Abstract
Mechanosensitive channels act as molecular transducers of mechanical force exerted on the membrane of living cells by opening in response to membrane bilayer deformations occurring in physiological processes such as touch, hearing, blood pressure regulation, and osmoregulation. Here, we determine the likely structure of the open state of the mechanosensitive channel of large conductance using a combination of patch clamp, fluorescence resonance energy transfer (FRET) spectroscopy, data from previous electron paramagnetic resonance experiments, and molecular and Brownian dynamics simulations. We show that structural rearrangements of the protein can be measured in similar conditions as patch clamp recordings while controlling the state of the pore in its natural lipid environment by modifying the lateral pressure distribution via the lipid bilayer. Transition to the open state is less dramatic than previously proposed, while the N terminus remains anchored at the surface of the membrane where it can either guide the tilt of or directly translate membrane tension to the conformation of the pore-lining helix. Combining FRET data obtained in physiological conditions with simulations is likely to be of great value for studying conformational changes in a range of multimeric membrane proteins.
INTRODUCTION
Mechanosensitive channels in bacteria form safety valves that protect cells from hypoosmotic shock, opening a wide pore under membrane tension to relieve excessive turgor pressure within the cell (Martinac, 2001; Strop et al., 2003; Perozo, 2006; Booth et al., 2007). The mechanosensitive channel of large conductance (MscL) is one of the best characterized membrane channels in both structural and functional terms. MscL channels respond to membrane tension (∼10–12 mN/m) (Berrier et al., 1996; Blount et al., 1996; Sukharev et al., 1999) and have a very high nonselective conductance (∼3 nS) (Sukharev et al., 1993; Cruickshank et al., 1997) so as to act as the last in a series of mechanisms designed to save osmotically challenged bacterial cells. A 3-D structure of the MscL homologue from Mycobacterium tuberculosis in a closed state shows the protein to be a homopentameric channel (Chang et al., 1998), with each subunit containing two membrane-spanning α helices (TM1 and TM2). The pore itself is lined by the TM1 helices and the external end of TM2, and it contains a tight constriction surrounded by hydrophobic residues known to be conserved across members of the MscL family (Oakley et al., 1999). An MscL homologue from Staphylococcus aureus was recently found to form functional tetrameric channels and was crystallized in a hypothesized intermediate open state with TM1 and TM2 tilted toward the plane of the membrane, but with a narrow constriction remaining in the pore (Liu et al., 2009).
A question of particular interest is how mechanosensitive channels sense mechanical force and how this is used to open the pore. Bacterial mechanosensitive channels directly sense membrane tension developed in the lipid bilayer alone (Martinac et al., 1990; Markin and Martinac, 1991; Moe and Blount, 2005), preserving their mechanosensitivity after reconstitution into artificial liposomes (Delcour et al., 1989; Häse et al., 1995; Berrier et al., 1996; Perozo et al., 2002b). This fact is underlined by the response of the channel to various amphipaths. In thin bilayers, the channel activates at lower pressures and intermediate states are stabilized. Insertion of the single-tailed lysophosphatidylcholine (LPC), on the other hand, greatly increases the open-channel probability for prolonged periods (Perozo et al., 2002a,b). Despite this knowledge, it is not clear which portions of the protein are used to sense membrane tension and how this is transferred to the pore, issues that an open-state structure may help to resolve. Given the large current known to pass through the pore in the open state, and the size of molecules that pass through the pore (Cruickshank et al., 1997), a significant conformation change must take place upon channel activation to create a large pore. Although a closed-state structure has been obtained from x-ray crystallography, it appears difficult to capture the open state of MscL obtained under membrane tension during crystallization. Furthermore, crystallization requires membrane proteins to be removed from their native environment, which may affect structure. Ideally, we would like to gain structural information under physiological conditions. In this direction, experiments using site-directed spin labeling (SDSL) electron paramagnetic resonance (EPR) spectroscopy of MscL reconstituted into artificial liposomes have been performed, indicating an open pore of >25 Å diameter (Perozo et al., 2002a). However, specific intersubunit distances could not be measured in the open state, as they are greater than the 15 Å that can be measured with the continuous wave (CW) EPR technique. Fluorescence resonance energy transfer (FRET) provides an alternative method of measuring inter-site distances over longer ranges (Wu and Brand, 1994; Gustiananda et al., 2004). Previous experiments using fluorescent labels on just one site of the protein have indicated that the diameter of the protein increased by 16 Å upon channel activation (Corry et al., 2005b).
The incomplete nature of the existing structural information means that questions remain as to the likely nature of the open state. Initially, it was suggested that the transmembrane helices end up side by side so that both TM1 and TM2 helices line the pore to accommodate the size of the open channel (Cruickshank et al., 1997; Chang et al., 1998; Batiza et al., 1999; Spencer et al., 1999; Yoshimura et al., 1999). SDSL CW EPR spectroscopy (Perozo et al., 2002a) and computational (Gullingsrud et al., 2001; Sukharev et al., 2001a,b) experiments, however, suggest that the open state may be lined mostly by TM1 helices, a feat that can only be achieved by a large tilting of the helices and a thinning of the protein in the plane of the membrane. The short N-terminal domain is known to be important to the function of the protein, as amino acid deletion or substitution in this region severely affects MscL function (Blount et al., 1996; Häse et al., 1997). It has been proposed that this domain functions as a second gate (Sukharev et al., 2001a,b; Betanzos et al., 2002; Anishkin et al., 2005); however, this is not supported by a recent study that used cysteine trapping of N-terminal mutant channels (Iscla et al., 2007). By conducting single cysteine mutations, site-directed fluorophore labeling, and FRET imaging of unilammelar bilayers formed by reconstituted liposomes, we have determined the structural rearrangements taking place at several regions within the protein. These data, along with existing SDSL EPR information, are used in computer simulations to quantify the conformational changes involved in channel opening. The demonstration of a successful application of site-directed fluorophore labeling FRET microscopy and simulation for obtaining structural information achieved here opens a new avenue for studying membrane proteins. An advantage of this technique is that measurements can be made in the same conditions as electrophysiological studies, and we are able to pick suitable sections of the sample (i.e., clearly unilamellar liposomes) rather than averaging over the entire sample. The resulting model yields information about the size and nature of conformational changes that lead to the open state, and shows how a highly conductive channel can be obtained with much more limited motion than previously suggested. Importantly, we are also able to clarify the role of the N-terminal domain, showing that it could either directly sense membrane tension and transfer this to the pore-lining helices or serve as an anchor that guides the TM1 to a greater tilt during MscL opening by maintaining its position along the membrane–cytoplasm interface, as suggested by Iscla et al. (2008).
