Abstract
Lipopolysaccharide (LPS) preconditioning provides neuroprotection against subsequent cerebral ischemic injury through activation of its receptor, Toll-like receptor 4 (TLR4). Paradoxically, TLR activation by endogenous ligands after ischemia worsens stroke damage. Here, we define a novel, protective role for TLRs after ischemia in the context of LPS preconditioning. Microarray analysis of brains collected 24 h after stroke revealed a unique set of upregulated genes in LPS-pretreated animals. Promoter analysis of the unique gene set identified an overrepresentation of type I interferon (IFN)-associated transcriptional regulatory elements. This finding suggested the presence of type I IFNs or interferon regulatory factors (IRFs), which upregulate interferon-stimulated genes. Upregulation of IFNβ was confirmed by real-time reverse transcription-PCR. Direct administration of IFNβ intracerebroventricularly at the time of stroke was sufficient for neuroprotection. TLR4 can induce both IFNβ and interferon-stimulated genes through its adapter molecule Toll/interleukin receptor domain-containing adaptor-inducing IFNβ (TRIF) and the IRF3 transcription factor. We show in oxygen glucose deprivation of cortical neurons, an in vitro model of stroke, that activation of TRIF after stroke reduces neuronal death. Furthermore, mice lacking IRF3 were not protected by LPS preconditioning in our in vivo model. Our studies constitute the first demonstration of the neuroprotective capacity of TRIF/IRF3 signaling and suggest that interferon-stimulated genes, whether induced by IFNβ or by enhanced TLR signaling to IRF3, are a potent means of protecting the brain against ischemic damage.
Introduction
It is increasingly clear that Toll-like receptor (TLR) signaling worsens stroke injury. Mice lacking TLR2 or TLR4 are less susceptible to damage in multiple models of cerebral ischemia (Cao et al., 2007; Lehnardt et al., 2007; Ziegler et al., 2007). TLRs are expressed by microglia, astrocytes, and endothelial cells and are activated by the damage-associated molecules HSP70 (TLR4) and HMGB1 (TLR2 and TLR4), present in the brain after ischemia (Kinouchi et al., 1993a,b; Faraco et al., 2007). TLR activation induces production of the inflammatory molecules tumor necrosis factor α (TNFα), IL1β, and inducible nitric oxide synthase and other cytotoxic mediators that increase tissue damage.
Although TLR4 activation after stroke exacerbates injury, activation of TLR4 before stroke protects the brain from damage. Systemic administration of lipopolysaccharide (LPS), a potent TLR4 ligand of bacterial origin, renders animals tolerant to injury in several models of cerebral ischemia (Tasaki et al., 1997; Rosenzweig et al., 2004; Hickey et al., 2007). LPS-induced tolerance to ischemic injury mirrors the phenomenon of LPS-induced tolerance to LPS. Initial exposure of macrophages to LPS induces proinflammatory TNFα, but, during subsequent exposure to LPS, TNFα production is reduced markedly as a result of disrupted signaling through the TLR4 adaptor molecule MyD88 (West and Heagy, 2002; Fan and Cook, 2004; Liew et al., 2005). Conversely, macrophages produce little interferon β (IFNβ) during initial exposure to LPS but enhance IFNβ production during secondary exposure (Broad et al., 2007), suggesting upregulated TLR4 signaling through the Toll/interleukin receptor domain-containing adaptor-inducing IFNβ (TRIF) adaptor molecule. Thus, pretreatment with LPS may cause cells to switch their dominant TLR4 signaling pathway.
TLR4 signaling through TRIF induces IFNβ via activation of the interferon regulatory factor IRF3. IFNβ, administered systemically, reduces ischemic brain damage (Liu et al., 2002; Veldhuis et al., 2003), likely through activation of interferon-stimulated genes (ISGs). IRF3 itself may have similar neuroprotective effects. IRF3 binds to interferon-stimulated response elements (ISREs) within gene promoters, increasing the expression of many ISGs to the same extent of that elicited by type I IFNs (Nakaya et al., 2001). Hence, activation of IRF3 may independently result in protection from ischemic stroke. Thus, enhanced TLR4 signaling to TRIF–IRF3–IFNβ would be expected to contribute to neuroprotection.
We propose that pretreatment or preconditioning with LPS changes the cellular environment such that subsequent activation of TLR4 increases signaling via TRIF to IRF3 and upregulates the neuroprotective cytokine IFNβ. Thus, in this way, LPS preconditioning may reprogram subsequent activation of TLR4 during ischemia, which leads to an increase in neuroprotective type I IFN signaling. Here we provide evidence for such reprogramming and its neuroprotective consequences.
Materials and Methods
Mice.
C57BL/6 mice (male, 8–12 weeks, ∼25 g) were purchased from The Jackson Laboratory. IFNβ knock-out mice were kindly provided by Dr. Leanderson (Lund University, Lund, Sweden). IRF3 knock-out mice were procured from RIKEN BioResource Center (Tsukuba, Japan). Both strains were backcrossed onto the C57BL/6 background for at least eight generations. All mice were housed in an American Association for Laboratory Animal Care-approved facility. Procedures were conducted according to Oregon Health and Science University, Institutional Animal Care and Use Committee, and National Institutes of Health guidelines.
LPS treatment.
Mice were given a 200 μl intraperitoneal injection of saline or LPS [0.2–1.0 mg/kg; Escherichia coli serotype 0111:B4; catalog #L2630, purified by phenol extraction, protein content <3% (Sigma)]. Each new lot of LPS was titrated to determine the optimal dose that confers neuroprotection in the particular strain of mouse being tested.
Middle cerebral artery occlusion.
Mice were anesthetized with 4% halothane and subjected to middle cerebral artery occlusion (MCAO) using the monofilament suture method described previously (Stevens et al., 2002). Briefly, a silicone-coated 8-0 monofilament nylon surgical suture was threaded through the external carotid artery to the internal carotid artery to block the middle cerebral artery and maintained intraluminally for 40, 45, or 60 min. Duration of occlusion was based on pilot studies performed to determine the time necessary to obtain an infarct size that is between 35 and 45% in the control groups of mice. It is well known that genetic background can influence ischemic outcome and thereby affect infarct size. The suture was then removed to restore blood flow. Cerebral blood flow (CBF) was monitored throughout surgery by laser Doppler flowmetry. The mean CBF during occlusion was between 10 and 17% of baseline in each of the studies presented. Mice that did not maintain a CBF drop within the norm of the group during the occlusion were excluded (<4% of all animals in the combined studies). Body temperature was maintained at 37°C with a thermostat-controlled heating pad. The survival rate for the MCAO procedure was >80%.
Infarct evaluation.
To visualize the region of infarction, 6 × 1 mm coronal midsections were placed in 1.5% 2,3,5 triphenyltetrazolium chloride (TTC) in 0.9% PBS and stained at 37°C for 15 min. The infarct size was determined from computer-scanned images of the hemispheres using NIH Image analyses. To account for edema within the infarct region, infarct area for each section was computed indirectly as follows: 100 × (contralateral hemisphere area − area of live tissue on ipsilateral hemisphere)/(contralateral hemisphere area) (Swanson et al., 1990).
Experimental design for gene expression studies.
C57BL/6 mice were divided into 10 groups with four animals per group: groups 1–3 received a saline injection and were killed at 3, 24, and 72 h, respectively. Groups 4–6 received an LPS injection and were killed at 3, 24, and 72 h, respectively. Groups 7 and 8 received a saline injection, followed 72 h later with a 45 min MCAO. Group 9 and 10 received an LPS injection, followed 72 h later with a 45 min MCAO. Groups 7 and 9 were killed at 3 h after start of occlusion with groups 8 and 10 killed 24 h after start of occlusion. At the time of the mice were killed, the animals were anesthetized and then perfused with heparinized saline. One group (n = 6) was included as unhandled controls. Under RNase-free conditions, a 1 mm section was removed (4 mm from rostral end) to determine the area of infarct based on TTC staining. The ipsilateral cortex region from the frontal 4 mm was isolated and snap frozen in liquid nitrogen.
RNA isolation.
Total RNA was isolated using the Qiagen RNeasy Lipid Mini kit. RNA from individual animals was hybridized to single arrays as described below.
GeneChip expression analyses.
Microarray assays were performed in the Affymetrix Microarray Core of the Oregon Health and Science University Gene Microarray Shared Resource. RNA samples were labeled using the NuGEN Ovation Biotin RNA Amplification and Labeling System_V1. Hybridization was performed as described in the Affymetrix technical manual with modifications as recommended for the Ovation labeling protocol. Labeled cRNA target was quality checked based on yield and size distribution. Quality-tested samples were hybridized to the MOE430 2.0 array. The array image was processed with Affymetrix GeneChip Operating Software. Arrays which did not meet empirically defined cutoffs within the core facility were remade and hybridized to fresh arrays. Data were normalized using the robust multichip average method (Irizarry et al., 2003). The normalized data were then analyzed using a two-way ANOVA model for each gene, using conditions and time as groups. Post hoc comparisons were made using the unhandled mice as a control group. p values were adjusted for multiple comparisons using the Hochberg and Benjamini method (Hochberg and Benjamini, 1990). Genes were considered significantly regulated if the adjusted p value was <0.05, and the fold change in regulation was greater than or equal to 2.
