Abstract
To better understand the interactions between catalysts and transition states during RNA strand cleavage, primary 18O kinetic isotope effects and solvent D2O isotope effects were measured to probe the mechanism of base-catalyzed 2'-O-transphosphorylation of the RNA dinucleotide 5'-UpG-3'. The observed 18O KIEs for the nucleophilic 2'-O and in the 5'-O leaving group at pH 14 are both large relative to reactions of phosphodiesters with good leaving groups, indicating that the reaction catalyzed by hydroxide has a transition state (TS) with advanced phosphorus-oxygen bond fission to the leaving group (18kLG = 1.034 ± 0.004) and phosphorous-nucleophile bond formation (18kNUC = 0.984 ± 0.004). A breakpoint in the pH dependence of the 2'-O-transphosphorylation rate to a pH independent phase above pH 13 has been attributed to the pKa of the 2'-OH nucleophile. A smaller nucleophile KIE is observed at pH 12 (18kNUC = 0.995 ± 0.004) that is interpreted as the combined effect of the equilibrium isotope effect (~1.02) on deprotonation of the 2′-hydroxyl nucleophile and the intrinsic KIE on the nucleophilic addition step (ca. 0.981). An alternative mechanism in which the hydroxide ion acts as a general base is considered unlikely given the lack of a solvent deuterium isotope effect above the breakpoint in the pH versus rate profile. These results represent the first direct analysis of the transition state for RNA strand cleavage. The primary 18O KIE results and the lack of a kinetic solvent deuterium isotope effect together provide strong evidence for a late transition state and 2'-O nucleophile activation by specific base catalysis.
Keywords: RNA, kinetic isotope effects, transphosphorylation, specific base, mass spectrometry, transition state
INTRODUCTION
Cleavage of RNA strands by nucleophilic attack of a ribose 2'-hydroxyl on the adjacent 3',5' phosphodiester to yield cyclic 2',3'-phosphate and 5'-hydroxyl products is catalyzed by numerous essential enzymes in biology with rate enhancements of up to 1010-fold1; 2. Extensive analyses of intra-and intermolecular phosphoryl transfer reactions reveal a complex energetic landscape for the reaction3; 4; 5; 6; 7; 8; 9; 10; 11; 12; 13. The overall addition-displacement mechanism can be concerted ANDN, or stepwise with formation of a phosphorane intermediate, depending on interactions with solution catalysts including acids, bases, and metal ions and their complexes4; 14; 15; 16; 17. RNA 2'-O-transphosphorylation necessarily involves protonation and deprotonation steps; however, it is exceedingly difficult to gain experimental insight into the trajectory and timing of these proton transfer steps, that is, whether they occur before, during or after transfer of the phosphoryl group. Understanding these details of mechanism is important because the 2'-O-transphosphorylation of RNA is catalyzed by both acid and base in solution5; 18; 19; 20; 21; 22; 23, and potential Bronsted acid and base functional groups are key components of phosphoryl transfer enzyme active sites24; 25; 26; 27; 28; 29. Defining the influence of interactions with acid and base catalysts on the reaction mechanisms in solution therefore provides an essential framework for understanding how enzymes achieve their characteristic attribute of tremendous rate enhancement.
In solution, the 2'-O-transphosphorylation reaction at pH 0–5 is believed to occur via a phosphorane intermediate, in which one or more of the non-bridging oxygens is protonated20; 23; 30. The pH vs. log rate profile displays an apparent slope of −2 between pH 1 and pH 2 and a slope of −1 between pH 2 and pH 55; 20; 31. Protonation of both the leaving group (pKa < 0) and one of the non-bridging oxygens (pKa ca. 2) is believed to account for the dependence of the rate constant on proton concentration over the acidic range. Under these conditions the phosphorane intermediate is sufficiently stable to undergo pseudorotation, resulting in isomerization to form the 2',5'-phosphodiester3; 9; 13; 20; 23; 32; 33. At pH 5–7 the cleavage reaction becomes pH independent, which is accounted for by the continued decrease in acid catalysis that is offset by increasing catalysis by hydroxide. Experimental and theoretical data suggest that under these conditions a dianionic oxyphosphorane transphosphorylation intermediate may form that is kinetically indistinguishable from a concerted displacement transition state and is too short-lived to undergo other processes such as protonation or pseudorotation5; 9; 23; 34; 35; 36; 37. Inversion of configuration of a phosphorothioate at the reactive phosphoryl supports a concerted mechanism under these conditions30. Above pH 13 the reaction becomes pH independent, with an apparent pKa of 13.5 presumed to reflect deprotonation of the 2'O nucleophile5; 19; 20; 21; 38; 39.
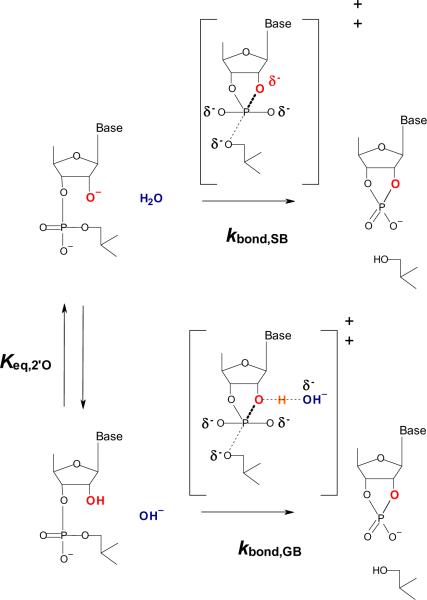
Pre-equilibrium deprotonation of the 2'-O nucleophile is considered to be the likely basis for the first order dependence of the cleavage rate constant on hydroxide ion and the loss of this dependence above pH 13.5 (Figure 1). In this mechanism, the rate constant for the reaction below the pKa increases with hydroxide ion concentration due to an increased fractional population of the reactive 2'-oxyanion nucleophile while above the pKa (indicated by Keq) the rate constant becomes independent of hydroxide concentration due to complete titration of the nucleophile, and the intrinsic rate constant for reaction from the 2'-oxyanion ground state is observed (kbond). An alternative model consistent with the data is that hydroxide ion acts as a general base, and that the proton bonded to the 2'-O position is transferred to hydroxide ion in the transition state. In this mechanism the rate constant for the reaction increases due to the increase in the concentration of the hydroxide ion catalyst, but above the breakpoint in the pH versus rate profile the increase in catalysis occurs because the effect of the increasing hydroxide ion concentration is offset by the corresponding decrease in the concentration of the reactive 2'-hydroxyl species. Direct experimental tests of mechanism at this level of detail are difficult, and ambiguity remains with respect to the identity of titrating groups giving rise to pH dependence and the charge distribution in the rate limiting transition state or states.
Figure 1.
General and specific base mechanisms of hydroxide ion catalyzed 2'-O-transphosphorylation. The conventional mechanism in which equilibrium deprotonation of the 2'-OH (Keq) occurs prior to nucleophilic attack and reaction with a rate constant governed by kbond,SB is shown as the upper pathway. An alternative mechanism in which reaction occurs from the 2'-OH ground state (kbond,GB) where a decrease in rate constant due to deprotonation of the nucleophile governed by Keq is offset by increasing hydroxide ion catalyst concentration is shown in the lower pathway.
