Abstract
The registration of images is a task that is at the core of many applications in computer vision. In computational neuroimaging where the automated segmentation of brain structures is frequently used to quantify change, a highly accurate registration is necessary for motion correction of images taken in the same session, or across time in longitudinal studies where changes in the images can be expected. This paper, inspired by Nestares and Heeger (2000), presents a method based on robust statistics to register images in the presence of differences, such as jaw movement, differential MR distortions and true anatomical change. The approach we present guarantees inverse consistency (symmetry), can deal with different intensity scales and automatically estimates a sensitivity parameter to detect outlier regions in the images. The resulting registrations are highly accurate due to their ability to ignore outlier regions and show superior robustness with respect to noise, to intensity scaling and outliers when compared to state-of-the-art registration tools such as FLIRT (in FSL) or the coregistration tool in SPM.
Keywords: image registration, robust statistics, inverse consistent alignment, motion correction, longitudinal analysis
1. Introduction
There is great potential utility for information extracted from neuroimaging data to serve as biomarkers, to quantify neurodegeneration, and to evaluate the efficacy of disease-modifying therapies. Currently, the accurate and reliable registration of images presents a major challenge, due to a number of factors. These include differential distortions that affect longitudinal time points in different ways; true, localized anatomical change that can cause global offsets in the computed registration, and the lack of inverse consistency in which the registration of multiple images depends on the order of processing, which can lead to algorithm-induced artifacts in detected changes. Thus, the development of an accurate, robust and inverse consistent method is a critical first step to quantify change in neuroimaging or medical image data in general.
Since the object of interest is typically located differently in each acquired image, accurate geometric transformations are necessary to register the input images into a common space. Approaches based on robust statistics are extremely useful in this domain, as they provide a mechanism for discounting regions in the images that contain true differences, and allow one to recover the correct alignment based on the remainder of the data. Inverse consistency is critical to avoid introducing bias into longitudinal studies. A lack of inverse consistency in registration is likely to bias subsequent processing and analysis, as documented in Yushkevicha et al. (2009). The goal of this work is thus to develop a robust and inverse consistent registration method for use in the analysis of neuroimaging data. The core application of this technique is intra-modality and intra-subject registration with important implications for:
Motion correction and averaging of several intra-session scans to increase the signal to noise ratio,
highly accurate alignment of longitudinal image data and
initial registration for higher-dimensional warps.
Although the remainder of this paper deals with neuroimaging data, the method can be used for other image registration task as well.
Highly accurate rigid registrations are of importance when averaging multiple scans taken within a session to reduce the influence of noise or subject motion. Since it is nearly impossible for a person to remain motionless throughout a 20 minutes scan, image quality can be increased by taking shorter scans and performing retrospective motion correction (Kochunov et al., 2006). Many common sequences are short enough to allow for several structural scans of the same modality within a session. Here even a slightly inaccurate registration will introduce additional artifacts into the final average and likely reduce the accuracy, sensitivity and robustness of downstream analysis.
Compared with cross-sectional studies, a longitudinal design can significantly reduce the confounding effect of inter-individual morphological variability by using each subject as his or her own control. As a result, longitudinal imaging studies are becoming increasingly common in clinical and scientific neuroimaging. Degeneration in subcortical structures and cortical gray matter is, for example, manifested in aging (Jack et al., 1997; Salat et al., 1999, 2004; Sowell et al., 2003, 2004), Alzheimer's disease (Dickerson et al., 2001; Thompson et al., 2003; Lerch et al., 2005), Huntington's disease (Rosas et al., 2002), multiple sclerosis (Sailer et al., 2003) and Schizophrenia (Thompson et al., 2001; Kuperberg et al., 2003; Narr et al., 2005) and has been useful towards understanding some of the major pathophysiological mechanisms involved in these conditions. As a result, in vivo cortical thickness and subcortical volume measures are employed as biomarkers of the evolution of an array of diseases, and are thus of great utility for evaluating the efficacy of disease-modifying therapies in drug trials. To enable the information exchange at specific locations in space, highly accurate and unbiased registrations across time are necessary. They need to be capable of efficiently dealing with change in the images, which can include true neurodegeneration, differential positioning of the tongue, jaws, eyes, neck, different cutting planes as well as session-dependent imaging distortions such as susceptibility effects.
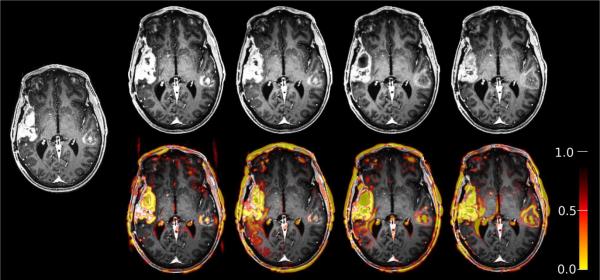
As an example see Figure 1 showing longitudinal tumor data (same slice of five acquisitions at different times, MPRAGE, 256 × 256 × 176, 1mm voxels) registered to the first time point (left) with the proposed robust method. The five time points are: 5 days prior to treatment start, 1 day prior, 1 day after treatment start, and 28, 56 days after treatment start. Despite of the significant change in these images the registration is highly accurate (verified visually in non-tumor regions). The bottom row depicts the outlier weights (red/yellow overlay), which are blurry regions of values between 0 (outlier) and 1 (regular voxel) that label differences in the images. In addition to the longitudinal change in tumor regions and consequential deformation (e.g. at the ventricles), the robust method also picks up differences in the scalp, eye region and motion artifacts in the background. In our robust approach the influence of these differences (or outliers) is reduced when constructing the registrations, while they have a detrimental influence on the final registration result in non robust methods.
Figure 1.
Robust registration of longitudinal tumor data (same slice of five acquisitions at different times). Left: target (first time point). Top row: aligned images. Bottom row: overlay of detected change/outlier regions (red/yellow). The outlier influence is automatically reduced during the iterative registration procedure to obtain highly accurate registrations of the remainder of the image; see also Fig. 6.
Statistically, robust parameter estimation has a history of supplying solutions to several computer vision problems (Stewart, 1999) as it is capable of estimating accurate model parameters in the presence of noise, measurement error (outliers) or true differences (e.g. change over time). The approach presented here is based on robust statistics and inspired by Nestares and Heeger (2000), who describe a robust multi-resolutional registration approach to rigidly register a set of slices to a full resolution image. Our approach, however, is designed to be inverse consistent to avoid introducing a bias. It also allows the calculation of an additional global intensity scale parameter to adjust for different intensity scalings that can be present especially in longitudinal data. A more complex intensity preprocessing is therefore not needed in most cases. Furthermore, we automatically estimate the single parameter of the algorithm that controls its sensitivity to outliers. This is a necessary addition, since a fixed parameter cannot adequately deal with different image intensity scales, which are common in MRI. In addition to the multi resolutional approach described in Nestares and Heeger (2000) we use moments for an initial coarse alignment to allow for larger displacements and situations where source and target may not overlap. Finally, we describe the registration of two full resolution images (instead of only a set of slices) and explain how both rigid and affine transformation models can be used in the symmetric algorithm. We demonstrate that our approach yields highly accurate registrations in brain regions and out-performs existing state-of-the-art registration algorithms.
The remainder of this paper is organized as follows. After discussing related work and introducing the theoretical background, such as robust statistics in Section 2, we present our symmetric registration model, different transformation models as well as intensity scaling in Section 3. Then we describe the registration algorithm in detail, taking care that the properties of the theory are carried over to the implementation (Section 4). We specifically focus on maintaining inverse consistency by resampling both images into a `half-way' space in intermediate steps as opposed to resampling the source at the estimated target location. This asymmetric sampling, which is commonly used, introduces a bias as the target image will not be resampled at all, and will thus be less smooth than the resampled source. In Section 5 we demonstrate the superiority of the proposed method over existing registration algorithms with respect to symmetry, robustness and accuracy on synthetic and real data as well as a motion correction application. The software implementing the presented robust registration is publicly distributed as part of the FreeSurfer (surfer.nmr.mgh.harvard.edu) software package as mri_robust_register.
2. Background
2.1. Related Work on Registration
Over the last 20 years methods for the registration of images (and in particular medical images) have been studied intensely (see e.g. Maintz and Viergever (1998); Maes et al. (1999); Hill et al. (2001) for surveys and comparisons). Many different applications domains exist for registration, including multimodal intra-subject registration, cross-subject volumetric registration, surface-based registration etc…, each of which require domain-specific approaches to maximize accuracy. Some of the most prominent intensity based algorithms are Cross-Correlation (Collins et al., 1995), Mutual Information (MI) (Maes et al., 1997, 1999; Wells et al., 1996), Normalized Mutual Information (NMI), and Correlation Ratio (CR) (Roche et al., 1998). Recently (Saad et al., 2009) found registration errors when comparing CR and MI and proposed a new cost function using a local Pearson correlation.
Intensity based methods consider information from the whole image and are often deemed to be more reliable and accurate than feature based methods (West et al., 1997, 1999). Driving the optimizations based on geometrically defined features such as points (Schönemann, 1931; Evans et al., 1989; Bookstein, 1991), edges (Nack, 1977; Kerwin and Yuan, 2001), contours (Medioni and Nevatia, 1984; Shih et al., 1997) or whole surfaces (Pelizzari et al., 1989; Fischl et al., 1999; Dale et al., 1999; Greve and Fischl, 2009) has the advantage of reducing computational complexity, but introduces reliability difficulties when extracting/placing the features. Furthermore, extracting surfaces is a complicated and time consuming process in itself and not feasible in cases where only an initial rigid registration is needed or for the purpose of averaging two structural scans from the same session. Additionally, hybrid approaches exist such as Greve and Fischl (2009), a surface based approach that additionally incorporates information derived from local intensity gradients. Note that a large body of work describes rigid registration in the Fourier domain, e.g. van der Kouwe et al. (2006); Bican and Flusser (2009); Costagli et al. (2009), but since we expect and wish to detect spatial outliers/change we operate in the spatial domain.
