomes protect ~150 bp of DNA. f-i, Topological analysis of H3- containing (f and g) and CENP-A-containing (h and i) nucleosomes. Analysis by gel electrophoresis in the absence (f and h) or presence (g and i) of chloroquine reveals that both H3-containing and CENP-A-containing nucleosomes wrap DNA in a left-handed manner.
To test the handedness of CENP-A-containing nucleosomes, we used well-established plasmid-based methodologies for nucleosome self-assembly and subsequent topological analysis that exploit the fact that conventional nucleosomes induce supercoils into closed, circular DNA templates19. Following standard nucleosome assembly reactions14,20, CENP-A-containing and H3-containing nucleosomes were assessed for histone content, protection from micrococcal nuclease digestion, and the extent and directionality of DNA wrapping (Fig. 4c). Nucleosomes were assembled by adding either (CENP-A/H4)2 or (H3/H4)2 heterotetramers and H2A/H2B heterodimers at a 1:2 (tetramer:dimer) molar ratio using a targeted nucleosome occupancy of 70% or 90% (calculated with a spacing of one nucleosome per 208 bp; Fig. S8). Following assembly, both H3- and CENP-A-containing nucleosomes contain equimolar amounts of each histone (Fig. 4d). Both types of nucleosomes protect ~150 bp of DNA (Fig. 4e). We noted that the band of protected DNA was slightly broader for CENP-A-containing nucleosomes than the tight band of protection in the case of H3-containing nucleosomes. This broadening is likely due to weakening of the interaction of CENP-A with the nucleosome entry/exit DNA resulting from the replacement of an arginine, that in H3 intercalates into the entry/exit DNA14, with lysine at CENP-A position 4921. For conventional nucleosomes, plasmid supercoiling indicates assembly and is assessed by increased mobility in agarose gels (one nucleosome induces ~1 supercoil22). The directionality of the supercoiling is readily determined by adding positive supercoils to the plasmids by including an intercalator (e.g. chloroquine) in the gel electrophoresis step. The intercalator reduces plasmid twist, thus compensatively increases writhe: plasmids that are originally negatively supercoiled (such as those induced by canonical nucleosome assembly; Fig. 4f) are relaxed by intercalator (Fig. 4g), while those that are positively supercoiled will accrue additional positive supercoils23. CENP-A nucleosomes induce supercoils (Fig. 4h) that are negatively supercoiled (Fig. 4i), in a manner indistinguishable from their conventional counterparts containing H3. (CENP-A/H4)2 heterotetramers can be assembled onto DNA24 and we found that they direct left-handed wrapping independently of the additional topological constraints incurred upon binding H2A/H2B dimers (Fig. S9), as do conventional (H3/H4)2 heterotetramers25.
For CENP-A to specify location of the centromere it must physically alter the chromatin into which it assembles. We have solved the structure of the sub-nucleosomal heterotetramer containing two copies each of CENP-A and H4 revealing physical features that mark centromeric chromatin. Strikingly, the three most pronounced physical changes—rotation at its tetramerization interface, extended positively charged bulge on its surface, and hydrophobic stitching to restrict conformational flexibility—are all mediated by amino acid substitutions relative to H3 that fall within the CATD (Fig. 3h), the region that has been shown to be sufficient to convert conventional H3 into a centromeric histone, in terms of its ability to track to centromeres and maintain centromere identity and function upon incorporation into centromeric nucleosomes2,9,26. Neither the L1 nor α2 helix alone is sufficient to convert H3 into a functional centromeric histone9,11,27, but rather act together to form a functional CATD. Thus, there is likely cooperation between the three distinct structural features encoded in this portion of CENP-A. Since the CATD mediates molecular recognition events prior to and after nucleosome assembly28,29 our physical studies contribute to current models of CENP-A directed self-propagation as a key component of the mechanism in the epigenetic inheritance of centromeres. Furthermore, our findings with purified components provide strong support to the notion that the octameric form of CENP-A-containing nucleosomes is a widely conserved feature of eukaryotic centromere identity and function. CENP-A alters the chromatin into which it assembles, not by switching handedness of DNA wrapping or disrupting the octameric nature of the nucleosome, but by altering nucleosome structure from within the folded histone core.