A more complete understanding of interplay between protonation and mechanism can be gained by analysis of the nucleophile and leaving group 18O kinetic isotope effects for RNA transphosphorylation and isomerization versus pH. Isotopic substitution of atoms undergoing chemical reactions affects reaction rates and equilibria8; 9; 40; 41; 42; 43. These kinetic and equilibrium isotope effects arise because heavier isotopes have lower zero point vibrational energies than their lighter counterparts. Differences in bond stiffness between the ground state and transition state result in differences in activation energy for the two different isotopologues and consequently differences in rate constant measured as kinetic isotope effects (KIEs, expressed as k16/k18). Analogously, these differences can alter the equilibrium distribution of heavy and light isotopes in reactants and products giving rise to equilibrium isotope effects (EIEs, expressed as K16/K18). For example, deprotonation of an alcohol such as the 2'-OH results in a less stiff bonding environment for the oxygen atom, which favors the lighter isotope, resulting in its enrichment in the oxyanion population44; 45. EIEs for deprotonation are therefore >1 and are referred to as “normal”. Similarly, KIEs on bond breakage as in the departure of an oxyanion leaving group are also normal. Increases in bond stiffness during the formation of the 2'O-P bond will tend to favor the heavier isotope and can result in KIEs that are <1, which are referred to as “inverse”.
A great advantage in interpreting KIEs for reactions involving RNA is the extensive experimental and conceptual framework derived from analyses of the KIEs of phosphodiester reactions9; 10; 11; 14; 16; 46; 47; 48; 49. Extensive studies of nucleophile, leaving group and non-bridging oxygen KIEs by Cleland and by Hengge demonstrate that these effects fall in the range of 0.93 – 1.07 (k16/k18) and provide important information on extents of bond formation and cleavage6; 11. Additionally, EIEs from ionizable protons influence the observed KIEs in a pH dependent manner; for example, the KIE for the non-bridging oxygen is different in the acid-catalyzed reaction versus the base-catalyzed reaction due in part to protonation10; 11. Therefore, the KIEs monitored for RNA reactions in solution should not only provide important new information on mechanism but also insight into proton transfer, and thus provide benchmarks for analysis of enzyme mechanism. Here, we report the first use of isotopically enriched [2'-18O] and [5'-18O] RNA dinucleotides to determine the nucleophile (18kNUC) and leaving group (18kLG) KIEs for specific base catalyzed 2'-O-transphosphorylation. The large magnitude for the nucleophile and leaving group KIEs are consistent with a late transition state with advanced 2'O-P bond formation and 5'O-P bond cleavage. A lower nucleophile KIE observed below the breakpoint in the pH dependence of the rate constant and the lack of a solvent deuterium isotope effect above the breakpoint provides evidence for nucleophilic activation by specific base catalysis.
EXPERIMENTAL SECTION
KIEs may be determined by analyzing as a function of reaction progress the change in the 16O/18O ratio of an RNA substrate of defined sequence that has been site-specifically enriched with 18O at either the 2'-O or 5'-O48. The larger rate constant for either the 18O- or 16O-containing RNA results in the progressive depletion of that isotopologue in the residual substrate population. Accomplishing this goal presents three key challenges: i. synthesis of populations of RNA enriched in 18O at the 2'-O or the 5'-O positions; ii. accurate and precise determination of reaction progress; and iii. precise determination of the 16O/18O ratio in the residual substrate molecules. The first challenge of obtaining 18O enriched RNA has been overcome by using [18O2]benzoic acid as a nucleophile to deliver the isotopic substitution to activated nucleoside ribose 2' or 5' carbons followed by hydrolysis of the benzoate ester50. The second requirement to precisely determine the fraction of reaction is accomplished by analytical HPLC. The third challenge of determining the isotope ratio in the substrate population is met by analyzing the 16O/18O ratio in UpG by tandem ESI-Q/TOF MS. These methods are described in the following sections.
Synthesis of [2'-18O] and [5'-18O] RNA
The 18O enriched RNA was synthesized using [18O2]benzoic acid as a nucleophile to deliver the isotopic substitution to activated nucleoside ribose 2' or 5' carbons followed by hydrolysis of the benzoate ester. Synthesis of [2'-18O] UpG is described in detail in Dai et al., 2008)50. To synthesize Up-[5'-18O]G the 2' and 3' hydroxyls of commercially available 5'-DMT-N-Bu-i-guanosine were protected with acetyl groups, and the 5'-DMT protecting group was subsequently removed by acid treatment (Scheme S1, Supplementary Information). The 5'-O was activated as the mesylate ester, which was subsequently treated with [2×18O2] benzoic acid to yield enrichment of the heavy isotopolog at the 5'O position. The hydroxyl protecting groups were removed to give the [5'-18O]-nucleoside. The [5'-18O]-nucleoside was coupled to commercially available uridine phosphoramidite to yield the RNA dinucleotide Up- [5'-18O]G.
RNA reaction kinetics
The 2'-O-transphosphorylation of 1–5 nmoles of UpG at pH 11–14 was carried out in 250–500 μL at 37°C at an ionic strength of 1 M adjusted to the desired reaction conditions using NaOH and NaCl giving a final RNA concentration of 2 –20 μM. Because the reaction does not generate additional protons, the only source comes from associated ammonium counter ions and the deprotonation of the guanosine and uridine nucleobases. Thus, the presence of low concentrations of RNA decreased the concentration of hydroxide by less than 10%. For the KIE determinations at pH 12 and 14 that contained 10 μM RNA, the presence of reactant decreased the concentration of hydroxide by 0.3% and 0.003%, respectively. At appropriate times after mixing substrate and base, aliquots were taken and neutralized with an equivalent of HCl followed by further dilution with 100–200 μL 0.1 M ammonium acetate pH 8.0. The reactant (UpG) and products (cUMP and G) were resolved by RP-HPLC on a 300 × 50 mm C18 column run isocratically at 1 mL/min of 0.1 M ammonium acetate and 4% acetonitrile. The fraction of reaction (F = [cUMP or G] / ([UpG] + [cUMP or G]) was quantified by integration of the chromatogram and the observed first order rate constant (kobs) determined by non linear least squares fitting (Origin). The values of kobs as a function of pH 10 – 14 in Figure 2C were fit to the rate equation for the base catalysis mechanisms shown in Figure 1 where,
| equation 1 |
which yielded an apparent pKeq of 13.45 ± 0.03 and a maximal kbond of 0.06 s−1. For KIE determinations the appropriate 18O-substituted substrate (either the 2', 5' or non-bridging oxygen) was introduced to an enrichment of ca. 50% and reacted as described above. The HPLC fractions containing the unreacted UpG were collected and dried under vacuum. The pellets were resuspended in 1 ml H2O, dried down 3–4 additional times, and finally resuspended in water at a concentration of 20–50 μM for subsequent isotopic analysis by MS.
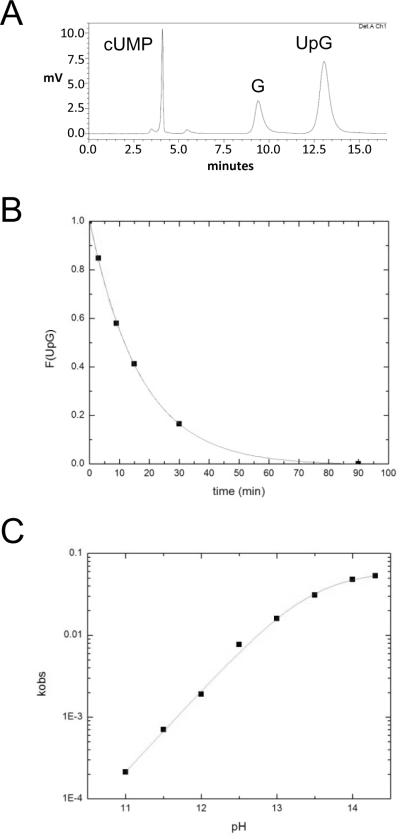
Figure 2.
