Abstract
Atomic force microscopy (AFM) is an emerging technique for a variety of uses involving the analysis of cells. AFM is widely applied to obtain information about both cellular structural and subcellular events. In particular, a variety of investigations into membrane proteins and microfilaments were performed with AFM. Here, we introduce applications of AFM to molecular imaging of membrane proteins, and various approaches for observation and identification of intracellular microfilaments at the molecular level. These approaches can contribute to many applications of AFM in cell imaging.
Keywords: membrane proteins; microfilaments; microscopy, atomic force
Introduction
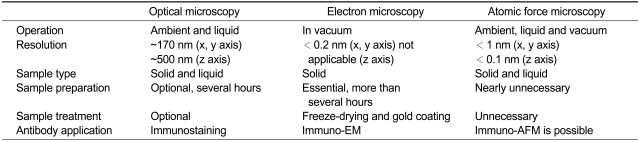
Optical microscopy has contributed greatly to various studies of biological materials, including cells and tissues (Yi et al., 2004; Lim et al., 2009). However, the resolution of optical microscopy, due to the diffraction limit of the light source, is not sufficient to visualize subcellular structures. The invention of electron microscopy (EM) advanced our insights into subcellular structures because it provided sub-nanometer resolution (Yingge et al., 2003; Ishikawa et al., 2005; Kadam et al., 2007). Although this microscopy is appropriate for visualization of subcellular structures, it requires complex sample preparation and must be operated in a vacuum environment.
Atomic force microscopy (AFM) has emerged as a key approach for molecular imaging of biological samples because it can provide high-resolution images that are comparable with those of electron microscopy with minimal sample damage during its preparation (Heuser and Kirschner, 1980; Svitkina et al., 1995; Small et al., 1999; Shahin and Barrera, 2008). AFM was found to have several advantages over other microscopic technologies (Table 1). AFM provides the real height profile of samples with sub-nanometer resolution. The resolution of AFM topographs surpasses optical microscopic images and is comparable to EM images (Ubbink and Schär-Zammaretti, 2005; Matsko, 2007). In addition, the process of sample preparation is far simpler than that of other microscopies. EM, a representative high-resolution imaging technique, requires processing of samples, such as cutting, detergent extraction, coating, and staining (Heuser and Kirschner, 1980; Svitkina et al., 1995; Small et al., 1999), and these treatments may cause damage to samples or result in artifacts (Franz, and Müller, 2005). On the other hand, because AFM does not require any additional treatment, biological samples can be imaged under nearly native conditions with minimal alteration or damage (Bonfiglio et al., 1995; Shahin and Barrera, 2008). Thus, AFM allows real-time observation of morphological changes caused by various stimuli with high resolution in a physiological environment. These advantages have attracted interests in AFM for cell imaging at remarkable resolution as an alternative imaging technique (Shahin and Barrera, 2008). In this review, we will introduce recent applications of AFM to imaging of cell membrane proteins and microfilaments, which are closely related with mechanical property and dynamics of cells.
Table 1.
Comparison between atomic force microscopy, optical microscopy, and electron microscopy in cell imaging.
Principles of AFM
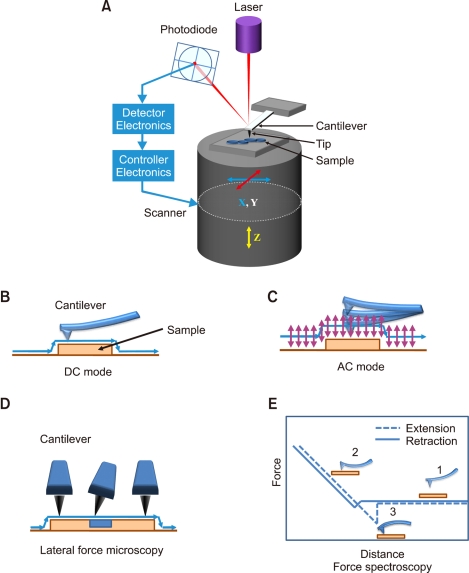
AFM uses ultra sharp tips mounted at the end of a flexible cantilever (tip radius 10-40 nm) as probes instead of an incident beam, as with optical microscopes (Zasadzinski et al., 1990; Hansma et al., 1994; Shahin and Barrera, 2008). The basic concept of AFM operation is reconstruction of three-dimensional maps by measuring the force between a sample and the sharp tip (Kirat et al., 2005). The cantilever is mounted on a piezoelectric scanner, which allows movement in either the horizontal (x, y) or vertical (z) dimension (Figure 1A). The tip is moved close to the sample surface and is raster scanned over the sample surface while moving vertically. AFM imaging can be classified into the following modes by the moving type of tips over the samples.
Figure 1.
Principle of operation and operation modes in AFM. (A) Schematics of atomic force microscopy principle. (B-E) Four widely used atomic force microscopy operation modes: (B) DC mode, (C) AC mode, (D) lateral force microscopy, and (E) Force spectroscopy. A typical force-distance curve is shown (E). The cantilever is not deflected yet (1). As the tip approaches to the surface, the cantilever bends upward by repulsive forces (2). As the tip retracts from the surface, the cantilever bends downward by attractive forces between the tip and sample (3).
