Abstract
Lymphangioleiomyomatosis (LAM) is characterized by cystic lung destruction caused by LAM cells (smooth-muscle-like cells) that have mutations in the tumor suppressor genes tuberous sclerosis complex (TSC) 1 or 2 and have the capacity to metastasize. Since chemokines and their receptors function in chemotaxis of metastatic cells, we hypothesized that LAM cells may be recruited by chemokine(s) in the lung. Quantification of 25 chemokines in bronchoalveolar lavage fluid from LAM patients and healthy volunteers revealed that concentrations of CCL2, CXCL1, and CXCL5 were significantly higher in samples from LAM patients than those from healthy volunteers. In vitro, CCL2 or MCP-1 induced selective migration of cells, showing loss of heterozygosity of TSC2 from a heterogeneous population of cells grown from explanted LAM lungs. Additionally, the frequencies of single-nucleotide polymorphisms in the CCL2 gene promoter region differed significantly in LAM patients and healthy volunteers (p = 0.018), and one polymorphism was associated significantly more frequently with the decline of lung function. The presence (i.e., potential functionality) of chemokine receptors was evaluated using immunohistochemistry in lung sections from 30 LAM patients. Expression of chemokines and these receptors varied among LAM patients and differed from that seen in some cancers (e.g., breast cancer and melanoma cells). These observations are consistent with the notion that chemokines such as CCL2 may serve to determine mobility and specify the site of metastasis of the LAM cell.
Metastatic cells migrate to specific sites distant from the primary tumor growth (1) and “home” to an appropriate environment described as “soil”, which is identified by specific soluble chemoattractants (2, 3). Metastasis of cells to specific organs appears to depend on soluble factors (e.g., chemokines) synthesized in the target tissue (3, 4), which bind to receptors on the surface of circulating cells. Anchored neoplastic cells undergo multiple genetic and/or biochemical changes that favor proliferation and tumor growth (1). Recruitment to a metastatic site depends on a cellular phenotype that includes detachment from adjacent cells and matrix, entrance to and survival in the lymphatic or hematological circulatory systems, and successful replication at the site of metastasis (1, 5).
Chemokines and their receptors have been implicated in neoplastic tumor metastasis. Four families of chemokines (C, CC, CXC, CX3C) are defined by locations of cysteines in their primary structures. They are produced by cells in response to infection and are responsible for the recruitment and homing of immune cells to sites of inflammation or injury (6). Many cells of nonimmune origin not only have chemokine receptors, but they also synthesize and secrete chemokines (7). Cancer cells, for example, exhibit a characteristic expression profile of chemokines and their receptors. Metastatic breast cancer cells express the chemokine receptors CXCR4 and CCR7, whereas malignant melanoma cells express CCR10, CXCR4, and CCR7. Thus, cells can, to some extent, be identified by their chemokine/chemokine receptor profiles, which appear to direct metastasis to specific organs (8).
Lymphangioleiomyomatosis (LAM),4 a rare multisystem disorder that affects primarily women of child-bearing age, is characterized by cystic lung destruction, leading to respiratory failure, recurrent pneumothoraces, chylous effusions, infiltration of the axial lymphatics (e.g., lymphangioleiomyomas), and abdominal tumors (e.g., renal angiomyolipomas) (9). LAM occurs sporadically or in association with tuberous sclerosis complex (TSC), an inherited disorder with variable penetrance, which results from mutations in TSC1 or TSC2 tumor-suppressor genes (10, 11). Mutations in the TSC2 gene, which encodes tuberin, are more common in LAM cells (10, 12, 13). Systemic dissemination of LAM cells in a metastatic process has been proposed (14, 15) and is consistent with the finding of LAM cells in renal, lymphatic, and lung lesions, which have identical TSC2 mutations (15), as well as the presence of LAM cells in blood and body fluids from LAM patients (16). These observations led us to investigate whether migration of LAM cells could be affected by chemokines.
Materials and Methods
Tissue samples and cell culture
The study group was comprised of 47 women from 22 to 59 years of age (mean 42 years) with documented LAM. Tissues from patients were used after informed consent was obtained in a study approved by the Institutional Review Board of the National Heart, Lung, and Blood Institute (protocol 95-H-0186). Tissues from which cells were grown were from LAM patients undergoing lung transplantation unless otherwise indicated.
Immunohistochemistry
For histological studies, tissues were fixed with buffered 10% formalin, embedded in paraffin, and sections (5 µm) were stained with H&E or reacted with mAb HMB-45 (Dako), which reacts with gp100 (diluted 1/25), the Pmel17 gene product, or Abs reactive with the indicated chemokine or chemokine receptor (Abs against CCR1, CCR3, CCR4, CCR5, CCR7, CCR8, CCR10, CXCR1, CXCR5, CXCR4, CCL27, and CCL28 from Imgenex, against CCR2 and CXCR6 from R&D Systems). Tissue sections were incubated with 0.3% hydrogen peroxide in methanol for 30 min at room temperature to block endogenous peroxidase activity. After washing with PBS, slides were incubated with anti-chemokine and chemokine receptor Abs (~10 µg/ml) for 2 h at room temperature. After washing, the EnVision+ systems (Dako) and Vectastain kit (Vector Laboratories) were used according to the manufacturers’ instructions to reveal reaction products. For controls, primary Abs were normal rabbit or mouse IgG. The intensity of staining was graded 0–3 for none, mild, moderate, or strong, respectively. The scoring was based on intensity relative to the negative control. Two pathologists scored each section in a blinded manner. The results reported are based on their consensus score. The percentage of patients represent samples that had an intensity score of 1–3 (see Figs. 2 and 4). A detailed list of patients is included in the supplemental material (supplemental Tables IS and IIS).5
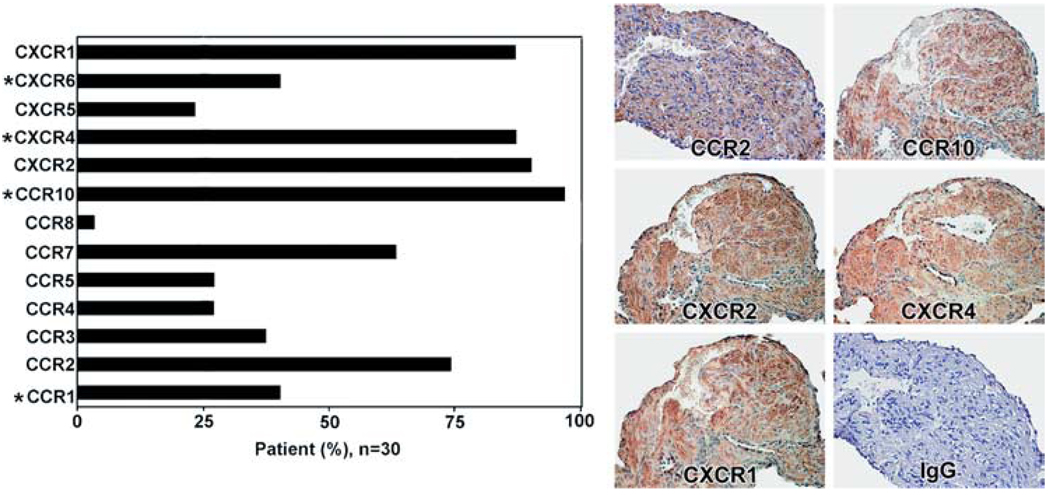
FIGURE 2.