MATERIALS AND METHODS
Expression, labeling, and reconstitution of MscL protein
The wild-type MscL protein contains no cysteine residues. Hence, site-directed mutagenesis (Agilent Technologies) was used to introduce cysteine at individual sites of interest in MscL that were subsequently labeled with Alexa Fluor maleimide dye (Invitrogen) specifically binding to cysteine. The MscL mutants were each cloned into pQE32 vector and expressed in Escherichia coli, followed by solubilization and purification via Ni-NTA affinity column, as described previously (Perozo et al., 2001, 2002b). The MscL protein mutant was then labeled according to the manufacturer’s instructions with equimolar concentrations of both Alexa Fluor 488 (AF488) and Alexa Fluor 568 (AF568) with a protein to each dye molar ratio of 1:5. Consequently, each of the five subunits had one label with the equal probability of any site being occupied by either AF488 or AF568, the functional protein having five fluorophores attached. For FRET analysis, the labeled protein was reconstituted into azolectin liposomes at a protein to lipid molar ratio of 1:250 (Corry et al., 2005b) and for patch clamp analysis with a protein to lipid ratio of 1:500 by weight (Häse et al., 1995).
Patch clamp analysis
Fluorophore-labeled channel mutants were tested for function in patch clamp analysis. Activity of the individual MscL mutants was recorded from single bilayer blisters in symmetrical solution (200 mM KCl, 40 mM MgCl2, and 5 mM HEPES buffer, pH 7.2/KOH), as reported previously (Häse et al., 1995). In control experiments, recording solution also contained 1 mM dithiothreitol (DTT) to prevent formation of disulfide bonds between cysteine residues, which would inhibit channel opening if fluorophores became detached. Gating and full channel openings were recorded as a result of membrane tension applied via negative hydrostatic pressure to the patch pipette. We also confirmed via patch clamp recordings that channel activity could be induced in the absence of suction by adding LPC to the bath solution (leading to insertion into the outer bilayer leaflet) at final concentrations between 2 and 3 µM. Typically, spontaneous channel gating occurred within 10 min after the addition of LPC, as reported previously (Perozo et al., 2002b). Additional control experiments were also performed with unlabeled mutants (supplemental material).
Fluorescence imaging and analysis
Liposomes containing reconstituted and labeled protein were imaged on a UV laser-scanning confocal microscope (MRC1000/1024; Bio-Rad Laboratories) using a water immersion objective lens (60×; NA1.2; Nikon) and the same recording solution as for the patch clamp studies. AF488 was excited at 488 nm with an argon laser and detected through a 522/535-nm bandpass filter, whereas AF568 was excited at 561 nm using a yellow laser and detected through a 585-nm long-pass filter.
FRET efficiencies were determined for individual liposomes using acceptor photobleaching before and after the addition of LPC. FRET data were analyzed using two methods. The first was a pixel by pixel analysis conducted using a modified version of a plug-in written by David Stepensky developed in ImageJ (Abramoff et al., 2004). In this, images of the donor and acceptor before and after acceptor bleaching were first aligned, and then background values obtained from a region away from the liposome in each image were subtracted before the resulting images were smoothed using a 3 × 3 mean filter. Regions of interest were then automatically selected using both donor and acceptor threshold values and at least 70% acceptor bleaching. Images were taken using acceptor excitation and emission channels before and after every bleaching cycle to monitor the degree of acceptor bleaching achieved. Corrections were then made to the intensity values of the donor after acceptor bleaching at each pixel to account for incomplete acceptor bleaching and bleaching of the donor during imaging:
in which Da’ is the corrected value of the donor after acceptor bleaching, Db and Da are the uncorrected donor values before and after acceptor bleaching, Ab and Aa are the acceptor values before and after acceptor bleaching, and C is a factor accounting for any undesired bleaching of the donor calculated for each liposome as described below. FRET efficiencies (E) were then calculated for each pixel in the region of interest using
The pixel by pixel analysis method is particularly sensitive to movement of the liposomes during imaging and could only be applied to a small proportion of the images collected. Thus, a more robust averaging method was also used in which regions of interest were specified manually, and background subtraction, donor correction, and FRET efficiency values were determined using the mean intensities in the region of interest, rather than for each pixel. For every liposome, an additional round of bleaching and imaging was performed after the acceptor was fully bleached to determine how the imaging and bleaching influenced the donor intensities. As every liposome bleached slightly differently depending primarily on its size, the donor bleaching correction for every liposome, C, could then be determined by comparing the donor intensity before and after the additional bleach.
An analysis of the orientations of the fluorophores relative to the membrane was performed with a method described previously (Corry, 2006). Using the fact that the exciting laser light was linearly polarized, the variation in emitted intensity of each fluorophore around spherical liposomes can be related to the so called “B factor” that comprises the average orientation of the fluorophores and their orientational mobility. Making such measurements on both donor and acceptor fluorophores allows the most likely orientation factor for transfer between them to be determined. This analysis method requires clean spherical liposomes, so only some of our images could be used.
Although the relationship between the fluorophore separation and transfer efficiency is well known for single donor–acceptor pairs, the situation is more complicated in our case, as each protein contains a random mix of five fluorophores. We use a Monte-Carlo scheme (using the program “exiFRET”) (Corry et al., 2005a) to relate the efficiency of energy transfer to the radius of the fluorophore pentamers, as shown in Fig. S1. Results for mutant M42C are taken from our previous publication (Corry et al., 2005b) and have been shown again here, as this illustrates one of the clearest examples of FRET.
Molecular and Brownian dynamics simulations
A model of the closed-state MscL channel from E. coli (Meyer, 2007) based on the crystal structure of the homologue from M. tuberculosis (Protein Data Bank accession no. 2OAR) was inserted into a POPC lipid bilayer and solvated in a 120 × 120 × 130–Å water box with 150 mM KCl (total of ∼155,000 atoms). Initially, the protein atoms were fixed while the lipid and water were minimized for 1,000 steps and then equilibrated for 50 ps. A harmonic constraint of 2 kcal/mol/Å was then placed on the protein α carbon, and equilibration proceeded for 20 ps before reducing the constraint to 1 kcal/mol/Å for a further 500 ps.
After equilibration, experimentally derived distances from FRET and EPR studies were enforced by including distance restraints between the Cα atoms on the associated residues. Because of the pentameric nature of the protein, each measurement yields 10 distance constraints: 5 to neighboring subunits and 5 to next nearest neighbors. In total, this yields 80 FRET-derived distance constraints and 640 EPR constraints. Distance constraints resulting from our FRET data were included as harmonic distance restraints with final force constants calculated as five times the inverse of the uncertainty in the FRET-derived distance data (thus force constants ranged from 0.2 to 2.9 kcal/mol/Å2). As the CW EPR measurements only provided lower bounds on the distance between residues within the subunits in the open state (Perozo et al., 2002a), these were included as lower bound constraints with force harmonic constants of 4.0 kcal/mol/Å2 when the distance between subunits was <18 and 0 when it was above. Both FRET and EPR restraints were slowly introduced during the simulations by gradually increasing the force constant over nine steps starting from one thousandth of the final value. We note that only distance changes between the closed and open states of the channel were used to minimize uncertainty arising from the orientation or size of the fluorophores. As noted above, we explicitly measure the orientation of the fluorophores to show that neither of these effects influences the distance changes seen in our data.