In the DC mode (contact mode), the most commonly used AFM mode, an AFM tip attached to the end of a flexible cantilever is in perpetual contact with the sample. When the AFM tip approaches to the sample surface, deflection of cantilever is induced by various forces between the tip and the sample according to the Hooke's law (Figure 1B). The AFM tip is raster scanned over the sample while the force applied to tip for keeping the constant deflection by feedback control. The DC mode AFM is very useful for imaging of samples with either hard surface or large variations in height. However, soft biological samples can be scraped or deformed by the frictional forces produced during scanning. Also, overloaded force applied on the cantilever may indent samples critically or wear out the sharp tip. In addition, AFM tip can be easily contaminated by adsorption of molecules detached from sample surface (Shahin and Barrera, 2008).
The AC mode AFM including non-contact and tapping was developed to overcome the problems related with friction, adhesion, and electrostatic forces (Zhong et al., 1993; Hansma et al., 1994). In the AC mode, the tip oscillates vertically near its near resonance frequency with amplitudes ranging between 20 and 100 nm and is in contact with the surface at lower part of its oscillation (Figure 1C) (Gadegaard, 2006; Shahin and Barrera, 2008). When the vertically oscillating tip comes closer to the surface of sample, the oscillation amplitude is reduced by interaction between the tip and the sample. AFM tip is raster scanned with keeping the oscillation amplitude. In this mode, since the orbit of cantilever oscillation is slightly distant from sample surface, samples are clearly imaged without any damage of sample surfaces. Thus, the AC mode AFM allows high-resolution imaging of sample surfaces including soft materials, poorly bounded molecules to a substrate (Dryden et al., 1999; Ellis et al., 1999; Hansma et al., 2003; Geisse et al., 2004).
The lateral force microscopy (LFM) is one of the imaging modes to indentify compositional differences of samples and to distinguish frictional characteristics of samples. In the LFM mode, operation mechanism is very similar to the DC mode AFM but the probe is scanned perpendicular to its length. During imaging in the LFM mode, cantilever will twist more or less depending on the frictional characteristics of the sample surface (Figure 1D). LFM is useful for differentiating components of heterogeneous surfaces since it can provide simultaneously topographic and frictional information of the sample surface. Furthermore, an applied use of LFM is chemical force microscopy, where the tip is modified with specific chemicals and scanned over the sample to investigate the character of various surfaces (e.g. functional group imaging) (Frisbie et al., 1994; Fiorini et al., 2001).
In force spectroscopy, the force between the AFM tip and samples is measured as a function of force vs. distance using Hooke's law, as the AFM tip approaches and retracts from the surface (Figure 1E). The attractive force between the tip and the sample surface may induce changes in the force-distance curve. The information of sample elasticity is obtained by analysis of these changes with appropriate theoretical models (Dupres et al., 2007). This approach is very useful in the study of mechanical properties of living cells and dynamic cellular events, including cellular signaling pathways and cell migration, because it can be performed under nearly physiological conditions (Grzywa et al., 2006; Lamontagne et al., 2008). This approach can be applied to molecular recognition on the cell surface by modification of AFM tips with cell adhesion proteins, such as cadherins (Baumgartner et al., 2000), integrins (Zhang et al., 2002; Li et al., 2003), selectins (Zhang et al., 2004) and bacterial adhesions (Dupres et al., 2005, 2007). Furthermore, force spectroscopy can be applied to force mapping by scanning the selected region, which can be compared to the topological imaging (Dupres et al., 2007; Heredia et al., 2007; Jung et al., 2009b).
Applications of AFM to imaging of membrane proteins
AFM has been widely applied to the imaging of small biological samples such as biomolecules (e.g., proteins and DNA) (Shin et al., 2005; Lesoil et al., 2010) and small organisms (e.g., viruses and bacteria) (Geisse et al., 2004; Kim et al., 2006), which are not visible under optical microscopy. Efforts to observe the ultra-structures of cells using AFM have gradually increased since its invention (Gould et al., 1990; Muller and Engel, 1999; Kreplak et al., 2007; Jaroslawski et al., 2007; Yu et al., 2007). Early studies of cells with AFM were focused on topological observation by high-resolution imaging. In this session, we introduce various applications of AFM to membrane biology.
