Abstract
Several single nucleotide polymorphism (SNP) genome-wide association studies (GWAS) have been completed in multiple sclerosis. Follow-up studies of the variants with the most promising rankings, especially when supplemented by informed candidate gene selection, have proven to be extremely successful. We report here the results of a multi-stage replication analysis of the putatively associated SNPs identified in the Wellcome Trust Case-Control Consortium non-synonymous SNP screen. In total the replication sample consisted of 3444 patients and 2595 controls. A combined analysis of the nsSNP screen and replication data provides evidence implicating a novel additional locus, rs3748816 in MMEL1 (OR=1.16, p=3.54×10−6) in multiple sclerosis susceptibility.
Keywords: Multiple Sclerosis, MMEL1, genetics
Introduction
Multiple sclerosis (MS) is considered to be an autoimmune disease of the central nervous system (CNS) that is characterised pathologically by inflammation, demyelination and axonal degeneration.1 Epidemiological evidence suggests that it results from unknown environmental factors acting on genetically susceptible individuals.2; 3
In the last two years the application of high-throughput genotyping methodologies has resulted in rapid progress in the identification of relevant susceptibility genes in MS. To date five Single Nucleotide Polymorphism (SNP) Genome-Wide Association Studies (GWAS) have been performed in MS; four based on indirectly screening common variation through linkage disequilibrium (LD)4–7 and the fifth based on directly typing non-synonymous coding SNPs (nsSNPs).8 It is this last study, performed as part of the Wellcome Trust Case Control Consortium (WTCCC) that has provided the foundation for the work reported here.
Individually none of the five MS GWAS thus far completed succeeded in identifying statistically unequivocal association at the screening stage (outside the expected association with the Major Histocompatibility Complex, MHC) although each provides an invaluable ranking of the variants tested. The extent to which the higher ranges of such rankings are enriched for genuine associations depends upon the power of the study and the prior odds that the variants tested are causative, or are in tight LD with such. Follow up studies of the variants with the most promising rankings, especially when supplemented by informed candidate gene selection, have proven to be extremely successful in MS. For example, by extending the typing of a modest number of the most promising variants identified in the WTCCC nsSNP screen we have already identified association with the tyrosine kinase 2 gene (TYK2) and MS.9 Similar highly focused replication efforts based on GWAS results have been equally successful in other complex genetic diseases such as celiac disease,10 type I diabetes11 and rheumatoid arthritis.12 In this article we report our most recent efforts to mine more deeply into the ranking generated in the nsSNP screen.8
Results
A two stage replication effort was employed in this study. In the first stage 1664 of the most promising markers identified in the nsSNP screen were typed in an independent set of 1038 cases and 1379 controls. These new data were combined with the original data from the nsSNP screen in order to provide a refined ranking. The results of this extension analysis are provided in Supplementary Table S1. In stage 2 of our replication effort we chose the 13 most promising SNPs identified from the extension analysis for further investigation in 2406 cases and 1216 controls. Five SNPs were found to have p < 10−4, however as rs601338 and rs602662 are in tight LD we only considered rs602662 further (the more strongly associated). Of the remaining 331 SNPs with a p < 0.05 we chose a further nine SNPs that were within candidate genes. As one of these SNPs (rs12912505) failed manufacture, in total we genotyped 12 SNPs.