A number of different registration methods are implemented in freely available software packages. The widely used registration tool FLIRT (Jenkinson et al., 2002), part of the FSL package (Smith et al., 2004), implements several intensity based cost functions such as standard least squares (LS), correlation ratio (CR) and mutual information (MI) as well as sophisticated optimization schemes to prevent the algorithms from being trapped in local minima. Another freely available and widely used registration tool is based on Collignon et al. (1995) and distributed within the SPM software package (Ashburner and Friston, 1999). In this paper, we use these two programs as standards to evaluate the accuracy and robustness of our technique.
Instead of applying a rigid or affine transformation model, more recent research in image registration has focused on nonlinear warps, which typically depend on an initial affine alignment. Non-linear models include higher-order polynomials (Woods et al., 1992, 1998), thin-plate splines (Bookstein, 1989, 1991), B-splines (Unser et al., 1993; Kostelec et al., 1998; Rueckert et al., 1999; Kybic et al., 2000), discrete cosine basis functions (Ashburner and Friston, 1997; Ashburner et al., 1997), linear elasticity (Navier-Stokes equilibrium) (Bajcsy and Kovavcivc, 1989; Gee et al., 1993) and viscous fluid approaches (Gee et al., 1993; Christensen et al., 1994). Specifically a method described in Periaswamy and Farid (2006) presents promising results. It is based on a linear model in a local neighborhood and employs the expectation/maximization algorithm to deal with partial data. Similar to our approach, it constructs a weighted least squares solution to deal with outlier regions, however, with an underlying globally non-linear (and usually asymmetric) transformation model.
Several inverse consistent approaches exist for nonlinear warps. Often both forward and backward warps are jointly estimated, e.g. (Christensen et al., 2001; Zeng and Chen, 2008). Others match at the midpoint (Beg and Kahn, 2007) or warp several inputs to a mean shape (Avants and Gee, 2004). Yeung et al. (2008) describe a post processing method to create a symmetric warp from the forward and backward warp fields.
While nonlinear methods are often capable of creating a perfect intensity match even for scans from different subjects (change information is stored in the deformation field), it is not trivial to model and adjust the parameters of these algorithms, in particular the trade-off between data matching and regularization. In addition, it is worth noting that perfect intensity matching does not guaranty accurate correspondence. These methods need to be designed to allow the warp enough freedom to accurately match the data while restricting the algorithm to force the warp to behave `naturally', for example preventing the merging of two gyri into one, or more simply to ensure smoothness and invertibility. Due to their robustness, transformation models with low degrees of freedom are generally better suited for tasks where no change (e.g. motion correction) or only little change (e.g. longitudinal settings) is expected. Furthermore, rigid or affine registrations are frequently used to initialize higher order warps. We therefore focus on highly accurate, low degrees of freedom, intensity based registrations in this work.
2.2. Robust Statistics
The field of robust statistics describes methods that are not excessively affected by outliers or other model violations. Classical methods rely heavily on assumptions that may not be met in real applications. Outliers in the data can have a large influence on the results. For example, the mean is influenced arbitrarily by a single outlier, while the median is robust and stays fixed even with outliers present. That is why robust parameter estimation plays an import role in computer vision applications (see e.g. Stewart (1999)).
A measure for robustness is the breakdown point that describes the fraction of incorrect (arbitrarily large) observations that can be present before the estimator produces an arbitrarily large result. The breakdown point of the mean is 0 while for the median it is 0.5, which is the maximum attainable, as for values above one half, it is impossible to distinguish between the correct and the contaminating distribution.
M-estimators are a generalization of maximum likelihood estimators (MLEs) and were introduced by Huber (1964). Instead of computing the estimator parameter θ minimizing for a family of probability density functions f of the observations x1 … xn as done for MLEs, Huber proposed to minimize any general function ρ:
| (1) |
The mean, for example, minimizes the sum of squared errors, so ρ(xi, θ) := (xi − θ)2 (where “:=” means “define”). The median can be understood as an M-estimator minimizing the sum of absolute errors ρ(xi, θ) := |xi − θ|. Since most commonly used ρ can be differentiated, the solution can be computed by finding the zeros to Σψ(xi, θ) with ψ(xi, θ) := ∂ρ(xi, θ)/∂θ. For most ρ and ψ no closed form solutions exist and iterative methods are used for the computations. Usually an iteratively reweighted least squares (IRLS) algorithm is performed (see next section).
A specific ρ used often in robust settings is the Tukey's biweight function (see Figure 2):
| (2) |
For small errors the biweight is similar to the squared error, but once a specific threshold c is reached it flattens out. Therefore large errors of outliers do not have an arbitrarily large influence on the result. Often the (scaled) derivative of ρ:
| (3) |
is referred to as the Tukey's biweight function, as it is used in the actual computations.
Figure 2.
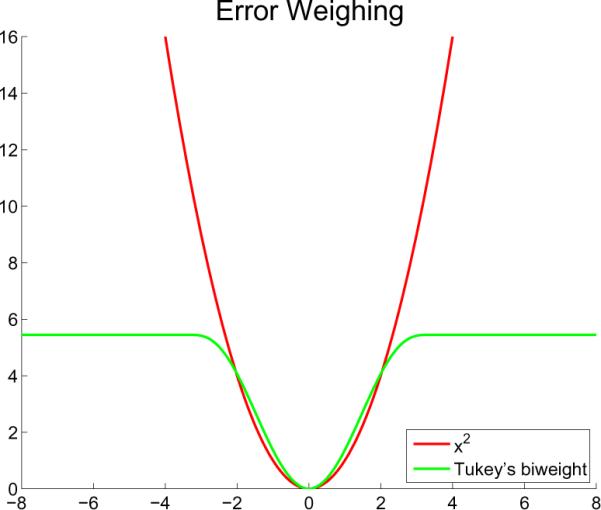
The robust Tukey's biweight function (green) limits the influence of large errors as opposed to the parabola (red).
To further highlight the difference between the robust and least squares approach Figure 3 depicts the distribution of the residuals after a successful registration (zoom-in into the histogram of residuals normalized by the number of voxels). For least-squares registration, the ideal residuals would be Gaussian noise, and in fact most residuals are around zero (the high peak there is cut off by the magnification). However, due to true differences in the images caused by distortion and anatomical change, larger residuals exist that cannot be explained by Gaussian noise models. These regions have extremely low probability under the Gaussian model (red curve in Fig. 3), which causes them to have a disproportionately large influence on the registration. As mentioned above, even a single large outlier can have an arbitrarily large effect on the result of the least-squares registration that is only optimal for zero-mean, unit variance Gaussian noise. Together with the residual distribution Fig. 3 shows two curves: where f (x) is either the parabola x2 (red) or the Tukey's biweight function ρ(x) (green). It can be seen that the parabola results in the Gaussian (red curve) and cuts off the tails significantly while the green function produced by the Tukey's biweight better models the larger residuals.
Figure 3.
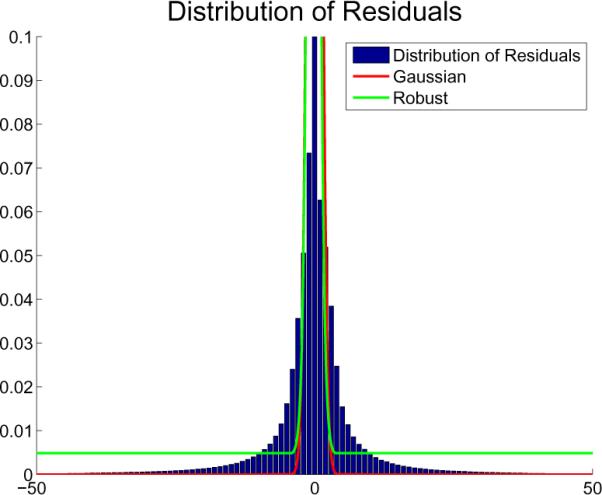
Distribution of residuals after successful registration together with the Gaussian (red) and robust (green) models (produced by the two functions from Fig. 2).
2.3. Iteratively Reweighted Least Squares
Consider a linear regression model with design matrix A and N observations in vector :
| (4) |
The M-estimator then minimizes the objective function
| (5) |
where vector is the i-th row of the matrix A. When using least-squares estimation we obtain the standard least squares linear regression solution, which can be solved directly. For a general ρ with derivative ψ := ρ′ one proceeds by differentiating the objective function (with respect to ) and by setting the partial derivatives to zero:
| (6) |
when setting the weights . These equations describe a weighted least squares problem that minimizes . Since the weights depend on the residuals ri, which in turn depend on the estimated coefficients (which depend on the weights), an iteratively reweighted least-squares algorithm is used. It selects an initial least-squares estimate (all weights equal to one), then calculates the residuals from the previous iteration and their weights, and then solves for a new weighted-least-squares estimate:
| (7) |
with the current weight matrix in iteration (j) (wi depends on the parameter vector ). These iterations are continued until a maximum number of iterations is reached or until the total squared error:
| (8) |
cannot be reduced significantly in the next iteration. It should be noted that the residuals are normalized before computing the weights in each step:
| (9) |
σ is a robust estimator for the standard deviation obtained by a scaled version of the median absolute deviation (MAD):
| (10) |
where the median is taken over all elements, i, j = 1, …, N.1 Fig. 4 shows a zoom-in of the distribution of residuals (blue) as presented in Fig. 3 of two images after successfull registration. Here also the distribution of the weighted residuals (wiri) is shown in green. It can be seen that the weights reduce the tail (large residuals) significantly.
Figure 4.
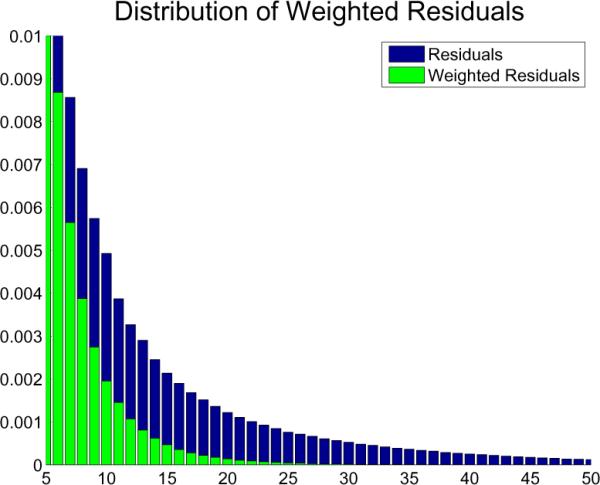
Zoom-in of the residual distribution of Fig. 3 with weighted residual distribution overlayed in green. It can be seen that the heavy tails are significantly reduced when using the robust weights.
