Abstract
Progressive accumulation of PrPSc, a hallmark of prion diseases, occurs when conversion of PrPC into PrPSc is faster than PrPSc clearance. Engulfment of apoptotic bodies by phagocytes is mediated by Mfge8 (milk fat globule epidermal growth factor 8). In this study, we show that brain Mfge8 is primarily produced by astrocytes. Mfge8 ablation induced accelerated prion disease and reduced clearance of cerebellar apoptotic bodies in vivo, as well as excessive PrPSc accumulation and increased prion titers in prion-infected C57BL/6 × 129Sv mice and organotypic cerebellar slices derived therefrom. These phenotypes correlated with the presence of 129Sv genomic markers in hybrid mice and were not observed in inbred C57BL/6 Mfge8−/− mice, suggesting the existence of additional strain-specific genetic modifiers. Because Mfge8 receptors are expressed by microglia and depletion of microglia increases PrPSc accumulation in organotypic cerebellar slices, we conclude that engulfment of apoptotic bodies by microglia may be an important pathway of prion clearance controlled by astrocyte-borne Mfge8.
Transmissible spongiform encephalopathies (TSEs) are fatal neurodegenerative disorders afflicting many mammals (Aguzzi, 2006). Prions, the infectious particles that cause TSEs, consist mostly of PrPSc, a β-sheet–rich higher-order aggregate of the membrane protein PrPC (Prusiner, 1982). TSE-affected brains display neuronal vacuolation and loss, microglial activation, astrogliosis, and deposition of PrPSc (Prusiner et al., 1983; Weissmann, 2004).
The molecular mechanisms underlying brain damage in prion diseases are not well understood. Grafting experiments of wild-type brain tissue into PrPC-deficient brains showed that the neuropathological changes only occurred in tissue expressing PrPC, even if proteinase K (PK)–resistant PrPSc was also detected in the surrounding Prnpo/o tissue (Brandner et al., 1996). These results indicate that neurotoxicity depends on PrPC expression by the target cells, whereas PrPSc does not appear to be intrinsically toxic. This notion was confirmed by neuron-specific ablation of Prnp (Mallucci et al., 2003) and in mice expressing anchorless PrP, which is converted into a protease-resistant isoform and forms amyloid plaques yet causes minimal neuronal damage (Chesebro et al., 2005).
Prion diseases exhibit frequent neuronal apoptosis (Liberski et al., 2004). Although inhibition of apoptosis by overexpressing Bcl-2 or ablating Bax did not affect the life expectancy of prion-inoculated mice (Steele et al., 2007), prion-infected brain cells may release membrane fragments even when undergoing nonapoptotic death. Furthermore, exosomes may be released by perfectly healthy cells (Théry et al., 2009) and may conceivably carry prion infectivity.
A trait common to each of these extracellular vesicles is the surface exposure of phosphatidyl serine (PS), which can be recognized by the secreted ligand, Mfge8 (milk fat globule epidermal growth factor 8; Patton and Keenan, 1975). By virtue of its affinity to PS, Mfge8 helps mediating the removal of apoptotic bodies (Hanayama et al., 2002). Phagocytic cells then bind Mfge8-opsonized apoptotic cells through αvβ3 and αvβ5 integrins. Mfge8 is secreted by some phagocytic cells, including immature DCs and thioglycolate-activated peritoneal macrophages, as well as nonhematopoietic cells, including mammary epithelial cells (Hanayama and Nagata, 2005) and follicular DCs (FDCs; Kranich et al., 2008). A recent study described a potential involvement of Mfge8 expressed by human astrocytes, microglia, and smooth muscle cells in the removal of Aβ plaques (Boddaert et al., 2007). A microarray screen also identified Mfge8 expression in mouse astrocytes (Cahoy et al., 2008). Another study claimed Mfge8 expression in vitro by the microglial cell line BV-2 (Fuller and Van Eldik, 2008).
In this study, we show by in situ RNA hybridization (ISH) and quantitative RT-PCR that Mfge8 is primarily expressed by subsets of astrocytes in the central nervous system (CNS). Furthermore, Mfge8 deficiency resulted in accelerated prion pathogenesis and enhanced PrPSc accumulation in the CNS and was accompanied by elevated numbers of apoptotic cerebellar granule cells. These results suggest that Mfge8 is required for the efficient removal of apoptotic cells in the CNS and possibly also for degradation of prions.
RESULTS
Mfge8-deficient mice show accelerated prion pathogenesis
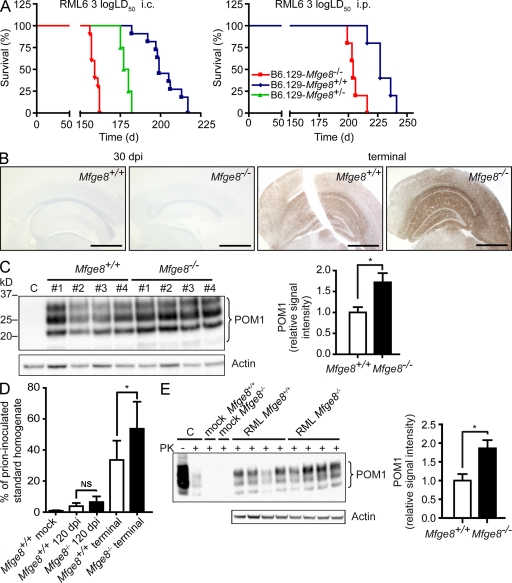
We inoculated Mfge8−/− mice (bred as intercrosses of the C57BL/6 and 129Sv mouse strains) i.c. (intracerebrally) or i.p. with RML6 (Rocky Mountain Laboratory strain, passage 6) prions (1,000 LD50 units). We monitored the mice for clinical signs of scrapie and defined the incubation period as the time until mice reached the terminal stage of disease. Mfge8−/− mice succumbed to scrapie much earlier than Mfge8+/+ mice. This acceleration was more pronounced after i.c. inoculation (∼40 d; Fig. 1 A, left) than after i.p. inoculation (∼20 d, Fig. 1 A, right), suggesting that it was caused by the absence of Mfge8 within the CNS rather than in extraneural compartments.
Figure 1.
