Abstract
Systemic lupus erythematosus (SLE) is characterized by high-avidity IgG antinuclear antibodies (ANAs) that are almost certainly products of T cell–dependent immune responses. Whether critical amino acids in the third complementarity-determining region (CDR3) of the ANA originate from V(D)J recombination or somatic hypermutation (SHM) is not known. We studied a mouse model of SLE in which all somatic mutations within ANA V regions, including those in CDR3, could be unequivocally identified. Mutation reversion analyses revealed that ANA arose predominantly from nonautoreactive B cells that diversified immunoglobulin genes via SHM. The resolution afforded by this model allowed us to demonstrate that one ANA clone was generated by SHM after a VH gene replacement event. Mutations producing arginine substitutions were frequent and arose largely (66%) from base changes in just two codons: AGC and AGT. These codons are abundant in the repertoires of mouse and human V genes. Our findings reveal the predominant role of SHM in the development of ANA and underscore the importance of self-tolerance checkpoints at the postmutational stage of B cell differentiation.
Systemic lupus erythematosus (SLE) is a multigenic systemic autoimmune disease with an immune complex–associated pathology that is prevalent among females of childbearing age. A hallmark of SLE is the appearance of serum antinuclear antibodies (ANAs) that appear to be products of T cell–dependent immunity, manifested by high-avidity binding, somatic mutations, and derivation from B cells that have undergone substantial clonal expansion (Shlomchik et al., 1987a, 1990; Tan, 1989). SLE-associated antibodies are frequently, but not exclusively, directed against histones, double-stranded DNA (dsDNA), histone–DNA complexes, and various ribonuclear proteins.
Considerable progress in dissecting SLE etiology and pathogenesis has come largely from mouse models with a genetic predisposition for spontaneous SLE-like disease (Theofilopoulos and Dixon, 1985). These models have been especially useful in identifying genetic contributions to specific disease manifestations and the role played by toll-like receptors (TLRs) in targeting the autoimmune response to specific TLR agonists (Fairhurst et al., 2006; Shlomchik, 2008). Spontaneous SLE-like disease in F1 hybrid mice between the NZB strain and several others bears striking resemblance to human SLE with respect to manifestations, genetics, and female gender bias. Genetic backcross studies involving NZB mice have identified a gene-rich interval at the distal end of the NZB chromosome 1 that is strongly associated with spontaneous autoantibody development (Vyse et al., 1997). This region is syntenic with a region of distal chromosome 1 associated with human SLE (Tsao et al., 1997).
A complementary body of work investigating immunological self-tolerance has identified several mechanisms of tolerance that are potentially breached in systemic autoimmunity. With respect to the B cell, these mechanisms involve anergy, receptor editing, clonal deletion, and a less well defined preplasma cell checkpoint (Goodnow et al., 2005; Culton et al., 2006; Nemazee, 2006). Studies of self-tolerance in B cells have relied heavily on mice carrying Ig transgenes that encode autoantibodies to artificial or natural self-antigens. In these models, self-tolerance is remarkably efficient, and autoantibody-encoding transgenes have, at most, a modest effect on development of ANAs (Brard et al., 1999; Mandik-Nayak et al., 1999; Yachimovich-Cohen et al., 2003; Steeves and Marion, 2004; Chen et al., 2006). When such transgenes are bred into autoimmune-prone strains, the autoreactive B cells that emerge are frequently oligoclonal and express edited receptors generated by endogenous Ig V gene rearrangements. Because it changes the unique identity of a B cell, receptor editing often obscures interpretations regarding the stages in B cell development when breaches in self-tolerance occur. Without this information, knowledge obtained from genetic mapping analyses, gene knockout studies, and other approaches cannot be fully interpreted in the context of disease etiology.
Many self-tolerance studies have focused on B cell developmental stages that precede immune activation, with the implicit assumption that autoreactive antecedents to disease-associated autoimmune B cells are generated in the bone marrow immediately after Ig gene recombination. Autoreactive cells generated in this manner would have to escape every self-tolerance checkpoint to participate in systemic autoimmunity (Goodnow et al., 2005). Alternatively, autoimmune B cells in SLE may be created by somatic hypermutation (SHM) in mature activated B cells responding to antigens in the periphery. Autoreactive B cells generated via this mutation-founder scenario would have to traverse fewer tolerance checkpoints before participating in the autoimmune process. To distinguish between these alternatives, somatic mutations within V region genes of autoimmune B cells must be identified and reverted to germline sequence so that their contribution to the specificity and affinity of the autoreactive B cell receptor (BCR) can be evaluated. Efforts to interpret results of such analyses have been confounded by cell sampling issues, uncertainty over whether a cell expresses one or multiple receptors, and difficulties in identifying somatic mutations. Defining mutations is problematic because of V gene polymorphisms and the presence of untemplated nucleotides in the heavy chain third complementarity-determining region (CDR3 [HCDR3]) that are added by terminal deoxynucleotidyl transferase (Tdt) during Ig gene recombination (Lafaille et al., 1989; Gilfillan et al., 1993). Yet defining mutations in HCDR3 is especially important because of its pivotal role in antigen binding, particularly with regard to nuclear antigens (Krishnan et al., 1996; Xu and Davis, 2000; Li et al., 2000; Guth et al., 2003). The HCDR3 caveat was common to all preceding studies and likely contributed to mixed interpretations regarding the role of SHM in the origin of ANA (Radic et al., 1989, 1993; Siminovitch et al., 1989; Xu and Davis, 2000; Wellmann et al., 2005; Mietzner et al., 2008).
In this study, we used a model of spontaneous autoimmunity in which all somatic mutations in V region genes of ANA-producing hybridomas could be unequivocally identified for reversion analyses. We found frequent unambiguous cases in which high-avidity autoimmune B cells were created from nonautoreactive precursors by SHM.
RESULTS
An SLE model for optimal identification of somatic mutations
Our objective was to determine whether B cells producing ANAs in SLE-like disease were derived from precursors that emerge in the bone marrow with an autoreactive BCR or from somatic mutants generated in the periphery. To this end, we generated a mouse in which all somatic mutations in monoclonal autoreactive antibodies could be unequivocally identified, even in HCDR3. This was accomplished by breeding two copies of a targeted null Tdt allele into an autoimmune-prone C57BL/6.NZB-Nba2 mouse (B6.Nba2; Gilfillan et al., 1993). The Nba2 interval spans ∼20 centimorgans of DNA from distal chromosome 1 of the NZB strain and confers upon aged mice a spontaneous autoimmunity with many of the classical hallmarks of SLE, including ANAs and a female gender bias (Rozzo et al., 2001). Notably, the chromosome 1–distal Nba2 interval is syntenic with a distal region of human chromosome 1 that is genetically associated with SLE (Tsao et al., 1997). As such, the Nba2 strain is considered an excellent model of spontaneous ANA development in human SLE. In Tdt-null mice, diversity at the borders of assembling gene segments is still created as a result of gain or loss of nucleotides, but the gains, referred to as P elements, occur through a templated process after the opening of sealed hairpins at the ends of coding segments (Lafaille et al., 1989). Prior studies established the feasibility of this approach by demonstrating ANA in Tdt-deficient autoimmune mice (Conde et al., 1998; Molano et al., 2003).
In the absence of Tdt, somatic mutations in HCDR3 of ANAs can be clearly defined. It is equally important that somatic mutations in VH, DH, JH, and JL gene segments used by such antibodies can also be unambiguously identified because the genomic sequence of the C57BL/6 strain is known. Heterozygous deficiencies in the Ig heavy chain (Igh) and kappa light chain (Igk) loci were also introduced because B cells may express both kappa and heavy chain alleles as a result of receptor editing or failed allelic exclusion (Chen et al., 1993; Gu et al., 1993). The heterozygous deficiencies limited expression to one heavy chain allele and made it possible to determine whether an autoreactive B cell hybridoma was expressing two light chains (κ + λ; Giachino et al., 1995; Li et al., 2002; Rezanka et al., 2005). The heterozygous Igh deficiency also assisted in distinguishing P-element additions from somatic mutations at the boundaries of the gene segments encoding HCDR3. As such, the genetic makeup of the B6.Nba2 Tdt−/− Igh+/− Igk+/− mouse allowed us to determine and reconstruct the original unmutated sequences of spontaneously arising ANAs before the advent of SHM.
Anti-chromatin antibodies in B6.Nba2 Tdt−/− Igh+/− Igk+/− mice
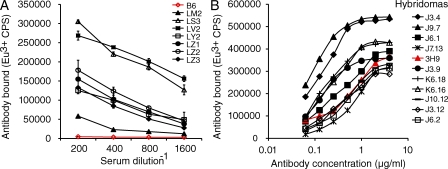
As is typical of the B6.Nba2 strain, a majority of aged female B6.Nba2 Tdt−/− Igh+/− Igk+/− mice developed high titers of serum anti-chromatin antibodies (Fig. 1 A). In most animals examined (6/8), these antibodies had already appeared by 5 mo of age. From one such spontaneously diseased mouse, we generated B cell hybridomas. In addition to anti-chromatin antibodies, this animal had substantial deposits of IgG in glomeruli of kidneys, as assessed by immunofluorescence (unpublished data). A screen of hybridoma supernatants for IgG antibodies directed against chromatin in a solid-phase europium-based immunoassay indicated that ∼5.7% (103/1,800) of them were positive. Representative results of a chromatin-binding assay using highly purified antibodies from several recloned hybridomas are shown in Fig. 1 B. The binding avidity of many of these antibodies was similar to, and sometimes exceeded, that of the prototypical ANA 3H9/Vκ4, which has served as a basis for numerous Ig transgene studies of tolerance and autoimmunity (Shlomchik et al., 1987a).