Transcriptional regulatory network analysis.
Using the Web-based program Promoter Analysis and Interaction Network Toolset (PAINT) version 3.5 (Vadigepalli et al., 2003), we examined the predicted regulatory elements associated with the unique gene regulation identified by microarray. In brief, using PAINT, we obtained the 5000 bp upstream sequence for the transcripts represented on the MOE430 Affymetrix gene chip (33,635 transcripts were identified with 5000 bp of upstream sequence). PAINT identified putative transcription factor binding sequences [transcriptional regulatory elements (TREs)] in these upstream sequences using the TRANSFAC PRO database version 10.4. This pool of genes and identified TREs was used as our reference comparison group. The statistical component of PAINT (false discovery rate adjusted p value set at ≤0.2) was used to determine the overrepresented TREs in individual gene clusters compared with the reference comparison group (i.e., uniquely expressed genes in LPS-preconditioned mice compared with 33,635 member reference group).
Intracerebral ventricular injection of IFNβ during MCAO.
Recombinant mouse (rm) IFNβ (Cell Sciences) or vehicle [artificial CSF (aCSF)] was injected into the left lateral ventricle as described previously (Meller et al., 2005). Injections (1 μl) of either rmIFNβ (200 U) or aCSF were administered immediately before and after surgery (60 min MCAO). Infarct volume was measured 24 h after stroke.
Quantitative real-time PCR for IFNβ.
RNA was treated with DNase and transcribed into cDNA using the Omniscript RT kit (Qiagen). Real-time reverse transcription-PCRs were performed using TaqMan PCR Master Mix (Applied Biosystems). For IFNβ, TaqMan Gene Expression Assay Mix for mouse IFNβ was used (Mm00439546_S1; Applied Biosystems). Primers and probe for β-actin were obtained from Integrated DNA Technologies: forward, 5′-AGAGGGAAATCGTGCGTGAC-3′; reverse, 5′-CAATAGTGATGACCTGGCCGT-3′; probe, CACTGCCGCATCCTCTTCCTCCC. Samples were run on an ABI-Prism 7700 (Applied Biosystems). Results were analyzed using Applied Biosystems sequence detection software. The relative quantitation of IFNβ was determined using the comparative cycle threshold (CT) method (2−ΔΔCT) described in Applied Biosystems User Bulletin #2. Results were normalized to β-actin and presented relative to unhandled mice. All reactions were performed in triplicate.
Oxygen glucose deprivation in vitro.
Primary mouse mixed cortical cultures were prepared from embryonic day 15 to 17 mouse fetuses. Cortices were dissected and dissociated with trypsin-EDTA (Invitrogen) and plated at a density of 4.5 × 105 cells/ml onto coverslips coated with poly-l-ornithine (15 mg/L). Cells were cultured in Neurobasal media (containing 4.5 g/L glucose; supplemented with Glutamax and B27-AO; Invitrogen) for 5 d before each experiment. Cultures consisted of ∼60% neurons (range, 53–66%) as determined by staining for neuronal-specific nuclear protein (Millipore Bioscience Research Reagents), with <5% astrocytes (GFAP+; Sigma) and <5% microglia (tomato lectin+; Vector Laboratories). Oxygen glucose deprivation (OGD) was performed by removal of the culture medium and replacement with Dulbecco's PBS (Invitrogen), followed by incubation in an anaerobic atmosphere of 85% N2, 10% CO2, and 5% H2 at 37°C for 3 h. The anaerobic conditions within the chamber were monitored using an electronic oxygen/hydrogen analyzer (Coy Laboratories). OGD was terminated by replacement of the exposure medium with Neurobasal medium (containing 4.5 g/L glucose; supplemented with Glutamax and B27-AO) and return of the cells to a normoxic incubator. Control plates were kept in the normoxic incubator during the OGD interval.
Cell death evaluation in vitro.
Cell death in vitro was examined 24 h after OGD by means of fluorescent, cell-permeable, DNA-binding dyes: propidium iodide (PI), as an indicator of cell death, and 4′,6-diamidino-2-phenylindole (DAPI), as an indicator of the total number of cells. Coverslips were incubated with PI (1.5 μg/ml; Sigma) for 5 min, washed with PBS, and fixed for 30 min in 10% Formalin. Coverslips were mounted on slides with Fluoromount-G mounting medium containing DAPI (Southern Biotechnology Associates). Stained cells were visualized with a fluorescent microscope (Leica) and analyzed using Metmorph7 software (Molecular Devices). The number of PI- and DAPI-stained cells were counted in two random fields of view on each coverslip, and percentage death was calculated as mean (PI)/(DAPI) × 100 per field of view. Each treatment was performed with triplicate coverslips within an experiment, and the entire experiment was repeated three or more times.
Results
Systemic administration of LPS induces an inflammatory response in the brain
As we have shown previously, systemic administration of LPS (0.2 mg/kg) given 3 d before MCAO substantially attenuates ischemic damage (Rosenzweig et al., 2004, 2007). To begin to elucidate possible mechanisms of neuroprotection, we isolated RNA from the cortex of LPS-treated and control mice at time points before MCAO. Using Affymetrix oligonucleotide microarrays, we identified 263 genes (228 increased, 35 decreased) significantly regulated 3 h after LPS injection and 176 genes (174 increased, 2 decreased) at 24 h after LPS treatment. However, within 72 h after LPS administration, most of the genomic changes had subsided to baseline with the exception of five differentially regulated genes that remained increased (Fig. 1A; Table 1). The saline-treated controls showed no statistically significant gene regulation at any time point when compared with the unhandled baseline group.
Figure 1.
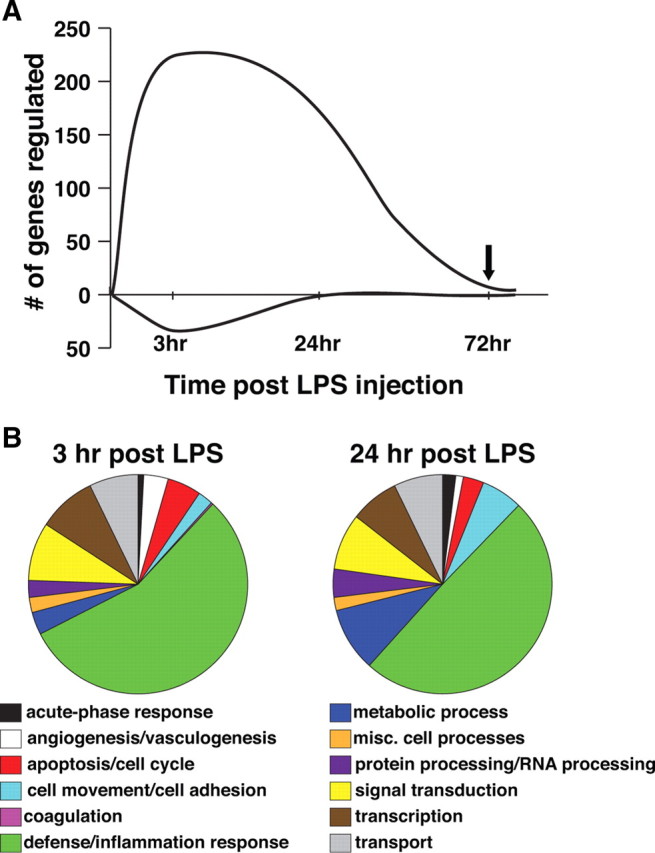
Systemic administration of LPS induces early gene regulation in the brain, consistent with an inflammatory response. C57BL/6 mice were administered saline or LPS (0.2 mg/kg, i.p.). At varying times (3, 24, and 72 h) after treatment, mice (n = 4 per time point) were killed, and cortical brain tissue was collected. RNA was isolated and hybridized to Affymetrix gene chips (MOE430). A, Graph represents the number of genes differentially regulated in LPS- or saline-treated mice compared with unhandled controls. Time of subsequent stroke is denoted with a black arrow. B, Putative biological functions were assigned to the regulated genes using available public databases and published literature.
Table 1.