Three separate simulations were conducted with membrane tensions of 0, 5, and 60 mN/m, each lasting either 13 ns (0 and 5 mN/m) or 18 ns (60 mN/m), using NAMD (Phillips et al., 2005) with the CHARMM27 all-atom force field (MacKerell et al., 1998) under a pressure of 1 atm and temperature of 298 K with a 1-fs time step. Pore radii were calculated with the program HOLE (Smart et al., 1993), and graphics were produced using VMD (Humphrey et al., 1996).
The conductance of the resulting open-channel structures was calculated using Brownian dynamics simulations (Li et al., 1998; Corry et al., 2002) in which the protein is taken as a rigid structure and the motion of individual ions is traced explicitly. Such simulations have been shown to produce accurate currents in a range of ion channels (Corry et al., 2001, 2002, 2004). Here, we use symmetric 300 mM KCl solution, and dielectric constants of 80 for the interior of the pore and baths and 2 for the protein were used when calculating the electrostatic forces on ions using Poisson’s equation. A 2-fs time step was used for ions inside the pore and 100 fs for ions in the baths.
Online supplemental material
The online supplemental material contains additional results and figures, including an examination of the importance of the labeling efficiency of the protein, more fluorescence images and analysis of the FRET data arising from them, and the orientations of the fluorophores. There are also patch recordings of each of the labeled mutants under hydrostatic pressure and additional simulation data, including the evolution of the protein and membrane structure during the simulations. An animation of ion conductance from Brownian dynamics simulations is also included. The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.200910376/DC1.
RESULTS and DISCUSSION
FRET and patch clamp recordings
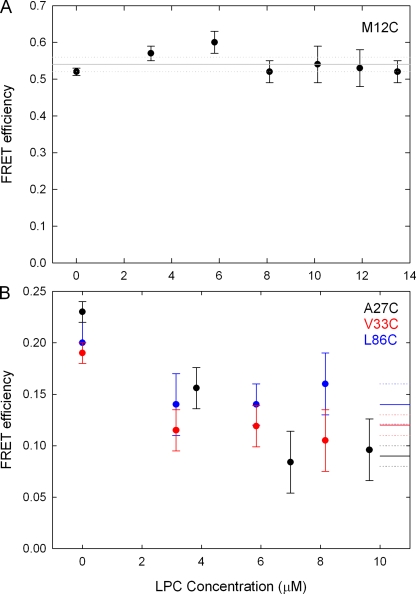
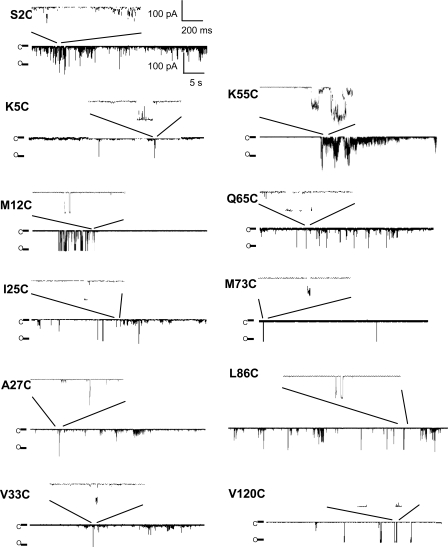
Fluorescence images of liposomes containing MscL labeled at different sites before and after the addition of LPC are shown in Fig. 1, and the increase in donor intensity after acceptor bleaching is a clear indication of the presence of FRET. Transfer efficiency values were determined by two methods (pixel by pixel and liposome average intensities), with the average difference between the two being <3%. A more detailed analysis of the pixel by pixel results is given in the supplemental material. A summary of the FRET results obtained is given in Table I, showing the large range of FRET efficiencies observed, whereas the locations of all the mutants studied is given in Fig. 2. The large FRET value for V120C is not unreasonable given the proximity of these sites in the cytoplasmic helices seen in the Tb-MscL crystal structure (nearest neighbors are only 13.5 Å apart). It is notable that although most of the mutants studied show a large change in FRET efficiency before and after the addition of LPC, this is not the case for the mutants M12C, G14C, I25C, and M73C, even at LPC concentrations as high as 13 µM (see Fig. 4). A similar lack of change between the closed and open states of M73C is seen in EPR data (unpublished data).
Figure 1.
Confocal fluorescence images of AF488- and AF568-labeled MscL protein reconstituted into liposomes. Images of the donor (AF488) are shown before (left column) and after (middle column) acceptor photobleaching. Images are colored by intensity according to the top scale bar. An image created from pixel by pixel analysis of the FRET efficiency is shown in the right column, colored according to the bottom scale bar. The mutant name and conditions are noted on the left, and the average FRET efficiency for the image is noted on the right. I, inner liposome; O, outer liposome.
Table I.
FRET efficiency values are indicated for liposomes imaged before and after the addition of LPC
| Mutant | FRET pre-LPC | FRET post-LPC | Δ Diameter FRET | Δ Diameter simulation |
| Å | Å | |||
| S2Ca | 0.22 ± 0.02 (24) | 0.11 ± 0.02 (30) | 22 ± 6 | 18.7 |
| K5Ca | 0.23 ± 0.01 (25) | 0.14 ± 0.02 (25) | 16 ± 4 | 17.5 |
| M12C | 0.45 ± 0.01 (30) | 0.46 ± 0.01 (36) | −1 ± 1 | 17.4 |
| G14C | 0.38 ± 0.02 (11) | 0.40 ± 0.02 (12) | −2 ± 3 | 22.9 |
| I25C | 0.16 ± 0.02 (20) | 0.15 ± 0.02 (34) | 2 ± 6 | 24.6 |
| A27Ca | 0.23 ± 0.01 (23) | 0.09 ± 0.01 (19) | 29 ± 4 | 25.9 |
| V33Ca | 0.19 ± 0.01 (21) | 0.12 ± 0.01 (26) | 14 ± 3 | 17.4 |
| M42Ca | 0.61 ± 0.01 (8) | 0.44 ± 0.01 (9) | 16 ± 2 | 15.1 |
| K55C | 0.06 ± 0.02 (18) | 0.03 ± 0.01 (16) | 23 ± 15 | 20.1 |
| Q65C | 0.09 ± 0.03 (25) | 0.05 ± 0.03 (17) | 18 ± 18 | 14.3 |
| M73C | 0.17 ± 0.02 (39) | 0.17 ± 0.02 (35) | 0 ± 5 | 14.1 |
| I86Ca | 0.21 ± 0.02 (46) | 0.14 ± 0.02 (34) | 12 ± 5 | 17.5 |
| M94Ca | 0.14 ± 0.01 (51) | 0.08 ± 0.01 (65) | 16 ± 5 | 20.1 |
| V120Ca | 0.78 ± 0.01 (28) | 0.66 ± 0.02 (49) | 14 ± 2 | 12.7 |
The number of liposomes used in the analysis is indicated in brackets in each case. The corresponding change in diameter at the location of the fluorophores calculated from the FRET data and that seen in the simulations (60 mN/m) are also indicated.
Residues used as restraints in the MD simulations.
Figure 2.
Location of mutants used to gain FRET data. Out of the 20 mutants selected, the 8 that gave the best FRET results (shown as blue side chains on the channel monomer) together with the location of each residue for which EPR data were used in the restrained simulation to create the open-state model (green spheres) are shown.
