Abstract
Mutations that affect splicing of precursor messenger RNAs play a major role in the development of hereditary diseases. Most splicing mutations have been found to eliminate GT or AG dinucleotides that define the 5′ and 3′ ends of introns, leading to exon skipping or cryptic splice-site activation. Although accurate description of the mis-spliced transcripts is critical for predicting phenotypic consequences of these alterations, their exact nature in affected individuals cannot often be determined experimentally. Using a comprehensive collection of exons that sustained cryptic splice-site activation or were skipped as a result of splice-site mutations, we have developed a multivariate logistic discrimination procedure that distinguishes the two aberrant splicing outcomes from DNA sequences. The new algorithm was validated using an independent sample of exons and implemented as a free online utility termed CRYP-SKIP (http://www.dbass.org.uk/cryp-skip/). The web application takes up one or more mutated alleles, each consisting of one exon and flanking intronic sequences, and provides a list of important predictor variables and their values, the overall probability of activating cryptic splice vs exon skipping, and the location and intrinsic strength of predicted cryptic splice sites in the input sequence. These results will facilitate phenotypic prediction of splicing mutations and provide further insights into splicing enhancer and silencer elements and their relative importance for splice-site selection in vivo.
Keywords: mutation, gene, splicing, cryptic splice site, exon skipping, RNA
Introduction
Removal of introns from precursor messenger RNA (pre-mRNA) by splicing is a critical step in eukaryotic gene expression. Splicing of human pre-mRNAs is mediated by conserved but highly degenerate sequences that include the MAG∣GURAGU consensus (M is A or C; R is purine; ∣ is the exon–intron boundary) at the 5′ splice site (ss) and the YAG∣R motif (Y is pyrimidine) at the 3′ss, which are preceded by an upstream poly-Y tract and the branch point sequence. In addition to these traditional sequences, accurate recognition of exons and introns by the spliceosome requires auxiliary elements in the pre-mRNA that repress or promote splicing, termed exonic or intronic splicing silencers (ESSs/ISSs) or enhancers (ESEs/ISEs). These signals are thought to act through combinatorial effects of RNA secondary structure1, 2 and/or numerous regulatory factors that bind to pre-mRNAs, including serine/arginine-rich (SR) proteins3, 4 and heterogeneous nuclear ribonucleoproteins.5, 6, 7, 8
Auxiliary splicing sequences have been characterized experimentally,3, 9, 10 computationally11, 12 or by a combination of the two approaches.13, 14, 15 Recent gene-specific16 and genome-wide17 comparisons showed that exonic sequences between authentic and aberrant splice sites that were activated by splice-site mutations have lower frequencies of ESEs and higher densities of ESSs than average exons. Conversely, intronic sequences between authentic and cryptic/de novo splice sites have more enhancers and less silencers than average introns.16, 17 Although the relative importance of various auxiliary signals in the development of aberrant splice-site activation in vivo is poorly understood, silencers, and putative octamer ESSs (PESSs)12 in particular, were identified as stronger predictors of aberrant splice-site activation than ESEs,16, 17, 18 consistent with a dominant role of repressive elements that keep highly abundant decoy splice-site signals in check.
Splicing mutations play a major role in the development of hereditary diseases and may represent up to ∼50% of disease-causing alterations in genes with a large number of introns.19, 20 This figure is likely to be an underestimate, because current mutation-screening policies are biased towards coding DNA, leaving aberrant splicing undetected in many cases, particularly de novo splice sites in introns. Pre-mRNA splicing can be impaired by mutations located anywhere in the gene, but most have been found in GT and AG dinucleotides that define 5′ and 3′ intron ends,21 reflecting their highest level of conservation among splice-site consensus sequences.22 Although accurate description of the resulting abnormal transcripts is important for predicting the degree of severity and the age of onset of both Mendelian and complex traits, RNA samples from affected individuals or their family members are often not available and functional splicing assays are costly and time-consuming. Computational prediction of these outcomes from genomic sequences would therefore provide a useful alternative, but such methods have not been available.