Imaging of membrane channels
AFM imaging has been performed in fixed or living cells under air or physiological environments after Gould et al. (1990) first demonstrated AFM images of dried red blood cells and bacteria. AFM has been used for the study of membrane channels because it allows observation of cell surfaces in their native environment at remarkable resolution (Jaroslawski et al. 2007; Yu et al., 2007; Muller, 2008; Stewart et al., 2010). Jaroslawski et al. (2007) have directly visualized gating of KirBac3.1 potassium channels by AFM imaging. KirVac3.1 is one member of a family of transmembrane potassium channels, and gating of Kir channels is spontaneous but modulated by intracellular molecules such as lipids (PIP2), G-proteins, nucleotides, and ions (H+, Ca2+, Mg2+). Yu et al. (2007) demonstrated that pH-induced conformational changes in connexin 26 hemichannels were modulated by aminosulfonate. They removed the upper layer of the gap junction channel by repetitive scanning AFM tip with loading of strong force. Stewart et al. (2010) revealed the tetrameric structure of the TRPM8 channel, which mediates the passage of cations across the plasma membrane, by AFM imaging. AFM imaging of two-dimensional crystals of Escherichia coli OmpF porins visualized voltage- and pH-induced gating of the porin, which facilitates the passage of small solutes (Muller and Engel, 1999). It was suggested that the bacteria protect themselves by these conformational changes of porins from drastic changes of the environment (Muller and Engel, 1999). Conformational changes of native membrane related to channel gating were shown by time-lapse AFM imaging (Muller and Engel, 1999; Jaroslawski et al., 2007; Yu et al., 2007).
Force spectroscopy of membrane proteins
Force spectroscopy with AFM, based on Hooke's law, has become a powerful tool for recognition of membrane proteins such as receptors. To probe membrane proteins in cells and to characterize the cell surface, the AFM tip needs to be modified with a specific chemical or ligand (Dufrene, 2000; Zhang et al., 2002; Li et al., 2003). Almqvist et al. (2004) studied the effect of membrane receptor clustering on local cell mechanics by mapping interaction forces between antibody-conjugated AFM tips and a vascular endothelial growth factor receptor. Jiang group (2009) reported distribution of Na+-K+ ATPase, a key transmembrane protein, in human red blood cells using AFM tips modified with antibody against Na+-K+ ATPase. This method was also applied to mapping of interactions between calcitonin and calcitonin receptor in osteocalst cells using AFM tips modified with calcitonin (Lehenkari et al, 2000). In addition, it was reported that the combined imaging of fluorescence, topography and recognition was applied to detect density, distribution, and localization of YFP-labeled CD1d molecules on α-galactosylceramide-loaded THP1 cells (Duman et al, 2010).
However, these approaches are limited to membrane proteins existed on the membrane surface of cells. Thus, Carnally et al. (2010) demonstrated interaction between sigma-1 receptors, which interact with a variety ion channel, and isolated acid-sensing ion channels using AFM. In addition, Kasai et al. (2010) applied AFM to observe tetrameric structure of ionotropic glutamate receptors after purification and reconstitution into lipid bilayers.
Various approaches to AFM imaging of microfilaments
One of the most active targets studied with AFM within cells is microfilaments. The microfilament plays an important role in many cellular processes such as physical and biochemical connection of the cell to external environment and spatial organization of cellular contents. It also generates coordinated forces that enable the cells to move and change their shape and generate (Fletcher and Mullins, 2010). In particular, the polymerization and depolymerization of microfilaments drive cytoplasmic organization, cell division, and cell motility (Azoury et al., 2009; Kueh and Mitchison, 2009). In addition, the association of microfilaments with the cell membrane plays a critical role in endocytosis and anchoring membrane proteins (Papakonstanti and Stournaras, 2008; Robertson et al., 2009). Thus, AFM was applied to reveal the mechanism of actin polymerization in vitro at the molecular scale (Ikawa et al, 2007). However, it was a big challenge to image intracellular microfilaments using AFM because AFM is limited to the topological analysis.
De-roofing and detergent extraction of the plasma membrane
Much work has been done to overcome the limits of topological analysis and to investigate intracellular structures by AFM imaging (Melling et al., 2003; Franz and Müller, 2005; Meller and Theiss, 2006). To remove the plasma membrane of cells, two approaches, de-roofing by short ultrasonic burst and detergent extraction, have been demonstrated (Heuser, 2000; Berdyyeva et al., 2005). The ultrastructural insight into the architecture of focal adhesions in de-roofed cells was provided by AFM topographs complemented with optical microscopy (Franz and Müller, 2005). Furthermore, this application offers accurate height information of focal adhesion and insight into the organization of microfilaments.
Several reports describe investigations of intracellular cytoskeletal structures after detergent extraction using the nonionic detergent Triton X-100 (Berdyyeva et al., 2005; Meller and Theiss, 2006). A method of permeabilization and embedding of lens epithelial cells was used to study the organization and distribution of intracellular proteins with AFM and confocal microscopy (Meller and Theiss, 2006). In this study, confocal microscopy was used to identify actin microfilaments by fluorescent labeling with rhodamine phalloidin, whereas AFM enabled the study of three-dimensional structures of microfilaments at the molecular level in fixed and living cells. Berdyyeva et al. (2005) described a novel application of AFM to directly visualize cytoskeletal fibers in human foreskin epithelial cells. In this approach, they used Triton X-100 to remove the membrane, soluble proteins, and organelles from the cell. After detergent extraction, the remaining cytoskeleton was directly imaged under either liquid or air-dried ambient conditions. While imaging in liquid has been used to observe the cytoskeleton near the top of cells, imaging under air was performed to visualize both surface filaments and the total volume of the cytoskeletal fibers.
