Abstract
The ability to recognize and react to specific environmental cues allows bacteria to localize to environments favorable to their survival and growth. Synthetic biologists have begun to exploit the chemosensory pathways that control cell motility to reprogram how bacteria move in response to novel signals. Reprogramming is often accomplished by designing novel protein or RNA parts that respond to specific small molecules not normally recognized by the natural chemosensory pathways. Additionally, cell motility and localization can be coupled to bacterial quorum sensing, potentially allowing consortia of cells to perform complex tasks.
Introduction
Bacteria use a variety of mechanisms to sense and respond to chemical signals in their environment. One of the best-studied sensing mechanisms is the chemotaxis system of E. coli, which controls bacterial movement and localization [1,2]. In E. coli, there are five transmembrane chemoreceptor proteins that recognize chemical signals such as amino acids and sugars. Ligand binding to chemoreceptors is coupled to a change in cell behavior via a series of cytosolic post-translational modifications involving six proteins (CheA, CheB, CheR, CheW, CheY, and CheZ) [1,2]. These signaling proteins work in concert to regulate the flagellar motor complex, which enables the cell to move up gradients of chemoattractants or down gradients of chemorepellents. The chemotactic response allows each individual in a population to direct its movement toward conditions that are favorable for its survival and growth.
While the chemotaxis response in E. coli governs the behavior of an individual cell, in many species of bacteria, cells use chemical signals to communicate with other members of a population. These quorum-sensing systems [3] are particularly interesting to synthetic biologists because they can potentially be engineered to allow heterogeneous populations, containing one or more strains or species of bacteria, to work together to perform complex tasks that a single strain would be unable to achieve on its own.
We will describe recent advances in the fields of protein engineering, RNA engineering, and synthetic biology directed toward reprogramming bacteria to recognize and follow novel small molecules, and in using these bacteria to accomplish specific tasks. Although many of these studies employ E. coli as a model system, we anticipate that the lessons learned can be extended to more advanced applications using additional bacterial species and more complex consortia.
Directed evolution to shift chemoreceptor ligand specificity
The first chemoreceptor mutants were discovered during efforts to elucidate the biochemical pathways that underlie E. coli chemotaxis [4]. Typically, cells were grown on semi-solid media containing various amino acids or sugars, and mutants that exhibited altered motility phenotypes were isolated and characterized. Cells that displayed altered motility in response to only a subset of chemoattractants were identified as chemoreceptor mutants [5].
Goulian and coworkers recognized that this motility assay could provide a powerful selection method for directing the evolution of endogenous chemoreceptor proteins to recognize new ligands [6]. Specifically, they tested the plasticity of the E. coli aspartate chemoreceptor (Tar) by evolving it to recognize new compounds, including cysteic acid, phenylalanine, or N-methyl-aspartate. Their motility selections also resulted in a Tar mutant strain that displayed improved chemotaxis toward glutamate and significantly reduced chemotaxis toward aspartate. Building upon this work, Goldberg et al. selected for Tar mutants that performed chemotaxis toward phenylacetic acid (PAA) [7**]. By adding a gene encoding penicillin acylase, an enzyme that hydrolyzes phenylacetyl glycine (PAG) to produce PAA, the authors created cells that could migrate up gradients of PAG. This reprogrammed strain and its use in a simple microbial consortium will be described further later in this review.
A major advantage of engineering the chemoreceptor proteins is that the intracellular signal transduction pathway of Che proteins remains in place, which allows cells to respond to new ligands on the same short time-scales (~200 ms [8]) that wild type cells do. Additionally, phenomena such as adaptation, in which a cell quickly alters its swimming behavior to changes in the concentration of a chemoattractant, but gradually returns to its pre-stimulus value, are presumably maintained in these engineered cells. However, one disadvantage of protein engineering approaches is that they most often involve making subtle changes on an existing protein scaffold, making it difficult to achieve dramatic changes in ligand specificity.