Here, we describe the development of a method that can distinguish exons that are skipped and exons that activate cryptic splice sites as a result of splicing mutations. The new procedure was capable of predicting the correct outcome in 72% of the cases and was implemented as an easy-to-use web application termed CRYP-SKIP, which is freely available at http://www.dbass.org.uk/cryp-skip/.
Materials and methods
To distinguish exon skipping and aberrant splice-site activation from pre-mRNA sequences, we set out to compare both traditional and auxiliary splicing elements between two well-defined groups of sequences. The first group (dataset termed EXSK) contained a set of 250 exons that were skipped as a result of disease-causing splicing mutations but did not activate cryptic splice sites in flanking exons or introns.17 We used the same ascertainment criteria17 to obtain 47 additional EXSK sequences from recently published reports (Supplementary Table 1). For the second group, we analyzed a total of 204 exonic sequences that sustained cryptic splice-site activation as a result of germ-line or somatic splicing mutations. This dataset (termed CR-E) is available from the updated Database of Aberrant Splice Sites (DBASS) maintained at http://www.dbass.org.uk23, 24 and includes all mutation-induced cryptic splice sites reported in peer-reviewed journals between 1981 and June 2008.
In each exonic sequence, we determined the location and strength of predicted (decoy) 3′ and 5′ss, and counts and densities of previously identified auxiliary splicing sequences. For decoy splice sites, we employed a neural network (NN) splice-site prediction algorithm and the NN Splice server (http://www.fruitfly.org/seq_tools/splice.html).25 ESSs discovered by a fluorescence-activated screen (FAS-ESSs)13 were computed using the FAS-ESS server (http://genes.mit.edu/fas-ess/). Putative enhancers and silencers obtained by comparing non-coding exons and pseudoexons or untranslated regions of intron-less genes (PEXSs) were identified with the PEXS algorithm12 (http://cubweb.biology.columbia.edu/pesx/). Scores for SF2/ASF, which was confirmed as the most important SR protein for aberrant splice-site activation in vivo,17 were computed using the updated matrix26 and a standard threshold implemented in the ESEFinder (version 3; http://rulai.cshl.edu/tools/ESE/).27 In addition, we employed a recently published set of 1131 and 708 exon and intron identity elements (EIEs and IIEs, respectively) and their Z-scores derived from DNA-strand asymmetry patterns.28 Finally, we examined both datasets using the Neighborhood Inference (NI) method29 that predicts the activity of splicing regulatory elements based on the local density of known sites in sequence space. For each potential predictor variable, we computed the total number of ESSs/ESEs per exon and, where applicable, the sum of their scores. Each count and score density was calculated for 100 nucleotides as described earlier.17
To model the relationship between the predictor variables and a dichotomous response (either EXSK or CR-E), we used multiple logistic regression to estimate the probability of cryptic splice-site activation (PCR-E; defined below for the final model) and exon skipping (1−PCR-E). Eighty per cent of EXSK (n=238) and CR-E (n=163) sequences were randomly chosen as a training set, whereas the remaining EXSK and CR-E sequences were used as a test set to validate the performance of our discrimination procedure. Competing models were compared by the likelihood ratio test and the likelihood-based Akaike's information criterion. The discrimination ability of each model was assessed using leave-one-out cross-validation with the training dataset. For data handling and statistical analysis, we employed the R-statistical software (http://www.r-project.org).30
Results
Algorithm
Median values of the predictor variables in EXSK and CR-E datasets and their distribution are shown in Table 1 and Supplementary Figure 1; full datasets are available in Supplementary Table 2. After comparing univariate models (Supplementary Table 3), we built an initial multivariate model using appropriately transformed predictor variables (Supplementary Figure 2). The NI and IIE score densities were omitted from this model, as they did not improve the model fit in the presence of the remaining variables (P-value of the likelihood ratio test was 0.83). Count densities were also left out, because they were highly correlated with the score densities and provided less information (for example, Pearson's correlation coefficients for EIEs and IIEs count/score densities were 0.86 and 0.94, respectively). The EIE score density was retained in the model despite its non-significance, as this predictor appeared to improve discrimination of EXSK and CR-E (Supplementary Table 4).