Genes differentially regulated 72 h after LPS preconditioning (time of stroke)
| Title | Symbol | Fold change |
|---|---|---|
| Serum amyloid A3 | Saa3 | 6.48 |
| Topoisomerase (DNA) II α | Top2a | 2.14 |
| UDP glucuronosyltransferase 2 family, polypeptide B37 | Ugt2b37 | 3.53 |
| RIKEN cDNA 4930440C22 gene | 2.24 | |
| RIKEN cDNA 4930554P06 gene | 2.35 |
Table 2.
Genes regulated 24 h after stroke only in LPS-preconditioned mice
| Symbol | LPS at 3 ha,b | LPS at 24 h | LPS at 72 h | LPS + MCAO at 3 h | LPS + MCAO at 24 h | |
|---|---|---|---|---|---|---|
| Apoptosis/cell cycle | ||||||
| CD274 antigen | Cd274 | 11.60 | NS | NS | NS | 2.48 |
| GLI pathogenesis-related 1 (glioma) | Glipr1 | NS | NS | NS | NS | 2.34 |
| Caspase 8 | Casp8 | NS | NS | NS | NS | 2.01 |
| Cell movement/cell adhesion | ||||||
| Filamin binding LIM protein 1 | Fblim1 | NS | NS | NS | NS | 2.44 |
| Selectin, endothelial cell | Sele | 1.98 | NS | NS | 1.66 | 2.37 |
| Neurogenic differentiation 4 | Neurod4 | NS | 1.60 | NS | NS | 2.33 |
| Kelch-like 6 (Drosophila) | Klhl6 | NS | NS | NS | NS | 2.18 |
| Glycoprotein (transmembrane) nmb | Gpnmb | NS | NS | NS | NS | 2.11 |
| Claudin 1 | Cldn1 | NS | NS | NS | NS | 2.03 |
| M-phase phosphoprotein 1 | Mphosph1 | NS | NS | NS | NS | 2.02 |
| PDZ and LIM domain 5 | Pdlim5 | NS | NS | NS | 1.73 | 2.01 |
| Protocadherin 20 | Pcdh20 | NS | NS | NS | NS | −2.20 |
| Protocadherin 21 | Pcdh21 | NS | NS | NS | NS | −2.37 |
| Coagulation | ||||||
| Coagulation factor V | F5 | NS | NS | NS | NS | 3.12 |
| Coagulation factor XIII, A1 subunit | F13a1 | NS | NS | NS | NS | 2.60 |
| Hepatocyte growth factor | Hgf | NS | NS | NS | NS | 2.60 |
| Protein S (α) | Pros1 | NS | NS | NS | NS | 2.41 |
| Defense response | ||||||
| Radical S-adenosyl methionine domain containing 2 | Rsad2, VIPERIN | 33.06 | 3.01 | NS | NS | 3.84 |
| Killer cell lectin-like receptor subfamily B member 1F | Klrb1f | NS | 4.77 | NS | NS | 3.77 |
| Interferon inducible GTPase 1 | Iigp1 | 23.33 | NS | NS | NS | 3.56 |
| CD52 antigen | Cd52 | NS | 2.37 | NS | NS | 3.33 |
| Fc receptor, IgG, high affinity I | Fcgr1 | NS | 2.77 | NS | NS | 3.09 |
| Interferon-induced protein with tetratricopeptide repeats 1 | Ifit1 | 19.84 | 4.03 | NS | NS | 3.06 |
| Guanylate nucleotide binding protein 3 | Gbp3 | 7.09 | 2.35 | NS | NS | 2.96 |
| SLAM family member 9 | Slamf9 | NS | 1.53 | NS | NS | 2.92 |
| Protein tyrosine phosphatase, receptor type, C | Ptprc, B220 | NS | 1.94 | NS | 1.58 | 2.65 |
| Transporter 1, ATP-binding cassette, subfamily B (MDR/TAP) | Tap1 | 3.42 | 1.78 | NS | NS | 2.65 |
| Histocompatibility 2, Q region locus 1 | H2-Q1 | 7.45 | 2.84 | NS | 2.48 | 2.60 |
| PYD and CARD domain containing | Pycard | NS | 2.11 | NS | NS | 2.58 |
| Lymphocyte cytosolic protein 2 | Lcp2 | 1.79 | NS | NS | NS | 2.51 |
| Fc receptor, IgE, high affinity I, γ polypeptide | Fcer1g | NS | 1.87 | NS | NS | 2.49 |
| 2′-5′ oligoadenylate synthetase-like 2 | Oasl2 | 5.05 | 3.87 | NS | NS | 2.48 |
| Phospholipid scramblase 2 | Plscr2 | 3.46 | NS | NS | NS | 2.47 |
| Interferon γ -induced GTPase | Igtp | 4.35 | 1.78 | NS | NS | 2.46 |
| Neutrophilic granule protein | Ngp | NS | NS | NS | NS | 2.44 |
| Interferon-induced transmembrane protein 6 | Ifitm6 | NS | NS | NS | NS | 2.44 |
| Complement component 1, q subcomponent, β polypeptide | C1qb | NS | 1.54 | NS | NS | 2.42 |
| Toll-like receptor 4 | Tlr4 | NS | NS | NS | NS | 2.42 |
| Myxovirus (influenza virus) resistance 1 | Mx1 | 11.10 | 2.43 | NS | NS | 2.34 |
| Lymphocyte cytosolic protein 1 | Lcp1 | NS | 1.60 | NS | NS | 2.29 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | Ddx58, RIG1 | 3.59 | 1.79 | NS | NS | 2.23 |
| Interleukin 1 β | Il1b | 4.69 | NS | NS | 2.08 | 2.20 |
| Leukocyte Ig-like receptor, subfamily B member 3 | Lilrb3 | NS | 1.54 | NS | NS | 2.13 |
| Histocompatibility 2, D region | H2-L | 1.55 | 2.03 | 1.63 | 1.61 | 2.13 |
| Stabilin 1 | Stab1 | NS | NS | NS | NS | 2.12 |
| Histocompatibility 2, K1, K region | H2-K1 | 2.01 | 2.38 | 1.74 | 1.86 | 2.11 |
| Protein tyrosine phosphatase, non-receptor type 6 | Ptpn6,Shp1 | NS | NS | NS | NS | 2.11 |
| E74-like factor 1 | Elf1 | NS | 1.98 | NS | 1.63 | 2.09 |
| Interferon-induced protein 35 | Ifi35 | 3.27 | 2.06 | NS | NS | 2.08 |
| SAM domain, SH3 domain and nuclear localization signals, 1 | Samsn1 | 1.91 | NS | NS | 1.54 | 2.08 |
| Histocompatibility 2, D region locus 1 | H2-D1 | NS | 2.05 | 1.62 | 1.60 | 2.08 |
| H-2 class I histocompatibility antigen, Q7 α chain precursor | QA-2 | 5.26 | 3.89 | NS | 3.08 | 2.08 |
| C-type lectin domain family 14, member a | Clec14a | −2.36 | NS | NS | NS | 2.07 |
| C-type lectin domain family 5, member a | Clec5a | NS | NS | NS | NS | 2.06 |
| Interferon-induced protein with tetratricopeptide repeats 3 | Ifit3 | 5.41 | 2.42 | NS | NS | 2.06 |
| Proteosome subunit, β type 8 | Psmb8 | 2.56 | 2.03 | NS | NS | 2.05 |
| Neutrophil cytosolic factor 1 | Ncf1 | 1.51 | NS | NS | NS | 2.03 |
| Lymphocyte antigen 6 complex, locus A | Ly6a | 1.79 | 1.88 | NS | NS | 2.01 |
| Signal peptide, CUB domain, EGF-like 1 | Scube1 | NS | NS | NS | NS | −2.03 |
| Corticotropin releasing hormone | Crh | NS | NS | NS | NS | −2.10 |
| Metabolic processes | ||||||
| Klotho | Kl | NS | NS | NS | NS | 2.48 |
| Hexokinase 2 | Hk2 | NS | NS | NS | NS | 2.26 |
| Ethanolamine kinase 1 | Etnk1 | NS | NS | NS | 2.89 | 2.24 |
| Carbonic anhydrase 13 | Car13 | NS | NS | NS | NS | 2.16 |
| Centromere protein A | Cenpa | NS | NS | NS | NS | 2.12 |
| Phosphodiesterase 3A, cGMP inhibited | Pde3a | NS | 2.19 | NS | NS | 2.08 |
| Folate receptor 1 (adult) | Folr1 | NS | NS | NS | NS | 2.04 |
| AMP deaminase 3 | Ampd3 | NS | NS | NS | NS | 2.02 |
| Miscellaneous cell processes | ||||||
| Schlafen 2 | Slfn2 | 4.00 | 2.18 | NS | NS | 3.35 |
| Calmodulin-like 4 | Calml4 | NS | NS | NS | NS | 2.50 |
| Ecotropic viral integration site 2b | Evi2b | NS | NS | NS | NS | 2.49 |
| Estrogen receptor 1 (α) | Esr1 | NS | NS | NS | 2.35 | 2.09 |
| Luc7 homolog (Saccharomyces / cerevisiae)-like | Luc7l | NS | NS | NS | NS | 2.04 |
| Protein/RNA processing | ||||||
| Ubiquitin specific peptidase 18 | Usp18 | 12.83 | 3.83 | NS | NS | 2.89 |
| RIKEN cDNA 5430435G22 gene | NS | NS | NS | NS | 2.75 | |
| Ribosomal protein L7 | Rpl7 | NS | 2.11 | NS | NS | 2.75 |
| Heat shock protein 8 | Hspb8 | NS | NS | NS | 1.70 | 2.42 |
| Serine (or cysteine) peptidase inhibitor, clade H, member 1 | Serpinh1 | NS | NS | NS | 1.70 | 2.33 |
| ST6 (α -N-acetyl-neuraminyl-2,3-β -galactosyl-1,3)-N-acetyl- galactosaminide α -2,6-sialyltransferase 2 | St6galnac2 | NS | NS | NS | NS | 2.31 |