Mfge8−/− mice show accelerated disease progression. (A) B6.129-Mfge8−/−, B6.129-Mfge8−/+, and B6.129-Mfge8+/+ were inoculated i.c. with 3 log LD50 (n = 10 for Mfge8−/−, n = 4 for Mfge8+/−, and n = 11 for Mfge8+/+) and i.p. with 3 log LD50 per group of RML6 prions. The mean incubation time after i.c. inoculation was significantly accelerated in Mfge8−/− mice (159 ± 2 d in Mfge8−/−, 179 ± 3 d in Mfge8+/−, and 202 ± 3.9 d in Mfge8+/+ mice; P < 0.0001, logrank). Differences in incubation time after i.p. inoculation were less pronounced (206 ± 6 d in Mfge8−/− and 229 ± 10 d in Mfge8+/+ mice) but still significant (P = 0.004, logrank test). (B) Stainings of brain sections after 30 dpi (left) or at terminal stage (right) using anti-PrP antibody SAF84. Bars, 1 mm. (C) Western blot of PK-digested brain homogenates from i.c. inoculated Mfge8+/+ and Mfge8−/− mice using anti-PrP antibody POM1 (C, uninoculated brain homogenate). Western blot for actin on non-PK–digested sample is shown below. Densitometric quantitation of POM1 signal normalized over actin expression. Bar graph shows mean relative signal intensity (n = 4; *, P = 0.03; Student’s t test). (D) MPA of brain homogenates from Mfge8+/+ and Mfge8−/− mice at 120 dpi or at terminal disease (n = 6–7 mice per group; *, P = 0.034; Student’s t test) indicated in percentages of RML6 prion–inoculated brain homogenate. Mfge8−/− mice sacrificed at 120 dpi showed a slight increase over Mfge8+/+ mice. (E) Mfge8+/+ and Mfge8−/− cerebellar slices were inoculated with 3 × 3 log LD50 RML6 prions or with noninfectious brain homogenate (mock), harvested after 5 wk, homogenized, and PK digested (C, brain homogenate from a terminal RML inoculated mouse used as positive control). PrPSc levels were analyzed by Western blotting using POM1 and normalized against actin (without PK digestion) by densitometry (bar graph; n = 4; *, P = 0.024; Student’s t test). Error bars represent SD. Histology, Western blot, and MPA results represent at least four independent experiments.
Because Mfge8 is highly expressed by FDCs (Kranich et al., 2008), which are important sites of extraneural prion replication (Aguzzi and Heikenwalder, 2005), we probed the splenic PrPSc content on PK-treated histoblots. Follicular PrPSc accumulation was similar in Mfge8+/+ and Mfge8−/− spleens (Fig. S1 A). Along with the accelerated pathogenesis of i.c. inoculated mice, this finding makes it unlikely that extraneural prion-modulating events caused the acceleration in Mfge8−/− mice. Therefore, we focused all further analyses on the interplay between Mfge8 and prions within the CNS.
Mfge8-deficient mice exhibit elevated levels of PrPSc
We reasoned that accelerated disease progression may result from enhanced accumulation of PrPSc, be it through increased production or through reduced degradation. This possibility was studied with four methodologies: qualitative assessment of PrP immunoreactivity in histological sections of terminally diseased brains (Fig. 1 B), quantitation of PrPSc chemiluminescence in Western blots of brain homogenates (Fig. 1 C), quantitation of aggregated PrP (PrPMPA) by the misfolded protein assay (MPA; Fig. 1 D; Lau et al., 2007), and infectivity titer determinations with the scrapie cell assay (SCA; Fig. S1 C). The MPA captures misfolded and aggregated proteins by precipitation with peptoid-coated magnetic beads; captured proteins are then disaggregated and quantitated by sandwich ELISA with anti-PrP antibodies (Polymenidou et al., 2008).
PrP immunoreactivity was much more intense in prion-infected Mfge8−/− brains than in Mfge8+/+ control brains (Fig. 1 B). Also, brains of terminally scrapie-sick Mfge8−/− mice displayed increased PK-resistant PrPSc and PrPMPA (P < 0.03 and P < 0.034, respectively; Fig. 1, C and D). This is noteworthy because Mfge8−/− mice attained terminal scrapie already at ∼160 d postinoculation (dpi), i.e., 40 d earlier than Mfge8+/+ mice. Because brain PrPSc accumulates exponentially over time, shorter incubation times typically go along with reduced PrPSc deposition and vice versa (Büeler et al., 1994). Therefore, we analyzed brain homogenates of preclinical Mfge8+/+ and Mfge8−/− mice sacrificed at 120 dpi by MPA. Already at this early time point, a tendency toward higher PrPSc levels was detected in Mfge8−/− brains, supporting the idea of accelerated PrPSc deposition (Fig. 1 D). PrPC concentrations, as measured by ELISA, did not differ between noninfected Mfge8+/+ and Mfge8−/− brain homogenates (Fig. S1 B), confirming that the increase in PrPSc was not caused by altered PrPC expression in Mfge8−/− mice.
Next, we asked whether the increased PrPSc deposition would lead to increased infectivity titers in Mfge8−/− brains. We compared brain homogenates from Mfge8+/+ and Mfge8−/− mice at terminal disease stage by SCA using the prion-susceptible neuroblastoma cell line PK1 (Klöhn et al., 2003). We observed a slight trend toward increased infectivity in Mfge8−/− brains (7.48 ± 0.07 TCI [infectious tissue culture] units and 7.30 ± 0.12 TCI units in Mfge8+/+ and Mfge8−/− brains, respectively), which, however, did not attain statistical significance (Fig. S1 C).
We also assessed PrPSc accumulation ex vivo using the prion organotypic slice culture assay (Falsig et al., 2008). Cerebellar Mfge8+/+ and Mfge8−/− slices were infected with 3,000 LD50 units of RML6 prions and harvested after 5 wk. Western blot analysis showed significantly higher (P < 0.024) PrPSc levels in Mfge8−/− brain slices (Fig. 1 E), confirming that increased prion accumulation in Mfge8−/− mice was entirely independent of any indirect effects caused by Mfge8 deficiency in extraneural compartments.
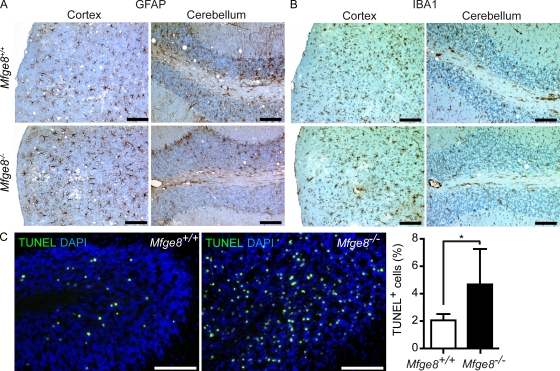
Next, we analyzed whether accelerated disease progression occurred concomitantly with additional pathological changes. The severity of astrogliosis, microgliosis (Fig. 2, A and B), and spongiosis (Fig. S1 D) at terminal disease was analyzed immunohistochemically. All of these parameters were similar in Mfge8+/+ and Mfge8−/− brains. Thus, the brain pathology of terminally scrapie-sick Mfge8−/− mice was as strong as in Mfge8+/+ mice, although terminal disease was reached 40 d earlier. We conclude that the processes leading to brain damage were also accelerated.
Figure 2.
Histopathological analysis of terminal Mfge8−/− brains. (A) Astroglia proliferation was analyzed by GFAP staining. (B) Microglial activation was visualized by IBA1 staining. (C) TUNEL staining on cerebellar cryosection of i.c. inoculated B6.129-Mfge8−/− (n = 5) and B6.129-Mfge8+/+ mice (n = 7). Sections were counterstained with DAPI. Bar graph shows quantitation of TUNEL+ cells from at least three cerebellar areas per mouse. Number of TUNEL+ cells was normalized against the number of DAPI+ nuclei. Graph shows percentage of TUNEL+ cells per area ± SD (*, P = 0.023; Student’s t test). Results summarize at least five independent experiments. Bars: (A and C) 100 µm; (B) 200 µm.
