Abstract
TAL1 (also known as SCL) is expressed in >40% of human T cell acute lymphoblastic leukemias (T-ALLs). TAL1 encodes a basic helix-loop-helix transcription factor that can interfere with the transcriptional activity of E2A and HEB during T cell leukemogenesis; however, the oncogenic pathways directly activated by TAL1 are not characterized. In this study, we show that, in human TAL1–expressing T-ALL cell lines, TAL1 directly activates NKX3.1, a tumor suppressor gene required for prostate stem cell maintenance. In human T-ALL cell lines, NKX3.1 gene activation is mediated by a TAL1–LMO–Ldb1 complex that is recruited by GATA-3 bound to an NKX3.1 gene promoter regulatory sequence. TAL1-induced NKX3.1 activation is associated with suppression of HP1-α (heterochromatin protein 1 α) binding and opening of chromatin on the NKX3.1 gene promoter. NKX3.1 is necessary for T-ALL proliferation, can partially restore proliferation in TAL1 knockdown cells, and directly regulates miR-17-92. In primary human TAL1-expressing leukemic cells, the NKX3.1 gene is expressed independently of the Notch pathway, and its inactivation impairs proliferation. Finally, TAL1 or NKX3.1 knockdown abrogates the ability of human T-ALL cells to efficiently induce leukemia development in mice. These results suggest that tumor suppressor or oncogenic activity of NKX3.1 depends on tissue expression.
T cell acute lymphoblastic leukemia (T-ALL) is a neoplastic disorder that occurs in 15% of pediatric and 25% of adult acute lymphoblastic leukemias. The TAL1/SCL gene (hereafter referred to as TAL1) is activated by chromosomal translocation, interstitial deletion, or unknown mechanisms in >40% of human T-ALLs (Bash et al., 1995; Ferrando et al., 2002, 2004; Armstrong and Look, 2005). This activation results in the production of a normal TAL1 protein ectopically expressed from the double-negative stage onwards during T-lymphopoiesis, whereas during normal T cell differentiation, TAL1 expression is restricted to the DN1-DN2 subset of immature CD4−/CD8− thymocytes (Herblot et al., 2000).
TAL1 encodes a class II basic helix-loop-helix (HLH [bHLH]) transcription factor that can activate or repress genes by forming E-box (CANNTG)–binding heterodimers with ubiquitous class I bHLH partners known as E-proteins, which include products of the E2A gene (E12 and E47), HEB, and E2-2 (Hsu et al., 1991, 1994a; Voronova and Lee, 1994). In hematopoietic cells, TAL1 regulates the transcriptional activities of its target genes through binding to an E-box/GATA motif and nucleation of a large complex that includes an E-protein, the LIM-only protein LMO2, GATA-1/2, Ldb1, and other associated transcription factors (Wadman et al., 1997; Lécuyer et al., 2002; Xu et al., 2003) or through its recruitment to regulatory sequences after interaction with GATA factors bound to DNA (Lahlil et al., 2004; Fujiwara et al., 2009; Yu et al., 2009). Functionally, TAL1 is a master regulatory protein of primitive and definitive hematopoiesis and in the development and maintenance of immature hematopoietic progenitors (Porcher et al., 1996; Robb et al., 1996; Brunet de la Grange et al., 2006; Souroullas et al., 2009; Lacombe et al., 2010). These TAL1 functions suggest that this transcription factor might have an important role during initiation or maintenance of T cell leukemia, but the transcriptional programs directly regulated by TAL1 in human T-ALL are still being elucidated.
The prevailing model of TAL1-induced leukemogenesis describes TAL1 as a transcriptional repressor through its heterodimerization with E-proteins and blocking the E2A, HEB, and/or E2-2 transcriptional activities (Park and Sun, 1998; O’Neil et al., 2004) and/or through a DNA binding–independent sequestration mechanism similar to that proposed for the Id family of HLH proteins (Benezra et al., 1990; Engel and Murre, 2001). This view is supported by the fact that E2A−/−, HEB−/−, and Id2–overexpressing transgenic mice exhibit a defect in T cell differentiation, characterized by a block at the transition of CD4/CD8 double-negative to double-positive thymocytes and by the emergence of T-ALL in surviving E2A-deficient mice or Id-overexpressing mice (Bain et al., 1997; Yan et al., 1997; Morrow et al., 1999). At the molecular level, TAL1 might exert its inhibitory effects on E-protein target genes by interfering with E2A-mediated recruitment of chromatin-remodeling complexes that facilitate transcription activation (Bradney et al., 2003) and/or by recruitment of corepressors such as mSin3A and HDAC1 (Huang and Brandt, 2000; O’Neil et al., 2004) or Brg1 and HDAC2 (Xu et al., 2006) on E2A, HEB, or E2-2 target genes.
In addition to the inhibition of class I bHLH functions, TAL1 might also exert its oncogenic effects through inappropriate gene activation. This possibility has been suggested by Ono et al. (1998), who have shown that TAL1 can induce the expression of the RALDH2 gene and is strengthened by the recent observation of a transcriptional regulatory network downstream of TAL1 in human T-ALL, showing that TAL1 might act as a repressor or an activator of target genes (Palomero et al., 2006). Interestingly, rather than requiring an E-box motif, activation of the RALDH2 gene by TAL1 occurred through its recruitment to a cryptic intronic promoter by DNA-bound GATA-3, although the relevance of this target gene for leukemogenesis remains unknown (Ono et al., 1998). A thorough understanding of additional genes directly activated by TAL1 in T-ALL will provide greater insight into the mechanisms by which TAL1 induces or maintains a leukemic phenotype.
To characterize genes directly or indirectly regulated by TAL1 in human T-ALL, we combined TAL1 knockdown by RNA interference and gene expression profiling in human T cell lines that express high or low levels of TAL1 protein. In addition to already known TAL1 target genes, we have identified the NKX3.1 gene as a potential TAL1 target gene in human T-ALL. NKX3.1 is a homeobox gene switched on during embryonic development of prostate tissue and then specifically expressed in the adult prostate epithelium (Shen and Abate-Shen, 2003). NKX3.1 is crucial for prostate stem cell maintenance (Wang et al., 2009), and its inactivation is one of the earliest events that occurs in prostate cancer initiation, thus defining NKX3.1 as a major tumor suppressor gene of prostate cancer (He et al., 1997; Abdulkadir et al., 2002; Magee et al., 2003). We show that TAL1 directly activates the transcription of the human NKX3.1 gene by binding to regulatory sequences that are not conserved in the mouse Nkx3.1 gene, suggesting differences between human TAL1-expressing T-ALL and mouse models of TAL1-mediated T-ALL. Finally, we document the role of NKX3.1 in human T-ALL both in vitro and in vivo.
RESULTS
TAL1 knockdown results in decreased proliferation of human T-ALL cell lines
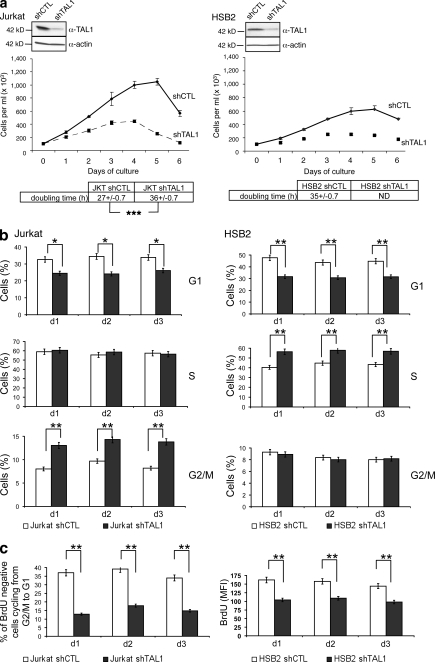
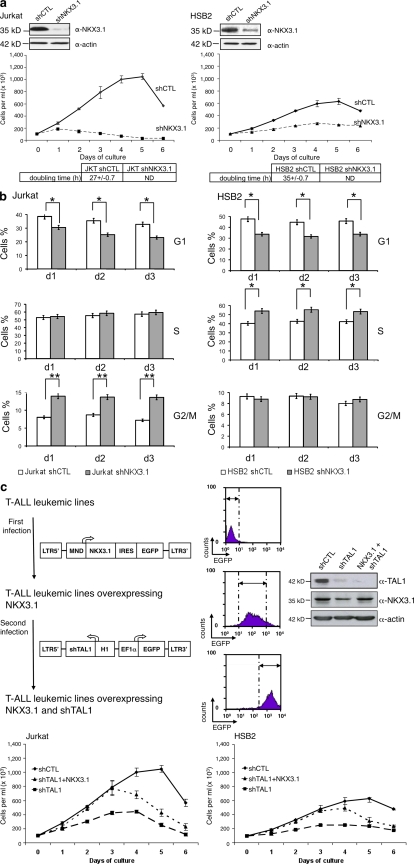
We decreased TAL1 protein level using a lentiviral delivery of shTAL1 in four human T-ALL cell lines (Jurkat, HSB2, CEM, and RPMI) that express high (Jurkat and RPMI) or low (HSB2 and CEM) levels of TAL1 protein (Fig. 1 a and Fig. S1 a). Growth curves of the four T-ALL cell lines showed that TAL1 knockdown impaired proliferation of these cell lines, but only when the cells were grown in low (<2.5%) concentrations of FCS (Fig. 1 a, bottom; Fig. S1 b; and not depicted for RPMI and CEM). These effects were obtained with another shTAL1, indicating little or no off-targets effects (unpublished data). To determine on which cellular functions TAL1 was acting, doubling time, apoptosis, and cell cycle of Jurkat and HSB2 cell lines expressing shCTL or shTAL1 were analyzed during the first 3 d of culture to avoid side effects caused by over-growth of leukemic cells. TAL1 knockdown lengthened the doubling time of cells (Fig. 1 a, bottom) but affected neither apoptosis nor necrosis (not depicted). Cell cycle analyses showed fewer cells in G1 phase and more cells in G2/M phase (Jurkat) or S phase (HSB2; Fig. 1 b). BrdU pulse chase labeling of Jurkat cells showed that TAL1 knockdown induced a partial blockage of the G2/M to G1 transition (Fig. 1 c, left), and diminished uptake of BrdU in HSB2 cells indicated a slow progression in S phase (Fig. 1 c, right). Thus, TAL1 knockdown resulted in an impaired proliferation under low serum conditions as the result of modifications of different phases of the cell cycle.
Figure 1.
TAL1 regulates proliferation of Jurkat and HSB2 T-ALL cell lines. (a, top) TAL1 protein in Jurkat and HSB2 cells that stably expressed shRNAs directed against human TAL1 (shTAL1) or hepatitis B virus (shCTL) was measured by immunoblotting. Actin is shown as a loading control. The data shown correspond to a representative experiment out of two performed. (middle) Growth curves of Jurkat and HSB2 cells expressing shTAL1 or shCTL. Graphs show number of cells ± SEM (n = 4 experiments). (bottom) doubling times of Jurkat and HSB2 cells expressing shCTL or shTAL1 analyzed during the exponential phase of the growth curves (***, P < 0.001). (b) Percentages of Jurkat (left) and HSB2 (right) cells expressing shCTL or shTAL1 in G1, S, and G2/M phases were determined at days 1, 2, and 3 of culture. Graphs show percentages of cells ± SEM (n = 4 experiments; *, P < 0.05; **, P < 0.01). (c, left) Percentages of BrdU-negative Jurkat cells expressing shCTL or shTAL1 cycling from G2/M to G1 were determined at days 1, 2, and 3 of culture. (right) BrdU mean fluorescence of HSB2 cells expressing shCTL or shTAL1 labeled with BrdU at days 1, 2, and 3 of culture. Graphs show percentages of cells ± SEM (n = 4 experiments; **, P < 0.01). MFI, mean fluorescence intensity.