| UDP-GlcNAc: β Gal β -1,3-N-acetylglucosaminyltransferase 5 | B3gnt5 | NS | NS | NS | NS | 2.27 |
| Phospholipase A2, group IVA | Pla2g4a | NS | NS | NS | NS | 2.27 |
| Z-DNA binding protein 1 | Zbp1 | 3.50 | 3.40 | NS | NS | 2.27 |
| A disintegrin-like and metallopeptidase with thrombospondin type 1 motif, 5 | Adamts5 | NS | NS | NS | NS | 2.13 |
| Translocating chain-associating membrane protein 2 | Tram2 | NS | NS | NS | NS | 2.08 |
| IMP4, U3 small nucleolar ribonucleoprotein | Imp4 | NS | NS | NS | NS | 2.08 |
| Ribosomal protein S25 | Rps25 | NS | NS | NS | NS | 2.08 |
| Ubiquitin-like, containing PHD and RING finger domains, 1 | Uhrf1 | NS | NS | NS | NS | 2.06 |
| cDNA sequence BC099439 | BC099439 | NS | NS | NS | NS | 2.05 |
| DnaJ (Hsp40) homolog, subfamily B, member 5 | Dnajb5 | NS | NS | NS | NS | −2.04 |
| Signal transduction | ||||||
| Receptor transporter protein 4 | Rtp4 | 3.83 | 3.46 | NS | NS | 2.92 |
| Component of Sp100-rs | Csprs | NS | 2.95 | NS | NS | 2.36 |
| Rho GTPase activating protein 30 | Arhgap30 | NS | NS | NS | NS | 2.33 |
| Adenylate cyclase 7 | Adcy7 | NS | NS | NS | NS | 2.28 |
| Guanine nucleotide binding protein, α 14 | Gna14 | NS | NS | NS | NS | 2.28 |
| Vomeronasal 1 receptor, A1 | V1ra1 | NS | 2.52 | NS | NS | 2.22 |
| Fibrinogen-like protein 2 | Fgl2 | NS | NS | NS | NS | 2.14 |
| Ras homolog gene family, member C | Rhoc | 1.66 | NS | NS | 1.55 | 2.12 |
| Pleckstrin homology, Sec7 and coiled/coil domains 4 | Pscd4 | NS | 1.52 | NS | NS | 2.07 |
| Pleckstrin homology domain containing, family G member 2 | Plekhg2 | NS | NS | NS | NS | 2.05 |
| Transcription | ||||||
| Reduced expression 2 | Rex2 | NS | NS | NS | NS | 2.28 |
| MyoD family inhibitor domain containing | Mdfic | NS | NS | NS | NS | 2.24 |
| Leucine rich repeat (in FLII) interacting protein 1 | Lrrfip1 | NS | NS | NS | 2.05 | 2.16 |
| Bromodomain adjacent to zinc finger domain 1A | Baz1a | NS | NS | NS | 2.28 | 2.05 |
| Ladybird homeobox 1 homolog (Drosophila) corepressor 1 | Lbxcor1 | NS | NS | NS | 1.87 | 2.04 |
| Annexin A11 | Anxa11 | NS | NS | NS | NS | 2.02 |
| Tripartite motif protein 30 | Trim30 | 8.15 | 2.13 | NS | NS | 2.02 |
| Inversin | Invs | NS | 2.11 | NS | NS | 2.02 |
| Transport | ||||||
| Transthyretin | Ttr | NS | NS | NS | NS | 12.74 |
| Translocator protein | Tspo | NS | 2.16 | NS | NS | 2.76 |
| Stanniocalcin 2 | Stc2 | NS | NS | NS | 1.97 | 2.64 |
| Transient receptor potential cation channel, subfamily M, member 3 | Trpm3 | NS | NS | NS | NS | 2.63 |
| Potassium voltage-gated channel, Isk-related subfamily, gene 2 | Kcne2 | NS | NS | NS | NS | 2.41 |
| Transferrin | Trf | NS | NS | NS | NS | 2.37 |
| Chloride channel calcium activated 1///chloride channel calcium activated 2 | Clca1///Clca2 | NS | NS | NS | NS | 2.26 |
| Mannose receptor, C type 1 | Mrc1 | NS | NS | NS | NS | 2.25 |
| Cysteine-rich hydrophobic domain 2 | Chic2 | NS | NS | NS | NS | 2.10 |
| Exocyst complex component 6 | Exoc6 | NS | NS | NS | NS | 2.10 |
| SEH1-like (S. cerevisiae) | Seh1l | NS | NS | NS | NS | 2.08 |
| Aquaporin 1 | Aqp1 | NS | NS | NS | NS | 2.06 |
| Poly (ADP-ribose) polymerase family, member 9 | Parp9 | 3.34 | 1.60 | NS | NS | 2.04 |
| RAB20, member RAS oncogene family | Rab20 | 1.98 | NS | NS | 1.95 | 2.04 |
| Triadin | Trdn | NS | NS | NS | NS | −2.04 |
| Solute carrier family 2 (facilitated glucose transporter), member 5 | Slc2a5 | NS | NS | NS | NS | −2.13 |
| Function unknown | ||||||
| RIKEN cDNA 1700040G22 gene | NS | 3.10 | NS | NS | 2.86 | |
| RIKEN cDNA 1500015O10 gene | NS | NS | NS | NS | 2.70 | |
| RIKEN cDNA 9330175E14 gene | 2.32 | 2.11 | NS | 1.62 | 2.62 | |
| BM241008 | NS | 2.28 | NS | NS | 2.61 | |
| RIKEN cDNA 1810053B01 gene | NS | NS | NS | NS | 2.57 | |
| RIKEN cDNA 1810032O08 gene | NS | NS | NS | NS | 2.50 | |
| Hypothetical LOC433632 | NS | NS | NS | NS | 2.44 | |
| RIKEN cDNA 3830408D24 gene | NS | NS | NS | NS | 2.38 | |
| Expressed sequence AI451617 | 3.29 | 2.95 | NS | NS | 2.34 | |
| Expressed sequence AU020206 | NS | 1.79 | NS | NS | 2.33 | |
| Expressed sequence AU045094 | NS | 2.54 | NS | NS | 2.32 | |
| RIKEN cDNA 2410131K14 gene | NS | NS | NS | NS | 2.29 | |
| Stefin A2 like 1 | Stfa2l1 | NS | NS | NS | NS | 2.29 |
| RIKEN cDNA 2810007J24 gene | NS | NS | NS | NS | 2.27 | |
| BG072508 | 2.54 | 3.00 | NS | NS | 2.27 | |
| Transcribed locus AW215795 | NS | 1.68 | NS | NS | 2.26 | |
| RIKEN cDNA 4632434I11 gene | NS | NS | NS | NS | 2.23 | |
| RIKEN cDNA 2610305J24 gene | NS | NS | NS | NS | 2.22 | |
| RIKEN cDNA 5430416N02 gene | NS | NS | NS | NS | 2.21 | |
| Predicted gene, EG240327 | 4.26 | 2.20 | NS | NS | 2.21 | |
| RIKEN cDNA 1110018M03 gene | NS | NS | NS | NS | 2.20 | |
| Similar to phospholipid scramblase 1 | LOC331000 | NS | 2.10 | NS | NS | 2.19 |
| cDNA sequence BC039210 | NS | NS | NS | NS | 2.18 | |
| RIKEN cDNA 4632432E15 gene | NS | NS | NS | NS | 2.16 | |
| RIKEN cDNA 1700010N08 gene | NS | 2.27 | NS | NS | 2.15 | |
| Expressed sequence AI447904 | 1.58 | 1.89 | NS | NS | 2.13 | |
| BB548602 | NS | NS | NS | NS | 2.13 | |
| RIKEN cDNA 2410006H16 gene | NS | NS | NS | NS | 2.12 | |
| Testis expressed gene 15 | Tex15 | NS | NS | NS | NS | 2.12 |
| AV227312 | NS | 1.66 | NS | NS | 2.10 | |
| BG068057 | NS | 2.43 | NS | NS | 2.10 | |
| Expressed sequence AU022436 | NS | NS | NS | NS | 2.09 | |
| Expressed sequence AU022479 | NS | NS | NS | 2.86 | 2.08 | |
| AV354139 | NS | NS | NS | NS | 2.08 | |
| BG068123 | NS | NS | NS | NS | 2.04 | |
| C85463 | NS | 2.40 | NS | NS | 2.03 | |
| RIKEN cDNA 2810026P18 gene | NS | NS | NS | NS | 2.02 | |
| RIKEN cDNA 4632409D06 gene | NS | 1.64 | NS | 2.60 | 2.02 | |
| Similar to EH-domain containing 2 | LOC673251 | NS | 1.52 | NS | 1.87 | 2.02 |
| BB485297 | 6.11 | NS | NS | NS | 2.01 | |
| Transcribed locus, BF148780 | NS | NS | NS | NS | 2.00 | |
| RIKEN cDNA E530001K10 gene | NS | NS | NS | NS | −2.01 | |
| LEM domain containing 1 | Lemd1 | NS | NS | NS | NS | −2.01 |
| Expressed sequence AI449310 | NS | NS | NS | NS | −2.05 | |
| AV118079 | NS | NS | NS | NS | −2.07 | |
| RIKEN cDNA 9630014M24 gene | NS | NS | NS | NS | −2.08 | |
| Transmembrane protein 107 | Tmem107 | NS | NS | NS | NS | −2.09 |
| RIKEN full-length enriched library, clone:D130065D02 | NS | NS | NS | NS | −2.10 | |
| RIKEN cDNA 1700040D17 gene | NS | NS | NS | −1.75 | −2.11 | |
| RIKEN cDNA 9630002A11 gene | NS | NS | NS | NS | −2.11 | |
| BE957247 | NS | NS | NS | NS | −2.19 | |
| BB335621 | NS | NS | NS | NS | −2.36 |
aFold change compared with unhandled baseline group.