Immunoreactive chemokine receptors in LAM nodules. Left, Immunoreactivity of patients (n = 30, except for CXCR6, n = 15; see supplemental Table IS) that reacted with Abs against the indicated receptors. CXCR6 was only determined on 15 patients. Asterisk (*) indicates the four differentially expressed receptors as shown in Fig. 1D. Right, Representative images of LAM cells reacted with anti-CCR2 Abs that did not react with vascular and alveolar cells or with anti-CCR10 Ab that reacted with proliferative LAM areas, but not with vascular, normal alveolar, or lymphatic-like structures. Abs against CXCR2 and CXCR4 reacted with vascular endothelial cells and nuclei of type II pneumocytes as well as LAM cells, which reacted also with anti-CXCR1 Ab that reacted only weakly with vascular endothelial cells. There was no reaction with normal rabbit IgG.
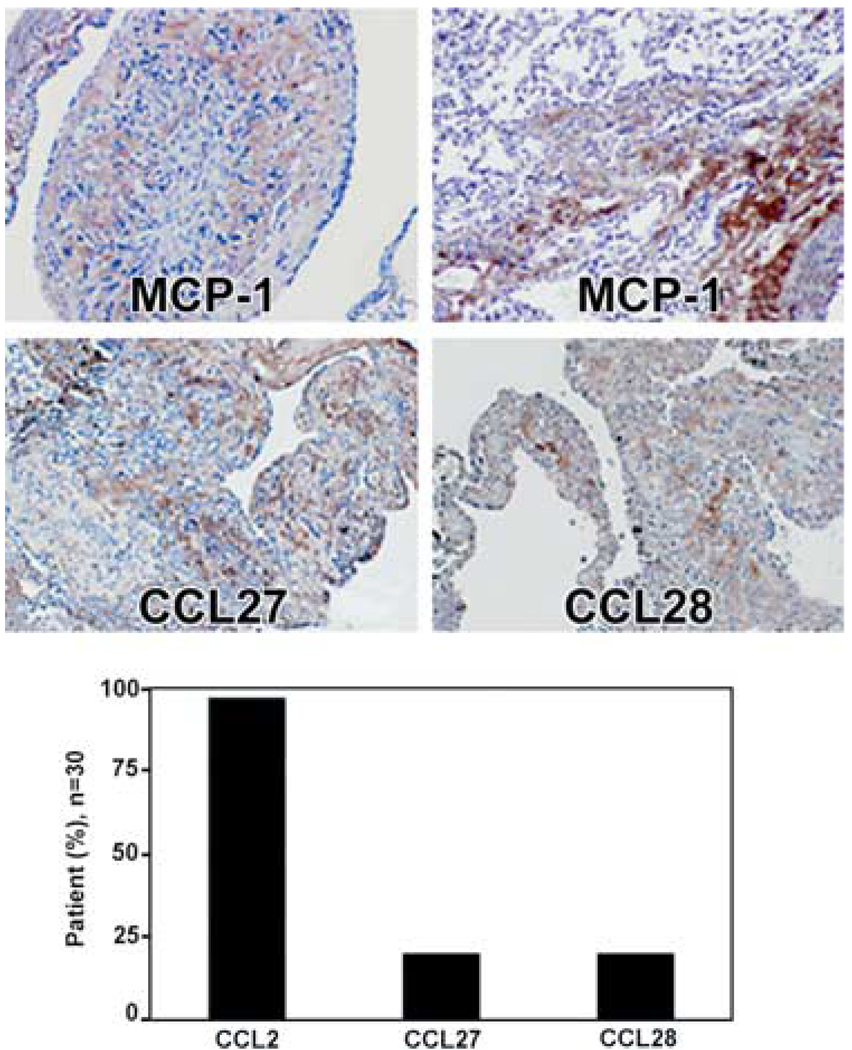
FIGURE 4.
Immunoreactivity of LAM cells to Abs against MCP-1/CCL2, CCL27, and CCL28. Upper, CCL2 immunoreactivity is seen in what appears to be extracellular matrix (not within LAM cells) of a LAM nodule as well as that of normal alveolar wall and outside of vascular structures. CCL27 and CCL28 immunoreactivity is seen in extracellular matrix area with only weak staining of LAM cells (magnification, ×100). Lower, Percentage of patients (n = 30) whose lung sections reacted with Abs against CCL2, CCL27, and CCL28.
Chemokine levels
Concentrations of cytokines, chemokines, or other biomarkers in bronchoalveolar lavage fluid (BALF) were measured using SearchLight proteome arrays (Pierce Biotechnology). Briefly, samples were diluted 1/5, 1/50, or 1/100 before a 1-h incubation on the array plates that were prespotted with capture Abs specific for each protein biomarker. Plates were emptied and washed three times before adding to each well a mixture of biotinylated detection Abs. After incubation for 30 min, plates were washed three times and incubated for 30 min with streptavidin-HRP. Plates were then washed before adding SuperSignal Femto chemiluminescent substrate (Pierce Biotechnology) and were immediately imaged using the SearchLight imaging system. Data were analyzed using ArrayVision software.
Concentrations of chemokines were measured using SearchLight proteome arrays. CXCL16, CCL27, and CCL28 were measured according to the manufacturer’s instructions (R&D Systems). Concentrations of the chemokines quantified by arrays of multiplex ELISAs and individual ELISAs were corrected for differences in lavage volumes using the epithelial lining fluid dilution factor.
Loss of heterozygosity (LOH) analysis
DNA extracted from cells that had migrated was amplified with 1× Taq buffer, 2.5 mM MgCl2, 300 µM dNTP mix, 33 µM random primers (15-mers; Operon Biotechnologies), and 5 U of TaqDNA polymerase (Continental Lab Products), with 50 cycles of 94°C for 1 min, 37°C for 2 min, ramping to 55°C for 3 min, and 55°C for 4 min. Five microliters of this mixture was then added to a PCR reaction with 1× Taq buffer, 1.5 mM MgCl2, 200 µM dNTP mix, and 1 U of TaqDNA polymerase. Samples were run on an ABI 3100 genetic analyzer (Applied Biosystems). The D16S3395 microsatellite marker on chromosome 16 was amplified with primer pair D16S3395 forward (5′-CTA ACC CTC AGC AGA GTT CTG-3′) and D16S3395 reverse (5′-Fam-CCT GGC AGT AAG TCC TGA AA-3′).