3. Robust Symmetric Registration
As described above, the first step in constructing a robust simultaneous alignment of several images into an unbiased common space for a longitudinal study or for motion correction, is to register two images symmetrically. To avoid any bias, the resulting registration must be inverse consistent, i.e., the same registration (inverse transformation) should be computed by the algorithm if the time points are swapped.
3.1. Symmetric Setup
We first describe our symmetric gradient based image registration setup. Instead of understanding the registration as a local shift of intensity values at specific locations from the source to the target, we transform both images: the source IS half way to the target IT and the target half way in the opposite direction towards the source. The residual at each voxel is
| (11) |
where is the intensity at voxel location is the local displacement from source to target and depends on the spatial parameters . This setup is symmetric in the displacement. We will explain later how an intensity scale parameter can be incorporated.
When applying a small additive change to the n parameters in vector we can write the result using a first order Taylor approximation
| (12) |
Since there is one such equation at each voxel, it is convenient to write this in matrix form (a row for each voxel):
| (13) |
We will call the design matrix containing the partial derivatives the A matrix. For N voxels and n parameters it is a N × n matrix. In the following we will simply refer to the residuals to be minimized as and the observations at the current location . Thus, equation (13) can be written .
The goal is to find the parameter adjustments that minimize Σρ(ri), which can be achieved with iteratively reweighted least squares (cf. Section 2.3). Choosing the Tukey's biweight function ρ will prevent the error from growing without bound. This will filter outlier voxels, and at the end of the iterative process we obtain the robust parameter estimate and the corresponding weights, which identify the regions of disagreement.
What remains is to set up the design matrix A, i.e. to compute the partial derivatives of (Eq. 11):
| (14) |
Here DI = (I1 I2 I3) denotes a row vector containing the partial derivatives of the image I in the three coordinate directions. The vector , the derivative of the displacement for each parameter pi, will be described in the following section. This formulation allows us to specify different transformation models (the ), that can easily be exchanged.
Note that common symmetric registration methods (Frackowiak et al., 2003) need to double the number of equations to set up both directions. They solve the forward and backward problem at the same time. In our approach this is not necessary, due to the symmetric construction detailed above. However, a symmetric setup like this is not su cient to guarantee symmetry. The full algorithm needs to be kept symmetric to avoid treating the source image differently from the target. Often, for example, the source is resampled to the target in each iteration, which introduces a bias. We describe below how to keep the algorithm symmetric by mapping both images into a halfway space to ensure that they are treated in the same manner, with both images being resampled into the symmetric coordinate system.
3.2. Transformation Model
This section describes some possible transformation models (for background see e.g. Frackowiak et al. (2003)). Depending on the application, different degrees of freedom (DOF) are allowed. For within subject registration, 6 DOF are typically used to rigidly align the images (translation and rotation) across different time points or within a session for the purpose of motion correction and averaging of the individual scans. To align images of different subjects to an atlas usually 12 DOF transforms (affine registrations) or higher dimensional warps are used. However, even in higher-dimensional approaches, a linear registration is often computed for initial alignment. In the next paragraphs we will describe how to implement a transformation model with up to 12 DOF.
Generally the displacement can be seen as a function of the n dimensional model parameter vector into (for a fixed location ). Here is assumed to be linear in the parameters (or it has to be linearized) and can be written as
| (15) |
where M can be seen as a 3 × n Jacobian matrix containing as columns the partials , needed in the construction of the design matrix A (see Eq. 14). In the following paragraphs we will compute these Jacobians M for the affine (MA) and the rigid (MRT) case. Note also that the displacement is not equivalent with the transformation T, but it is the amount of which a location is displaced, so .
The affine 12 DOF displacement is given by a translation vector and a 3 × 3 matrix:
| (16) |
It is straightforward to construct a transformation matrix (in homogeneous coordinates) from these parameters:
| (17) |
For the rigid case, we can restrict this transform, to only allow rotation and translation. However, for small rotation it is more convenient to use the cross product to model the displacement of a rotation around the vector (p4, p5, p6)T by its length in radians:
| (18) |
Note that this model is used to compute the values p4…p6 in each step. It is not used to map the voxels to the new location as small amounts of stretching could accumulate. To construct the transformation, only the translation and the rotation around the vector (p4, p5, p6)T by its length are considered. With , , and (a unit quaternion) we obtain the transformation matrix T:
| (19) |
After specifying the displacement model, we can plug it into equation (14) and obtain the matrix equation:
| (20) |
3.3. Intensity Scaling
Images can differ in both geometry and intensity in longitudinal settings. If the assumption that a set of images share an intensity scale is violated, many intensity based registration algorithm can exhibit degraded accuracy. Often a pre-processing stage such as histogram matching (Mishra et al., 1995; Nestares and Heeger, 2000) is employed. An alternative to preprocessing the images is to utilize a similarity measure that is insensitive to scalings of intensity such as mutual information or entropy. Due to difficulties when estimating geometric and intensity changes simultaneously only a few exceptions such as Woods et al. (1992, 1998); Ashburner and Friston (1997); Ashburner et al. (1997); Periaswamy and Farid (2006) incorporate explicit models of intensity differences obviating the need for complex intensity pre-processing.
We can easily incorporate a global intensity scale parameter s into our model in a symmetric fashion. First the intensity scale factor is applied to both source and target to adjust their intensities to their geometric mean:
| (21) |
Recall that the additive spacial displacement was kept symmetric by adding half the displacement to the source and half of the negative displacement to the target, to move both to towards a common half way space. The intensity scale factor is multiplicative, so instead of simply multiplying the source image's intensities by s we scale them by and the target by to map both images to their intensity (geometric) mean. This keeps the residual function symmetric with respect to the intensity scaling factor in addition to the symmetric displacement setup.
For the approximation, the corresponding partial derivative is added in the Taylor approximation:
| (22) |
Thus, in order to incorporate intensity scaling, one simply appends s to the parameter vector and attaches a column to matrix A, containing the partial derivative of the vector with respect to s:
| (23) |
4. Registration Algorithm
The algorithm consists of the following steps:
Initialize Gaussian Pyramid: by subsampling and smoothing the images.
Initialize Alignment: compute a coarse initial alignment using moments at the highest resolution.
Loop Resolutions: iterate through pyramid (low to high resolution):
-
Loop Iterations: on each resolution level iterate registration to obtain best parameter estimate. For each iteration step:
-
(a)
Symmetry: take the current optimal alignment, map and resample both images into a half way space to maintain symmetry.
-
(b)
Robust Estimation: construct the overdetermined system (Eq. 20) and solve it using iteratively reweighted least squares to obtain a new estimate for the parameters.
-
(a)
Termination: If the difference between the current and the previous transform is greater than some tolerance, iterate the process at this resolution level up to a maximal number of iterations (Step 4), otherwise switch to the next higher resolution (Step 3).
The above algorithm will be described in more detail in the following sections.
4.1. Gaussian Pyramid (Step 1)
Since the Taylor based registration can only estimate small displacements, it is necessary to employ a multiresolution approach (Roche et al., 1999; Hellier et al., 2001), together with an initial alignment (see next section). As described in Nestares and Heeger (2000) we construct a Gaussian pyramid, bisecting each dimension on each level until the image size is approximately 163. We typically obtain about 5 resolution levels with a standard adult field-of-view (FOV) for an MRI image that is approximately 1mm isotropic (i.e. an FOV of 256mm). First a standard Gaussian filter (5-tab cubic B-Spline approximation)
| (24) |
is applied in each direction of the image, which is then subsampled to the lower resolution. These pyramids (source and target) need to be constructed only once for the entire process.
4.2. Initial Alignment (Step 2)
In order to speed up the registration and increase its capture range, an initial coarse alignment is constructed using moments. Geometric moments have proven to be an efficient tool for image analysis (Del Bimbo, 1999). For a grayscale image with pixel intensities I(x1, x2, x3), the raw image moments Mijk are calculated by
| (25) |
where i, j, k are the exponents of the coordinates x1, x2, x3 respectively (taking the values 0 or 1 in the following equation). The centroid of an image can be derived from the raw moments:
| (26) |
We compute the translation needed to align the centroids and use it by default as an initial transformation to ensure overlapping images when starting the robust registration algorithm. Furthermore, it is possible to use central moments as defined below to compute an initial rotational alignment. For full head images with possibly different cropping planes, such a rotational pre-alignment can be very inaccurate and should therefore only be used when aligning skull stripped images. Central moments are defined translation invariant by using the centroid (Eq. 26):
| (27) |
The covariance matrix of the image I can now be defined using :
| (28) |
The eigenvectors of the covariance matrix correspond to the three principal axes of the image intensities (ordered according to the corresponding eigenvalues). These axes are then aligned for two images. Care needs to be taken to keep the correct orientation. This is achieved by flipping the first eigenvector if the system has left-handed orientation. Even if both systems are right-handed, it can still happen that two of the axes are pointing in the opposite direction, which can be detected and fixed by projecting each axis onto its corresponding axis in the other image and flipping it if necessary. If the angle between the corresponding axes is too large, the correct orientation cannot be determined without additional information and the initial rotational alignment is not performed. Note that initial moment based orientation alignment was never necessary and therefore not used in any of our tests, since head MRI images are usually oriented similarly.
4.3. Loops (Step 3)
There are three nested loops in the registration algorithm: the different resolutions of the pyramid (step 3), several iterations on each level (remapping the images (step 4), and finally the iteratively reweighted least squares algorithm for the robust parameter estimation (inside step 4(b), see Section 2.3). Note, when switching form a lower to a higher resolution in step 3, the translational parameters need to be adjusted (scaled by 2) when given in voxel coordinates.
4.4. Registration (Step 4)
On each resolution level there are several iterations of the resampling and robust parameter estimation as explained next.