bNS, Not significantly regulated; false discovery rate adjusted p value >0.05.
Interferon transcriptional regulatory elements are associated with LPS preconditioning
We identified TREs associated with the unique gene regulation detected in the LPS and saline preconditioned animals using the Web-based program PAINT version 3.5. We compared the TREs identified in the cluster of genes uniquely increased in LPS-preconditioned mice 24 h after stroke (158 genes) (Fig. 2) to a reference cluster consisting of ∼33,000 transcripts from the MOE430 gene chip. This allowed the determination of overrepresented TREs associated with the genes in the preconditioned cluster. We performed the same comparison using the cluster of genes uniquely increased in the saline-pretreated mice 24 h after stroke (128 genes) (Fig. 2). Analysis of the LPS-preconditioned group identified 14 TREs with an adjusted p value <0.2, whereas the saline-pretreated cluster revealed only five overrepresented TREs (Table 3). Five of the 14 identified TREs in the LPS-preconditioned cluster are interferon associated [IRF (V$IRF_Q6 and V$IRF_Q6_01), IRF8, ISRE, and IRF7], and two are nuclear factor-κB (NFκB) components (cRel, NF-kappaBp65). A network depiction of interactions between the identified TREs and the genes in the LPS-preconditioned cluster is displayed in Figure 3. The interferon-associated TREs (in red) are linked to a substantial number of the genes shown (60%; 76 of 127). In fact, a large number of the genes with identified IFN TREs have been reported in the literature to be induced by type I interferons (Fig. 4, red asterisks). NFκB regulatory elements were also overrepresented; these sequences were found on 54 of the 127 genes (42%) (Fig. 3, blue), with 30 of those also sharing IFN TREs (Fig. 4). It has been reported recently that cRel directly binds to the promoter and regulates several ISG genes during IFN stimulation (Wei et al., 2008). Thus, cRel/NFκB may play an integral role in the interferon fingerprint associated with LPS preconditioning. Collectively, 79% (100 of 127) of the genes induced 24 h after stroke in LPS-preconditioned mice contain a regulatory sequence for IFN or NFκB (Fig. 4). The predominance of the type I interferon signature prompted us to pursue the possible role of enhanced type I interferon signaling in LPS-induced neuroprotection.
Figure 2.

LPS preconditioning induces a unique set of genes in response to MCAO. C57BL/6 mice were preconditioned with LPS (0.2 mg/kg) or saline 72 h before MCAO (45 min). At 3 or 24 h after MCAO, mice (n = 4 per time point) were killed, and the ipsilateral cortical brain tissue was collected. RNA was isolated and hybridized to Affymetrix gene chips (MOE430). Venn diagram showing the number of genes differentially regulated in each condition compared with unhandled controls. Arrows indicate the direction of regulation.
Table 3.
TREs identified as significantly overrepresented in genes induced 24 h after stroke in LPS-preconditioned mice
| Transcriptional regulatory element | Adjusted p values for overrepresentation |
|
|---|---|---|
| LPS group | Saline group | |
| c-Rel/V$CREL_01 | 0.00 | >1.00 |
| IRF/V$IRF_Q6 | 0.00 | >1.00 |
| IRF/V$IRF_Q6_01 | 0.01 | >1.00 |
| NF-kappaB (p65)/V$NFKAPPAB65_01 | 0.01 | 0.86 |
| RREB-1/V$RREB1_01 | 0.01 | >1.00 |
| IRF-8/V$ICSBP_Q6 | 0.03 | 0.99 |
| NF-Y/V$NFY_Q6 | 0.03 | >1.00 |
| ISRE/V$ISRE_01 | 0.07 | >1.00 |
| STAT5B (homodimer)/V$STAT5B_01 | 0.07 | >1.00 |
| IRF-7/V$IRF7_01 | 0.15 | >1.00 |
| COMP1/V$COMP1_01 | 0.17 | 0.37 |
| Freac-3/V$FREAC3_01 | 0.17 | >1.00 |
| Muscle TATA box/V$MTATA_B | 0.17 | >1.00 |
| Ik1/V$IK1_01 | 0.199 | 0.64 |
| S8/V$S8_01 | >1.00 | 0.00 |
| E2/V$E2_01 | 0.69 | 0.0024 |
| C/EBPβ /V$CEBPB_02 | >1.00 | 0.13 |
| HNF-1/V$HNF1_C | >1.00 | 0.13 |
| Myogenin/NF-1/MYOGNF1_01 | 0.94 | 0.19 |
Figure 3.
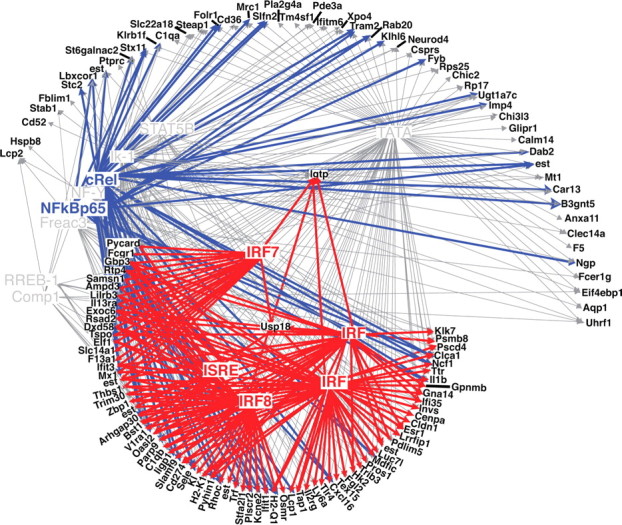
Interferon- and NFκB-related TREs identified in the majority of genes increased after stroke in LPS-preconditioned mice. Hypothetical gene–TRE network showing the relationship of the identified TREs to the genes increased after stroke in LPS- preconditioned mice. Genes are depicted in black, interferon-associated TREs are represented in red, NFκB-associated TREs are in blue, and other TREs are in gray. p value threshold set at 0.2.
Figure 4.
Genes induced in LPS-preconditioned mice after stroke with identified interferon- or NFκB-related TREs. Matrices depicting the occurrence of IFN and NFκB regulatory sites within the genes induced 24 h after stroke in LPS-preconditioned mice. Genes are listed down the right side, with TREs listed across the top. Identification of a TRE within the 5′ sequence of a gene is denoted with a red box. Red asterisks show genes induced by type I interferons as reported in the published literature.