Chemotaxis assays
Chemotaxis was assayed in 96- or 24-well plates (Chemicon International) according to the manufacturer’s instructions, using, respectively, 4 × 104 or 1 × 106 cells per well. Cells in serum-free medium were placed in the upper chambers. Lower chambers contained medium without or with 10% FBS, without or with CCL2 (PrepoTech), CCL27 (R&D Systems), or CCL28 (R&D Systems) and, as indicated, 4 µM cytochalasin D. After incubation of plates (~18 h), cells that migrated through filters were detached and lysed. DNA in lysates was quantified with CyQuant GR Dye (fluorescence at 485/538 nm). To assess LOH for TSC2, cells that had migrated were detached and washed with PBS before DNA was extracted using the QIAamp DNA Mini kit (Qiagen).
Cultured cell lines
Pulmonary artery smooth muscle cells (PASM) (Clonetics) and Malme-3M malignant melanoma cells (American Type Culture Collection) were grown according to each supplier’s instructions.
Sectioning and staining for Affymetrix oligonucleotide microarray
Snap-frozen lung tissue specimens were embedded in OCT compound (Electron Microscopy Sciences), and serial 10-µm sections were cut from frozen sections with a cryotome (Shandon Lipshaw). Sections were collected on RNase-free microscope slides, immediately placed on dry ice, and stored at −80°C. Histology was monitored before laser capture microdissection (LCM). To minimize degradation of RNA, a single tissue section was thawed for microdissection. To prepare and stain sections for LCM, the HistoGene LCM frozen section staining kit (Arcturus Engineering) was used according to the manufacturer’s instructions.
Laser capture microdissection
The PixCell II LCM system (Arcturus Engineering), which incorporates an Olympus IX-50 microscope, was used to collect LAM nodules. LAM cells were captured (10,000 shots per cap for gene microarray) on a CapSure Macro Cap (Arcturus Engineering) with a 30-µm laser beam setting (pulse, 40 mW; duration, 9 ms). The cap with captured cells was cleared of nonspecifically adhering tissue using a CapSure clean-up pad (Arcturus Engineering). Images were acquired using the PixCell II image archiving workstation (Arcturus Engineering) at ×10 power.
Affymetrix oligonucleotide microarray
The human U133A chip containing 22,238 genes (Affymetrix) was used for oligonucleotide microarray. The very small amount of RNA recovered from LCM samples was purified using PicoPure RNA isolation kit (Arcturus Engineering) according to the manufacturer’s instructions. The quantity and quality of purified RNA were measured using Nano-Chip and Pico-Chip systems (Agilent Technologies). Biotin-labeled cRNA (Enzo Clinical Labs) from 20 to 100 ng of RNA was used in a two-round amplification protocol of RiboAmp OA RNA amplification kit (Arcturus Engineering). Purified cRNA (20 µg) was fragmented; 15 µg was used to prepare the hybridization mixture for standard format GeneChip microarray. As a quality control, both of the cRNAs before and after fragmentation were separated by electrophoresis in 1% denatured agarose gel. Hybridization quality and array performance were assessed by using the Affymetrix GeneChip eukaryotic hybridization control kit and test 3 chip (Affymetrix).
Microarray data analysis
Microarray data were prepared by eliminating arrays with high scale factors. Remaining arrays were analyzed using Affymetrix MAS 5.1 to make the present/absent call and compute array performance. The present/absent calls and expression estimates were imported into GeneSpring 6.1 for analyses. Replicates were averaged to identify gene expression levels that were indicative of biological variation and not variation due to measurement processes. Due to the number of replicates, the global error model was used to estimate measurement and sample-to-sample variation. Significance of intergroup differences was evaluated by the Mann-Whitney U test. The data were deposited on Gene Expression Omnibus (GEO), according to the minimum information about a microarray experiment criteria (GEO no. GSE12027).
CCL2 polymorphisms
Genomic DNA was prepared from whole blood with the PureGene kit (Gentra Systems), following manufacturer’s directions, and fragments of DNA containing polymorphisms were amplified by PCR with primers 5′-CCG AGA TGT TCC CAG CAC AG-3′ and 5′-CTG CTT TGC TTG TGC CTC TT-3′ for A(−2578)G and 5′-TGC TGA TAT GAC TAA GCC AGG AGA-3′ and 5′-ATC AGG GGA AAC CTC TCT CTG ATC-3′ for A(−2136)T. Genotypes were determined by restriction fragment polymorphism analysis, using PvuII for A(−2578)G and Bc1I for A(−2136)T.
LAM patients were matched to healthy volunteers 1:1 based on ethnicity, gender, and age (±5 years) (National Heart, Lung, and Blood Institute protocol 96-H-100). A 1:1 case-control study was designed to compare the CCL2 promoter genotypes in LAM patients and controls by using multinomial logistic regression in the statistical software SPSS. To correlate the lung function (e.g., forced expiratory volume in 1 s (FEV1) and diffusion capacity of carbon monoxide (DLCO) with different polymorphisms of CCL2, linear regression analyses (SPSS) were performed to assess the relationships between the phenotypes and CCL2 genotypes within the LAM population.
Results
To assess the potential role of chemokines in LAM cell migration, we compared amounts of 25 different chemokines in BALF from LAM patients and healthy, age-matched female volunteers (Table I). Concentrations of CCL2, CCL19, CXCL1, CXCL5, CXCL11, CXCL16, and CXCL12 differed significantly between the two groups (Table I). These data are consistent with the hypothesis that chemokines may play a role in the motility and the recruitment of LAM cells from other sites to the lung. Among those chemokines, CCL2, CXCL1, and CXCL5 were significantly higher in patients with pulmonary LAM.
Table I.