Figure 4.
FRET efficiency versus LPC concentration. (A) For mutant M12C, the measured FRET efficiency shows no change upon the addition of LPC. (B) For mutants A27C, V33C, and L86C, the FRET efficiency decreases upon the addition of LPC, but it does not decrease further when LPC concentration is >6 µM. The average post-LPC FRET values and associated uncertainties (as reported in Table II) are represented by the solid and dashed lines on the right-hand side of the figure.
To help understand these results, patch clamp recordings were undertaken to see whether the mutants are still functional, as shown in Fig. 3. We confirmed that the fluorophore-labeled MscL mutants were functional by the application of negative hydrostatic pressure to the patch pipette (Fig. S4), finding that channels are active for all labeled mutants except G14C. Similarly, all mutants investigated in this study except G14C can be activated by the addition of LPC to the excised liposome patch (Fig. 3). Activation by pressure and LPC of the mutant M42C has been confirmed previously (Corry et al., 2005b). For comparison, we have also tested unlabeled A27C and V120C mutants by negative pressure and LPC (Fig. S5). In contrast to the fluorophore-labeled mutant channels, only channels gating at subconducting levels were observed in recordings obtained from unlabeled A27C channels in the absence of DTT, whereas activity of fully open channels could be recorded upon the addition of 25 mM DTT to the bath solution (Fig. S5 A). Similarly, a majority of the patches containing unlabeled V120C channels exhibited activity of fully open channels in the presence of DTT, and only a few patches exhibited such activity in the absence of DTT (Fig. S5 B). Similar to labeled channels the unlabeled channels could be activated by LPC with or without DTT being present (Fig. S5).
Figure 3.
Representative recording traces for fluorophore-labeled channel mutants activated with LPC in the absence of pressure. For each mutant, a longer recording trace and an inset with the enlarged recording trace is shown. Note the different scale bars for the initial traces and for the inset. C, the closed state of one MscL channel; O, the open state of one MscL channel.
The lack of change in FRET efficiency for the mutants M12C, G14C, I25C, and M73C could be explained by many factors. The low open probabilities of the channel upon the addition of LPC witnessed in the patch clamp studies for M12C, G14C, and M73C would limit the associated FRET changes. It is possible that there is little structural change in the protein when moving between the closed and open states at these positions, but this is unlikely, particularly for I25C that sits at the narrowest part of the pore in the closed state and has been shown in SDSL EPR experiments to undergo significant motion during the channel activation (Perozo et al., 2002a). Therefore, it may be that either the fluorophores are confined within the pore when attached to this site and thus cannot accurately report on the structural change of the protein, or that the open probability of the channel is low for these cases when FRET measurements are made.
An important consideration for these experiments is to ensure that we have very high open probabilities when making our post-LPC FRET measurements. Seven lines of evidence support the conclusion that high open probabilities are being achieved: (1) FRET data were collected at several different LPC concentrations, and results were only used for the open state once the efficiency showed no change upon increasing LPC concentration, as shown for four mutants in Fig. 4. This ensures that we obtain the maximum open probability possible using LPC. (2) We note that the FRET measurements are not made under the same LPC concentrations as the patch clamp recording shown in Fig. 3. LPC concentration in patch clamp experiments had to be lower, corresponding to lower channel open probability, because during the patch clamp experiments, liposome patches are under residual stress due to the gigaohm seal formation, which causes the bilayer to break easier in the presence of LPC. An advantage of the FRET measurement in liposomes is that the bilayer is not stressed and can tolerate high LPC concentrations (>25% LPC molar ratio with PC (Perozo et al., 2002b). The open probability during FRET measurements is, therefore, larger than that seen in the patch recording of Fig. 3, as the EPR data show an increasing open probability with increasing LPC insertion in the liposomes. (3) Perhaps most conclusively, EPR experiments conducted under similar conditions indicate that spin coupling between labeled residues (including those in the most constricted portions of the channel) are eliminated at these high LPC concentrations (Perozo et al., 2002b). As spin coupling is clearly observed under resting conditions, the lack of coupling observed after the addition of LPC is only possible if the closed-channel probability is too small to detect. The considerations of open probability in the present study are the same as those in recent EPR experiments (Perozo et al., 2002a,b; Vásquez et al., 2008). (4) There is electrophysiological evidence for continuous long opening of MscL in the presence of low concentration of LPC after membrane stretch was released (Fig. 4 a in Perozo et al., 2002a). (5) The diameter changes measured using FRET are large and consistent with changes expected for an open state, in agreement with the results from molecular modeling, SDSL EPR, and electrophysiological studies (Cruickshank et al., 1997; Sukharev et al., 2001a,b; Perozo et al., 2002a). Our model indicates an 18-nm2 in-plane protein expansion, which agrees well with experimental measurements of 20.4 ± 4.8 nm2 (Chiang et al., 2004). In addition, the size of the pore found in our proposed open-channel models described below and the likely conductance of these (determined from Brownian dynamics simulation) are consistent with measured values for the fully open channel. Although the presence of closed channels during our FRET measurements could mean that we underestimate the structural changes involved in activation, if our FRET values involved a significant proportion of closed channels we would expect to see less structural change. (6) Occasionally, results were obtained with a proteoliposome within another proteoliposome. After the addition of LPC, the distribution of FRET efficiencies for outer and inner liposome has clearly been distinguishable, such that the outer liposome is affected by the presence of LPC (i.e., the channels are in their open state), whereas the inner liposome remains unaffected by LPC (i.e., the channels remain closed; e.g., M42C mutant in Fig. 1). These results strongly indicate that a substantial change in FRET efficiency occurs in these situations due to the insertion of LPC into the outer liposome (see supplemental material). (7) Finally, a 3-D x-ray structure of SaMscL showing a tetrameric channel in an intermediate open state (Liu et al., 2009) is consistent with the MscL open-channel structure presented in this study as discussed below.
The changes in FRET efficiency seen in the remaining cases upon adding LPC most likely represent underlying structural changes of the MscL protein resulting in the fluorophores on each subunit moving farther from one another when the channel is opened. Although it is possible that the changes could (at least in part) be caused by alteration in the relative orientations of the fluorophores or a change in the position of the dye head group relative to the site of attachment, this can be excluded by analyzing the observed orientation of the fluorophore transition moments following the procedure developed previously (Corry et al., 2006) (see supplemental material). As the measured orientations of the dyes are almost identical before and after the addition of LPC, this data also shows that the use of a value for the orientation factor (κ2) close to 2/3 is well justified. The measured FRET efficiencies can be related to distances within the protein, provided that the pentameric nature of the protein and labeling probabilities are taken into account (Corry et al., 2005a). The change of radius or diameter upon channel activation is less likely to be influenced by any systematic errors in the FRET measurements and is less sensitive to the value of the κ2 (and R0) than is the absolute radius of the channel. This change in diameter is shown for each mutant in Table I.