Reduced TAL1 expression in Jurkat or HSB2 T cell lines is associated with up- or down-regulation of a common set of genes that includes the NKX3.1 gene
To identify TAL1 target genes involved in the proliferation of T-ALL cell lines, we performed gene expression profiling of Jurkat and HSB2 cell lines expressing shTAL1 or shCTL. We isolated RNA from Jurkat and HSB2 cells after 2 or 3 d of culture and used them to interrogate Affymetrix DNA microarrays representing 30,000 known human genes. Transcription profiles were compared, and a common set of genes for which expressions were dependent on TAL1 in both cell lines was found (Fig. S2; deposited in ArrayExpress under accession no. E-MEXP-2197). Among these genes, we found already known TAL1 target genes such as RAG1, TM4SF2, or RALDH2 (Ito et al., 2003; Ono et al., 1998; Soulier et al., 2005), thus validating the cell lines used to identify TAL1 target genes in human T-ALL.
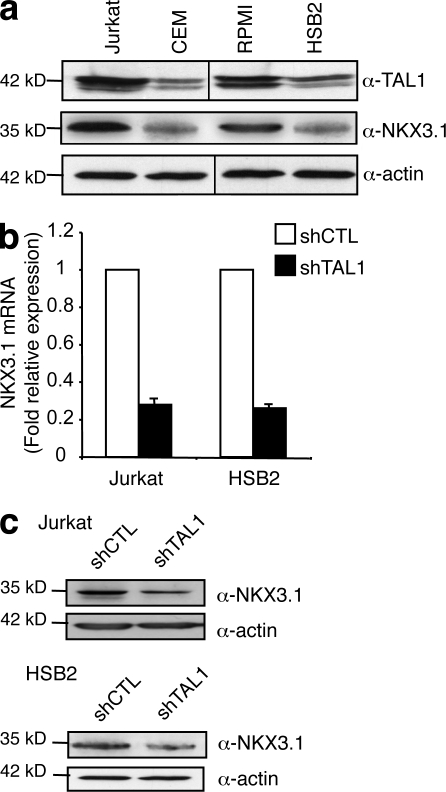
Among potential TAL1 target genes, we focused our study on the NKX3.1 gene as (a) this gene is not expressed in normal human thymocytes but is ectopically expressed in primary T-ALL samples expressing TAL1 (Soulier et al., 2005), (b) NKX2.5, another member of the NKX homeobox protein family, is activated by chromosomal translocations in a small number of primary human T-ALL and T cell lines (Nagel et al., 2003; Przybylski et al., 2006), and (c) NKX3.1 is involved in the maintenance of prostate stem cells and is a tumor suppressor gene of prostate cancer (Magee et al., 2003; Wang et al., 2009). NKX3.1 protein level correlated with TAL1 protein level in the four T-ALL cell lines studied (Fig. 2 a), and NKX3.1 messenger RNA (mRNA) and NKX3.1 protein levels were three- to fourfold decreased when TAL1 expression was decreased in Jurkat and HSB2 cells using shTAL1 constructs (Fig. 2, b and c). These results indicated that NKX3.1 gene expression was dependent on the presence of TAL1 in T cell lines and suggested a possible function of the NKX3.1 protein in TAL1-mediated proliferation of human T-ALL.
Figure 2.
NKX3.1 mRNA and protein levels are dependent on TAL1 expression. (a) TAL1 and NKX3.1 protein in high (Jurkat and RPMI) and low (CEM and HSB2) TAL1-expressing T cell lines was measured by immunoblotting. Actin is shown as a loading control. The data shown correspond to one representative experiment out of two performed. Vertical black lines indicate that intervening lanes have been spliced out. (b) NKX3.1 mRNA was determined by quantitative RT-PCR normalized to GAPDH mRNA in shCTL- or shTAL1-expressing T cell lines. Error bars indicate SEM (n = 4 experiments). (c) NKX3.1 protein was measured by Western blotting using nuclear extracts from the indicated T cell lines. Actin is shown as a loading control. The data shown correspond to one representative experiment out of three performed.
The human NKX3.1 gene is directly activated by TAL1 in T-ALL cell lines through regulatory sequences that are not conserved in the mouse Nkx3.1 gene
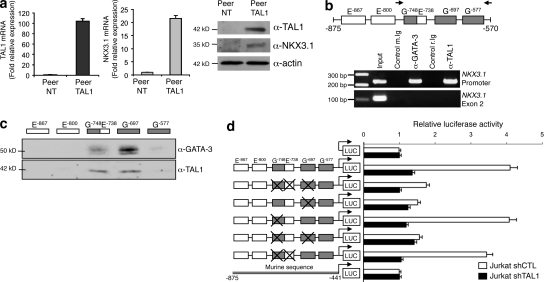
To determine whether TAL1 could directly activate transcription of the NKX3.1 gene, we first expressed TAL1 in a human T cell line, Peer, which expresses neither TAL1 nor NKX3.1. Ectopic expression of TAL1 in Peer cells resulted in the transcriptional activation of the endogenous NKX3.1 gene and expression of the NKX3.1 protein (Fig. 3 a), indicating that the NKX3.1 gene is a potential direct target of TAL1. Chromatin immunoprecipitation (ChIP) showed that TAL1 and GATA-3, the only member of the GATA family expressed in T cells, were specifically bound in vivo to the −875/−570 region of the human NKX3.1 gene promoter (Fig. 3 b). This region contains multiple E-boxes and GATA binding sites (Fig. S3 a), and using oligonucleotides that cover all E-boxes and GATA binding sites present in the −875/−570 human NKX3.1 promoter region, we found that TAL1 and GATA-3 can bind an E-box located at −738 and/or two GATA binding sites at −748 and −697 (Fig. 3 c). To identify the relative contributions of these three regulatory motifs to the transcriptional activity of the NKX3.1 gene promoter, the −977/−482 sequence and associated mutants at positions −748/−738 and/or −697 were cloned upstream of a minimal promoter that drives luciferase expression and used in transient transfection assays. This promoter region could activate transcription in Jurkat cells (Fig. 3 d) but not in NIH3T3 cells (not depicted), and this activation was dependent on TAL1 expression (Fig. 3 d). Transcriptional activities of the different mutants of the −977/−482 promoter region pinpointed the GATA-3–TAL1 binding site at −697 as a major regulatory site that mediated the transcriptional activity of this NKX3.1 promoter region in Jurkat cells (Fig. 3 d). Interestingly, the two regulatory sequences that can bind TAL1 in the human NKX3.1 gene are not conserved in the mouse gene (Fig. S3 a), and as a consequence, no transcriptional activity of the corresponding mouse Nkx3.1 promoter region could be detected in Jurkat cells (Fig. 3 d). These results suggest why increased Nkx3.1 mRNA levels are not found in gene expression profiling of mouse TAL1–LMO1 leukemic cells (Fig. S3 b).
Figure 3.
TAL1 directly activates NKX3.1 gene expression in human T cell lines. (a) Peer, a T-ALL cell line which expresses neither TAL1 nor NKX3.1, was transduced with lentiviral particles containing TAL1 cDNA. Expression of TAL1 (left) and NKX3.1 (right) mRNA was studied in Peer cells (Peer NT) or in Peer cells expressing TAL1 (Peer TAL1) by quantitative PCR normalized to GAPDH mRNA. Error bars indicate SEM (n = 4 experiments). (b) The human −875/−570 NKX3.1 promoter region (+1 is the ATG start codon) contains E-boxes (open boxes) and GATA binding sites (gray boxes). Arrows indicate positions of oligonucleotides used for PCR amplification. Jurkat cell chromatin extract was immunoprecipitated with GATA-3, TAL1 antibodies, or control-matched Ig (m., mouse; r., rabbit). Immunoprecipitated DNA was amplified to detect the −790/−570 NKX3.1 promoter or exon 2 sequences. Input is amplification of Jurkat cell DNA. The results shown are representative of four independent experiments. (c) Jurkat cell nuclear extract was incubated with biotin-labeled oligonucleotides that covered E-boxes and/or GATA binding sites present in the −875/−570 NKX3.1 promoter region. Proteins bound to the different oligonucleotides were analyzed by Western blot using GATA-3 or TAL1 antibodies. The data shown correspond to one representative experiment out of three performed. (d) Jurkat cells expressing shCTL or shTAL1 were transfected with plasmids containing wild-type or mutated −977/−482 sequence of the NKX3.1 gene promoter cloned upstream of a minimal promoter that drives the firefly luciferase (LUC) gene. The mutated sequences, indicated by crosses, abolished GATA-3 and/or TAL1 binding sites. The corresponding mouse Nkx3.1 gene promoter region was cloned upstream of the same reporter gene and assayed in Jurkat cells. Transfections were normalized using the dual-luciferase reporter assay system, and expression of Renilla luciferase provides an internal control. Data are the means ± SEM (n = 4 experiments).
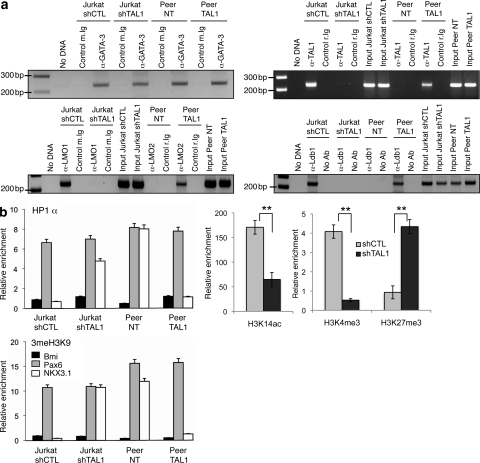
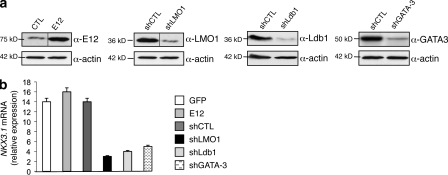
At the chromatin level, GATA-3 was bound to the NKX3.1 promoter in T cell lines expressing or not NKX3.1 (Fig. 4 a, top left). Expression of TAL1 led to its recruitment by GATA-3 (Fig. 4 a, top right) and to the formation of a transcriptional complex containing LMO1/2 and Ldb1 proteins on the NKX3.1 promoter (Fig. 4 a, bottom). Recruitment (respectively suppression) of this complex on the NKX3.1 promoter was associated with suppression (respectively recruitment) of HP1-α (heterochromatin protein 1 α) binding and histone H3 trimethylation on lysine 9, which are hallmarks of gene repression (Fig. 4 b, left). The effects of TAL1 knockdown on activating (H3K14ac or H3K4me3) or repressive (H3K27me3) histone marks further strengthened a TAL1-dependent chromatin structure of the NKX3.1 gene promoter in the Jurkat cell line (Fig. 4 b, right). The NKX3.1 gene activation is similar to the TAL1-mediated RALDH2 gene activation in human T-ALL (Ono et al., 1998). We found TAL1-dependent chromatin modifications of the RALDH2 gene similar to the ones found on the NKX3.1 promoter (Fig. S4), indicating that TAL1 might regulate the chromatin structure of a subset of genes already marked by GATA-3. As the GATA-3–TAL1 interaction nucleates a transcriptional complex containing Ldb1 and LMO1 in Jurkat cells, the expression of GATA-3, Ldb1, and LMO1 was knocked down after transduction of Jurkat cells by lentiviruses that could express short hairpin RNAs (shRNAs) targeting the corresponding mRNAs (Fig. 5 a), and the NKX3.1 mRNA level was quantified in the transduced cell line. NKX3.1 mRNA level was not affected by overexpression of E12, indicating that NKX3.1 gene expression was not dependent on E2A protein level but decreased when the protein level of any member of the TAL1-containing complex decreased (Fig. 5 b). Altogether, these results indicated that ectopic expression of TAL1 in human T-lymphoid cells could activate the transcription of the human gene NKX3.1 at its chromosomal locus via TAL1 recruitment by GATA-3 already bound to the NKX3.1 promoter and formation of a nuclear complex that can eliminate repression of the NKX3.1 gene.