4.4.1. Half Way Space (Step 4a)
The registration model (Eq. 11) is designed to maintain symmetry in the algorithm, however we must also ensure that all steps are performed similarly for both images. Therefore it is not sufficient to map the source to the target in each iteration and re-estimate the new parameters. In such a setup only the source would be resampled at (or close to) the target location while the target would not go through the resampling process. In order to avoid this asymmetry, which can introduce biases due to the arbitrary specification of source and target, we propose to resample both images to the half way space in each iteration step.
For a given transformation T from source to target the half way maps are constructed by approximating the square root of the matrix T (here T is again assumed to be a 4 × 4 matrix in homogeneous coordinates). For a positive definite matrix T (we don't allow reflections and projections) there exists exactly one positive definite matrix with . For its computation we use the Denman-Beavers square root iteration (Denman and Beavers, 1976; Cheng et al., 2001): Let Y0 = T and Z0 = I, where I is the identity matrix. The iteration is defined by
| (29) |
The matrix Yk converges quadratically to the square root , while Zk converges to its inverse, . Once has been approximated, the source image is mapped to and the target to (to ensure both get resampled at the same location). For the resampling process tri-linear interpolation is used, although other interpolation algorithms can easily be employed. Note that to maintain symmetry the square root iteration should only be stopped when the largest element of is sufficiently small.
4.4.2. Robust Estimation (Step 4b)
To set up the robust estimation problem (Eq. 20), the partial derivatives and a smoothed version of both images need to be computed. Smoothing is used to prevent the algorithm from being trapped in a local minimum. For smoothing we apply a Gaussian kernel in each image direction (Nestares and Heeger, 2000):
| (30) |
The smoothed derivatives can be computed by applying
| (31) |
in the direction of the derivative and the smoothing kernel in the two other directions. Once the image derivatives DI are computed, the matrix A and vector can be constructed (see Eq. 20). If the matrix gets too large, it is often sufficient to subsample the image at the highest resolution and only select every second voxel. As the derivatives and intensity information are selected from the high resolutional image the result will still be more accurate than stopping at the previous lower resolution level in the Gaussian pyramid. For further improvement stochastic sampling algorithms can be employed to avoid aliasing. Subsampling specific regions more densely than others (e.g. depending on gradients, edges or the outlier weights) is also likely to improve accuracy. Our tests, however, show very accurate results even with the simple subsampling algorithm.
Once the system has been constructed, the iteratively reweighted least squares algorithm (Section 2.3) is employed to compute the new parameters and weights. For this reason, the saturation parameter c of the Tukey's biweight functions must be specified. In Nestares and Heeger (2000) a constant saturation value c = 4.685 is recommend (suggested for Gaussian noise in Holland and Welsch (1977)). However, a fixed value cannot adjust well to different image contrast types and SNR levels, such as non-linear deformations or larger intensity differences. In these cases it can happen that the registration fails as too many voxels are considered outliers. Therefore in order to reduce the number of detected outliers particularly in the brain, it is necessary to find a less sensitive (i.e. larger) value in these cases. The user can always adjust this parameter according to the specific image situation. For full head scans, however, we developed a method that automatically estimates the sensitivity parameter. It also works remarkably well in brain-only registrations. For full head images, a global limit on the number of outlier voxels will not be a good measure, as large outlier regions especially at the skull, jaw and neck should be permitted. The following outlier measure uses a Gaussian to weigh voxels at the center of the image more strongly than voxels further away (see also Figure 5):
| (32) |
where di is the distance of voxel i to the center. W is zero iff (if and only if) all weights wi are one (meaning no outliers). A large W means that many voxels in the center of the image are labeled outlier. In that case the saturation is automatically incremented and W recomputed until W < Wthresh. All of this can be computed quickly on a lower resolution level (we choose the third highest level, i.e. for a 2563 image this is 643). The threshold Wthresh will be discussed and determined in Section 5.4 below. Note that in situations with significant outliers in the center, a global unweighted threshold can be used instead or the sensitivity parameter can be adjusted manually.
Figure 5.
Gaussian filter at the center .
4.5. Termination (Step 5)
In order to measure how much a new parameter estimate differs from the last iteration, the root mean square (RMS) deviation of the two corresponding transformations is computed. This measure will also be used to assess the quality of a registration when compared to some ground truth. The RMS deviation measures the average difference of voxel displacements inside a spherical volume for two given affine transformations and , where M1, M2 are two 3 × 3 linear transformation matrices and t1, t2 the corresponding 3×1 translation vectors. The RMS error for a spherical volume with radius r is then given by:
| (33) |
where tr is the trace (see Jenkinson (1999) for the derivation). An average displacement error is used as a quality measure for a transformation instead of, for example, the maximum displacement because it depends on all voxels contained in the sphere instead of possibly only a single voxel. The misalignment of a single voxel is not very important if the rest of the image is aligned accurately. While a translation has an equally strong effect everywhere, a rotation, for example, shifts voxels different distances depending on the distance to the rotation center. For a translation of 0.1mm (and 1mm3 voxels) both maximum displacement and average displacement are the same ERMS = 0.1. Such a displacement can easily be seen on the screen when switching between the images. Even ERMS of 0.05 and below can be noticed when magnifying the images. These displacements, however, are too small to visualize in a printed version of the image (e.g. checkerboard).
In this work the RMS error is measured on the transformations defined in RAS (right, anterior, superior) coordinates with the origin located approximately at the center of the image. The radius of the spherical volume is set to r = 100 which corresponds to 100mm, enough to include the full brain. The iterations of the parameter estimation are usually terminated once ERMS < 0.01 i.e. the average displacement consecutive estimates is below 0.01mm, which is very restrictive. To avoid long runtimes in ill conditioned cases, a maximum number of iterations can also be specified by the user (the default is 5).
5. Results
This section presents results quantifying the accuracy and robustness of the robust registration in comparison to other commonly used methods. As mentioned above, the robust registration is capable of ignoring outlier regions. This can be verified when checking the weights during a successful registration, as shown in Figure 6. The top images show the (enhanced) differences between the target and registered source. The regions that contain the strongest differences are correctly detected as outliers as can be seen in Figure 6 (bottom), where the weights are overlayed (red/yellow regions). Note that the weights range from 0 to 1 and are blurry (because they are computed on smoothed images), so they can blur in from neighboring slices.
Figure 6.
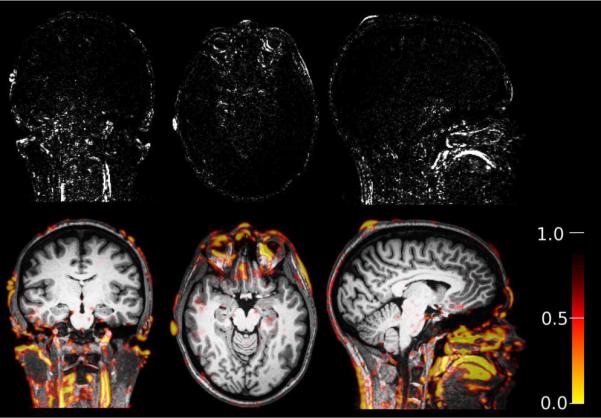
The red/yellow regions (bottom row) are detected as outlier regions during the registraion procedure of this Multiecho MPRAGE test-retest data. Their influence is automatically reduced. It can be seen that the detected outliers agree with the non-rigid differences after successful registration (top row) located mainly in the neck, eye, scalp and jaw/tongue region; see also Fig. 1.
Figure 7 (left) is an example of an image that is misaligned using FLIRT with the mutual information similarity function. The visible edges in the brain regions indicate an alignment error. Figure 7 (right) shows the differences when using the robust registration, where clearly less strong edges are visible. The remaining residuals are due to resampling and noise. Figure 7 bottom shows a magnification of the target (red) and registered source (green) on top of each other. The red and green edges (left) at the ventricle and temporal lobe indicate misalignment while yellow regions are accurately aligned (right). The difference between the two transforms here is ERMS = 0.88, almost one voxel on average.
Figure 7.
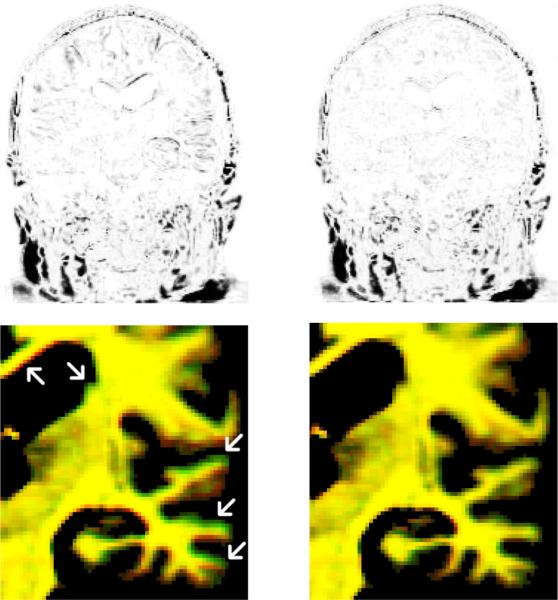
Difference after alignment. Left: FLIRT MI (the visible structures in the brain indicate misalignment). Right: Robust method (accurate alignment, residual differences due to noise and resampling). The top shows the difference images and the bottom a zoom-in into the aligned target (red) and source (green). A good alignment should be yellow (right) while the inaccurate registration shows misaligned red and green edges (left).
In the following sections we analyze the performance of different registration tools. We use the RMS deviation of two transformations as described in Section 4.5 to quantify the distance (the error) of a computed transformation with respect to some ground truth transformation. For the following tests we use a set of 14 healthy subjects each with two scans 14 days apart. The images are MPRAGE T1 weighted full head scans (on Siemens Sonata 1.5T) and are resampled to 2563 voxels each with 1mm side length (original dimensions 256×256×128 with 1mm ×1mm ×1.33mm voxels).
