Abstract
Regulatory T cells (T reg cells) constitute a population of CD4+ T cells that limits immune responses. The transcription factor Foxp3 is important for determining the development and function of T reg cells; however, the molecular mechanisms that trigger and maintain its expression remain incompletely understood. In this study, we show that mice deficient for the Ets-1 transcription factor (Ets-1−/−) developed T cell–mediated splenomegaly and systemic autoimmunity that can be blocked by functional wild-type T reg cells. Spleens of Ets-1−/− mice contained mostly activated T cells, including Th2-polarized CD4+ cells and had reduced percentages of T reg cells. Splenic and thymic Ets-1−/− T reg cells expressed low levels of Foxp3 and displayed the CD103 marker that characterizes antigen-experienced T reg cells. Thymic development of Ets-1−/− T reg cells appeared intrinsically altered as Foxp3-expressing cells differentiate poorly in mixed fetal liver reconstituted chimera and fetal thymic organ culture. Ets-1−/− T reg cells showed decreased in vitro suppression activity and did not protect Rag2−/− hosts from naive T cell–induced inflammatory bowel disease. Furthermore, in T reg cells, Ets-1 interacted with the Foxp3 intronic enhancer and was required for demethylation of this regulatory sequence. These data demonstrate that Ets-1 is required for the development of natural T reg cells and suggest a role for this transcription factor in the regulation of Foxp3 expression.
Ets-1 is the founding member of a family of winged helix-turn-helix transcription factors that was initially found as part of a fusion protein in the E26 avian erythroblastosis virus (Leprince et al., 1983; Nunn et al., 1983). The Ets domain, which is shared by all ETS proteins, specifically recognizes DNA sequences that contain a GGAA/T core element (Nye et al., 1992). Ets-1 is involved in multiple biological processes such as hematopoiesis, angiogenesis, or tumor progression (Dittmer, 2003). Analyses of mice bearing an Ets-1–deficient lymphoid system have shed light on the complex functions performed by this transcription factor. Ets-1–deficient (Ets-1−/−) mice have impaired pre–B cell receptor–dependent stage of B cell maturation, decreased response to B cell receptor cross-linking, and compromised development of marginal zone and B1a B cells (Eyquem et al., 2004b). Furthermore, lack of functional Ets-1 allows the massive development of plasma cells, which is associated with hypersecretion of IgM and production of autoantibodies (Bories et al., 1995; Wang et al., 2005). In addition, we have shown that Ets-1−/− mice displayed impaired expansion of double-positive (DP) thymocytes and defective allelic exclusion at TCR-β locus, suggesting a role of Ets-1 in pre-TCR function. Maturation of peripheral T cells also requires functional Ets-1 as mice carrying a hypomorphic mutation of this transcription factor displayed impaired Th1 T cell fate and enhanced Th17 differentiation (Grenningloh et al., 2005; Moisan et al., 2007).
Autoimmune disease is a consequence of the generation of self-reactive T cells. Although a key mechanism whereby dysregulated T cell responses are avoided in large part through intrathymic deletion of self-reactive clones, additional mechanisms are also critical. Among those is active regulation by a subset of CD4+ T cells, called regulatory T cells (T reg cells), which is characterized by the transcription factor Foxp3 (Fontenot et al., 2003; Hori et al., 2003; Khattri et al., 2003). T reg cells possess robust immunosuppressive activities, and their absence or impaired function results in lymphoproliferation and multiorgan autoimmunity in mice and IPEX (immunodysregulation polyendocrinopathy enteropathy X linked) in humans (Ochs et al., 2007).
Most T reg cells are generated in the thymus where the synergistic action of several pathways downstream of the TCR, costimulatory molecules, and cytokine receptors is required for the active transcription of Foxp3 and the differentiation of thymocytes into the T reg cell lineage. First, self-antigens induce or favor the differentiation of CD4+CD8+ thymocytes into T reg cells through TCR- and CD28-dependent signals (Bensinger et al., 2001; Aschenbrenner et al., 2007). The second pathway involves IL-2/IL-15 signaling as Foxp3+ T reg cell development in Il2rβ−/−, Il2rγ−/−, and Il2−/− × Il15−/− mice is dramatically impaired (Burchill et al., 2007). More recently, TGF-β was also shown to be required for the generation of thymic T reg cells (Liu et al., 2008). In the periphery, several mechanisms contribute to increase or sustain T reg cell numbers. Conversion of naive CD4+ T cells into Foxp3-expressing T reg cells (iTreg) has been shown to occur through the action of TGF-β in the absence of other proinflammatory cytokines (Chen et al., 2003). TGF-β is thought to be involved in the maintenance of peripheral T reg cells as reduced numbers of Foxp3+ T reg cells have been reported in both Tgfβ1−/− mice and in mice with a T cell–specific deletion of Tgfβr2 (Marie et al., 2005, 2006; Li et al., 2006). Furthermore, using sophisticated reconstitution mouse models, CD28 and IL-2 signaling were shown to participate in peripheral T reg cell homeostasis, in addition to their role in thymic development (Tai et al., 2005).
In mice, transcription factors acting downstream of the TCR and cytokine receptors contribute to drive and maintain expression of Foxp3 by interactions with specific regulatory sequences (Huehn et al., 2009; Hori, 2010). TCR activation results in the binding of CREB (cyclic adenosine monophosphate–responsive element–binding protein) to an intronic enhancer element (hereafter referred to as CNS2) in the Foxp3 gene (Kim and Leonard, 2007). The STAT5 transcription factor, which is activated by the IL-2–Jak3 signaling pathway, plays a key role by directly activating the expression of the Foxp3 locus through two main specific elements, CNS1 and CNS2 (Burchill et al., 2007, 2008; Yao et al., 2007). TIEG1 (TGF-β–inducible early gene 1; also known as KFl10) was shown to bind to the Foxp3 promoter and to cooperate with itchy E3 ubiquitin protein ligase homologue (ITCH) to induce Foxp3 expression (Venuprasad et al., 2008). In addition, the TGF-β–induced transcription factor SMAD3 (mothers against decapentaplegic homologue 3) was reported to control the activity of a specific Foxp3 intronic enhancer element in cooperation with NFAT (Tone et al., 2008). Interestingly, stable up-regulation of Foxp3 expression is correlated with epigenetic events such as DNA demethylation that are lacking in Foxp3-positive in vitro TGF-β–activated T cells (Polansky et al., 2008). Thus, identification of factors that contribute to the regulation of the Foxp3 gene appears critical to better define the T reg cell compartment. In this study, we investigated the function of Ets-1 in the development of T reg cells and further analyzed the role of these cells in the development of autoimmunity in mice.
RESULTS
Mice lacking Ets-1 display splenomegaly, constitutive T cell activation, and spontaneous Th2 cytokine expression
In the C57BL/6 background, complete inactivation of the Ets-1 locus leads to >98% perinatal mortality (Eyquem et al., 2004a), whereas in mixed 129sv/C57BL/6 background, ∼12% of the mice reach adulthood. 6-wk-old viable Ets-1−/− mice had dramatically enlarged spleens (Fig. 1 A) containing an approximately fourfold higher total cell number than WT littermate controls (Table S1). This increase was essentially accounted for by B cells (fourfold) and a high side scatter heterogeneous population (Fig. S1 A) that included eosinophils (not depicted).
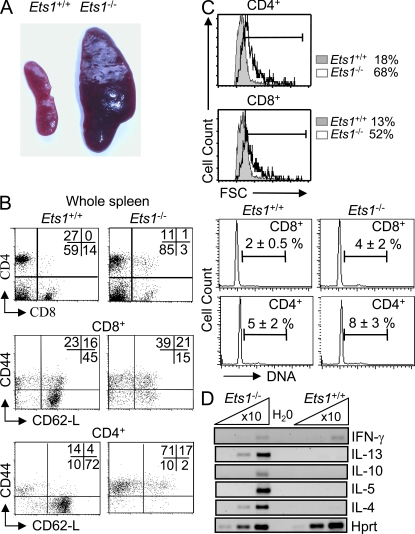
Figure 1.
Splenomegaly and Th2-activated T cells in Ets-1−/− mice. (A) Spleens removed from 5–6-wk-old Ets-1−/− and WT (Ets-1+/+) littermate controls. Results are representative of at least five animals. (B) Spleen cell populations from Ets-1−/− and Ets-1+/+ mice were stained with the indicated antibodies and analyzed by flow cytometry. Numbers indicate the percentages of cells falling in each quadrant. Dot plots are gated on the indicated population. (C, top) Forward light scatter (FSC) of CD4+ or CD8+ gated spleen cell populations from Ets-1−/− and Ets-1+/+ littermate controls. (bottom) DNA content analysis of sorted CD8 and CD4 spleen cells from Ets-1−/− and Ets-1+/+ littermate controls. Numbers indicate percentages of cells within gates. (D) Semiquantitative RT-PCR analysis of cytokines. cDNA from freshly purified CD4+ spleen cells from Ets-1+/+ and Ets-1−/− mice was 10-fold serially diluted and subjected to semiquantitative RT-PCR analysis using oligonucleotides corresponding to the indicated genes. Results are representative of at least three independent experiments.
In contrast, the number of T cells was reduced twofold as compared with controls (Table S1), and the reduction affected both CD4 and CD8 subsets (Fig. 1 B). Most CD4+ and CD8+ Ets-1−/− splenocytes were large (Fig. 1 C), and an increased percentage had an activated phenotype with up-regulated levels of CD44 and down-regulation of CD62-L (Fig. 1 B). Notably, cells with a naive phenotype (CD62-LhighCD44low) represented at best 2% and 15%, respectively, of the CD4 and CD8 T cell populations (Fig. 1 B). Furthermore, the percentages of CD69- and CD95-expressing cells were significantly increased in Ets-1−/− T cells (Fig. S1 B). Moreover, DNA content analysis revealed a 1.5–2-fold increase of the percentage of cycling cells (Fig. 1 C).