Methods Summary
The CENP-A/H4 complex was crystallized using the hanging-drop vapor diffusion method and hexagonal crystals of trypsinized and engineered N-terminal truncated complexes appeared in 5-7 days. Crystallographic data were collected at Argonne National Laboratory and Advanced Light Source and one chain of H3 and one chain of H4 from the crystal structure of nucleosome (PDB ID 1KX515) were used as a search model for molecular replacement. Initial models were completed through several cycles of manual model rebuilding and refinement. Nucleosome assembly was performed using a standard approach for generating conventional nucleosome arrays20. We used pUC19 plasmid DNA as the template and calculated maximum nucleosome occupancy based on one nucleosome occupying 208 base pairs of DNA. Histone content of nucleosomes was analyzed by SDS-PAGE and coomassie blue staining. Supercoiling-based topological analysis was performed and the products were run on a 0.8% agarose gel and post-stained with ethidium bromide. Partial micrococcal nuclease digestion was performed and the products analyzed on a 1.5% agarose gel and post-stained with ethidium bromide. HeLa cells were cultured in DMEM containing 10% newborn calf serum, transfected with the indicated plasmids using Effectene (Qiagen, Valencia, CA), and processed for immunofluorescence two days after transfection as described elsewhere2. YFP-CENP-A constructs were described elsewhere9. Mutations were introduced using the Quickchange system (Stratagene, La Jolla, CA) and the plasmids were sequenced. For each sample, images were collected at 0.2 μm z-sections that were subsequently deconvolved using identical parameters. The z-stacks were then projected as single two-dimensional images.
Methods
Protein Purification and Partial Proteolysis
Recombinant human versions of all sub-nucleosomal histone complexes were produced as described2,28 except that the 6-X-His tag on H2A was cleaved by Precission protease (GE Healthcare, Piscataway, NJ) prior to dimer reconstitution with untagged H2B. For partial trypsinization experiments, the indicated complexes (1 mg/ml each) were incubated with sequence grade trypsin (Roche) at the indicated concentration for 4, 6, or 18 hr, in buffer containing 20 mM sodium phosphate (pH 6.9), 2 M NaCl, and 5 mM β-mercaptoethanol. The products were analyzed by SDS-PAGE and coomassie blue staining.
Protein Crystallization and Structure Determination
For crystals obtained through partial proteolysis, the CENP-A/H4 complex (7 mg/ml) in 20 mM phosphate buffer (pH 6.9) containing 500 mM NaCl was incubated with 1 pg/ml trypsin at 4°C for four days. Trypsinization was stopped by addition of equimolar phenylmethanesulphonyl fluoride. For crystallization, the protein solution (3.5 mg/ml) was mixed with the reservoir solution (2 M ammonium sulfate, 0.1 M HEPES [pH 7.9]) at a 3:1 ratio (3+1 μL) and the crystallization experiment was performed using the hanging-drop vapor diffusion method at 22°C. Hexagonal crystals appeared after one week and reached maximal dimensions of 80 × 80 × 100 μm. A crystal was cryoprotected in four steps each including a 10 s incubation in mother liquor supplemented with 5%, 10%, 15% and 20% ethylene glycol followed by freezing in liquid N2. The crystals were generally very sensitive to manipulation and exhibited severe decay due to radiation damage (high Rsym). Data were collected at GM/CA-CAT beamline 23-ID-D at APS.
Crystals of higher resolution were obtained using an engineered version, lacking the first 59 N-terminal residues of CENP-A and the first 19 N-terminal residues of H4 that was purified as described for the full length proteins. A protein solution (3.3 mg/ml) was mixed 1:1 with reservoir solution (0.17 M ammonium sulfate, 0.085 M sodium cacodylate [pH 6.5], 25.5% PEG-8000, 15% glycerol) in hanging drops and incubated at 22°C. Hexagonal crystals appeared after 5 days. Crystals were deep frozen in liquid nitrogen without further treatment. Data were collected at 8.2.2 beamline at ALS.
All data processing was done with the XDS program package30. One chain of H3 and one chain of H4 from the crystal structure of nucleosome (PDB ID 1KX515) (or the structure of trypsinized (CENP-A/H4)2 in the case of the engineered truncated version) were used as a search model for molecular replacement with PHASER31. The initial model was completed through several cycles of manual model rebuilding in COOT32 and refinement in Refmac533 for structure obtained with partially proteolyzed proteins, while high-resolution structure obtained with engineered proteins was subjected to automatic model rebuilding using program ARP/wARP34. In addition to protein and water moieties, sulfate or phosphate molecules were also modeled and are bound at the sites where protein-DNA contacts are made in the corresponding positions in canonical nucleosomes14. The structural figures were all prepared in Pymol (www.pymol.org).