Figure 4.
TAL1 expression modifies the chromatin structure of the NKX3.1 gene promoter. (a) Jurkat shCTL, Jurkat shTAL1, Peer NT, and Peer TAL1 cell chromatin extracts were immunoprecipitated with GATA-3 (top left), TAL1 (top right), LMO1/2 (bottom left), and Ldb1 (bottom right) antibodies, control-matched Ig, or no antibody (no Ab). Immunoprecipitated DNA was amplified to detect the −790/−570 NKX3.1 promoter region as in Fig. 3 b. The data shown correspond to one representative experiment out of two performed. (b, left) HP1-α, me3H3K9 antibodies, or control Ig was used to immunoprecipitate the same chromatin extracts as in panel a. NKX3.1, Pax6 (repressed gene), and Bmi (activated gene) promoter sequences were detected by quantitative PCR on the immunoprecipitated DNA. (right) H3K14ac, H3K4me3, and H3K27me3 antibodies or control Ig was used to immunoprecipitate chromatin extracts from Jurkat shCTL and Jurkat shTAL1. NKX3.1 promoter sequence was detected by quantitative PCR on the immunoprecipitated DNA. Data are the means ± SEM (n = 3 experiments; **, P < 0.01).
Figure 5.
LMO1, Ldb1, and GATA-3 proteins are necessary for NKX3.1 expression in Jurkat cells. (a) shRNAs directed against LMO1, Ldb1, and GATA-3 decreased the corresponding protein level compared with cells expressing an shCTL. Jurkat cells were also transduced with a lentivirus that allowed overexpression of the E12 or GFP protein. Nuclear extracts from transduced Jurkat cells were analyzed by Western blotting using the indicated antibodies. Actin is shown as a loading control. The data shown correspond to one representative experiment out of two performed. Vertical black lines indicate that intervening lanes have been spliced out. (b) NKX3.1 mRNA was monitored by quantitative RT-PCR normalized to GAPDH mRNA in Jurkat cells transduced with lentiviral vectors that expressed GFP, E12 cDNA, or control shRNA or shRNA directed against LMO1, Ldb1, or GATA-3. Data are the means ± SEM (n = 4 experiments).
NKX3.1 is required for the proliferation of TAL1-expressing T cell lines
NKX3.1-deficient mice develop prostatic hyperplasia, and NKX3.1 has been shown to regulate cell cycle exit during luminal cell regeneration, indicating that NKX3.1 has an antiproliferative effect in the prostate (Lei et al., 2006). As the NKX3.1 gene is activated by TAL1 in human T cell lines, we studied the role of NKX3.1 in the cellular proliferation of Jurkat and HSB2 cell lines expressing shNKX3.1. NKX3.1 protein level was decreased using two different shNKX3.1 lentiviral constructs (Fig. 6 a, top; and not depicted). NKX3.1 knockdown impaired proliferation of Jurkat and HSB2 cells cultured in low (<2.5%) FCS concentrations (Fig. S5 a), increased doubling time, and did not affect apoptosis/necrosis of the cells (Fig. 6 a, bottom; and not depicted). NKX3.1 protein level was more reduced with shNKX3.1 than with shTAL1 (Fig. S5 b), indicating why the effects on cell growth of NKX3.1 knockdown could be more dramatic than TAL1 knockdown. In Jurkat and HSB2 cells, NKX3.1 knockdown also had effects on the cell cycle similar to those of TAL1; i.e., it diminished the percentage of cells in G1 phase and increased the percentage of cells in G2/M phase (Jurkat) or in S phase (HSB2; Fig. 6 b). Finally, constitutive expression of NKX3.1 in shTAL1-expressing Jurkat and HSB2 cells (Fig. 6 c, top) rescued the proliferative deficiency associated with low TAL1 expression but did not prevent cell death (Fig. 6 c, bottom).
Figure 6.
NKX3.1 mediates proliferating effects of TAL1 in human T cell lines. (a, top) NKX3.1 protein in Jurkat and HSB2 cells that stably expressed shRNAs directed against human NKX3.1 (shNKX3.1) or human hepatitis B virus (shCTL) was measured by immunoblotting. Actin is shown as a loading control. The data shown correspond to one representative experiment out of two performed. (middle) Growth curves of Jurkat and HSB2 cells expressing shNKX3.1 or shCTL. Graphs show number of cells ± SEM (n = 4 experiments). (bottom) Doubling times of Jurkat and HSB2 cells expressing shCTL or shNKX3.1 analyzed during the exponential phase of the growth curves. (b) Percentages of Jurkat (left) and HSB2 (right) cells expressing shCTL or shNKX3.1 in G1, S, and G2/M phases were determined at days 1, 2, and 3 of culture. Graphs show percentages of cells ± SEM (n = 3 experiments; *, P < 0.05; **, P < 0.01). (c, top left) Jurkat and HSB2 cells were transduced with viral particles containing human NKX3.1 cDNA followed by an IRES/EGFP under the control of the MND promoter. These cells were then transduced with viral particles containing the EF1-α–EGFP construct and shTAL1 under the control of the H1 promoter. Each transduction was 100% efficient as shown by FACS analysis of EGFP expression. Dashed lines and arrows determine the fluorescent population studied. (top right) Western blot analysis of TAL1 and NKX3.1 expression in shCTL- and shTAL1-expressing Jurkat cells. “NKX3.1 + shTAL1” defined the NKX3.1-expressing Jurkat cells transduced with the shTAL1 lentivirus. Actin is shown as a loading control. The data shown correspond to one representative experiment out of two performed. (bottom) Growth curves of Jurkat and HSB2 cells expressing shCTL, shTAL1, or shTAL1 and NKX3.1. Graphs show number of cells ± SEM (n = 4 experiments).
To identify common target genes regulated by TAL1 and NKX3.1 knockdown, we performed gene expression profiling of Jurkat and HSB2 T cell lines expressing shNKX3.1, shTAL1, or shCTL. We isolated RNAs from cells after 2 d of culture and used them to interrogate Affymetrix DNA microarrays. Transcription profiles were compared, and we characterized a common set of genes possibly involved in leukemic cell proliferation and for which expressions were dependent on TAL1 and NKX3.1 expressions (Fig. S6 a; deposited in ArrayExpress under accession no. E-MEXP-2197). Most genes have an expression dependent on TAL1 and/or NKX3.1, whereas few genes have an expression dependent only on NKX3.1, indicating NKX3.1 as a mediator of TAL1-regulated pathways (Fig. S6 b).
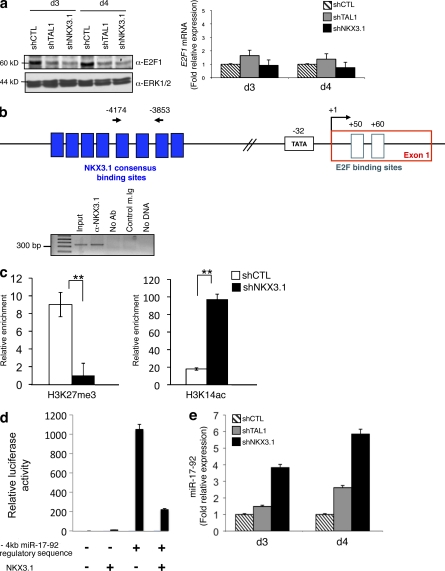
MicroRNAs (miRNAs) are expressed in human T-ALL (Chen et al., 2010), and the oncogenic 17-92 cluster of miRNAs is of high importance in human hematopoietic cancers (Mavrakis et al., 2010). Furthermore, miR-17-92 regulates expression of E2F1 and thus of the cell cycle (Nagel et al., 2009), and miR-19, a member of the miR-17-92 cluster, is sufficient to promote leukemogenesis in Notch1-induced T-ALL (Mavrakis et al., 2010). As NKX3.1 knockdown results in growth inhibition, we studied E2F1 expression in Jurkat cells expressing shCTL, shTAL1, or shNKX3.1 and showed a decreased E2F1 protein level without any significant decrease of E2F1 mRNA level (Fig. 7 a). NKX3.1 could bind a miR-17-92 gene region located 4 kb 5′ from the transcription start site (Fig. 7 b). This binding is associated with increased (vs. decreased) binding of histone H3 marked with repressive label (vs. activating label; H3K27me3 vs. H3K14ac) on this regulatory sequence (Fig. 7 c), and NKX3.1 could repress transcriptional activity of the −4-kb regulatory sequence of the miR-17-92 gene (Fig. 7 d). Finally, quantitative PCR analysis of pri-miR-17-92 showed that its expression level inversely correlated with TAL1 and NKX3.1 expressions (Fig. 7 e). These results identified miR-17-92 as a direct target of NKX3.1 in the Jurkat cell line. Altogether, these results showed that NKX3.1 could mediate the proliferative effects of TAL1 in T cell lines through a direct regulation of a specific set of genes.
Figure 7.
NKX3.1 directly regulates miR-17-92 promoter activity. (a, left) E2F1 protein in shCTL-, shTAL1-, and shNKX3.1-expressing Jurkat cells at days 3 and 4 of culture was measured by immunoblotting. ERK1/2 is shown as a loading control. The data shown correspond to one representative experiment out of three performed. (right) E2F1 mRNA was determined by quantitative RT-PCR normalized to GAPDH mRNA in the same T cell lines at days 3 and 4 of culture. Error bars indicate SEM (n = 4 experiments). (b, top) Schematic organization of the miR-17-92 promoter region. The transcriptional start site (+1), TATA box (−32), and exon 1 and E2F binding sites (+50 and +60) are indicated. The cluster of NKX3.1 consensus binding sites located 4 kb upstream of the transcription start site is shown by blue boxes. Location of the primer set used in the ChIP analysis is shown by arrows. (bottom) Jurkat cell chromatin extract was immunoprecipitated with NKX3.1 antibody or control Ig. Immunoprecipitated DNA was amplified to detect a 300-bp region located at −4 kb in the miR-17-92 promoter region. Input is amplification of Jurkat cell DNA. The data shown correspond to one representative experiment out of two performed. (c) H3K27me3 and H3K14ac antibodies or control Ig was used to immunoprecipitate chromatin extracts from Jurkat shCTL or Jurkat shNKX3.1. The −4-kb miR-17-92 regulatory sequence was detected by quantitative PCR on the immunoprecipitated DNA. Data are the means ± SEM (n = 3 experiments; **, P < 0.01). (d) Jurkat cells were transfected with a plasmid containing the −4-kb miR-17-92 regulatory sequence that binds NKX3.1 cloned upstream of a promoter that drives the firefly luciferase gene with or without an NKX3.1 cDNA sequence. Transfections were normalized using the dual-luciferase reporter assay system. Data are the means ± SEM (n = 4 experiments). (e) miR-17-92 RNA was determined by quantitative RT-PCR normalized to 36B4 RNA in shCTL, shTAL1, or shNKX3.1 Jurkat cells at days 3 and 4 of culture. Error bars indicate the SEM (n = 4 experiments).