Analysis by semiquantitative RT-PCR of the transcription status of several cytokines in freshly isolated Ets-1−/− CD4+ T cells revealed high expression levels of IL-4, IL-5, IL-10, and IL–13 but not IFN-γ (Fig. 1 D). Consistently, upon in vitro stimulation, secretion of IL-4 and IL-10 was markedly increased, whereas IL-2 and IFN-γ were barely detected (Fig. S2).
These results show that inactivation of the Ets-1 transcription factor induces splenomegaly and the development of activated T cells with CD4+ cells expressing high levels of Th2 cytokines. These features were associated with severe naive T cell lymphopenia.
Ets-1–deficient T cells trigger clearance of marginal zone B cells, B cell activation, and elevated serum levels of IgG1 and IgE
In line with previous descriptions (Eyquem et al., 2004b), histological analysis of Ets-1−/− spleens showed a marked abnormal structure with loss of B cell follicles and marginal zones (Fig. S3). Viable mutant mice contained high numbers of activated B cells, a slight increase in the percentage of cycling cells (unpublished data), and high serum titers of IgM, as well as elevated levels of auto antibodies (Eyquem et al., 2004b).
To specifically assess a direct role of mutant T cells in the B cell immune disorder of Ets-1−/− mice, CD3-ε–deficient mice were engrafted with Ets-1−/− (KO/CD3) or WT Ets-1+/+ (WT/CD3) fetal thymi under the kidney capsule. 5–6 wk after graft, both KO/CD3 and WT/CD3 mice had reconstituted CD4 and CD8 splenic T cell compartments (Fig. 2 A). Similar to Ets-1−/− mice, the KO/CD3 chimeras had splenomegaly (see next paragraph). Although spleens from WT/CD3 bear a significant subset (∼33%) of naive CD44lowCD62-L+CD4+ T cells, KO/CD3 chimeras, like Ets-1−/− mice, displayed increased percentages of activated CD44highCD4+ cells and very few CD44lowCD62-L+CD4+ splenocytes (Fig. 2 B).
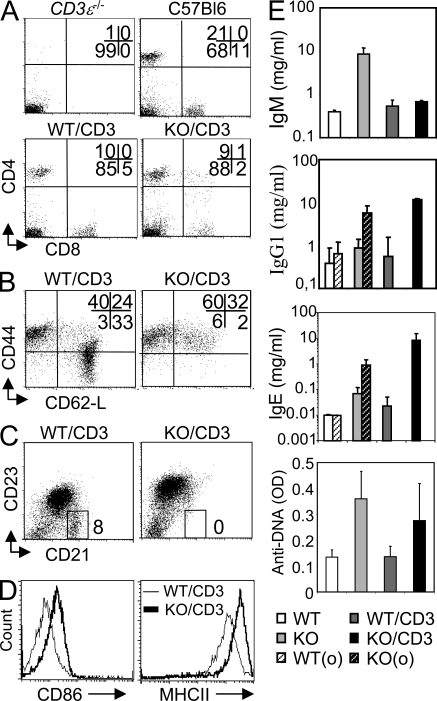
Figure 2.
Ets-1−/− T cells induce B cell activation and IgG1 and IgE hypergammaglobulinemia. Spleen cell population from CD3ε−/−, WT (C57BL/6), CD3ε−/− engrafted with WT embryonic thymus (WT/CD3), and CD3ε−/− engrafted with Ets-1−/− embryonic thymus (KO/CD3) were analyzed by flow cytometry. (A) Dot plots are gated on whole splenocytes. (B) Dot plots are gated on CD4+ spleen cells. Numbers represent the percentage of cells falling in each quadrant. (C) Dot plots are gated on B220+ cells. Percentages of cells falling into the indicated gate are shown. (D) CD86 and MHC class II molecules on the surface of gated B220+ spleen cells from CD3ε−/− chimera mice engrafted with WT (WT/CD3) thymus correspond to thin lines, and mice engrafted with Ets-1−/− (KO/CD3) thymus correspond to thick lines. These results are representative of three independent experiments. (E) Serum Ig levels of WT or Ets-1−/− (KO) mice or CD3ε−/− recipient mice engrafted with WT (WT/CD3) or Ets-1−/− (KO/CD3) embryonic thymus. Ig levels were measured 6 or 8 wk after transfer when indicated (0). ELISA was used to detect anti-DNA antibodies in the serum of mice. Values are indicated as OD. Results are representative of four independent experiments. Error bars indicate standard deviation.
Whereas WT/CD3 spleens had a normal frequency of marginal zone (CD21+CD23−) B cells, this population was undetectable in KO/CD3 chimeras (Fig. 2 C). In these mice, the CD86 and MHC II on B cells was significantly higher than in control chimeras (Fig. 2 D), indicating that most B cells were in an activated state.
WT/CD3 mice had normal levels of IgM, IgG1, and IgE (Fig. 2 E). In KO/CD3 chimeras, IgM titers were similar to controls, but IgG1 and IgE titers were 10-fold increased (Fig. 2 E and not depicted). Furthermore, elevated concentrations of anti-DNA antibodies could be specifically detected in KO/CD3 chimeras (Fig. 2 E). These results demonstrate that except for IgM levels, Ets-1−/− T cells trigger all the features of B cell dysregulation observed in Ets-1−/− mice.
WT T reg cells inhibit Ets-1−/− T cell–mediated splenomegaly and B cell autoimmune disorders
Enhanced in vivo T cell activation can originate from impaired development or loss of function of naturally occurring T reg cells. To test whether a similar process occurs in Ets-1−/− mice, we generated Ets-1/CD3 chimeras that received (or not) WT Ly5.1 CD25+CD4+ (hereafter referred to as donor T reg cells) at the time of fetal thymus transplantation.
As expected, 5–6 wk after graft, KO/CD3 chimera mice had enlarged spleens compared with WT/CD3 controls (Fig. 3 A). Strikingly, injection of donor T reg cells in KO/CD3 chimeras was sufficient to block development of splenomegaly (Fig. 3 A). Interestingly, in these chimeras, donor T reg cells represented 50–60% of the CD4+ T cell population as compared with 13–22% in WT/CD3 controls (Fig. S4 A). In addition, Ets-1−/− CD4+ T cells represented only 2% of total spleen cells in KO/CD3 chimeras that received T reg cells compared with 7% in the absence of donor T reg cells, suggesting that donor T reg cells might inhibit expansion of Ets-1−/− T cells. Notably, the CD4 T cell compartment of KO/CD3 chimeras that received donor T reg cells was also devoid of naive (CD44lowCD62-L+) cells (Fig. S4 A). To further investigate the function of WT T reg cells on Ets-1−/− T cells, CD45.2+CD4+ was sorted from WT/CD3 or KO/CD3 chimeras that had received (or not) CD45.1 WT T reg cells. Real-time RT-PCR analysis showed a 12-fold reduction of IL-4 expression in Ets-1−/− CD4+ splenocytes that were isolated from mice containing WT T reg cells compared with those that did not receive suppressive cells (Fig. S4 B).
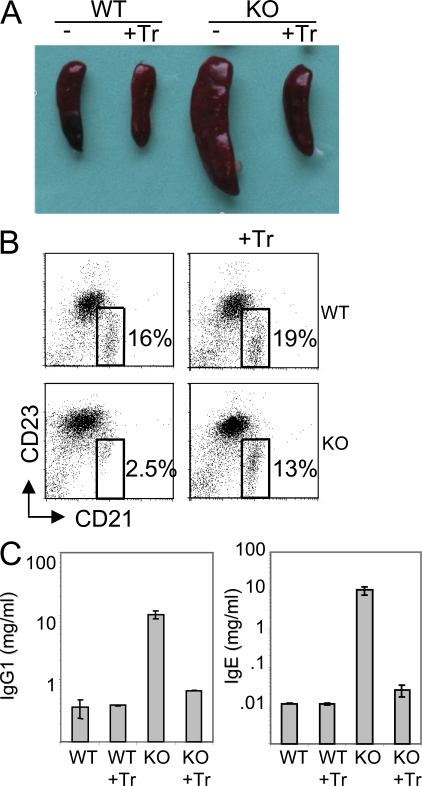
Figure 3.
WT T reg cells inhibit immune defects triggered by Ets-1−/− T cells. (A) Spleens removed from 5–6-wk-old CD3ε−/− mice engrafted with WT embryonic thymus and CD3ε−/− engrafted with Ets-1−/− embryonic thymus (KO). +Tr indicates that WT Ly5.1 CD4+CD25+ cells were injected on the day of thymus engraftment. (B) Dot plots show B220+ gated spleen cells from CD3ε−/− mice engrafted with thymus alone (left) or thymus plus WT CD4+CD25+ (right). The percentage of marginal zone B cells (boxed) is indicated in each dot blot. Results are representative of three independent experiments. (C) Serum IgG1 and IgE in mice shown in A. Results are representative of three animals of each type. Error bars indicate standard deviation.
Furthermore, whereas no CD21+CD23− marginal zone B cells were detected in the spleen of KO/CD3 chimeras, injection of WT T reg cells clearly prevented clearance of this B cell subset (Fig. 3 B). Injection of WT T reg cells also prevented IgG1 and IgE hypergammaglobulinemia in KO/CD3 chimeras (Fig. 3 C). These results demonstrate that Ets-1−/− T cells are responsive to T reg cell suppression and point toward a defective T reg cell compartment in Ets-1–deficient mice.