Concentration of chemokines in BALF from patients with LAM (n = 29) and normal healthy volunteers (NV) (n = 25)a
| Chemokine | NV | LAM |
|---|---|---|
| CCL1 | 360 ± 80 | 240 ± 40 |
| CCL2 | 4,710 ± 1,050 | 12,710 ± 3,430* (p = 0.033) |
| CCL3 | 1,510 ± 340 | 1,470 ± 230 |
| CCL4 | 1,290 ± 360 | 1,730 ± 270 |
| CCL5 | 420 ± 90 | 610 ± 90 |
| CCL7 | 280 ± 60 | 210 ± 40 |
| CCL8 | 420 ± 90 | 290 ± 60 |
| CCL11 | 1,050 ± 380 | 490 ± 100 |
| CCL13 | 270 ± 90 | 230 ± 60 |
| CCL17 | 290 ± 60 | 290 ± 50 |
| CCL19 | 1,730 ± 550 | 550 ± 140* (p = 0.044) |
| CCL20 | 860 ± 380 | 680 ± 170 |
| CCL22 | 560 ± 220 | 460 ± 80 |
| CCL21 | 1,520 ± 390 | 1,760 ± 410 |
| CCL27b | ND | ND |
| CCL28b | ND | ND |
| XCL1 | 2,080 ± 490 | 2,300 ± 630 |
| CXCL1 | 23,960 ± 5,000 | 44,900 ± 7,410* (p = 0.024) |
| CXCL5 | 4,470 ± 1,350 | 10,180 ± 2,440* (p = 0.047) |
| CXCL8 | 1,820 ± 270 | 2,630 ± 360 |
| CXCL9 | 3,710 ± 840 | 3,790 ± 650 |
| CXCL10 | 1,900 ± 580 | 1,260 ± 270 |
| CXCL11 | 640 ± 160 | 270 ± 70* (p = 0.043) |
| CXCL12 | 13,910 ± 5,110 | 2,170 ± 660* (p = 0.026) |
| CXCL16 | 230 ± 44 | 123 ± 16* (p = 0.033) |
Means of concentrations of the chemokines determined by arrays of multiplex ELISAs were corrected for differences in bronchoalveolar lavage volumes using the epithelial lining fluid dilution factor and are reported as pg/ml with the exception of CXCL16 (ng/ml; NV (n = 22); LAM (n = 27)).
Significant; ND, nondetectable.
Determined by individual ELISA.
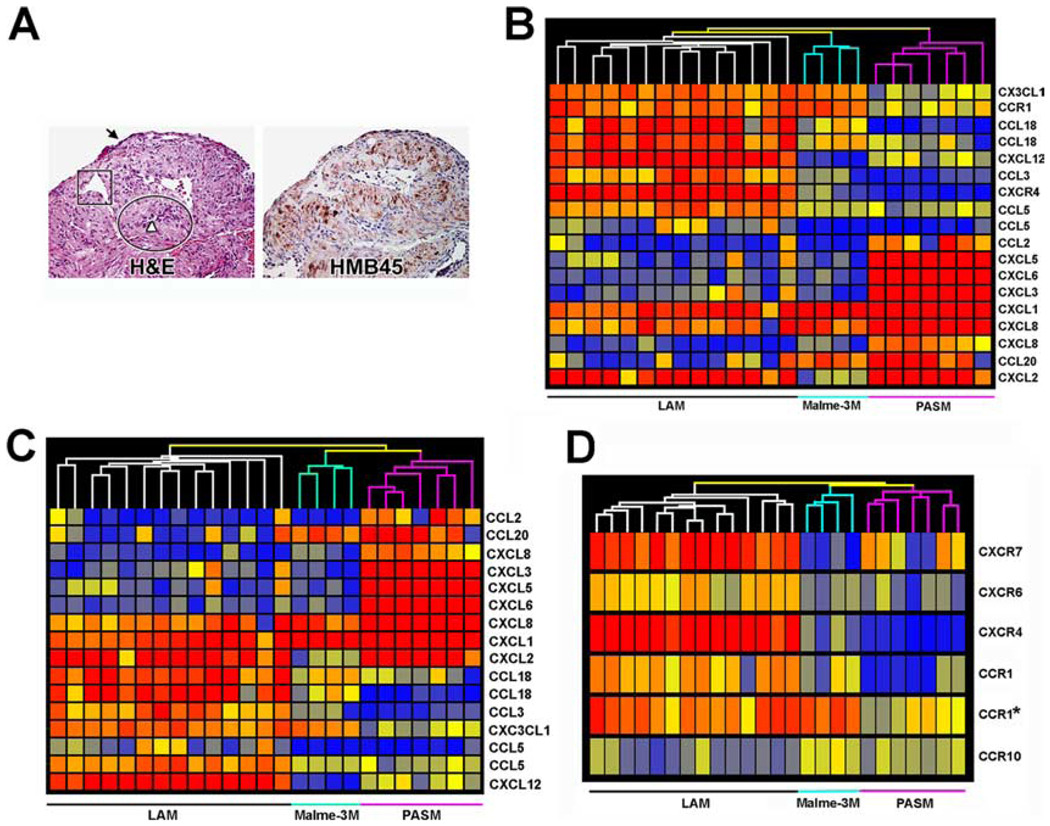
Nodular structures in the lungs of LAM patients contain smooth muscle-like cells, termed LAM cells, which were both spindle-shaped (Fig. 1A) and epithelioid (17). LAM cells were identified by immunoreactivity with the mAb HMB-45 (18, 19), which recognizes gp100, encoded by the Pmel17 gene (20).
FIGURE 1.
Gene expression profile of microdissected LAM cells. A, Serial sections (magnification, ×200) of lung from one LAM patient, representative of samples from 30 patients. Left, Stained with H&E. Arrow indicates type II pneumocytes; a proliferative area of LAM cells is shown in the oval; square outlines vascular structure in close proximity to the proliferating LAM cells; arrowhead indicates lymphatic-like structures. Right, Reactivity of LAM cells with mAb HMB-45. B, Hierarchical cluster diagram of 18 probe sets differentially expressed among the 79 probe sets that represent chemokines and chemokine receptors. C, Among the 18 probe sets, 13 chemokines clustered LAM cells from PASM and Malme-3M. D, Five chemokine receptors, that is, CXCR7 (212977_at), CXCR6 (206974_at), CXCR4 (217028_at), CCR1 (205098_at and 205099_s_at), and CCR10 (220565_at), differentiate gene expression in LAM cells from PASM and Malme-3M cells. The probes used for clustering are indicated in Table II.
These nodular structures were microdissected for comparison with PASM and Malme-3M melanoma cells. These cells were chosen because LAM cells appear phenotypically similar to smooth muscle cells and contain premelanosomal structures found in melanocytes and melanoma cells (21). Although several studies have compared biological aspects of airway smooth muscle cells with putative LAM cells (22), it has been difficult to define an experimental control; however, our initial experiments that compared the expression of ~4000 genes from microdissected LAM nodules with established cell lines including airway smooth muscle cells showed that PASM were closely related to LAM cells (data not shown). Our initial studies showed that Malme-3M contained melanosomes in their cytoplasm as well as reactivity with HMB-45 similar to that of cells grown from LAM lungs (21, 23). In contrast, other melanoma cell lines, A2058 and CHL1, reacted weakly with the HMB-45 and contained lesser amounts of this splice form of the PMel17 gene product (21). The relatively high level of gp100 in Malme-3M led us to use it as a cell line control.