5.1. Inverse Consistency
Since this algorithm is intended to compute inverse consistent registrations, we need to verify experimentally that the final transforms are exactly inverses of each other when switching source and target. For each subject we register the image from time point 2 to the time point 1 and vice versa while comparing the obtained transforms of several common registration algorithms. We compare the robust registration (with different parameter settings) with the FLIRT registrations (Jenkinson et al., 2002) from the FSL suite (Smith et al., 2004) using different cost functions: standard least squares [FLIRT-LS], correlation ratio [FLIRT-CR], mutual information [FLIRT-MI]. Furthermore, we compare with a registration tool from the SPM software (Ashburner and Friston, 1999) based on Collignon et al. (1995) [SPM]. The robust variants are: [Robust] robust rigid registration (no intensity scale), [Robust-I] with intensity scaling, and [Robust-I-SS] with additional sub-sampling at the highest resolution. We also include our implementation with standard least squares [LS] instead of the robust Tukey's biweight error function to see the effect of the robust estimation with no other differences in the algorithm. In Figure 8 the RMS deviation of the forward and inverse backward transforms are computed and compared for different image types as used in the FreeSurfer software package: full head scans (orig), intensity normalized images (T1) and normalized skull stripped images (norm).
Figure 8.
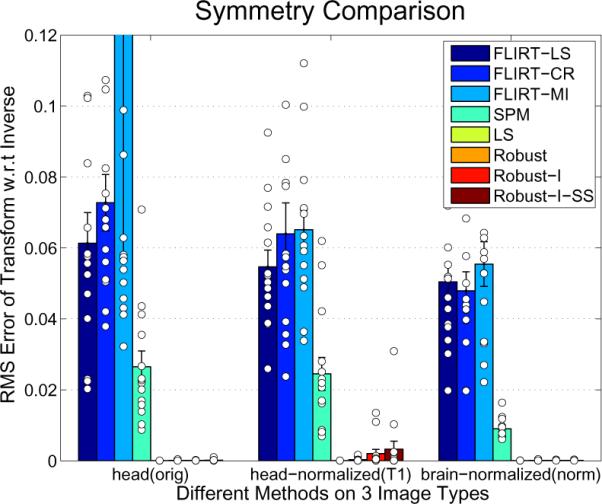
Comparison of inverse consistency using different methods: FLIRT LS: least squares, CR: correlation ratio, MI: mutual information, SPM, LS (our implementation with least squares instead of robust error function), Robust registration, Robust-I (+intensity scaling) and Robust-I-SS (subsampling on the highest resolution). The white circles represent the individual registrations.
The FLIRT registrations perform similarly. The higher mean in the mutual information method on the orig images is due to a single outlier (ERMS = 2.75). It can be seen that the robust registration methods are extremely symmetric, even with intensity scaling switched on, adding another degree of freedom and a higher chance for numerical instabilities. Also our non-robust method [LS] with the standard least squares error function is perfectly symmetric in all cases. This test, however, does not tell us anything about the accuracy of the registration.
5.2. Tests Using Synthetic Data
In this section we present results using images that were transformed, intensity scaled and otherwise manipulated with known transformations, which then can be used as ground truth. We compare how well several registration algorithms perform on the same test set of the 14 MPRAGE T1 weighted full head scans (Siemens Sonata 1.5T) of the same healthy subjects. The registration methods are the same as in the previous section (FLIRT, SPM and robust registration).
A random rigid transformation (rotation, translation) was computed for each image. The parameters were chosen in a way that reflects possible (large) head movements in a scanner: 50mm translation in a random direction together with a random rotation of 25 degrees around an arbitrary axis with the origin at the center of the image. The maximum displacement of a corner of theses image was between 130mm and 140mm. The parameters were chosen, so that all methods can find approximate solutions. For larger transformations [SPM] was no longer capable of recovering the correct registrations at all, while the robust methods performed perfectly (not shown) in tests up to 100mm translation and 40 degrees rotation. These transformations move the images apart so that there is almost no overlap, furthermore parts of the face, skull, neck and jaw can be cropped because they are mapped outside the field of view. The robust approach can deal well with this kind of partial matching. Moreover, we believe that, due to the multiresolution algorithm and the initial moment based alignment, even larger transformations will be recovered accurately.
For the synthetic registration comparison, the transform that represents half the random translation and rotation is used to map and resample each image at the target location and the inverse is applied to map and resample each image at the source location. This ensures that both images (source and target) will be resampled and do not move outside the field of view as easily. An accurate registration from source to target needs to be close to the original random transform. The accuracy is measured using the RMS deviation (see Section 4.5) of the ground truth and the computed transformation matrix. Four different tests were performed. In all cases random rigid motion was applied (as described above):
Only-Motion: only random rigid motion.
Noise: significant Gaussian noise was added with σ = 10 (Figure 9 middle).
Outlier-Boxes: 80 boxes (each box 303 voxel) were created and copied from a random location to another random location within the same image, with 40 boxes each in source and target (Figure 9 right).
Intensity: global intensity scaling (±5%) was performed.
Figure 9.
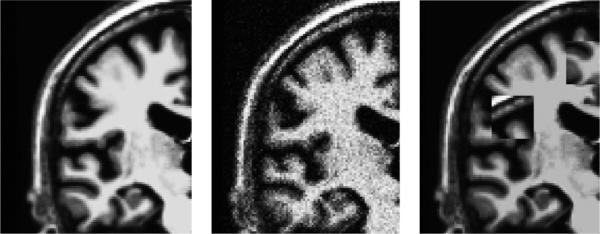
Close-ups of test images: original (left) with Gaussian noise σ = 10 (middle) and with outlier boxes (right).
The results of this experiment are given in Figure 10. It can be seen that the robust version outperforms the three different FLIRT similarity functions in all tests. [SPM] yields similar accuracy, but fails completely for larger transforms (not shown). The robust registration shows almost no influence of the outlier boxes since these are accurately detected as such. There is only little influence of noise. However, when the global intensity scale is different in the two images, the robust registration methods needs 7 DOF (one additional intensity scale parameter: [Robust-I]) to maintain accuracy, because it strongly depends on similar intensity levels. This underlines the importance of incorporating automatic intensity scaling into the robust registration method. Subsampling on the highest resolution in the robust registration [Robust-I-SS] leads to a significant reduction in memory and run time, but still yields the same registration accuracy in these tests. The simple non-robust implementation LS performs poorly in most cases.
Figure 10.
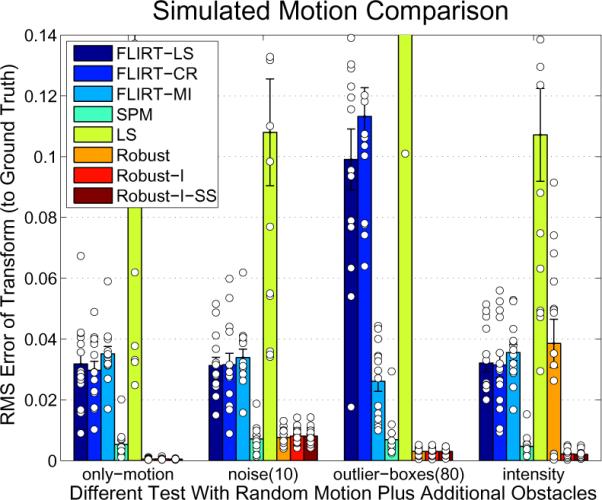
Accuracy of different methods (see Fig. 8). The four different tests are: random rigid motion, additional Gaussian noise (σ = 10mm), 80 boxes of outlier data and intensity scaling.
It should be noted that the FLIRT methods produce a few individual registrations with low accuracy when outliers or noise are present (as can be seen by checking the scatter data, the small circles in Figure 10, some are too large and not shown). The SPM method on the other hand produces quite accurate results in most test cases. However, as mentioned above it fails completely for larger transformations.
5.3. Tests Using Real Data
In contrast to the simulations with available ground truth transformations we do not know the correct registration in advance in typical registration paradigms. Therefore we need to establish a different performance metric. This can be achieved by registering the skull stripped and intensity normalized images of a test-retest study (two time points) with different registration methods. These registrations are highly accurate as the images contain only brains of healthy normals and only small changes in the brain are involved (e.g. noise etc.). In these well behaved situations the registration of these brain images computed by the different algorithms deviate from each other only by small amounts. The goal here is to find registrations of the corresponding full head images that are as close as possible to the brain-only, intensity normalized registrations.
The group chosen for this test is the same as described above. This test will be more noisy as the `ground truth' is already defined inaccurately. Figure 11 (left) shows the distances of all other methods to the SPM registration of the skull stripped normalized image (norm). It can be seen that compared to the full head registrations, the norm registrations are on a similar low level for all methods (SPM has of course zero distance to itself). SPM has been chosen to construct the ground truth registration of the norm images, as it performed more accurately than the FLIRT methods in the previous tests. We did not choose a robust registration to establish the ground truth to not favor our method. However, we tested establishing the `ground truth' with any other method which leads to very similar results and almost exactly the same plots.
Figure 11.
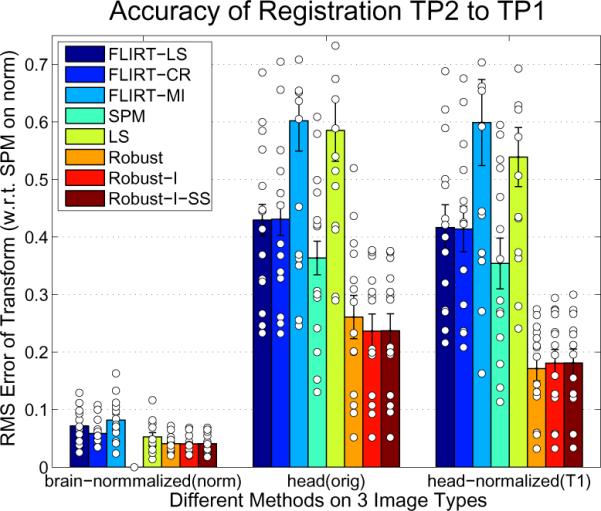
Accuracy of different methods (see Fig. 8) with respect to SPM (on the norm images).
The results on the full head (orig) image (Figure 11 middle) and intensity normalized full head (T1) image (Figure 11 right) evidence behavior that is similar to the previous tests. SPM performs (here only slightly) better than the FLIRT methods, while the robust registration yields the most accurate results. As expected for the orig images intensity scaling [Robust-I] improves the registrations further, while for the normalized T1 images it is not necessary. Again subsampling [Robust-I-SS] on the highest resolution reaches the same accuracy, indicating that the expensive iterations on the highest resolution level can be avoided.