Figure 1.
Chromatin-binding IgG antibodies in B6.Nba2 Tdt−/− Igh+/− Igk+/− mice. (A) Serum titers of IgG anti-chromatin antibodies from B6.Nba2 Tdt−/− Igh+/− Igk+/− mice (8–10 mo old). Red line, representative serum from a nonautoimmune C57BL/6 mouse. Standard errors are shown. (B) Chromatin binding assay with purified monoclonal antibodies generated from a spontaneously autoimmune B6.Nba2 Tdt−/− Igh+/− Igk+/− mouse (female, 12 mo old). A prototypical ANA, 3H9/Vκ4, is shown in red. Bound antibodies were detected in a solid-phase europium (Eu3+)-based fluoroimmunometric assay (as counts per second). The figure shows one of two experiments with similar results.
Heavy and light chain V region genes were sequenced for 30 of the hybridomas using a reverse transcriptase PCR procedure. The sequences were queried against entries in several databases, most notably the National Center for Biotechnology Information C57BL/6 genome Build 36.0, to identify corresponding germline V gene segments used by each hybridoma. Unambiguous matches for VH, DH, JH, and Jk gene segments were found in every case and permitted somatic mutations to be identified clearly (Supplemental data). In a few cases, the heterozygous deficiency at the Igh locus also aided in the identification of mutations located at the gene segment boundaries. For example, we were able to infer that a thymidine nucleotide in the VH/D boundary in hybridomas J6.1 and J5.5 (lineage 4; Fig. 2) was added in a templated manner (P element) during V gene segment assembly. Had it been inserted instead via SHM, the preceding sequence would have been shifted out of the proper translational reading frame by 1 base and unable to encode a BCR, which is required for B cell survival.
Figure 2.
HCDR3 sequences for clones subjected to mutation-reversion analysis. Germline-encoded DH sequences are shown above and germline-encoded VH and JH sequences are shown below hybridoma (shaded) sequences. Somatic mutations are underlined. The blue lowercase t in J6.1 is a templated nucleotide (P element) added during VH(D)JH recombination. In lineage #4, somatic mutations that eliminated Arg codons are explicitly indicated for both members.
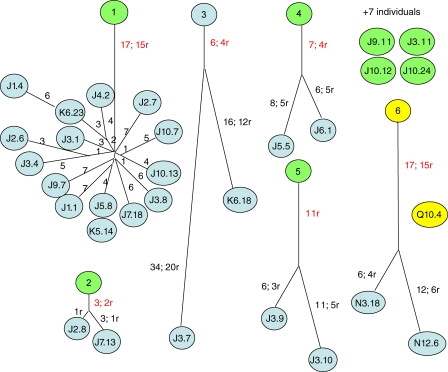
V region gene sequences revealed that the 30 hybridomas represented 12 antecedent B cell clones. Seven clones were represented by one hybridoma each. Four clones were represented by two hybridomas each, and one large clone was represented by 15 hybridomas (Fig. 3). This indicated that half of the chromatin-reactive hybridomas that were sampled represented one large clone. Large lineages of autoimmune B cells in spontaneous SLE are common, as they have been seen in several other studies (Shan et al., 1994; Colombo et al., 2000; Zhang et al., 2001).
Figure 3.
Dendrograms of multimember lineages. Numbers of somatic mutations are indicated along each branch, where r denotes an amino acid replacement and red indicates a mutation shared by all members of the lineage. The length of a branch is proportional to the number of somatic mutations. Clones from the first animal subjected to reversion are colored green, and those from a second autoimmune mouse are in yellow. Four of seven clones from the first animal that were represented by single hybridomas and subjected to reversion are shown.
Structural evidence that autoreactivity was generated via SHM
Several key observations emerged from the sequence analyses to support the idea that the chromatin reactivity of these antibodies was generated by SHM. First, every antibody was encoded by somatically mutated V region genes, carrying from 2 to 24 aa replacements (Table I). Second, each multimember lineage was defined by shared somatic mutations producing amino acid replacements among all clone members (Fig. 3). This is a prediction of the mutation-founder scenario. In the large lineage #1, for example, all members shared 17 somatic mutations, 15 of which produced amino acid replacements. A strong selection event apparently drove the proliferation of a single mutated B cell relative to its siblings at or near the time it had acquired the 17th somatic mutation. At this point, we infer that the cell was strongly autoreactive because an engineered antibody containing only these shared mutations bound strongly to chromatin (unpublished data). Third, 11 of 12 clones carried somatic mutations producing arginine (Arg) residues, and in four of the five multimember lineages, there was at least one mutation producing a shared Arg replacement among clone members (Table I and Supplemental data). Arg residues are often critical for antinuclear specificities (Radic and Weigert, 1994; Kalsi et al., 1996; Krishnan et al., 1996; Jang et al., 1998; Li et al., 2000; Rahman et al., 2001; Tanner et al., 2001; Guth et al., 2003; Haley et al., 2004). Studies with DNA-binding proteins have shown that their flexible side chains can interact via hydrogen bonds with bases in the major and minor grooves of DNA or form salt bridges and hydrogen bonds with the phosphodiester backbone (Steitz, 1990; Luscombe et al., 2001). One of the two shared arginine mutations in lineage #1 occurred in HCDR3 at the boundary of the DFL16.1 gene segment and the JH1 gene segment (Fig. 2). Had this clone been derived from a mouse with a functional gene for Tdt, we would not have been able to draw this conclusion with confidence. However, although HCDR3 arginines are often associated with affinity for nucleic acids, this is not always the case (Rahman et al., 2001). In HCDR3 of lineage #4, a pair of Arg codons sustained nonsynonymous somatic mutations. In hybridoma J5.5, a VH Arg codon near the boundary with the D gene segment was mutated to a Thr codon (Fig. 2). In the other hybridoma of lineage #4 (J6.1), a mutation at the beginning of the D gene segment produced an Arg to Ser codon conversion (Fig. 2).
Table I.
Summary of hybridoma V region genes
| Hybridoma | Vκ germline genes and shared mutations; shared replacementsa | Mutations: total; replacements | VH germline genes, isotype, and shared mutations; shared replacements | Mutations: total; replacements | Unanimously shared Arg mutations |
| Lineage #1 (mouse 1) | |||||
| J1.1 | Vκ CR-1/Jκ2 unanimously shared mutation10; 8r | 15; 9r | VH7183.9.15/ DFL16.1/ JH1 γ2b unanimously shared mutations 7r | 11; 10r | 0 Vκ; 2 VH |
| J1.4 | 13; 9r | 13; 11r | 0 Vκ; 2 VH | ||
| J2.6 | 14; 9r | 9; 8r | 0 Vκ; 2 VH | ||
| J2.7 | 14; 10r | 10; 8r | 0 Vκ; 2 VH | ||
| J3.1 | 12; 8r | 8; 7r | 0 Vκ; 2 VH | ||
| J3.4 | 10; 8r | 13; 9r | 0 Vκ; 2 VH | ||
| J3.8 | 11; 9r | 11; 8r | 0 Vκ; 2 VH | ||
| J4.2 | 10; 8r | 11; 9r | 0 Vκ; 2 VH | ||
| J5.8 | 14; 9r | 9; 8r | 0 Vκ; 2 VH | ||
| K5.14 | 14; 9r | 9; 8r | 0 Vκ; 2 VH | ||
| J7.18 | 14; 8r | 13; 9r | 0 Vκ; 2 VH | ||
| J9.7 | 13; 8r | 12; 10r | 0 Vκ; 2 VH | ||
| J10.7 | 10; 8r | 13; 9r | 0 Vκ; 2 VH | ||
| J10.13 | 11; 8r | 11; 7r | 0 Vκ; 2 VH | ||
| K6.23 | 13; 9r | 7; 7r | 0 Vκ; 2 VH | ||
| Lineage #2 (mouse 1) | |||||
| J2.8b | Vκ ai4/ Jκ1 unanimously shared mutations 1r | 2; 2r | VH3609.12.174/ DSP2.2/JH1 γ2c/γ2b unanimously shared mutations 2; 1r | 2; 1r | 0 Vκ; 0 VH |
| J7.13b | 1; 1r | 5; 2r | 0 Vκ; 0 VH | ||
| Lineage #3 (mouse 1) | |||||
| J3.7 | Vκ CR-1/Jκ1 unanimously shared mutations 3; 2r | 19; 12r | VHJ558.59.155/DSP2.2/JH3 γ1 unanimously shared mutations 3; 2r | 21; 12r | 0 Vκ; 1 VHc |
| K6.18 | 11; 7r | 11; 9r | 0 Vκ; 1 VH | ||
| Lineage #4 (mouse 1) | |||||
| J5.5 | Vκ 23-43 /Jκ5 unanimously shared mutations 1r | 5; 4r | VHQ52.2.4/DST4/ JH2 γ2c Unanimously shared mutations 6; 3r | 10; 5r | 1 Vκ; 0 VH |
| J6.1 | 4; 3r | 9; 6r | 1 Vκ; 0 VH | ||
| Lineage #5 (mouse 1) | |||||
| J3.9 | Vκ ai4/ Jκ2 unanimously shared mutations 6r | 9; 8r | VHJ558.26.116/DQ52/JH3 γ1 unanimously shared mutations 5r | 8; 6r | 3 Vκ; 1 VH |
| J3.10 | 7; 7r | 15; 9r | 3 Vκ; 1 VH | ||
| Single clones (mouse 1) | |||||
| J3.11 | Vκ ai4/ Jκ5 | 11; 7r | VHJ558.26.116/DSP2.5/ JH2 γ2c | 25; 15r | 1 Vκ; 0 VH |
| J3.12 | Vκ aa4/ Jκ1 | 2; 0r | VH7183.20.37/DSP2.x/JH4 γ2c | 13; 11r | 0 Vκ; 1 VH |
| J6.2 | Vκ 23-39/Jκ2 | 17; 9r | VHJ558.79.184/DST4/JH4 γ2c | 16; 9r | 1 Vκ; 1 VHd |
| J9.11 | Vκ 23-45/Jκ4 | 1; 1r | VHJ558.75.177/DSP2x/JH1 γ2c | 1; 1r | 1 Vκ; 0 VH |
| J10.12 | Vκ19-25/Jκ1 | 7; 5r | VH3609.12.174/ DSP2.x/JH1 γ2c | 4; 4r | 0 Vκ; 1 VH |
| J10.24 | Vκ ai4/Jκ2 | 8; 4r | VHJ558.26.116/DSP2.2/JH2 γ1 | 3; 3r | 1 Vκ; 0 VH |
| K6.16 | Vκ 23-43/Jκ2 | 10; 6r | VHJ588.2.88/ DFL16.1/JH2 γ1 | 18; 11r | 1 Vκ; 1 VH |
| Lineage #6 (mouse 2) | |||||
| N3.18 | Ai4/Jk2 | 7; 4r | VHJ558.26.116/DSP2.5/JH4 γ2c | 15;13r | 1 Vκ; 0 VH |
| N12.6 | Unanimously shared mutations 3; 1r | 13; 6r | Unanimously shared mutations 14; 12r | 15;13r | 1 Vκ; 0 VH |
| Single clones (mouse 2) | |||||
| Q10.4 | 23-43/Jk5 | 4; 3r | VHJ558.26.116/DFL16.1/JH1 γ2c | 2; 1r | 1 Vκ; 0 VH |
Empty cells in the second and fourth columns indicate the same information as the cell above.