Determination of reaction progress, rate constants and pH dependence of the 2'-O-transphosphorylation reaction of 3'-UpG-5'. A. Separation of the reactant (UpG) from the products (cUMP and G) by RP-HPLC. Reaction mixtures were injected onto a 300 × 50 mm C18 column and eluted isocratically at 1 ml/min of 0.1 M ammonium acetate and 4% acetonitrile. The chromatogram shows absorbance at 260 nm. B. A characteristic time course used to determine reaction progress and the rate constants for the reaction. The reaction progress as determined by integration of the chromatogram is plotted as a function of time after addition of base. C. A plot of kobs determined using RP-HPLC as depicted in panels A and B versus pH. The line is a fit of the data to the mechanism shown in Scheme 1 and gives an apparent pKa of 13.45 ± 0.03.
Quantitative determination of 16O/18O ratios in UpG by electrospray ionization quadrupole / time of flight tandem mass spectrometry (ESI-Q/TOF)
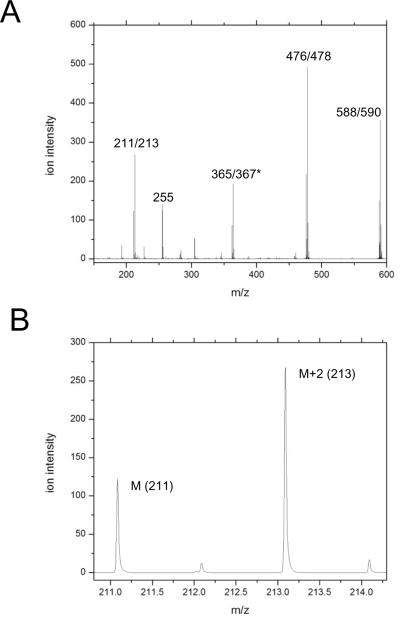
Determination of the substrate 16O/18O ratio in UpG isolated by RP-HPLC was accomplished by tandem MS using an ABI QStar electrospray ionization quadrupole time-of-flight tandem mass spectrometer in negative ion mode. The sample was injected at 1−0.5 μL/min using an ion voltage of −2200. The entire deprotonated molecular ion isotopic cluster of the substrate with monoisotopic m/z 588 was isolated using low resolution in the initial quadrupole stage and fragmented by inert gas collision at a collision energy of 35. In the second stage and the resulting fragments at 100–600 m/z were resolved by TOF MS. To be described in more detail in Anderson et al., in prep, we observe that fragmentation of the monoisotopic ion yields high intensity product ions at m/z 476 and 211 that result from loss of the uridine nucleobase, and from ribosephosphate, respectively as shown in Figure 3A. Both species contain the enriched 2'-O and 5'-O and were sufficiently abundant for precise quantitation of 16O/18O ratios in the residual substrate population with 10–15 minutes of continuous data acquisition. The second TOF-MS dimension produces highly enhanced signal to noise as demonstrated in Figure 3 for the 211/213 ion pair. The absence of background noise and the exclusion of contaminants due to the high mass resolution of the TOF result in a precision of 0.5% or better as assessed by analytical dilution of isotopic standards. Fitting the m/z data to a series of Gaussian peaks was used to quantify the abundance of the two isotopologues and determine the 16O/18O ratio51, alternatively with complete baseline separation of the isotopic peaks and the absence of apparent contaminants the Analyst integration routine produced equivalent results.
Figure 3.
Determination of 16O/18O ratios in the residual UpG reactant by whole molecule mass spectrometry. A. ESI-Q/TOF tandem mass spectrum of [2'-18O]UpG. The TOF spectrum from fragmentation of the deprotonted molecular m/z 589 UpG ion. Ions at m/z 476/478 and m/z 211/213 that are used for quantitation of the 16O/18O ratio are indicated. B. The resolution of the M and M+2 isotopologues for the m/z 211/213 ion.
Determination of kinetic isotope effects by fitting isotopologue ratio versus reaction progress data
The KIEs reported in Table 1 were calculated from plots of ln(18O/16O) versus F by fitting to:
| equation 2 |
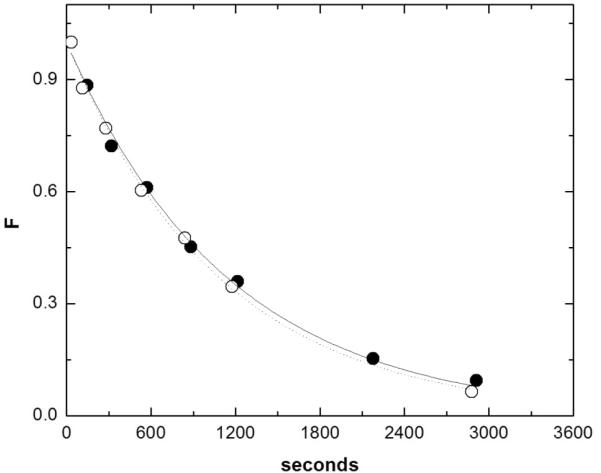
where 18k is the isotope effect, F is the fraction of substrate consumed as determined from integration of the HPLC chromatogram, and I is the initial 18O/16O ratio in the unreacted substrate52. Each experiment was performed in duplicate and the intensity data for both the 211/213 and 476/478 ions were analyzed, yielding four determinations of the KIE for each experiment. The KIEs and errors reported in Table 1 are averages and standard deviations of six to ten different determinations coming from at least three independent experiments. The solvent D2O effect on 2'-O-transphosphorylation of 5'-UpG-3' was determined at pH 14. The kD was measured by dissolving NaOH in 99.9% D2O or H2O to a concentration of 1M hydroxide (deuteroxide). 500 μL of either solution was used to resuspend 1 nmole of UpG and thus the contribution of adventitious protons to the D2O reaction was exceedingly small. Kinetic analyses of reactions run in H2O (filled symbols) or D2O (open symbols) were run in parallel. Comparison of the rate constants yields a solvent D2O effect (kH/kD) of 0.95.
Table 1.
KIE Results for Intramolecular Transphosphorylation Reactionsa
| 18kLG | 18kNUC | 18kNUC(corr)b | Reference | |
|---|---|---|---|---|
| UpG (pH 14) | 1.034 ± 0.004 | 0.984 ± 0.003 | 0.981 | |
| UpG (pH 12) | 1.037 ± 0.002 | 0.996 ± 0.002 | 0.981 | |
| uridine-3'-m-nitrobenzyl phosphate | 1.0272 | NDc | 66 | |
| uridine-3'-p-nitrophenol phosphate | 1.0059 | ND | 46 | |
| hydroxypropyl-p-nitrophenol phosphate | 1.0064 | 1.0327 | 1.0079 | 1 |
Standard errors are shown for KIEs determined in this study. The observed nucleophile KIEs (18knuc) reflect contributions from the effect for deprotonation and the effect for nucleophilic attack, as discussed in the text.
The intrinsic nucleophile KIEs were obtained by correcting the observed values for the calculated EIE for deprotonation of hydroxypropyl-p-nitrophenol phosphate.
ND not determined
RESULTS
The precise determination of reaction progress by separation of the reactants and products by RP-HPLC and integration of the chromatogram is illustrated in Figure 2A & B. The first order rate constants for reaction at different pHs were determined under constant ionic strength (I = 1 M). The dependence of the 2'-O-transphosphorylation reaction rate constant on hydroxide ion concentration (pH) under the conditions used for isotope effect measurements is shown in Figure 2C. As observed previously, the log of the rate constant increases linearly with pH between ca. 11 – 13, and at pH values greater than 13 the rate constant becomes independent of pH5; 53. Fitting the data to the rate equation for the reaction in Figure 1 (equation 1) gives a pKa of 13.45 ± 0.03, which is close to the value of 13.5 for hydrolysis of an RNA linkage within a DNA oligonucleotide determined under similar temperature and ionic strength conditions19.