Increased prevalence of apoptotic cells in the cerebellum of Mfge8−/− mice
Beyond accelerated prion disease, Mfge8−/− mice have no obvious CNS defects. However, prion infections lead to significant apoptosis in the CNS, and the role of Mfge8 in the removal of apoptotic cells is well established (Hanayama et al., 2002, 2004; Kranich et al., 2008). If the absence of Mfge8 were to impair the removal of prion-infected apoptotic neurons, this might contribute to the rise in PrPSc in Mfge8−/− mice and could explain their shortened incubation time.
Therefore, we determined the prevalence of apoptotic cells by terminal transferase dUTP nick end labeling (TUNEL) in at least three cerebellar regions of five to seven Mfge8+/+ and Mfge8−/− mice. The mean frequency of TUNEL+ cells per region was normalized against the total number of DAPI+ nuclei in each region of interest. We detected an approximately twofold increase of TUNEL+ cells in cerebelli of terminally sick Mfge8−/− mice (P = 0.023; Fig. 2 C). In other brain areas, TUNEL+ cells were rare, and no difference was detected between Mfge8−/− and Mfge8+/+ mice (unpublished data). This finding suggests that Mfge8 is involved in the removal of prion-damaged cells in the cerebellum. The paucity of apoptotic cells in other areas implies that factors other than Mfge8 ensure rapid removal of dead cells there.
Astrocytes express Mfge8
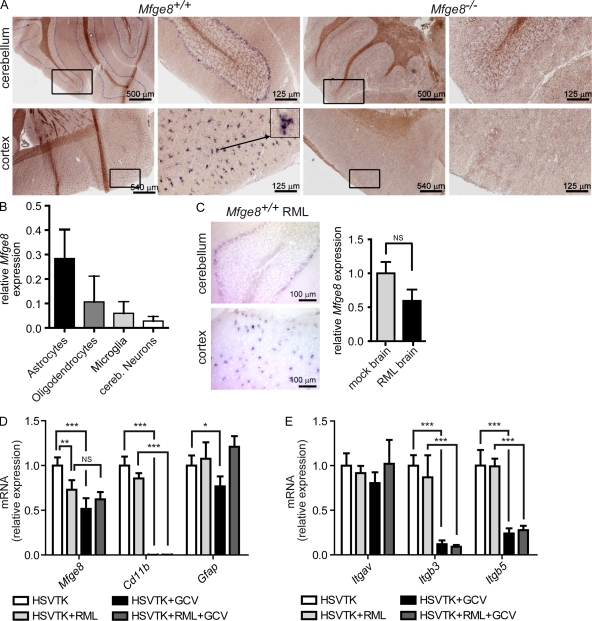
The cellular source of Mfge8 in the CNS is not well characterized. Human and mouse astrocytes, microglia, and smooth muscle cells (Boddaert et al., 2007; Fuller and Van Eldik, 2008) were reported to produce Mfge8, suggesting that microglia in vivo may also be a source of Mfge8. As we did not detect Mfge8 in the CNS by immunohistochemistry using anti-Mfge8 antibodies (unpublished data), we focused on RNA expression analysis as a more reliable tool for detecting the source of secreted proteins such as Mfge8. We performed ISHs using an Mfge8 RNA probe and found Mfge8 transcription in the cerebellum at the watershed between internal granular and molecular layers. Mfge8-expressing cells lacked the characteristic morphology of Purkinje neurons. In view of their localization and morphology, Bergman glia (a specialized type of astrocytes) were deemed the most likely source of Mfge8 (Fig. 3 A, top). We also found cortical Mfge8-expressing cells with a stellate appearance indicative of astrocytes (Fig. 3 A, bottom). To validate the presumption that Mfge8 is expressed by astrocytes, we quantified Mfge8 messenger RNA (mRNA) expression by quantitative RT-PCR on cultured CNS cells enriched for distinct subpopulations. We found sustained Mfge8 expression in cultures enriched for astrocytes (purity of the astrocyte population, >95%; unpublished data) but little Mfge8 mRNA in cultures enriched for oligodendrocytes (purity, ∼85%) or for cerebellar granule neurons (purity, ∼95%). Conversely, we detected only minute amounts of Mfge8 RNA in cultured microglia (purity, >95%), although this is the main phagocytic cell type in the CNS (Fig. 3 B), and the microglial cell line BV-2 was reported to express Mfge8 (Fuller and Van Eldik, 2008). Because explanted microglia tend to dedifferentiate and to become activated, we wondered whether microglia up-regulate Mfge8 expression upon activation. Therefore, we performed quantitative RT-PCR and ISH on brains of terminally prion-sick mice, which displayed an extremely high density of activated microglia. Even here, the Mfge8 expression pattern was similar to that of healthy mice (Fig. 3 C, left). In fact, the overall amount of Mfge8 RNA was slightly reduced in RML-inoculated mice as compared with mice inoculated with noninfected brain homogenate (Fig. 3 C, right). Combination of immunofluorescence to visualize microglia using IB4 (isolectin B4) and ISH for Mfge8 failed to show colocalization (Fig. S2) of IB4 and Mfge8. These results negate a major contribution of microglia to the availability of Mfge8 within the CNS, be it in health or in disease.
Figure 3.
Analysis of the cellular source of Mfge8. (A) ISH for Mfge8 on brain cryosections. Mfge8+ cells (dark purple) were found in cerebellar (top) and cortical areas (bottom). Low and high magnification photographs of the same cerebellar (top) and cortical (bottom) areas are shown. Boxes indicate areas shown in higher magnifications. The arrow indicates the cell shown in high magnification. (B) Expression of Mfge8 in cultured astrocytes, microglia, oligodendrocytes, and neurons was quantified by quantitative RT-PCR. Mfge8 expression in each subset relative to total brain is shown (n = 3). (C) ISH for Mfge8 in prion-inoculated Mfge8+/+ mice at terminal stage (left). Right panel shows relative Mfge8 mRNA expression in mock- and RML-inoculated brains as assessed by quantitative RT-PCR (n = 3). (D) Quantitative RT-PCR quantitation of Mfge8, Cd11b, and Gfap expression in cerebellar slice cultures from untreated CD11b-HSVTK mice (HSVTK), prion-inoculated untreated CD11b-HSVTK mice (HSVTK + RML), GCV-treated CD11b-HSVTK mice (HSVTK + GCV), and prion-inoculated GCV-treated CD11b-HSVTK mice (HSVTK + RML + GCV; Student’s t test; n = 4). Uninfected HSVTK + GCV slices compared with HSVTK slices have a slight but significant reduction of Mfge8 (***, P = 0.0007). No difference in expression in HSVTK + RML + GCV slices compared with HSVTK + RML slices (NS, P = 0.1566). HSVTK + RML slices compared with HSVTK show significant down-regulation (**, P = 0.0082). CD11b expression is down-regulated by almost 3 logs in GCV -treated slices, confirming efficient depletion of microglia (HSVTK + GCV: ***, P < 0.0001; HSVTK + RML + GCV: ***, P < 0.0001). Gfap expression is slightly reduced after GC treatment (HSVTK + GCV: *, P = 0.0255). (E) Analysis of integrin αv (Itgav), β3 (Itgb3), and β5 (Itgb5) expression by quantitative RT-PCR (Student’s t test; n = 4). No significant changes in Itgav expression were found. Itgb3 and Itgb5 were down-regulated in GCV-treated compared with nontreated slices (Itgb3 HSVTK + GCV: ***, P < 0.0001; Itgb3 HSVTK + RML + GCV: ***, P = 0.0007; Itgb5 HSVTK + GCV: ***, P = 0.0002; Itgb5 HSVTK + RML + GCV: ***, P < 0.0001). Error bars represent SD. ISH and quantitative RT-PCR results are shown representative of 3 and 12 independent experiments, respectively.