Controlling cell motility and localization with synthetic RNA parts
Topp and Gallivan took an alternate approach to engineering bacterial taxis [9**]. Using a synthetic riboswitch, which is an RNA molecule that controls gene expression in a ligand-dependent fashion absent other protein cofactors [10,11], the authors engineered E. coli to follow a gradient of the small molecule theophylline [9**]. Specifically, they used a riboswitch to activate the expression of one of the six E. coli Che proteins—CheZ, a phosphatase that regulates the tumbling frequency of E. coli. In the absence of ligand, CheZ was not expressed, and the cells tumbled in place, while addition of the ligand resulted in the production of CheZ, allowing the cells to move. The engineered cells displayed a ‘pseudotactic’ response to theophylline, in which the diffusion coefficient of the cells was proportional to the ligand concentration. Because the diffusion coefficient of the cells was greatly reduced in the absence of the ligand, the engineered cells were able to precisely localize to a chemical signal, as migration away from the ligand caused the cells to stop moving.
One advantage of using a synthetic riboswitch to control motility is that molecular recognition is achieved using an RNA aptamer that can be selected in vitro, without the need for a preexisting scaffold. Thus, it is possible to develop riboswitches that recognize ligands that are significantly different in structure than native chemoattractants. Using a motility-based selection [12], Sinha et al. developed a synthetic riboswitch that recognizes atrazine, one of the most heavily used herbicides in the US [13**]. The riboswitch allowed cells to migrate in the presence of atrazine. By adding an atrazine chlorohydrolase gene that leads to atrazine breakdown, the authors engineered cells that could ‘seek and destroy’ this pollutant (Figure 1). However, because the riboswitch operates intracellularly, and atrazine is not particularly cell-permeable, this system only responds to relatively high (>100 μM) concentrations of atrazine, even though the aptamer is capable of binding atrazine at much lower concentrations [13**]. Additionally, because synthetic riboswitches activate protein expression, rather than modifying existing proteins, their response times are longer than engineered chemoreceptors. Nevertheless, the ability to engineer synthetic riboswitches to recognize completely novel compounds, and the ability to introduce riboswitches into new organisms with minimal modification [14], are important advantages.
Figure 1.
A) The structure of atrazine. B) E. coli were engineered to follow atrazine. When cells were plated at the center of a Petri dish containing atrazine, they moved toward the edges of the plate. C) When a gene that allowed the cells to catabolize atrazine was added, the cells formed concentric rings. The dark regions between the rings are areas where atrazine was degraded.
Manipulating the motility and localization of cells within a consortia
Many of the systems described thus far involve a single strain of genetically engineered bacteria. However, such populations are typically unstable when transplanted from the laboratory to the field [15]. Outside of the laboratory, mixed bacterial populations (consortia) are far more common, in part because they are more resistant to an array of environmental conditions [16]. Additionally, a consortium of engineered cells may be better suited to accomplish a complex task than any single engineered strain. Key to the creation of a synthetic consortium is the ability to coordinate the behavior of the mixed population of cells, such that members of the consortia can co-localize.
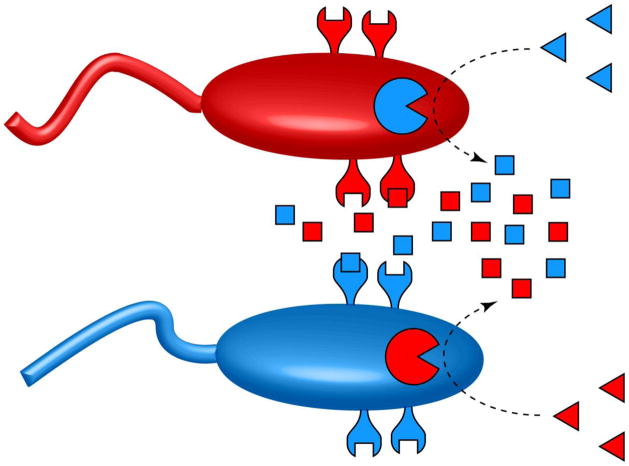
Building upon their previous chemoreceptor engineering work, Goldberg et al. engineered two strains of E. coli, each of which migrates toward a chemical signal that can be produced only by its partner strain [7**]. Specifically, the researchers engineered one strain to express the wild-type aspartate chemoreceptor and penicillin acylase, while its partner strain was engineered to express a PAA-responsive mutant chemoreceptor and asparaginase II, an enzyme that hydrolyzes asparagine to aspartate (Figure 2). When plated as homogeneous populations on semi-solid agar, neither strain was motile. However, when the two strains were combined, the consortium possessed all the components needed to migrate up gradients that contain both PAG and asparagine. Because neither strain is motile unless its partner strain and both chemical signals are present, this consortium acts as a Boolean ‘AND’ gate. The ability to engineer consortia that can perform logical tasks will be critical for developing microbial communities that can be safely deployed in the field.