Table 1. Median values of potential predictors of the splicing outcome.
| Group | Mutation outcome | Number of sequences | Exon length | PESS density | NN splice density of 5′ss | SF2/ASF score density | FAS-ESS density (hex2) | NI score density | EIE score density | IIE score density |
|---|---|---|---|---|---|---|---|---|---|---|
| Training set | EXSK | 238 | 116 | 1.22 | 0.03 | 10.02 | 3.46 | 30.31 | 440.83 | 313.16 |
| CR-E | 163 | 154 | 0.65 | 0.14 | 12.36 | 3.06 | 36.86 | 460.18 | 274.81 | |
| Test set | EXSK | 59 | 102 | 1.01 | 0.02 | 10.52 | 3.24 | 31.69 | 449.46 | 316.58 |
| CR-E | 41 | 131 | 0.69 | 0.10 | 12.23 | 3.45 | 37.41 | 431.27 | 251.75 | |
| All | EXSK | 297 | 111 | 1.09 | 0.03 | 10.12 | 3.42 | 31.19 | 443.11 | 313.70 |
| CR-E | 204 | 148 | 0.65 | 0.14 | 12.30 | 3.08 | 37.29 | 453.41 | 273.06 |
Abbreviations: EXSK, exon skipping; CR-E, cryptic splice-site activation.
Predictor variables are defined in the text.
On the basis of our final multivariate model, we define PCR-E as:
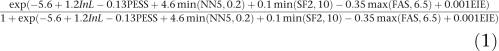 |
where L is exon length (in nucleotides), PESS is the PESS density, NN5 is the density of decoy 5′ss, SF2 is the SF2/ASF score density, FAS is the FAS-ESS hex2 density and EIE is the EIE score density (Supplementary Table 2). Coefficient estimates of the final model, their standard errors and significance are shown in Table 2.
Table 2. Multiple logistic regression table.
| Predictor variable | Coefficient estimate | Standard error | P-valuea | OR (95% CI) |
|---|---|---|---|---|
| Intercept | −5.56 | 2.08 | 0.010 | – |
| Log(length) | 1.22 | 0.26 | <0.001 | 3.40 (2.05, 5.64) |
| PESS density | −0.13 | 0.06 | 0.030 | 0.88 (0.77, 0.99) |
| min(NN5, 0.2) | 4.64 | 1.33 | <0.001 | 103.22 (7.61, 1400.40) |
| min(SF2, 10) | 0.10 | 0.05 | 0.035 | 1.11 (1.00, 1.22) |
| max(FAS-ESS, 6.5) | −0.35 | 0.20 | 0.032 | 0.70 (0.47, 1.04) |
| EIE score | 0.0010 | 0.00079 | 0.203 | 1.00 (9.99, 1.00) |
Abbreviations: OR, odds ratio; CI, confidence intervals.
Likelihood ratio test.
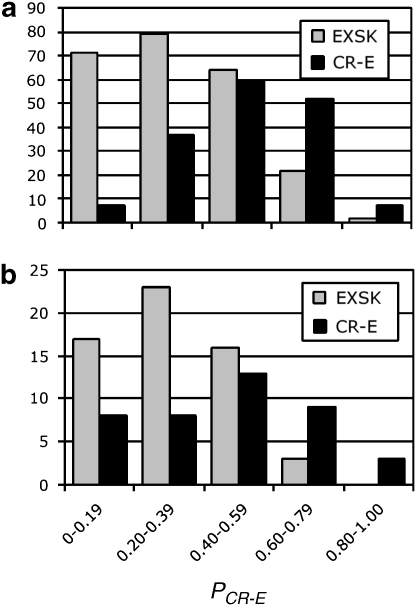
We next evaluated our discrimination procedure using an independent set of exons. Figure 1 shows the PCR-E distribution computed separately for the training and independent set. In the independent dataset, 54% of CR-E sequences had the PCR-E value >0.5, whereas 85% of EXSK sequences had the estimated PCR-E value ≤0.5. Conversely, 72% of exons with the PCR-E value ≤0.5 underwent exon skipping, whereas 71% of exons with the PCR-E value >0.5 sustained cryptic splice-site activation. Taken together, 72/100 (72%) sequences in the test set were correctly classified.