Amino acid replacements due to somatic mutation.
J2.8 is γ2c; J7.13 is γ2b.
Members of this clone share an Arg replacement in VH CDR1 but as a result of different base substitutions (mutations).
Arg codon may be the result of either a somatic mutation in CDR3 or a P-element addition.
Finally, few of the clones showed signs of light chain receptor editing (Gay et al., 1993; Tiegs et al., 1993). None of the antibodies expressed a lambda light chain, and only 3 of the 12 clones used distal Jk5 or Jk4 gene segments (Table I). This is approximately the frequency expected for clones attempting to negotiate a productive rearrangement. Collectively, these structural observations were consistent with the idea that many of the chromatin-reactive B cells were derived from nonautoreactive B cells in secondary lymphoid tissues via the process of SHM.
A large autoreactive lineage derived from a nonautoreactive B cell
To test the mutation-founder idea conclusively, we reverted the 15 nonsynonymous mutations shared by all members of the large lineage #1 to regenerate codons expressed by the original B cell as it emerged in the bone marrow before SHM. These included the HCDR3 somatic mutations, as illustrated in Fig. 2 (top sequence). The primers used to construct the template for reversion mutagenesis were located distally with respect to the variable gene coding sequences to accurately recreate all features of the unmutated (and mutated) variable genes, including the leader sequences. This was done because mutations in leader sequences can affect antibody binding (Ping et al., 1993). The reverted constructs were transfected in the context of an IgG2b genomic expression vector into SP2/0 cells to produce antibody, which was affinity purified and treated with DNase and 1 M NaCl to remove potential contaminating nuclear antigens as previously described (Guth et al., 2003). This was done because unpurified ANAs from culture supernatants can display altered binding specificity and avidity profiles as a result of contaminating nuclear antigens that may form a bridge between the antibody and an intended target antigen in immunoassays.
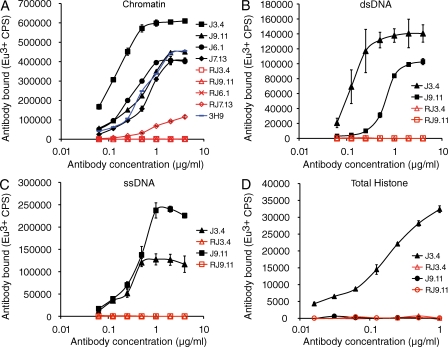
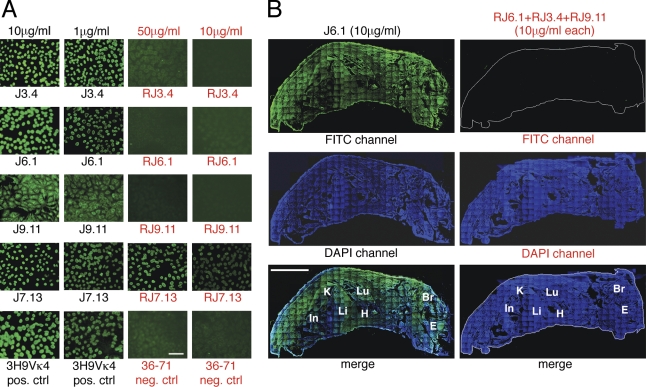
Binding immunoassays revealed that the revertant Ab of lineage #1 lost all measurable affinity for chromatin, as well as for various nuclear antigen subspecificities (Fig. 4). The revertant also failed to demonstrate detectable antinuclear activity against fixed HEp-2 cells by immunofluorescence, even when used at a substantially higher concentration than that of the original mutant antibody (Fig. 5 A). Finally, the revertant failed to bind detectably any tissue antigen in frozen sections of whole neonatal mice, as assessed by immunofluorescence (Fig. 5 B). To validate our IgG2b transfection vector and procedure, we also reconstructed the original mutant antibody and expressed it in SP2/0 cells. The mutant Ab generated in this way retained its original activity, as expected (unpublished data). On the basis of these results, we conclude that the largest chromatin-reactive clone was derived from a normal B cell whose antinuclear specificity was created by SHM.
Figure 4.
Chromatin binding assays for monoclonal antibodies and engineered revertants lacking somatic mutations. Standard errors are shown. R and red font color denote germline revertants. Two of the original monoclonal antibodies (J3.4 and J9.11) demonstrated binding activity against several chromatin components as shown in B–D. All antibodies were purified by a stringent affinity method designed to remove potentially contaminating nuclear antigens (Guth et al., 2003). A prototypical ANA, 3H9/Vκ4, is shown in blue in A. Bound antibodies were detected in a solid-phase europium (Eu3+)-based fluoroimmunometric assay (as counts per second). One of three experiments with similar results is shown.
Figure 5.
Immunofluorescence staining of HEp-2 cells and whole frozen sections of neonatal mouse with monoclonal antibodies and engineered revertants without somatic mutations. (A) Stains of fixed HEp-2 cells. R and red font color denote revertant. Note higher concentrations used for revertant antibodies. 3H9/Vκ4 and 36–71 served as positive and negative controls, respectively. The experiment was performed three times. Bar, 100 µm. (B) Stains of whole frozen sections of neonatal mice. Mutant antibody J6.1 served as positive control. Experimental section was stained with a mixture of revertant antibodies (RJ3.4, RJ9.11, and RJ6.1), each at a concentration equivalent to that of the positive control J6.1 mAb. Bound positive antibodies were detected with an FITC-coupled sheep anti–mouse IgG (γ-chain specific). Sections were counterstained with DAPI (blue) to highlight organs (In, intestine; Li, liver; K, kidney; Lu, lung; H, heart; Br, brain; E, eye). One of two experiments with similar results is shown. Bar, 3 mm.
Other autoreactive clones created by SHM
To determine whether mutation-generated autoreactivity in the large clone was the exception or the rule, we interrogated additional independent clones by reversion analysis. As a rigorous test, we initially chose three clones with characteristics suggesting that they might have emerged with antinuclear BCR directly from the bone marrow. Lineage #2 was chosen because no Arg replacement mutations were shared by its two members. Lineage #4 was selected because its two members used a Jk5 gene segment, suggesting that the clone may have exhausted options for receptor editing in attempts to replace an autoreactive BCR. A third clone (J9.11) was chosen because its antibody V genes contained the fewest number of nonsynonymous somatic mutations (only 2) among the 30 clones that were examined. In addition, it also used a distal Jk4 gene segment. All antibodies were purified as described for lineage #1 and tested in various binding assays. Despite their structural features, each of these clones sustained dramatic losses in autoreactivity activity upon mutation reversion, completely in two cases and ∼50-fold in the third (Figs. 4 and 5). In addition, neither the revertants nor the original mutant mAb bound to cardiolipin detectably (Fig. S1). As with lineage #1, the transfection and expression methods were validated by recreating and expressing the three mutants in the context of the IgG2b expression vector. In each case, autoreactivity was preserved.