Expression of chemokines and their receptors genes (Table II) were assessed using microarrays. The gene probes of Table II were selected according to the chemokine and chemokine receptor genes defined by the International Union of Pharmacology (IUPHAR) (www.iuphar.org/about_intro.html) (24 –26). In these microarrays, 79 Affymetrix probe sets were associated with chemokines and their receptors, representing a total of 60 genes (Table II). The discrepancy in the number of probes and number of genes arises from the design of microarrays, which include different probes to quantify a single assigned gene. Although the most direct measurement of the transcript products is determined by the antisense probes, indicated with “_at” after the identification number, other related measurements of the genes include determination of alternative polyadenylated transcripts or alternative splice forms indicated with “_s_at”. The probe number followed by “_x_at” designates probe sets for multiple transcripts of related genes. Thus, it is important to identify the type of probe used for the specific measurement of a gene or related genes.
Table II.
Normalized expression level of genes for chemokines and their receptorsa
| Gene | GenBank | Probe Set | LAM | Malme-3M | PASM |
|---|---|---|---|---|---|
| CX3CL1b,c | U84487 | 823_at | 2.69 | 2.21 | 0.69 |
| CCL18b,c | Y13710 | 32128_at | 4.73 | 0.91 | 0.07 |
| XCR1 | NM_005283 | 221468_at | 0.12 | 0.21 | 0.14 |
| CCL24 | NM_002991 | 221463_at | 0.09 | 0.12 | 0.15 |
| CCR10d | NM_016602 | 220565_at | 0.46 | 0.87 | 0.67 |
| CCRL1 | NM_016557 | 220351_at | 0.54 | 0.29 | 0.59 |
| CXCL14 | NM_004887 | 218002_s_at | 0.68 | 0.12 | 0.17 |
| CXCR3 | Z79783 | 217119_s_at | 0.06 | 0.08 | 0.05 |
| CXCR4c,d | AJ224869 | 217028_at | 11.44 | 0.43 | 0.08 |
| CCL13 | Z77651 | 216714_at | 0.19 | 0.10 | 0.12 |
| CCL2b,c | S69738 | 216598_s_at | 0.18 | 0.10 | 1.87 |
| CXCL5 | BG166705 | 215101_s_at | 0.07 | 0.07 | 0.14 |
| CXCL5b,c | AK026546 | 214974_x_at | 0.44 | 0.26 | 31.00 |
| XCL2 | NM_003175 | 214567_s_at | 0.09 | 0.06 | 0.05 |
| SCYA8 | AI984980 | 214038_at | 0.97 | 0.27 | 0.38 |
| CXCR7d | AI817041 | 212977_at | 4.76 | 0.15 | 0.89 |
| CXCR4 | AF348491 | 211919_s_at | 2.11 | 1.63 | 1.69 |
| GPR30 | U58828 | 211829_s_at | 0.05 | 0.04 | 0.05 |
| IL8b,c | AF043337 | 211506_s_at | 0.17 | 0.33 | 2.09 |
| CXCR6 | U73531 | 211469_s_at | 0.23 | 0.31 | 0.29 |
| CCRL2 | AF015524 | 211434_s_at | 0.31 | 0.11 | 0.06 |
| CXCL11 | AF002985 | 211122_s_at | 0.16 | 0.14 | 0.29 |
| CMKLR1 | U79526 | 210659_at | 0.95 | 0.52 | 0.56 |
| CCL23 | U58913 | 210549_s_at | 0.14 | 0.10 | 0.09 |
| CCL23 | U58913 | 210548_at | 0.21 | 0.19 | 0.15 |
| CCL15 | AF031587 | 210390_s_at | 0.15 | 0.53 | 0.14 |
| CXCL11 | AF030514 | 210163_at | 0.14 | 0.07 | 0.10 |
| CCL11 | D49372 | 210133_at | 0.85 | 0.75 | 0.51 |
| CCL19 | U88321 | 210072_at | 0.48 | 0.14 | 0.09 |
| CCL18b,c | AB000221 | 209924_at | 4.47 | 1.26 | 0.54 |
| CXCL2b,c | M57731 | 209774_x_at | 9.68 | 0.57 | 13.78 |
| CXCL12b,c | U19495 | 209687_at | 8.82 | 0.15 | 0.76 |
| CXCR4 | L01639 | 209201_x_at | 1.82 | 1.55 | 1.07 |
| CCR4 | NM_005508 | 208376_at | 0.47 | 0.35 | 0.55 |
| CCR3 | NM_001837 | 208304_at | 0.13 | 0.13 | 0.15 |
| CCL7 | NM_006273 | 208075_s_at | 0.15 | 0.20 | 1.05 |
| CCR8 | NM_005201 | 208059_at | 0.42 | 0.75 | 0.81 |
| CCL27 | NM_006664 | 207955_at | 0.07 | 0.23 | 0.18 |
| CCL17 | NM_002987 | 207900_at | 0.10 | 0.08 | 0.10 |
| CCL22 | NM_002990 | 207861_at | 0.27 | 0.24 | 0.35 |
| CXCL5 | NM_002994 | 207852_at | 0.06 | 0.06 | 0.19 |
| CXCL3b,c | NM_002090 | 207850_at | 0.42 | 0.18 | 27.06 |
| CCR2 | NM_000648 | 207794_at | 0.27 | 0.41 | 0.48 |
| CXCR3 | NM_001504 | 207681_at | 0.23 | 0.25 | 0.26 |
| CMKLR1 | NM_004072 | 207652_s_at | 0.15 | 0.09 | 0.22 |
| CCL1 | NM_002981 | 207533_at | 0.14 | 0.11 | 0.13 |
| CCR9 | AF145439 | 207445_s_at | 0.51 | 0.63 | 0.62 |
| CCL16 | NM_004590 | 207354_at | 0.24 | 0.16 | 0.41 |
| IL8RA | NM_000634 | 207094_at | 0.22 | 0.39 | 0.40 |
| IL8RB | NM_001557 | 207008_at | 0.08 | 0.06 | 0.05 |
| CCR5 | NM_000579 | 206991_s_at | 0.60 | 0.13 | 0.10 |
| CCL25 | NM_005624 | 206988_at | 0.15 | 0.28 | 0.30 |
| CCR6 | NM_004367 | 206983_at | 0.22 | 0.25 | 0.36 |