Figure 2.
An engineered cell consortium consisting of two interacting strains that produce a mutually interdependent chemotactic response. The red cell is sensitive to molecules that are produced by the blue cell, and the blue cell is sensitive to compounds produced by the red cell.
Cirino and coworkers took a different approach to engineering motile cell consortia. They asked whether a ‘sender’ strain of E. coli could induce motility in a separate ‘receiver’ E. coli strain. The sender strain was engineered to constitutively synthesize an acyl homoserine lactone (AHL) signaling molecule [17*]. The ‘receiver’ strain included the Plux/LuxR system from Vibrio fischeri, which was used to couple the presence of the AHL signaling molecule to the expression of a protein required for flagellar rotation (MotB). Only when the sender strain was plated close to the receiver strain did the concentrations of AHL rise to sufficient levels to induce motility in the receiver strain. We anticipate that this simple, yet elegant, system can be further elaborated to display more complex functions
Synthetic consortia can also mimic the behaviors of natural populations, and in doing so, provide excellent model systems to study microbial ecology. Balagaddé et al. engineered a chemically mediated ecosystem consisting of two distinct E. coli populations that communicate through an engineered quorum-sensing pathway [18]. This pathway was designed to create a predator-prey relationship between the two populations. The result is a synthetic ecosystem that requires a specific balance to ensure the survival of both predator and prey.
In later studies, Song and colleagues used this synthetic system to analyze how cell motility and long-range diffusion of chemical signals combine to affect the biodiversity of an ecosystem [19*]. The authors seeded predator and prey populations at different distances from one another. They found that when the distances between the predator and prey were either much greater or much less than the diffusion limit of the predator chemical signal (which kills the prey), cell motility was a minor factor in determining biodiversity. However, when the populations of predator and prey were seeded slightly beyond the limits of diffusion of the predator chemical signal, decreased motility allowed for greater biodiversity because the prey remained segregated from the predator and both strains were able to survive. This work nicely supplemented earlier studies that focused primarily on the contributions of motility to microbial biodiversity [20,21]. Future studies that employ model systems consisting of reprogrammed cells will continue to improve our understanding of population dynamics and the effects of various environmental conditions on both natural and synthetic bacterial consortia.
Medicinal applications of engineered bacteria that localize to a specific signal
Engineering bacteria to recognize specific molecules and respond by performing preprogrammed tasks is still a very new field, but the sophistication of these molecular logic systems is increasing rapidly. As an example, Voigt and coworkers reprogrammed E. coli to invade cancer cells depending on the local environment [22**]. By placing a gene that allows for cell invasion, the inv gene, under the control of naturally occurring promoters that respond to conditions such as hypoxia or to cell density, the authors controlled where and when cell invasion occurred. This proof-of-principle can be expanded upon to engineer bacteria that identify and localize to areas of an organism that produce a specific small molecule or protein of interest. One can imagine that additional reprogramming of these bacteria could allow for activation of a signal that indicates when and where the engineered bacteria have found high concentrations of the specific molecule it was designed to seek out.
Conclusion and Perspective
A variety of synthetic constructs, such as engineered chemoreceptors and riboswitches, have been combined with naturally-occurring genetic circuits to reprogram bacteria to recognize and follow novel ligands. The additional ability to engineer cell consortia provides new opportunities for encoding complex functions that no single strain would be able to accomplish on its own. However, there are many hurdles that stand between these early laboratory models and systems that can be deployed in the field. Notably, engineered systems tend to be less robust than their natural counterparts. Thus, systems that work well in the carefully controlled environment of a Petri dish may perform unpredictably in a more complex environment, and in some cases, may be outcompeted by native organisms that are already well adapted. Making significant progress in these areas will likely require contributions from disciplines that operate on length-scales ranging from molecules to ecosystems, and many levels in between. As synthetic biology matures as a field, we expect to see clever solutions to these problems that will produce tangible advances in biotechnology and biomedicine.