Constrained molecular dynamics
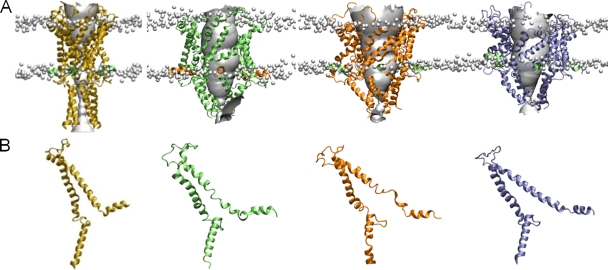
Apart from the four mutants described above, the remaining results suggest a consistent diameter change of >14 Å for all parts of the protein studied. To determine how these structural changes are likely to influence the overall structure of the protein, they were incorporated into molecular dynamics simulations. An initial model of the pore based on the crystal structure of Tb-MscL (Meyer, 2007) has a very small minimum radius, as shown in Fig. 5 (and see supplemental material). However, when the results of our FRET measurements and previous SDSL EPR data are included as 720 individual distance restraints in the simulation, very large conformational changes take place, creating a wide pore in each of the conditions tested. It should be noted that the experimental restraints are not aimed at reproducing forces experienced by the protein during activation, but rather act as a structure-refinement tool to make the protein adopt a conformation consistent with the experimental data. The resulting structures obtained with 0, 5, and 60 mN/m of tension on the membrane are indicated in Fig. 5, along with the initial closed state of the channel. In all cases the TM1 and TM2 helices are seen to move outward to reveal a 28-Å diameter pore. Note that an open-channel pore >25 Å is indicated by SDSL EPR experiments (Perozo et al., 2002a). Molecular modeling suggests a pore of 30–37 Å (Sukharev et al., 2001a,b), whereas electrophysiological experiments gave an estimate of a diameter of the pore of ∼30 Å (Cruickshank et al., 1997). Collectively, these results indicate that FRET experiments in this study yielded a pore of a fully open channel. The current–voltage relationship calculated on each of the structures using Brownian dynamics simulations gives a conductance of between 3.0 and 3.5 nS (see supplemental material), similar to the currents measured in patch clamp studies, suggesting also that the pore is wide enough to represent the open state of the channel (Häse et al., 1995, 1997).
Figure 5.
Simulation studies incorporating EPR and FRET data. (A) The closed state (yellow) and model open states of the protein obtained under membrane tension of 0 (green), 5 (orange), and 60 mN/m (blue), respectively, viewed in the plane of the membrane. The location of the lipid phosphate head groups is indicated by the gray balls to give an indication of the position of the membrane. (B) The structure of a single subunit of each structure is indicated.
The introduction of tension does have a significant effect on the final structure of the protein found in the simulations. As described in the supplemental material, the application of tension decreases the thickness of the membrane, which favors a greater tilting of the transmembrane helices. Notably, in the absence of membrane tension, some unraveling of the intracellular end of the TM1 helix is seen during the simulations. This is most likely an artifact created by forcing the protein to adopt a new conformation in a limited time frame. When a tension of 60 mN/m is applied to the membrane, this unraveling is mostly eliminated. This suggests that transition to the open state is more favorable under tension with a slightly thinner membrane, consistent with experimental data showing increased activation by membrane stretch in bilayers formed from shorter lipids (Martinac and Hamill, 2002; Perozo et al., 2002b). To check if the protein remains in the same open state in the absence of the experimental restraints, we removed the restraints and continued the simulations for an additional 5 ns. As described in the supplemental material, no significant change in the structure is seen in this time, and the radius of the pore remains relatively constant. If the tension is removed in addition to the experimental restraints, however, the channel begins to shrink, as seen in Fig. S9).
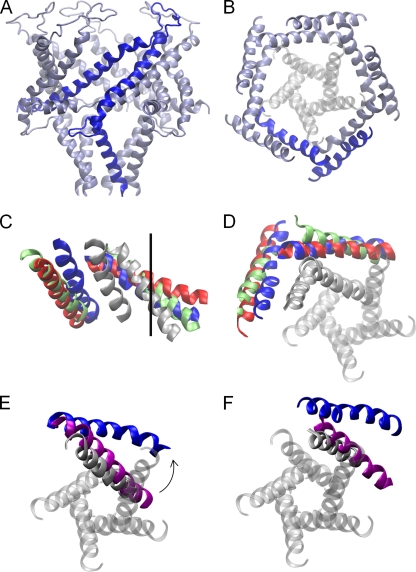
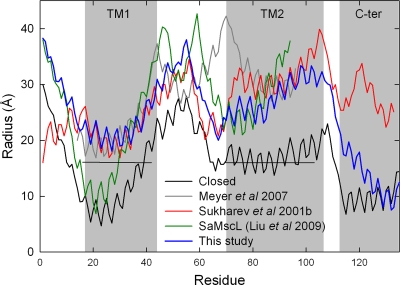
The structural change taking place between the closed state of the pore and our open-channel models found under tension is significant, as can be seen in Fig. 6. The TM1 and TM2 helices maintain their relationship to one another, but both move outward in a radial direction, which is consistent with the results of Betanzos et al. (2002), who showed that cross-linking TM1 to TM2 did not abolish MscL opening. The result of this is that the full length of TM1 lines the pore along with the extracellular half of TM2, yielding a configuration consistent with lipid and water accessibility data from SDSL EPR, but without substantial rotation of TM1. This conformational arrangement is somewhat different to recent hypotheses that suggest the open state of the pore to be lined principally by TM1 (Gullingsrud et al., 2001; Sukharev et al., 2001a,b; Perozo et al., 2002a; Meyer, 2007). To illustrate this difference, we superimpose our open-state model on previous models in Fig. 6 (C and D). All open-state models have a similar pore radius, and all show TM1 and TM2 tilting toward the plane of the membrane by around 20°. But, in our case the TM2 helix lies closer to the pore than in previous suggestions, which can also be seen by plotting the radial position of each residue as in Fig. 7, a configuration supported by our FRET data. The 18-nm2 in-plane protein expansion in our model obtained under 60 mN/m of tension agrees well with experimental measurements of 20.4 ± 4.8 nm2 (Chiang et al., 2004).
Figure 6.
Open-channel structure. The symmetrized open-channel model obtained from the simulations conducted with 60 mN/m of tension are shown, viewed from in the plane of the membrane (A) and just the TM helices from above the membrane (B), with one subunit highlighted for clarity. The location of TM1 in the closed-state crystal structure is shown in gray. Comparisons with previous open-channel models are shown viewed from in (C) and above (D) the plane of the membrane. In these, TM1 and TM2 of just one subunit of the closed structure (gray), our open-state model (blue), and the models of Meyer (2007) (green) and Sukharev et al. (2001b) (red) are shown. The location of the pore axis is marked by the black line. Comparisons of the positions of TM1 (E) and TM2 (F) in our open-state model and the hypothesized intermediate SaMscL structure (purple). TM1 of all five subunits of the closed structure is shown in D–F for clarity.
Figure 7.
Comparisons of open-channel models. The radius of the protein at the position of the α-carbon atom of each residue is shown for the closed-state model of the protein (Chang et al., 1998) (black), the open-state model developed here (blue), the previously proposed open-state models (gray) of Meyer (2007) and (red) Sukharev et al. (2001b), and the SaMscL crystal structure (green) (Liu et al., 2009). The location of the EPR lower bound distance restraints included in the simulations is shown by the horizontal lines.