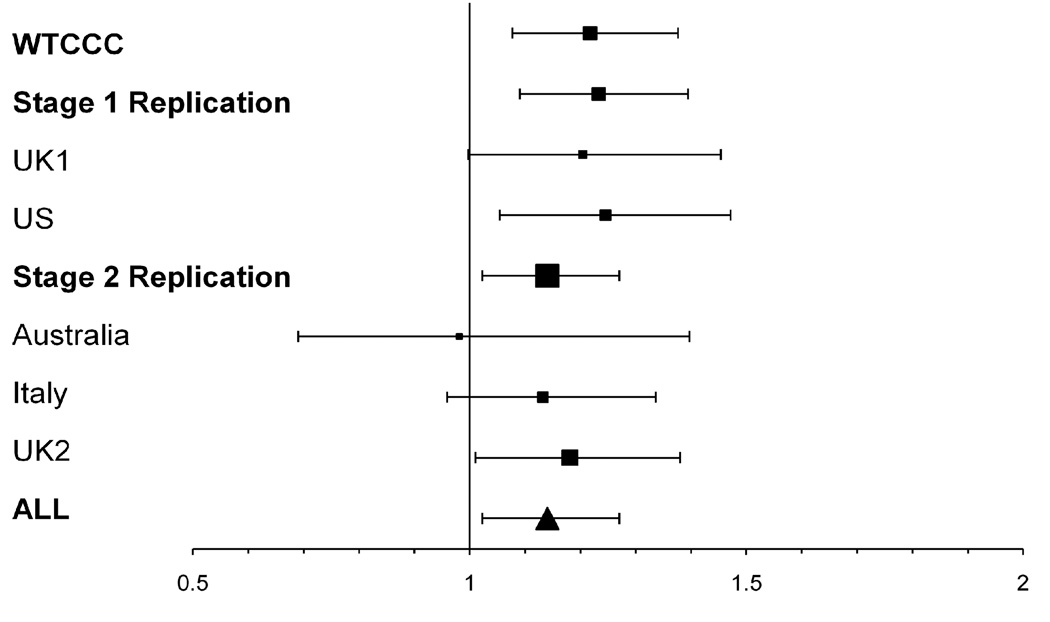
The results of the 12 SNPs analysed in both replication stages are shown in Table 2 along with the combined analysis of all available data from the nsSNP screen, stage 1 and stage 2 replication efforts. The population specific results for the top 12 SNPs are shown in Supplementary Table S2. Using the Breslow-Day test, some evidence for heterogeneity was shown for one SNP (rs379707). A random effects model was therefore applied for the analysis of this SNP, though it did not alter the Odds Ratio (OR) (data not shown). In the combined analysis of all available data, the most significant association was with rs3748816 which lies in the membrane metalloendopeptidase-like 1 gene (MMEL1) (p=3.54×10−6, OR=1.16, 95% CI=1.09–1.24). The modest power in the second stage limits the extent to which the evidence for association is increased by this typing. However the result for rs3748816 remains nominally significant even if corrected for the number of tests performed in the original screen. A detailed plot of the effect size of this variant within MMEL1 by country of ascertainment is shown in Figure 1.
Table 2.
Results from the replication analysis of the most promising 12 SNPs
| SNP | Locus | Chr | Risk Allele |
WTCCC nsSNP Screena |
Replication Stage1b |
Replication Stage 2c |
Replication Analysis Combined |
Combined Analysisd | RAF Case |
RAF Control |
Breslow Day Test |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-value | p-value | p-value | p-value | p-value | Odds Ratio (CI 95%) | p-value | ||||||
| rs3748816 | MMEL1 | 1 | T | 1.63×10−3 | 1.34×10−3 | 0.0178 | 3.57×10−4 | 3.54×10−6 | 1.16 (1.09–1.24) | 0.696 | 0.660 | 0.650 |
| rs2304497 | ACLY | 17 | A | 0.0187 | 9.40×10−4 | 0.389 | 1.11×10−3 | 9.76×10−5 | 1.21 (1.10–1.33) | 0.898 | 0.877 | 0.540 |
| rs4801853 | C19orf48 | 19 | C | 3.18×10−3 | 0.0130 | 0.0744 | 3.24×10−3 | 1.13×10−4 | 1.14 (1.07–1.22) | 0.741 | 0.712 | 0.634 |
| rs930557 | MCPH1 | 8 | G | 4.64×10−3 | 0.395 | 0.254 | 0.0279 | 5.87×10−4 | 1.15 (1.06–1.24) | 0.203 | 0.181 | 0.292 |
| rs602662 | FUT2 | 19 | C | 0.0151 | 6.95×10−4 | 0.738 | 0.0229 | 1.40×10−3 | 1.10 (1.04–1.17) | 0.493 | 0.467 | 0.681 |
| rs2043211 | CARD8 | 19 | T | 0.1514 | 0.0592 | 0.160 | 0.0311 | 0.0111 | 1.09 (1.02–1.16) | 0.332 | 0.314 | 0.742 |
| rs2066807 |
STAT2- IL23A |
12 | C | 0.0583 | 0.0637 | 0.640 | 0.107 | 0.0187 | 1.16 (1.03–1.31) | 0.941 | 0.931 | 0.344 |
| rs10489990 | CD207 | 2 | A | 3.43×10−3 | 0.0461 | 0.400 | 0.460 | 0.0340 | 1.07 (1.01–1.14) | 0.363 | 0.347 | 0.0845 |
| rs2034310 | CD300LF | 17 | T | 0.0223 | 0.0246 | 0.532 | 0.197 | 0.0444 | 1.09 (1.00–1.18) | 0.176 | 0.166 | 0.285 |
| rs40401 | IL3 | 5 | A | 0.153 | 0.0239 | 0.465 | 0.280 | 0.0827 | 1.07 (0.99–1.15) | 0.223 | 0.213 | 0.227 |
| rs2290610 | IL5RA | 3 | A | 0.0990 | 0.0587 | 0.0369 | 0.834 | 0.423 | 1.03 (0.96–1.09) | 0.637 | 0.634 | 0.0511 |
| rs379707 | FYB | 5 | T | 0.0257 | 0.169 | 0.0163 | 0.458 | 0.697 | 1.01 (0.95–1.08) | 0.696 | 0.695 | 0.00741 |
Chr = chromosome; RAF = risk allele frequency
Results shown in bold are in the opposite direction to the association identified in the WTCCC nsSNP screen
The WTCCC nsSNP screen included 975 cases and 1466 controls from the UK population
Replication Stage 1 includes a total of 1038 cases and 1379 controls from the UK and US population