To further address whether microglia might constitute a source of Mfge8, cerebellar slice cultures were prepared from CD11b-HSVTK mice (Heppner et al., 2005), from which microglia can be depleted by administering ganciclovir (GCV; Falsig et al., 2008). The efficiency of depletion was assessed by quantitative RT-PCR for CD11b, which is exclusively expressed in the brain by microglia. CD11b expression was reduced by almost 3 logs (Fig. 3 D), confirming that microglia were virtually absent after the addition of GCV. Mfge8 expression was slightly reduced after microglia depletion (Fig. 3 D, HSVTK + GCV). This suggests that in organotypic slices, which show prodigious levels of microglial activation, some Mfge8 may be microglia borne (Falsig et al., 2008). Conversely, GCV-treated slices displayed a slight reduction in Gfap (Glial fibrillary acidic protein) expression, suggesting that astrocytes were also affected by GCV to some degree. Thus, the decrease in Mfge8 expression might represent a consequence of marginal astrocyte depletion. Alternatively, the depletion of microglia may quench astrocytic transcription of Mfge8 indirectly, e.g., through the depletion of microglia-borne cytokines. Expression of neuron-specific Nefh (neurofilament heavy chain) and oligodendrocyte-specific Mbp (myelin basic protein) was not affected by GCV treatment (unpublished data). Consistent with the in vivo results (Fig. 3 C, right), we also observed a slight reduction of Mfge8 expression in prion-infected slice cultures (Fig. 3 D, HSVTK + RML). The aforementioned findings support the notion that most or all Mfge8 present in the CNS is not produced by microglia but rather by subsets of astrocytes encompassing cerebellar Bergman glia.
We next analyzed the expression of αvβ3 and αvβ5 integrins in CD11b-HSVTK slices, receptors that were shown to be required for Mfge8-dependent engulfment of apoptotic cells (Hanayama et al., 2002). Although quantitative RT-PCR analysis of the αv subunit (Itgav) was unchanged in microglia-depleted slices, expression of the β3 (Itgb3) and β5 (Itgb5) subunit was decreased by ∼90% and 75%, respectively (Fig. 3 E). Although microglia are not the major source of Mfge8, they are the major cell type expressing the Mfge8 receptors αvβ3 and αvβ5. Thus, our findings strongly suggest that microglia are required for removal of apoptotic prion-infected cells.
The influence of Mfge8 on prion pathogenesis is modulated by quantitative trait loci
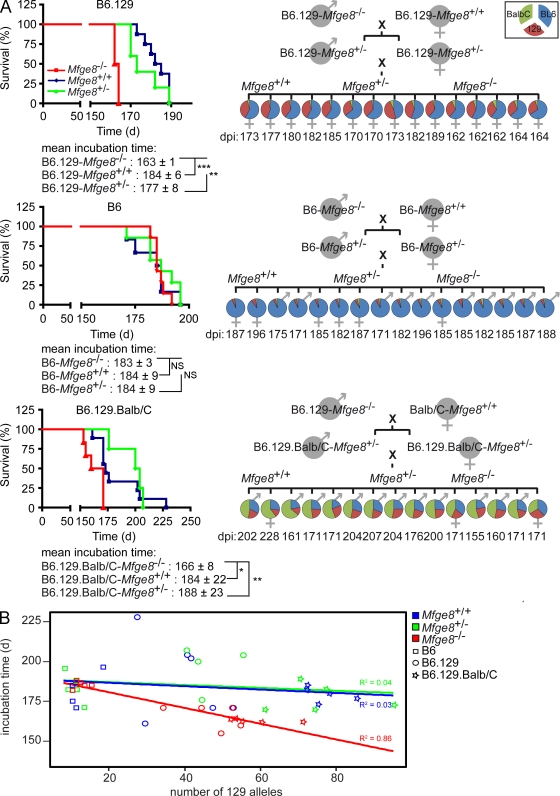
We then tested the reproducibility of the aforementioned findings using RML5 prions (derived from a different passage of the same inoculum as RML6) and also repeated the inoculations using Mfge8−/− mice on a different genetic background. Our initial inoculation experiments with RML6 were conducted using the originally published Mfge8−/− strain (Hanayama et al., 2004) bred on a mixed C57BL/6 × 129Sv background (B6.129-Mfge8−/−). This resulted in a 40-d acceleration of the disease. Inoculations of B6.129-Mfge8−/− with 3 log LD50 of RML5 resulted again in a strongly accelerated incubation time. In the latter experiment, the mean difference was 20 d (Fig. 4 A, top left) and was, again, highly statistically significant (P = 0.0002). The variation of the latency between B6.129-Mfge8−/− inoculated with RML5 and RML6 (Fig. 1 A) is likely to reflect differences between the infectivity titers of the two inocula.
Figure 4.
Inoculation of Mfge8−/− mice on different genetic backgrounds. (A) Mfge8+/+, Mfge8+/−, and Mfge8−/− mice on B6.129 (top), C57BL/6 (middle), and B6.129.Balb/C (bottom) background were inoculated i.c. with 3 log LD50 of RML5 prions (n = 5–9 per group). Significantly accelerated incubation times between Mfge8−/− and Mfge8+/+ or Mfge8+/− mice were only seen in B6.129 and B6.129.Balb/C backgrounds (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Right panel summarizes the STR analysis of inoculated mice. Pie charts show the percentage of B6 (blue), Balb/C (green), and 129Sv (red) alleles per mouse. Each chart represents one mouse. Only alleles that could unambiguously be assigned to one of the three different genetic backgrounds are represented in the pie charts. (B) Linear regression analysis comparing incubation time with number of 129Sv alleles. No correlation was seen between incubation time and number of 129Sv alleles in Mfge8+/+ and Mfge8+/− mice. However, there was a strong correlation in Mfge8−/− mice. Results represent at least five independent experiments.
When we inoculated Mfge8−/− mice that had been backcrossed to the C57BL/6 background for 10 generations (B6-Mfge8−/−) with 3 log LD50 RML5, we did not observe any acceleration of pathogenesis. B6-Mfge8−/−, B6-Mfge8+/−, and B6-Mfge8+/+ mice all succumbed to disease after a time period of ∼180 d (Fig. 4 A, middle left). Accordingly, the PrPSc levels and the density of apoptotic granule cells were not altered in B6-Mfge8−/− mice (Fig. S3, A and B). These results suggest that the effect of Mfge8 on the course of prion infections can be overridden by unidentified genetic modifiers that are polymorphic between the B6 and 129Sv strains. To gain more insights into the distribution of these modifiers, we crossed B6.129-Mfge8−/− mice to a third genetic background (Balb/C). Inoculation of the resulting F2 generation littermates (B6.129.Balb/C-Mfge8−/−, -Mfge8+/−, and -Mfge8+/+) showed that pathogenesis was again accelerated by the removal of Mfge8 (Fig. 4 A, bottom left). The variability of incubation times was increased, most likely because of the increased genetic heterogeneity of the mouse collectives.