Figure 1.
PCR-E distribution of exonic sequences that underwent cryptic splice-site activation or exon skipping. (a) Training set; (b) independent set. All exonic sequences, the intrinsic strength of their authentic and cryptic splice sites, underlying mutations and their phenotypic consequences are described in the online Database of Aberrant Splice Sites. Their PCR-E values were determined as in Equation (1). Each column shows the number of EXSK (gray) or CR-E (black) events that had the PCR-E value in the interval shown on the x-axis.
The dependence of the PCR-E on predictor variables can be described in terms of the odds of a cryptic splice-site activation (OCR-E), defined as PCR-E/(1−PCR-E). Assuming the above model, a 33% increase in exon length would increase the estimated OCR-E by 42% if the values of the remaining predictors are kept unchanged. The same increase in the estimated OCR-E would require a rise in NN5 by 0.076, or in SF2 by 3.4, or in EIE by 346.6, with the values of the remaining predictors fixed. Conversely, a 42% decrease in the estimated OCR-E would result from an increase in PESS by 4.1 or from an increase in FAS density by 1.54, again without changing the values of the remaining five predictors. The OCR-E value was not much influenced by NN5 higher than 0.2, SF2 higher than 10 and FAS lower than 6.5 (Supplementary Figure 2). Together, these results illustrate how the values of some predictors influence the odds of cryptic splice-site activation, with practical implications for predicting aberrant splice-site activation ab initio.
To further validate the performance of the algorithm using experimental data, we compared a previously observed outcome of splicing mutations in the RB1 gene 31 with PCR-E values for each exon (Supplementary Table 5). This comparison showed that 13/14 (93%) exons that were skipped as a result of mutation were correctly predicted by the CRYP-SKIP algorithm. The only exception was exon 25, in which the predicted cryptic 5′ss activation (ATG/GTATGT in the middle of the exon) was not observed, despite a relatively high PCR-E value of 0.64. A failure to activate this splice site in vivo (mutation IVS25+1G>A31) may be explained by the small size of reduced exonic segment (33 nt), which may need additional splicing enhancer elements in flanking intronic sequences.
Finally, we calculated PCR-E values for 43 243 constitutively spliced human exons32 and for a set of 1909 alternatively spliced exons that are conserved between mouse and humans.33 As expected, the average PCR-E values were higher in the former group (0.39 vs 0.34, t-test, P<10−15), suggesting that alternatively spliced or weakly included exons are less likely to sustain cryptic splice-site activation when their authentic site is mutated. This is probably attributable mainly to a smaller average exon size of the latter group (139 vs 128 nt).
CRYP-SKIP
Our final regression model was incorporated in a new algorithm termed CRYP-SKIP, which was implemented as a common gateway interface script on a public server available at http://www.dbass.org.uk/cryp-skip/ or http://cryp-skip.img.cas.cz. The algorithm determines the exon length for each submitted sequence and performs a search for PESSs with the Z-score cut-off value of −2.6212, FAS-ESSs hex2 set13 and EIEs.28 For the analysis of decoy splice sites and SF2/ASF scores, the script (programmed in Perl) interacts with the NN splice server and the ESEFinder, respectively. The web application computes count and score densities of these elements and employs the logistic regression model as described above to calculate the PCR-E value (Equation (1)) for each submitted sequence.