Reduced Foxp3 expression in Ets-1−/− CD4+ T cells
Based on the aforementioned observations, we studied the thymic and peripheral T reg cell compartments of Ets-1−/− mice. As previously reported (Bories et al., 1995; Eyquem et al., 2004a), the thymus of these mice had 10–12 times lower cell numbers, and although CD4+8+ thymocytes represented around 50% of the cells, the percentage of CD4+ single-positive (SP) thymocytes was comparable to age-matched littermates (Fig. 4 A). Within this subset, cells expressing Foxp3 were detected in mutant mice at frequencies similar to those of littermates. However, the amounts of Foxp3 were clearly lower in Ets-1−/− thymocytes (Fig. 4 A), whereas CD25 was expressed at normal levels (not depicted).
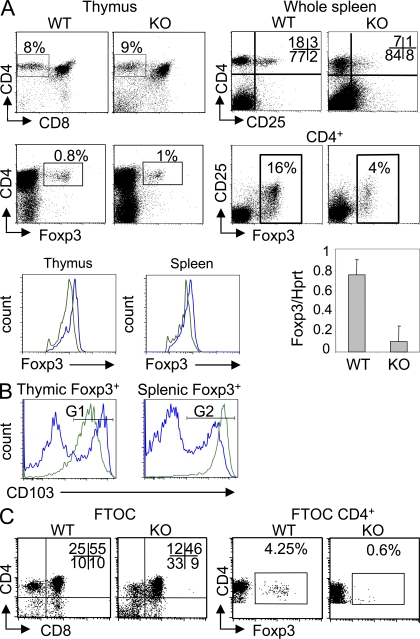
Figure 4.
Impaired development of T reg cells in Ets-1−/− mice. (A) Thymus and spleen cells from viable adult Ets-1−/− (KO) mice and WT littermate controls were analyzed by flow cytometry. Numbers indicate the percentage of the gated population. Histograms show Foxp3 expression in gated CD4+Foxp3+ cells from Ets-1+/+ (blue line) and Ets-1−/− (green line) mice. Bar graph depicts a real-time RT-PCR analysis of Foxp3 expression in sorted CD4+CD25+ cells from spleens of Ets-1−/− (KO) or Ets-1+/+ littermate controls (WT). Results are expressed as a Foxp3/Hprt ratio. Error bars indicate standard deviation. (B) Histograms show CD103 expression in gated Foxp3+ cells from Ets-1+/+ (blue line) and Ets-1−/− (green line) mice. The gates G1 and G2 indicate CD103-positive populations. The results in A and B are representative of at least five independent experiments. (C) FTOCs from Ets-1−/− mice and Ets-1+/+ littermates were analyzed by flow cytometry. Dot plots are gated as indicated. Numbers indicate the percentage of the gated population and are representative of six independent experiments.
In the spleen, the percentage of Foxp3+ cells among Ets-1−/− CD4+ splenocytes was fourfold lower than in controls (Fig. 4 A). The expression level of Foxp3 and the quantity of Foxp3 transcripts in sorted Ets-1−/− CD25+CD4+ cells were also decreased (Fig. 4 A). In contrast, expression levels of CD25 within Foxp3+ were equivalent in mutants and controls (Fig. 4 A).
Because in Ets-1−/− mice ∼98% of the peripheral CD4+ T cells were activated, we determined the frequency of Ets-1−/− Foxp3+CD4+ splenocytes that expressed CD103, a marker which is normally up-regulated in activated T reg cells infiltrating nonlymphoid tissues (Sather et al., 2007). As shown in Fig. 4 B, 85% of T reg cells expressed CD103 (gate G2), compared with 28% in control and, on average, at significantly higher levels. In the thymus, 44% of Ets-1+/+ Foxp3+ cells were CD103+ and expressed high levels of this marker (Fig. 4 B, gate G1). Strikingly, 83% of Ets-1−/− Foxp3+ thymocytes expressed CD103, although at lower levels. Given the very high frequency of CD103+ peripheral T reg cells and the fact that this marker is up-regulated on homeostatically generated peripheral T reg cells (Feuerer et al., 2010), these results raised the possibility that the majority of thymic Ets-1−/− Foxp3+CD103+ cells might have come from the periphery. This would imply that thymic production of T reg cells is significantly reduced in Ets-1−/− mice.
To investigate thymic development of T reg cells in a peripheral T cell free system, we performed fetal thymus organ cultures (FTOCs) with E15 thymi of Ets-1−/− and WT mice. At day 7 of culture, WT thymic lobes had fully developed (9 × 105 ± 105 cells/thymus), including 45–65% of DPs (median = 50%; n = 6) and 7–25% of CD4 SP. Foxp3+ cells were readily identified and represented 2.5–4.5% of CD3+4+8− thymocytes (Fig. 4 C). In contrast, Ets-1−/− lobes developed poorly (2.5 × 105 ± 105 cells/thymus) with DPs ranging from 10 to 50% (median = 25%; n = 6) and 3 to 12% for CD4 SPs. Within this subset, Foxp3+ cells represented <0.6%, the cells expressing very low amounts of the Foxp3 protein. Altogether, the results show that, despite the normal frequency of thymic Foxp3-expressing cells in adults, the development of Foxp3+ T cell is significantly compromised in Ets-1−/− mice.
Codevelopment of WT and Ets-1−/− fetal liver cells does not alter the development of WT T reg cells nor rescues normal generation of Ets-1−/− T reg cells
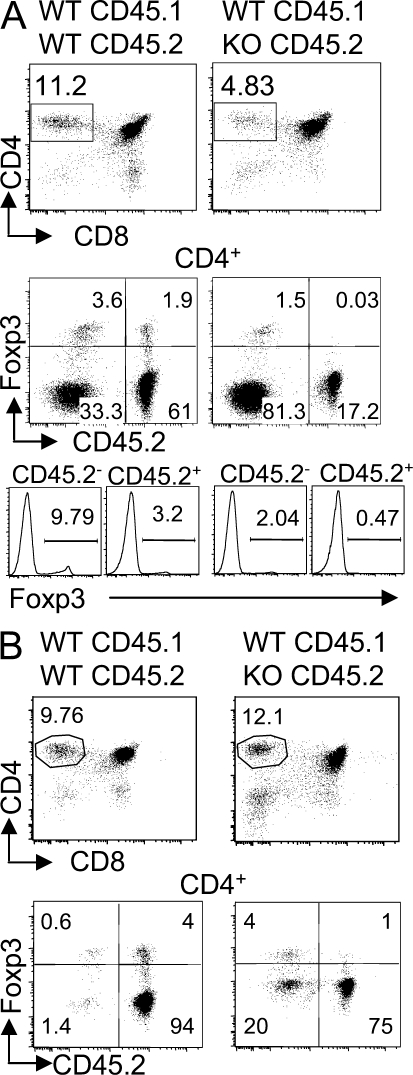
To evaluate the cell-intrinsic role of Ets-1 in T reg cells, we cotransferred various ratios of WT (CD45.1+) and Ets-1−/− (CD45.2+) fetal liver cells into irradiated Rag−/−γc−/− mice. In 1:1 WT/KO chimera, analyzed 1 mo after transfer when T cells were still poorly detected in peripheral blood, the frequency of Ets-1−/− cells in CD4 SP was only 4.83% (Fig. 5 A). Strikingly, 2.04% of WT (CD45.2−) CD4+ SP cells expressed Foxp3 at normal levels, but only 0.47% of the Ets-1−/− (CD45.2+) counterpart stained for Foxp3 and expressed very low levels of this transcription factor (Fig. 5 A). The deficit of Ets-1−/− T reg cells appeared even more pronounced when gating on thymocytes expressing high level of Foxp3 (Fig. 5 A, middle). In contrast, in 1:1 WT/WT chimeras, normal Foxp3+ cells were readily detected in the CD4 SP derived from both donors. To investigate the development of T reg cells in the presence of an excess of Ets-1−/− cells, we analyzed 1:20 WT/KO chimera 2 mo after transfer. As shown in Fig. 5 B, although >75% of the CD4+ SPs was derived from the Ets-1−/− background, the development of WT Foxp3+ thymocytes was not impaired and represented ∼80% of the T reg cell subset. This deficit of Ets-1−/− CD4+Foxp3+ cells, which resulted in an increased percentage of Ets-1−/− CD4+Foxp3− SP cells among the CD45.2 subset, appeared significant in our series of chimera mice (Fig. S5). Collectively, these results show that the thymic environment in these chimeras is capable of efficiently supporting normal but not Ets-1−/− T reg cell development and point toward an intrinsic role for Ets-1 in this process.
Figure 5.
Impaired development of Ets-1−/− Foxp3+ thymocytes in mixed fetal liver reconstituted mice. (A) Thymus cell populations from Rag−/−γc−/− mice reconstituted with 1:1 mixed CD45.1+ WT plus CD45.2+ WT or CD45.2+ Ets-1−/− fetal liver cells were analyzed 1 mo after transfer. The gated cell population is indicated above each dot plot. Histograms show Foxp3 expression within CD4+ SP thymocytes gated on CD45.2− or CD45.2+ cells. Results are representative of three independent mice. In WT/WT chimera, the percentage of Foxp3+ among CD4+CD8−CD45.2− populations was not significantly different from the percentage of Foxp3+ among CD4+CD8−CD45.2+ cells (P = 0.2262), whereas in the WT/KO mice, the difference was significant (P = 0.03826). (B) Thymus cell populations from Rag−/−γc−/− mice reconstituted with 1:20 mixed CD45.1+ WT plus CD45.2+ WT or CD45.2+ Ets-1−/− fetal liver cells were analyzed by flow cytometry 2 mo after transfer. Results are representative of three independent mice. In WT/WT chimera, the percentage of Foxp3+ among CD4+CD8−CD45.2− populations was not significantly different from the percentage of Foxp3+ among CD4+CD8−CD45.2+ cells (P = 0.0571), whereas in the WT/KO mice, the difference was significant (P = 0.00684). Numbers indicate the percentage of the gated populations.