Replication Stage 2 includes a total of 2406 cases and 1216 controls from the Australia, Italy and the UK population
The combined analysis includes all data from the WTCCC nsSNP screen, replication stage 1 and replication stage 2
Figure 1. Odds ratio for the rs3744816(T) allele in MMEL1.
The area of the box is proportional to the number of cases included in the study. The error bars indicate the 95% confidence intervals. The population specific odds ratios are shown for reference.
Discussion
By systematically extending the analysis of the putatively associated SNPs identified in our nsSNP GWAS we provide suggestive evidence implicating an additional locus (rs3748816 in MMEL1) involved in MS susceptibility. The SNP rs3748816 is located in exon 15 of MMEL1 with the protective ancestral “C” allele coding for the polar hydrophilic amino acid Threonine (ACG) and the risk “T” allele coding a non-polar hydrophobic Methionine (ATG) at amino acid position 509. This change is predicted to be “probably damaging” based on the program Polymorphism Phenotyping (PolyPhen). MMEL1, which is also known as Neprilysin 2 (NEP2), belongs to the M13 family of metalloendopeptidases. Unlike other members of this family that are ubiquitously expressed, MMEL1 expression is limited to the testis and CNS. In rodents, the largest isoform is predominantly expressed in testis, cleavage of this isoform producing a secreted soluble enzyme. The second isoform that is 23 amino acids shorter is a membrane-associated protein highly expressed in the CNS.13 Little data exists though as to the exact function of the MMEL1 gene. As with other members of the metalloendopeptidase family, MMEL1 appears to be involved in neuropeptide degradation,13 and a role in Alzheimer’s disease has also been suggested as it is involved in the degradation of beta-amyloid.14
The associated SNP lies within a region of extensive LD on chromosome 1 encompassing the MMEL1 gene and just extending to the tumour necrosis factor receptor superfamily 14 gene (TNFRSF14). A SNP within TNFRSF14 (rs2234167) that lies on the boundary of this LD block was typed in the first replication stage but did not show any evidence for significance in the combined analysis of the original nsSNP screen and stage 1 replication data (p=0.2514). The LD between rs3748816 and rs2234167, as determined from the 4858 individuals that were typed for both markers in stage 1 of the study, is low (D’=1, r2=0.083). Considerable work will be required to fine map this signal to ascertain if the signal is due to association with TNFRSF14 or MMEL1.
Two recent GWAS in rheumatoid arthritis (RA) also identified an association with the MMEL1 gene.12; 15 The SNPs tested in the RA screens (rs3890745 and rs10910099) were not analysed in the current study. Both SNPs though are in strong LD with rs3748816 (r2=0.962 and 0.924 respectively based on HapMap data) with the major allele increasing susceptibility in both of the RA screens and in our study. Growing evidence suggests that there is an overlap in susceptibility factors between autoimmune diseases,16; 17 yet it remains to be determined if the association with MMEL1 in MS and RA is due to the same variant. Further work is required to fully establish the function of this gene and hence its potential role in MS disease pathogenesis.