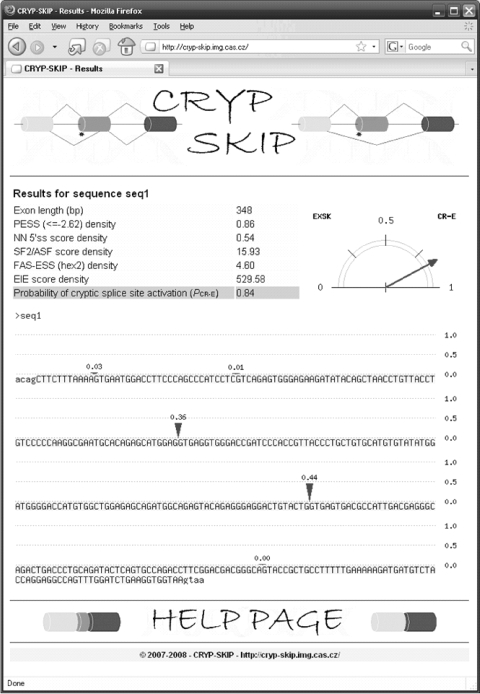
CRYP-SKIP users submit a DNA sequence (FASTA format) consisting of one exon (in upper case) and ∼100 nt of flanking intervening sequences (in lower case). Pairs of wild-type and mutated sequences or multiple FASTA sequences are permitted, as long as the total sequence does not exceed a limit of 4000 bp. The server output is a single page with a summary table containing a list of predictors, their calculated values and PCR-E, which is graphically shown as a pointer next to the table (Figure 2). PCR-E takes values between 0 and 1, with higher values speaking in favor of cryptic splice-site activation and lower values in favor of exon skipping (Figure 1). Finally, CRYP-SKIP shows predicted cryptic splice sites as vertical marks in the input sequence; their size reflects the relative intrinsic strength of decoy splice sites and sums to the PCR-E value of the submitted exon.
Figure 2.
A screenshot of the CRYP-SKIP output. PCR-E is shown on the right as a pointer balancing between exon skipping (EXSK) and cryptic splice-site activation (CR-E). Values of each predictor variable used in the regression model are summarized in a table on the left. The last row of each table shows the numerical value of PCR-E for each input sequence. Exonic sequences (highlighted in light blue) are in upper case and flanking introns are in lower case. Predicted aberrant splice sites are shown as arrows, with their size reflecting their relative predicted strength (scale 0–1 on the right). The sum of their sizes equals the PCR-E value for the output sequence. The color reproduction of this figure is available on the html full text version of the manuscript.
Discussion
CRYP-SKIP is a comprehensive computational tool that predicts whether inactivation of authentic splice sites by mutation is more likely to result in exon skipping or aberrant splice-site activation. As the two events represent the vast majority of pathogenic transcripts induced by splice-site mutations, the algorithm will facilitate prediction of aberrant splicing outcomes for most splicing mutations in human genes. Because cis-acting splicing signals and spliceosome components are generally well conserved in higher eukaryotes, the same algorithm should discriminate the two aberrant splicing outcomes in other mammalian or vertebrate species as well, although this remains to be tested.
Our model is based on a comprehensive sample of carefully selected and well-documented dichotomous events that gives us a unique opportunity to study splice-site selection in vivo as opposed to in vitro experiments. This approach should be instrumental when addressing the question why a particular decoy splice-site signal was selected by the spliceosome despite having a lower intrinsic strength than similar signals in the vicinity that were not recognized. Both CR-E and EXSK datasets are likely to expand in future, which will be facilitated by a more widespread use of RNA-based mutation screening and by more rigorous characterization of aberrant transcripts at the nucleotide level in affected individuals. Published reports should include estimates of the relative amounts of RNAs transcribed from mutated alleles and also evidence for adequate separation of RT-PCR products using polyacrylamide gel electrophoresis as many cryptic splice sites are activated just a few nucleotides away from authentic sites. Finally, because ins/del polymorphisms represent an important and underappreciated source of disease-associated cryptic splice sites or pseudoexon activation, particularly in repetitive sequences (Meili et al,34), DNA-based mutation screening of disease genes should employ and further develop methods capable of detecting structural variants.