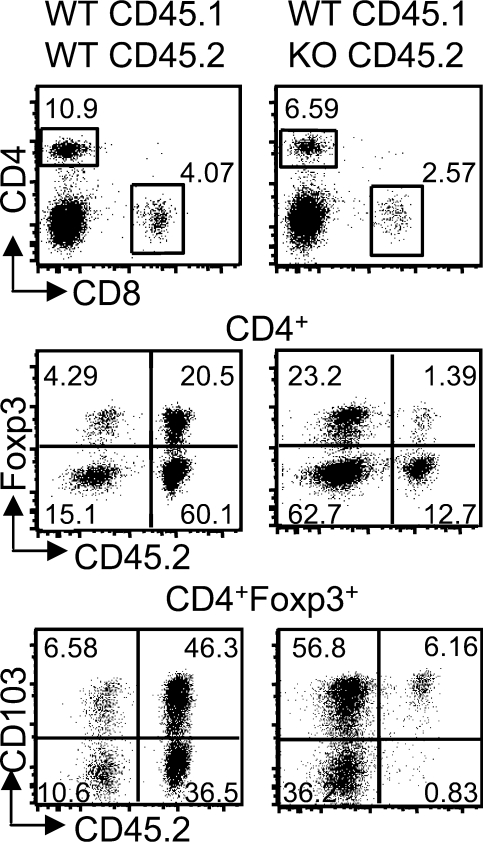
Phenotype of peripheral Ets-1−/− CD4+ cells in mixed fetal liver chimeras
As described in the previous section, 75% of CD4+ SP thymocytes in 1:20 WT/KO chimera were derived from the mutated background (Fig. 5 B). However, in the spleen of these animals, Ets-1−/− cells represented <15% of CD4+ T cells (Fig. 6), indicating that conventional mutated CD4+ T cells have a significant competitive deficit compared with WT cells. Within the CD4+ subset, the majority of Foxp3+ was derived from the WT. The deficit of Ets-1−/− CD4+Foxp3+ cells, and consequently the increased percentage of Ets-1−/− CD4+Foxp3− splenocytes, was significant in our series of chimera mice (Fig. S6). Furthermore, although around 60% of WT T reg cells were CD103+, ∼88% of Ets-1−/− Foxp3+CD4+ expressed CD103 (Fig. 6). In contrast, ∼50% of the very few Ets-1−/− Foxp3+CD4+ thymocytes expressed CD103 as compared with 54% in the WT counterparts (Fig. S7 A). Thus, in these lymphopenic chimeras, the severe quantitative deficit of Ets-1−/− thymic T reg cells correlated with a significantly lower frequency of antigen-experienced T reg cells in the thymus than in the periphery. This reinforced our hypothesis that in viable Ets-1−/− mice, recirculating Foxp3+ Ets-1−/− cells were masking a quantitative deficit in the development of thymic T reg cells.
Figure 6.
Defective Ets-1−/− T reg cells in the spleen of mixed chimera mice. Spleen cell populations from Rag−/−γc−/− mice reconstituted with a 1:20 mixture of CD45.1+ WT plus CD45.2+ WT or CD45.2+ Ets-1−/− fetal liver cells were analyzed by flow cytometry 2 mo after transfer. The same chimeras as in Fig. 5 B are shown. The gated cell population is indicated above each dot plot. Numbers indicate the percentage of the gated populations. Results are representative of three independent experiments. In spleens of WT/WT chimera, the percentage of Foxp3+ among CD4+CD45.2− populations was not significantly different from the percentage of Foxp3+ among CD4+CD45.2+ cells (P = 0.2262), whereas in the WT/KO mice, the difference was significant (P = 0.03826).
We next investigated the activation status of Ets-1−/− CD4+ and CD8+ T cells in the spleen of fetal liver chimera mice. As described for Ets-1 viable mice, in chimeras that only received Ets-1−/− fetal liver cells, >80% of CD4 cells were CD44highCD62-L−, and <1% had a naive phenotype (CD62-L+CD44low; Fig. S7). In contrast, in mixed 1:20 WT/KO chimera, the percentages of CD62-L+CD44high and CD62-L+CD44low within Ets-1−/− CD4+ and CD8+ subsets were markedly increased (Fig. S7). Although the frequency of CD44high cells did not change, the percentage of cells that expressed CD62-L was reduced by half. Interestingly, a high percentage of WT T cells in chimera mice did not express a typical naive phenotype (CD62-L+CD44low), suggesting that our lymphopenic host mouse model may lead to the underestimation of the frequency of naive T cells (Fig. S7). Although spleens from 1:20 chimera contained a significant percentage of Ets-1−/− T cells, none of them had elevated spleen cells numbers nor displayed splenomegaly (Fig. S7 B and not depicted). In contrast, mice reconstituted only with Ets-1−/− fetal liver cells (KO chimera) were affected by both defects. Altogether, these results show that the presence of WT cells in mixed chimera is sufficient to control spleen cell numbers but to only partially rescue Ets-1−/− naive CD4 and CD8 splenocytes.
In vitro TGF-β–mediated up-regulation of Foxp3 is not abrogated in Ets-1−/− peripheral T cells
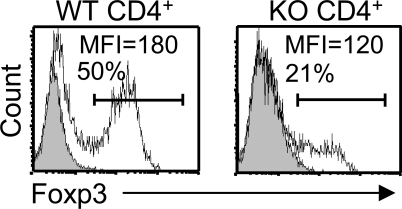
Next, we investigated whether Ets-1−/− CD4+ T cells could up-regulate Foxp3 after in vitro activation. CD4+CD62-L+ splenocytes were FACS sorted from Ets-1−/− and WT mice and activated with anti-CD3 in IL-2– and TGF-β–containing medium. 48 h after activation, the percentage of Foxp3-expressing cells was lower in Ets-1−/− (21%) than in WT cultures (50%; Fig. 7). We should note that, as shown in Fig. 1 B, 90% of the splenic CD4+CD62-L+ T cell population of Ets-1−/− mice also expressed high levels of CD44, as compared with 5% in the control population. Therefore, it is not excluded that the differences observed are caused by a differential susceptibility to anti-CD3/IL-2 and TGF-β treatment among CD4 T cell subsets rather than a specific effect caused by Ets-1 inactivation. These results indicate that inactivation of the Ets-1 transcription factor reduces but does not abrogate the capacity of Ets-1−/− CD4+ T cells to up-regulate Foxp3 in vitro.
Figure 7.
In vitro development of Ets-1−/− T reg cells. FACS analysis of Foxp3 expression in in vitro stimulated naive WT (WT CD4+) or Ets-1−/− CD4+ (KO CD4+) sorted spleen cells. Histograms show the staining of anti-Foxp3 and isotype control (gray) antibodies. Mean fluorescent intensity (MFI) and the percentage of cells in the indicated gate are shown. Results represent four independent experiments.
Ets-1−/− T reg cells show decreased in vitro and in vivo suppressive activity
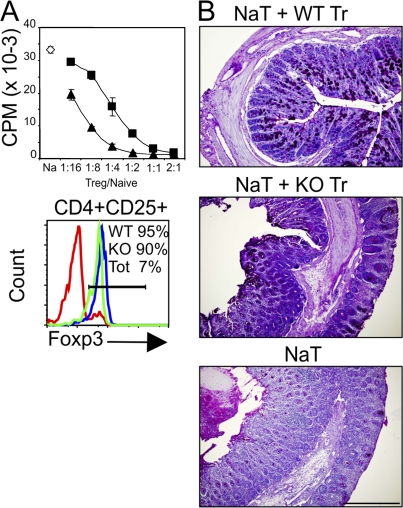
To examine the in vitro suppressive function of CD4+CD25+ spleen cells that develop in the Ets-1−/− background, naive T cells (CD4+CD62-L+CD25−) were sorted from WT C57BL/6 mice and were activated in vitro in the presence of various numbers of sorted 90% Foxp3-expressing T reg cells (CD4+CD25high) from Rag/Ets-1+/+ or Rag/Ets-1−/− chimera mice. At high concentrations, both WT and mutant T reg cells inhibited anti-CD3–induced T cell proliferation (Fig. 8 A). However, the suppressive activity of mutant T reg cells was less efficient as the same level of inhibition was achieved by four- to fivefold fewer WT T reg cells (Fig. 8 A). Thus, Ets-1−/− cells have a decreased in vitro suppressive activity.
Figure 8.
Impaired suppressive activity of Ets-1−/− T reg cells. (A) WT naive T cells (CD4+CD25−CD62-L+) were stimulated with α-CD3 and APC in the presence of CD4+CD25+ cells from WT (closed triangles) or Ets-1−/− (closed squares) Rag−/− chimeric mice at the indicated ratios. The open diamond indicates naive T cells alone. Error bars indicate the standard deviation of triplicate values. Histograms show Foxp3 expression in sorted T reg cells from Ets-1+/+ (WT; blue) and Ets-1−/− (KO; green) mice used for the in vitro suppression assay. Sorted CD4+ spleen cells (Tot; red) were stained as a control. Numbers indicate the percentage of cells falling into the gate indicated by the horizontal bar. Results are representative of three experiments. (B) Development of colitis 7 wk after adoptive cell transfer. Histological appearance of colon in representative Rag2−/− recipient mice 7 wk after transfer WT naive T cells (NaT) alone or with CD4+CD25+ from WT (WT Tr) or Ets-1−/− (KO Tr) mice. Sections of colon from individual mice were imbedded in formalin and stained with hematoxylin and eosin. Results are representative of four different mice of each type. Bar, 100 µm.