Acknowledgments
We would like to thank members of the Gallivan lab for helpful comments and discussions. This work was supported by the NIH (GM074070 to JPG).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Baker MD, Wolanin PM, Stock JB. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 2.Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 3.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adler J. Chemotaxis in bacteria. J Supramol Struct. 1976;4:305–317. doi: 10.1002/jss.400040302. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong JB, Adler J, Dahl MM. Nonchemotactic mutants of Escherichia coli. J Bacteriol. 1967;93:390–398. doi: 10.1128/jb.93.1.390-398.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derr P, Boder E, Goulian M. Changing the specificity of a bacterial chemoreceptor. J Mol Biol. 2006;355:923–932. doi: 10.1016/j.jmb.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 7**.Goldberg SD, Derr P, DeGrado WF, Goulian M. Engineered single- and multi-cell chemotaxis pathways in E. coli. Mol Syst Biol. 2009;5:283. doi: 10.1038/msb.2009.41. Creates a simple consortium in which each of two strains is needed to produce the ligand that allows the other strain to swim. Thus, both strains are required for motility of the consortium in the presence of the attractants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segall JE, Manson MD, Berg HC. Signal processing times in bacterial chemotaxis. Nature. 1982;296:855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- 9**.Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J Am Chem Soc. 2007;129:6807–6811. doi: 10.1021/ja0692480. First use of a synthetic riboswitch to control bacterial motility. The bacteria are capable of moving only when theophylline is present, localizing to areas with higher concentrations of theophylline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 12.Topp S, Gallivan JP. Random walks to synthetic riboswitches--a high-throughput selection based on cell motility. Chembiochem. 2008;9:210–213. doi: 10.1002/cbic.200700546. [DOI] [PubMed] [Google Scholar]
- 13**.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat Chem Biol. 2010 doi: 10.1038/nchembio.369. Creates bacteria containing a novel synthetic atrazine riboswitch, which directs the bacteria to seek and destroy atrazine, an environmental pollutant. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Topp S, Reynoso CM, … 2010.
- 15.Chao WL, Feng RL. Survival of genetically engineered Escherichia coli in natural soil and river water. J Appl Bacteriol. 1990;68:319–325. doi: 10.1111/j.1365-2672.1990.tb02881.x. [DOI] [PubMed] [Google Scholar]
- 16.Burmolle M, Webb JS, Rao D, Hansen LH, Sorensen SJ, Kjelleberg S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol. 2006;72:3916–3923. doi: 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Weiss LE, Badalamenti JP, Weaver LJ, Tascone AR, Weiss PS, Richard TL, Cirino PC. Engineering motility as a phenotypic response to LuxI/R-dependent quorum sensing in Escherichia coli. Biotechnol Bioeng. 2008;100:1251–1255. doi: 10.1002/bit.21862. Using a quorum-sensing pathway, the authors link motility of a ‘receiver’ E. coli strain to the production of an AHL molecule by a second ‘sender’ strain. [DOI] [PubMed] [Google Scholar]
- 18.Balagadde FK, Song H, Ozaki J, Collins CH, Barnet M, Arnold FH, Quake SR, You L. A synthetic Escherichia coli predator-prey ecosystem. Mol Syst Biol. 2008;4:187. doi: 10.1038/msb.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Song H, Payne S, Gray M, You L. Spatiotemporal modulation of biodiversity in a synthetic chemical-mediated ecosystem. Nat Chem Biol. 2009;5:929–935. doi: 10.1038/nchembio.244. By employing a synthetic model ecosystem, the authors elucidate what role motility plays in biodiversity while also demonstrating how a predator-prey system can limit bacterial localization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerr B, Riley MA, Feldman MW, Bohannan BJ. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 21.Reichenbach T, Mobilia M, Frey E. Mobility promotes and jeopardizes biodiversity in rock-paper-scissors games. Nature. 2007;448:1046–1049. doi: 10.1038/nature06095. [DOI] [PubMed] [Google Scholar]
- 22**.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. Bacteria were successfully reprogrammed to invade mammalian cells under specific conditions using natural gene control elements. This work highlights potential future applications for bacteria that can localize to specific environmental cues. [DOI] [PubMed] [Google Scholar]