NKX3.1 expression, regulation, and function in primary human T-ALL
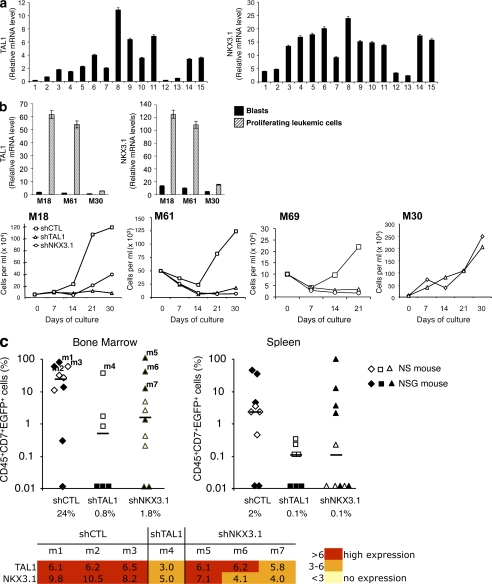
As cell lines may not reliably reflect patient samples, TAL1 and NKX3.1 mRNA levels were quantified in human primary circulating T-ALL blasts, and these quantifications showed a correlation between TAL1 and NKX3.1 expressions (Fig. 8 a). T-ALLs proliferate in bone marrow and other organs, but 90% of the circulating blasts are in the G0 phase and do not proliferate (Fig. S7 a, left). Therefore, to study any role of TAL1 and NKX3.1 in proliferation of leukemic cells, we used the previously described MS5-DL1 mouse stromal cell line, which allows activation, maintenance, and proliferation of primary T-ALL leukemic cells in vitro (Fig. S7 a [right panel] and b; Armstrong et al., 2009). Four human T-ALL primary samples that express (M18, M61, and M69) or do not express (M30) TAL1 were grown on MS5-DL1. TAL1 and NKX3.1 mRNA levels were 10 times higher in M18 and M61 leukemic proliferating cells than in M18 or M61 circulating blasts, and these mRNAs were not detected in M30 proliferating cells (Fig. 8 b, top). These increased mRNA levels were mostly related to proliferation and not to activation of the Notch signaling pathway, as shown by the effect of DAPT, an inhibitor of this pathway, on TAL1 and NKX3.1 mRNA levels (Fig. S7 c) and on Ki67-positive leukemic cells (Fig. S7 d). Decreased TAL1 or NKX3.1 expression abrogated proliferation of M18, M61, and M69 but not of M30 leukemic cells, indicating that these two proteins were necessary for proliferation of primary TAL1-expressing T-ALL leukemic cells (Fig. 8 b, bottom). Decreased TAL1 or NKX3.1 expression also impaired leukemia development in vivo as leukemia development from shTAL1- or shNKX3.1-expressing M18 leukemic cells was limited compared with control cells after transplantation into immunodeficient mice (Fig. 8 c, top). Furthermore, NKX3.1 or TAL1 expression was detected in the leukemias that developed in mice, indicating that only blasts with little inhibition of TAL1 or NKX3.1 could escape from the proliferation defect and were selected and responsible for T-ALL leukemia development (Fig. 8 c, bottom). These results show that TAL1 and NKX3.1 are involved in efficient development of human T-ALL in recipient immunodeficient mice.
Figure 8.
NKX3.1 is expressed in human TAL1-expressing T-ALL cells and is necessary for in vitro proliferation and in vivo development of human T-ALL. (a) TAL1 and NKX3.1 mRNA levels in human T-ALL blasts at diagnosis. Quantitative RT-PCR analysis of TAL1 and NKX3.1 mRNA was performed using RNA prepared from blasts obtained from 15 human T-ALL. Relative expressions were normalized to β2m mRNA. Error bars represent means ± SEM (n = 4 experiments). (b, top) TAL1 and NKX3.1 mRNA levels were analyzed by quantitative RT-PCR using RNA extracted from TAL1-expressing (M18 and M61) or non-TAL1–expressing (M30) T-ALL leukemic cells at diagnosis (black bars) or after 3 d of culture on the MS-5 cell line expressing the DL1 Notch ligand (MS5-DL1; hatched bars). Relative expressions were normalized to β2m mRNA level. Error bars indicate SEM (n = 4 experiments). (bottom) Growth curves of M18, M61, M69, and M30 primary T-ALL expressing shCTL, shTAL1, or shNKX3.1 co-cultured with MS5-DL1 for 21 d (M69) or 1 mo. (c) M18 leukemic cells expressing shCTL, shTAL1, or shNKX3.1 were injected into NS and NSG mice. (top) percentages of human leukemic transduced (CD45+CD7+EGFP+) cells detected in bone marrow and spleen of mice at 6 (NSG) and 12 (NS) wk after transplantation. Median engraftment levels (horizontal bars) are indicated under the diagram. (bottom) Quantitative RT-PCR analyses of TAL1 and NKX3.1 mRNA were performed using RNA from CD45+CD7+EGFP+ cells isolated from mice that developed leukemia (m1–m7). Relative expression levels were normalized to β2m mRNA level and represented by a color scale (n = 3 experiments).
DISCUSSION
The class II bHLH factor TAL1 is aberrantly expressed in 60% of children and in 45% of adults with T-ALL, but the mechanisms that mediate its oncogenic potential remain unclear (O’Neil and Look, 2007). Using RNA interference to knock down TAL1 expression in human T-ALL cell lines that express low or high levels of TAL1 protein, we showed that TAL1 regulates proliferation in these cell lines. These results are in accordance with a previously described role for TAL1 in promoting the aberrant growth of Jurkat cells without any effect on apoptosis (Palomero et al., 2006). The effects observed on Jurkat and HSB2 cell proliferation shown in this article are particularly drastic as we used lentiviruses that transduce 100% of cells and consequently eliminate any potential side effects, such as affecting clonal selection. Moreover, the cell culture conditions used (i.e., 1% FCS) allowed us to detect this drastic effect of TAL1 knockdown on proliferation, indicating that growth factors present in the serum can complement some of the oncogenic effects of TAL1.
To study the TAL1 network that regulates the proliferation of T-ALL cell lines, we performed gene expression profiling using oligonucleotide microarrays after 2 or 3 d of culture of Jurkat or HSB2 cell lines expressing or not TAL1. We focused our study on a set of genes whose transcriptional activity was dependent on TAL1 expression in both cell lines. Some of these genes, like Six6, RAG2, or RALDH2, have been shown to be specifically expressed in TAL1-expressing primary T-ALL blasts (Soulier et al., 2005), indicating that our screening might be more relevant to human T-ALL biology than to cell line–associated secondary events. No obvious metabolic pathway regulated by TAL1 expression could be deduced from the analysis of the set of genes characterized in the two cell lines, and our results indicated that TAL1 might act as an activator or a repressor in human T-ALL, a feature previously suggested (Hsu et al., 1994b; Palomero et al., 2006).
Among the genes whose expression depends on TAL1, we focused our study on NKX3.1, a member of the NKX family of homeobox genes that is specifically expressed in the prostate epithelium during prostate development and in adulthood. Only one member of the NKX family, NKX2.5, has already been characterized as a master oncogene in human T-ALL. NKX2.5 is activated only by T-ALL–specific rearrangements (Przybylski et al., 2006) and is expressed in two T cell lines, Peer and CCFR-CEM (Nagel et al., 2003). In accordance with previous studies, we did not find any expression of NKX2-5 in Jurkat, HSB2, or RPMI T cell lines but found NKX2.5 expression in CCFR-CEM cells (Nagel et al., 2003; Przybylski et al., 2006). As TAL1 knockdown (respectively expression) did not affect NKX2.5 expression in CCFR-CEM cells (respectively Peer; unpublished data), we did not study any further NKX2.5 roles in T cell lines and focused our research on NKX3.1. We showed that the NKX3.1 gene is directly activated by TAL1 in T-ALL through a GATA-3–mediated recruitment of a TAL1–LMO–Ldb1 complex on regulatory sequences of the NKX3.1 gene. This recruitment is associated with suppression of the NKX3.1 gene repression at the chromatin level. This peculiar gene activation in human T-ALL is similar to the TAL1-mediated RALDH2 gene activation in T-ALL (Ono et al., 1998) and pinpoints GATA-3 as an important component of TAL1 function in human T cell leukemogenesis. During erythropoiesis, a TAL1–LMO2–Ldb1–E2A complex can distinguish active from repressive GATA-1 binding sites as this complex is only present at activating elements and is not associated with repressive GATA-1 binding sites. In human T-ALL, TAL1–E2A direct binding to DNA is often associated with repression of target genes after recruitment of corepressors such as LSD1 or HP1-α (Wen et al., 2005; Hu et al., 2009), but we now showed that the recruitment of the TAL1–LMO–Ldb1 complex by GATA-3 could relieve the repression of NKX3.1 and RALDH2 genes by antagonizing HP1-α function. Altogether, these results indicated a similarity between TAL1 functions in erythropoiesis and T cell leukemogenesis with a repressive or activating function of TAL1 depending on its recruitment to DNA. Finally, as for the RALDH2 gene, the regulatory sequences that mediated TAL1 recruitment on the NKX3.1 gene promoter are not conserved between human and mouse, which suggests why the Nkx3.1 gene is not activated in mouse models of TAL1-mediated T-ALL. Tissue-specific transcriptional regulation is markedly different between human and mouse. A recent study has shown that >50% of four tissue-specific transcription factor binding sites are species specific, and it is thus not surprising to find specific gene activations in human T-ALL that cannot be mimicked by mouse models (Odom et al., 2007). Together with works on the transcriptional networks downstream of TAL1 in human T-ALL (Rangarajan et al., 2004; Soulier et al., 2005), our study indicates that mouse models of human T-ALL might recapitulate only part of human T cell leukemogenesis. Species-specific differences have already been described for transformation of rodent cells compared with human cells (Rangarajan et al., 2004). Rodent cells seemed to need fewer events to be transformed as two signaling pathways are required for tumorigenic conversion of normal mouse fibroblasts, whereas perturbation of six pathways is required for the transformation of human fibroblasts (Rangarajan et al., 2004). Our results indicated that TAL1-mediated leukemogenesis might also require less signaling pathways in mouse than in human and shows the importance of paralleled experiments in mouse and human leukemia and of mouse models of human leukemia.