As a final test, we reverted the somatic mutations in six additional ANA-producing clones. Two of these were from a second 8-mo-old autoimmune female mouse with serum anti-chromatin IgG, although of a lower titer than previous mouse. Only ∼0.5% of the hybridomas (4/832) from this animal was initially scored positive for IgG anti-chromatin. One of these clones was represented by two hybridomas (Fig. 3). The other 4 clones were from the original set of 12 derived from the first animal. In every case, the germline revertants lost all detectable autoreactivity as assessed in solid-phase binding assays against chromatin, in immunofluorescence assays against fixed HEp-2 cells, and in immunofluorescence assays against whole frozen sections of neonatal mice (Figs. S1–4). It is clear from these analyses that SHM frequently created antinuclear clones from nonautoreactive precursors; in 9 of 10 clones, SHM was responsible for all detectable autoreactivity and in the 10th clone for almost all of it.
The precursor to an ANA-producing clone generated by VH gene replacement
The absence of Tdt in these autoimmune mice enabled us to define the origins of nucleotides at junctional boundaries of assembled V, D, and J gene segments encoding the ANA in our panel. Clone Q10.4 was unusual because it had 8 bases at the VH/D boundary that were derived neither from the corresponding VHJ558.26.116 and DFL16.1 gene segments nor from a P-element addition (Fig. 6 A). To determine if this short sequence was a residual footprint left by a VH gene replacement event (Kleinfield et al., 1986; Reth et al., 1986; Zhang, 2007), we searched 1.2 megabases of B6 genomic DNA located downstream of the VHJ558.26.116 gene for this sequence (GAGACAGA). This region of DNA included all VH genes located between VHJ885.26.116 and the D gene segment cluster, any one of which was a potential candidate for replacement by a secondary rearrangement involving VHJ558.26.116. The GAGACAGA sequence was found 60 times. For each of the 60 hits, we extracted the preceding 300 bases and performed an Ig BLAST search to determine if they encoded a VH gene in any reading frame. This analysis produced one VH gene (VH12.1.78) that contained the GAGACAGA sequence precisely at its 3′ end. This VH gene also contained a cryptic heptamer recombination signal sequence located 3 bases immediately 5′ of GAGACAGA. When the translational reading frame of VH12.1.78 was extended through the recombined VH/D junction, it terminated in two consecutive stop codons within the DFL16.1 gene segment (Fig. 6 B). These observations reveal that the Q10.4 ANA clone had traversed a circuitous pathway to autoimmunity. It had originally carried a nonproductive VH12.1.78/DFL16.1/JH1 gene in which VH12.1.78 was subsequently replaced by VHJ558.26.116 to generate a productive heavy chain gene. After Vk gene rearrangement, it is possible that the original B cell was autoreactive and underwent receptor editing, as suggested by the use of Jk5. Alternatively, several Vk recombination events terminating in a Vk-Jk5 exon may have been required to produce the first functional kappa gene. Either way, subsequent SHM then generated the autoreactive nuclear specificity in this cell. To our knowledge, this is the first demonstration of an endogenous VH gene replacement that corrected a naturally occurring nonproductive V/D/J rearrangement.
Figure 6.
Functional Igh gene for the Q10.4 hybridoma created by VH replacement. (A) The Q10.4 VH/D/JH boundary sequence illustrating an 8-base sequence remaining from the original VH12.1.78 rearrangement in red. (B) The original VH12.1.78 rearrangement showing the nonproductive translational reading frame ending in consecutive stop codons (red). The cryptic heptamer recombination signal sequence within VH12.1.78 is shown in blue. The 8-base sequence of VH12.1.78 that is destined to remain after replacement by VHJ558.26.116 is also shown in red.
A propensity for arginine mutations at specific serine codons
Many studies have indicated the important role of Arg residues in antinuclear activity (Radic and Weigert, 1994; Kalsi et al., 1996; Krishnan et al., 1996; Jang et al., 1998; Li et al., 2000; Rahman et al., 2001; Tanner et al., 2001; Guth et al., 2003; Haley et al., 2004). In agreement with this, when the single Arg replacement mutation in the J9.11 clone (Vk CDR1) was reverted to a germline serine codon, there was nearly a complete loss of anti-chromatin activity in the resultant antibody (Fig. S5). Arg mutations can also impart positive charges to antibodies, and there are studies reporting that cationic ANAs are prone to deposit in the kidney and induce pathology (Ebling and Hahn, 1980; Dang and Harbeck, 1984). Although several of our antibody V regions with Arg mutations had relatively high calculated isoelectric points (pI = 9.1–9.4), we did not see evidence of pathology when they were injected (as hybridomas) into irradiated BALB/c mice (unpublished data).
A close inspection of all Arg mutations within our panel of hybridomas revealed that a majority of them occurred at AGC and AGT serine codons. 40% of independent Arg mutations in VH and Vk genes expressed by our hybridoma panel were at AGC (Table II). This is likely a result of the fact that AGC is the most intrinsically favored triplet target of SHM and because AGC can mutate to an arginine codon by any 1 of 3 different single base changes (Smith et al., 1996). In addition, the AGC codon is well represented in Ig V genes of our hybridoma panel; it is present in the Vk and VH genes at approximately two and three times the frequency expected for random codon use, respectively. Although AGT is not a prefered target of SHM, it is also well represented in the VH and Vk genes of our panel, at 2.2 and 6.6× the frequency expected for random codon use (Table II). Moreover, the combined frequencies of AGC and AGT in CDR1 and CDR2 are even higher, at 4.3 (VH) and 7.1 (Vk)× that expected. This is reminiscent of an observation reported by Radic and Weigert (1994) that among the six serine codons, AGC and AGT were overrepresented in CDR of a sample of ANA V genes. Collectively, mutations at AGC and AGT accounted for 66% (20/30) of all Arg mutations in the V genes of our panel.
Table II.
Frequent Arg mutations at AGC and AGT codons in V genes
| V genes | Arg mutations at AGC | Arg mutations at AGT | AGC codons in V genesa | AGT codons in V genes | AGC+AGT CDR1 and CDR2 | AGC+AGT FR1 and FR2 |
| % | % | % | % | % | % | |
| VH | 41.7 | 16.7 | 3.7 | 2.0 | 10.8 | 1.8 |
| Vκ | 38.9 | 33.3 | 3.0 | 6.1 | 17.9 | 1.9 |
This table refers to anti-chromatin antibodies in this study.
AGC and AGT codon usage among 52,926 mouse genomic codons is 1.6 and 0.92%, respectively (GenBank: http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=10092).
These observations prompted us to analyze codon use by all C57BL/6 Ig VH and Vk genes listed in the Ig Blast public database (http://www.ncbi.nlm.nih.gov/igblast/). This analysis revealed that AGC was used ∼1.9× more frequently in VH genes and 1.6× more frequently in Vk genes than expected on the basis of either random codon use or actual codon use by other mouse genes. AGT use by mouse V genes was even higher (Table III). Finally, similar results for both codons were seen when all human VH and Vk and Vλ genes in the BLAST Ig database were analyzed. The results were nearly the same when only one allele (listed first in the database) for each of the human V genes was analyzed. Collectively, these observations support the idea that SHM frequently creates nuclear-reactive B cell clones from normal B cell clones and that this occurs by mutations that convert AGC and AGT serine codons to arginine codons.
Table III.
Frequent use of AGC and AGT codons in antibody V genes
| V genes | AGC codon use | AGT codon use | ||
| All mouse V genesa | All human V genesb | All mouse V genes | All human V genes | |
| % | % | % | % | |
| VH | 3.1 | 3.4 | 1.67 | 2.2 |
| Vκ | 2.6 | 3.9 | 5.5 | 4.7 |
| Vλ | 2.4 | 3.4 | 2.4 | 2.4 |
This table shows V gene sequences taken directly from Ig BLAST (http://www.ncbi.nlm.nih.gov/igblast/). A majority of the Ig BLAST sequences were from the international ImMunoGeneTics information system (IMGT; http://www.imgt.org).
AGC and AGT codon usage among 52,926 mouse genomic codons is 1.6 and 0.92%, respectively (GenBank: http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=10092).
AGC and AGT codon usage among 40,662,582 human genomic codons is 1.95 and 1.21%, respectively (GenBank: http://www.kazusa.or.jp/codon/cgi-bin/showcodon.cgi?species=9606).
DISCUSSION
The results of this study provide conclusive evidence that SHM generates spontaneous autoreactive B cells that participate in systemic autoimmunity. By generating ANA-producing hybridomas from mice with a homozygous genetic deficiency in Tdt and heterozygous deficiencies at the Igh and Igk loci, we were able to unequivocally identify all of the somatic mutations in V region genes of spontaneous ANA-producing B cells and to test them functionally in reversion analyses. The requirement of somatic mutations for antinuclear activity was absolute in 9 of 10 clones analyzed and included a large lineage comprising approximately half of the sampled B cells. A predominant fraction of mutations producing Arg codons occurred at AGC and AGT serine codons, which are abundant in V genes and able to mutate to an Arg codon by 3 alternative base changes. AGC is also the most intrinsically mutable codon substrate for SHM. Considering the importance of Arg residues in antinuclear activity, these findings collectively support the idea that SHM routinely generates ANA-specific B cells during T cell–dependent immunity.