| CCR2 | NM_000647 | 206978_at | 1.25 | 0.92 | 1.14 |
| CXCR6d | NM_006564 | 206974_at | 1.26 | 0.51 | 0.41 |
| CCL13 | NM_005408 | 206407_s_at | 0.52 | 0.18 | 0.54 |
| PF4 | NM_002619 | 206390_x_at | 0.39 | 0.93 | 1.70 |
| XCL1 | U23772 | 206366_x_at | 0.33 | 0.15 | 0.22 |
| XCL1 | NM_002995 | 206365_at | 0.16 | 0.52 | 0.30 |
| CCR7 | NM_001838 | 206337_at | 1.54 | 0.50 | 0.83 |
| CXCL6b,c | NM_002993 | 206336_at | 0.33 | 0.30 | 64.72 |
| CX3CR1 | U20350 | 205898_at | 0.86 | 0.16 | 0.10 |
| CCL20b,c | NM_004591 | 205476_at | 0.40 | 2.90 | 12.33 |
| CCL15 | NM_004166 | 205392_s_at | 3.36 | 0.77 | 1.13 |
| CXCL13 | NM_006419 | 205242_at | 0.34 | 0.25 | 0.09 |
| HM74 | NM_006018 | 205220_at | 0.73 | 0.54 | 0.50 |
| CCL3b,c | NM_002983 | 205114_s_at | 2.20 | 0.41 | 0.17 |
| CCR1d | NM_001295 | 205099_s_at | 1.59 | 0.65 | 0.17 |
| CCR1c,d | AI421071 | 205098_at | 2.72 | 3.28 | 0.89 |
| CCL5b,c | NM_002985 | 204655_at | 2.30 | 0.77 | 0.69 |
| CCL21 | NM_002989 | 204606_at | 1.05 | 0.41 | 0.46 |
| CXCL10 | NM_001565 | 204533_at | 2.47 | 1.50 | 1.41 |
| CXCL1b,c | NM_001511 | 204470_at | 4.40 | 11.08 | 144.50 |
| CCL4 | NM_002984 | 204103_at | 0.82 | 0.35 | 0.13 |
| CXCL9 | NM_002416 | 203915_at | 2.61 | 1.03 | 1.28 |
| CX3CL1 | NM_002996 | 203687_at | 0.32 | 0.23 | 0.12 |
| CXCL12 | NM_000609 | 203666_at | 2.42 | 1.34 | 1.29 |
| CXCL8b,c | NM_000584 | 202859_x_at | 2.39 | 3.49 | 118.70 |
| CCL5b,c | M21121 | 1405_i_at | 0.59 | 0.05 | 0.07 |
For each gene, the normalization was performed with Affymetrix 5.1 before importion into GeneSpring and scaling the mean intensity of each sample to 1. Data from 14 different samples (patients) with sporadic LAM, 4 different cultures of Malme-3M, and 8 different cultures of pulmonary artery smooth muscle cells were analyzed.
Probes used for clustering in Fig. 1C.
Probes used for clustering in Fig. 1B.
Probes used for clustering in Fig. 1D.
Among those 79 probes, 18 differed significantly, representing 15 genes (false discovery rate of 0.001), and they formed distinct clusters in nonsupervised analysis (Fig. 1B). Of those genes, 13 encode chemokines and only two (CCR1 and CXCR4) correspond to chemokine receptors (Fig. 1B). Chemokine genes also distinguish LAM cells from Malme-3M and PASM with a false discovery rate of 0.001 (Fig. 1C). The two chemokine receptors (CCR1 and CXCR4) were not sufficient to group the different samples. However, statistical analysis showed that with a false discovery rate of 0.01, five chemokine receptors represented with six probes (Fig. 1D) allowed for clustering of the different cell types. Among the five genes, four encoded chemokine receptors genes (CCR1, CXCR4, CXCR6, and CCR10) that are present in tissue sections of LAM patients (Fig. 2). To determine the strength of the clustering, we took the most informative set of genes (CXCR7 (212977_at), CXCR6 (206974_at), CXCR4 (217028_at), CCR1 (205098_at and 205099_s_at), and CCR10 (220565_at) and using a resampling method (10,000 times) it was determined that in 100% of the cases the LAM samples were correctly clustered.
Thus, as had been found for breast cancer and melanoma cells (8), LAM cells were characterized by a specific pattern of chemokine receptor gene expression that distinguished them from smooth muscle and melanoma cells. Additionally, the production of chemokines by LAM cells suggests an inflammatory response (27) (Table I).
More than 50% of smooth-muscle-like LAM cells from 30 LAM patients reacted with Abs against the chemokine receptors CCR2, CCR7, CCR10, CXCR2, CXCR4, and CXCR1 (Fig. 2, left panel). The presence of seven other chemokine receptors was detected in <50% of the samples. These findings are consistent with a potential role for chemokine receptors in LAM cell function. We failed to detect a clustering of patient subgroups based on chemokine receptor immunoreactivity (data not shown). Heterogeneity of reaction with antichemokine receptor Abs was found among LAM lung nodules. Because receptors CXCR1, CXCR2, CXCR4, CCR2, CCR7, and CCR10 were present more frequently than the others in LAM cells within lung nodules, we hypothesized that chemokines such as CCL2, CCL27, and/or CCL28 might influence LAM cell migration.