Imaging in semi-in situ environment
De-roofing and detergent extraction to remove the plasma membrane may cause damage to samples during their preparation (Svitkina et al., 1995; Small et al., 1999; Berdyyeva et al., 2005). Recently, we reported a semi-in situ system through photodynamic therapy (PDT) to study the ultrastructure of microfilaments without additional treatment and under near-native conditions (Jung et al., 2009a). PDT is an anticancer modality that uses photosensitizers preferentially accumulated in cancer cells (Almeida et al, 2004; Ferreira et al, 2004; Tsai et al, 2005; Uzdensky et al, 2005; Lim et al., 2009). Subsequent activation of photosensitizer in target cells or tissues by light of an appropriate wavelength causes a cascade of biological events through various photophysical pathways, which induce production of reactive oxygen species, elevation of intracellular Ca2+, activation of caspases, and translocation of apoptosis-inducing factor, and ultimately result in cell death through apoptosis or necrosis (Almeida et al, 2004; Ferreira et al, 2004; Tsai et al, 2005; Yoo et al., 2009). In bladder cancer cells, PDT induced conversion of intracellular into extracellular microfilaments, and the extracellular microfilaments formed by PDT was subjected to AFM (Jung et al., 2009a). AFM imaging of the microfilaments showed a stranded helical structure. This approach enabled us to investigate microfilaments at the molecular level under a semi-in situ environment.
On-stage labeling and imaging
Although AFM provides ultrastructural information at the molecular level, it is difficult to identify intracellular biomolecules because the technique is limited to the analysis of surface topology. To overcome this limitation, a combination of AFM and confocal microscopy has been recognized as an alternative method for the investigation of biological materials with high resolution (Franz and Müller, 2005; Sharma et al., 2005; Duman et al, 2010). It would be both convenient and useful to directly install AFM on a confocal microscope, but there may be compatibility issues between the two. Thus, a tandem imaging method was introduced to allow comparative analysis of AFM and confocal microscopic images obtained in the same scanning field without co-installation of two pieces of imaging equipment (Jung et al., 2009a). This approach is very useful for both identification and high-resolution imaging of certain proteins in a complex mixture such as cells.
Although intracellular components can be identified by the combinatory or tandem imaging method, an alternative method is required to identify ultrastructural components of cells, which are not detectable with confocal microscopy due to its limited resolution. On-stage labeling and imaging, based on a previous report (Jung et al., 2009a), can provide be a solution to overcome this limitation. In this new approach, pre-imaging, labeling, and post-imaging are sequentially performed in aqueous solution without moving samples on the scanner tube of AFM. A coverslip with fixed cells is mounted on a scanner tube and AFM imaging is performed (pre-imaging). After pre-imaging, the fluid cell is perfused with a labeling molecule in a buffer solution to probe target components of cells. Various molecules, including antibodies, toxins, peptides or conjugated nanoparticles, can be used to label the target molecules. After washing with the buffer solution, AFM imaging is performed at the same position (post-imaging). Figure 2 shows a possible schematic of on-stage labeling and imaging for the identification of microfilaments by AFM. This new approach can be used for the simultaneous identification and analysis of ultrastructural components of cells at the molecular level, which is not likely with other approaches. In addition, this on-stage approach will be more useful when it is used in parallel with the hybrid AFM-confocal microscopy, because on-stage labeling can be evaluated with confocal microscopy before on-stage imaging is performed with AFM. Furthermore, this on-stage labeling and imaging method has a potential to be developed into immuno-AFM.
Figure 2.
Identification of microfilaments by on-stage labeling/imaging. Samples mounted on the scanner tube are sequentially imaged in aqueous solution by AFM before (pre-imaging) and after (post-labeling) labeling with molecular probes including antibodies. Line profiles from pre-imaging (from a to b) and post-imaging (from a' to b') are compared.
High-speed AFM imaging
The image acquisition time in AFM might be too long to analyze biomolecular processes in real time because they generally occur on a millisecond or less (Ando et al., 2008a). Recently, improvement of the AFM tip and feedback control system has resulted in the development of high-speed AFM. The imaging rate of high-speed AFM is more than 30 frames per second and it enabled researchers to study dynamic biomolecular processes such as protein-protein interactions (Ando et al., 2008a, 2008b) and DNA-enzyme interactions (Hansma et al., 2006). For example, Gilmore's group revealed single-molecule dynamics of the DNA-enzyme complexes with high-speed AFM (Gilmore et al., 2009). Ando's group used high-speed AFM for studying dynamic behavior of actin and myosin V (Ando et al., 2008b). In addition, high-speed AFM can allow investigators to trace morphological changes of cells after external stimulus in real time. Thus, high-speed AFM has a strong potential for visualization of microfilament dynamics in cells.
Conclusion
AFM is becoming a powerful technique for the ultrastructural study of biological samples because it can provide high-resolution images without causing damage during sample preparation and scanning. Recently, efforts towards the investigation of membrane proteins and microfilaments using AFM have increased to study dynamic events within cells. Various membrane channels were successfully analyzed by the topological imaging of AFM, and the force spectroscopy was very useful in force mapping of membrane receptors. Although microfilaments were successfully imaged by AFM through additional treatments such as de-roofing or detergent extraction, cell shrinkage using external stimuli such as PDT can be an alternative method for the investigation of intracellular structures using AFM.