A further three markers showed evidence for association in our final analysis with p-values < 0.001. While the evidence for association is not definitive, further analysis in larger samples is probably merited for all three markers. The strongest signal of these three markers was with rs2304497 in the ATP-citrate lyase (ACLY) gene. Rs2304497 lies in exon 4 of ACLY and changes a glutamic acid (Glu) to aspartic acid (Asp) at amino acid position 175, a change that is predicted to be “benign” (polyPhen). ACLY lies within the same LD block as the promoter for the 2′,3′-cyclic-nucleotide 3′-phosphodiesterase (CNP) gene on chromosome 17q, a gene which promotes microtubule assembly18 and in mice has been shown to be essential for axonal survival.19 The next strongest association was with rs4801853 in the c19orf48 which lies within a gene dense region on chromosome 19q13.33. In the initial analysis of our nsSNP screen and the stage 1 replication samples, six SNPs within this region were associated with MS at p <0.05. We only chose the most significantly associated SNP to study further. Amongst the genes in this region is a cluster of the kallikrein genes. Kallikreins are a family of serine proteases, of which KLK6 has been shown to be upregulated in MS plaques20 and both KLK1 and KLK6 have been implicated in degeneration and disease progression in MS.21 KLK1 and KLK3 have also been associated with susceptibility to Systemic Lupus Erythematosis (SLE).22 Finally, the association with the rs930557 marker in the Microcephalin (MCPH1) gene may be the result of uncorrected population stratification as this SNP is under strong positive selection in European and Asian populations.23
At the time the WTCCC nsSNP screen was designed an attempt was made to include all known coding variants, however, the 12,374 SNPs included in the screen represent only a small fraction of such variants. The tendency for nsSNPs to have a lower allele frequency also means that even those that have been tested will probably not have been examined to a level where effects can be excluded. For example, in the combined analysis of our nsSNP screen and stage 1 replication data, at a 5% risk allele frequency we only have 53% power to identify a variant with an OR of 1.2 at a p-value < 0.05,24 making it likely that genuinely associated low frequency variants may have been missed. Furthermore, by screening only nsSNPs, other variations that may be involved in disease such as intronic or copy number variations will not have been captured. As with other genuine associations seen in complex genetic diseases, the associated variants we have identified in this and other studies have a relatively limited effect on an individual’s risk, but together the growing number of validated associations is helping to build a greater understanding of the mechanisms of disease pathogenesis.
In summary, we have found suggestive evidence for an association with rs3748816 in the MMEL1 gene. Independent replication and functional analysis will be necessary to fully confirm this locus as a genuine susceptibility gene in MS.
Materials and Methods
Samples
The first replication stage included subjects recruited through the University of Cambridge, UK; Brigham and Women’s Hospital in Boston, USA; and the University of California San Francisco, USA. Healthy unrelated controls were also included from both US sites and from the 1958 birth cohort in the UK. Controls were selected in order to optimise the matching of gender and age. The second replication stage included additional samples recruited through the University of Cambridge, UK; John Hunter Hospital Newcastle, Australia; and the University of Eastern Piedmont, Novara Italy. All samples were collected with informed written consent and appropriate local ethical approval. All patients satisfied recognised clinical criteria for the diagnosis of MS.25; 26 Subjects were chosen with all four main types of MS and varying disability. Full demographic features are provided in Table 1. The individuals included in the original WTCCC non-synonymous screen are described elsewhere.8
Table 1.