It is also interesting to compare our open-state model with the recent SaMscL crystal structure that is hypothesized to represent an intermediate open state of the channel (Liu et al., 2009). TM1 is tilted at a similar angle to the membrane in both structures, but although the intracellular end of the helix forms a constriction to the pore in the SaMscL structure, in our case it is farther from the channel axis, resulting in a much wider pore (Fig. 6 E). TM2 also sits farther from the pore axis in our structure, as required to accommodate the new position of TM1 (Fig. 6 F). Our model is consistent with the suggestion of Liu et al. (2009) that opening the pore from the intermediate state involves a rotation of the TM helices. Starting from the SaMscL structure and pivoting the TM1 and TM2 helices of the adjacent subunit as a rigid block by 20° about an axis perpendicular to the membrane passing through Gly48 places the TM helices in almost exactly the same position seen in our open-state model as shown in the supplemental material. In support of our model, such a rotation of the SaMscL structure places TM2 closer to the pore than in previous suggestions.
All our open-state models involve a much smaller conformational change from the closed state than either of the previous models. To illustrate this, we align both one subunit and the complete pentamer of each model with the closed state and calculate the magnitude of the structural changes required to open each. Although the presence of membrane tension creates a greater tilting of the TM helices and thus a greater structural change upon activation, this is less than seen in the previous models as shown in Table II. Because our simulations start from the closed state of the pore, this may bias the results toward open conformations that are similar to the closed state of the pore. Previous restraint-driven simulations (Perozo et al., 2002a) sample a greater range of possible structures, but they do not evaluate the energy of all atomic interactions within the protein and environment as done here. Although certainly not conclusive, the fact that a stable and wide pore can be achieved with more limited structural changes than that seen in some previous models lends credence to the open-channel structure presented here. Further simulations that can combine all the atomic interactions used here with greater conformational sampling and experimental data may yield the most conclusive model of the open state.
Table II.
The tilt in the TM helices is shown for each of the open-state models as well as the magnitude of structural change required to move from the closed state to each open state
| Model | Tilt TM1 | Tilt TM2 | RMSD in subunit | RMSD in pentamer |
| Å | Å | |||
| Closed | 37.2 | 34.4 | ||
| 0 dyn/cm | 52.5 | 39.0 | 8.1 | 11.5 |
| 5 dyn/cm | 56.3 | 42.4 | 9.9 | 12.0 |
| 60 dyn/cm | 59.8 | 47.0 | 8.9 | 12.7 |
| Meyer (32) | 51.2 | 54.5 | 12.6 | 17.7 |
| Sukharev (27) | 71.5 | 56.3 | 9.5 | 33.2 |
The structural change is determined by aligning either a single subunit to a subunit in the closed structure, or the whole pentamer to the closed pentameric structure, and presented as a root mean-square deviation (RMSD).
A recent study using electrostatic interactions and disulfide cross-linking to probe the channel suggests that specific residue pairs that do not interact in the closed state of the pore do interact in the open state (G26/I92 and V23/I96), something that requires a rotation of the TM1 helix relative to TM2 (Li et al., 2009). The interpretation of these experiments is complicated due to the competition of electrostatic interactions between the charged sites within the TM1 helix and interactions between TM1 and TM2, as well as to whether the addition of charged groups in proximal locations assists rotation. Similar rotations have been suggested in the models based on EPR studies (Perozo et al., 2002a) and MTS accessibility data (Bartlett et al., 2004), as well as data based on intragenic suppression of the V23A and G26S mutations (Li et al., 2004). Further support for such TM1 helix rotation has been provided by a study using metal-binding sites generated in the channel pore (Iscla et al., 2004). In contrast, an open-channel model supported by disulfide cross-linking has been proposed that does not involve TM1 rotation (Betanzos et al., 2002), and no such rotation is seen in the recent S. aureus crystal structure (Liu et al., 2009). Our FRET data are in agreement with the open diameter of the channel estimated from EPR studies (∼69 Å compared with 70 Å) as well as that of the channel pore estimated from EPR and molecular sieving experiments (28 Å compared with >25 Å). Our models, however, show little rotation of TM1 relative to TM2. Rather, the pairs of TM1–TM2 helices appear to rotate as a rigid body about an axis perpendicular to the membrane.
The N terminus remains in a stable helical configuration along the membrane water interface in all our simulations, with a nonpolar face of the helix pointing into the membrane and a polar face pointing out as can be seen in Fig. 5, consistent with the estimated distances and recently refined MscL crystal structure (Steinbacher et al., 2007; Iscla et al., 2008). This result helps resolve contention about the role of this domain, as in this position it could not function as a second gate but may act to anchor the protein to the surface of the lipid bilayer. More interestingly, this interaction of the N terminus with the membrane could serve a role in sensing membrane tension and pressure gradients. The direct connection between this region and the intracellular end of the pore-lining TM1 means that a radial force on the N terminus may directly be transferred into an increased tilt in the TM1 helix relative to the membrane by conferring to the N terminus a role of an anchor that guides the TM1 to a greater tilt, especially upon membrane thinning (Iscla et al., 2008). A thinning of the protein in the plane of the membrane and pore widening is apparent in Fig. 5. The reduced hydrophobic thickness of the protein, possibly in conjunction with strain in the TM1 helix seen to some extent in our simulations, may act as a spring to return the pore to a resting state once tension on the N terminus is removed. This hypothesized role is consistent with the increased pressure required to open N-terminal deletion mutants (Blount et al., 1996; Häse et al., 1997) that would reduce the transduction of membrane tension to TM1. FRET data from mutant V120C indicate a widening of the C-terminal α-helical bundle; however, there is a large degree of scatter in the FRET data at this position, suggesting variability in the conformation of this domain, which has also been implied to participate in the channel gating (Yoshimura et al., 2008). In all our simulations, this comes apart at the top to create a larger entrance to the pore, but the lower end of the bundle remains intact and may help to prevent very large solutes entering the pore, as suggested previously (Anishkin et al., 2003).
The method of combining data from FRET confocal microscopy obtained in liposomes with simulations has enabled us to determine a likely open-state structure of MscL in a natural lipid environment. Broad similarities of our structures with previous experiments (Cruickshank et al., 1997; Sukharev et al., 2001a,b; Perozo et al., 2002b; Anishkin et al., 2003; Anishkin and Sukharev, 2004; Iscla et al., 2008; Yoshimura et al., 2008), and close agreement with the hypotheses based on the recently reported SaMscL structure (Liu et al., 2009), support the applicability of this method, but the ability to measure inter-site separations over longer distances than in previous studies has allowed us to suggest different locations for some parts of the protein. A similar method has been used to study the conformational distributions in flexible prion peptides (Gustiananda et al., 2004); however, the use of FRET to gain detailed structural information for multimeric membrane proteins has required different approaches to protein labeling, control of the protein state, and careful analysis of the orientations, geometries, and number of fluorescent probes. We believe that the methodology demonstrated here provides an excellent tool for studying the structures of membrane proteins in natural environments.