In the future, it will be desirable to extend this tool to ab initio prediction of mutation-induced cryptic splice sites in flanking intronic sequences. Location of aberrant splice sites in introns is not symmetrical, reflecting the more complicated pattern of 3′ss organization compared with the 5′ss.23, 35 The variability of traditional splicing signals at the 3′ss (branch site, polypyrimidine tracts, 3′YAG and upstream AG exclusion zones) from one intron to another is considerable, and cooperative assembly of spliceosomal complexes at these signals is further confounded by local secondary structure and by multiple, distant or non-canonical branch points. In addition, auxiliary splicing sequences have been studied less in introns than in exons, although some of them have recently been characterized in more detail, such as short G-rich repeats.36, 37, 38 The extended datasets should provide more power for building robust models that include additional predictors, which did not give significant P-values in the analyzed sample and/or did not significantly improve our model, including decoy 3′ss, RESCUE-ESE11 and NI.29 Future efforts should also be facilitated by a more comprehensive dissection of cooperative interactions between splicing signals upstream of 3′ss, including distant branch sites. Thus, prediction algorithms discriminating the two aberrant RNA outcomes from DNA sequences are likely to be further improved, ultimately leading to better understanding of splice-site selection in vivo and more accurate characterization of human mutations and their phenotypic consequences.
Acknowledgments
We thank Sarah Ennis and Nicholas Maniatis for useful discussions and critical reading of the manuscript. This work was funded by a Grant from the JDRF (2008-47) to IV.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Hertel KJ, Maniatis T. SR proteins are ‘locators' of the RNA splicing machinery. Curr Biol. 1999;9:R6–R7. doi: 10.1016/s0960-9822(99)80032-3. [DOI] [PubMed] [Google Scholar]
- Ghetti A, Pinol-Roma S, Michael WM, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Xing J, Dreyfuss G. The hnRNP F protein: unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J Biol Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Mol Cell Biol. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Androphy EJ, Singh RN. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Kangsamaksin T, Chao MS, Banerjee JK, Chasin LA. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol Cell Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A, Ram O, Amit M, et al. Comparative analysis identifies exonic splicing regulatory sequences–the complex definition of enhancers and silencers. Mol Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Wimmer K, Roca X, Beiglbock H, et al. Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5′ splice-site disruption. Hum Mutat. 2007;28:599–612. doi: 10.1002/humu.20493. [DOI] [PubMed] [Google Scholar]
- Kralovicova J, Vorechovsky I. Global control of aberrant splice site activation by auxiliary splicing sequences: evidence for a gradient in exon and intron definition. Nucleic Acids Res. 2007;35:6399–6413. doi: 10.1093/nar/gkm680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Xiao X, Van Nostrand E, Burge CB. General and specific functions of exonic splicing silencers in splicing control. Mol Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ars E, Serra E, Garcia J, et al. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- Teraoka SN, Telatar M, Becker-Catania S, et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am J Hum Genet. 1999;64:1617–1631. doi: 10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. Human Gene Mutation. Oxford: BIOS Scientific Publishers; 1993. [Google Scholar]
- Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorechovsky I. Aberrant 3′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2006;34:4630–4641. doi: 10.1093/nar/gkl535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Chivers MC, Kralovicova J, et al. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucletide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet. 2006;15:2490–2508. doi: 10.1093/hmg/ddl171. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li WH, Krainer AR, Zhang MQ. RNA landscape of evolution for optimal exon and intron discrimination. Proc Natl Acad Sci USA. 2008;105:5797–5802. doi: 10.1073/pnas.0801692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Shomron N, Yeo GW, Schneider A, Xiao X, Burge CB. Inference of splicing regulatory activities by sequence neighborhood analysis. PLoS Genet. 2006;2:e191. doi: 10.1371/journal.pgen.0020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RDC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing: ISBN 3-900051-07-0; 2007. [Google Scholar]
- Houdayer C, Dehainault C, Mattler C, et al. Evaluation of in silico splice tools for decision-making in molecular diagnosis. Hum Mutat. 2008;29:975–982. doi: 10.1002/humu.20765. [DOI] [PubMed] [Google Scholar]
- Carmel I, Tal S, Vig I, Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004;10:828–840. doi: 10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Bakowski MA, Morris Q, et al. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Meili D, Kralovicova J, Zagalak J, et al. Disease-causing mutations improving the branch site and polypyrimidine tract: pseudoexon activation of LINE-2 and antisense Alu lacking the poly(T)-tail Hum Mutat 2009(in press). [DOI] [PubMed]
- Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989;3:2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- McCullough AJ, Berget SM. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralovicova J, Vorechovsky I. Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J Immunol. 2006;176:2381–2388. doi: 10.4049/jimmunol.176.4.2381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.