Determination of the substrate 16O/18O ratio in the residual unreacted UpG population was accomplished by tandem Q/TOF MS of negative ions generated by direct infusion using an electrospray source (Anderson et al., in prep). The UpG dinucleotide gives rise to the expected m/z 588 unprotonated molecular ion (M) (Figure 3A). The UpG ion population was isolated by quadrupole MS and subsequently fragmented by inert gas collision. The resulting fragments were resolved by a second dimension of time of flight mass spectrometry. Fragmentation yields high intensity product ions at m/z of 476, 365, 255 and 211. The m/z 255 product ion is a contaminant, presumably palmitate, also present in background samples.
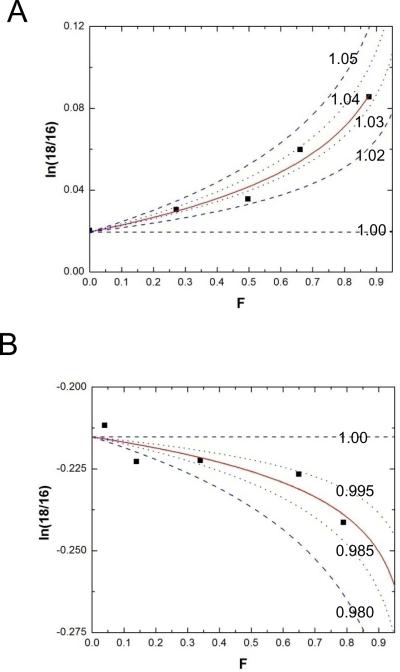
In reactions containing a mixture of natural abundance UpG and Up[5'-18O]G, the fraction of the residual substrate containing the 18O label clearly increases due to the faster reaction of the lighter 16O isotope (Figure 4A). The lower activation energy for the lighter isotopologue is indicative of a normal value for the leaving group isotope effect, 18kLG. In contrast, the population of molecules in which the 2'-O is enriched in 18O label shows the opposite result. The change in isotope ratio clearly shows accumulation of the 16O labeled molecules indicative of a larger rate constant for the heavier 18O containing molecules and therefore an inverse 18kNUC (Figure 4B). As described in the Experimental Section, individual KIEs measurements were determined from plots of ln(18O/16O) versus F (e.g. Figure 4A & B)52. The values for 18kNUC and 18kLG observed at pH 12 and 14 are shown in Table 1. Fits of the data from individual experiments to equation 2 (Experimental Section) gave standard errors of <0.5%. The opposite observed enrichments of the 5'-18O and 2'-18O populations of UpG indicate that the observed effects are not due to adventitious enrichment of one of the isotopes on sample workup. KIEs determined from two different ions (m/z 486/488 and m/z 211/213) gave the same KIE within 0.5%, providing a check on internal accuracy.
Figure 4.
Determination of observed KIEs by analysis of ln(18O/16O) versus F data. A. Determination of 18kLG by quantification of the change in 18O/16O as a function of reaction progress (filled symbols). The red line is a fit of the data to equation 1 yielding a KIE of 1.032. The blue lines represent simulations that span the range of observable 18O KIEs. B. Determination of 18kNUC. The data and fit to equation 1 are indicated as in panel A. The blue lines represent simulations of KIEs of unity to 0.980 inverse for comparison.
To better understand the extent to which proton transfer occurs in the TS above the breakpoint in the pH versus rate profile the kinetic solvent deuterium isotope effect for UpG 2'-O-transphosphorylation at pH 14 was measured. Solvent deuterium effects have been reported for a variety of phosphoryl transfer reactions28; 54; 55; 56. Intermolecular reactions show a significant effect at pH values of 1256 and below, while the solvent D2O effect above this range has not been reported. The solvent deuterium isotope for the base-catalyzed 2'-O-transphosphorylation of UpG at pH 14 was determined by direct comparison of the reaction rates in H2O and D2O (Figure 5). Conversion of the substrate was monitored by RP-HPLC and integration of the chromatogram as described above. The difference in the rate constants between H2O and D2O was less than 10% (0.055 min−1 and 0.052 min−1, respectively).
Figure 5.
Determination of the solvent D2O effect on 2'-O-transphosphorylation of 5'-UpG-3' at pH 14. Kinetic analyses of reactions in H2O (filled symbols) or D2O (open symbols) were run in parallel. Comparison of the rate constants yield a solvent D2O effect (kH/kD) of 0.95.
DISCUSSION
The results reported here provide the first analysis of the mechanism and transition state for RNA strand cleavage by 2'-O-transphosphorylation by analysis of primary KIEs. Consideration of the magnitude of the observed effects provides insight into the role of hydroxide in catalyzing the reaction. Interpretation of KIEs reported here also provides increased understanding of the changes in protonation state that occur during the reaction. Additionally, comparison of the values for RNA to the extensive mechanistic framework established for reactions of phosphodiesters by KIE analyses provides new insight into the transition state charge distribution.
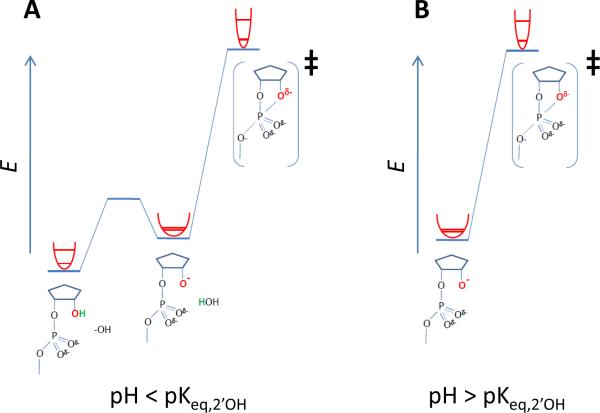
First, it is important to consider whether the observed KIEs are consistent with a general or specific base mechanism for catalysis by hydroxide. The conventional model for the observed pH-rate profile for RNA hydrolysis is depicted in the top pathway in Figure 1 and in reaction coordinate diagrams in Figure 6. In this specific base catalysis model the kinetically observed pKa at pH 13.5 is due to the deprotonation of the 2'-OH. Based on this mechanism, at pH higher than pKa the 18kobs should reflect 18kbond. At lower pH, the ground state changes from the alkoxide to the hydroxyl. As illustrated in Figure 6A, this results in a stiffer ground state, and consequently, predicts that a more normal KIE, in this case that is closer to unity, should be observed. A precise understanding of the contribution from bond formation requires knowledge of the EIE for deprotonation. No experimental primary 18O-EIE for the deprotonation of an aliphatic alcohol has been published. However, normal 18O EIEs in the range of 1–2% on deprotonation for several compounds have been reported57. The 18Keq for deprotonation of nitrophenol is reported as 1.015358 and values of 1.019-1.014 for phosphoric acid and glucose phosphate has been measured59. An EIE of 1.045 for deprotonation of water was determined by Green and Taube by analyzing the equilibrium between solutions of NaOH and H2O vapor60. However, as pointed out by Hengge and colleagues61 this value includes secondary contributions from water molecules that solvate hydroxide that will also be normal and therefore the intrinsic value for deprotonation will be smaller, but still normal. Recently, Humphry et al.45 estimated from ab initio calculations an EIE of 1.0245 for deprotonation of the hydroxyl of 2-hydroxypropyl-p-nitrophenyl phosphate.