Acknowledgments
We thank Paul Rohde, Corinne Vallet, Dr. Andrew Battle, and Maryrose Constantine for technical assistance.
This project was supported by grants from the Australian Research Council (to B. Corry, P. Rigby, and B. Martinac), the Australian National Health and Medical Research Council (to B. Martinac and B. Corry), and an award under the merit allocation scheme on the National Computational Infrastructure at the ANU and considerable supercomputer time from iVEC. The authors acknowledge the facilities and the scientific and technical assistance of the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy, Characterisation & Analysis, The University of Western Australia, a facility funded by The University, State, and Commonwealth Governments.
Lawrence G. Palmer served as editor.
Footnotes
Abbreviations used in this paper:
- AF488
- Alexa Fluor 488
- AF568
- Alexa Fluor 568
- CW
- continuous wave
- DTT
- dithiothreitol
- EPR
- electron paramagnetic resonance
- FRET
- fluorescence resonance energy transfer
- LPC
- lysophosphatidylcholine
- MscL
- mechanosensitive channel of large conductance
- SDSL
- site-directed spin labeling
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. 2004. Image processing with ImageJ. Biophotonics International. 11:36–42 [Google Scholar]
- Anishkin A., Sukharev S. 2004. Water dynamics and dewetting transitions in the small mechanosensitive channel MscS. Biophys. J. 86:2883–2895 10.1016/S0006-3495(04)74340-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anishkin A., Gendel V., Sharifi N.A., Chiang C.S., Shirinian L., Guy H.R., Sukharev S. 2003. On the conformation of the COOH-terminal domain of the large mechanosensitive channel MscL. J. Gen. Physiol. 121:227–244 10.1085/jgp.20028768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anishkin A., Chiang C.S., Sukharev S. 2005. Gain-of-function mutations reveal expanded intermediate states and a sequential action of two gates in MscL. J. Gen. Physiol. 125:155–170 10.1085/jgp.200409118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett J.L., Levin G., Blount P. 2004. An in vivo assay identifies changes in residue accessibility on mechanosensitive channel gating. Proc. Natl. Acad. Sci. USA. 101:10161–10165 10.1073/pnas.0402040101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batiza A.F., Rayment I., Kung C. 1999. Channel gate! Tension, leak and disclosure. Structure. 7:R99–R103 10.1016/S0969-2126(99)80061-6 [DOI] [PubMed] [Google Scholar]
- Berrier C., Besnard M., Ajouz B., Coulombe A., Ghazi A. 1996. Multiple mechanosensitive ion channels from Escherichia coli, activated at different thresholds of applied pressure. J. Membr. Biol. 151:175–187 10.1007/s002329900068 [DOI] [PubMed] [Google Scholar]
- Betanzos M., Chiang C.S., Guy H.R., Sukharev S. 2002. A large iris-like expansion of a mechanosensitive channel protein induced by membrane tension. Nat. Struct. Biol. 9:704–710 10.1038/nsb828 [DOI] [PubMed] [Google Scholar]
- Blount P., Sukharev S.I., Moe P.C., Schroeder M.J., Guy H.R., Kung C. 1996. Membrane topology and multimeric structure of a mechanosensitive channel protein of Escherichia coli. EMBO J. 15:4798–4805 [PMC free article] [PubMed] [Google Scholar]
- Booth I.R., Edwards M.D., Black S., Schumann U., Miller S. 2007. Mechanosensitive channels in bacteria: signs of closure? Nat. Rev. Microbiol. 5:431–440 10.1038/nrmicro1659 [DOI] [PubMed] [Google Scholar]
- Chang G., Spencer R.H., Lee A.T., Barclay M.T., Rees D.C. 1998. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science. 282:2220–2226 10.1126/science.282.5397.2220 [DOI] [PubMed] [Google Scholar]
- Chiang C.S., Anishkin A., Sukharev S. 2004. Gating of the large mechanosensitive channel in situ: estimation of the spatial scale of the transition from channel population responses. Biophys. J. 86:2846–2861 10.1016/S0006-3495(04)74337-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.H., Allen T.W., Kuyucak S. 2002. Conducting-state properties of the KcsA potassium channel from molecular and Brownian dynamics simulations. Biophys. J. 82:628–645 10.1016/S0006-3495(02)75427-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry B. 2006. An energy-efficient gating mechanism in the acetylcholine receptor channel suggested by molecular and Brownian dynamics. Biophys. J. 90:799–810 10.1529/biophysj.105.067868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry B., Allen T.W., Kuyucak S., Chung S.H. 2001. Mechanisms of permeation and selectivity in calcium channels. Biophys. J. 80:195–214 10.1016/S0006-3495(01)76007-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry B., Hoyles M., Allen T.W., Walker M., Kuyucak S., Chung S.H. 2002. Reservoir boundaries in Brownian dynamics simulations of ion channels. Biophys. J. 82:1975–1984 10.1016/S0006-3495(02)75546-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry B., O’Mara M., Chung S.H. 2004. Conduction mechanisms of chloride ions in ClC-type channels. Biophys. J. 86:846–860 10.1016/S0006-3495(04)74160-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry B., Jayatilaka D., Rigby P. 2005a. A flexible approach to the calculation of resonance energy transfer efficiency between multiple donors and acceptors in complex geometries. Biophys. J. 89:3822–3836 10.1529/biophysj.105.069351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry B., Rigby P., Liu Z.W., Martinac B. 2005b. Conformational changes involved in MscL channel gating measured using FRET spectroscopy. Biophys. J. 89:L49–L51 10.1529/biophysj.105.072009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry B., Jayatilaka D., Martinac B., Rigby P. 2006. Determination of the orientational distribution and orientation factor for transfer between membrane-bound fluorophores using a confocal microscope. Biophys. J. 91:1032–1045 10.1529/biophysj.106.080713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank C.C., Minchin R.F., Le Dain A.C., Martinac B. 1997. Estimation of the pore size of the large-conductance mechanosensitive ion channel of Escherichia coli. Biophys. J. 73:1925–1931 10.1016/S0006-3495(97)78223-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcour A.H., Martinac B., Adler J., Kung C. 1989. Modified reconstitution method used in patch-clamp studies of Escherichia coli ion channels. Biophys. J. 56:631–636 10.1016/S0006-3495(89)82710-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullingsrud J., Kosztin D., Schulten K. 2001. Structural determinants of MscL gating studied by molecular dynamics simulations. Biophys. J. 80:2074–2081 10.1016/S0006-3495(01)76181-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustiananda M., Liggins J.R., Cummins P.L., Gready J.E. 2004. Conformation of prion protein repeat peptides probed by FRET measurements and molecular dynamics simulations. Biophys. J. 86:2467–2483 10.1016/S0006-3495(04)74303-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häse C.C., Le Dain A.C., Martinac B. 1995. Purification and functional reconstitution of the recombinant large mechanosensitive ion channel (MscL) of Escherichia coli. J. Biol. Chem. 270:18329–18334 10.1074/jbc.270.31.18329 [DOI] [PubMed] [Google Scholar]