To investigate whether differential expression of Mfge8 or its receptors αvβ3 and αvβ5 integrins might contribute to the observed background effect, we analyzed their mRNA levels by quantitative RT-PCR on whole brain cDNA from B6 and B6.129 Mfge8+/+ mice (Fig. S4 A). Expression levels of Mfge8, Itgb3, and Itgb5 were similar in all mice analyzed, yet one specific primer pair annealing to the 3′ end of the Itgb3 open reading frame yielded dramatically different quantitative RT-PCR reads between the two strains (unpublished data). This idiosyncrasy suggested the existence of polymorphisms between the B6 and 129Sv alleles of the Itgb3 gene leading to differential annealing and artifactual quantitative RT-PCR results. Therefore, we analyzed the microsatellite marker D11Mit128, which is located within 1 cM from the Itgb3 locus and discriminates between B6 and 129Sv allelotypes, in prion-inoculated mice (Fig. S4 B). We found that only Mfge8−/− mice homozygous for the 129Sv allele of D11Mit128 showed accelerated prion pathogenesis (Fig. S4 B, red box plots). Whenever a B6 allele of D11Mit128 was present, the presence or absence of Mfge8 failed to alter the incubation time (Fig. S4 B, blue and green box plots).
These results suggest that Mfge8/αvβ3 integrin–dependent phagocytosis is affected by strain-specific Itgb3 allotypes. Therefore, we sequenced the open reading frame of Itgb3 of 129Sv, C57BL/6, and Balb/C Mfge8+/+ mice. We found several strain-specific single nucleotide polymorphisms, yet none of these resulted in codon changes (Fig. S4 C). Thus, the linkage to D11Mit128 may point to noncoding polymorphisms that may affect splicing, mRNA stability, or posttranscriptional control by microRNAs.
We then performed a whole genome linkage analysis of all inoculated mice with 209 short tandem repeats (STRs; mean genetic distance, ∼7 cM; 13.5 Mbp) polymorphic between B6, 129Sv, or Balb/C mice (Table S1). Alleles that were shared by two different mouse strains were counted separately (B6 + 129Sv, B6 + Balb/C, or 129Sv + Balb/C), and all remaining alleles were regarded as noninformative (Table I). The percentages of B6 (blue), Balb/C (green), and 129Sv (red) alleles were summarized in pie charts (Fig. 4 A, right). Analysis of covariance was performed to analyze the independent effects of Mfge8 deficiency and genetic background on incubation time. In crossbred mice, the Mfge8 genotype significantly correlated with the incubation time of prion disease, even after elimination of possible genetic confounders by adjusting the contribution of each genetic background. The number of 129Sv alleles also influenced the incubation time (Table I). Linear regression analysis revealed a correlation of the incubation time and the amount of 129Sv alleles only in Mfge8−/− mice (R2 = 0.86; Fig. 4 B). Therefore, Mfge8 deficiency accelerates the prion pathogenesis, but only in the genetic context of the 129Sv strain.
Table I.
Statistical analysis of STR results
| Source | F | P-value |
| Number of B6 alleles and Mfge8 genotype | ||
| Number of B6 alleles | 0.4 | 0.53 |
| Mfge8 genotype | 4.2 | 0.021 |
| Number of 129Sv alleles and Mfge8 genotype | ||
| Number of 129Sv alleles | 6.3 | 0.016 |
| Mfge8 genotype | 5.6 | 0.007 |
| Number of Balb/C alleles and Mfge8 genotype | ||
| Number Balb/C alleles | 0.6 | 0.44 |
| Mfge8 genotype | 4.1 | 0.023 |
| Number of B6/129Sv alleles and Mfge8 genotype | ||
| Number of B6/129Sv alleles | 5.7 | 0.022 |
| Mfge8 genotype | 5.3 | 0.009 |
| Number of B6/Balb/C alleles and Mfge8 genotype | ||
| Number of B6/Balb/C alleles | 0.3 | 0.61 |
| Mfge8 deficiency | 4.2 | 0.022 |
| Number of 129Sv/Balb/C alleles and Mfge8 genotype | ||
| Number of 129Sv/Balb/C alleles | 0.1 | 0.81 |
| Mfge8 deficiency | 4.2 | 0.022 |
| Number of unknown alleles and Mfge8 genotype | ||
| Number of unknown alleles | 0.9 | 0.34 |
| Mfge8 deficiency | 3.5 | 0.039 |
Summary of results from analysis of covariance for incubation time with Mfge8 genotype as factor and number of alleles corresponding to the various genetic backgrounds as covariates. The between-subjects factors are Mfge8+/+ (n = 16), Mfge8+/− (n = 14), and Mfge8−/− (n = 16). The dependent variable is incubation time. F-values correspond to (1,42) and (2,42) degrees of freedom for the covariates and the genotype, respectively.
DISCUSSION
Many factors control incubation times of prion diseases after i.p. inoculation, including components of the immune system (Mabbott et al., 1998; Aguzzi, 2003), depletion of FDCs (Montrasio et al., 2000), and manipulations of the complement system (Klein et al., 2001; Mabbott et al., 2001). However, interference with prion replication in the CNS is more difficult, and there are only few interventions known to alter the incubation time of prion diseases after i.c. inoculation. Even fewer factors have been found to accelerate pathogenesis after i.c. administration of prions (LaCasse et al., 2008; Spinner et al., 2008). Mice expressing a mutant TLR4 (Toll-like receptor 4) defective in intracellular signaling were reported to experience accelerated disease after prion inoculation, implying that absence of TLR4 might impair microglia activation and consequently prion clearance. However, there was no difference in PrPSc levels at terminal disease stage in this model (Spinner et al., 2008). Even in mice engineered to overexpress PrPC, which have drastically shortened incubation times, the accelerated prion replication does not lead to elevated PrPSc levels (Fischer et al., 1996). In contrast, Mfge8−/− mice displayed enhanced PrPSc deposition despite accelerated disease progression. These findings strongly suggest that these mice suffer from decreased prion clearance.
Why does the absence of Mfge8 result in elevated levels of PrPSc? Upon prion infection, neurons may experience progressive damage and ultimately undergo apoptosis; during this process, they may secrete apoptotic PS-coated bodies containing prions (Ashok and Hegde, 2006; Leblanc et al., 2006). We suggest that astrocyte-derived Mfge8 opsonizes these prion-laden apoptotic bodies by binding to PS exposed on their surface. Microglia, the main phagocytic cell type of the CNS, express the Mfge8 receptors, αvβ3 and αvβ5 integrin heterodimers. Because microglia antagonize prion replication (Falsig et al., 2008), it appears plausible that Mfge8-opsonized, prion-laden apoptotic bodies are taken up and degraded by microglia. Because clearance of apoptotic bodies is inefficient in the absence of Mfge8, prions may persist in the brain and exacerbate PrPSc accumulation, thereby accelerating the disease.
Whereas the brain concentration of PrPSc was dramatically increased in the absence of Mfge8, prion infectivity titers were altered to a much smaller extent. Because prion titers are likely to reflect the number of individual PrP aggregates capable of nucleating further PrP, these findings suggest that the defective clearance caused by the absence of Mfge8 allows primarily for the elongation of PrPSc seeds rather than for their numerical expansion.