To determine the importance of Ets-1 in T reg cell in vivo function, we used the mouse model of inflammatory bowel disease (IBD) by adoptively transferring WT naive T cells with or without cotransferring WT or Ets-1−/− T reg cells into Rag2−/− recipient mice. As expected, recipient mice that received WT naive CD4+CD25−CD62-L+ T cells (pathogenic) alone developed severe colitis 6–7 wk after cell transfer, with thickening of the colon, loss of normal, pellet-shaped stools, and marked mononuclear cellular infiltrate (Fig. 8 B). Rag2−/− mice that were cotransferred with WT CD4+CD25+ T reg cells and WT naive T cells were protected from developing colitis with normal pellet-shaped stools and normal mucosal structure. In marked contrast, Ets-1−/− T reg cells did not prevent the development of colitis after being cotransferred with WT naive T cells, and mice showed features of colitis, including thickening of the colon and mononuclear cellular infiltrate (Fig. 8 B).
This result demonstrates that Ets-1−/− T reg cells do not protect from naive T cell–induced IBD. This could be because of decreased suppressive function (as shown in vitro), although decreased survival of mutant T reg cells both in vivo and in vitro cannot be excluded.
Ets-1 binds Foxp3 transcriptional regulatory sequences and controls their methylation status
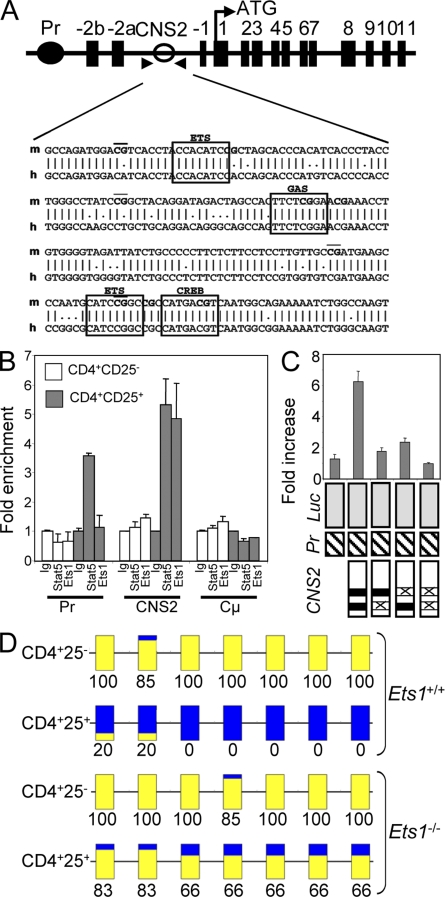
To further characterize the function of Ets-1 in T reg cells, we analyzed Foxp3 transcriptional regulatory sequences. A previous study has identified an enhancer element (CNS2) located between exon −2b and −1 of Foxp3 that contains T reg cell–specific STAT5- and CREB-binding sites (Kim and Leonard, 2007). Furthermore, this region displays a CpG island that appears to be specifically demethylated in T reg cells. Analysis of human and mouse enhancer regions with TFSEARCH software (http://www.cbrc.jp/research/db/TFSEARCH.html) confirmed the presence of two conserved ETS-binding sites located near STAT5/CREB-interacting sites that were previously suggested by Floess et al. (2007; Fig. 9 A). Ets-1 binding to the native Foxp3 gene in mouse primary cells was assessed using sorted CD4+CD25− and CD4+CD25bright splenocytes and chromatin immunoprecipitation (ChIP) with anti–Ets-1, anti-STAT5, or irrelevant (Ig) antibodies. Subsequent real-time PCR amplification of the Foxp3 gene surrounding putative binding sites in the CNS2 region showed expected STAT5 but also significant Ets-1 binding in T reg cells but not in naive cells (Fig. 9 B). Conversely, analysis of the Foxp3 promoter region did not reveal significant Ets-1 binding (Fig. 9 B). As a control, we assessed a DNA segment of the IgH constant region (Cµ) that is not expressed in T cells and found neither STAT5 nor Ets-1 binding. To investigate the functional significance of ETS-binding sites on the activity of CNS2, the minimal enhancer region (including the two ETS-binding sites) was cloned into the pGL3-promoter vector, and luciferase activity was measured into transfected Jurkat T cells. Whereas the intact enhancer exhibited an approximately sixfold increase in activity in response to PMA plus ionomycin, mutation of either ETS-binding site (or both) dramatically reduced CNS2 induced function (Fig. 9 C). Thus, ETS-binding sites are required for the enhancer function of the CNS2 region. To further investigate how Ets-1 binding impacted the Foxp3 locus, the methylation status of CNS2 was analyzed by bisulphite sequencing on DNA isolated from male T cells (see Materials and methods). Interestingly, striking differences between WT and Ets-1−/− CD4+CD25bright T cells could be observed. CpG motifs within our amplified sequence displayed a high degree of methylation (100%) within conventional CD4+CD25− T cells from both genotypes (Fig. 9 D). However, whereas the amplified region was unmethylated within WT T reg cells, CpG dinucleotides in this region were largely methylated in Ets-1−/− T reg cell (Fig. 9 D). This result demonstrated that Ets-1 is essential for epigenetic modulations associated with Foxp3 expression.
Figure 9.
Ets-1 is involved in epigenetic changes at the Foxp3 locus. (A) Schematic representation of the mouse Foxp3 gene. Exons are shown as black boxes and are labeled. The enhancer region (CNS2) containing the CpG island and the promoter region (Pr) are indicated as open and closed circles, respectively. Comparison of mouse (m) and human (h) sequences of CNS2 is shown. CpG are underlined, and potential Stat (GAS), CREB, and Ets-1–binding sites are boxed. (B) Sorted splenic CD25brightCD4+ and CD25−CD4+ T cells were treated with IL-2 for 1 h. ChIP was performed using normal rabbit serum (Ig), anti-STAT5 (Stat5), or anti–Ets-1 (Ets1) antibodies. Quantification of immunoprecipitated DNA fragments was performed by real-time PCR using primers for CNS2, the promoter region, and the irrelevant IgH Cµ. Values were normalized to corresponding input control and are expressed as fold enrichment relative to normal rabbit serum for each experiment. Results are representative of three individual experiments. (C) The conserved Ets-binding sites (indicated as gray boxes) in CNS2 are essential for PMA plus ionomycin–induced Foxp3 enhancer activity. Mutations (shown as X) in the Ets-binding sites were introduced into the Foxp3 reporter construct. Constructs were transected into Jurkat cells, followed by no stimulation or stimulation with PMA plus ionomycin for 17 h. Luciferase activity was normalized to cotransfected β-galactosidase. Bar graphs show the fold increase of normalized luciferase activities between the treated versus untreated conditions. Results are representative of four independent experiments. (B and C) Error bars indicate standard deviation. (D) Methylation pattern of Foxp3 CNS2 in CD25brightCD4+ and CD25−CD4+ T cells derived from male Ets-1−/− or Ets-1+/+ mice. The BiQ Analyzer was used to analyze the sequence traces from 10 independent clones from each amplified bisulfite-treated DNA. The methylation status of individual CpG motifs within the amplified region is colored in yellow (methylated) or blue (not methylated). Numbers indicate the percentage of methylation at CpG sites. Results are representative of three individual experiments.
DISCUSSION
The data presented in this study show that Ets-1 is required for normal development and function of T reg cells and that defects in this cell subset were responsible for some of the immunological disorders in Ets-1−/− mice. Viable young mutant animals had reduced numbers of T reg cells in the spleen, but the frequency in the thymus appeared normal. In both sites, Ets-1−/− T reg cells had an unusual phenotype in that they expressed CD103 (Fig. 4 B), a marker typical of cells that experienced antigen under certain inflammatory conditions (Huehn et al., 2004; Suffia et al., 2005). This raised the possibility that the majority of thymic Ets-1−/− T reg cells were antigen-experienced recirculating cells, thereby masking an important quantitative deficit in thymic development of these cells. This was indeed the case, as supported by the very low frequency of thymic T reg cells in FTOCs (Fig. 4 C), in 5-d-old newborns (not depicted), and in mixed WT/KO chimeras analyzed at relative early time points (Figs. 5 and 6).
Ets-1−/− mice have a severe phenotype that affects other lymphocyte lineages. In such context, the T reg cell deficit could be secondary to T reg cell–independent abnormalities, such as the T cell lymphopenia, the hyperactivation status of conventional T cells, and dysfunctions resulting thereof. As demonstrated in this study using mixed WT/KO fetal liver chimeras, normal T reg cell development was achieved in a thymic environment even with an excess of Ets-1−/− cells. In contrast, the same environment did not rescue the deficit of T reg cell development of Ets-1−/− precursors, ruling out a potential role of extrinsic factors, e.g., IL-2 or TGF-β, in this deficit. These results strongly supported a T reg cell–intrinsic role of Ets-1.
Normal T reg cells were shown to prevent Ets-1−/− T cell–mediated splenomegaly and B cell dysfunctions observed in viable mutants. This block correlated with decreased accumulation of Ets-1−/− T cells, drastic reduction in the capacity of mutant CD4+ T cells to produce IL-4, and significant decrease of the proportion of CD44highCD62-L− cells to the benefit of activated/memory cells coexpressing high levels of these two molecules (Fig. S7). Moreover, cells with a naive phenotype (CD44lowCD62-Lhigh) were now clearly detected, but their frequency was still significantly lower than in controls. Thus, Ets-1−/− conventional T cells were highly susceptible to key T reg cell–mediated negative signals, but the failure to accumulate high numbers of naive cells could indicate that Ets-1−/− conventional CD4 T cells did not respond to T reg cell–mediated signals inhibiting activation or that they were in an intrinsic state of activation caused by the lack of Ets-1.