- Häse C.C., Le Dain A.C., Martinac B. 1997. Molecular dissection of the large mechanosensitive ion channel (MscL) of E. coli: mutants with altered channel gating and pressure sensitivity. J. Membr. Biol. 157:17–25 10.1007/s002329900212 [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38: 27–28 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Iscla I., Levin G., Wray R., Reynolds R., Blount P. 2004. Defining the physical gate of a mechanosensitive channel, MscL, by engineering metal-binding sites. Biophys. J. 87:3172–3180 10.1529/biophysj.104.049833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iscla I., Levin G., Wray R., Blount P. 2007. Disulfide trapping the mechanosensitive channel MscL into a gating-transition state. Biophys. J. 92:1224–1232 10.1529/biophysj.106.090316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iscla I., Wray R., Blount P. 2008. On the structure of the N-terminal domain of the MscL channel: helical bundle or membrane interface. Biophys. J. 95:2283–2291 10.1529/biophysj.107.127423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.C., Hoyles M., Kuyucak S., Chung S.H. 1998. Brownian dynamics study of ion transport in the vestibule of membrane channels. Biophys. J. 74:37–47 10.1016/S0006-3495(98)77764-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wray R., Blount P. 2004. Intragenic suppression of gain-of-function mutations in the Escherichia coli mechanosensitive channel, MscL. Mol. Microbiol. 53:485–495 10.1111/j.1365-2958.2004.04150.x [DOI] [PubMed] [Google Scholar]
- Li Y., Wray R., Eaton C., Blount P. 2009. An open-pore structure of the mechanosensitive channel MscL derived by determining transmembrane domain interactions upon gating. FASEB J. 23:2197–2204 10.1096/fj.09-129296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Gandhi C.S., Rees D.C. 2009. Structure of a tetrameric MscL in an expanded intermediate state. Nature. 461:120–124 10.1038/nature08277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A.D., Jr, Bashford D., Bellott M., Dunbrack R.L., Jr, Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- Markin V.S., Martinac B. 1991. Mechanosensitive ion channels as reporters of bilayer expansion. A theoretical model. Biophys. J. 60:1120–1127 10.1016/S0006-3495(91)82147-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B. 2001. Mechanosensitive channels in prokaryotes. Cell. Physiol. Biochem. 11:61–76 10.1159/000047793 [DOI] [PubMed] [Google Scholar]
- Martinac B., Hamill O.P. 2002. Gramicidin A channels switch between stretch activation and stretch inactivation depending on bilayer thickness. Proc. Natl. Acad. Sci. USA. 99:4308–4312 10.1073/pnas.072632899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B., Adler J., Kung C. 1990. Mechanosensitive ion channels of E. coli activated by amphipaths. Nature. 348:261–263 10.1038/348261a0 [DOI] [PubMed] [Google Scholar]
- Meyer G.R. 2007. Electron paramagnetic resonance spectroscopy and computer modelling studies of the structure of prokaryotic mechanosensitive channels. PhD Thesis. University of Queensland, Queensland, Australia: 132 pp [Google Scholar]
- Moe P., Blount P. 2005. Assessment of potential stimuli for mechano-dependent gating of MscL: effects of pressure, tension, and lipid headgroups. Biochemistry. 44:12239–12244 10.1021/bi0509649 [DOI] [PubMed] [Google Scholar]
- Oakley A.J., Martinac B., Wilce M.C. 1999. Structure and function of the bacterial mechanosensitive channel of large conductance. Protein Sci. 8:1915–1921 10.1110/ps.8.10.1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo E. 2006. Gating prokaryotic mechanosensitive channels. Nat. Rev. Mol. Cell Biol. 7:109–119 10.1038/nrm1833 [DOI] [PubMed] [Google Scholar]
- Perozo E., Kloda A., Cortes D.M., Martinac B. 2001. Site-directed spin-labeling analysis of reconstituted Mscl in the closed state. J. Gen. Physiol. 118:193–206 10.1085/jgp.118.2.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo E., Cortes D.M., Sompornpisut P., Kloda A., Martinac B. 2002a. Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature. 418:942–948 10.1038/nature00992 [DOI] [PubMed] [Google Scholar]
- Perozo E., Kloda A., Cortes D.M., Martinac B. 2002b. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat. Struct. Biol. 9:696–703 10.1038/nsb827 [DOI] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart O.S., Goodfellow J.M., Wallace B.A. 1993. The pore dimensions of gramicidin A. Biophys. J. 65:2455–2460 10.1016/S0006-3495(93)81293-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer R.H., Chang G., Rees D.C. 1999. ‘Feeling the pressure’: structural insights into a gated mechanosensitive channel. Curr. Opin. Struct. Biol. 9:448–454 10.1016/S0959-440X(99)80063-3 [DOI] [PubMed] [Google Scholar]
- Steinbacher S., Bass R., Strop P., Rees D.C. 2007. Structures of the prokaryotic mechanosensitive channels MscL and MscS. Mechanosensitive Ion Channels, Part A., Simon S., Hamill O., Benos D., Elsevier Academic Press, Inc., San Diego: 1–24 [Google Scholar]
- Strop P., Bass R., Rees D.C. 2003. Prokaryotic mechanosensitive channels. Adv. Protein Chem. 63:177–209 [DOI] [PubMed] [Google Scholar]
- Sukharev S.I., Martinac B., Arshavsky V.Y., Kung C. 1993. Two types of mechanosensitive channels in the Escherichia coli cell envelope: solubilization and functional reconstitution. Biophys. J. 65:177–183 10.1016/S0006-3495(93)81044-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev S.I., Sigurdson W.J., Kung C., Sachs F. 1999. Energetic and spatial parameters for gating of the bacterial large conductance mechanosensitive channel, MscL. J. Gen. Physiol. 113:525–540 10.1085/jgp.113.4.525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev S., Betanzos M., Chiang C.S., Guy H.R. 2001a. The gating mechanism of the large mechanosensitive channel MscL. Nature. 409:720–724 10.1038/35055559 [DOI] [PubMed] [Google Scholar]
- Sukharev S., Durell S.R., Guy H.R. 2001b. Structural models of the MscL gating mechanism. Biophys. J. 81:917–936 10.1016/S0006-3495(01)75751-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vásquez V., Sotomayor M., Cordero-Morales J., Schulten K., Perozo E. 2008. A structural mechanism for MscS gating in lipid bilayers. Science. 321:1210–1214 10.1126/science.1159674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Brand L. 1994. Resonance energy transfer: methods and applications. Anal. Biochem. 218:1–13 10.1006/abio.1994.1134 [DOI] [PubMed] [Google Scholar]
- Yoshimura K., Batiza A., Schroeder M., Blount P., Kung C. 1999. Hydrophilicity of a single residue within MscL correlates with increased channel mechanosensitivity. Biophys. J. 77:1960–1972 10.1016/S0006-3495(99)77037-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K., Usukura J., Sokabe M. 2008. Gating-associated conformational changes in the mechanosensitive channel MscL. Proc. Natl. Acad. Sci. USA. 105:4033–4038 10.1073/pnas.0709436105 [DOI] [PMC free article] [PubMed] [Google Scholar]