Increased levels of IFNβ after stroke in LPS-preconditioned mice
The increase in interferon-inducible genes and overrepresentation of interferon-associated TREs suggested that IFNβ may be present in the brain cortex after stroke in LPS-preconditioned mice. Using real-time PCR, we examined the levels of IFNβ transcript in the brain after stroke in LPS-preconditioned and saline-treated mice. IFNβ levels were increased after stroke in the preconditioned and non-preconditioned mice compared with unhandled controls (Fig. 5). However, levels in LPS-preconditioned mice were ninefold higher at 3 h (LPS treated, 59.4 ± 22 vs saline treated, 6.7 ± 3; p < 0.001) and 3.5-fold higher at 24 h (LPS treated, 45.3 ± 23 vs saline treated, 11.7 ± 6; p < 0.001) after stroke. We examined levels of IFNβ just before MCAO (72 h after injection) to confirm that the increase in IFNβ after stroke was independent of any residual increase of IFNβ resulting from the preconditioning LPS injection. Levels of IFNβ in LPS- and saline-treated mice were statistically equivalent to unhandled controls (1.49 ± 1.4 and 0.74 ± 0.6, respectively) (data not shown). Thus, after stroke, mice preconditioned with LPS mount a more robust IFNβ response to ischemic injury.
Figure 5.
Increased levels of IFNβ after MCAO in LPS-preconditioned mice. Real-time PCR analysis was performed on RNA derived from the cortices after MCAO (3 and 24 h) of mice preconditioned with either LPS or saline. Data were normalized to β-actin. Results are presented as fold increase relative to unhandled controls. n = 3–4 mice per group; data are group means ± SEM; ***p < 0.001 by two-way ANOVA with Bonferroni's post hoc test.
IFNβ does not play a role in the endogenous response of the brain to ischemia
IFNβ after stroke in non-preconditioned mice was increased 6.7-fold over unhandled mice (Fig. 5). Although not as robust as in LPS-preconditioned mice, this does suggest that IFNβ may play a protective role in the endogenous response to stroke. To determine whether the increase after stroke alone could be protective, IFNβ knock-out mice were subjected to 40 min MCAO, followed by 72 h of reperfusion. Wild-type and IFNβ knock-out mice displayed infarcts of similar size (38.5 ± 2 vs 39.5 ± 1%; p = 0.7). Thus, IFNβ itself does not appear to play a critical role in the usual response by the brain to ischemia.
IFNβ protects against ischemic injury
To determine whether the more robust increase in IFNβ (ninefold) (Fig. 5) after stroke in LPS-preconditioned mice would effect ischemia, we tested the effect of exogenous IFNβ administration in the brain after MCAO. We injected C57BL/6 mice intracerebroventricularly with recombinant mouse IFNβ immediately before and after MCAO and measured infarct size 24 h later. Animals treated with IFNβ showed a significant reduction in infarct volume versus vehicle-treated mice (31.9 ± 4 vs 49.4 ± 2%; p < 0.001). This result supports the notion that increased expression of IFNβ within the brain would confer protection from ischemic injury.
TRIF-mediated signaling is protective in ischemia modeled in vitro
The increase in IFNβ and type I interferon-associated genes in response to stroke in the LPS-preconditioned mice mirrors the secondary response to LPS in classic endotoxin tolerance and supports a possible reprogramming of the TLR response to stroke, resulting in a TRIF-mediated event. To determine whether signaling via TRIF after stroke would induce protection, we used the synthetic TLR3 ligand Poly I:C (InvivoGen), which signals exclusively via a TRIF-dependent pathway. We reasoned that activation of this pathway at the time of ischemia would provide acute protection from ischemic damage. To test this, mixed cortical cultures from mice were exposed to OGD for 3 h, followed by treatment with varying doses of Poly I:C and subsequently returned to normoxic conditions. Twenty-four hours later, cell death was determined. Acute Poly I:C treatment after OGD significantly reduced OGD-mediated cell death at all three doses tested (Fig. 6). Thus, signaling via TRIF after ischemia provides protection against damage in vitro, which suggests that TRIF-mediated signaling in the brain after stroke could reduce ischemic injury.
Figure 6.
TRIF signaling after oxygen glucose deprivation induces neuroprotection. Mixed cortical cultures were exposed to 3 h OGD, followed by treatment with increasing doses of Poly I:C. Cell death was assessed by PI staining performed 24 h after. Mean ± SEM are shown; ***p < 0.001 versus media-treated OGD control. n = 2–4 individually repeated experiments. Poly I:C treatment at the highest dose without exposure to OGD did not induce cell death (data not shown).
IRF3 is required for LPS-induced protection from brain ischemia
To further explore the TRIF–IRF3 pathway, we tested whether IRF3 is a critical effector of LPS-induced ischemic protection. First, we determined whether IRF3 is involved in the endogenous response by the brain to stroke. IRF3 knock-out mice were subjected to 40 min MCAO, followed by 72 h of reperfusion. IRF3 knock-out mice displayed infarcts of similar size to wild-type mice (41.2 ± 5 vs 43.4 ± 4%; p = 0.8). Thus, IRF3 does not play a critical role in the usual response by the brain to ischemia. Next we determined whether IRF3 is a required effector of LPS-induced tolerance to ischemia. IRF3 knock-out mice were pretreated with LPS (0.4 mg/kg) 72 h before 40 min MCAO and killed 24 h later. Figure 7 shows that IRF3 knock-out mice fail to be preconditioned with LPS (17.3 vs 53.2% reduction). Hence, IRF3 is required for the protective effects of LPS pretreatment.
Figure 7.
IRF3 is an essential mediator of LPS preconditioning. IRF3 knock-out and wild-type mice were pretreated with LPS (0.4 mg/kg) or saline 72 h before 40 min MCAO. Infarct volume was measured 24 h after surgery using TTC staining. Data shown are group means ± SEM; *p < 0.05 by two-way ANOVA with Bonferroni's post hoc test; n = 7–10 per group.
Discussion
We propose a molecular model of LPS-induced neuroprotection from ischemic injury wherein systemic LPS preconditioning reprograms TLR4 signaling in response to stroke, directing it toward a neuroprotective pathway. Administration of systemic LPS induces an early inflammatory genomic response in the brain that has receded by 72 h. However, the response to stroke in these LPS-preconditioned mice is altered, and a new pattern of gene regulation is induced as early as 3 h after exposure to ischemia. The genomic changes in the brain in response to LPS suggest a possible activation of brain TLRs before the stroke, which go on to respond with an altered signaling pathway after activation by the stroke event. The fact that IFN regulatory elements are overrepresented among the unique genes induced after stroke in LPS- preconditioned animals suggests that TLR signaling in this setting may be altered. To pursue this, we examined the possibility that increased signaling via the TRIF-dependent pathway could serve as a neuroprotective mechanism associated with LPS preconditioning.
The induction of IFNβ is a hallmark of the TRIF-dependent TLR4 cascade (Biswas et al., 2007). In accordance with this, we found that LPS preconditioning increased IFNβ within the brain 3 and 24 h after stroke. We then tested whether the increase in IFNβ was required to confer neuroprotection. Mice lacking IFNβ displayed infarcts of similar size to wild-type mice, suggesting that endogenous IFNβ does not protect the brain from ischemic injury. However, exogenous administration of IFNβ intracerebroventricularly at the time of stroke conferred significant protection against ischemic damage, indicating that local upregulation of this cytokine may be neuroprotective. Our in vitro studies using modeled ischemia showed that activation of the TRIF–IRF3 pathway with the TLR3 ligand Poly I:C after OGD reduced neuronal death, thus supporting the notion that TRIF-dependent signaling can provide protection to ischemic injury.
The role of the TRIF–IRF3 cascade in the natural response of the brain to stroke appears relatively minor because mice lacking IRF3 displayed infarcts of similar size to wild-type mice. However, there appears to be a critical role for IRF3 in LPS preconditioning because IRF3-deficient mice could not be protected from ischemic injury in the setting of LPS preconditioning. Because these mice are deficient in IRF3 during the induction of tolerance as well as after the ischemic event, we cannot rule out a requirement for IRF3 during the induction phase as well as the resolution phase. In preliminary studies, we found that Poly I:C preconditioning is protective in stroke (our unpublished observation), which suggests that TRIF signaling may be sufficient for the induction of tolerance. In models of endotoxin–endotoxin (LPS) tolerance in which previous exposure to low-dose endotoxin provides protection against subsequent high-dose exposure, there is substantial support for the notion that TRIF–IRF3 signaling is required. Biswas et al. (2007) showed that endotoxin tolerance was induced in MyD88-deficient mice but not in TRIF- or IRF3-deficient mice (Biswas et al., 2007). Thus, although TRIF-dependent signaling is likely involved in the induction of tolerance, the enhancement of IRF3-dependent genes after stroke in LPS-preconditioned mice and the protective response elucidated with Poly I:C after OGD supports a role for the TRIF–IRF3 pathway in the protective phenotype as well.