Although this issue is difficult to assess given the lymphopenic context of Ets-1−/− and mixed WT/KO chimeras, our experiments also provided evidence suggestive of cell intrinsic defects in conventional Ets-1−/− CD4 T cells, which provide possible explanations for the distinct spectrum of immune disorders in these mice as compared with other T reg cell–defective models with partial or complete impaired Foxp3 function. First, these cells had reduced growth potential and/or survival in vitro (Bories et al., 1995; Muthusamy et al., 1995). This could explain the T cell lymphopenia, despite the severe T reg cell deficit, instead of uncontrolled T cell expansion. Second, they showed enhanced Th2 polarization but poor capacity to produce IFN-γ (Fig. 1 and Fig. S2) or to induce IBD in Rag2-deficient hosts (not depicted). Together with the absence of IBD in viable Ets-1−/− mice, these results suggest that inactivation of Ets-1 might negatively impact Th1 polarization, which is in line with the finding that T-bet function was markedly impaired in Ets-1 mutated T cells, stimulated under Th1-skewing conditions (Grenningloh et al., 2005). Collectively, these results suggest that Ets-1 inactivation also affects conventional T cells in a cell-intrinsic manner and provide an explanation for the absence of massive T cell expansions and enhanced Th2 polarization in mutant animals.
Ets-1 may regulate Foxp3 expression trough several nonmutually exclusive ways. It could have an indirect effect by regulating the transcription of genes encoding direct regulators of Foxp3 or by controlling expression of signaling molecules acting upstream of Foxp3 (i.e., IL-2R or TCR). However, our finding that Ets-1 binds to the CNS2 region and that the enhancer activity of this element relies on ETS-binding sites rather supports a more direct effect. Histone modification and DNA demethylation have been associated with stable expression of Foxp3 in both thymic and peripherally generated T reg cells (Floess et al., 2007; Polansky et al., 2008). In this context, inactivation of nuclear factors such as Runx–CBF-β that are involved in chromatin remodeling processes markedly impaired the T reg cell compartment (Kitoh et al., 2009; Klunker et al., 2009; Rudra et al., 2009). Interestingly, Runx–CBF-β−/− and Ets-1−/− mice both displayed T reg cell deficit associated with hyper IgE secretion and enhanced Th2 polarization (Kitoh et al., 2009). Thus, as the Runx–CBF-β complex, Ets-1 could contribute to trigger the epigenetic changes required for stable Foxp3 expression. This hypothesis is strengthened by the highly methylated status of the CNS2 region in splenic Ets-1−/− T reg cells that correlated with the reduced level of Foxp3 transcripts and proteins (Figs. 4 and 9 D). However, although CBF-β was required to maintain high levels of Foxp3 in the periphery, unlike Ets-1−/− mice, CBF-β–deficient mice had normal thymic T reg cell development (Kitoh et al., 2009; Rudra et al., 2009). This suggests that Ets-1 function may not be restricted to the context of such epigenetic modifier nuclear complexes.
We report in this study that functional Ets-binding sites within the CNS2 region are located near sequences occupied by STAT5 and CREB nuclear factors in T reg cells (Fig. 9 A; Kim and Leonard, 2007). Interestingly, Ets-1 has been shown to interact with STAT5 in T cells that were activated in vitro in the presence of IL-2 and, upon those conditions, to bind DNA target sites (Rameil et al., 2000). Thus, during thymic T reg cell development, Ets-1–STAT5 complexes could interact with Foxp3 regulatory sequences, thereby ensuring optimal expression of the gene. Furthermore, upon cellular activation, phosphorylated Ets-1 and CREB are both able to associate with CBP/p300, two proteins know to carry histone acetyl transferase activities (Chrivia et al., 1993; Yang et al., 1998; Foulds et al., 2004). Thus, for thymocytes that experience strong TCR/CD28 and IL-2 signals, Ets-1 activity could be required to efficiently target a large complex of nuclear factors to the Foxp3 intronic enhancer. However, gene-targeted deletion of the CNS2 region was shown to impair heritable Foxp3 expression in dividing cells but, unlike Ets-1 inactivation, not to affect the development of thymic T reg cells (Zheng et al., 2010). These data suggest that Ets-1 could interact with additional regulatory sequences within the Foxp3 locus.
The CNS1 sequence appears to be essential for TGF-β–mediated Foxp3 induction in conventional T cells but dispensable for T reg cell thymic development. We show in this study that TGF-β could induce Foxp3 expression in Ets-1−/− T cells in vitro (Fig. 7), and because we failed to detect Ets-1 binding to the Foxp3 promoter (Fig. 9 B), it seems unlikely that neither the CNS1 nor the promoter would be required for Ets-1–mediated regulation of the Foxp3 gene. An additional region, CNS3, was shown to increase the frequency of T reg cells generated in the thymus and in the spleen, but its deletion left the levels of Foxp3 unaffected (Zheng et al., 2010). Thus, Ets-1 may not act through a single regulatory region but rather interact at several sites, and only from these combined activities would proper expression and epigenetic modification at the Foxp3 locus be achieved.
Altogether, our data support a model in which Ets-1 participates in a nucleoprotein complex whose function would be regulated by TCR and/or γ-chain signaling pathways to induce and lock up Foxp3 expression during the development of thymic T reg cells. These results identify Ets-1 as a key regulator of the immune system and of the biology of T reg cells.
MATERIALS AND METHODS
Mice.
Mice carrying the inactivated ets-1 gene have been previously described (Bories et al., 1995; Eyquem et al., 2004a). All mice used in this study, including WT C57BL/6 mice, were maintained in our specific pathogen-free breeding facility (Departement d'Experimentation Animal, Institut Universitaire d'Hématologie, Saint Louis, France) and were sacrificed for analysis between 5 and 6 wk of age. All mouse experiments were subject to approval by the Institut Universitaire d'Hémathologie Institutional Animal Care and Use Committee.
Flow cytometry and cell sorting.
Single cell suspensions were stained with antibodies according to standard procedures and analyzed on a FACScalibur or a Canto flow cytometer (BD). FACS results were analyzed with CellQuestpro (BD) or FlowJo software (Tree Star, Inc.). The following antibodies were purchased from BD: anti–CD8-α (53-6.7), anti-CD4 (L3T4), anti-CD25 (7D4), anti–CD3-ε (clone 145-2C11), anti-CD44 (IM7), anti-CD69 (H1.2F3), anti-CD95 (Jo2), anti-CD95L (MFL3), anti–CD62-L (MEL-14), anti-CD103 (M290), anti-CD45.2 (104), anti-CD23 (B3B4), anti-CD21 (7G6), anti-CD86 (GL1), and anti–I-A/I-E (M5/114.15.2). mAbs were conjugated with FITC, phycoerythrin, biotin or allophycocyanin, Pacific blue, streptavidin PerCP-Cy5.5, and streptavidin Cy-Chrome.
Cell sorting was performed on a FACSvantage SE (BD), and in all cases, the purity of the sorted cells was estimated at >99%. FACS DNA content analysis was performed as described previously (Lagresle et al., 2002). For intracellular staining of Foxp3, single cell suspensions from spleens and thymi were stained with anti-CD4, anti-CD8, and anti-CD25 or anti-CD3. Subsequent intracellular staining for Foxp3-expressing cells was performed using the phycoerythrin anti-Foxp3 staining kit (eBioscience) according to the manufacturer's instructions.
In vitro differentiation of naive T cells into Foxp3-expressing cells.
Sorted CD4+CD25−CD62-L+ T cells and irradiated Rag2−/− spleen cells (1:3 ratio) were cultured in RPMI medium containing 10% fetal calf serum, 5 µg/ml soluble anti-CD3 (2C11), 3 ng/ml TGF-β1, and 20 U/ml mouse recombinant IL-2. Foxp3 expression was measured after 72 h of activation.
In vitro suppression activity.
Sorted naive WT CD4+CD25− T cells (1.5 × 104 cells) were set up in triplicates in 96-well round-bottom plates (Costard) with 105 irradiated Rag2−/− spleen cells and mixed or not with variable numbers of FACS sorted WT or Ets-1−/− CD4+CD25high T reg cells to obtain the indicated ratios (T reg cells/naives) in medium containing 5 µg/ml anti-CD3. Intracellular FACS analysis of sorted CD4+CD25high T reg cells revealed Foxp3 expression in >90% of the cells. Proliferation was assessed after 3 d by pulsing the cells with 1 µCi/well [3H] thymidine (GE Healthcare) for the last 16 h of culture. Cells were harvested onto filter membranes, and the amount of incorporated [3H] thymidine was measured with a liquid scintillation counter Wallac1409 (PerkinElmer). Data are representative of three independent experiments.
In vivo cell transfer.
All mice used for chimera were in a C57BL/6 background. CD45.2/CD45.1 Rag2−/−γc−/− chimera mice were generated as previously described (Eyquem et al., 2004b). For T cell reconstitution of CD3-ε–deficient mice, thymi were isolated from 18.5 pc embryos and grafted under the kidney capsule of CD3ε−/− mice. 5–6 wk after transfer, recipient chimera mice were sacrificed for analysis. For T reg cell transfers, spleen cell suspensions were stained with anti–CD25-biotin antibody (7D4), followed by incubation with strepavidin-coupled MicroBeads (Miltenyi Biotec). The cells were passed twice through a MACS separation column (Miltenyi Biotec) to obtain a >95% pure CD25+CD4+ population. 5 × 105 purified CD25+CD4+ cells were injected i.v. into Rag2−/− recipient mice. 5 × 105 of electronically sorted CD4+CD25−CD62-L+ naive T cells were coinjected in the indicated mice.
PCR assays.