Figure 6.
Reaction coordinate diagrams for specific base mechanism of hydroxide ion catalyzed 2'-O-transphosphorylation showing the changes in bond stiffness at the 3'O position below (A) and above (B) its pKa (pKeq,2'O). Equilibrium deprotonation of the 2'-OH (at eq,2'O) occurs prior to nucleophilic attack and reaction to form the rate limiting transition state. A two dimensional projection of the relative changes in bonding environment for the 2'-O are depicted as hyperbolas for the protonated and deprotonated ground states and the transition state. The lower line in each hyperbola represents the zeropoint vibrational energy of the heavy (18O) isotope and the upper line represents the zeropoint vibrational energy of the light (16O) isotope). For the reaction of the 2'-oxyanion in B the stiffer bonding environment in the transition state due to formation of the new 2'O-P bond results in an inverse 18kNUC. For the reaction below the pKeq,2'O depicted in A, the ground state bonding environment is stiffer due to protonation, and as a consequence, there is less of a change in overall bond stiffness between the ground state and transition state, resulting in an observed 18kNUC that is closer to unity (less inverse).
Using the rate equation for the specific base mechanism shown in Figure 1, the observed 18kNUC values at pH 14 and 12, and an estimated 18Keq for deprotonation of 1.024545, the intrinsic 18kbond for RNA 2'-O-transphorphorylation can be calculated using the following expression:
| equation 3 |
where 18kNUC is the experimentally observed KIE, k16, and k18 are the rate constants for reaction of the two oxygen isotopologues, k16,bond and k18,bond are the intrinsic rate constants for reaction from the 2' oxyanion ground state, K16,eq and K18,eq are the equilibrium constants for deprotonation of the 2'-16O and 2'-18O, respectively. Assuming that the equilibrium isotope effect on deprotonation (18Keq) is 1.0245 as estimated for the deprotonation of the 2'-hydroxyl of hydroxypropyl-p-nitrophenol phosphate45, one can use the full equation to calculate the intrinsic 18kbond from the observed values for 18kNUC(obs) at pH 12 and pH 14. Here, the K18,eq is obtained by applying the EIE calculated for deprotonation of the hydroxyl group of 2-hydroxypropyl-p-nitrophenyl phosphate to the experimentally determined K16,eq from Figure 2C. For values of 18kNUC that we observe at both pH values, the estimated 18kbond from this method is 0.981. This correspondence is a good check on internal consistency and indicates that our measurement of KIE and estimation of 18Keq are reasonable.
For the reaction at pH 14 the only significant change in the bonding environment for the 2'-O is the formation of the new bond to phosphorus assuming that the reaction proceeds through attack of the 2'-oxyanion nucleophile. The inverse 18kNUC values observed at both pH 12 and 14 reflect advanced formation of this bond in the rate-limiting transition state. At pH 12 the transition state is essentially unchanged, but in the ground state the 2'-O is bonded to a proton, which results in a stiffer bonding environment compared to the 2'-oxyanion (Figure 6). As discussed above, the magnitude of the effect of this stiffer ground state environment on the observed nucleophile KIE is determined by the EIE for deprotonation.
An alternative model is that hydroxide ion acts as a general base, and that the proton bonded to the 2'-O position is transferred in the transition state (Figure 1, lower pathway). As in the specific base mechanism (Figure 1, upper pathway) the ground state changes as the pH becomes greater than 13.5. In the general base mechanism the rate constant for the reaction increases from pH 11–13 due to the increase in the concentration of the hydroxide ion catalyst. Above the breakpoint in the pH versus rate profile the increase in catalysis due to the increase in hydroxide ion concentration is offset by the corresponding decrease in the concentration of the protonated 2'-hydroxyl species, which remains the state from which nucleophilic attack occurs. Thus, the same pH dependence of the rate constant is observed as in the specific base. While the 2'-oxyanion is clearly a better nucleophile, which suggests the specific base mechanism would be favored, the general base mechanism cannot be excluded based on this consideration alone since electrostatic repulsion may limit the degree to which it can adopt the appropriate in-line conformation for the reaction compared to a protonated 2'-hydroxyl.
In the case of the general base mechanism the observed 18kNUC is still defined by the magnitudes of 18kbond and 18Keq, however, the intrinsic 18kbond is observed below rather than above the observed kinetic pKa. That is, above the pKa the observed KIE must be corrected to account for equilibrium protonation of the 2'-oxyanion ground state. Using the rate equation for the mechanism shown in the lower pathway in Figure 1, the intrinsic 18kbond can be calculated in a similar manner:
| equation 4 |
In this hypothetical general base mechanism the estimated intrinsic 18kbond is 0.995, which is significantly more normal than the value obtained for the 18kbond for the specific base mechanism (0.984). In this mechanism the ground state bonding environment is stiffer due to protonation, and as a consequence, there is less of a change in overall bond stiffness between the ground state and transition state, resulting in an observed 18kNUC that is closer to unity (less inverse). Thus, the primary 18O KIEs alone do not distinguish between these two mechanisms, although the specific base mechanism appears more likely given the much greater nucleophilicity of the 2'-oxyanion.
Evidence in favor of the specific base mechanism comes from consideration of the kinetic solvent D2O isotope effects for the 2'-O-transphosphorylation reaction and comparisons with effects from other intermolecular phosphoryl transfer reactions. It is well established that there are significant solvent D2O effects on the base catalyzed intermolecular phosphoryl transfer. Effects of 4.9 and 7.2 were reported for reactions of 1-(5'-O-methyl-β-D-ribofuranoyl)-2-methylbenzimidazole 2'-phenylphosphate and 2'-(2-methoxylethyl)phosphate, respectively62. A similar value of 4.0 was reported for the alkaline cleavage of 2-hydroxypropyl-p-nitrophenyl phosphate63. These large effects are primarily attributable to the difference in the deprotonation of the attacking nucleophile. Importantly, lower solvent D2O effects are observed for reactions of fully ionized oxyanion nucleophiles. Values of 0.9 for attack of hydroxide on the p-nitrophenyl ester of 3'-thymidine monophosphate55; 0.8 for cis-4-hydroxytetrahydrofuran 3-phenylposphate64, and 1.0 with dialkyl 2-carboxyphenyl phosphates65 have been reported. Thus, the observed effects suggest a primary contribution from proton transfer when the reaction occurs from the neutral oxygen ground state which is absent when the reaction occurs from the 2'-oxyanion ground state. This interpretation also explains the observed attenuation of the solvent D2O effect at high pH for the cleavage of p-nitrophenol ester of 3'-UMP by a highly active Zn(II) complex56. To confirm this result for 2'-O-transphosphorylation of UpG under the conditions used to monitor primary 18O KIEs here, we analyzed the solvent D2O effect at pH 14 (Figure 4). As suggested by previous results there is essentially no solvent deuterium KIE for the reaction consistent with no proton transfer in the transition state. This result is incompatible with the general base mechanism, which predicts a solvent effect should be observable both above and below the kinetic pKa at 13.5. Thus, we conclude that the primary and solvent KIE data together support the conventional model in which the 2'-O undergoes equilibrium deprotonation to yield the 2'-oxyanion prior to nucleophilic attack.