We postulate that LPS preconditioning against ischemic injury results in redirected TLR4 signaling that resembles endotoxin tolerance. Cells made tolerant to endotoxin (LPS) are known to suppress the proinflammatory MyD88–TNFα pathway by upregulating pathway inhibitors, namely IRAK-M, Tollip, Ship1, and Trim30α, among others (Lang and Mansell, 2007; Shi et al., 2008), which results in decreased inflammatory cytokine responses during secondary exposure to TLR4 ligands. Inhibition of these pathways shunts subsequent TLR4 signaling down the TRIF–IRF3–IFNβ pathway and results in enhanced production of IFNβ (Bagchi et al., 2007). Similarly, LPS preconditioning may upregulate inflammatory pathway inhibitors in the brain that shunt subsequent TLR signaling down the TRIF–IRF3–IFNβ pathway. Thus, in the setting of ischemia, release of endogenous TLR ligands would be expected to lead to TLR signaling that is shunted down the TRIF–IRF3–IFNβ pathway and result in upregulation of IFNβ.
Our data support a model of redirected TLR4 signaling after stroke in preconditioned animals. Unlike control animals, LPS- preconditioned mice demonstrate a significant upregulation of IFNβ- and IFN-associated genes after stroke, which, based on promoter region analyses, are likely produced via the TLR4–TRIF–IRF3 pathway. Hirotani et al. (2005) have shown in endotoxin tolerance, using TRIF-deficient mice, that induction of type I interferon-associated genes after a secondary challenge is exclusively dependent on TRIF (Hirotani et al., 2005). Together, these data suggest that the type I IFN “fingerprint” is generated downstream of TLR4–TRIF–IRF3 and supports the concept of TLR4 reprogramming.
The potential for the reprogrammed TLR response to be protective is evinced by the TRIF-mediated neuronal protection in our in vitro model and via the protective effect of IFNβ directly administered in the brain. Others have shown that systemic administration of IFNβ reduces tissue damage in both a rat and rabbit model of ischemic stroke (Liu et al., 2002; Veldhuis et al., 2003). In these studies, the protective effects of IFNβ may have been mediated through leakage across the blood–brain barrier (BBB) after stroke wherein IFNβ may have then acted directly on the brain parenchyma. A recent study by Maier et al. (2006) in which BBB integrity is believed to be preserved failed to show attenuation of ischemic brain injury after systemic administration of IFNβ (Maier et al., 2006). Thus, as with our data, these results support the notion that the neuroprotective effects of IFNβ occur centrally.
We show that IRF3 is required for LPS-induced neuroprotection and that this is most likely through its induction of IFNβ and other ISGs. Two potential ISG modulators of the protective effect, Trim30 and Ifit1, were identified in our microarray analysis. Trim30 has been shown to negatively regulate LPS-induced TNFα and IL6 expression via inhibition of TLR4-induced NFκB activation (Shi et al., 2008). In the brain, we found increased mRNA levels of Trim30 at 3 and 24 h after LPS treatment and at 24 h after stroke in LPS-preconditioned mice. This induction could play a role in the suppression of inflammatory cytokines in the brain after stroke. Although Shi et al. (2008) reported that Trim30 expression depends on NFκB, the promoter sequence also contains an ISRE site (Fig. 4). In addition, we find induction of Ifit1 after LPS administration and again 24 h after stroke in LPS-preconditioned mice. The closely related gene Ifit2 has been shown recently to suppress the LPS-mediated induction of TNFα and IL6 (Berchtold et al., 2008), although it has not been directly linked to suppression of TLR signaling. The induction of these two ISG genes and IFNβ in LPS-preconditioned mice support the hypothesis of a reprogrammed TLR response to stroke resulting in a neuroprotective state.
Our data suggest that systemic administration of LPS reprograms TLR4-expressing cells within the brain. TLR4 is widely expressed in the brain (Lehnardt et al., 2002; Olson and Miller, 2004; Chakravarty and Herkenham, 2005), and many studies have shown that peripheral LPS induces a proinflammatory response within the brain (Chen et al., 2005; Qin et al., 2008). However, it is unclear whether LPS crosses the BBB and/or whether it induces peripheral cytokines, which in turn, cross into the brain. Recent evidence suggests that systemic LPS elicits TLR4 signaling in the brain independent of peripheral cytokine responses (Chakravarty and Herkenham, 2005; Gosselin and Rivest, 2008). However, other researchers have failed to find LPS within the brain parenchyma after systemic administration (Singh and Jiang, 2004). It is clear that LPS binds to cerebral endothelial cells (Singh and Jiang, 2004; Verma et al., 2006). Because these cells are an interface between the systemic circulation and the brain parenchyma, they may help integrate information from both compartments. Hence, reprogramming of TLR4 may occur within the cerebral endothelium.
In summary, we have shown that LPS preconditioning reprograms the cellular response to stroke and causes a type I IFN response, with a critical and protective role for IRF3. These reprogramming events may exemplify endogenous processes that protect the brain against additional injury and suggest that LPS preconditioning fundamentally changes the cellular response to stroke. This is the first demonstration that a preconditioning stimulus results in an interferon fingerprint after the ischemic event and the first report of a neuroprotective role for TRIF–IRF3 signaling after injury.
Footnotes
This work was supported by National Institutes of Health Grant R01 NS050567 (M.P.S.-P.). We thank Jo-Lynn Boule, Eric Tobar, Delfina Homen, Tao Yang, and Dr. Nikola Lessov for excellent technical support and Dr. Roger Simon for constructive discussion. Microarray assays were performed in the Affymetrix Microarray Core of the Oregon Health & Science University Gene Microarray Shared Resource.
References
- Bagchi A, Herrup EA, Warren HS, Trigilio J, Shin HS, Valentine C, Hellman J. MyD88-dependent and MyD88-independent pathways in synergy, priming, and tolerance between TLR agonists. J Immunol. 2007;178:1164–1171. doi: 10.4049/jimmunol.178.2.1164. [DOI] [PubMed] [Google Scholar]
- Berchtold S, Manncke B, Klenk J, Geisel J, Autenrieth IB, Bohn E. Forced IFIT-2 expression represses LPS induced TNF-alpha expression at posttranscriptional levels. BMC Immunol. 2008;9:75. doi: 10.1186/1471-2172-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, Bist P, Dhillon MK, Kajiji T, Del Fresno C, Yamamoto M, Lopez-Collazo E, Akira S, Tergaonkar V. Role for MyD88-independent, TRIF pathway in lipid A/TLR4-induced endotoxin tolerance. J Immunol. 2007;179:4083–4092. doi: 10.4049/jimmunol.179.6.4083. [DOI] [PubMed] [Google Scholar]
- Broad A, Kirby JA, Jones DE. Toll-like receptor interactions: tolerance of MyD88-dependent cytokines but enhancement of MyD88-independent interferon-beta production. Immunology. 2007;120:103–111. doi: 10.1111/j.1365-2567.2006.02485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao CX, Yang QW, Lv FL, Cui J, Fu HB, Wang JZ. Reduced cerebral ischemia-reperfusion injury in Toll-like receptor 4 deficient mice. Biochem Biophys Res Commun. 2007;353:509–514. doi: 10.1016/j.bbrc.2006.12.057. [DOI] [PubMed] [Google Scholar]
- Chakravarty S, Herkenham M. Toll-like receptor 4 on nonhematopoietic cels sustains CNS inflammation during endotoxemia, independent of systemic cytokines. J Neurosci. 2005;25:1788–1796. doi: 10.1523/JNEUROSCI.4268-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Zhou H, Beltran J, Malellari L, Chang SL. Differential expression of cytokines in the brain and serum during endotoxin tolerance. J Neuroimmunol. 2005;163:53–72. doi: 10.1016/j.jneuroim.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Fan H, Cook JA. Molecular mechanisms of endotoxin tolerance. J Endotoxin Res. 2004;10:71–84. doi: 10.1179/096805104225003997. [DOI] [PubMed] [Google Scholar]