We are confident that all functionally relevant binding was lost in nine of the clones examined because multivalent immunoassays are so sensitive that they can reveal binding interactions that are below the threshold required for immune recruitment. In the arsonate (Ars) model, for example, a canonical antibody can be readily detected in immunoassays with protein conjugates of sulfanilic acid (Sulf), even though immunization with Sulf-carrier protein does not recruit cells producing this antibody (Fish et al., 1989). Clone J7.13 is the one exception that retained a small measure of antinuclear activity after reversion, raising the possibility that it escaped all tolerance checkpoints in B cell development. Alternatively, the avidity of the germline antecedent of J7.13 for self-antigen may be physiologically irrelevant and insufficient to induce a tolerant phenotype. It is noteworthy that clone J7.13 used a Jk1 gene segment. This could reflect insufficient avidity for self-antigen to induce receptor editing or a defect in receptor editing in B6.Nba2 mice. Distinguishing between these can only be determined by generating mice that carry an unmutated version of this BCR and comparatively analyzing the resultant B cells in B6 and B6.Nba2 mice.
Our approach avoided caveats that previously obscured complete identification of somatic mutations in disease-associated autoreactive B cells. Chief among these was the undefined nature of unmutated CDR3 sequences in antecedents to autoimmune clones. This was resolved by an absence of Tdt in our mice. The high resolution imparted by a Tdt deficiency is underscored by the fact that it enabled us to define, for the first time, an endogenous VH gene replacement that corrected a naturally generated nonproductive V/D/J gene. The subset of prior studies that interpreted somatic mutations as being critical to ANA, particularly those involving human ANA, did not have the benefit of genetic ablations to define the role of somatic mutations in CDR3 and of secondary rearrangements to autoreactivity. Nevertheless, these preceding studies framed an important issue and provided the first evidence that somatic mutations were required for the generation of disease-associated ANA (Radic et al., 1989, 1993; Siminovitch et al., 1989; Xu and Davis, 2000; Wellmann et al., 2005; Lambrianides et al., 2007; Mietzner et al., 2008). Whether the conclusion of our study applies to autoantibodies in other autoimmune diseases is unknown. It is noteworthy, however, that in young children that progress to type I diabetes, the first anti-insulin antibodies detected in serum are high-avidity IgGs (Palmer et al., 1983; Yu et al., 2000). This may be an indication that their insulin specificities are generated by SHM.
In testing the relevance of somatic mutations to the antinuclear activity of a given antibody, we took the precaution of eliminating all of them. This was done because preservation or improvement of a germline-determined affinity might require several somatic mutations, whereas a subset of the same mutations might be destructive. One somatic mutation might be functionally destructive if acquired before, but beneficial if acquired after, another mutation. The hybridoma sampling approach was also an important component of our approach. Many studies have demonstrated a strong concordance between serum antibodies and mAb produced by hybridomas sampled in physiological and disease settings (Reth et al., 1978; Griffiths et al., 1984; Wysocki et al., 1986; Shlomchik et al., 1987b). However, the relationship between serum ANA and receptors expressed by B cells bearing markers of memory is not as well defined. Cells sorted for memory markers might include a tolerant category, as suggested by the high frequency of antinuclear BCR specificities observed in such cells isolated from nonautoimmune individuals (Tiller et al., 2007) and by the observation that in mouse models of immunity to foreign antigens, autoreactivity tends to be purged during memory B cell development (Notidis et al., 2002; Guay et al., 2004).
The patterns and frequencies of somatic mutations in our clones indicated that autoimmune B cells were generated at both early and late stages after induction of SHM. For example, some of the mutant antinuclear B cells carried few mutations, whereas members of the large lineage shared 17 somatic mutations, indicating strong selection at or near the time that the last shared mutation was acquired. SHM normally diversifies antibody genes during physiological responses to foreign immunogen within the microenvironment of the germinal center (GC). Therefore, the most straightforward interpretation of our results is that the mutant antinuclear B cells were generated de novo at this time (Apel and Berek, 1990; Jacob et al., 1991). Although we favor this idea, it is conceivable that SHM and selection are not properly regulated and occur elsewhere in B6.Nba2 mice. SHM outside of the GC has been reported for rheumatoid-factor B cells in one model of autoimmunity (William et al., 2002). Either way, our results support the counterintuitive conclusion that a developing B cell emerging from the bone marrow with a normal nonautoreactive BCR may be more dangerous than one arising with an autoreactive BCR.
In autoimmune-prone mice that express transgenic antinuclear BCR, the autoimmune clones that ultimately arise in disease often express altered BCRs that are products of receptor editing. One possible explanation is that upon receptor editing, the precursors either replaced one autoreactive BCR with another or were unable to dilute the autoreactive BCR by expressing a second benign heavy or light chain gene. Our findings suggest an alternative possibility. Editing may have been functionally successful, thus permitting the B cells to survive early self-tolerance checkpoints, become engaged in an immune response, and undergo SHM to become autoreactive. A mature B cell that gains autoreactivity during immunity has to escape fewer tolerance checkpoints than one that emerges in the bone marrow with an autoreactive BCR.
Several observations support the idea that SHM frequently creates autoreactive B cells during the GC reaction. The varying numbers of shared somatic mutations in our lineages indicate that mutations creating antinuclear BCR can apparently be acquired at early or late time points after the commencement of SHM. In addition, there appears to be a bias favoring somatic mutations that generate Arg codons because of an abundance of specific serine codons in antibody V genes that have an intrinsic bias to mutate to Arg codons or are preferred targets of SHM. Nevertheless, previous studies have demonstrated one or more tolerance checkpoints at, or after, the GC phase of B cell differentiation (Hande et al., 1998; Culton et al., 2006). This raises an intriguing question: if ANA-specific B cells are frequently generated in the GC and must subsequently be censored, what is the major benefit of self-tolerance checkpoints during development of the preimmune B cell repertoire?
One possibility to consider is that early censorship offers the added benefit of preventing autoreactive B cells from competing with normal B cells for physical and functional niches in the immune system. As such, central tolerance in the preimmune repertoire may ensure that it is populated with B cells that are most likely to generate successful progeny if recruited into an immune response. From this perspective, receptor editing and deletion of autoreactive B cells in the preimmune repertoire may be viewed as quality control measures that ultimately enhance immune responses. The existence of receptor editing as a potential salvage operation supports the idea that generating a functional B cell repertoire is a difficult or energetically expensive prospect, otherwise deletion should suffice. This idea is also supported by microsequence substrate preferences of the SHM mechanism, which has an intrinsic target bias to limit structural damage to the V region (Dörner et al., 1997; Kepler, 1997; Shapiro et al., 2002). This quality control idea leads to the prediction that defects in receptor editing and deletion of autoreactive B cells in the preimmune repertoire would result in compromised humoral immunity.
MATERIALS AND METHODS
Mice.
B6.Nba2 congenic mice were described previously and were provided by S. Rozzo and B. Kotzin (University of Colorado Health Sciences Center; Denver, CO; Rozzo et al., 2001). Tdt-deficient mice were provided by D. Mathis (Harvard University, Cambridge, MA; Gilfillan et al., 1993). Kappa gene-deficient mice were provided by D. Huszar (AstraZeneca R&D Boston, Waltham, MA; Chen et al., 1993) and heavy chain gene–deficient mice (Gu et al., 1993) were purchased from The Jackson Laboratory. All loci were previously bred onto a B6 genetic background and intercrossed to generate B6.Nba2 Tdt −/− IgH+/− Igk+/− mice in the Biological Resource Center at National Jewish Health. All animals were handled according to a protocol approved by the Institutional Animal Care and Use Committee of National Jewish Health. Genotyping was performed by PCR with primers shown in Table S1.
Hybridoma production.
Hybridomas were generated from a 12-mo-old B6.Nba2 Tdt −/− IgH+/− Igk+/− spontaneously autoimmune mouse using the SP2/0/ mIL-6 fusion partner (Harris et al., 1992). Hybridoma culture supernatants were tested for the production of IgG antibodies in a Eu+-linked fluoroimmunoassay using a goat anti–mouse IgG H-chain–specific antibody (SouthernBiotech) as described in Antibody binding assays. Hybridomas that scored positive were cloned by limiting dilution. Heavy chain isotypes of monoclonal antibodies were determined by ELISA using an isotyping kit (SouthernBiotech).
Sequencing hybridoma V region genes.