SAXS
SAXS data were collected at APS, beamline BIO-CAT ID-18 on protein samples in the range of 1-3 mg/ml in buffer containing 20 mM sodium phosphate (pH 6.9), 1 M NaCl, 1 mM EDTA, 8 mM DTT, and 2.4 % glycerol. Engineered tailless versions of proteins, lacking the first 59 N-terminal residues (CENP-A) or the first 60 N-terminal residues (H3) and the first 19 N-terminal residues of H4, were used in SAXS experiments to investigate the solution behavior of the histone fold domains. 3-10 1 s exposures were captured for each complex and used to generate averaged scattering profiles. Initial analysis and data reduction were performed in the program PRIMUS35. P(r) functions and the Dmax values were calculated with the program GNOM36. Theoretical intensities for high-resolution models were obtained using CRYSOL37 with PDB files containing only protein atoms. Dmax values were determined empirically by using different Dmax in increments of 5 Å and choosing values where P(r) most closely approaches zero while giving plausible curves. SASREF (ATSAS program package) was used to model CENP-A/H4 and H3/H4 dimer relationships within each tetramer that most closely match the SAXS data. Calculations were performed for each heterotetramer, using CENP-A/H4 or H3/H4 dimers in P2 symmetry. Two restraints were imposed at the CENP-A/CENP-A or H3/H3 interface (e.g. preserving 8 Å distance between backbone atoms of residues at the dimer-dimer interface [Leu112/Leu112’ and His115/His115’ in CENP-A/CENP-A’ or Cys110/Cys110’ and His113/His113’ in H3/H3’). Similar results were obtained with a single restraint. All SASREF models obtained with SAXS data for (CENP-A/H4)2 are similar to each other as well as to its corresponding crystal structures. SASREF models obtained for (H3/H4)2 SAXS data occasionally resulted in extreme rotation of the dimerization interface in a manner that precludes formation of the 4-helix bundle, and these models were discarded. For each tetramer, 10 of the high-resolution SASREF models were converted to bead models using SITUS38 and averaged using DAMAVER39. The resulting envelopes were displayed in UCSF Chimera40. Docking of ribbon diagrams into the averaged envelopes was performed by SUPCOMB (ATSAS program package).
Nucleosome Assembly
Nucleosome assembly was performed using a standard approach for generating conventional nucleosome arrays20. We used pUC19 plasmid DNA (purified by two successive CsCl gradient steps) as the template. The maximum nucleosome occupancy of pUC19 was calculated based on one nucleosome occupying 208 base pairs of DNA. The molar quantities of histones required for 70% and 90% nucleosome occupancy of pUC19 were calculated for both CENP-A-containing nucleosomes (assuming one [CENP-A/H4]2 tetramer and two [H2A/H2B] dimers) and H3-containing nucleosomes (one [H3/H4]2 tetramer and two [H2A/H2B] dimers). Histones and pUC19 were mixed in TE buffer containing 2 M NaCl, followed by stepwise dialysis in TE buffer to lower ionic strength (1 M NaCl, 0.75 M NaCl, and finally 2.5 mM NaCl). Histone content of nucleosomes was analyzed by SDS-PAGE and coomassie blue staining.
DNA Topology Assays
Purification of topoisomerase I ND423 and supercoiling-based topological analysis were performed as previously described41. Briefly, 5μg of either naked pUC19 DNA, CENP-A-containing nucleosomes (90% occupancy), or H3-containing nucleosomes (90% occupancy) were relaxed with topoisomerase I in a buffer comprised of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 0.1 mM EDTA, 50 mg/ml BSA, and 0.5 mM DTT for 30 min at 30°C. Reactions were terminated with addition of a stoppage buffer comprised of 20 mM EDTA (pH 8), 0.2 M NaCl, 1% (w/v) SDS, and 0.25 mg/ml glycogen. DNA was then deproteinized with 50 mg/ml proteinase K for 30 min at 37°C, and purified by phenol/chloroform extraction. The products were run on a 0.8% agarose gel and post-stained with ethidium bromide.
Micrococcal Nuclease Protection
Partial micrococcal nuclease digestion was performed as previously described41. Briefly, 5 μg of either naked pUC19 DNA, CENP-A-containing nucleosomes (90% occupancy), or H3-containing nucleosomes (90% occupancy) were digested with 7.5 units of micrococcal nuclease (Roche) in a buffer comprised of 0.75 mM HEPES (pH 7.6), 10 mg/ml PEG 8000, 50 mM KCl, 10 mM CaCl2, and 0.5% glycerol for 10 min at 37°C. Reactions were terminated with addition of stoppage buffer and prepared as described above, and then analyzed on a 1.5% agarose gel and post-stained with ethidium bromide.