The aforementioned model implies a multitiered control of phagocytosis in the brain, whereby astrocytes are the source of Mfge8 and microglia are the executioners of phagocytosis. In this model, microglia would operate at highest efficiency only when armed by astrocyte-borne Mfge8. In the absence of the latter, removal of apoptotic cells, and of any prions that might be contained therein, would be impaired. We have discovered an analogous collaboration in germinal centers of lymphoid organs between FDCs and tingible body macrophages. There, FDCs produce Mfge8 and thereby license tingible body macrophages to engulf apoptotic cells (Kranich et al., 2008). In the CNS, an additional level of complexity may be added by the finding that astrocytes express markers of phagocytic pathways, including the Mfge8 receptor αvβ5 (Barres, 2008; Cahoy et al., 2008), and may have phagocytic potential of their own.
The regulation of phagocytosis is complex, and several independent, redundant pathways of phagocytosis are known. Mfge8 may not be the only mediator of engulfment, and some hitherto unidentified factors may, in certain situations, compensate for its absence. Strong evidence for the latter scenario comes from our observation that the acceleration of prion pathogenesis seen in Mfge8−/− mice is crucially dependent on the genetic background of mice. Although the Mfge8 genotype was the major determinant of incubation time, one or more additional loci (potentially including the Mfge8 receptors) act as important modifiers. Because these loci appear to be polymorphic between the 129Sv and the C57BL/6 strains of mice, it may be possible to identify them through larger-scale analyses of prion pathogenesis in F2 crosses between said mouse strains.
MATERIALS AND METHODS
Mice.
Mfge8−/− mice were donated by S. Nagata (Graduate School of Medicine, Kyoto University, Sakyo-ku, Kyoto, Japan) and were on a mixed (C57BL/6 × 129Sv)F1 (Hanayama et al., 2004) or C57BL/6 background. B6.129.Balb/C-Mfge8−/− mice were generated in our facility by intercrossing B6.129-Mfge8−/− mice with Balb/C mice (Harlan Laboratories). As controls, C57BL/6 (Harlan Laboratories) or F1 offsprings of crossings between C57BL/6 and 129Sv mice (Charles River) were used. For microglia depletion experiments, CD11b-HSVTK mice were used (Heppner et al., 2005). All experiments were performed in accordance with Swiss federal legislation and were approved by the Veterinäramt des Kantons Zurich (Veterinary office of the Canton Zurich).
Inoculations.
Mice were inoculated i.p. with 100 µl of brain homogenate diluted in PBS with 5% BSA and containing 3 log LD50 units of the Rocky Mountain Laboratories scrapie strain (passage 5 and 6, thus called RML5 and RML6). For i.c. inoculations, 30 µl of inoculum with 3 log LD50 units was administered. Scrapie was diagnosed according to clinical criteria (ataxia, kyphosis, priapism, and hind leg paresis). Mice were sacrificed on the day of onset of terminal clinical signs of scrapie.
Histoblot, Western blot, ELISA, histology, TUNEL assay, and ISH.
Detection of splenic PrPSc accumulation by histoblot (Taraboulos et al., 1992), detection of PrPC and PrPSc by Western blot (Lau et al., 2007), and quantitation of PrPC by sandwich ELISA (Polymenidou et al., 2008) were performed as described previously.
Immunohistochemistry for SAF84 (1:200), GFAP (1:1,000), and IBA1 (1:1,000) was performed on paraffin sections and detected with diaminobenzidine (Sigma-Aldrich). Tissue was treated with formic acid to inactivate prion infectivity. Sections were incubated in Ventana buffer, and stains were performed on an immunohistochemistry robot (NEXEX; Ventana Instruments) using an IVIEW DAB Detection kit (Ventana Instruments).
TUNEL stainings were performed on cryosections with the ApopTaq Plus Fluorescein In Situ Apoptosis Detection kit (Millipore) according to the manufacturer’s instructions. ISH using an Mfge8 riboprobe was performed as described previously (Kranich et al., 2008). In selected experiments, slides were subsequently stained with IB4–Alexa Fluor 647 (Invitrogen) and DAPI (Sigma-Aldrich).
MPA.
Brain homogenate was diluted in TBS with 0.1% Triton X-100 and subjected to immunoprecipitation using magnetic beads coupled to the peptoid PSR1 (Lau et al., 2007). Samples were incubated 1 h at 37°C under permanent agitation and then washed and denatured with 0.1 N NaOH. After neutralization (0.3 M Na2H2PO4), samples were placed on a magnet, and supernatant was transferred to POM19 (Polymenidou et al., 2005)-coated ELISA plates. After incubation (1 h at 37°C), plates were washed, and POM2-AP (Polymenidou et al., 2005) was added. After incubation with substrate (enhance chemiluminescence; GE Healthcare), plates were read in a luminometry reader (Lumiscan Ascent; Thermo Fisher Scientific).
SCA.
SCA was performed as described previously (Klöhn et al., 2003) with minor adaptations. Prion-susceptible neuroblastoma N2aPK1 cells were inoculated with Mfge8+/+ and Mfge8−/− brain homogenate in 96-well plates for 3 d. Prion infectivity titers were determined after cells were split three times 1:3 and three times 1:10. For this, 25,000 cells from each well were filtered onto the membrane of an ELISPOT plate, treated with PK, and denatured. Individual infected (PrPSc positive) cells were detected by ELISA using the anti-PrP antibody POM1 (Polymenidou et al., 2005). The number of TCI units per aliquot was calculated from the proportion of negative to total wells using the Poisson equation.
Cell-enriched primary cultures.
Mixed glia cultures were prepared from cerebra of pups (1–2 d postpartum) as previously described (Giulian and Baker, 1986). Purity of microglia was determined by staining with IB4 and was typically >95%. Purity of oligodendrocyte precursors and astrocytes was determined by NG2 and GFAP staining and was found to typically exceed 85% and 95%, respectively. Cerebellar granule neurons were cultured from 7-d-old mice as previously published (Leist et al., 1997). Cultures consisted of ∼95% cerebellar granule neurons and <5% astrocytes, as determined by staining for βIII-tubulin and GFAP.
Quantitative RT-PCR.
Total RNA from brain or cultured cells was isolated using TRIZOL (Invitrogen) and subjected to cDNA synthesis using QuantiTect Reverse Transcription kit (QIAGEN) according to the manufacturer’s instruction. Quantitative real-time PCR was performed on a 7900HT Fast Real-Time PCR system (Applied Biosystems). Expression levels were normalized using Gapdh (glyceraldehyde-3-phosphate dehydrogenase).