Variable genes of the hybridomas were cloned with RT-PCR and 5′ rapid amplification of cDNA ends (RACE) using sets of constant region primers and anchor primers. Total RNA was isolated from the hybridomas with RNeasy (QIAGEN), and first-strand cDNA was synthesized with Superscript III RT (Invitrogen) using an oligo (dT) primer. First-strand cDNA was purified using QIAquick (QIAGEN), and a poly (dG) tail was added to 3′ ends with Tdt (Invitrogen) at 37°C for 20 min in the presence of 0.4 mM dGTP. First-round 5′ RACE PCR was performed on 5 µl of tailed cDNA using high-fidelity Phusion DNA polymerase (Finnzymes) with anchor primer 1 and a heavy chain or light chain constant region outer primer (Table S1) under the following conditions: an initial incubation at 98°C for 1 min; then 18 cycles at 98°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final incubation at 72°C for 10 min. Second-round PCR was performed on 1 µl of first-round PCR product with anchor primer 2 and a nested constant region heavy chain or light chain primer under the following conditions: an initial incubation at 98°C for 1 min; then 20 cycles of 98°C for 30 s, 60°C for 30 s, and 72°C for 1 min; and a final incubation at 72°C extension for 10 min. Heavy chain PCR products were extracted from 2% agarose gels and inserted into the pCR4-TOPO vector (Invitrogen). Light chain PCR products were purified with QIAquick and digested with restriction enzymes PflFI or PflmI (New England Biolabs, Inc.) to disrupt the rearranged Vk21-12 gene transcribed by the SP2/0 fusion partner (only three other kappa genes have both restriction sites). After 2% agarose gel electrophoresis of the PCR products, the uncut bands were extracted and inserted into pCR-4-TOPO for sequencing. DNA sequencing was performed with BigDye terminator mixtures in conjunction with a 3100 capillary sequencer (Applied Biosystems) at the University of Colorado Cancer Center DNA Sequencing and Analysis Core Facility (Aurora, CO).
Sequence analysis.
To identify germline precursors of expressed hybridoma VH and Vk gene segments, the latter were aligned against the entire B6 genome reference sequence (National Center for Biotechnology Information build 36) using mouse Blast (http://www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=10090) and Ig Blast (www.ncbi.nlm.nih.gov/projects/igblast). JH, JD, and Jk gene segment use and somatic mutations therein were determined manually by comparing hybridoma sequences to B6 germline correlates. VH and Vk gene designations in Table I were taken from Johnston et al. (2006) and from Brekke and Garrard (2004), respectively. Heavy and light chain sequences were combined into single files for each member of a clone and then aligned and analyzed with the CustalW analysis function of the program MacVector (v7.2.2) to generate lineage dendrograms.
Site-directed mutagenesis and expression of antibodies.
The Ig variable genes were amplified from genomic DNA and cloned into the pCR-4-TOPO using sense primers that hybridized upstream of promoter elements identified in Brekke and Garrard (2004) and Johnston et al. (2006) and antisense primers that hybridized to intronic sequences immediately 3′ of expressed JH or Jk gene segments (Table I). Resulting plasmids were used as templates for germline primers (Table S1) in a multiple-site mutagenesis protocol (Sawano and Miyawaki, 2000), modified by increasing the denaturation temperature to 98°C, using Phusion DNA polymerase in PCR and by transforming TOP10 E. coli (Invitrogen). The original mutated VH-D-JH or revertant VH-D-JH genes were excised with EcoR1 and HindIII and ligated into a genomic IgG2b expression vector as previously described (Zhang et al., 2001). Light chain fragments were cloned into the EcoRI site of a genomic expression vector containing both the intronic and 3′ kappa enhancer elements. SP2/0/mIL-6 cells were cotransfected with paired heavy and light chain genomic constructs. Mycophenolic acid–resistant transfectomas were screened for the production of IgG antibody. Positive transfectomas were cloned by limiting dilution in 96-well trays. Heavy and light chain pairing was confirmed in a sandwich europium assay, using microtiter trays coated with 1 µg/ml goat anti–mouse kappa and developed with 0.5 µg/ml biotin-labeled goat anti–mouse IgG Ab as described in Antibody binding assays.
Antibody binding assays.
Antibodies were purified by two rounds of affinity purification and treatments with DNase I and NaCl to remove associated chromatin as previously described (Guth et al., 2003). Antibody purity was assessed by SDS-PAGE. To test for dsDNA binding, 96-well microtiter plates (Microlon high-binding; Greiner Bio-One Inc.) were coated with 0.01% poly-L-lysine (Sigma-Aldrich) followed by 10 µg/ml dsDNA overnight at 4°C. 96-well trays were directly coated (no poly-lysine) with the following: calf thymic chromatin; a mixture of 10 µg/ml of total histones H1, H2A, H2B, H3, and H4 (Roche); and 10 µg/ml ssDNA or cardiolipin in ethanol. Control plates were coated with blocking buffer (PBS with 2 mg/ml BSA, 1 mg/ml gelatin, 0.05% Tween-20, and 0.01% thimerosol) alone. With the exception of the streptavidin-europium binding and release steps, all subsequent steps were performed in blocking buffer with 1 mM EDTA to prevent nonspecific adherence of antibody and detection reagents and degradation of DNA, as previously described (Guth et al., 2003). Experimental mAb at defined concentrations was added to the plates and detected with 0.5 µg/ml biotin-labeled goat anti–mouse IgG followed by 50 ng/ml streptavidin-conjugated Eu3+ (PerkinElmer). Eu3+ was detected with a time-resolved fluorometer (VICTOR2; PerkinElmer) as previously described (Guth et al., 2003).
Immunofluorescence assays.
mAbs were tested for autoreactivity against fixed HEp-2 cells (Bio-Rad Laboratories) and against frozen sections of a whole neonatal mouse adhered to ProbeOn Plus microscope slides (Thermo Fisher Scientific). Slides were incubated with mAb for 30 min, washed with PBS for 5 min, and incubated with FITC-labeled sheep anti–mouse IgG antibody (1/200; Sigma-Aldrich) for 30 min. For frozen sections, slides were covered with mounting medium containing DAPI (Vector Laboratories). Pictures were taken with an inverted microscope (Axiovert 200M; Carl Zeiss, Inc.) at 200× (HEp-2) or 100× (frozen sections) magnification. A montage of images taken of frozen sections was assembled using Slidebook 4.1 software (Intelligent Imaging Innovations Inc.).
Online supplemental material.
Fig. S1 shows an anti-cardiolipin binding assay performed with ANA and engineered revertants. Fig. S2 shows an anti-chromatin binding assay with ANA and engineered revertants. Fig. S3 shows immunofluorescent staining of HEp-2 cells with six ANAs and engineered revertants not represented in Fig. 5 A. Fig. S4 shows immunofluorescent staining of whole frozen sections of a neonatal mouse with six revertants not analyzed in Fig. 5 B. Fig. S5 demonstrates that a single kappa Arg mutation is required for high-avidity binding by the J9.11 clone.Table S1 is a composition of all primers used for amplification and cloning throughout the project. Supplemental data shows the sequences of the mutated antibody V regions that were subjected to reversion analysis. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092712/DC1.
Acknowledgments
The authors thank Drs. Tim Manser, John Cambier, Ed Palmer, and Michael Holers for their helpful critique and suggestions, Drs. Diane Mathis, Dennis Huszar, Brian Kotzin, and Stephen Rozzo for the Tdt−/−, Igk −/− mice, and B6.Nba2 mice, respectively, and Dr. Judith Spiegel for proof reading the manuscript.
This work was supported by grants from the National Institutes of Health #R01AI033613, #R01AI073945, #T32 AI007405, and #R03 AI088408.
The authors have no conflicting financial interest.