What could be the role of NKX3.1, a tumor suppressor gene in human prostate cancer, in human T-ALL? To address this question, we first inhibited NKX3.1 gene expression in the different TAL1+/T-ALL cell lines studied and found a near failure of proliferation when the NKX3.1 protein was down-regulated. These results indicated that NKX3.1 could mediate some of the proliferative effects of TAL1 in T-ALL and complementation experiments of TAL1 knockdown by overexpression of NKX3.1 confirmed this hypothesis. During the prostate cancer initiation, NKX3.1 blocks some of the effects of PTEN loss and inhibits AKT phosphorylation (Lei et al., 2006). In human T-ALL, PTEN is frequently deleted (Maser et al., 2007), and thus the TAL1-mediated activation of NKX3.1 might block some of the effects of PTEN loss during TAL1-mediated T cell leukemogenesis. However, in Jurkat cells, which harbor a mutated PTEN protein, NKX3.1 knockdown did not result in a diminished AKT phosphorylation (unpublished data), indicating that NKX3.1 regulated the proliferation of these cells using other pathways. The oncogenic function of NKX3.1 in human T-ALL might also be related to the role of NKX3.1 in cellular proliferation of LNCaP, a hormone-sensitive model of prostate cancer (Possner et al., 2008). As NKX3.1 might have different roles during prostate cancer progression, including a role in proliferation at the early steps of prostate carcinogenesis, our results indicated that NKX3.1 might only have proliferative functions in human T-ALL. To determine the metabolic pathways regulated by NKX3.1 and TAL1 in human T-ALL, we performed and analyzed gene expression profiling and found a common set of genes for which transcription was dependent on TAL1 and NKX3.1 expression in Jurkat and HSB2. Among these genes, two subsets could be related to the growth curves previously obtained. The first one contains genes known to be involved in the T cell signaling and/or cellular proliferation (Fli1, Vav3, or EiF4E3), and the second one contains genes involved in calcium metabolism (calnexin and calumenin) that regulates T cell proliferation (Michalak et al., 2002; Medyouf et al., 2007). In addition to mRNAs whose levels were dependent on NKX3.1 expression, we found that miR-17-92 was directly regulated by NKX3.1. miR-19, a member of the miR-17-92 cluster, has been recently shown to promote leukemogenesis in a mouse Notch1-induced T-ALL (Mavrakis et al., 2010), and activation of miR-17-92 by NKX2-5 suppresses apoptosis via reduction of E2F1 (Nagel et al., 2009). Our results indicated that NKX3.1 might be a possible link between TAL1 and the miR-17-92 transcriptional regulation in human T-ALL, but further studies are needed to precisely define the role of NKX3.1 on mature miRNA expression (particularly miR-19, derived from miR-17-92).
Patient-derived T-ALL cell lines may not totally reflect the original patient samples. To further explore the roles of TAL1 and NKX3.1 in leukemia development, we used patient samples. We first studied TAL1 and NKX3.1 expression levels in primary samples from T-ALL patients and showed a correlation between NKX3.1 and TAL1 gene expressions, a result which is in accordance with previous data (Soulier et al., 2005). However, as we observed that blood-derived T-ALL blasts are mostly quiescent cells, we switched to a cell culture system we recently showed to allow activation and/or proliferation of T-ALL leukemic cells as well as preservation of T-ALL leukemia–initiating cell activity (Armstrong et al., 2009). In this context, we found that TAL1 and NKX3.1 gene expressions were increased >10 times in leukemic cells recovered after culture in comparison with uncultivated T-ALL blasts. These results suggested that caution should be exercised when using gene expression profiling of blood-derived T-ALL blasts, especially when studying genes involved in the activation or proliferation of T-ALL cells. In the context of culture-activated leukemic cells, we found that the Notch pathway did not significantly regulate TAL1 and NKX3.1 gene expression, indicating that the TAL1–NKX3.1 and Notch pathways might cooperate in the development of human T-ALL. We finally showed that NKX3.1 expression is an important component of the development of human T-ALL in vivo as NKX3.1 knockdown in human T-ALL samples limited leukemia burden in immunodeficient mice. These results strengthen the importance of NKX3.1 gene activation by TAL1 during human T cell leukemogenesis. Further experiments are needed to establish whether NKX3.1 and TAL1 participate in the maintenance and/or proliferation of T-ALL leukemia–initiating cell, as we have recently demonstrated for Notch (Armstrong et al., 2009).
In conclusion, the results described in this study characterize NKX3.1 as the first gene directly activated by TAL1 in human T-ALL and involved in the proliferation of human T-ALL in vitro and in vivo. These findings extend the current view of TAL1-mediated T leukemogenesis, e.g., interference with the transcriptional regulation elicited by the tumor suppressor gene E2A, and may define new pathways that could be used for the treatment of T-ALL.
MATERIALS AND METHODS
Cell lines, T-ALL samples, and culture conditions.
Jurkat, HSB2, and Peer cells were maintained in RPMI 1640 containing 10% FCS (Invitrogen). Cells were grown in 10, 5, 2.5, and 1% FCS for the growth curve analysis, and viable cells were counted every day by trypan blue exclusion. Primary T-ALL samples were obtained and characterized (Armstrong et al., 2009) at diagnosis after informed consent of all patients’ parents, in accordance with national ethics rules, and cultured with mouse stromal MS5 cells as described previously (Armstrong et al., 2009).
Lentiviral vectors and cell transduction of cell lines and T-ALL primary samples.
Oligonucleotides 5′-GTTCAGCCATCAGAAGTAC-3′ (respectively 5′-TGTGTCACTGAATATCAAC-3′) were designed in the coding region of the human NKX3.1 gene and used for PCR amplification. The 230-bp DNA fragment obtained was purified and cloned in the pSuper plasmid 3′ of the polIII H1 promoter. H1-shRNANKX3.1 DNA fragment was then subcloned in pTRIP/ΔU3–EF1-α–EGFP, where the GFP coding sequence is under the control of the EF1-α promoter (TRIP/ΔU3–EF1-α–EGFP; Sirven et al., 2001). The lentiviral vectors pTRIP/ΔU3–EF1-α–EGFP encoding an shTAL1 or a control shCTL directed against the human hepatitis B virus have been described previously (Lazrak et al., 2004; Brunet de la Grange et al., 2006), and we also used another shTAL1 (5′-GCAACATTGTTCACCTTTG-3′) to avoid off-target effects. The human TAL1 coding sequence cloned under the EF1-α promoter (TRIP/ΔU3–EF1-α–TAL1) has been described previously (Ravet et al., 2004). The human NKX3.1 coding sequence was cloned into the pTRIPΔU3-Mnd-IRES-EGFP after PCR amplification using primers (forward) 5′-GCGGGTGCATTCAGGCCAAGG-3′ and (reverse) 5′-GGTGTGCACACCTCTTGCTCC-3′ and sequenced. Concentrated VSV/G pseudotyped shRNA and cDNA lentiviral vectors were produced as described previously (Ravet et al., 2004) and added to T-ALL cell lines cultured at a P24 concentration of 0.5 µg/ml twice at 24-h intervals. T-ALL samples were transduced with unconcentrated G/IL-7SUx envelope pseudotyped vectors (Verhoeyen et al., 2003) according to Gerby et al. (2010). In brief, T-ALL samples were spinoculated for 2 h in the presence of plastic-bound human DL1, 5 µg/ml protamine sulfate, and G/IL-7SUx pseudotyped lentiviral vectors, and transduction was continued for a further 48 h. EGFP+ T-ALL primary cells were sorted (FACSAria; BD) and used in functional assays.
Cell cycle assay.
Cells (106) were labeled with 10 µM BrdU for 45 min at days 1, 2, and 3 of culture. Cells were washed to remove BrdU and put in chase medium (RPMI 1640 and 1% FCS) or immediately fixed. After a chase period of 2 h, cells were fixed with 70% ethanol and stored overnight at −20°C. After washing and centrifugation, cells were treated with 10 µg/ml RNase A for 20 min at 37°C. DNA was denatured with 2 N HCL/0.5% Triton X-100 for 30 min at 37°C to recover BrdU epitope and rinsed in PBS. Cells were incubated with FITC-conjugated mouse anti-BrdU antibody (1:10; BD) for 1 h at room temperature, washed in PBS, and then incubated with 20 µg/ml propidium iodide before acquisition on the flow cytometer (Mow Flow Cytometer [Beckman Coulter] and FACSCalibur [BD]). Data were analyzed with FlowJo software (Tree Star, Inc.) for bivariate analysis of DNA content versus BrdU.
Oligonucleotide pull-down.
Cells were washed with PBS and incubated for 10 min at 4°C in buffer A (10 mM Hepes, pH 7.6, 3 mM MgCl2, 10 mM KCl, 5% glycerol, and 0.5% NP-40) containing proteinase inhibitor cocktail (Roche). After centrifugation, nuclear pellets were resuspended in buffer B (10 mM Hepes, pH 7.6, 3 mM MgCl2, 300 mM KCl, 5% glycerol, and 0.5% NP-40) and incubated for 30 min at 4°C. For oligonucleotide pull-down assays, nuclear extracts from Jurkat cells were incubated with 1 µg of the indicated double-strand biotin-labeled oligonucleotide at 4°C overnight in buffer containing 10 mM Hepes, pH 7.6, 3 mM MgCl2, 150 mM KCl, 5% glycerol, and 0.5% NP-40. DNA–protein complexes were pelleted using streptavidin beads (GE Healthcare), and the beads were then washed three times with buffer A, resuspended in Laemmli buffer, and subjected to immunoblotting analysis using antibodies against TAL1 or GATA-3 (Santa Cruz Biotechnology, Inc.).
Biotin-labeled oligonucleotides used for the oligonucleotide pull-down were as follows: E−867, 5′-biotin-TACATGTTAACAGTTGGTAATCTGCA-3′; E−800, 5′-biotin-GCAGTTTTTGCATTTGTCCTGGCCTA-3′; G−748/E−738, 5′-biotin-GATGGTATGTCATGTGTCTGGGGAGG-3′; G−697, 5′-biotin-GGTTATTGCGGATAAAGGAACCAC-3′; and G−577, 5′-biotin-TGAGGTCGTAGATATTGCAGATCT-3′.
Immunoblotting analysis.
Total proteins from 1–2 × 106 T-ALL cell lines were extracted with 1× Laemmli buffer. Proteins were separated by 12% SDS-PAGE and transferred onto nitrocellulose membrane (Schleicher & Schuell). Membranes were incubated in Tris-buffered saline with 5% nonfat milk containing antibodies against TAL1 (3BTL73 mouse monoclonal antibody provided by D. Mathieu [Centre National de la Recherche Scientifique Unité Mixte de Recherche 5535, Montpellier, France] or Santa Cruz Biotechnology, Inc.), LMO1 (Abcam), Ldb1, GATA-3, E2F1, or NKX3.1 (Santa Cruz Biotechnology, Inc.). Normalization was performed using β-actin (clone AC-15; Sigma-Aldrich) or ERK1/2 antibodies (Cell Signaling Technology). After staining with secondary antibodies, the proteins were detected with Supersignal West Pico chemiluminescent substrate (Thermo Fisher Scientific). Hybridization signals were quantified by using the GS-800 calibrated densitometer with Quantity One 1D analysis software (Bio-Rad Laboratories).
ChIP.