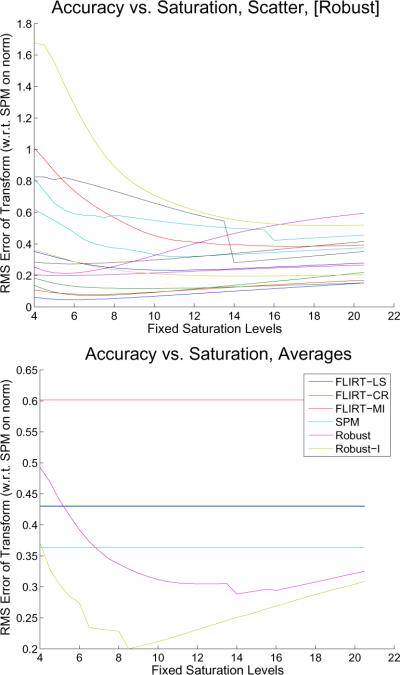
5.4. Parameter Estimation
As described in Section 4.4.2 a fixed saturation level c cannot be recommended for all image types. The value c = 4.685 from Nestares and Heeger (2000) will lead to erroneous registrations in many common settings. Figure 12 (top) shows the accuracy of each robust registration of the orig images plotted vs. the selected saturation level. For some subjects the optimal registration is reached at c ≈ 6 while other need a higher value c ≈ 15. For the normalized T1 images or for [Robust-I] (with intensity scaling enabled) the results look similar (not shown), however with individual minima spread between c = 4 and c = 9. When using a fixed saturation level for all registrations, c ≈ 14 is optimal for [Robust] with an average RMS error of slightly below 0.3 and c = 8.5 is optimal for [Robust-I]. Even with a fixed saturation, both robust methods are on average better than the other non-robust registration methods (cf. Figure 12 bottom).
Figure 12.
Top: Accuracy of [Robust] for each individual subject. Bottom: Mean accuracy of the methods, where [Robust] and [Robust-I] depend on the saturation level (fixed across all subjects). It can be seen (bottom) that [Robust] reaches its minimal average registration error at the fixed saturation level of c = 14 and [Robust-I] at c = 8.5. For most fixed saturation levels, both methods perform better on average than FLIRT or SPM (note, the averages of [FLIRT-LS] and [FLIRT-CR] almost coincide, compare with Fig. 11 middle).
For [Robust] without intensity scaling, a relative high saturation value (c = 14) is particularly necessary to compensate for the differences in image intensity. Lower values might label too many voxels outlier due to the intensity differences or non-linearities, resulting in misaligned images (see Figure 13 for an example). Instead of manually inspecting the outliers and registrations while determining an optimal saturation setting per image, we introduce the center focused weight measure W (Eq. 32) for full head images to indicate when too many outliers are detected in brain regions and to adjust the sensitivity accordingly. Figure 13 (bottom row) shows the same image registration, where the automatic parameter estimation results in less detected outliers and a successful alignment.
Figure 13.
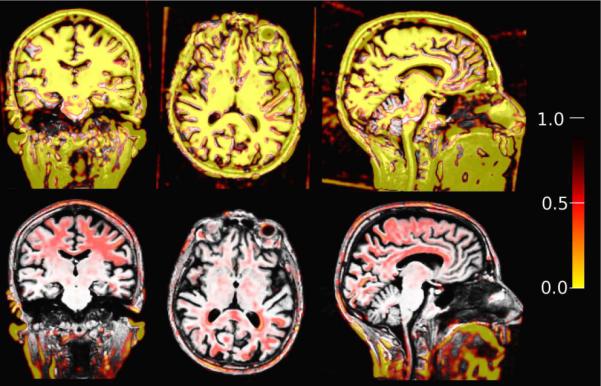
Top: Fixed low saturation of c = 4.685 (high outlier sensitivity) in a registration with intensity differences and non-linearities results in too many outlier and consequently in misalignment. Bottom: Automatic sensitivity estimation adjusts to a higher saturation value (low outlier sensitivity) to register the images successfully. The detected outlier regions are labeled red/yellow.
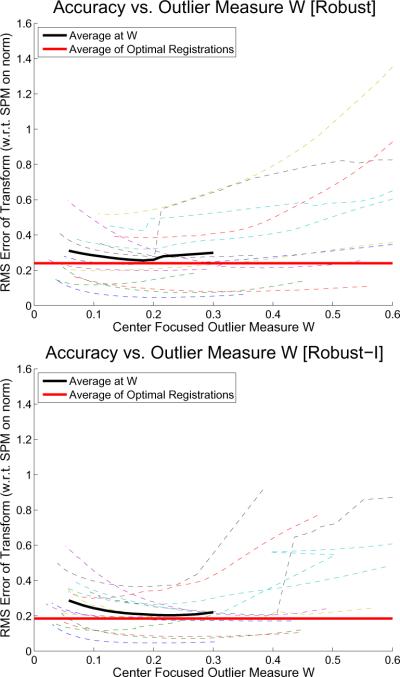
We will now determine an optimal W for the automatic saturation estimation. Figure 14 presents scatter plots of registration accuracies [Robust] and [Robust-I] on the full head (orig) images here plotted versus W. The horizontal red line shows the average minimum error when choosing the individual saturation that leads to the best registration for each subject (with respect to the ground truth). The automatic saturation estimation can almost reach this optimum by fixing the center focused weight measure W around 0.2 (see the black curve showing the average of W between 0.05 and 0.3). Additionally, W is quite robust since the average (black dashed curve) is relatively flat. Ensuring a W around 0.2 for the tested image types in the automatic saturation estimation leads to registrations that are almost as accurate as when taking the optimal result per subject (which is of course not know a priori).
Figure 14.
Registration accuracy for each subject depending on center focused weight W (Robust top, Robust-I bottom). Red horizontal line: averaging best registration per subject. Black curve: average performance at specific W. Dashed curves: individual subject's results.
5.5. Application: Motion Correction
Frequently several images of a given scan type are acquired within a session and averaged in order to increase SNR. These images are not perfectly aligned due to small head movements in the scanner (for some groups of patients there can be even large differences in head location, due to uncontrolled motion) and need to be registered first. Since not only noise but other differences such as jaw movement or motion artifacts are prevalent in these images, a robust registration method should be optimally suited to align the images while discounting these outlier regions. It can be expected that except for noise, brain tissue and other rigid parts of the head will not contain any significant differences (except rigid location changes). A misalignment of the within-session scans will of course affect the average image negatively and can reduce the accuracy of results generated by downstream processes. Therefore highly accurate registrations for motion correction are the first step, for example, towards detecting subtle morphometric differences associated with disease processes or therapeutic intervention.
To test the robust registration for this type of within-session motion correction, the two scans of the first session in the longitudinal data set presented above were selected. The second scan was registered to the first with the different registration methods. It was then resampled at the target location (first scan) and an average image was created. Since these within-session scans should show no change in brain anatomy, it can be expected that the difference between scan 1 and aligned scan 2 in brain regions will be very small and mainly be due to noise (and of course scan 2 will be smoother due to resampling). Therefore a larger difference in the brain-region between the registered images implies misalignment, most likely due to image differences elsewhere (e.g. jaw, neck, eyes) or less likely due to non-linear differences between the two scans. The gradient non-linearities will badly influence all rigid registrations similarly, while possible non-brain outlier regions will influence the employed methods differently. Therefore we will evaluate the performance of full head registration only within the brain mask.2
We first quantify the registration error and compute the sum of squared errors (SSE) of the intensity values in scan I1 and aligned/resampled scan :
| (34) |
where the sum is taken over all brain voxels . The brain masks to specify brain regions were created automatically for each subject with the FreeSufer software package and visually inspected to ensure accuracy.
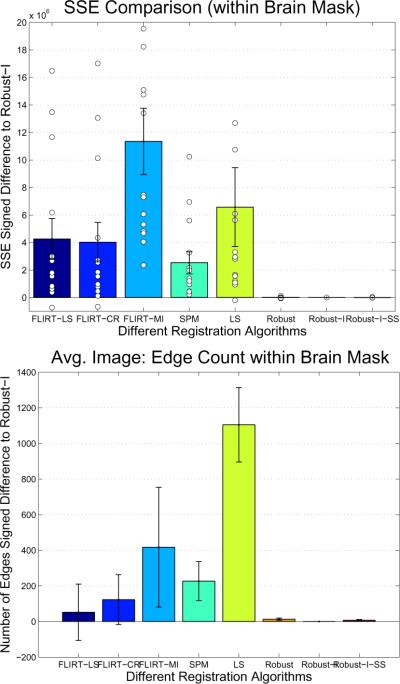
The SSE measure quantifies the intensity differences of the two images after alignment within the brain. For a perfect registration these differences should be small as they only measure noise, non-linearities and global intensity scaling (all of these should be small as the two images are from the same scan session). Figure 15 (top) shows the signed difference of SSE with respect to the result of the method [Robust-I]. The robust methods perform best on average, while [FLIRT-LS], [FLIRT-CR] and [LS] yield a better results (lower SSE) only in one single instance (white circles with negative value). To test significance of these results, we applied a Wilcoxon signed rank test (Wilcoxon, 1945) for each algorithm with respect to [Robust-I] to test if the median of the pairwise differences is equal to zero (null hypothesis). This is similar to the t-test on the pairwise differences, without the assumption of normally distributed data. We found that all non-robust methods show significant differences from [Robust-I] at a p < 0.001 while the null hypothesis cannot be rejected within the robust methods, as expected, since their performance is basically the same.
Figure 15.
Error of motion correction task in brain region for different registration methods (top: sum of squared errors comparison, bottom: edge count of average image). Both plots show the signed difference to Robust-I.
In order to test if differences can be detected in the resulting average images, we count the number of edges. Correctly aligned images should minimize the number of edges since all edges will be aligned, while misalignment increases the edge count. The edges were detected by scanning the x component of the gradient (using the Sobel filter) in the x directions and counting local maxima above a threshold of 5. Figure 15 (bottom) shows that the misalignment increases edge count on average when compared to [Robust-I]. However, due to the large variance the FLIRT results are not significant. [SPM] is significantly different at level p = 0.058 and [LS] at the p < 0.001 significance level in the Wilcoxon signed rank test.