Gapdh forward (5′-CCACCCCAGCAAGGAGACT-3′), Gapdh reverse (5′-GAAATTGTGAGGGAGATGCT-3′), Mfge8 forward (5′-ATATGGGTTTCATGGGCTTG-3′), Mfge8 reverse (5′-GAGGCTGTAAGCCACCTTGA-3′), Integrin αv forward (5′-CACCAGCAGTCAGAGATGGA-3′), Integrin αv reverse (5′-TGCCTTGCTGAATGAACTTG-3′), Integrin β3 forward (5′-TGACATCGAGCAGGTGAAAG-3′), Integrin β3 reverse (5′-GAGTAGCAAGGCCAATGAGC-3′), Integrin β5 forward (5′-GGTTTCGGGTCTTTTGTTGA-3′), Integrin β5 reverse (5′-CCGATCTTCTCCTTGCAGAC-3′), CD11b forward (5′-CGGAAAGTAGTGAGAGAACTGTTTC-3′), CD11b reverse (5′-CTTATAATCCAAGGGATCACCGAATTT-3′), Gfap forward (5′-CACGAACGAGTCCCTAGAGC-3′), Gfap reverse (5′-GAATGGTGATGCGGTTTTCT-3′), and Mbp (available from GenBank/EMBL/DDBJ under accession no. NM_010777) expression was detected using the commercially available QuantiTect primer assay (QIAGEN).
Organotypic slice cultures and microglia depletion.
Organotypic cerebellar brain slices were prepared from 10–11-d-old pups, prion infected with 10−4 dilution of RML6 prions, and maintained according to previously published protocols (Falsig et al., 2008). 5 wk postinoculation, slices were harvested, and the PrPSc content of PK-treated samples was analyzed by Western blotting with anti-PrP antibody POM1. Microglia were depleted from organotypic brain slice cultures as previously described (Falsig et al., 2008).
STR analysis.
Whole genome STR analysis was performed using fluorescently labeled primers (FAM, VIC, NED; all from Applied Biosystems; Table S1). Genomic DNA was amplified by PCR, denatured, and sequenced on a 3130xl sequencer (Applied Biosystems). Analysis, allele calling, binning, and calibration of various mouse strains were performed manually and in combination with an in-house–developed software.
Statistical analysis.
Continuous data are presented as mean ± SD and were compared between groups using the unpaired Student’s t test. Incubation times were analyzed using the Kaplan-Meier method and compared between groups using the logrank test. The relation between incubation time and genetic background was analyzed using linear regression. The combined effect of Mfge8 genotype and genetic background on incubation time was analyzed using analysis of covariance with Mfge8 genotype as factor and the number of alleles corresponding to different genetic backgrounds as covariates. P-values <0.05 were considered statistically significant. SPSS 13 software (SPSS Inc.) was used for statistical analyses.
Online supplemental material.
Fig. S1 shows PrPSc accumulation in spleen, PrPC quantitation, and analysis of spongiosis. Fig. S2 shows that IB4+ microglia do not express Mfge8. Fig. S3 demonstrates that PrPSc levels and the amount of TUNEL+ apoptotic cells in the cerebellum of terminally sick mice are not changed between B6-Mfge8−/− and B6-Mfge8+/+ mice. Fig. S4 depicts Mfge8 and integrin expression levels in the brain, the correlation between the D11Mit128 allotype, and incubation time and Itgb3 single nucleotide polymorphisms in mice of different genetic backgrounds. Table S1, included as a separate PDF file, shows an overview of primers used for STR analysis. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092401/DC1.
Acknowledgments
We thank Shigekazu Nagata for providing Mfge8−/− mice, Burkhart Seifert for statistical analyses, and Mario Nuvolone, Caihong Zhu, Bei Li, Cristobál Tostado, Mirzet Delic, Winfried Förderer, Marianne König, André Wethmar, and Norbert Wey for technical assistance.
The Aguzzi laboratory is supported by grants of the Ernst Jung Foundation, the Stammbach Foundation, the European Union (LUPAS, PRIORITY), the Swiss National Science Foundation, a Sinergia grant from the Swiss National Science Foundation, and the National Center of Competence in Research on Neural Plasticity and Repair. A. Aguzzi is a recipient of an Advanced Grant of the European Research Council.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- CNS
- central nervous system
- dpi
- day postinoculation
- FDC
- follicular DC
- GCV
- ganciclovir
- ISH
- in situ RNA hybridization
- MPA
- misfolded protein assay
- mRNA
- messenger RNA
- PK
- proteinase K
- PS
- phosphatidyl serine
- SCA
- scrapie cell assay
- STR
- short tandem repeat
- TSE
- transmissible spongiform encephalopathy
- TUNEL
- terminal transferase dUTP nick end labeling
References
- Aguzzi A. 2003. Prions and the immune system: a journey through gut, spleen, and nerves. Adv. Immunol. 81:123–171 10.1016/S0065-2776(03)81004-0 [DOI] [PubMed] [Google Scholar]
- Aguzzi A. 2006. Prion diseases of humans and farm animals: epidemiology, genetics, and pathogenesis. J. Neurochem. 97:1726–1739 10.1111/j.1471-4159.2006.03909.x [DOI] [PubMed] [Google Scholar]
- Aguzzi A., Heikenwalder M. 2005. Prions, cytokines, and chemokines: a meeting in lymphoid organs. Immunity. 22:145–154 10.1016/j.immuni.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Ashok A., Hegde R.S. 2006. Prions and retroviruses: an endosomal rendezvous? EMBO Rep. 7:685–687 10.1038/sj.embor.7400749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres B.A. 2008. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 60:430–440 10.1016/j.neuron.2008.10.013 [DOI] [PubMed] [Google Scholar]
- Boddaert J., Kinugawa K., Lambert J.C., Boukhtouche F., Zoll J., Merval R., Blanc-Brude O., Mann D., Berr C., Vilar J., et al. 2007. Evidence of a role for lactadherin in Alzheimer’s disease. Am. J. Pathol. 170:921–929 10.2353/ajpath.2007.060664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner S., Isenmann S., Raeber A., Fischer M., Sailer A., Kobayashi Y., Marino S., Weissmann C., Aguzzi A. 1996. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 379:339–343 10.1038/379339a0 [DOI] [PubMed] [Google Scholar]
- Büeler H., Raeber A., Sailer A., Fischer M., Aguzzi A., Weissmann C. 1994. High prion and PrPSc levels but delayed onset of disease in scrapie-inoculated mice heterozygous for a disrupted PrP gene. Mol. Med. 1:19–30 [PMC free article] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A., et al. 2008. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28:264–278 10.1523/JNEUROSCI.4178-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., Raymond L., Favara C., Baron G., Priola S., et al. 2005. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science. 308:1435–1439 10.1126/science.1110837 [DOI] [PubMed] [Google Scholar]
- Falsig J., Julius C., Margalith I., Schwarz P., Heppner F.L., Aguzzi A. 2008. A versatile prion replication assay in organotypic brain slices. Nat. Neurosci. 11:109–117 10.1038/nn2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M., Rülicke T., Raeber A., Sailer A., Moser M., Oesch B., Brandner S., Aguzzi A., Weissmann C. 1996. Prion protein (PrP) with amino-proximal deletions restoring susceptibility of PrP knockout mice to scrapie. EMBO J. 15:1255–1264 [PMC free article] [PubMed] [Google Scholar]