Identification of ultrastructural components of cells is one of the big challenges to be resolved. AFM force mapping will be helpful in identification of subcellular components under physiological conditions or in fixed cells. On-stage labeling and imaging approach can also provide simultaneous ultrastructural images and identification of molecules such as proteins in cells.
Acknowledgements
This work was supported in part by the Korea Research Foundation through the Basic Research Program (2008-05943), and by the Korean Ministry of Education, Science and Technology (the Regional Core Research Program/Medical & Bio-Materials Research Center). We would like to thank the Korea Basic Science Institute (Chuncheon Center) for help with facilities.
Abbreviations
- AFM
atomic force microscopy
- EM
electron microscopy
- LFM
lateral force microscopy
References
- 1.Almeida RD, Manadas BJ, Carvalho AP, Duarte CB. Intracellular signaling mechanisms in photodynamic therapy. Biochim Biophys Acta. 2004;1704:59–86. doi: 10.1016/j.bbcan.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Almqvist N, Bhatia R, Primbs G, Desai N, Banerjee S, Laly R. Elasticity and adhesion force mapping reveals real-time clustering of growth factor receptors and associated changes in local cellular rheological properties. Biophys J. 2004;86:1753–1762. doi: 10.1016/S0006-3495(04)74243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ando T, Uchihashi T, Kodera N, Miyagi A, Nakakita R, Yamashita H, Sakashita M. High-speed atomic force microscopy for studying the dynamic behavior of protein molecules at work. Jpn J Appl Phys. 2008a;45:1897–1903. [Google Scholar]
- 4.Ando T, Uchihashi T, Kodera N, Yamamoto D, Miyagi A, Taniguchi M, Yamashita H. High-speed AFM and nano-visualization of biomolecular processes. Pflugers Arch. 2008b;456:211–225. doi: 10.1007/s00424-007-0406-0. [DOI] [PubMed] [Google Scholar]
- 5.Azoury J, Verlhac MH, Dumont J. Actin filaments: key players in the control of asymmetric divisions in mouse oocytes. Biol Cell. 2009;101:69–76. doi: 10.1042/BC20080003. [DOI] [PubMed] [Google Scholar]
- 6.Baumgartner W, Hinterdorfer P, Ness W, Raab A, Vestweber D, Schindler H, Drenckhahn D. Cadherin interaction probed by atomic force microscopy. Proc Natl Acad Sci USA. 2000;97:4005–4010. doi: 10.1073/pnas.070052697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berdyyeva T, Woodworth CD, Sokolov I. Visualization of cytoskeletal elements by the atomic force microscope. Ultramicroscopy. 2005;102:189–198. doi: 10.1016/j.ultramic.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Bonfiglio A, Parodi MT, Tonini GP. Subcellular details of early events of differentiation induced by retinoic acid in human neuroblastoma cells detected by atomic force microscopy. Exp Cell Res. 1995;216:73–79. doi: 10.1006/excr.1995.1009. [DOI] [PubMed] [Google Scholar]
- 9.Carnally SM, Johannessen M, Henderson RM, Jackson MB, Edwardson JM. Demonstration of a direct interaction between sigma-1 receptors and acid-sensing ion channels. Biophys J. 2010;98:1182–1191. doi: 10.1016/j.bpj.2009.12.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dryden DT, Davies GD, Martin I, Powell LM, Murray NE, Ellis DJ, Berge T, Edwardson JM, Henderson RM. The assembly of the EcoKI type I DNA restriction/modification enzyme and its interaction with DNA. Biochem Soc Trans. 1999;27:691–696. doi: 10.1042/bst0270691. [DOI] [PubMed] [Google Scholar]
- 11.Dufrene YF. Direct characterization of the physicochemical properties of fungal spores using functionalized AFM probes. Biophys J. 2000;78:3286–3291. doi: 10.1016/S0006-3495(00)76864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duman M, Pfleger M, Zhu R, Rankl C, Chtcheglova LA, Neundlinger I, et al. Improved localization of cellular membrane receptors using combined fluorescence microscopy and simultaneous topography and recognition imaging. Nanotechnology. 2010;21:115504–115510. doi: 10.1088/0957-4484/21/11/115504. [DOI] [PubMed] [Google Scholar]
- 13.Dupres V, Menozzi FD, Locht C, Clare BH, Abbott NL, Cuenot S, Bompard C, Raze D, Dufrene YF. Nanoscale mapping and functional analysis of individual adhesins on living bacteria. Nat Methods. 2005;2:515–520. doi: 10.1038/nmeth769. [DOI] [PubMed] [Google Scholar]