Centromere localization
HeLa cells were cultured in DMEM containing 10% newborn calf serum, transfected with the indicated plasmids using Effectene (Qiagen, Valencia, CA), and processed for immunofluorescence two days after transfection as described elsewhere2. YFP-CENP-A constructs were described elsewhere9. Mutations were introduced using the Quickchange system (Stratagene, La Jolla, CA) and the plasmids were sequenced to verify the coding sequence of mutant versions of CENP-A. Digital images were captured using Leica LAF software by a Hamamatsu Orca AG charge-coupled device camera mounted on a Leica DMI6000B inverted microscope. For each sample, images were collected at 0.2 μm z-sections that were subsequently deconvolved using identical parameters. The z-stacks were then projected as single two-dimensional images and assembled using Adobe Photoshop (version 9) and Illustrator (version 12).
Supplementary Material
Acknowledgments
We thank D. Cleveland for plasmids and steadfast encouragement to pursue a physical understanding of the centromere, S. Wood for generating cleaved histone H2A, K. Gupta for help with collecting data and technical suggestions, K. Ferguson, G. Van Duyne, M. Lemmon, J. Shorter, L. Jansen, D. Foltz, J. Shah, D. Alvarado, K. Moravcevic, and T. Panchenko for helpful discussions and comments on the manuscript. This work was supported by the NIH research grant GM82989, a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, and a Rita Allen Foundation Scholar Award to B.E.B. N.S. is supported by a postdoctoral fellowship from the American Cancer Society and E.A.B. has been supported by the Penn Structural Biology Training Grant (NIH GM08275) and a predoctoral fellowship from the American Heart Association.
Footnotes
Author Contributions N.S. designed and performed experiments, solved and refined the structures, analyzed data, and wrote the manuscript, E.A.B. and D.J.R. performed experiments and analyzed data, and B.E.B. directed the project, designed and performed experiments, analyzed data, and wrote the manuscript.
References
- 1.Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 3.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 4.Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith MM. Centromeres and variant histones: what, where, when and why? Curr Opin Cell Biol. 2002;14:279–285. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- 8.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 9.Black BE, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 10.Erhardt S, et al. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shelby RD, Vafa O, Sullivan KF. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J Cell Biol. 1997;136:501–513. doi: 10.1083/jcb.136.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Black BE, Brock MA, Bédard S, Woods VL, Cleveland DW. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc Natl Acad Sci U S A. 2007;104:5008–5013. doi: 10.1073/pnas.0700390104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 15.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 16.Altaf M, et al. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X, et al. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat Struct Mol Biol. 2008;15:1122–1124. doi: 10.1038/nsmb.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Englander SW. Hydrogen exchange and mass spectrometry: A historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lusser A, Kadonaga JT. Strategies for the reconstitution of chromatin. Nat Methods. 2004;1:19–26. doi: 10.1038/nmeth709. [DOI] [PubMed] [Google Scholar]
- 20.Carruthers LM, Tse C, Walker KP, Hansen JC. Assembly of defined nucleosomal and chromatin arrays from pure components. Meth Enzymol. 1999;304:19–35. doi: 10.1016/s0076-6879(99)04004-5. [DOI] [PubMed] [Google Scholar]
- 21.Conde e Silva N, et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 22.Simpson RT, Thoma F, Brubaker JM. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 23.Esposito F, Sinden RR. Supercoiling in prokaryotic and eukaryotic DNA: changes in response to topological perturbation of plasmids in E. coli and SV40 in vitro, in nuclei and in CV-1 cells. Nucleic Acids Res. 1987;15:5105–5124. doi: 10.1093/nar/15.13.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci U S A. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bina-Stein M, Simpson RT. Specific folding and contraction of DNA by histones H3 and H4. Cell. 1977;11:609–618. doi: 10.1016/0092-8674(77)90078-2. [DOI] [PubMed] [Google Scholar]
- 26.Bassett EA, et al. Epigenetic centromere specification directs Aurora B accumulation but is insufficient to efficiently correct mitotic errors. J Cell Biol. 2010 Jul 26;190(2):177–85. doi: 10.1083/jcb.201001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vermaak D, Hayden HS, Henikoff S. Centromere Targeting Element within the Histone Fold Domain of Cid. Mol Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foltz DR, et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carroll CW, Silva MCC, Godek KM, Jansen LET, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 31.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Dodson EJ, Winn M, Ralph A. Collaborative Computational Project, number 4: providing programs for protein crystallography. Meth Enzymol. 1997;277:620–633. doi: 10.1016/s0076-6879(97)77034-4. [DOI] [PubMed] [Google Scholar]
- 34.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konarev PV, Volkov VV, Sokolova AV, Koch MH, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 36.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 37.Svergun D, Barberato C, Koch MHJ. CRYSOL – a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 38.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF, et al. UCSF Chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Fyodorov DV, Kadonaga JT. Chromatin assembly in vitro with purified recombinant ACF and NAP-1. Meth Enzymol. 2003;371:499–515. doi: 10.1016/S0076-6879(03)71037-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.