Demographic features of affected individuals
| Samples | Cases | Controls | Average Age at Onset |
EDSS % | Disease Course % | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <3 | 3 to <6 | ≥6 | Unkn | RR | SP | PP | PR | Unkn | ||||
| WTCCC nsSNP screen | 975 | 1466 | 30.3 | 27.2 | 24.3 | 48.1 | 0.4 | 54.6 | 32.0 | 13.0 | 0.0 | 0.4 |
| Replication Stage 1 | 1038 | 1379 | ||||||||||
| UK1 | 369 | 764 | 32.3 | 35.8 | 17.6 | 43.6 | 3.0 | 54.2 | 26.3 | 15.5 | 0.0 | 3.8 |
| US | 669 | 615 | 33.9 | 53.0 | 23.8 | 23.3 | 0 | 49.9 | 17.9 | 10.2 | 3.0 | 19.0 |
| Replication Stage 2 | 2406 | 1216 | ||||||||||
| Australia | 159 | 120 | 34.4 | 33.8 | 38.9 | 26.8 | 0.6 | 63.9 | 18.4 | 5.1 | 2.5 | 10.1 |
| Italy | 829 | 635 | 32.5 | 47.1 | 25.2 | 16.6 | 11.2 | 69.0 | 11.3 | 8.7 | 1.4 | 9.6 |
| UK2 | 1418 | 461 | 30.0 | 28.2 | 27.3 | 42.5 | 2.0 | 57.9 | 31.0 | 8.8 | 0.0 | 2.3 |
EDSS = Expanded Disability Status Scale; RR = Relapse Remitting; SP = Secondary Progressive; PP = Primary Progressive; PR – Progressive Relapsing; Unkn = Unknown
Markers
In the original WTCCC nsSNP screen a carefully developed set of over 15,000 nsSNPs were typed in five UK DNA sets: controls from the 1958 birth cohort (1958BC) and patient samples from individuals with MS, ankylosing spondylitis (AS), autoimmune thyroid disease (ATD) and breast cancer (BC). After the application of stringent quality control the primary MS specific component of the analysis (MS vs 1958BC) yielded reliable data from 12,374 SNPs in up to 975 MS cases and 1466 controls. Two additional analyses were conducted using data from the other conditions considered in the project on the basis that for most variants these samples would represent reasonable “controls” in an analysis of susceptibility to MS. In the first of these additional analyses the MS data was compared with that from all other samples considered (MS vs 1958BC+AS+AITD+BC) and in the second (an “immune” based analysis) the three autoimmune conditions were compared with the non-autoimmune samples (MS+AS+AITD vs 1958BC+BC). We used the following criteria to select markers for inclusion in our current follow-up effort:
P-value ranked within the top 1% of any of the three MS relevant analyses (n=303).
Having p<0.1 in both the main nsSNP analysis (MS vs 1958BC) and in the parallel screen performed by the International Multiple Sclerosis Genetics Consortium (IMSGC)4 (n=99). This obviously only applied to those variants that were included in both studies.
Any marker with a minor allele frequency of >5% and p<0.2 in the main nsSNP analysis (MS vs 1958BC) (n=1506).
After excluding the 18 SNPs we had previously investigated 9 these criteria identified a total of 1908 SNPs worthy of further follow up, of which 1853 were successfully genotyped. Following the application of stringent quality control measures (see below), 1664 of these gave reliable data.
Genotyping/Quality Control - stage 1
Genotyping was performed using the Illumina Infinium 60K BeadChip platform.27 The Illumina Infinium protocol involves amplification of genomic DNA, fragmentation of this DNA, hybridization of the DNA on the BeadChip, extension on the BeadChip, and imaging to read the chip. Samples were excluded if the genotyping efficiency was low (<95%). Similarly SNPs were excluded if the genotyping success rate was <95% or there was evidence of significant deviation from Hardy-Weinberg equilibrium (HWE) (p<0.001).
Genotyping/Quality Control - stage 2
The 13 SNPs considered in stage 2 of the replication were genotyped using TaqMan® Assays-On-Demand (Applied Biosystems). One of the SNPs however failed in the manufacturing process (rs12912505) so in total 12 SNPs were analysed. Genotyping was completed on a 7900 Sequence Detection System and plots generated and analysed using Sequence Detection System Software v2.1. All 12 SNPs fulfilled quality control requirements with a genotyping success rate >95% and Hardy-Weinberg equilibrium p>0.001. In this stage samples were excluded if the genotyping success was <50%. This resulted in the exclusion of a total of 48 samples, 14 from the Italian dataset (2 cases, 12 controls) and 34 from the UK dataset (15 cases, 19 controls) which resulted in the final analysis of a total of 2406 cases and 1216 controls in the second stage replication effort.
Statistical analysis
Data quality checking and statistical analysis was completed using PLINK v 1.05. As the results of several populations were combined, we analysed the data using a Cochran-Mantel-Haenzel test treating each population as a separate stratum and tested for heterogeneity between populations using the Breslow-Day statistic (see Table 2).