Footnotes
Abbreviations used:
- ANA
- antinuclear antibody
- BCR
- B cell receptor
- CDR3
- third complementarity-determining region
- dsDNA
- double-stranded DNA
- GC
- germinal center
- HCDR3
- heavy chain CDR3
- SHM
- somatic hypermutation
- SLE
- systemic lupus erythematosus
- Tdt
- terminal deoxynucleotidyl transferase
- TLR
- toll-like receptor
References
- Apel M., Berek C. 1990. Somatic mutations in antibodies expressed by germinal centre B cells early after primary immunization. Int. Immunol. 2:813–819 10.1093/intimm/2.9.813 [DOI] [PubMed] [Google Scholar]
- Brard F., Shannon M., Prak E.L., Litwin S., Weigert M. 1999. Somatic mutation and light chain rearrangement generate autoimmunity in anti–single-stranded DNA transgenic MRL/lpr mice. J. Exp. Med. 190:691–704 10.1084/jem.190.5.691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekke K.M., Garrard W.T. 2004. Assembly and analysis of the mouse immunoglobulin kappa gene sequence. Immunogenetics. 56:490–505 10.1007/s00251-004-0659-0 [DOI] [PubMed] [Google Scholar]
- Chen C., Li H., Tian Q., Beardall M., Xu Y., Casanova N., Weigert M. 2006. Selection of anti-double-stranded DNA B cells in autoimmune MRL-lpr/lpr mice. J. Immunol. 176:5183–5190 [DOI] [PubMed] [Google Scholar]
- Chen J., Trounstine M., Kurahara C., Young F., Kuo C.C., Xu Y., Loring J.F., Alt F.W., Huszar D. 1993. B cell development in mice that lack one or both immunoglobulin kappa light chain genes. EMBO J. 12:821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M., Dono M., Gazzola P., Roncella S., Valetto A., Chiorazzi N., Mancardi G.L., Ferrarini M. 2000. Accumulation of clonally related B lymphocytes in the cerebrospinal fluid of multiple sclerosis patients. J. Immunol. 164:2782–2789 [DOI] [PubMed] [Google Scholar]
- Conde C., Weller S., Gilfillan S., Marcellin L., Martin T., Pasquali J.L. 1998. Terminal deoxynucleotidyl transferase deficiency reduces the incidence of autoimmune nephritis in (New Zealand Black x New Zealand White)F1 mice. J. Immunol. 161:7023–7030 [PubMed] [Google Scholar]
- Culton D.A., O’Conner B.P., Conway K.L., Diz R., Rutan J., Vilen B.J., Clarke S.H. 2006. Early preplasma cells define a tolerance checkpoint for autoreactive B cells. J. Immunol. 176:790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang H., Harbeck R.J. 1984. The in vivo and in vitro glomerular deposition of isolated anti-double-stranded-DNA antibodies in NZB/W mice. Clin. Immunol. Immunopathol. 30:265–278 10.1016/0090-1229(84)90061-8 [DOI] [PubMed] [Google Scholar]
- Dörner T., Brezinschek H.P., Brezinschek R.I., Foster S.J., Domiati-Saad R., Lipsky P.E. 1997. Analysis of the frequency and pattern of somatic mutations within nonproductively rearranged human variable heavy chain genes. J. Immunol. 158:2779–2789 [PubMed] [Google Scholar]
- Ebling F., Hahn B.H. 1980. Restricted subpopulations of DNA antibodies in kidneys of mice with systemic lupus. Comparison of antibodies in serum and renal eluates. Arthritis Rheum. 23:392–403 10.1002/art.1780230402 [DOI] [PubMed] [Google Scholar]
- Fairhurst A.M., Wandstrat A.E., Wakeland E.K. 2006. Systemic lupus erythematosus: multiple immunological phenotypes in a complex genetic disease. Adv. Immunol. 92:1–69 10.1016/S0065-2776(06)92001-X [DOI] [PubMed] [Google Scholar]
- Fish S., Zenowich E., Fleming M., Manser T. 1989. Molecular analysis of original antigenic sin. I. Clonal selection, somatic mutation, and isotype switching during a memory B cell response. J. Exp. Med. 170:1191–1209 10.1084/jem.170.4.1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay D., Saunders T., Camper S., Weigert M. 1993. Receptor editing: an approach by autoreactive B cells to escape tolerance. J. Exp. Med. 177:999–1008 10.1084/jem.177.4.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachino C., Padovan E., Lanzavecchia A. 1995. κ+λ+ dual receptor B cells are present in the human peripheral repertoire. J. Exp. Med. 181:1245–1250 10.1084/jem.181.3.1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan S., Dierich A., Lemeur M., Benoist C., Mathis D. 1993. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 261:1175–1178 10.1126/science.8356452 [DOI] [PubMed] [Google Scholar]
- Goodnow C.C., Sprent J., Fazekas de St Groth B., Vinuesa C.G. 2005. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature. 435:590–597 10.1038/nature03724 [DOI] [PubMed] [Google Scholar]
- Griffiths G.M., Berek C., Kaartinen M., Milstein C. 1984. Somatic mutation and the maturation of immune response to 2-phenyl oxazolone. Nature. 312:271–275 10.1038/312271a0 [DOI] [PubMed] [Google Scholar]
- Gu H., Zou Y.R., Rajewsky K. 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 73:1155–1164 10.1016/0092-8674(93)90644-6 [DOI] [PubMed] [Google Scholar]
- Guay H.M., Panarey L., Reed A.J., Caton A.J. 2004. Specificity-based negative selection of autoreactive B cells during memory formation. J. Immunol. 173:5485–5494 [DOI] [PubMed] [Google Scholar]
- Guth A.M., Zhang X., Smith D., Detanico T., Wysocki L.J. 2003. Chromatin specificity of anti-double-stranded DNA antibodies and a role for Arg residues in the third complementarity-determining region of the heavy chain. J. Immunol. 171:6260–6266 [DOI] [PubMed] [Google Scholar]
- Haley J., Mason L.J., Nagl S., Giles I., Latchman D.S., Isenberg D.A., Rahman A. 2004. Somatic mutations to arginine residues affect the binding of human monoclonal antibodies to DNA, histones, SmD and Ro antigen. Mol. Immunol. 40:745–758 10.1016/j.molimm.2003.10.018 [DOI] [PubMed] [Google Scholar]
- Hande S., Notidis E., Manser T. 1998. Bcl-2 obstructs negative selection of autoreactive, hypermutated antibody V regions during memory B cell development. Immunity. 8:189–198 10.1016/S1074-7613(00)80471-9 [DOI] [PubMed] [Google Scholar]
- Harris J.F., Hawley R.G., Hawley T.S., Crawford-Sharpe G.C. 1992. Increased frequency of both total and specific monoclonal antibody producing hybridomas using a fusion partner that constitutively expresses recombinant IL-6. J. Immunol. Methods. 148:199–207 10.1016/0022-1759(92)90173-Q [DOI] [PubMed] [Google Scholar]
- Jacob J., Kelsoe G., Rajewsky K., Weiss U. 1991. Intraclonal generation of antibody mutants in germinal centres. Nature. 354:389–392 10.1038/354389a0 [DOI] [PubMed] [Google Scholar]
- Jang Y.J., Sanford D., Chung H.Y., Baek S.Y., Stollar B.D. 1998. The structural basis for DNA binding by an anti-DNA autoantibody. Mol. Immunol. 35:1207–1217 10.1016/S0161-5890(98)00095-9 [DOI] [PubMed] [Google Scholar]
- Johnston C.M., Wood A.L., Bolland D.J., Corcoran A.E. 2006. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J. Immunol. 176:4221–4234 [DOI] [PubMed] [Google Scholar]
- Kalsi J.K., Martin A.C., Hirabayashi Y., Ehrenstein M., Longhurst C.M., Ravirajan C., Zvelebil M., Stollar B.D., Thornton J.M., Isenberg D.A. 1996. Functional and modelling studies of the binding of human monoclonal anti-DNA antibodies to DNA. Mol. Immunol. 33:471–483 10.1016/0161-5890(95)00138-7 [DOI] [PubMed] [Google Scholar]
- Kepler T.B. 1997. Codon bias and plasticity in immunoglobulins. Mol. Biol. Evol. 14:637–643 [DOI] [PubMed] [Google Scholar]
- Kleinfield R., Hardy R.R., Tarlinton D., Dangl J., Herzenberg L.A., Weigert M. 1986. Recombination between an expressed immunoglobulin heavy-chain gene and a germline variable gene segment in a Ly 1+ B-cell lymphoma. Nature. 322:843–846 10.1038/322843a0 [DOI] [PubMed] [Google Scholar]
- Krishnan M.R., Jou N.T., Marion T.N. 1996. Correlation between the amino acid position of arginine in VH-CDR3 and specificity for native DNA among autoimmune antibodies. J. Immunol. 157:2430–2439 [PubMed] [Google Scholar]
- Lafaille J.J., DeCloux A., Bonneville M., Takagaki Y., Tonegawa S. 1989. Junctional sequences of T cell receptor gamma delta genes: implications for gamma delta T cell lineages and for a novel intermediate of V-(D)-J joining. Cell. 59:859–870 10.1016/0092-8674(89)90609-0 [DOI] [PubMed] [Google Scholar]
- Lambrianides A., Giles I., Ioannou Y., Mason L., Latchman D.S., Manson J.J., Isenberg D.A., Rahman A. 2007. Arginine mutation alters binding of a human monoclonal antibody to antigens linked to systemic lupus erythematosus and the antiphospholipid syndrome. Arthritis Rheum. 56:2392–2401 10.1002/art.22743 [DOI] [PubMed] [Google Scholar]
- Li Z., Schettino E.W., Padlan E.A., Ikematsu H., Casali P. 2000. Structure-function analysis of a lupus anti-DNA autoantibody: central role of the heavy chain complementarity-determining region 3 Arg in binding of double- and single-stranded DNA. Eur. J. Immunol. 30:2015–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li H., Weigert M. 2002. Autoreactive B cells in the marginal zone that express dual receptors. J. Exp. Med. 195:181–188 10.1084/jem.20011453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe N.M., Laskowski R.A., Thornton J.M. 2001. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 29:2860–2874 10.1093/nar/29.13.2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandik-Nayak L., Seo S.J., Sokol C., Potts K.M., Bui A., Erikson J. 1999. MRL-lpr/lpr mice exhibit a defect in maintaining developmental arrest and follicular exclusion of anti–double-stranded DNA B cells. J. Exp. Med. 189:1799–1814 10.1084/jem.189.11.1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietzner B., Tsuiji M., Scheid J., Velinzon K., Tiller T., Abraham K., Gonzalez J.B., Pascual V., Stichweh D., Wardemann H., Nussenzweig M.C. 2008. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc. Natl. Acad. Sci. USA. 105:9727–9732 10.1073/pnas.0803644105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molano I.D., Redmond S., Sekine H., Zhang X.K., Reilly C., Hutchison F., Ruiz P., Gilkeson G.S. 2003. Effect of genetic deficiency of terminal deoxynucleotidyl transferase on autoantibody production and renal disease in MRL/lpr mice. Clin. Immunol. 107:186–197 10.1016/S1521-6616(03)00035-4 [DOI] [PubMed] [Google Scholar]