Immunoprecipitation conditions were determined to ensure that the amount of antibody and percentage of cross-linking agent did not limit the ChIP yields. Cells (40 × 106) from the indicated cell lines were cross-linked for 10 min at 37°C with 1% formaldehyde in RPMI medium containing 10% FCS, washed twice with ice-cold PBS, and lysed in 2 ml of ChIP lysis buffer for 10 min (50 mM Tris, pH 8.0, 10 mM EDTA, and 1% SDS). Chromatin fragments between 250 and 3,000 bp were obtained by sonication with a Sonifier 450 (Branson).
5 µg of antibodies against HP1-α (Euromedex), H3K9me3 (Millipore), H3K14ac (Millipore), H3K4me3 (Millipore), and H3K27me3 (Millipore) was used to precipitate 1 µg of fragmented chromatin. Immunoprecipitations were performed in buffer containing 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and a protease inhibitor cocktail (Roche) at 4°C overnight. A negative control without antibody or with control Ig was run simultaneously with the samples. Immunocomplexes were collected by adsorption to protein G agarose/Salmon Sperm DNA beads (Millipore). Washing and elution of immunocomplexes were performed, cross-linking was reversed for 5 h at 65°C, and samples were then treated with proteinase K (Invitrogen). Immunoprecipitated DNA was extracted with phenol-chloroform-isoamyl alcohol. The Bmi, Pax6, NKX3.1, RALDH2, and miR-17-92 promoters were assessed for the presence of histone modifications using quantitative PCR. Amplification was performed using power SYBR green PCR Master mix (Applied Biosystems) supplemented with 0.5 µM of specific primer pairs and 100 ng DNA in a final volume of 20 µl. Amplification conditions included an initial denaturation at 95°C for 10 min, followed by 35 cycles at 95°C for 15 s and 60°C for 60 s. Data were normalized to input signal and reported to IgG values ± SD.
4 µg α-LMO1 (Abcam), 2 µg α-LMO2, 2 µg α-Ldb1, 5 µg α-TAL1, 5 µg α–GATA-3, and 5 µg α-NKX3.1 antibodies (Santa Cruz Biotechnology, Inc.) were used to precipitate 30 µg of fragmented chromatin. Immunoprecipitations were performed as described in the previous paragraphs, and PCR amplifications of eluted DNA from immunocomplexes were performed in 20 µl of reaction mixture using the specific primers for the NKX3.1, RALDH2, and miR-17-92 promoters. Amplification conditions included an initial denaturation at 95°C for 5 min, followed by 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The amplified DNA was separated on 2% agarose gels and visualized with ethidium bromide.
The sequences of the primers used were as follows: NKX3.1 promoter forward, 5′-AACGCCTTCCATCCGTCTGG-3′; NKX3.1 promoter reverse, 5′-TGGTGCAAACTCAGATCTGC-3′; NKX3.1 exon 2 forward, 5′-CCTCCCTGGTCTCCGTGTA-3′; NKX3.1 exon 2 reverse, 5′-TGTCACCTGAGCTGGCATTAC-3′; −4-kb miR-17-92 regulatory sequence forward, 5′-CCAGATAGCAAAGATATAACAG-3′; −4-kb miR-17-92 regulatory sequence reverse, 5′-CTTATATGAATCACGACTGAC-3′; RALDH2 intron 2 forward, 5′-AGAGTGTTCCCTGTCTATAATCCAGCC-3′; RALDH2 intron 2 reverse, 5′-TGTTAGAGAGGAAGAGGCACAACTGA-3′; RALDH2 exon 6 forward, 5′-AACTTCCCCCTGCTGATGTTTGCC-3′; and RALDH2 exon 6 reverse, 5′-CTTGATGAGGGCTCCCATGTAGAGTGC-3′.
For quantitative PCR, the primers used were as follows: NKX3.1 promoter forward, 5′-ATCATCTCTGTTCCACTTTCG-3′; NKX3.1 promoter reverse, 5′-TAACCTGTTAACTTTCCTTCC-3′; Pax6 promoter forward, 5′-GTCCGGTGCCTTGAACCAT-3′; Pax6 promoter reverse, 5′-GCGCAACTACCGCCTCTAAA-3′; Bmi promoter forward, 5′-CCAGTCTAGTGCATGCCTTCTTAA-3′; Bmi promoter reverse, 5′-CAAGCCAGCGACGCAGT-3′; miR-17-92 promoter forward, 5′-CCAGATAGCAAAGATATAACAG-3′; and miR-17-92 promoter reverse, 5′-GTAACCTTGTTTGGGAAGACAC-3′.
Real-time quantitative RT-PCR.
Total RNA was isolated from 5 × 105 cells using 1 ml TRIZOL reagent (Invitrogen) or the mirVana miRNA isolation kit (Applied Biosystems). RNA was reverse transcribed using random hexamers and the Superscript RT kit (Invitrogen) according to the manufacturer’s instructions. The cDNA reactions were diluted 1:10 in water and used as template in real-time PCR reactions using the power SYBR green PCR Master mix or the TaqMan Universal PCR MasterMix No AmpErase UNG (Applied Biosystems; supplemented with 0.5 µM specific probe for the specific primer pairs if necessary) in the 7300 fast real-time PCR system (Applied Biosystems). The cycle at which a particular sample reaches an arbitrary threshold fluorescent level (Ct) is indicative of the input quantity of the template. The raw data were obtained in terms of Ct values that refer to the PCR cycle number during exponential amplification at which the product crosses an arbitrary threshold. To adjust for variations in the amount of RNA, the Ct values for each gene were normalized against the Ct values for the housekeeping genes, GAPDH, β2m, or 36B4. The primer sequences used for real-time quantitative PCR were the following: GAPDH forward, 5′-GGGAAACTGTGGCGTGAT-3′; GAPDH reverse, 5′-GGAGGAGTGGGTGTCGCTGTT-3′; β2m forward, 5′-CACAGCCCAAGATAGTTAAGT-3′; β2m reverse, 5′-CCAGCCCTCCTAGAGC-3′; NKX3.1 forward, 5′-CCTCCCTGGTCTCCGTGTA-3′; NKX3.1 reverse, 5′-TGTCACCTGAGCTGGCATTAC-3′; TAL1 forward, 5′-TCTGAAGCAAGGCGGTGGAC-3′; TAL1 reverse, 5′-GGAAGACCGTGCCGTCTTCA-3′; Hes1 forward, 5′-CAACACGACACCGGATAAACC-3′; Hes1 reverse, 5′-CCAGAATGTCCGCCTTCC-3′; Deltex forward, 5′-TTCTGACTTCAGGAGCGAAAG-3′; Deltex reverse, 5′-TGCCCACTCCCAACGA-3′; E2F1 forward, 5′-CCCAACTCCCTCTACCCTTGA-3′; E2F1 reverse, 5′-TCTGTCTCCCTCCCTCACTTTC-3′; miR-17-92 forward, 5′-CAGTAAAGGTAAGGAGAGCTCAATCTC-3′; miR-17-92 reverse, 5′-CATACAACCACTAAGCTAAAGAATAATCTGA-3′; miR-17-92 probe (200 nM final), FAM-TGGAAATAAGATCATCATGCCCACTTGAGAC-TAMRA; 36B4 forward 5′- GATGCCCAGGGAAGACAG-3′; 36B4 reverse 5′-TCTGCTCCCACAATGAAACAT-3′; and 36B4 probe (50 nM final), FAM-GACCTGGA-TAMRA.
Microarray processing.
Quality of RNA samples was assessed using RNA 6000 Nano chips (Agilent Technologies), and quantity was assessed by measuring the absorbance at 260 nm with ND-1000 (NanoDrop). cRNA was prepared by the PartnerChip Company according to the protocols of the manufacturer (Affymetrix). In brief, 2 µg of RNA samples was used to generate first-strand cDNA using a T7–oligo (dT) primer and the Superscript II RT (Invitrogen). Second-strand synthesis was achieved using a cocktail of enzymes from Escherichia coli (DNA ligase and DNA polymerase I), RNase H, and T4 DNA polymerase. Clean-up of double-stranded cDNA was performed using the GeneChip Sample Cleanup Module (Affymetrix). In vitro transcription was performed in the presence of T7 RNA polymerase and a biotinylated nucleotide analogue/ribonucleotide mix for cRNA amplification and biotin labeling (GeneChip IVT Labeling kit; Affymetrix). Resulting biotinylated cRNAs were cleaned up using the GeneChip Sample Cleanup Module and quantified by absorbance measurement at 260 nm (NanoDrop). Starting with 5 µg of total RNA yielded between 50 and 80 µg of purified cRNA. Then, 20 µg of cRNA from samples was incubated at 94°C for 35 min in a fragmentation buffer to be reduced to a mean size of ∼35–200 nt and finally added to the hybridization buffer. Fragmented cRNAs were hybridized on HG-U133 Plus 2.0 array (Affymetrix) for 16 h at 45°C together with internal hybridization controls (bioB, bioC, bioD, cre, and oligonucleotide B2). The washing and staining procedure was performed in the Fluidics Station 450 (Affymetrix). Probe arrays were exposed to 10 washes in nonstringent wash buffer A (6× saline–Na phosphate–EDTA and 0.01% Tween 20) at 30°C, followed by 6 washes in stringent buffer B (100 mM 2-[4-morpholino]ethanesulfonic acid, 0.1 M [Na+], and 0.01% Tween 20) at 50°C. The biotinylated cRNAs were stained with 10 µg/ml of a streptavidin-phycoerythrin conjugate (SAPE) for 5 min at 35°C and washed again with nonstringent buffer A (10 washes at 30°C). An antibody amplification step was added using 0.1 mg/ml goat IgG and 3 µg/ml biotinylated antistreptavidin antibody followed by an additional SAPE stain (5 min at 35°C for each step). Finally, arrays were washed 15 times in nonstringent buffer A at 35°C before scanning in GeneChip Scanner 3000 (Affymetrix).
Microarray analyses were performed at the Institut de Génétique et de Biologie Moléculaire et Cellulaire French resource and by PartnerChip. HG-U133 Plus 2.0 array (Affymetrix) contains ∼54,600 human probe sets corresponding to ∼22,400 UniGene clusters. CEL files containing raw intensities were produced using GCOS (GeneChip operating software; Affymetrix). Data were normalized using GeneChip robust multi-array average, which generates intra- and interchip normalizations in a single step. Microarray data were submitted to ArrayExpress (deposited under accession no. E-MEXP-2197).
Promoter in silico analysis.
NKX3.1 promoter sequence alignments were generated using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Mouse (promoter ID 55970) and human (promoter ID 41156) sequences were obtained from the Transcriptional Regulator Element Database (http://rulai.cshl.edu/cgi-bin/TRED/tred.cgi?process=searchPromForm). The c13orf25 gene contains a polycistronic cluster of miRNAs (miR-17–92), and its promoter has been previously described (Ota et al., 2004; Woods et al., 2007).
Promoter reporter constructs, transient transfections, and luciferase assays.