- 14.Dupres V, Verbelen C, Dufrene YF. Probing molecular recognition sites on biourfaces using AFM. Biomaterials. 2007;28:2393–2402. doi: 10.1016/j.biomaterials.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Ellis DJ, Dryden DT, Berge T, Edwardson JM, Henderson RM. Direct observation of DNA translocation and cleavage by EcoKI endonuclease using atomic force microscopy. Nat Struct Biol. 1999;6:15–17. doi: 10.1038/4882. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira SDRM, Tedesco AC, Sousa G, Zangaro RA, Silva NS, Pacheco MTT, et al. Direct observation of DNA translocation and cleavage by the EcoKI endonuclease using atomic force microscopy. Lasers Med Sci. 2004;18:207–212. [Google Scholar]
- 17.Fiorini M, Mckendry R, Cooper MA, Rayment T, Abell C. Chemical force microscopy with active enzymes. Biophys J. 2001;80:2471–2476. doi: 10.1016/S0006-3495(01)76215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fletcher DA, Mullins RD. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franz CM, Müller DJ. Analyzing focal adhesion structure by atomic force microscopy. J Cell Sci. 2005;118:5315–5323. doi: 10.1242/jcs.02653. [DOI] [PubMed] [Google Scholar]
- 20.Frisbie CD, Rozsnyai LF, Noy A, Wrighton MS, Lieber CM. Functional group imaging by chemical force microscopy. Science. 1994;265:2071–2074. doi: 10.1126/science.265.5181.2071. [DOI] [PubMed] [Google Scholar]
- 21.Gadegaard N. Atomic force microscopy in biology: technology and techniques. Biotech Histochem. 2006;81:87–97. doi: 10.1080/10520290600783143. [DOI] [PubMed] [Google Scholar]
- 22.Geisse NA, Cover TL, Henderson RM, Edwardson JM. Targeting of Helicobacter pylori vacuolating toxin to lipid raft membrane domains analysed by atomic force microscopy. Biochem J. 2004;381:911–917. doi: 10.1042/BJ20031719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilmore JL, Suzuki Y, Tamulaitis G, Siksnys V, Takeyasu K, Lyubchenko YL. Single-molecule dynamics of the DNA-EcoRII protein complexes revealed with high-speed atomic force micrscopy. Biochemistry. 2009;48:10492–10498. doi: 10.1021/bi9010368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gould SAC, Drake B, Prater CB, Weisenhorn AL, Manne S, Hansma HG, et al. From atoms to integrated circuit chips, blood cells, and bacteria with the atomic force microscope. J Vac Sci Technol A. 1990;8:369–373. [Google Scholar]
- 25.Grzywa EL, Lee AC, Lee GU, Suter DH. High-resolution analysis of neuronal growth cone morphology by comparative atomic force and optical microscopy. J Neurobiol. 2006;66:1529–1543. doi: 10.1002/neu.20318. [DOI] [PubMed] [Google Scholar]
- 26.Hansma HG, Oroudjev E, Baudrey S, Jaeger L. TectoRNA and 'kissing-loop' RNA: atomic force microscopy of self-assembling RNA structure. J Microsc. 2003;212:273–279. doi: 10.1111/j.1365-2818.2003.01276.x. [DOI] [PubMed] [Google Scholar]
- 27.Hansma PK, Cleveland JP, Radmacher M, Walters DA, Hillner PE, Benzanilla M, et al. Tapping mode atomic force microscopy in liquids. Appl Phys Lett. 1994;64:1738–1740. [Google Scholar]
- 28.Hansma PK, Schitter G, Fantner GE, Prater C. High-speed atomic force microscopy. Science. 2006;314:601–602. doi: 10.1126/science.1133497. [DOI] [PubMed] [Google Scholar]
- 29.Heredia A, Bui CC, Suter U, Young P, Schäffer TE. AFM combines functional and morphological analysis of peripheral myelinated and demyelinated nerve fibers. Neuroimage. 2007;37:1218–1226. doi: 10.1016/j.neuroimage.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Heuser J. The production of 'cell cortices' for light and electron microscopy. Traffic. 2000;1:545–552. doi: 10.1034/j.1600-0854.2000.010704.x. [DOI] [PubMed] [Google Scholar]
- 31.Heuser JE, Kirschner MW. Filament organization revealed in platinum replicas of freeze-dried cytoskeletons. J Cell Biol. 1980;86:212–234. doi: 10.1083/jcb.86.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikawa T, Hoshino F, Watanabe O, Li Y, Pincus P, Safinya CR. Molecular scale imaging of F-actin assemblies immobilized on a photopolymer. Phys Rev Lett. 2007;98:018101. doi: 10.1103/PhysRevLett.98.018101. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa H, Hashinoto S, Tanno M, Ishikawa T, Tanaka T, Shimono M. Cytoskeleton and surface structures of cells directly attached to the tooth in the rat junctional epithelium. J Periodontal Res. 2005;40:354–363. doi: 10.1111/j.1600-0765.2005.00815.x. [DOI] [PubMed] [Google Scholar]
- 34.Jaroslawski S, Zadek B, Ashcroft F, Venien-Bryan C, Scheuring S. Direct visualization of KirBac3.1 potassium channel gating by atomic force microscopy. J Mol Biol. 2007;374:500–505. doi: 10.1016/j.jmb.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 35.Jiang J, Hao X, Cai M, Shan Y, Shang X, Tang Z, et al. Localization of Na+-K+ ATPases in quasi-native cell membranes. Nano Lett. 2009;9:4489–4493. doi: 10.1021/nl902803m. [DOI] [PubMed] [Google Scholar]