The magnitude of the nucleophile and leaving group KIEs provide information on differences in bonding between the ground state and transition state. However, interpretation of nucleophile KIEs are subject to further complexity in addition to changes in protonation, since the magnitude of 18kbond is the product of two factors, the temperature-independent factor (TIF) and the temperature-dependent factor (TDF)66. The TIF is also referred to as the imaginary frequency ratio because the force constant for this motion is negative, which in turn gives an imaginary vibrational frequency (ω = (k/m)1/2)67. The TIF reflects the extent to which the labeled atom participates in the reaction coordinate. Because the imaginary frequency of the lighter isotope is always larger, this term always makes a normal (>1) contribution to the observed isotope effect. The TDF reflects differences in vibrations (bonding) of the labeled atom in the ground state compared to the transition state. The increase in bond stiffness due to formation of the new O-P bond therefore results in an inverse contribution to the observed KIE. For early transition states in which the contribution from formation of the new bond to phosphorus is small, the observed nucleophile KIE can be normal due to a larger contribution from the TIF. Examples of normal observed isotope effects for oxygen nucleophiles in phosphoryl transfer reactions include concerted displacement of a good leaving group like nitrophenol45; 55; 68 or displacement of sulfur from a phosphorothioate ester61. In contrast, because there is no TIF for a stepwise mechanism in which leaving group displacement is rate limiting, the observed nucleophile KIE can assume a large inverse value as reported for hydrolysis of a metal complexed phosphodiester (0.937)17. An more inverse 18kNUC for nucleophilic attack is reported for the reaction of hydroxypropyl-p-nitrophenol phosphate catalyzed by dinuclear zinc(II) catalyst indicating an altered, later transition state for the catalyzed reaction (0.9874 versus 1.0079 for the catalyzed and uncatalyzed reactions, respectively)45. In this context, the estimated inverse 18kbond value reported here for concerted 2'-O-transphosphorylation reaction of RNA (0.981) indicates that the inverse contribution from the TDF is dominant and strongly suggests that nucleophile bond formation is advanced in the transition state.
The large, normal value of 18kLG (1.034) reported here for the RNA 2'-O-transhosphorylation reaction catalyzed by specific base reflects a large decrease in bond order resulting in a less stiff vibrational environment for the 5'O in the transition state. By comparison, the magnitude of the leaving group KIE for 2'-O-transphosphorylation of uridine-3'-p-nitrophenyl phosphate is small46; 49 (ca. 18kLG = 1.006) consistent with an early TS as observed for solvent hydrolysis of phosphodiesters with good leaving groups10. The RNA reaction mechanism is likely to be different because the 5'-O leaving group has much higher pKa than nitrophenol (7.1 for p-nitrophenol compared to >13 for a ribose hydroxyl19). Indeed, a large leaving group KIE (18kLG = 1.027) was observed for reactions of uridine-3'-m-nitrobenzyl (pKa 14.9)69. These data indicate that for the RNA reaction the P-5'O bond is being broken and the 5'-O is departing as the alkoxide. Protonation of the leaving group in the TS would act to stiffen the bonding environment and offset the loss of bonding to the phosphorous atom, resulting in a KIE that is closer to unity. Computational studies by Warshel also suggest greater nucleophilic participation in reaction coordinate motion in order to expel leaving groups with higher pKa7, consistent with the Hammond-Leffler postulate70; 71 and supported by the large inverse intrinsic nucleophile KIE of 0.981 as discussed above. Together the experimental and computational data indicate a “late” or product-like TS for RNA reaction with its relatively unreactive 5' ribose hydroxyl leaving group (pKa 14).
CONCLUSIONS
The cleavage of RNA chains by attack of the adjacent 2'-OH is universal in biology and has been intensively studied since the 1950's, yet this is the first characterization of RNA cleavage with primary isotope effects capable of directly informing on the extent of bond cleavage in the transition state. Comparing the results reported here with this literature and KIE measurements of model phosphoryl transfer reactions reveals that significant differences exist in the transition states for concerted phosphoryl transfer reactions and that the high pKa of the leaving group coupled with the enforced geometry of cleavage can significantly alter the transition state for the reaction. The KIE data indicate a concerted mechanism in which bond formation to the nucleophile and bond cleavage from the leaving group are both advanced in a “late” or product-like transition state. The nucleophile isotope effects implicate equilibrium deprotonation of the 2'O prior to nucleophilic attack, and thus catalysis by hydroxide occurs by a specific base mechanism. Additionally, the general methods used in these studies provide the opportunity to determine both nucleophile and leaving group KIEs on reactions involving RNA itself and will provide important insights into mechanisms employed by other chemical catalysts. Additionally, these methods provide a means for investigating enzyme-catalyzed mechanisms of phosphoryl transfer with physiologically relevant substrates.
Supplementary Material
ACKNOWLEDGEMENT
The authors gratefully acknowledge the support of the Dr. Mark Chance and the CWRU Center for Proteomics and Bioinformatics. We thank Dr. Masaru Miyagi for expert instruction and advice on mass spectrometric analysis of nucleotides. The anonymous reviewers are also gratefully acknowledged for comments which improved the clarity and content of the manuscript. This work was supported by grants NIH GM079647, NIH GM056740 to MEH, NSF 00543 to VEA and NIH AI081987 to JAP.
REFERENCES
- 1.Mildvan AS. Mechanisms of signaling and related enzymes. Proteins. 1997;29:401–16. [PubMed] [Google Scholar]
- 2.Wolfenden R. Thermodynamic and extrathermodynamic requirements of enzyme catalysis. Biophys Chem. 2003;105:559–72. doi: 10.1016/s0301-4622(03)00066-8. [DOI] [PubMed] [Google Scholar]
- 3.Westheimer FH. Pseudo-rotation in the hydrolysis of phosphate esters. Acc Chem Res. 1968;1:70–78. [Google Scholar]
- 4.Kirby AJ, Jencks WP. The reactivity if nucleophilic reagents toward the p-nitrophenyl phosphate dianion. J Am Chem Soc. 1965;87:3209–3216. [Google Scholar]
- 5.Oivanen M, Kuusela S, Lonnberg H. Kinetics and mechanism for the cleavage and isomerization of phosphodiester bonds of RNA by Bronsted acids and bases. Chem Rev. 1998;98:961–990. doi: 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- 6.Cleland WW, Hengge AC. Enzymatic mechanisms of phosphate and sulfate transfer. Chem Rev. 2006;106:3252–78. doi: 10.1021/cr050287o. [DOI] [PubMed] [Google Scholar]