A 494-bp DNA fragment of the human NKX3.1 promoter spanning nt −977 to −482 relative to the start codon was amplified using the primers 5′-ATGCCTCGAGATCATCTCTGTTCCACTTTCG-3′ (forward) and 5′-GCATTCTAGAGATGAGCACGCAGTCACTGC-3′ (reverse). A 434-bp DNA fragment of the mouse NKX3.1 promoter spanning nt −875 to −441 relative to the start codon was amplified using the primers 5′-ATGCCTCGAGACCTGGCTGTCCAAGAAATCC-3′ (forward) and 5′-GCATTCTAGAGAAGGTCCAAGTAGTATGAC-3′ (reverse). The fragments were sequenced and cloned upstream from a TATA box sequence (5′-TCTAGAGGGTATATAATGGATCTAAGTAAGCTT-3′), and the resulting DNA fragments were cloned upstream from the firefly luciferase-coding sequence in pGL2b (Promega). Mutations (base substitution) of the E-box E−738 and the GATA motifs G−748 and G−697 in the human promoter were performed using the QuikChange II site-directed mutagenesis kit (Agilent Technologies). The binding site sequences 5′-GATG-3′ (G−748), 5′-GATA-3′ (G−697), and 5′-CATGTG-3′ (E−738) were, respectively, mutated to 5′-GCCC-3′, 5′-GCCC-3′, and 5′-CCTCGG-3′ sequences. Jurkat cells or Jurkat shTAL1 (107) were electroporated in a volume of 150 µl of PBS-Hepes (10 mM, pH 7.4) by a single pulse of 200 V at 960 µF with a Gene Pulser apparatus (Bio-Rad Laboratories). 10 µg of reporter gene plasmid and 2 µg of pRL-TK plasmid (Promega) for normalization of transfection efficiency were used. Cells were stimulated with 1 µM ionomycin and 50 ng/ml PMA (Sigma-Aldrich) and harvested 24 h after transfection. Luciferase activities were determined using the dual-luciferase reporter assay system as indicated by the manufacturer (Promega).
A 1612-bp DNA fragment of the human miR-17-92 promoter spanning nt −4840 to −3228 relative to the transcriptional start site was amplified using the primers 5′-TACTGAGGTACCAAGTGCCCCCAGAAAGGCAA-3′ (forward) and 5′-TACTGACTCGAGCAAAGACCATAATCATTTAACCTG-3′ (reverse). The resulting DNA fragment was cloned into KpnI and XhoI sites of the pGL3-control vector (Promega) upstream from the firefly luciferase–coding sequence, sequenced, and cotransfected with 100 ng pcDNA3-NKX3.1 and Renilla luciferase vector (Promega) into HEK 293 cells. Luciferase activities were determined using the dual-luciferase reporter assay system as indicated by the manufacturer (Promega).
Animals.
NOD.CB17-Prkdc(scid) (abbreviated NOD/scid; NS) and NOD.Cg-Prkdc(scid)Il2rg(tm1Wjll)/SzJ (abbreviated NOD/scid/IL-2Rγ null; NSG; both from The Jackson Laboratory) were housed in pathogen-free animal facilities at the Commissariat à l’Energie Atomique et aux Energies Alternatives, Fontenay-aux-Roses, France. Mice were sublethally irradiated at 3 Gy (IBL 637 CisBio International, France; dose rate 0.61 Gy/min) and anesthetized with isoflurane before i.v. injection of human leukemic transduced (CD45+CD7+EGFP+; 5 × 104/mouse) cells. All experimental procedures were performed in compliance with French Ministry of Agriculture regulations (animal facility registration number A920322) for animal experimentation. Mice were euthanized when they reached endpoints set to meet accepted animal care guidelines. For analysis of the development of leukemia, human leukemic cells in bone marrow, spleen, and thymus were stained with conjugated mouse anti–human-specific monoclonal antibodies PE-CD45 and PC5-CD7 (Beckman Coulter).
Statistical analyses.
Statistical significance of compared measurements was determined using the Student’s two-tailed t test. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 were considered statistically significant. Data are presented as means ± SEM.
Online supplemental material.
Fig. S1 shows TAL1 protein measured by immunoblot in different T-ALL cell lines and demonstrates serum-dependent effects of TAL1 knockdown on Jurkat and HSB2 growth. Fig. S2 shows that reduced TAL1 expression in Jurkat and HSB2 T cell lines is associated with up- and down-regulation of a common set of genes. Fig. S3 shows the sequence alignment of human and mouse NKX3.1 gene promoter regions and analysis of Nkx3.1 and Nkx2.9 mRNA expression levels performed by global gene expression in transgenic mice. Fig. S4 shows the fixation of GATA-3 and TAL1 on the regulatory sequence located in the second intron of the RALDH2 gene associated with activating (H3K14ac) or repressive (H3K27me3 or H3K9me3) histone marks. Fig. S5 shows serum-dependent effects of NKX3.1 knockdown on Jurkat and HSB2 growth and NKX3.1 protein measurement by immunoblot in shCTL, shTAL1, and shNKX3.1 T-ALL cell lines. Fig. S6 shows a classification of the TAL1 and/or NKX3.1 target genes in the Jurkat T cell line. Fig. S7 shows that T-ALL blasts in peripheral blood are in G0 phase and cycle in the MS-5 stroma cell line and that the effects of NKX3.1 in primary cells are not caused by the Notch pathway. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100745/DC1.
Acknowledgments
We thank Dr. Daniel Mathieu for human TAL1 antibody and Philippe Kastner for unpublished results in microarrays. We are grateful to Julien Calvo for expert assistance on quantitative PCR. We acknowledge Linhda Couvelard, Sylvain Baulande, and Pascal Soularue (PartnerChip) for their help in the analysis of microarrays.
S. Kusy was supported by fellowships from the Commissariat à l’Energie Atomique et aux Energies Alternatives and from the Association pour la Recherche sur le Cancer (ARC). B. Gerby was supported by a studentship from the Ligue Nationale contre le Cancer (LNCC) and Société Francaise d’Hématologie. N. Goardon was supported by studentships from the ARC and the Société Française d’Hématologie. F. Ferri was supported by a fellowship from Domaine Intérêt Majeur Stem Pole. D. Gérard was supported by a fellowship from the ARC. F. Armstrong was supported by fellowships from LNCC and Centre de Ressources de Modèles Expérimentaux de Cancer. This project was supported by grants from the Institut National de la Santé et de la Recherche Médicale, ARC, LNCC, Association Laurette Fugain, Institut National du Cancer, Cancéropole Ile de France, and Agence Nationale pour la Recherche.
The authors declare that they have no competing financial interests.
Footnotes
Abbreviations used:
- bHLH
- basic HLH
- ChIP
- chromatin immunoprecipitation
- HLH
- helix-loop-helix
- miRNA
- microRNA
- mRNA
- messenger RNA
- shRNA
- short hairpin RNA
- T-ALL
- T cell acute lymphoblastic leukemia
References
- Abdulkadir S.A., Magee J.A., Peters T.J., Kaleem Z., Naughton C.K., Humphrey P.A., Milbrandt J. 2002. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol. Cell. Biol. 22:1495–1503 10.1128/MCB.22.5.1495-1503.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong F., Brunet de la Grange P., Gerby B., Rouyez M.C., Calvo J., Fontenay M., Boissel N., Dombret H., Baruchel A., Landman-Parker J., et al. 2009. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood. 113:1730–1740 10.1182/blood-2008-02-138172 [DOI] [PubMed] [Google Scholar]
- Armstrong S.A., Look A.T. 2005. Molecular genetics of acute lymphoblastic leukemia. J. Clin. Oncol. 23:6306–6315 10.1200/JCO.2005.05.047 [DOI] [PubMed] [Google Scholar]
- Bain G., Engel I., Robanus Maandag E.C., te Riele H.P., Voland J.R., Sharp L.L., Chun J., Huey B., Pinkel D., Murre C. 1997. E2A deficiency leads to abnormalities in alphabeta T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 17:4782–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bash R.O., Hall S., Timmons C.F., Crist W.M., Amylon M., Smith R.G., Baer R. 1995. Does activation of the TAL1 gene occur in a majority of patients with T-cell acute lymphoblastic leukemia? A pediatric oncology group study. Blood. 86:666–676 [PubMed] [Google Scholar]
- Benezra R., Davis R.L., Lockshon D., Turner D.L., Weintraub H. 1990. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 61:49–59 10.1016/0092-8674(90)90214-Y [DOI] [PubMed] [Google Scholar]
- Bradney C., Hjelmeland M., Komatsu Y., Yoshida M., Yao T.P., Zhuang Y. 2003. Regulation of E2A activities by histone acetyltransferases in B lymphocyte development. J. Biol. Chem. 278:2370–2376 10.1074/jbc.M211464200 [DOI] [PubMed] [Google Scholar]
- Brunet de la Grange P., Armstrong F., Duval V., Rouyez M.C., Goardon N., Romeo P.H., Pflumio F. 2006. Low SCL/TAL1 expression reveals its major role in adult hematopoietic myeloid progenitors and stem cells. Blood. 108:2998–3004 10.1182/blood-2006-05-022988 [DOI] [PubMed] [Google Scholar]
- Chen J., Odenike O., Rowley J.D. 2010. Leukaemogenesis: more than mutant genes. Nat. Rev. Cancer. 10:23–36 10.1038/nrc2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I., Murre C. 2001. The function of E- and Id proteins in lymphocyte development. Nat. Rev. Immunol. 1:193–199 10.1038/35105060 [DOI] [PubMed] [Google Scholar]
- Ferrando A.A., Neuberg D.S., Staunton J., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., et al. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 1:75–87 10.1016/S1535-6108(02)00018-1 [DOI] [PubMed] [Google Scholar]
- Ferrando A.A., Herblot S., Palomero T., Hansen M., Hoang T., Fox E.A., Look A.T. 2004. Biallelic transcriptional activation of oncogenic transcription factors in T-cell acute lymphoblastic leukemia. Blood. 103:1909–1911 10.1182/blood-2003-07-2577 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., O’Geen H., Keles S., Blahnik K., Linnemann A.K., Kang Y.A., Choi K., Farnham P.J., Bresnick E.H. 2009. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell. 36:667–681 10.1016/j.molcel.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerby B., Armstrong F., de la Grange P.B., Medyouf H., Calvo J., Verhoeyen E., Cosset F.L., Bernstein I., Amselem S., Boissel N., et al. 2010. Optimized gene transfer into human primary leukemic T cell with NOD-SCID/leukemia-initiating cell activity. Leukemia. 24:646–649 10.1038/leu.2009.235 [DOI] [PubMed] [Google Scholar]
- He W.W., Sciavolino P.J., Wing J., Augustus M., Hudson P., Meissner P.S., Curtis R.T., Shell B.K., Bostwick D.G., Tindall D.J., et al. 1997. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 43:69–77 10.1006/geno.1997.4715 [DOI] [PubMed] [Google Scholar]
- Herblot S., Steff A.M., Hugo P., Aplan P.D., Hoang T. 2000. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat. Immunol. 1:138–144 10.1038/77819 [DOI] [PubMed] [Google Scholar]