- 36.Jung SH, Park JY, Yoo JO, Shin IC, Kim YM, Ha KS. Identification and ultrastructural imaging of photodynamic therapy-induced microfilaments by atomic force microscopy. Ultramicroscopy. 2009a;109:1428–1434. doi: 10.1016/j.ultramic.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Jung YJ, Park YS, Yoon KJ, Kong YY, Park JW, Nam HG. Molecule-level imaging of Pax6 mRNA distribution in mouse embryonic neocortex by molecular interaction force microscopy. Nucl Acids Res. 2009b;37:e10. doi: 10.1093/nar/gkn965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadam K, D'souza S, Natraj U. Spatial distribution of actin and tubulin in human sperm nuclear matrix-intermediate filament whole mounts-a new paradigm. Microsc Res Tech. 2007;70:589–598. doi: 10.1002/jemt.20438. [DOI] [PubMed] [Google Scholar]
- 39.Kasai N, Ramanujan CS, Fujimoto I, Shimada A, Ryan JF, Torimitsu K. AFM observation of single, functioning ionotropic glutamate receptors reconstituted in lipid bilayers. Biochim Biophys Acta. 2010;1800:655–661. doi: 10.1016/j.bbagen.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Kim HS, Jung SH, Kim SH, Suh IB, Kim WJ, Jung JW, et al. High-Throughput analysis of mumps virus and the virus-specific monoclonal antibody on the arrays of a cationic polyelectrolyte with a spectral SPR biosensor. Proteomics. 2006;6:6426–6432. doi: 10.1002/pmic.200600432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirat KE, Burton I, Dupres V, Dufrene YF. Sample preparation procedures for biological atomic force microscopy. J Microsc. 2005;218:199–207. doi: 10.1111/j.1365-2818.2005.01480.x. [DOI] [PubMed] [Google Scholar]
- 42.Kreplak L, Wang H, Aebi U, Kong XP. Atomic force microscopy of mammalian urothelial surface. J Mol Biol. 2007;374:365–373. doi: 10.1016/j.jmb.2007.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kueh HY, Mitchison TJ. Structural plasiticity in actin and tubulin polymer dynamics. Science. 2009;325:960–963. doi: 10.1126/science.1168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamontagne CA, Cuerrier CM, Grandbois M. AFM as a tool to probe and manipulate cellular processes. Pflugers Arch. 2008;456:61–70. doi: 10.1007/s00424-007-0414-0. [DOI] [PubMed] [Google Scholar]
- 45.Lehenkari PP, Charras GT, Nytak A, Horton MA. Adapting atomic force microscopy for cell biology. Ultramicroscopy. 2000;82:289–295. doi: 10.1016/s0304-3991(99)00138-2. [DOI] [PubMed] [Google Scholar]
- 46.Lesoil C, Nonaka T, Sekiguchi H, Osada T, Miyata M, Afrin R, et al. Molecular shape and binding force of Mycoplasma mobile's leg protein Gli349 revealed by an AFM study. Biochem Biophys Res Commun. 2010;391:1312–1317. doi: 10.1016/j.bbrc.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 47.Li F, Redick SD, Erickson HP, Moy VT. Force measurements of the α5β1 integrin-fibronectin interaction. Biophys J. 2003;84:1252–1262. doi: 10.1016/S0006-3495(03)74940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim YC, Yoo JO, Park D, Kang G, Hwang BM, Kim YM, et al. Antitumor effect of photodynamic therapy with chlorin-based photosensitizer DH-II-24 in colorectal carcinoma. Cancer Sci. 2009;100:2431–2436. doi: 10.1111/j.1349-7006.2009.01326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsko NB. Atomic force microscopy applied to study macromolecular content of embedded biological material. Ultramicroscopy. 2007;107:95–105. doi: 10.1016/j.ultramic.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 50.Meller K, Theiss C. Atomic force microscopy and confocal laser scanning microscopy on the cytoskeleton of permeabilised and embedded cells. Ultramicroscopy. 2006;106:320–325. doi: 10.1016/j.ultramic.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Melling M, Karimian-Teherani D, Behnam M, Mostler S. Mophological study of the healthy human oculomotor nerve by atomic force microscopy. Neuroimage. 2003;20:795–801. doi: 10.1016/S1053-8119(03)00359-8. [DOI] [PubMed] [Google Scholar]
- 52.Muller DJ, Engel A. Voltage and pH-induced channel closure of porin OmpF visualized by atomic force microscopy. J Mol Biol. 1999;285:1347–1351. doi: 10.1006/jmbi.1998.2359. [DOI] [PubMed] [Google Scholar]
- 53.Muller DJ. AFM: a nanotool in membrane biology. Biochemistry. 2008;47:7986–7998. doi: 10.1021/bi800753x. [DOI] [PubMed] [Google Scholar]