- 7.Rosta E, Kamerlin SC, Warshel A. On the interpretation of the observed linear free energy relationship in phosphate hydrolysis: a thorough computational study of phosphate diester hydrolysis in solution. Biochemistry. 2008;47:3725–35. doi: 10.1021/bi702106m. [DOI] [PubMed] [Google Scholar]
- 8.Harris ME, Cassano AG. Experimental analyses of the chemical dynamics of ribozyme catalysis. Curr Opin Chem Biol. 2008;12:626–39. doi: 10.1016/j.cbpa.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleland WW, Hengge AC. Mechanisms of phosphoryl and acyl transfer. FASEB Journal. 1995;9:1585–94. doi: 10.1096/fasebj.9.15.8529838. [DOI] [PubMed] [Google Scholar]
- 10.Hengge AC, Aleksandra TE, Cleland WW. Studies of transition-state structures of phosphoryl transfer reactions of phosphodiesters of p-nitrophenol. J Am Chem Soc. 1995;117:5919–5926. [Google Scholar]
- 11.Hengge AC. Isotope effects in the study of phosphoryl and sulfuryl transfer reactions. Acc Chem Res. 2002;35:105–12. doi: 10.1021/ar000143q. [DOI] [PubMed] [Google Scholar]
- 12.Kirby AJ, Younas M. The reactivity of phosphate esters. Reactions of diesters with nucleophiles. J Chem Soc B. 1970:1165–1172. [Google Scholar]
- 13.Lonnberg H, Stromberg R, Williams A. Compelling evidence for a stepwise mechanism of the alkaline cyclisation of uridine 3'-phosphate esters. Org Biomol Chem. 2004;2:2165–7. doi: 10.1039/b406926a. [DOI] [PubMed] [Google Scholar]
- 14.Rawlings J, Cleland WW, Hengge AC. Metal-catalyzed phosphodiester cleavage: secondary 18O isotope effects as an indicator of mechanism. J Am Chem Soc. 2006;128:17120–5. doi: 10.1021/ja065931a. [DOI] [PubMed] [Google Scholar]
- 15.Humphry T, Forconi M, Williams NH, Hengge AC. Altered mechanisms of reactions of phosphate esters bridging a dinuclear metal center. J Am Chem Soc. 2004;126:11864–9. doi: 10.1021/ja047110g. [DOI] [PubMed] [Google Scholar]
- 16.Grzyska PK, Czyryca PG, Purcell J, Hengge AC. Transition state differences in hydrolysis reactions of alkyl versus aryl phosphate monoester monoanions. J Am Chem Soc. 2003;125:13106–11. doi: 10.1021/ja036571j. [DOI] [PubMed] [Google Scholar]
- 17.Humphry T, Forconi M, Williams NH, Hengge AC. An altered mechanism of hydrolysis for a metal-complexed phosphate diester. J Am Chem Soc. 2002;124:14860–1. doi: 10.1021/ja027671c. [DOI] [PubMed] [Google Scholar]
- 18.Davis AM, Hall AD, Williams A. Charge description of base-catalyzed alcoholysis of aryl phosphodiesters: A ribonuclease model. J Am Chem Soc. 1988;110:5105–5108. [Google Scholar]
- 19.Li Y, Breaker RR. Kinetics of RNA Degradation by Specific Base Catalysis of Transesterification Involving the 2∋-Hydroxyl Group. J Am Chem Soc. 1999;121:5364–5372. [Google Scholar]
- 20.Jarvinen P, Oivanen M, Lonnberg H. Interconversion and phosphoester hydrolysis of 2',5'- and 2',3'-dinucleoside monophosphates: Kinetics and mechanism. J Org Chem. 1991;56:5396–5401. [Google Scholar]
- 21.Koike T, Inoue Y. STRUCTURE AND REACTIVITY OF OLIGONUCLEOTIDES. PART I KINETICS OF THE NON-ENZYMATIC TRANSPHOSPHORYLATION OF ADENYLYL-(3′–5′)-ADENOSINE 3′-PHOSPHATE AND OTHER DINUCLEOTIDES. Chemistry Letters. 1972;7:569–572. [Google Scholar]
- 22.Lipkin D, Talbert PT, Cohn M. The Mechanism of the Alkaline Hydrolysis of Ribonucleic Acids. J Am Chem Soc. 1954;76:2871–2872. [Google Scholar]
- 23.Breslow R. Kinetics and mechanism in RNA cleavage. Proc Natl Acad Sci U S A. 1993;90:1208–11. doi: 10.1073/pnas.90.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedor MJ. Comparative enzymology and structural biology of RNA self-cleavage. Annu Rev Biophys. 2009;38:271–99. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 25.Hamilton CS, Spedaliere CJ, Ginter JM, Johnston MV, Mueller EG. The roles of the essential Asp-48 and highly conserved His-43 elucidated by the pH dependence of the pseudouridine synthase TruB. Arch Biochem Biophys. 2005;433:322–34. doi: 10.1016/j.abb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Polgar L. The catalytic triad of serine peptidases. Cell Mol Life Sci. 2005;62:2161–72. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richard JP. The enhancement of enzymatic rate accelerations by Bronsted acid-base catalysis. Biochemistry. 1998;37:4305–9. doi: 10.1021/bi972655r. [DOI] [PubMed] [Google Scholar]
- 28.Schowen KB, Limbach HH, Denisov GS, Schowen RL. Hydrogen bonds and proton transfer in general-catalytic transition-state stabilization in enzyme catalysis. Biochim Biophys Acta. 2000;1458:43–62. doi: 10.1016/s0005-2728(00)00059-1. [DOI] [PubMed] [Google Scholar]
- 29.Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–91. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 30.Oivanen M, Ora M, Almer H, Stromberg R, H L. Hydrolytic reactions of the diastereomeric phosphoromonothioate analogs of uridylyl(3',5')uridine: Kinetics and mechanism of desulfurization, phosphoester hydrolysis, and transesterification to the 2',5'-isomers. J Org Chem. 1995;60:5620–5627. [Google Scholar]
- 31.Emilsson GM, Nakamura S, Roth A, Breaker RR. Ribozyme speed limits. Rna. 2003;9:907–18. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez CS, Faza ON, Gregersen BA, de Lera AR, York DM. Pseudorotation of natural and chemically modified biological phosphoranes: implications for RNA catalysis. Chemphyschem. 2004;5:1045–9. doi: 10.1002/cphc.200400091. [DOI] [PubMed] [Google Scholar]
- 33.Cook RM, Turley PC, Deibert CE, Fierman AH, Haake P. Pentacoordinate Intermediates and the Rate-Determining Step in Displacement at Phosphorus. J Am Chem Soc. 1972;94:9260–9261. [Google Scholar]
- 34.Perreault DM, Anslyn EV. Angew. Chem. 1997;109:470–490. [Google Scholar]
- 35.Lim C, Karplus M. Nonexistence of dianionic phosphorane intermediates in an ab initio study of the base-catalyzed hydrolysis of ehtylene phosphate. J Am Chem Soc. 1990;112:5872–5873. [Google Scholar]
- 36.Davies JE, Doltsinis NL, Kirby AJ, Roussev CD, Sprik M. Estimating pKa values for pentaoxyphosphoranes. J Am Chem Soc. 2002;124:6594–9. doi: 10.1021/ja025779m. [DOI] [PubMed] [Google Scholar]
- 37.Lopez X, Schaefer M, Dejaegere A, Karplus M. Theoretical evaluation of pK(a) in phosphoranes: implications for phosphate ester hydrolysis. J Am Chem Soc. 2002;124:5010–8. doi: 10.1021/ja011373i. [DOI] [PubMed] [Google Scholar]
- 38.Lui X, Reese CB. Tetrahedron Letters. 1995:3413–3416. [Google Scholar]
- 39.Acharya S, Foldesi A, Chattopadhyaya J. The pK(a) of the internucleotidic 2'-hydroxyl group in diribonucleoside (3'-->5') monophosphates. J Org Chem. 2003;68:1906–10. doi: 10.1021/jo026545o. [DOI] [PubMed] [Google Scholar]
- 40.Hengge AC. Isotope effects in the study of enzymatic phosphoryl transfer reactions. FEBS Lett. 2001;501:99–102. doi: 10.1016/s0014-5793(01)02638-2. [DOI] [PubMed] [Google Scholar]