RT-PCR reactions were performed in a 50-µl reaction containing the indicated amounts of genomic DNA, 2 ng/µl of each primer, 0.2 µM deoxy-nucleoside triphosphate, 2 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl, pH 8.8, 0.1% Triton X-100, and 1 U/50 µl Taq polymerase. Reactions were performed for 4 min at 95°C; 35 cycles of 1 min at 95°C, 1 min at 57°C, and 1.5 min at 72°C; and 5 min at 72°C.
Real-time RT-PCR was performed using the LightCycler FastStart kit (Roche; Espeli et al., 2006) on a LightCycler system. Amounts of RNA were calculated by reference to a log-linear standard regression curve. This curve was constructed from the number of cycles necessary to detect product accumulation after amplification of 2 µl of serial dilutions of a standard cDNA solution. The calculation of the relative concentration was performed using the LightCycler software (Roche).
Primers used were hypoxanthine-guanine phosphoribosyltransferase (HPRT) forward, 5′-CACAGGACTAGAACACCTGC-3′; HPRT reverse, 5′-GCTGGTGAAAAGGACCTCT-3′; IL-2 forward, 5′-CCTGAGCAGGATGGAGAATTACA-3′; IL-2 reverse, 5′-GCCTTATGTGTTGTAAGCAGGAGG-3′; IL-4 forward, 5′-TGACGGCACAGAGCTATTGA-3′; IL-4 reverse, 5′-ATGGTGGCTCAGTACTACGA-3′; IL-5 forward, 5′-ATGAGGCTTCCTGTCCCTACTC-3′; IL-5 reverse, 5′-GTCACCATGGAGCAGCTCAGCC-3′; IL-10 forward, 5′-GGTTGCCAAGCCTTATCGGA-3′; IL-10 reverse, 5′-ACCTGCTCCACTGCCTTGCT-3′; IL-13 forward, 5′-GGTGCCAAGATCTGTGTCTCTCC-3′; IL-13 reverse, 5′-GAGATGCCCAGGGATGGTCTCC-3′; IFN-γ forward, 5′-GCTCTGAGACAATGAACGCT-3′; IFN-γ reverse, 5′-AAAGAGATAATCTGGCTCTGC-3′; Foxp3 forward, 5′-CCAAGGATCCTACCCACTGCTGG-3′; and Foxp3 reverse, 5′-CCCAGAGGTGCCTCCGCACTGC-3′.
Plasmid constructs.
The minimal CNS2 region of the Foxp3 gene was PCR amplified and cloned into pGL3-Promoter (Promega) to yield pFoxp3CNS2. PCR primers used were 5-EFoxp, 5′-CACACAGTAAGAAGGTGGATCCATGC-3′; and 3-EFoxp 5′-CTGGGCTGGCCAGCCAGCTTCCTGCACTGTC-3′.
The site-directed mutagenesis kit (Agilent Technologies) was used to mutate the GGA motif into CCA in the targeted ETS sites. Primers for mutagenesis were as follows: FwMt E1, 5′-ACGTCACCTACCTCTAGGGCTAGCACCCAC-3′; RvMt E1, 5′-GTGGGTGCTAGCCCTAGAGGTAGGTGACGT-3′; FwMt E2, 5′-AGCCCAATCCATGGGGCCGCCATGACG-3′; and RvMt E2, 5′-CGTCATGGCGGCCCCATGGATTGGGCT-3′. The integrity of all the CNS2 regions inserted in the pGL3-Promoter vector was established by sequencing.
Transient transfections and luciferase assays.
Jurkat cells were transiently transfected with 1 µg of the indicated pGL3-luciferase plasmids using electroporation with the Cell Line Nucleofector kit V (Lonza). For each transfection, 0.5 µg of β-galactosidase–expressing plasmid (Invitrogen) was added for normalization. Activation and luciferase assays were performed as previously described (Kim and Leonard, 2007).
ChIP assays.
ChIP assays were performed using the Imprint ChIP kit (Sigma-Aldrich) as previously described (Eyquem et al., 2002). CD4+CD25bright and CD4+CD25− cells were sorted from spleens and stimulated with 100 U/ml IL-2 for 1 h. Formaldehyde (final concentration 1%) was then added to cross-link proteins and DNA. The cell lysates were sonicated and immunoprecipitated with normal rabbit serum (Millipore), α-STAT5 (Santa Cruz Biotechnology, Inc.), and α–Ets-1 (Santa Cruz Biotechnology, Inc.). The immunoprecipitated DNA was eluted and amplified by real-time SYBR green PCR using an ABI 7300 (Applied Biosystems). Values were normalized to serial dilution of corresponding input controls and are expressed as fold enrichment relative to normal rabbit serum for each experiment. The sequence-specific primers used for amplification of the Foxp3 gene are CNS2, 5′-ATCTGGCCAAGTTCAGGTTG-3′ and 5′-GGAACTGGTGTGACTGTGTGA-3′; and Foxp3 promoter, 5′-CACTCAGAGACTCGCAGCAG-3′ and 5′-GGGGTAGTGCTCTGTCTCCA-3′. Primers for IgH Cµ irrelevant region are 5′-GGCCTCGCAGATGAGTTTAG-3′ and 5′-GTGCCCATTCCAGGTAAGAA-3′.
Detection of serum Ig.
Concentrations of serum polyclonal IgM, IgG1, IgE, and IgG2b were determined using isotype-specific ELISA (SouthernBiotech). The concentrations were determined by comparing test sample dilution series with isotype control standards (SouthernBiotech). For IgE detection, the capturing antibody was anti-IgE (R35-72; BD). For detection of autoantibodies, the capturing agent used was double-stranded sheared salmon sperm DNA (Eppendorf) at 10 µg/ml.
Bisulphite sequencing.
Genomic DNA was isolated from electronically sorted male T cells as previously described (Eyquem et al., 2002). Sodium bisulphite treatment of genomic DNA was performed using the QIAGEN kit according the manufacturer's protocol. In a subsequent PCR amplification, uracils were replicated as thymidines. PCRs were performed on Eppendorf thermocyclers in a final volume of 25 µl containing 1× PCR buffer, 1 U Taq DNA polymerase (Promega), 200 µM deoxy-nucleoside triphosphate, 12.5 pmol each of forward and reverse primers, and 7 ng of bisulphate-treated genomic DNA. The amplification conditions were 95°C for 15 min and 40 cycles of 95°C for 1 min, 55°C for 45 s, and 72°C for 1 min, and a final extension step of 10 min at 72°C. PCR products were purified and cloned into Topo vectors. Plasmid from individual clones was purified and sequenced in both directions applying the PCR primers and the ABI Big Dye Terminator v1.1 cycle sequencing chemistry (Applied Biosystems), followed by capillary electrophoresis on an ABI 3100 genetic analyzer (Applied Biosystems). Trace files were interpreted using BiQ Analyzer (Max-Planck-Institut, Saarbrucken, Germany), which allows for quantification of methylation signals. For each sample, both PCR amplification and sequencing were repeated once. The following primers (5′ to 3′ direction) were used for both PCR amplification of bisulphite-converted genomic DNA and sequence reactions: Amp 2 forward, 5′-ATTTGAATTGGATATGGTTTGT-3′; and reverse, 5′-AACCTTAAACCCCTCTAACATC-3′.
Online supplemental material.
Fig. S1 shows size and phenotypic analysis of splenic T cells from Ets-1−/− mice. Fig. S2 shows in vitro cytokine secretion by WT and Ets-1−/− T cells. Fig. S3 depicts the organization of spleens from Ets-1+/+ and Ets-1−/− mice. Fig. S4 shows that Ets-1−/− T cells are responsive to suppression from WT T reg cells. Figs. S5 and S6 show an intrinsic defect of Ets-1−/− thymic T reg cell development and splenic T reg cells, respectively. Fig. S7 shows that WT cells partially rescue Ets-1−/− naive CD4 and CD8 splenocytes. Table S1 shows the numbers of B and T cells (CD4+ and CD8+) in spleens from Ets-1+/+ and Ets-1−/− mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092153/DC1.
Acknowledgments
We thank M. Goodhardt and R.S. Allan for criticism of the manuscript.
This work was supported by a grant from the Ligue Nationale Contre le Cancer (RS 06/75-16). K. Chemin and E. Mouly were supported by fellowships from the Fondation pour la Recherche Médicale, and O. Burlen-defranoux and A. Bandeira were supported by a grant from the Association pour la Recherche sur le Cancer (ARC #1007).
The authors have no conflicting financial interests.