- 54.Papakonstanti EA, Stournaras C. Cell responses regulated by early reorganization of actin cytoskeleton. FEBS Lett. 2008;582:2120–2127. doi: 10.1016/j.febslet.2008.02.064. [DOI] [PubMed] [Google Scholar]
- 55.Robertson AS, Smythe E, Ayscough KR. Functions of actin in endocytosis. Cell Mol Life Sci. 2009;66:2049–2065. doi: 10.1007/s00018-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shahin V, Barrera NP. Providing unique insight into cell biology via atomic force microscopy. Int Rev Cytol. 2008;265:227–252. doi: 10.1016/S0074-7696(07)65006-2. [DOI] [PubMed] [Google Scholar]
- 57.Sharma A, Anderson KI, Muller DJ. Actin microbridges characterized by laser scanning confocal and atomic force microscopy. FEBS Lett. 2005;579:2001–2008. doi: 10.1016/j.febslet.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 58.Shin M, Song M, Rhee JH, Hong Y, Kim YJ, Seok YJ, et al. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Eσ70 as a cofactor for looping. Genes Dev. 2005;19:2388–2398. doi: 10.1101/gad.1316305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Small J, Rottner K, Hahne P, Anderson KI. Visualising the actin cytoskeleton. Microsc Res Tech. 1999;47:3–17. doi: 10.1002/(SICI)1097-0029(19991001)47:1<3::AID-JEMT2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 60.Stewart AP, Egressy K, Lim A, Edwardson JM. AFM imaging reveals the tetrameric structure of the TRPM8 channel. Biochem Biophys Res Commun. 2010;394:383–386. doi: 10.1016/j.bbrc.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 61.Svitkina TM, Verkhovsky AB, Borisy GG. Improved procedure for electron microscopic visualization of the cytoskeleton of cultured cells. J Struct Biol. 1995;115:290–303. doi: 10.1006/jsbi.1995.1054. [DOI] [PubMed] [Google Scholar]
- 62.Tsai JC, Wu CL, Chen HF, Chen CT. Reorganization of cytoskeleton induced by 5-aminolevulinic acid-mediated photodynamic therapy and its correlation with mitochondrial dysfunctioin. Lasers Surg Med. 2005;36:398–408. doi: 10.1002/lsm.20179. [DOI] [PubMed] [Google Scholar]
- 63.Ubbink J, Schär-Zammaretti P. Probing bacterial interactions: integrated approaches combining atomic force microscopy, electron microscopy and biophysical techniques. Micron. 2005;36:293–320. doi: 10.1016/j.micron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 64.Uzdensky A, Kolpakova E, Juzeniene A, Juzenas P, Moan J. The effect of sub- lethal ALA-PDT on the cytoskeleton and adhesion of cultured human cancer cells. Biochim Biophys Acta. 2005;1722:43–50. doi: 10.1016/j.bbagen.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 65.Yi SJ, Choi HJ, Yoo JO, Yuk JS, Jung HI, Lee SH, et al. Arachidonic acid activates tissue transglutaminase and stress fiber formation via intracellular reactive oxygen species. Biochem Biophys Res Commun. 2004;325:819–826. doi: 10.1016/j.bbrc.2004.10.122. [DOI] [PubMed] [Google Scholar]
- 66.Yingge Z, Xia J, Lan S. The relations between neurite development and the subcellular structures of hippocampal neuron somata. J Struct Biol. 2003;144:327–336. doi: 10.1016/j.jsb.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 67.Yoo JO, Lee CH, Hwang BM, Kim W, Kim YM, Ha KS. Regulation of intracellular Ca2+ in the cytotoxic response to photodynamic therapy with a chlorin-based photosensitizer. J Porphyr Phthalocyanines. 2009;13:811–817. [Google Scholar]
- 68.Yu J, Bippes CA, Hand GM, Muller DJ, Sosinsky GE. Aminosulfonate modulated pH-induced conformational changes in connexin26 hemichannels. J Biol Chem. 2007;282:8895–8904. doi: 10.1074/jbc.M609317200. [DOI] [PubMed] [Google Scholar]
- 69.Zasadzinski JA, Helm CA, Longo ML, Weisenhorn AL, Gould SA, Hansma PK. Scanning tunneling microscopy and atomic force microscopy of biological surfaces. Ann N Y Acad Sci. 1990;589:476–491. [Google Scholar]
- 70.Zhang X, Wojcikiewicz E, Moy VT. Force spectroscopy of the leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction. Biophys J. 2002;83:2270–2279. doi: 10.1016/S0006-3495(02)73987-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang X, Bogorin DF, Moy VT. Molecular basis of the dynamic strength of the sialyl Lewis X-selectin interaction. Chem Phys Chem. 2004;5:175–182. doi: 10.1002/cphc.200300813. [DOI] [PubMed] [Google Scholar]
- 72.Zhong Q, Inniss D, Kjoller K, Elings VB. Fractured polymer/silica fiber surface studied by tapping mode atomic force microscopy. Surf Sci. 1993;290:688–692. [Google Scholar]