- Fuller A.D., Van Eldik L.J. 2008. MFG-E8 regulates microglial phagocytosis of apoptotic neurons. J. Neuroimmune Pharmacol. 3:246–256 10.1007/s11481-008-9118-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulian D., Baker T.J. 1986. Characterization of ameboid microglia isolated from developing mammalian brain. J. Neurosci. 6:2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R., Nagata S. 2005. Impaired involution of mammary glands in the absence of milk fat globule EGF factor 8. Proc. Natl. Acad. Sci. USA. 102:16886–16891 10.1073/pnas.0508599102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanayama R., Tanaka M., Miwa K., Shinohara A., Iwamatsu A., Nagata S. 2002. Identification of a factor that links apoptotic cells to phagocytes. Nature. 417:182–187 10.1038/417182a [DOI] [PubMed] [Google Scholar]
- Hanayama R., Tanaka M., Miyasaka K., Aozasa K., Koike M., Uchiyama Y., Nagata S. 2004. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 304:1147–1150 10.1126/science.1094359 [DOI] [PubMed] [Google Scholar]
- Heppner F.L., Greter M., Marino D., Falsig J., Raivich G., Hövelmeyer N., Waisman A., Rülicke T., Prinz M., Priller J., et al. 2005. Experimental autoimmune encephalomyelitis repressed by microglial paralysis. Nat. Med. 11:146–152 10.1038/nm1177 [DOI] [PubMed] [Google Scholar]
- Klein M.A., Kaeser P.S., Schwarz P., Weyd H., Xenarios I., Zinkernagel R.M., Carroll M.C., Verbeek J.S., Botto M., Walport M.J., et al. 2001. Complement facilitates early prion pathogenesis. Nat. Med. 7:488–492 10.1038/86567 [DOI] [PubMed] [Google Scholar]
- Klöhn P.C., Stoltze L., Flechsig E., Enari M., Weissmann C. 2003. A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc. Natl. Acad. Sci. USA. 100:11666–11671 10.1073/pnas.1834432100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranich J., Krautler N.J., Heinen E., Polymenidou M., Bridel C., Schildknecht A., Huber C., Kosco-Vilbois M.H., Zinkernagel R., Miele G., Aguzzi A. 2008. Follicular dendritic cells control engulfment of apoptotic bodies by secreting Mfge8. J. Exp. Med. 205:1293–1302 10.1084/jem.20071019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCasse R.A., Striebel J.F., Favara C., Kercher L., Chesebro B. 2008. Role of Erk1/2 activation in prion disease pathogenesis: absence of CCR1 leads to increased Erk1/2 activation and accelerated disease progression. J. Neuroimmunol. 196:16–26 10.1016/j.jneuroim.2008.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A.L., Yam A.Y., Michelitsch M.M., Wang X., Gao C., Goodson R.J., Shimizu R., Timoteo G., Hall J., Medina-Selby A., et al. 2007. Characterization of prion protein (PrP)-derived peptides that discriminate full-length PrPSc from PrPC. Proc. Natl. Acad. Sci. USA. 104:11551–11556 10.1073/pnas.0704260104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc P., Alais S., Porto-Carreiro I., Lehmann S., Grassi J., Raposo G., Darlix J.L. 2006. Retrovirus infection strongly enhances scrapie infectivity release in cell culture. EMBO J. 25:2674–2685 10.1038/sj.emboj.7601162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M., Volbracht C., Kühnle S., Fava E., Ferrando-May E., Nicotera P. 1997. Caspase-mediated apoptosis in neuronal excitotoxicity triggered by nitric oxide. Mol. Med. 3:750–764 [PMC free article] [PubMed] [Google Scholar]
- Liberski P.P., Sikorska B., Bratosiewicz-Wasik J., Gajdusek D.C., Brown P. 2004. Neuronal cell death in transmissible spongiform encephalopathies (prion diseases) revisited: from apoptosis to autophagy. Int. J. Biochem. Cell Biol. 36:2473–2490 10.1016/j.biocel.2004.04.016 [DOI] [PubMed] [Google Scholar]
- Mabbott N.A., Farquhar C.F., Brown K.L., Bruce M.E. 1998. Involvement of the immune system in TSE pathogenesis. Immunol. Today. 19:201–203 10.1016/S0167-5699(98)01253-5 [DOI] [PubMed] [Google Scholar]
- Mabbott N.A., Bruce M.E., Botto M., Walport M.J., Pepys M.B. 2001. Temporary depletion of complement component C3 or genetic deficiency of C1q significantly delays onset of scrapie. Nat. Med. 7:485–487 10.1038/86562 [DOI] [PubMed] [Google Scholar]
- Mallucci G., Dickinson A., Linehan J., Klöhn P.C., Brandner S., Collinge J. 2003. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 302:871–874 10.1126/science.1090187 [DOI] [PubMed] [Google Scholar]
- Montrasio F., Frigg R., Glatzel M., Klein M.A., Mackay F., Aguzzi A., Weissmann C. 2000. Impaired prion replication in spleens of mice lacking functional follicular dendritic cells. Science. 288:1257–1259 10.1126/science.288.5469.1257 [DOI] [PubMed] [Google Scholar]
- Patton S., Keenan T.W. 1975. The milk fat globule membrane. Biochim. Biophys. Acta. 415:273–309 [DOI] [PubMed] [Google Scholar]
- Polymenidou M., Stoeck K., Glatzel M., Vey M., Bellon A., Aguzzi A. 2005. Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet Neurol. 4:805–814 10.1016/S1474-4422(05)70225-8 [DOI] [PubMed] [Google Scholar]
- Polymenidou M., Moos R., Scott M., Sigurdson C., Shi Y.Z., Yajima B., Hafner-Bratkovic I., Jerala R., Hornemann S., Wuthrich K., et al. 2008. The POM monoclonals: a comprehensive set of antibodies to non-overlapping prion protein epitopes. PLoS One. 3:e3872 10.1371/journal.pone.0003872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S.B. 1982. Novel proteinaceous infectious particles cause scrapie. Science. 216:136–144 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- Prusiner S.B., McKinley M.P., Bowman K.A., Bolton D.C., Bendheim P.E., Groth D.F., Glenner G.G. 1983. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 35:349–358 10.1016/0092-8674(83)90168-X [DOI] [PubMed] [Google Scholar]
- Spinner D.S., Cho I.S., Park S.Y., Kim J.I., Meeker H.C., Ye X., Lafauci G., Kerr D.J., Flory M.J., Kim B.S., et al. 2008. Accelerated prion disease pathogenesis in Toll-like receptor 4 signaling-mutant mice. J. Virol. 82:10701–10708 10.1128/JVI.00522-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele A.D., King O.D., Jackson W.S., Hetz C.A., Borkowski A.W., Thielen P., Wollmann R., Lindquist S. 2007. Diminishing apoptosis by deletion of Bax or overexpression of Bcl-2 does not protect against infectious prion toxicity in vivo. J. Neurosci. 27:13022–13027 10.1523/JNEUROSCI.3290-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Jendroska K., Serban D., Yang S.L., DeArmond S.J., Prusiner S.B. 1992. Regional mapping of prion proteins in brain. Proc. Natl. Acad. Sci. USA. 89:7620–7624 10.1073/pnas.89.16.7620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Ostrowski M., Segura E. 2009. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9:581–593 10.1038/nri2567 [DOI] [PubMed] [Google Scholar]
- Weissmann C. 2004. The state of the prion. Nat. Rev. Microbiol. 2:861–871 10.1038/nrmicro1025 [DOI] [PubMed] [Google Scholar]