6. Conclusion
In this work a robust registration method based on Nestares and Heeger (2000) is presented, with additional properties such as initial coarse alignment, inverse consistency, sensitivity parameter estimation and global intensity scaling. Automatic intensity scaling is necessary for the method to function when global intensity differences exist. Similarly the automatic estimation of the saturation parameter avoids misalignment in specific image situations where a fixed value potentially ignores too many voxels.
The presented method outperforms commonly used state-ofthe-art registration tools in several tests, and produces results that are optimally suited for motion correction or longitudinal studies, where images are taken at different points in time. Local differences in these images can be very large due to movement or true anatomical change. These differences will influence the registration result, if a statistically non-robust approach is employed. In contrast, the robust approach presented here maintains high accuracy and robustness in the presence of noise, outlier regions and intensity differences.
The symmetric registration model together with the `half-way' space resampling ensure inverse consistency. If an unbiased average of two images is needed, it is easily possible to resample both, target and source, at the `half-way' location and perform the averaging in this coordinate system. Furthermore, these registrations can be employed to initialize nonlinear warps without introducing a bias. Robust registration has been successfully applied in several registration tasks in our lab, including longitudinal processing and motion correction. The software is freely available within the FreeSurfer package as the mri_robust_register tool.
Future research will extend the presented registration to more than two images and incorporate these algorithms into a longitudinal processing stream, where more than two time points may be involved. In those settings instead of simply registering all images to the first time point, it is of interest to create an unbiased template image and simultaneously align all input images in order to transfer information at a specific spatial location across time. Similar to the idea in (Avants and Gee, 2004) it is possible to estimate the unbiased (intrinsic) mean image and the corresponding transforms iteratively based on the pairwise registration algorithm described in this paper.
7. Acknowledgements
Support for this research was provided in part by the National Center for Research Resources (P41 RR14075, and the NCRR BIRN Morphometric Project BIRN002, U24 RR021382), the National Institute for Biomedical Imaging and Bioengineering (R01 EB006758), the National Institute on Aging (R01 AG022381,U54 AG024904), the National Institute for Neurological Disorders and Stroke (R01 NS052585, R01 NS042861, P01 NS058793). Additional support was provided by The Autism & Dyslexia Project funded by the Ellison Medical Foundation. The authors would like to thank Dr. Greg Sorensen for kindly supplying the tumor sample data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The constant is a necessary bias correction. The MAD alone estimates the 50% interval ω around the median rm of the distribution of r: P(|r − rm| ≤ ω) = 0.5. Under normality ω = 0.6745 σ ⇒ σ = 1.4826 ω.
In some applications it might be better to compute registrations on skull stripped brains directly. However automatic skull stripping is a complex procedure, and frequently needs the user to verify all slices manually. Furthermore, in some situations it makes sense to keep the skull, for example, when registering to a Talairach space with skull to estimate intracranial content, which depends on head size rather than brain size. Finally even skull stripped images can contain significant differences, for example in longitudinal data or simply because different bits of non-brain are included, so that the robust registration is still the best choice.
References
- Ashburner J, Friston K. Multimodal image coregistration and partitioning – a unified framework. NeuroImage. 1997;6(3):209–217. doi: 10.1006/nimg.1997.0290. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston K. Nonlinear spatial normalization using basis functions. Human Brain Mapping. 1999;7(4):254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Neelin P, Collins DL, Evans A, Friston K. Incorporating prior knowledge into image registration. NeuroImage. 1997;6(4):344–352. doi: 10.1006/nimg.1997.0299. [DOI] [PubMed] [Google Scholar]
- Avants B, Gee JC. Geodesic estimation for large deformation anatomical shape averaging and interpolation. NeuroImage. 2004;23(1):139–150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Bajcsy R, Kovavcivc S. Multiresolution elastic matching. Computer Vision Graphics and Image Processing. 1989;46(1):1–21. [Google Scholar]
- Beg MF, Kahn A. Symmetric Data Attachment Terms for Large Deformation Image Registration. IEEE Transactions on Medical Imaging. 2007;26(9):1179–1189. doi: 10.1109/TMI.2007.898813. [DOI] [PubMed] [Google Scholar]
- Bican J, Flusser J. 3D Rigid registration by cylindrical phase correlation method. Pattern Recognition Letters. 2009;30:914–921. [Google Scholar]
- Bookstein FL. Principal warps: Thin-plate splines and the decomposition of deformations. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1989;11:567–585. [Google Scholar]
- Bookstein FL. Thin-plate splines and the atlas problem for biomedical images. IPMI '91: Proceedings of the 12th International Conference on Information Processing in Medical Imaging; Springer-Verlag; 1991. pp. 326–342. [Google Scholar]
- Cheng SH, Higham NJ, Kenney CS, Laub AJ. Approximating the logarithm of a matrix to specified accuracy. SIAM Journal on Matrix Analysis and Applications. 2001;22(4):1112–1125. [Google Scholar]
- Christensen GE, Rabbitt RD, Miller MI. 3D brain mapping using a deformable neuroanatomy. Physics in Medicine and Biology. 1994;39(3):609–618. doi: 10.1088/0031-9155/39/3/022. [DOI] [PubMed] [Google Scholar]
- Christensen GE, Johnson HJ. Consistent Image Registration. IEEE Transactions on Medical Imaging. 2001;20(7):568–582. doi: 10.1109/42.932742. [DOI] [PubMed] [Google Scholar]
- Collignon A, Maes F, Delaere D, Vandermeulen D, Suetens P, Marchal G. Information Processing in Medical Imaging. Kluwer; 1995. Automated multi-modality image registration based on information theory; pp. 263–274. [Google Scholar]
- Collins D, Holmes C, Peters T, Evans A. Automatic 3-D model-based neuroanatomical segmentation. Human Brain Mapping. 1995;3(3):190–208. [Google Scholar]
- Costagli M, Waggoner RA, Ueno K, Tanaka K, Cheng K. Correction of 3D rigid body motion in fMRI time series by independent estimation of rotational and translational effects in k-space. NeuroImage. 2009;45:749–757. doi: 10.1016/j.neuroimage.2008.12.067. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis: I. segmentation and surface reconstruction. NeuroImage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Del Bimbo A. Visual information retrieval. Morgan Kaufmann Publishers Inc.; San Francisco: 1999. [Google Scholar]
- Denman ED, Beavers AN. The matrix sign function and computations in systems. Applied Mathematics and Computation. 1976;2(1):63–94. [Google Scholar]
- Dickerson B, Goncharova I, Sullivan M, Forchetti C, Wilson R, Bennett DA, Beckett L, deToledo Morrell L. MRI-derived entorhinal and hippocampal atrophy in incipient and very mild alzheimer's disease. Neurobiological Aging. 2001;22:747–754. doi: 10.1016/s0197-4580(01)00271-8. [DOI] [PubMed] [Google Scholar]
- Evans A, Marrett S, Collins D, Peters T. Anatomical-functional correlative analysis of the human brain using three-dimensional imaging systems. Proceedings of the SPIE - The International Society for Optical Engineering. 1989;1092:264–274. [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis: Ii: Inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Frackowiak R, Friston K, Frith C, Dolan R, Price C, Zeki S, Ashburner J, Penny W. Human Brain Function. 2nd Edition Academic Press; 2003. [Google Scholar]
- Gee JC, Gee JC, Reivich M, Reivich M, Bajcsy R, Bajcsy R. Elastically deforming a three-dimensional atlas to match anatomical brain images. Computer Assisted Tomography. 1993;17(2):225–236. doi: 10.1097/00004728-199303000-00011. [DOI] [PubMed] [Google Scholar]
- Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. NeuroImage. 2009;48(1):63–72. doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellier P, Barillot C, Memin E, Perez P. Hierarchical estimation of a dense deformation field for 3-D robust registration. IEEE Transactions on Medical Imaging. 2001;20(5):388–402. doi: 10.1109/42.925292. [DOI] [PubMed] [Google Scholar]
- Hill DLG, Batchelor PG, Holden M, Hawkes DJ. Medical image registration. Physics in Medicine and Biology. 2001;46(3):R1–R45. doi: 10.1088/0031-9155/46/3/201. [DOI] [PubMed] [Google Scholar]
- Holland PW, Welsch RE. Robust regression using iteratively reweighted least-squares. Communications in Statistics - Theory and Methods. 1977;6(9):813–827. [Google Scholar]
- Huber PJ. Robust estimation of a location parameter. Annals Math. Statist. 1964;35:73–1001. [Google Scholar]
- Jack CR, Petersen RC, Xu YC, Waring SC, O'Brien PC, Tangalos EG, Smith GE, Ivnik RJ, Kokmen E. Medial temporal atrophy on mri in normal aging and very mild alzheimer's disease. Neurology. 1997;49(3):786–790. doi: 10.1212/wnl.49.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M. Tech. Rep. TR99MJ1. Oxford Center for Functional Magnetic Resonance Imaging of the Brain (FMRIB); 1999. Measuring transformation error by RMS deviation. [Google Scholar]
- Jenkinson M, Bannister PR, Brady JM, Smith SM. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Kerwin W, Yuan C. Active edge maps for medical image registration. Medical Imaging 2001: Image Processing. 2001;4322:516–526. SPIE. [Google Scholar]