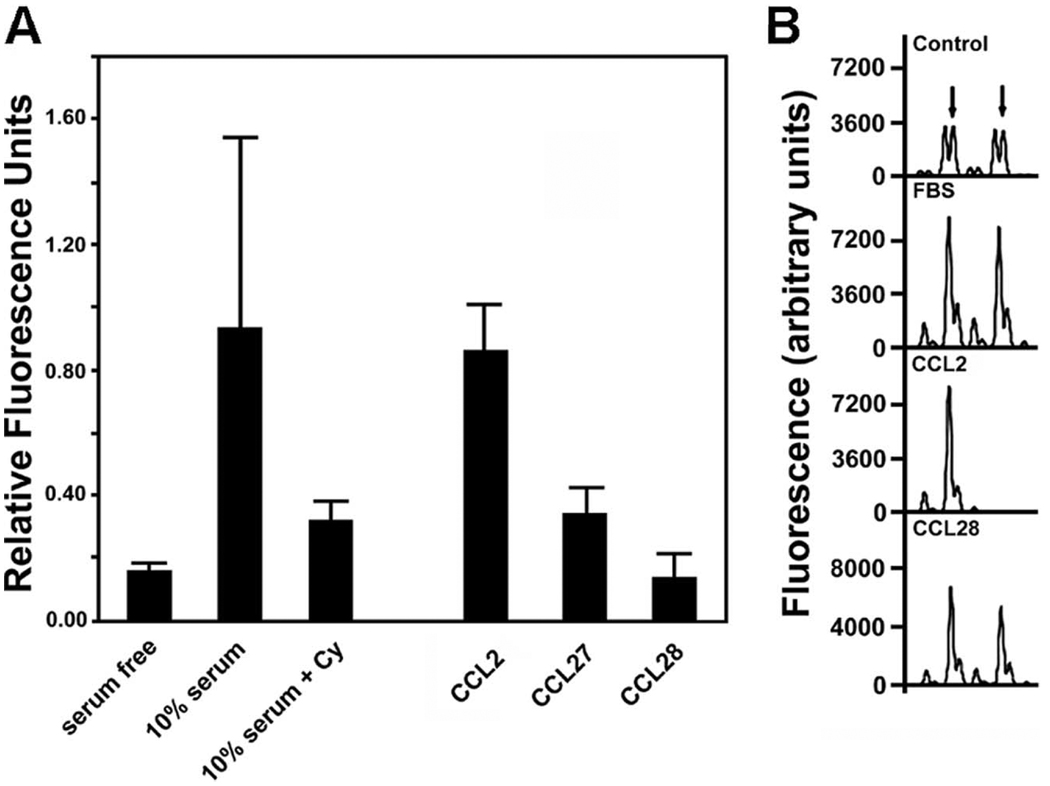
Of note, we reported previously that levels of CCL2 are influenced by loss of TSC2 function (28). To demonstrate LAM cell migration in response to CCL2, heterogeneous populations of cells grown from explanted lungs of LAM patients undergoing lung transplantation that contained ~20% LAM cells (determined by fluorescence in situ hybridization analyses) were used. Chemotaxis in response to chemokines and other agonists was quantified in Boyden chambers (Fig. 3A). Migration of cells in response to FBS was 4–5-fold that in the absence of serum, and it was successfully blocked by inhibition of cytoskeletal function with cytochalasin D, confirming serum-dependent migration (29, 30). As shown in Fig. 3, in the absence of serum, CCL2 significantly enhanced migration (p = 0.041), whereas CCL27 (p = 0.172) and CCL28 (p = 0.833) were without significant effect. Additionally, cell migration in the presence of fractalkine/CX3CL1 and stromal cell-derived factor (SDF)-1α/β/CXCL12 did not differ from that of unstimulated cells (data not shown).
FIGURE 3.
Chemotactic migration and loss of TSC2 heterozygosity by cells grown from explanted lungs of patients with LAM. After incubation for 24 h without serum, the heterogeneous populations of cells were incubated in Boyden chambers where medium in lower chambers contained no additions (serum-free = control), 10% serum without or with 4 µM cytochalasin D (Cy), or CCL2, CCL27, or CCL28 (each 100 ng/ml). A, Amounts of cells (relative fluorescence units) that had migrated through filters in 18–20 h are reported as means ± SD of values from triplicate incubations in five experiments. B, Cells that had migrated through 24-well Boyden chambers were detached and DNA was extracted for analysis of the D16S3395 microsatellite marker on chromosome 16. In the top (control) chromatogram, the amplified alleles are from the DNA of a heterogeneous population; arrows indicate two alleles of TSC2. Two alleles were also seen in DNA from cells that migrated in response to FBS or in the presence of CCL28, but only one was observed among cells that responded to CCL2. Results were similar with cells from two patients.
Because a heterogeneous population of cultured cells from LAM lungs was used in these studies, we evaluated loss of heterozygosity for TSC2, a hallmark of LAM cells, among cells that had migrated under the different conditions. Cells from two patients that migrated in response to CCL2 had loss of TSC2 heterozygosity, whereas those that migrated in the presence of serum, CCL28 (Fig. 3B), or CCL27 did not. Thus, LAM cells appeared to be selectively mobilized by CCL2.
As CCL2 was one of the factors secreted by cells with dysfunctional TSC2 (28), we investigated its presence in LAM lung specimens from 30 patients, using Abs against CCL2, CCL27, and CCL28 (Fig. 4). CCL2 immunoreactivity was observed on what appears to be the surface of LAM nodules from ~70% of patients, although only ~10% of those tissues reacted with Abs against CCL27 or CCL28 (Fig. 4). We were unable to detect CCL27 or CCL28 by ELISA in BALF of LAM patients (Table I).
Frequencies of two polymorphisms in the promoter of the CCL2 gene were compared in LAM patients and age- and sex-matched healthy volunteers (Table III). Both differed significantly in the two groups with frequency of AA at positions −2578 and −2136 greater in LAM patients than in volunteers (Table III). Combining genotypes of the two promoter polymorphisms resulted in greater statistical significance (p = 0.002) of the difference between the LAM patients and the healthy volunteers, with 36% of LAM patients homozygous for AA at both promoter positions vs 22% of the control population.
Table III.
Distribution of the CCL2 gene polymorphismsa
| Single Nucleotide Polymorphism |
No. (%) of Samples | ||
|---|---|---|---|
| LAM | Normal volunteer | ||
| A(−2578)Gb | |||
| AA | 120 (57.1) | 103 (49.0) | |
| AG | 81 (38.6) | 86 (41.0) | |
| GG | 9 (4.3) | 21 (10.0) | |
| A(−2136)Tc | |||
| AA | 140 (70.7) | 115 (58.1) | |
| AT | 51 (25.8) | 77 (38.9) | |
| TT | 7 (3.5) | 6 (3.0) | |
| A(−2578)G | A(−2136)Td | ||
| AA | AA | 70 (35.9) | 43 (22.1) |
| AT | 36 (18.5) | 49 (25.1) | |
| TT | 7 (3.6) | 6 (3.1) | |
| AG | AA | 61 (31.3) | 55 (28.2) |
| AT | 13 (6.7) | 24 (12.3) | |
| TT | 0 | 0 | |
| GG | AA | 8 (4.1) | 15 (7.7) |
| AT | 0 | 3 (1.5) | |
| TT | 0 | 0 | |
Samples from 210 LAM patients and 210 healthy-matched volunteers were analyzed.
p = 0.018.
p = 0.036.
p = 0.002.
To explore a functional role of these polymorphisms, we correlated them with rate of decline in lung function (DLCO and FEV1), finding that the rate of decline in FEV1 correlated with the polymorphism A(−2578)G (p = 0.028) with a more rapid rate of decline in those homozygous for the A allele (Table IV). For the patients with A allele, a trend toward more rapid decline in DLCO was not quite significant (p = 0.058). As might be expected, given its role in disease progression, this genotype appears to be present in individuals who had been diagnosed at a younger age (p = 0.054). All data are consistent with the conclusion that the CCL2 gene and protein may be genetic modifiers in the development of LAM.