- Nemazee D. 2006. Receptor editing in lymphocyte development and central tolerance. Nat. Rev. Immunol. 6:728–740 10.1038/nri1939 [DOI] [PubMed] [Google Scholar]
- Notidis E., Heltemes L., Manser T. 2002. Dominant, hierarchical induction of peripheral tolerance during foreign antigen-driven B cell development. Immunity. 17:317–327 10.1016/S1074-7613(02)00392-8 [DOI] [PubMed] [Google Scholar]
- Palmer J.P., Asplin C.M., Clemons P., Lyen K., Tatpati O., Raghu P.K., Paquette T.L. 1983. Insulin antibodies in insulin-dependent diabetics before insulin treatment. Science. 222:1337–1339 10.1126/science.6362005 [DOI] [PubMed] [Google Scholar]
- Ping J., Schildbach J.F., Shaw S.Y., Quertermous T., Novotny J., Bruccoleri R., Margolies M.N. 1993. Effect of heavy chain signal peptide mutations and NH2-terminal chain length on binding of anti-digoxin antibodies. J. Biol. Chem. 268:23000–23007 [PubMed] [Google Scholar]
- Radic M.Z., Weigert M. 1994. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annu. Rev. Immunol. 12:487–520 10.1146/annurev.iy.12.040194.002415 [DOI] [PubMed] [Google Scholar]
- Radic M.Z., Mascelli M.A., Erikson J., Shan H., Shlomchik M., Weigert M. 1989. Structural patterns in anti-DNA antibodies from MRL/lpr mice. Cold Spring Harb. Symp. Quant. Biol. 54:933–946 [DOI] [PubMed] [Google Scholar]
- Radic M.Z., Mackle J., Erikson J., Mol C., Anderson W.F., Weigert M. 1993. Residues that mediate DNA binding of autoimmune antibodies. J. Immunol. 150:4966–4977 [PubMed] [Google Scholar]
- Rahman A., Haley J., Radway-Bright E., Nagl S., Low D.G., Latchman D.S., Isenberg D.A. 2001. The importance of somatic mutations in the V(lambda) gene 2a2 in human monoclonal anti-DNA antibodies. J. Mol. Biol. 307:149–160 10.1006/jmbi.2000.4491 [DOI] [PubMed] [Google Scholar]
- Reth M., Hämmerling G.J., Rajewsky K. 1978. Analysis of the repertoire of anti-NP antibodies in C57BL/6 mice by cell fusion. I. Characterization of antibody families in the primary and hyperimmune response. Eur. J. Immunol. 8:393–400 10.1002/eji.1830080605 [DOI] [PubMed] [Google Scholar]
- Reth M., Gehrmann P., Petrac E., Wiese P. 1986. A novel VH to VHDJH joining mechanism in heavy-chain-negative (null) pre-B cells results in heavy-chain production. Nature. 322:840–842 10.1038/322840a0 [DOI] [PubMed] [Google Scholar]
- Rezanka L.J., Kenny J.J., Longo D.L. 2005. Dual isotype expressing B cells [kappa(+)/lambda(+)] arise during the ontogeny of B cells in the bone marrow of normal nontransgenic mice. Cell. Immunol. 238:38–48 10.1016/j.cellimm.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Rozzo S.J., Allard J.D., Choubey D., Vyse T.J., Izui S., Peltz G., Kotzin B.L. 2001. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 15:435–443 10.1016/S1074-7613(01)00196-0 [DOI] [PubMed] [Google Scholar]
- Sawano A., Miyawaki A. 2000. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28:E78 10.1093/nar/28.16.e78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan H., Shlomchik M.J., Marshak-Rothstein A., Pisetsky D.S., Litwin S., Weigert M.G. 1994. The mechanism of autoantibody production in an autoimmune MRL/lpr mouse. J. Immunol. 153:5104–5120 [PubMed] [Google Scholar]
- Shapiro G.S., Aviszus K., Murphy J., Wysocki L.J. 2002. Evolution of Ig DNA sequence to target specific base positions within codons for somatic hypermutation. J. Immunol. 168:2302–2306 [DOI] [PubMed] [Google Scholar]
- Shlomchik M.J. 2008. Sites and stages of autoreactive B cell activation and regulation. Immunity. 28:18–28 10.1016/j.immuni.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Shlomchik M.J., Aucoin A.H., Pisetsky D.S., Weigert M.G. 1987a. Structure and function of anti-DNA autoantibodies derived from a single autoimmune mouse. Proc. Natl. Acad. Sci. USA. 84:9150–9154 10.1073/pnas.84.24.9150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomchik M.J., Marshak-Rothstein A., Wolfowicz C.B., Rothstein T.L., Weigert M.G. 1987b. The role of clonal selection and somatic mutation in autoimmunity. Nature. 328:805–811 10.1038/328805a0 [DOI] [PubMed] [Google Scholar]
- Shlomchik M., Mascelli M., Shan H., Radic M.Z., Pisetsky D., Marshak-Rothstein A., Weigert M. 1990. Anti-DNA antibodies from autoimmune mice arise by clonal expansion and somatic mutation. J. Exp. Med. 171:265–292 10.1084/jem.171.1.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siminovitch K.A., Misener V., Kwong P.C., Song Q.L., Chen P.P. 1989. A natural autoantibody is encoded by germline heavy and lambda light chain variable region genes without somatic mutation. J. Clin. Invest. 84:1675–1678 10.1172/JCI114347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.S., Creadon G., Jena P.K., Portanova J.P., Kotzin B.L., Wysocki L.J. 1996. Di- and trinucleotide target preferences of somatic mutagenesis in normal and autoreactive B cells. J. Immunol. 156:2642–2652 [PubMed] [Google Scholar]
- Steeves M.A., Marion T.N. 2004. Tolerance to DNA in (NZB x NZW)F1 mice that inherit an anti-DNA V(H) as a conventional micro H chain transgene but not as a V(H) knock-in transgene. J. Immunol. 172:6568–6577 [DOI] [PubMed] [Google Scholar]
- Steitz T.A. 1990. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q. Rev. Biophys. 23:205–280 10.1017/S0033583500005552 [DOI] [PubMed] [Google Scholar]
- Tan E.M. 1989. Antinuclear antibodies: diagnostic markers for autoimmune diseases and probes for cell biology. Adv. Immunol. 44:93–151 10.1016/S0065-2776(08)60641-0 [DOI] [PubMed] [Google Scholar]
- Tanner J.J., Komissarov A.A., Deutscher S.L. 2001. Crystal structure of an antigen-binding fragment bound to single-stranded DNA. J. Mol. Biol. 314:807–822 10.1006/jmbi.2001.5178 [DOI] [PubMed] [Google Scholar]
- Theofilopoulos A.N., Dixon F.J. 1985. Murine models of systemic lupus erythematosus. Adv. Immunol. 37:269–390 10.1016/S0065-2776(08)60342-9 [DOI] [PubMed] [Google Scholar]
- Tiegs S.L., Russell D.M., Nemazee D. 1993. Receptor editing in self-reactive bone marrow B cells. J. Exp. Med. 177:1009–1020 10.1084/jem.177.4.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Tsuiji M., Yurasov S., Velinzon K., Nussenzweig M.C., Wardemann H. 2007. Autoreactivity in human IgG+ memory B cells. Immunity. 26:205–213 10.1016/j.immuni.2007.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao B.P., Cantor R.M., Kalunian K.C., Chen C.J., Badsha H., Singh R., Wallace D.J., Kitridou R.C., Chen S.L., Shen N., et al. 1997. Evidence for linkage of a candidate chromosome 1 region to human systemic lupus erythematosus. J. Clin. Invest. 99:725–731 10.1172/JCI119217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyse T.J., Rozzo S.J., Drake C.G., Izui S., Kotzin B.L. 1997. Control of multiple autoantibodies linked with a lupus nephritis susceptibility locus in New Zealand black mice. J. Immunol. 158:5566–5574 [PubMed] [Google Scholar]
- Wellmann U., Letz M., Herrmann M., Angermüller S., Kalden J.R., Winkler T.H. 2005. The evolution of human anti-double-stranded DNA autoantibodies. Proc. Natl. Acad. Sci. USA. 102:9258–9263 10.1073/pnas.0500132102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- William J., Euler C., Christensen S., Shlomchik M.J. 2002. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 297:2066–2070 10.1126/science.1073924 [DOI] [PubMed] [Google Scholar]
- Wysocki L., Manser T., Gefter M.L. 1986. Somatic evolution of variable region structures during an immune response. Proc. Natl. Acad. Sci. USA. 83:1847–1851 10.1073/pnas.83.6.1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.L., Davis M.M. 2000. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity. 13:37–45 10.1016/S1074-7613(00)00006-6 [DOI] [PubMed] [Google Scholar]
- Yachimovich-Cohen N., Fischel R., Bachar N., Yarkoni Y., Eilat D. 2003. Autoimmune NZB/NZW F1 mice utilize B cell receptor editing for generating high-affinity anti-dsDNA autoantibodies from low-affinity precursors. Eur. J. Immunol. 33:2469–2478 10.1002/eji.200324025 [DOI] [PubMed] [Google Scholar]
- Yu L., Robles D.T., Abiru N., Kaur P., Rewers M., Kelemen K., Eisenbarth G.S. 2000. Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proc. Natl. Acad. Sci. USA. 97:1701–1706 10.1073/pnas.040556697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Smith D.S., Guth A., Wysocki L.J. 2001. A receptor presentation hypothesis for T cell help that recruits autoreactive B cells. J. Immunol. 166:1562–1571 [DOI] [PubMed] [Google Scholar]
- Zhang Z. 2007. VH replacement in mice and humans. Trends Immunol. 28:132–137 10.1016/j.it.2007.01.003 [DOI] [PubMed] [Google Scholar]