Supplementary Material
Acknowledgements
We thank the many multiple sclerosis patients and healthy individuals who participated in this study and the International Multiple Sclerosis Genetics Consortium (IMSGC, https://www.imsgc.org/) for providing the network through which we were able to collaborate. This work was supported by the Medical Research Council (G0700061) and MRC programme grant U.1052.00.012.00001.01, the National Institute of Health (R01 NS049477 and NS032830) and the Cambridge NIHR Biomedical Research Centre. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113. We acknowledge use of DNA from the British 1958 Birth Cohort collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. JLM was supported by a grant from the National Multiple Sclerosis Society (RG4201-A-1). PLD is a Harry Weaver Neuroscience Scholar of the National MS Society. L.B. is supported by a PhD Lagrange Fellowship. SD is supported by a FISM grant (2008/R/11).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372(9648):1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 2.Hogancamp WE, Rodriguez M, Weinshenker BG. The epidemiology of multiple sclerosis. Mayo Clin Proc. 1997;72(9):871–878. doi: 10.4065/72.9.871. [DOI] [PubMed] [Google Scholar]
- 3.Hauser SL, Oksenberg JR. The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron. 2006;52(1):61–76. doi: 10.1016/j.neuron.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 4.International Multiple Sclerosis Genetics Consortium (IMSGC) Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357(9):851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 5.Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18(4):767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ANZgene. Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41(7):824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 7.Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J, et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 2008;40(12):1402–1403. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]
- 8.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ban M, Goris A, Lorentzen AR, Baker A, Mihalova T, Ingram G, et al. Replication analysis identifies TYK2 as a multiple sclerosis susceptibility factor. Eur J Hum Genet. 2009;17(10):1309–1313. doi: 10.1038/ejhg.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40(4):395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39(7):857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40(10):1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ouimet T, Facchinetti P, Rose C, Bonhomme MC, Gros C, Schwartz JC. Neprilysin II: A putative novel metalloprotease and its isoforms in CNS and testis. Biochem Biophys Res Commun. 2000;271(3):565–570. doi: 10.1006/bbrc.2000.2664. [DOI] [PubMed] [Google Scholar]
- 14.Huang JY, Bruno AM, Patel CA, Huynh AM, Philibert KD, Glucksman MJ, et al. Human membrane metallo-endopeptidase-like protein degrades both beta-amyloid 42 and beta-amyloid 40. Neuroscience. 2008;155(1):258–262. doi: 10.1016/j.neuroscience.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barton A, Thomson W, Ke X, Eyre S, Hinks A, Bowes J, et al. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet. 2008;40(10):1156–1159. doi: 10.1038/ng.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Multiple Sclerosis Genetics Consortium (IMSGC) The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun. 2009;10(1):11–14. doi: 10.1038/gene.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fung EY, Smyth DJ, Howson JM, Cooper JD, Walker NM, Stevens H, et al. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun. 2009;10(2):188–191. doi: 10.1038/gene.2008.99. [DOI] [PubMed] [Google Scholar]
- 18.Bifulco M, Laezza C, Stingo S, Wolff J. 2',3'-Cyclic nucleotide 3'-phosphodiesterase: a membrane-bound, microtubule-associated protein and membrane anchor for tubulin. Proc Natl Acad Sci U S A. 2002;99(4):1807–1812. doi: 10.1073/pnas.042678799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, et al. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33(3):366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- 20.Scarisbrick IA, Blaber SI, Lucchinetti CF, Genain CP, Blaber M, Rodriguez M. Activity of a newly identified serine protease in CNS demyelination. Brain. 2002;125(Pt 6):1283–1296. doi: 10.1093/brain/awf142. [DOI] [PubMed] [Google Scholar]
- 21.Scarisbrick IA, Linbo R, Vandell AG, Keegan M, Blaber SI, Blaber M, et al. Kallikreins are associated with secondary progressive multiple sclerosis and promote neurodegeneration. Biol Chem. 2008;389(6):739–745. doi: 10.1515/BC.2008.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu K, Li QZ, Delgado-Vega AM, Abelson AK, Sanchez E, Kelly JA, et al. Kallikrein genes are associated with lupus and glomerular basement membrane-specific antibody-induced nephritis in mice and humans. J Clin Invest. 2009;119(4):911–923. doi: 10.1172/JCI36728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans PD, Gilbert SL, Mekel-Bobrov N, Vallender EJ, Anderson JR, Vaez-Azizi LM, et al. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309(5741):1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- 24.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 25.McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50(1):121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 26.Poser CM, Paty DW, Scheinberg L, McDonald WI, Davis FA, Ebers GC, et al. New diagnostic criteria for multiple sclerosis: guidelines for research protocols. Ann Neurol. 1983;13(3):227–231. doi: 10.1002/ana.410130302. [DOI] [PubMed] [Google Scholar]
- 27.Steemers FJ, Chang W, Lee G, Barker DL, Shen R, Gunderson KL. Whole-genome genotyping with the single-base extension assay. Nat Methods. 2006;3(1):31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.