- Kochunov P, Lancaster JL, Glahn DC, Purdy D, Laird AR, Gao F, Fox P. Retrospective motion correction protocol for high-resolution anatomical mri. Human Brain Mapping. 2006;27(12):957–962. doi: 10.1002/hbm.20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostelec P, Weaver J, Healy DJ. Multiresolution elastic image registration. Medical Physics. 1998;25(9):1593–1604. doi: 10.1118/1.598403. [DOI] [PubMed] [Google Scholar]
- van der Kouwe A, Benner T, Dale A. Real-time rigid body motion correction and shimming using cloverleaf navigators. Magnetic Resonance in Medicine. 2006;56(5):1019–1032. doi: 10.1002/mrm.21038. [DOI] [PubMed] [Google Scholar]
- Kuperberg GR, Broome M, McGuire PK, David AS, Eddy M, Ozawa F, Goff D, West WC, Williams S, van der Kouwe A, Salat D, Dale AM, Fischl B. Regionally localized thinning of the cerebral cortex in Schizophrenia. Archives of General Psychiatry. 2003;60:878–888. doi: 10.1001/archpsyc.60.9.878. [DOI] [PubMed] [Google Scholar]
- Kybic J, Thevenaz P, Nirkko A, Unser M. Unwarping of unidirectionally distorted EPI images. IEEE Transactions on Medical Imaging. 2000;19(2):80–93. doi: 10.1109/42.836368. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Pruessner JC, Zijdenbos A, Hampel H, Teipel SJ, Evans AC. Focal decline of cortical thickness in Alzheimer's disease identified by computational neuroanatomy. Cerebral Cortex. 2005;15:955–1001. doi: 10.1093/cercor/bhh200. [DOI] [PubMed] [Google Scholar]
- Maes F, Collignon A, Vandermeulen D, Marchal G, Suetens P. Multimodality image registration by maximization of mutual information. IEEE Transactions on Medical Imaging. 1997;16(2):187–198. doi: 10.1109/42.563664. [DOI] [PubMed] [Google Scholar]
- Maes F, Vandermeulen D, Suetens P. Comparative evaluation of multiresolution optimization strategies for multimodality image registration by maximization of mutual information. Medical Image Analysis. 1999;3:373–386. doi: 10.1016/s1361-8415(99)80030-9. [DOI] [PubMed] [Google Scholar]
- Maintz J, Viergever M. A survey of medical image registration. Medical Image Analysis. 1998;2(1):1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- Medioni G, Nevatia R. Matching images using linear features. Pattern Analysis and Machine Intelligence. 1984;6(6):675–685. doi: 10.1109/tpami.1984.4767592. [DOI] [PubMed] [Google Scholar]
- Mishra D, Chan AK, Chui CK. Histogram equalization, image registration, and data fusion for multispectral images. Proceedings of the SPIE - The International Society for Optical Engineering. 1995;2496:1025–1031. [Google Scholar]
- Nack M. Rectification and registration of digital images and the effect of cloud detection. Proceedings of Machine Processing of Remotely Sensed Data. 1977:12–23. [Google Scholar]
- Narr KL, Bilder RM, Toga AW, Woods RP, E. RD, Szeszko PR, Robinson D, Sevy S, Gunduz-Bruce H, Wang Y-P, Deluca H, Thompson P. Mapping cortical thickness and gray matter concentration in first episode Schizophrenia. Cerebral Cortex. 2005;15:708–719. doi: 10.1093/cercor/bhh172. [DOI] [PubMed] [Google Scholar]
- Nestares O, Heeger DJ. Robust multiresolution alignment of MRI brain volumes. Magnetic Resonance in Medicine. 2000;43(5):705–715. doi: 10.1002/(sici)1522-2594(200005)43:5<705::aid-mrm13>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Pelizzari CA, Chen GTY, Spelbring DR, Weichselbaum RR, Chen C-T. Accurate three-dimensional registration of CT, PET, and/or MR images of the brain. Journal of Computer Assisted Tomography. 1989;13(1):20–26. doi: 10.1097/00004728-198901000-00004. [DOI] [PubMed] [Google Scholar]
- Periaswamy S, Farid H. Medical image registration with partial data. Medical Image Analysis. 2006;10(3):452–464. doi: 10.1016/j.media.2005.03.006. Special Issue on The Second International Workshop on Biomedical Image Registration (WBIR'03) [DOI] [PubMed] [Google Scholar]
- Roche A, Malandain G, Ayache N, Prima S. Towards a better comprehension of similarity measures used in medical image registration. MICCAI '99: Proceedings of the Second International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer-Verlag; 1999. pp. 555–566. [Google Scholar]
- Roche A, Malandain G, Pennec X, Ayache N. Proceedings MICCAI'98. Vol. 1496 of LNCS. Springer Verlag; 1998. The correlation ratio as a new similarity measure for multimodal image registration; pp. 1115–1124. [Google Scholar]
- Rosas HD, Liu AK, Hersch S, Glessner M, Ferrante RJ, Salat D, van der Kouwe A, Jenkins BG, Dale AM, Fischl B. Regional and progressive thinning of the cortical ribbon in Huntington's disease. Neurology. 2002;5(5):695–701. doi: 10.1212/wnl.58.5.695. [DOI] [PubMed] [Google Scholar]
- Rueckert D, Sonoda L, Hayes I, Hill DLG, Leach M, Hawkes DJ. Nonrigid registration using free-form deformations: Application to breast MR images. IEEE Transactions on Medical Imaging. 1999;18(8):400–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Saad ZS, Glen DR, Chen G, Beauchamp MS, Desai R, Cox RW. A new method for improving functional-to-structural mri alignment using local pearson correlation. NeuroImage. 2009;44(3):839–848. doi: 10.1016/j.neuroimage.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailer M, Fischl B, Salat D, Tempelmann C, Schönfeld M, Busa E, Bodammer N, Heinze H-J, Dale AM. Focal thinning of the cerebral cortex in multiple sclerosis. Brain. 2003;126(8):1734–1744. doi: 10.1093/brain/awg175. [DOI] [PubMed] [Google Scholar]
- Salat D, Buckner R, Snyder A, Greve DN, Desikan R, Busa E, Morris J, Dale AM, Fischl B. Thinning of the cerebral cortex in aging. Cerebral Cortex. 2004;14:721–730. doi: 10.1093/cercor/bhh032. [DOI] [PubMed] [Google Scholar]
- Salat D, Kaye J, Janowsky J. Prefrontal gray and white matter volumes in healthy aging and alzheimer disease. Archives of Neurology. 1999;56(3):338–44. doi: 10.1001/archneur.56.3.338. [DOI] [PubMed] [Google Scholar]
- Schönemann P. A generalized solution of the orthogonal procrustes problem. Psychometrika. 1931;31(1):1–10. [Google Scholar]
- Shih W, Lin W, Chen C. Contour-model-guided nonlinear deformation model for intersubject image registration. Proceedings of the SPIE - The International Society for Optical Engineering. 1997;3034:611–620. [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23:208–219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Peterson BS, Thompson P, Welcome SE, Henkenius AL, Toga AW. Mapping cortical changes across the human life span. Nature Neuroscience. 2003;6:309–315. doi: 10.1038/nn1008. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Thompson P, Leonard CM, Welcome SE, Kan E, Toga AW. Longitudinal mapping of cortical thickness and brain growth in normal children. J Neuroscience. 2004;24(38):8223–8231. doi: 10.1523/JNEUROSCI.1798-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CV. Robust parameter estimation in computer vision. SIAM Reviews. 1999;41:513–537. [Google Scholar]
- Thompson P, Hayashi KM, Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer S, Doddrell DM, Toga AW. Dynamics of gray matter loss in alzheimer's disease. J Neuroscience. 2003;23(3):994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P, Vidal CN, Giedd J, Gochman P, Blumenthal J, Nicolson R, Toga AW, Rapoport J. Mapping adolescent brain change reveals dynamic wave of accelerated gray matter loss in very early onset Schizophrenia. Proceedings of National Academy of Sciences USA. 2001;98(20):11650–11655. doi: 10.1073/pnas.201243998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unser M, Aldroubi A, Gerfen C. A multiresolution image registration procedure using spline pyramids. SPIE Conference on Mathematical Imaging: Wavelet Applications in Signal and Image Processing.1993. pp. 160–170. [Google Scholar]
- Wells W, Viola P, Atsumi H, Nakajima S, Kikinis R. Multi-modal volume registration by maximization of mutual information. 1996. [DOI] [PubMed] [Google Scholar]
- West J, Fitzpatrick JM, Wang MY, Dawant BM, Maurer CJ, Kessler RM, Maciunas RJ. Retrospective intermodality registration techniques for images of the head: surface-based versus volume-based. IEEE Transactions on Medical Imaging. 1999;18(2):144–150. doi: 10.1109/42.759119. [DOI] [PubMed] [Google Scholar]
- West J, Fitzpatrick JM, Wang MY, Dawant BM, Maurer CR, Kessler RM, Maciunas RJ, Barillot C, Lemoine D, Collignon A, Maes F, Sumanaweera TS, Harkhess B, Hemler PF, Hill DLG, Hawkes DJ, Studholme C, Maintz JBA, Viergever MA, Mal G, Pennec X, Noz ME, Maguire GQ, Pollack M, Pelizzari CA, Robb RA, Hanson D, Woods RP. Comparison and evaluation of retrospective intermodality brain image registration techniques. Journal of Computer Assisted Tomography. 1997;21(4):554–566. doi: 10.1097/00004728-199707000-00007. [DOI] [PubMed] [Google Scholar]
- Wilcoxon F. Individual comparisons by ranking methods. Biometrics. 1945;1:80–83. [Google Scholar]
- Woods RP, Cherry S, Mazziotta J. Rapid automated algorithm for aligning and reslicing PET images. Computer Assisted Tomography. 1992;16(4):620–633. doi: 10.1097/00004728-199207000-00024. [DOI] [PubMed] [Google Scholar]
- Woods RP, Grafton S, Holmes C, Cherry S, Mazziotta J. Automated image registration: I. general methods and intrasubject, intramodality validation. Computer Assisted Tomography. 1998;22(1):139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Yeung SK, Tang CK, Shi PC, Pluim JPW, Viergever MA, Chung ACS, Shen HC. Enforcing stochastic inverse consistency in non-rigid image registration and matching. Proceedings CVPR'08. 2008:8. [Google Scholar]
- Yushkevicha PA, Avantsa BB, Dasa SR, Plutab J, Altinaya M, Craigea C. Bias in estimation of hippocampal atrophy using deformation-based morphometry arises from asymmetric global normalization: An illustration in ADNI 3 tesla MRI data. NeuroImage. 2009 doi: 10.1016/j.neuroimage.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Chen Y. Bioinformatics Research and Applications, ISBRA'08, 293–304, Lecture Notes in Computer Science 4983. Springer; 2008. Accurate Inverse Consistent Non-rigid Image Registration and Its Application on Automatic Re-contouring. [Google Scholar]