- Hsu H.L., Cheng J.T., Chen Q., Baer R. 1991. Enhancer-binding activity of the tal-1 oncoprotein in association with the E47/E12 helix-loop-helix proteins. Mol. Cell. Biol. 11:3037–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.L., Wadman I., Baer R. 1994a. Formation of in vivo complexes between the TAL1 and E2A polypeptides of leukemic T cells. Proc. Natl. Acad. Sci. USA. 91:3181–3185 10.1073/pnas.91.8.3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H.L., Wadman I., Tsan J.T., Baer R. 1994b. Positive and negative transcriptional control by the TAL1 helix-loop-helix protein. Proc. Natl. Acad. Sci. USA. 91:5947–5951 10.1073/pnas.91.13.5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Li X., Valverde K., Fu X., Noguchi C., Qiu Y., Huang S. 2009. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc. Natl. Acad. Sci. USA. 106:10141–10146 10.1073/pnas.0900437106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Brandt S.J. 2000. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol. Cell. Biol. 20:2248–2259 10.1128/MCB.20.6.2248-2259.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito E., Honma R., Imai J., Azuma S., Kanno T., Mori S., Yoshie O., Nishio J., Iwasaki H., Yoshida K., et al. 2003. A tetraspanin-family protein, T-cell acute lymphoblastic leukemia-associated antigen 1, is induced by the Ewing’s sarcoma-Wilms’ tumor 1 fusion protein of desmoplastic small round-cell tumor. Am. J. Pathol. 163:2165–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe J., Herblot S., Rojas-Sutterlin S., Haman A., Barakat S., Iscove N.N., Sauvageau G., Hoang T. 2010. Scl regulates the quiescence and the long-term competence of hematopoietic stem cells. Blood. 115:792–803 10.1182/blood-2009-01-201384 [DOI] [PubMed] [Google Scholar]
- Lahlil R., Lécuyer E., Herblot S., Hoang T. 2004. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol. Cell. Biol. 24:1439–1452 10.1128/MCB.24.4.1439-1452.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazrak M., Deleuze V., Noel D., Haouzi D., Chalhoub E., Dohet C., Robbins I., Mathieu D. 2004. The bHLH TAL-1/SCL regulates endothelial cell migration and morphogenesis. J. Cell Sci. 117:1161–1171 10.1242/jcs.00969 [DOI] [PubMed] [Google Scholar]
- Lécuyer E., Herblot S., Saint-Denis M., Martin R., Begley C.G., Porcher C., Orkin S.H., Hoang T. 2002. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 100:2430–2440 10.1182/blood-2002-02-0568 [DOI] [PubMed] [Google Scholar]
- Lei Q., Jiao J., Xin L., Chang C.J., Wang S., Gao J., Gleave M.E., Witte O.N., Liu X., Wu H. 2006. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 9:367–378 10.1016/j.ccr.2006.03.031 [DOI] [PubMed] [Google Scholar]
- Magee J.A., Abdulkadir S.A., Milbrandt J. 2003. Haploinsufficiency at the Nkx3.1 locus. A paradigm for stochastic, dosage-sensitive gene regulation during tumor initiation. Cancer Cell. 3:273–283 10.1016/S1535-6108(03)00047-3 [DOI] [PubMed] [Google Scholar]
- Maser R.S., Choudhury B., Campbell P.J., Feng B., Wong K.K., Protopopov A., O’Neil J., Gutierrez A., Ivanova E., Perna I., et al. 2007. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 447:966–971 10.1038/nature05886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis K.J., Wolfe A.L., Oricchio E., Palomero T., de Keersmaecker K., McJunkin K., Zuber J., James T., Khan A.A., Leslie C.S., et al. 2010. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat. Cell Biol. 12:372–379 10.1038/ncb2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medyouf H., Alcalde H., Berthier C., Guillemin M.C., dos Santos N.R., Janin A., Decaudin D., de Thé H., Ghysdael J. 2007. Targeting calcineurin activation as a therapeutic strategy for T-cell acute lymphoblastic leukemia. Nat. Med. 13:736–741 10.1038/nm1588 [DOI] [PubMed] [Google Scholar]
- Michalak M., Lynch J., Groenendyk J., Guo L., Robert Parker J.M., Opas M. 2002. Calreticulin in cardiac development and pathology. Biochim. Biophys. Acta. 1600:32–37 [DOI] [PubMed] [Google Scholar]
- Morrow M.A., Mayer E.W., Perez C.A., Adlam M., Siu G. 1999. Overexpression of the Helix-Loop-Helix protein Id2 blocks T cell development at multiple stages. Mol. Immunol. 36:491–503 10.1016/S0161-5890(99)00071-1 [DOI] [PubMed] [Google Scholar]
- Nagel S., Kaufmann M., Drexler H.G., MacLeod R.A. 2003. The cardiac homeobox gene NKX2-5 is deregulated by juxtaposition with BCL11B in pediatric T-ALL cell lines via a novel t(5;14)(q35.1;q32.2). Cancer Res. 63:5329–5334 [PubMed] [Google Scholar]
- Nagel S., Venturini L., Przybylski G.K., Grabarczyk P., Schmidt C.A., Meyer C., Drexler H.G., Macleod R.A., Scherr M. 2009. Activation of miR-17-92 by NK-like homeodomain proteins suppresses apoptosis via reduction of E2F1 in T-cell acute lymphoblastic leukemia. Leuk. Lymphoma. 50:101–108 10.1080/10428190802626632 [DOI] [PubMed] [Google Scholar]
- O’Neil J., Look A.T. 2007. Mechanisms of transcription factor deregulation in lymphoid cell transformation. Oncogene. 26:6838–6849 10.1038/sj.onc.1210766 [DOI] [PubMed] [Google Scholar]
- O’Neil J., Shank J., Cusson N., Murre C., Kelliher M. 2004. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer Cell. 5:587–596 10.1016/j.ccr.2004.05.023 [DOI] [PubMed] [Google Scholar]
- Odom D.T., Dowell R.D., Jacobsen E.S., Gordon W., Danford T.W., MacIsaac K.D., Rolfe P.A., Conboy C.M., Gifford D.K., Fraenkel E. 2007. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 39:730–732 10.1038/ng2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., Fukuhara N., Yoshie O. 1998. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Mol. Cell. Biol. 18:6939–6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota A., Tagawa H., Karnan S., Tsuzuki S., Karpas A., Kira S., Yoshida Y., Seto M. 2004. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 64:3087–3095 10.1158/0008-5472.CAN-03-3773 [DOI] [PubMed] [Google Scholar]
- Palomero T., Odom D.T., O’Neil J., Ferrando A.A., Margolin A., Neuberg D.S., Winter S.S., Larson R.S., Li W., Liu X.S., et al. 2006. Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia. Blood. 108:986–992 10.1182/blood-2005-08-3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.T., Sun X.H. 1998. The Tal1 oncoprotein inhibits E47-mediated transcription. Mechanism of inhibition. J. Biol. Chem. 273:7030–7037 10.1074/jbc.273.12.7030 [DOI] [PubMed] [Google Scholar]
- Porcher C., Swat W., Rockwell K., Fujiwara Y., Alt F.W., Orkin S.H. 1996. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 86:47–57 10.1016/S0092-8674(00)80076-8 [DOI] [PubMed] [Google Scholar]
- Possner M., Heuser M., Kaulfuss S., Scharf J.G., Schulz W., Hermann-Ringert R., Thelen P. 2008. Functional analysis of NKX3.1 in LNCaP prostate cancer cells by RNA interference. Int. J. Oncol. 32:877–884 [PubMed] [Google Scholar]
- Przybylski G.K., Dik W.A., Grabarczyk P., Wanzeck J., Chudobska P., Jankowski K., von Bergh A., van Dongen J.J., Schmidt C.A., Langerak A.W. 2006. The effect of a novel recombination between the homeobox gene NKX2-5 and the TRD locus in T-cell acute lymphoblastic leukemia on activation of the NKX2-5 gene. Haematologica. 91:317–321 [PubMed] [Google Scholar]
- Rangarajan A., Hong S.J., Gifford A., Weinberg R.A. 2004. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 6:171–183 10.1016/j.ccr.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Ravet E., Reynaud D., Titeux M., Izac B., Fichelson S., Roméo P.H., Dubart-Kupperschmitt A., Pflumio F. 2004. Characterization of DNA-binding-dependent and -independent functions of SCL/TAL1 during human erythropoiesis. Blood. 103:3326–3335 10.1182/blood-2003-05-1689 [DOI] [PubMed] [Google Scholar]
- Robb L., Elwood N.J., Elefanty A.G., Köntgen F., Li R., Barnett L.D., Begley C.G. 1996. The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 15:4123–4129 [PMC free article] [PubMed] [Google Scholar]
- Shen M.M., Abate-Shen C. 2003. Roles of the Nkx3.1 homeobox gene in prostate organogenesis and carcinogenesis. Dev. Dyn. 228:767–778 10.1002/dvdy.10397 [DOI] [PubMed] [Google Scholar]
- Sirven A., Ravet E., Charneau P., Zennou V., Coulombel L., Guétard D., Pflumio F., Dubart-Kupperschmitt A. 2001. Enhanced transgene expression in cord blood CD34(+)-derived hematopoietic cells, including developing T cells and NOD/SCID mouse repopulating cells, following transduction with modified trip lentiviral vectors. Mol. Ther. 3:438–448 10.1006/mthe.2001.0282 [DOI] [PubMed] [Google Scholar]
- Soulier J., Clappier E., Cayuela J.M., Regnault A., García-Peydró M., Dombret H., Baruchel A., Toribio M.L., Sigaux F. 2005. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood. 106:274–286 10.1182/blood-2004-10-3900 [DOI] [PubMed] [Google Scholar]
- Souroullas G.P., Salmon J.M., Sablitzky F., Curtis D.J., Goodell M.A. 2009. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell. 4:180–186 10.1016/j.stem.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeyen E., Dardalhon V., Ducrey-Rundquist O., Trono D., Taylor N., Cosset F.L. 2003. IL-7 surface-engineered lentiviral vectors promote survival and efficient gene transfer in resting primary T lymphocytes. Blood. 101:2167–2174 10.1182/blood-2002-07-2224 [DOI] [PubMed] [Google Scholar]
- Voronova A.F., Lee F. 1994. The E2A and tal-1 helix-loop-helix proteins associate in vivo and are modulated by Id proteins during interleukin 6-induced myeloid differentiation. Proc. Natl. Acad. Sci. USA. 91:5952–5956 10.1073/pnas.91.13.5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman I.A., Osada H., Grütz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145–3157 10.1093/emboj/16.11.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kruithof-de Julio M., Economides K.D., Walker D., Yu H., Halili M.V., Hu Y.P., Price S.M., Abate-Shen C., Shen M.M. 2009. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 461:495–500 10.1038/nature08361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Huang S., Pack S.D., Yu X., Brandt S.J., Noguchi C.T. 2005. Tal1/SCL binding to pericentromeric DNA represses transcription. J. Biol. Chem. 280:12956–12966 10.1074/jbc.M412721200 [DOI] [PubMed] [Google Scholar]
- Woods K., Thomson J.M., Hammond S.M. 2007. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 282:2130–2134 10.1074/jbc.C600252200 [DOI] [PubMed] [Google Scholar]
- Xu Z., Huang S., Chang L.S., Agulnick A.D., Brandt S.J. 2003. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell. Biol. 23:7585–7599 10.1128/MCB.23.21.7585-7599.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Meng X., Cai Y., Koury M.J., Brandt S.J. 2006. Recruitment of the SWI/SNF protein Brg1 by a multiprotein complex effects transcriptional repression in murine erythroid progenitors. Biochem. J. 399:297–304 10.1042/BJ20060873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Young A.Z., Soares V.C., Kelley R., Benezra R., Zhuang Y. 1997. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol. Cell. Biol. 17:7317–7327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Riva L., Xie H., Schindler Y., Moran T.B., Cheng Y., Yu D., Hardison R., Weiss M.J., Orkin S.H., et al. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell. 36:682–695 10.1016/j.molcel.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]