- Faraco G, Fossati S, Bianchi ME, Patrone M, Pedrazzi M, Sparatore B, Moroni F, Chiarugi A. High mobility group box 1 protein is released by neural cells upon different stresses and worsens ischemic neurodegeneration in vitro and in vivo. J Neurochem. 2007;103:590–603. doi: 10.1111/j.1471-4159.2007.04788.x. [DOI] [PubMed] [Google Scholar]
- Gosselin D, Rivest S. MyD88 signaling in brain endothelial cells is essential for the neuronal activity and glucocorticoid release during systemic inflammation. Mol Psychiatry. 2008;13:480–497. doi: 10.1038/sj.mp.4002122. [DOI] [PubMed] [Google Scholar]
- Hickey EJ, You X, Kaimaktchiev V, Stenzel-Poore M, Ungerleider RM. Lipopolysaccharide preconditioning induces robust protection against brain injury resulting from deep hypothermic circulatory arrest. J Thorac Cardiovasc Surg. 2007;133:1588–1596. doi: 10.1016/j.jtcvs.2006.12.056. [DOI] [PubMed] [Google Scholar]
- Hirotani T, Yamamoto M, Kumagai Y, Uematsu S, Kawase I, Takeuchi O, Akira S. Regulation of lipopolysaccharide-inducible genes by MyD88 and Toll/IL-1 domain containing adaptor inducing IFN-beta. Biochem Biophys Res Commun. 2005;328:383–392. doi: 10.1016/j.bbrc.2004.12.184. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Gautier L, Cope LM. An R Package for oligonucleotide array statistical analysis. In: Parmigiani G, Irizarry RA, Zeger SL, editors. The analysis of gene expression data: methods and software. New York: Springer; 2003. [Google Scholar]
- Kinouchi H, Sharp FR, Hill MP, Koistinaho J, Sagar SM, Chan PH. Induction of 70-kDa heat shock protein and hsp70 mRNA following transient focal cerebral ischemia in the rat. J Cereb Blood Flow Metab. 1993a;13:105–115. doi: 10.1038/jcbfm.1993.13. [DOI] [PubMed] [Google Scholar]
- Kinouchi H, Sharp FR, Koistinaho J, Hicks K, Kamii H, Chan PH. Induction of heat shock hsp 70 mRNA and HSP70 kDa protein in neurons in the “penumbra” following focal cerebral ischemia in the rat. Brain Res. 1993b;619:334–338. doi: 10.1016/0006-8993(93)91630-b. [DOI] [PubMed] [Google Scholar]
- Lang T, Mansell A. The negative regulation of Toll-like receptor and associated pathways. Immunol Cell Biol. 2007;85:425–434. doi: 10.1038/sj.icb.7100094. [DOI] [PubMed] [Google Scholar]
- Lehnardt S, Lachance C, Patrizi S, Lefebvre S, Follett PL, Jensen FE, Rosenberg PA, Volpe JJ, Vartanian T. The toll-like receptor TLR4 is necessary for lipopolysaccharide-induced oligodendrocyte injury in the CNS. J Neurosci. 2002;22:2478–2486. doi: 10.1523/JNEUROSCI.22-07-02478.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnardt S, Lehmann S, Kaul D, Tschimmel K, Hoffmann O, Cho S, Krueger C, Nitsch R, Meisel A, Weber JR. Toll-like receptor 2 mediates CNS injury in focal cerebral ischemia. J Neuroimmunol. 2007;190:28–33. doi: 10.1016/j.jneuroim.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. doi: 10.1038/nri1630. [DOI] [PubMed] [Google Scholar]
- Liu H, Xin L, Chan BP, Teoh R, Tang BL, Tan YH. Interferon beta administration confers a beneficial outcome in a rabbit model of thromboembolic cerebral ischemia. Neurosci Lett. 2002;327:146–148. doi: 10.1016/s0304-3940(02)00371-3. [DOI] [PubMed] [Google Scholar]
- Maier CM, Yu F, Nishi T, Lathrop SJ, Chan PH. Interferon-beta fails to protect in a model of transient focal stroke. Stroke. 2006;37:1116–1119. doi: 10.1161/01.STR.0000208214.46093.d5. [DOI] [PubMed] [Google Scholar]
- Meller R, Stevens SL, Minami M, Cameron JA, King S, Rosenzweig H, Doyle K, Lessov NS, Simon RP, Stenzel-Poore MP. Neuroprotection by osteopontin in stroke. J Cereb Blood Flow Metab. 2005;25:217–225. doi: 10.1038/sj.jcbfm.9600022. [DOI] [PubMed] [Google Scholar]
- Nakaya T, Sato M, Hata N, Asagiri M, Suemori H, Noguchi S, Tanaka N, Taniguchi T. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem Biophys Res Commun. 2001;283:1150–1156. doi: 10.1006/bbrc.2001.4913. [DOI] [PubMed] [Google Scholar]
- Olson JK, Miller SD. Microglia initiate central nervous system innate and adaptive immune responses through multiple TLRs. J Immunol. 2004;173:3916–3924. doi: 10.4049/jimmunol.173.6.3916. [DOI] [PubMed] [Google Scholar]
- Qin L, He J, Hanes RN, Pluzarev O, Hong JS, Crews FT. Increased systemic and brain cytokine production and neuroinflammation by endotoxin following ethanol treatment. J Neuroinflammation. 2008;5:10. doi: 10.1186/1742-2094-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig HL, Lessov NS, Henshall DC, Minami M, Simon RP, Stenzel-Poore MP. Endotoxin preconditioning prevents the cellular inflammatory response during ischemic neuroprotection in mice. Stroke. 2004;35:2576–2581. doi: 10.1161/01.STR.0000143450.04438.ae. [DOI] [PubMed] [Google Scholar]
- Rosenzweig HL, Minami M, Lessov NS, Coste SC, Stevens SL, Henshall DC, Meller R, Simon RP, Stenzel-Poore MP. Endotoxin preconditioning protects against the cytotoxic effects of TNFa after stroke: a novel role for TNFa in LPS-ischemic tolerance. J Cereb Blood Flow Metab. 2007;27:1663–1674. doi: 10.1038/sj.jcbfm.9600464. [DOI] [PubMed] [Google Scholar]
- Shi M, Deng W, Bi E, Mao K, Ji Y, Lin G, Wu X, Tao Z, Li Z, Cai X, Sun S, Xiang C, Sun B. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat Immunol. 2008;9:369–377. doi: 10.1038/ni1577. [DOI] [PubMed] [Google Scholar]
- Singh AK, Jiang Y. How does peripheral lipopolysaccharide induce gene expression in the brain of rats? Toxicology. 2004;201:197–207. doi: 10.1016/j.tox.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Stevens SL, Bao J, Hollis J, Lessov NS, Clark WM, Stenzel-Poore MP. The use of flow cytometry to evaluate temporal changes in inflammation. Brain Res. 2002;932:110–119. doi: 10.1016/s0006-8993(02)02292-8. [DOI] [PubMed] [Google Scholar]
- Swanson RA, Morton MT, Tsao-Wu G, Savalos RA, Davidson C, Sharp FR. A semiautomated method for measuring brain infarct volume. J Cereb Blood Flow Metab. 1990;10:290–293. doi: 10.1038/jcbfm.1990.47. [DOI] [PubMed] [Google Scholar]
- Tasaki K, Ruetzler CA, Ohtsuki T, Martin D, Nawashiro H, Hallenbeck JM. Lipopolysaccharide pre-treatment induces resistance against subsequent focal cerebral ischemic damage in spontaneously hypertensive rats. Brain Res. 1997;748:267–270. doi: 10.1016/s0006-8993(96)01383-2. [DOI] [PubMed] [Google Scholar]
- Vadigepalli R, Chakravarthula P, Zak DE, Schwaber JS, Gonye GE. PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. OMICS. 2003;7:235–252. doi: 10.1089/153623103322452378. [DOI] [PubMed] [Google Scholar]
- Veldhuis WB, Derksen JW, Floris S, Van Der Meide PH, De Vries HE, Schepers J, Vos IM, Dijkstra CD, Kappelle LJ, Nicolay K, Bär PR. Interferon-beta blocks infiltration of inflammatory cells and reduces infarct volume after ischemic stroke in the rat. J Cereb Blood Flow Metab. 2003;23:1029–1039. doi: 10.1097/01.WCB.0000080703.47016.B6. [DOI] [PubMed] [Google Scholar]
- Verma S, Nakaoke R, Dohgu S, Banks WA. Release of cytokines by brain endothelial cells: a polarized response to lipopolysaccharide. Brain Behav Immun. 2006;20:449–455. doi: 10.1016/j.bbi.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Wei L, Fan M, Xu L, Heinrich K, Berry MW, Homayouni R, Pfeffer LM. Bioinformatic analysis reveals cRel as a regulator of a subset of interferon-stimulated genes. J Interferon Cytokine Res. 2008;28:541–551. doi: 10.1089/jir.2007.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MA, Heagy W. Endotoxin tolerance: a review. Crit Care Med. 2002;30:S64–S73. [PubMed] [Google Scholar]
- Ziegler G, Harhausen D, Schepers C, Hoffmann O, Röhr C, Prinz V, König J, Lehrach H, Nietfeld W, Trendelenburg G. TLR2 has a detrimental role in mouse transient focal cerebral ischemia. Biochem Biophys Res Commun. 2007;359:574–579. doi: 10.1016/j.bbrc.2007.05.157. [DOI] [PubMed] [Google Scholar]