Author contributions: E. Mouly, A. Bandeira, and J.-C. Bories designed the research and analyzed the data. E. Mouly, K. Chemin, O. Burlen-defranoux, M. Chopin, L. Mesnard, M. Leite-de-Moraes, and H.V. Nguyen performed experiments. A. Bandeira had major contributions to the work. A. Bandeira and J.-C. Bories wrote the manuscript.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- DP
- double positive
- FTOC
- fetal thymus organ culture
- HPRT
- hypoxanthine-guanine phosphoribosyltransferase
- IBD
- inflammatory bowel disease
- SP
- single positive
References
- Aschenbrenner K., D'Cruz L.M., Vollmann E.H., Hinterberger M., Emmerich J., Swee L.K., Rolink A., Klein L. 2007. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat. Immunol. 8:351–358 10.1038/ni1444 [DOI] [PubMed] [Google Scholar]
- Bensinger S.J., Bandeira A., Jordan M.S., Caton A.J., Laufer T.M. 2001. Major histocompatibility complex class II–positive cortical epithelium mediates the selection of CD4+25+ immunoregulatory T cells. J. Exp. Med. 194:427–438 10.1084/jem.194.4.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bories J.C., Willerford D.M., Grévin D., Davidson L., Camus A., Martin P., Stéhelin D., Alt F.W. 1995. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 377:635–638 10.1038/377635a0 [DOI] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vogtenhuber C., Blazar B.R., Farrar M.A. 2007. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 178:280–290 [DOI] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vang K.B., Moon J.J., Chu H.H., Lio C.W., Vegoe A.L., Hsieh C.S., Jenkins M.K., Farrar M.A. 2008. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 28:112–121 10.1016/j.immuni.2007.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jin W., Hardegen N., Lei K.J., Li L., Marinos N., McGrady G., Wahl S.M. 2003. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 198:1875–1886 10.1084/jem.20030152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia J.C., Kwok R.P., Lamb N., Hagiwara M., Montminy M.R., Goodman R.H. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 365:855–859 10.1038/365855a0 [DOI] [PubMed] [Google Scholar]
- Dittmer J. 2003. The biology of the Ets1 proto-oncogene. Mol. Cancer. 2:29 10.1186/1476-4598-2-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli M., Rossi B., Mancini S.J., Roche P., Gauthier L., Schiff C. 2006. Initiation of pre-B cell receptor signaling: common and distinctive features in human and mouse. Semin. Immunol. 18:56–66 10.1016/j.smim.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Eyquem S., Lagresle C., Fasseu M., Sigaux F., Bories J.C. 2002. Disruption of the lineage restriction of TCR beta gene rearrangements. Eur. J. Immunol. 32:3256–3266 [DOI] [PubMed] [Google Scholar]
- Eyquem S., Chemin K., Fasseu M., Bories J.C. 2004a. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc. Natl. Acad. Sci. USA. 101:15712–15717 10.1073/pnas.0405546101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyquem S., Chemin K., Fasseu M., Chopin M., Sigaux F., Cumano A., Bories J.C. 2004b. The development of early and mature B cells is impaired in mice deficient for the Ets-1 transcription factor. Eur. J. Immunol. 34:3187–3196 10.1002/eji.200425352 [DOI] [PubMed] [Google Scholar]
- Feuerer M., Hill J.A., Kretschmer K., von Boehmer H., Mathis D., Benoist C. 2010. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc. Natl. Acad. Sci. USA. 107:5919–5924 10.1073/pnas.1002006107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floess S., Freyer J., Siewert C., Baron U., Olek S., Polansky J., Schlawe K., Chang H.D., Bopp T., Schmitt E., et al. 2007. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 5:e38 10.1371/journal.pbio.0050038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot J.D., Gavin M.A., Rudensky A.Y. 2003. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4:330–336 10.1038/ni904 [DOI] [PubMed] [Google Scholar]
- Foulds C.E., Nelson M.L., Blaszczak A.G., Graves B.J. 2004. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol. Cell. Biol. 24:10954–10964 10.1128/MCB.24.24.10954-10964.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh R., Kang B.Y., Ho I.C. 2005. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med. 201:615–626 10.1084/jem.20041330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S. 2010. c-Rel: a pioneer in directing regulatory T-cell lineage commitment? Eur. J. Immunol. 40:664–667 10.1002/eji.201040372 [DOI] [PubMed] [Google Scholar]
- Hori S., Nomura T., Sakaguchi S. 2003. Control of regulatory T cell development by the transcription factor Foxp3. Science. 299:1057–1061 10.1126/science.1079490 [DOI] [PubMed] [Google Scholar]
- Huehn J., Siegmund K., Lehmann J.C., Siewert C., Haubold U., Feuerer M., Debes G.F., Lauber J., Frey O., Przybylski G.K., et al. 2004. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J. Exp. Med. 199:303–313 10.1084/jem.20031562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huehn J., Polansky J.K., Hamann A. 2009. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat. Rev. Immunol. 9:83–89 10.1038/nri2474 [DOI] [PubMed] [Google Scholar]
- Khattri R., Cox T., Yasayko S.A., Ramsdell F. 2003. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 4:337–342 10.1038/ni909 [DOI] [PubMed] [Google Scholar]
- Kim H.P., Leonard W.J. 2007. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J. Exp. Med. 204:1543–1551 10.1084/jem.20070109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitoh A., Ono M., Naoe Y., Ohkura N., Yamaguchi T., Yaguchi H., Kitabayashi I., Tsukada T., Nomura T., Miyachi Y., et al. 2009. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 31:609–620 10.1016/j.immuni.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Klunker S., Chong M.M., Mantel P.Y., Palomares O., Bassin C., Ziegler M., Rückert B., Meiler F., Akdis M., Littman D.R., Akdis C.A. 2009. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J. Exp. Med. 206:2701–2715 10.1084/jem.20090596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagresle C., Gardie B., Eyquem S., Fasseu M., Vieville J.C., Pla M., Sigaux F., Bories J.C. 2002. Transgenic expression of the p16(INK4a) cyclin-dependent kinase inhibitor leads to enhanced apoptosis and differentiation arrest of CD4-CD8- immature thymocytes. J. Immunol. 168:2325–2331 [DOI] [PubMed] [Google Scholar]
- Leprince D., Gegonne A., Coll J., de Taisne C., Schneeberger A., Lagrou C., Stehelin D. 1983. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature. 306:395–397 10.1038/306395a0 [DOI] [PubMed] [Google Scholar]
- Li M.O., Sanjabi S., Flavell R.A. 2006. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 25:455–471 10.1016/j.immuni.2006.07.011 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang P., Li J., Kulkarni A.B., Perruche S., Chen W. 2008. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat. Immunol. 9:632–640 10.1038/ni.1607 [DOI] [PubMed] [Google Scholar]
- Marie J.C., Letterio J.J., Gavin M., Rudensky A.Y. 2005. TGF-β1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J. Exp. Med. 201:1061–1067 10.1084/jem.20042276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie J.C., Liggitt D., Rudensky A.Y. 2006. Cellular mechanisms of fatal early-onset autoimmunity in mice with the T cell-specific targeting of transforming growth factor-beta receptor. Immunity. 25:441–454 10.1016/j.immuni.2006.07.012 [DOI] [PubMed] [Google Scholar]
- Moisan J., Grenningloh R., Bettelli E., Oukka M., Ho I.C. 2007. Ets-1 is a negative regulator of Th17 differentiation. J. Exp. Med. 204:2825–2835 10.1084/jem.20070994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy N., Barton K., Leiden J.M. 1995. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 377:639–642 10.1038/377639a0 [DOI] [PubMed] [Google Scholar]
- Nunn M.F., Seeburg P.H., Moscovici C., Duesberg P.H. 1983. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 306:391–395 10.1038/306391a0 [DOI] [PubMed] [Google Scholar]
- Nye J.A., Petersen J.M., Gunther C.V., Jonsen M.D., Graves B.J. 1992. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 6:975–990 10.1101/gad.6.6.975 [DOI] [PubMed] [Google Scholar]
- Ochs H.D., Gambineri E., Torgerson T.R. 2007. IPEX, FOXP3 and regulatory T-cells: a model for autoimmunity. Immunol. Res. 38:112–121 10.1007/s12026-007-0022-2 [DOI] [PubMed] [Google Scholar]
- Polansky J.K., Kretschmer K., Freyer J., Floess S., Garbe A., Baron U., Olek S., Hamann A., von Boehmer H., Huehn J. 2008. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 38:1654–1663 10.1002/eji.200838105 [DOI] [PubMed] [Google Scholar]
- Rameil P., Lécine P., Ghysdael J., Gouilleux F., Kahn-Perlès B., Imbert J. 2000. IL-2 and long-term T cell activation induce physical and functional interaction between STAT5 and ETS transcription factors in human T cells. Oncogene. 19:2086–2097 10.1038/sj.onc.1203542 [DOI] [PubMed] [Google Scholar]
- Rudra D., Egawa T., Chong M.M., Treuting P., Littman D.R., Rudensky A.Y. 2009. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 10:1170–1177 10.1038/ni.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sather B.D., Treuting P., Perdue N., Miazgowicz M., Fontenot J.D., Rudensky A.Y., Campbell D.J. 2007. Altering the distribution of Foxp3+ regulatory T cells results in tissue-specific inflammatory disease. J. Exp. Med. 204:1335–1347 10.1084/jem.20070081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suffia I., Reckling S.K., Salay G., Belkaid Y. 2005. A role for CD103 in the retention of CD4+CD25+ Treg and control of Leishmania major infection. J. Immunol. 174:5444–5455 [DOI] [PubMed] [Google Scholar]
- Tai X., Cowan M., Feigenbaum L., Singer A. 2005. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat. Immunol. 6:152–162 10.1038/ni1160 [DOI] [PubMed] [Google Scholar]
- Tone Y., Furuuchi K., Kojima Y., Tykocinski M.L., Greene M.I., Tone M. 2008. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9:194–202 10.1038/ni1549 [DOI] [PubMed] [Google Scholar]
- Venuprasad K., Huang H., Harada Y., Elly C., Subramaniam M., Spelsberg T., Su J., Liu Y.C. 2008. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat. Immunol. 9:245–253 10.1038/ni1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., John S.A., Clements J.L., Percy D.H., Barton K.P., Garrett-Sinha L.A. 2005. Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. Int. Immunol. 17:1179–1191 10.1093/intimm/dxh295 [DOI] [PubMed] [Google Scholar]
- Yang C., Shapiro L.H., Rivera M., Kumar A., Brindle P.K. 1998. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol. Cell. Biol. 18:2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z., Kanno Y., Kerenyi M., Stephens G., Durant L., Watford W.T., Laurence A., Robinson G.W., Shevach E.M., Moriggl R., et al. 2007. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 109:4368–4375 10.1182/blood-2006-11-055756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Josefowicz S., Chaudhry A., Peng X.P., Forbush K., Rudensky A.Y. 2010. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 463:808–812 10.1038/nature08750 [DOI] [PMC free article] [PubMed] [Google Scholar]