- 41.Cleland WW, Cook PF. Enzyme Kinetics and Mechanism. Garland Publishers; London and New York: 2007. [Google Scholar]
- 42.Buddenbaum WE, Shiner VJ. Computation of Isotope Effects on Equilibria and Rates. In: Cleland WW, O'Leary MH, Northrop DB, editors. Isotope Effects on Enzyme-Catalyzed Reactions. University Park Press; London: 1977. pp. 1–36. [Google Scholar]
- 43.Bigeleisen J. Correlation of kinetic isotope effects with chemical bondingin three-center reactions. Pure Appl Chem. 1964;8:217–223. [Google Scholar]
- 44.Kolmodin K, Luzhkov VB, Aqvist J. Computational study of the influence of solvent on (16)O/(18)O equilibrium isotope effects in phosphate deprotonation reactions. J Am Chem Soc. 2002;124:10130–5. doi: 10.1021/ja012669r. [DOI] [PubMed] [Google Scholar]
- 45.Humphry T, Iyer S, Iranzo O, Morrow JR, Richard JP, Paneth P, Hengge AC. Altered transition state for the reaction of an RNA model catalyzed by a dinuclear zinc(II) catalyst. J Am Chem Soc. 2008;130:17858–66. doi: 10.1021/ja8059864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hengge AC, Bruzik KS, Tobin AE, Cleland WW, Tsai MD. Kinetic Isotope Effects and Stereochemical Studies on a Ribonuclease Model: Hydrolysis Reactions of Uridine 3'-Nitrophenyl Phosphate. Bio-Organic Chemistry. 2000;28:119–133. doi: 10.1006/bioo.2000.1170. [DOI] [PubMed] [Google Scholar]
- 47.Feng G, Tanifum EA, Hengge AC, Williams NH. Mechanism and transition state structure of aryl methylphosphonate esters doubly coordinated to a dinuclear cobalt(III) center. J Am Chem Soc. 2009;131:12771–9. doi: 10.1021/ja904134n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cleland WW. Use of isotope effects to elucidate enzyme mechanisms. CRC Crit Rev Biochem. 1982;13:385–428. doi: 10.3109/10409238209108715. [DOI] [PubMed] [Google Scholar]
- 49.Rishavy MA, Hengge AC, Cleland WW. Lanthanide Catalyzed Cyclization of Uridine 3'-p-Nitrophenyl Phosphate. Bio-Organic Chemistry. 2000;28:283–292. doi: 10.1006/bioo.2000.1179. [DOI] [PubMed] [Google Scholar]
- 50.Dai Q, Frederiksen JK, Anderson VE, Harris ME, Piccirilli JA. Efficient synthesis of [2'–18O]uridine and its incorporation into oligonucleotides: a new tool for mechanistic study of nucleotidyl transfer reactions by isotope effect analysis. J Org Chem. 2008;73:309–11. doi: 10.1021/jo701727h. [DOI] [PubMed] [Google Scholar]
- 51.Cassano AG, Wang B, Anderson DR, Previs S, Harris ME, Anderson VE. Inaccuracies in selected ion monitoring determination of isotope ratios obviated by profile acquisition: nucleotide 18O/16O measurements. Anal Biochem. 2007;367:28–39. doi: 10.1016/j.ab.2007.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cook PF, Cleland WW. Enzyme Kinetics and Mechanism. Garland Science; New York: 2007. Isotope effects as a probe of mechanism; pp. 253–324. [Google Scholar]
- 53.Breaker RR, Emilsson GM, Lazarev D, Nakamura S, Puskarz IJ, Roth A, Sudarsan N. A common speed limit for RNA-cleaving ribozymes and deoxyribozymes. Rna. 2003;9:949–57. doi: 10.1261/rna.5670703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakano S, Bevilacqua PC. Proton inventory of the genomic HDV ribozyme in Mg(2+)-containing solutions. J Am Chem Soc. 2001;123:11333–4. doi: 10.1021/ja0166850. [DOI] [PubMed] [Google Scholar]
- 55.Cassano AG, Anderson VE, Harris ME. Evidence for direct attack by hydroxide in phosphodiester hydrolysis. J Am Chem Soc. 2002;124:10964–5. doi: 10.1021/ja020823j. [DOI] [PubMed] [Google Scholar]
- 56.Yang MY, Iranzo O, Richard JP, Morrow JR. Solvent deuterium isotope effects on phosphodiester cleavage catalyzed by an extraordinarily active Zn(II) complex. J Am Chem Soc. 2005;127:1064–5. doi: 10.1021/ja044617i. [DOI] [PubMed] [Google Scholar]
- 57.Rishavy MA, Cleland WW. 13C,15N, and 18O equilibrium isotope effects and fractionation factors. Can. J. Chem. 1999;77:967–977. [Google Scholar]
- 58.Hengge AC, Hess RA. Concerted of stepwise mechanism for acyl transfer reactions of p-nitrophenol acetate? Transition state structures from kinetic isotope effects. J Am Chem Soc. 1994;116:11256–11263. [Google Scholar]
- 59.Knight WB, Weiss PM, Cleland WW. Determination of equilibrium 18O isotope effects on the deprotonation of phosphate and phosphate esters and the anomeric effect on deprotonation of glucose-6-phosphate. J Am Chem Soc. 1986;108:2759–2761. [Google Scholar]
- 60.Green M, Taube H. Isotopic fractionation in the OH--H2O exchange reaction. J. Phys. Chem. 1963;67:1565–6. [Google Scholar]
- 61.Iyer S, Hengge AC. The effects of sulfur substitution for the nucleophile and bridging oxygen atoms in reactions of hydroxyalkyl phosphate esters. J Org Chem. 2008;73:4819–29. doi: 10.1021/jo8002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Virtanen A, Polari L, Valila M, Mikkola S. Kinetic solvent deuterium isotope effect in transesterification of RNA models. J Phys Org Chem. 2005;18:385–397. [Google Scholar]
- 63.Bonfa L, Gatos F, Mancin F, Tecilla P, Tonellato U. The Ligand Effect on the Hydrolytic Reactivity of Zn(II) Complexes toward Phosphate Diesters. Inorg Chem. 2003;42 doi: 10.1021/ic034139x. [DOI] [PubMed] [Google Scholar]
- 64.Usher DA, Richardson DI, Jr., Oakenfull DG. Models of Ribonuclease Action. 11. Specific Acid, Specific Base, and Neutral Pathways for Hydrolysis of a Nucleotide Diester Analog. J Am Chem Soc. 1970;92:4699–4712. doi: 10.1021/ja00718a037. [DOI] [PubMed] [Google Scholar]
- 65.Bruice TC, Blasko A, Arasasingham RD, Kim J-S. Participation of two carboxyl groups in phosphodiester hydrolysis. 2. A kinetic, isotopic and 31P NMR study of the hydrolysis of a phosphodiester with carboxyl groups fixed in an attack conformation. J Am Chem Soc. 1995;117:12070–12077. [Google Scholar]
- 66.Melander L, Saunders WH. Reaction Rates of Isotopic Molecules. Wiley Interscience; New York: 1987. [Google Scholar]
- 67.Lewars E. Computaional Chemistry: Introduction to the theory and application of molecular and quantum mechanics. Kluwer Academic Publishers; Norwell, MA: 2003. [Google Scholar]
- 68.Cassano AG, Anderson VE, Harris ME. Analysis of solvent nucleophile isotope effects: evidence for concerted mechanisms and nucleophilic activation by metal coordination in nonenzymatic and ribozyme-catalyzed phosphodiester hydrolysis. Biochemistry. 2004;43:10547–59. doi: 10.1021/bi049188f. [DOI] [PubMed] [Google Scholar]
- 69.Gerratana B, Sowa GA, Cleland WW. Characterization of the transition state structures and mechanisms for the isomerization and cleavage reactions of 3'-m-nitrobenzyl phosphate. J Am Chem Soc. 2000;122:12615–12621. [Google Scholar]
- 70.Hammond GS. A correlation of reaction rates. J Am Chem Soc. 1955;77 [Google Scholar]
- 71.Leffler JE. Parameters fr the description of transition states. Science. 1953;117:340–341. doi: 10.1126/science.117.3039.340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.