Table IV.
Correlation of the CCL2 gene polymorphisms with lung functiona
| Polymorphism | Age | Age of Diagnosis | Rate of Decline in FEV1 (%/Year) | Rate of Decline in DLCO (%/Year) |
|---|---|---|---|---|
| A(−2578)G | ||||
| AA | 49.8 ± 0.9 (109) | 39.8 ± 0.8 (105)b | 3.6 ± 0.4 (99)c | 3.7 ± 0.4 (99)d |
| AG | 51.5 ± 1.3 (73) | 41.6 ± 1.3 (74) | 1.9 ± 0.4 (71) | 2.6 ± 0.4 (71) |
| GG | 51.9 ± 2.3 (7) | 46.6 ± 2.1 (7) | 3.3 ± 2.0 (7) | 2.4 ± 0.4 (7) |
| A(−2136)T | ||||
| AA | 51.1 ± 0.9 (133) | 41.1 ± 0.8 (129) | 2.7 ± 0.4 (121) | 2.9 ± 0.3 (121) |
| AT | 49.3 ± 1.3 (49) | 40.2 ± 1.3 (51) | 3.3 ± 0.6 (50) | 4.1 ± 0.6 (50) |
| TT | 49.0 ± 4.0 (6) | 39.0 ± 4.2 (6) | 2.1 ± 0.5 (6) | 2.8 ± 0.4 (6) |
Linear regression analyses (SPSS) were performed to assess the relationship between the phenotypes and CCL2 genotypes within the LAM population. Ages were as of 2005. Data are means ± SEM (no. of patients).
p = 0.054.
p = 0.028.
p = 0.058.
Discussion
Chemokines, which induce migration of leukocytes and other cells, are considered to be synthesized largely by cells of the immune system, resulting in cell recruitment to a site of inflammation as part of an inflammatory response (27). The function of chemokines produced by noninflammatory cells, such as smooth muscle cells, is less clear but they may play a role in recruitment of cells in health as well as disease (7) and affect stroma cells (31). Since inflammatory processes appear to be required for recruitment of cancer cells (32), and LAM nodules apparently lack inflammatory cells (33), we propose that production of chemokines in this situation is a response to different stimuli.
Hypoxia enhances the levels of chemokine receptors (34, 35), including CXCR4 that is relatively abundant in LAM cells (Figs. 1 and 2). Patients with LAM may experience hypoxia with exercise (36), which could augment the synthesis of chemokine and chemokine receptors. HIFs (hypoxia-inducible factors) are master transcription factors that modulate the expression of several genes during hypoxia (37). Loss of TSC2 function, which is characteristic of LAM cells, through the activation of mammalian target of rapamycin (mTOR) can increase HIF-1α (38), which could influence directly the transcription of genes (37) such as those for CXCR4 (39) and SDF-1 (40). Thus, expression of chemokines and their receptors in LAM could be regulated, in part, by hypoxia, and thus is relevant to LAM patients with moderate or severe disease who may experience desaturation with exercise.
Gene expression of chemokines are regulated at the transcriptional level largely by binding of sequence-specific transcription factors such as IFN regulatory factors (41) and GATA transcription factor (42). Binding of these factors to specific sequences may be altered by a single nucleotide difference, as may be the case for the CCL2 polymorphisms, which could contribute through modulation of chemokine gene expression. Two of the several CCL2 gene polymorphisms are in the promoter and are reported to have multiple effects on CCL2 levels. For the A(−2578)G polymorphism, the G allele correlated with higher levels of CCL2 in IL-1β-stimulated mononuclear cells than did the A allele (43, 44), whereas individuals with the A allele had higher plasma levels of CCL2 than did those with the G allele (45). There are other reports of no correlation between this polymorphism and plasma CCL2 concentrations (46, 47). LAM patients had a significantly greater frequency of AA homozygosity at both sites than did age-matched healthy volunteers, suggesting that CCL2 might be a modifier gene for the development or diagnosis of LAM. The AA homozygotes of the A(−2578)G polymorphism had somewhat higher (p = 0.16) concentrations of CCL2 in BALF than did AG heterozygotes or GG homozygotes (data not shown). Perhaps greater production of CCL2 in the lungs of individuals homozygous for the A allele of A(−2578)G results in conditions for favorable LAM cell localization and proliferation. The association of the A(−2578)G polymorphism with a rate of lung function decline as measured by FEV1 suggests that CCL2 could be a disease modifier. We had reported that cells with dysfunctional TSC2 produced large amounts of CCL2 (28), and CCL2 production by LAM cells, in which TSC2 is dysfunctional, could enhance mobilization of cells that respond to this chemokine, including LAM cells.
The high expression of inflammatory chemokines in bronchoalveolar lavage from LAM patients (CCL2, CXCL1, and CXCL5; Table I) and the production of chemokines of mRNA by cells from LAM nodules (Table II and Fig. 1) suggest the involvement of inflammation in the pathogenesis of LAM. Interestingly, CCL2 and its receptor are highly expressed in a dominant negative transgenic mouse model of Tsc2 (48). Furthermore, expression of CXCL1 by breast cancer cells (MDA-MB-231) confers on them metastatic potential (49). Finally, growth-regulated oncogene 1 (Gro-1)/CXCL5 has been implicated in tumor progression by causing senescence of fibroblasts that could be associated with ovarian cancer (50). In summary, the highly expressed chemokines in the BALF of LAM patients could play roles in cell motility, affecting metastasis and/or influencing the LAM microenvironment by altering LAM cell-stroma interactions.
LAM has not been defined as an inflammatory disease; however, hyperplastic type II pneumocytes (51) and the presence of bronchiolitis in 47 of 74 (~60%) patients with patients with LAM (52) suggest that both are associated with inflammatory responses (53). LAM cell proliferation clearly could be influenced by molecules that participate in acute or chronic inflammation.
Renal angiomyolipomas were postulated to be a primary source of lung LAM cells (54). Anchoring of LAM cells in the lung may require their transit via the lymphatic and/or cardiovascular circulation (54). Identification of LAM cells in blood and other body fluids (e.g., chylous pleural effusions) of LAM patients (16) suggested that the cells could be disseminated in a fashion similar to that of neoplastic cells (1). Chemokines might affect not only cell mobilization but also their homing (8, 55, 56). Reported studies on the effects of chemokines on cancer cell mobilization were performed on well-characterized homogeneous cell populations (e.g., MDA-MB-231) (8). Here, we show that the chemokine CCL2 selectively attracts LAM cells, as defined by TSC2 LOH, from heterogeneous cell populations.
Chemokine receptors including CCR5 (57) and CXCR4 (58) have been described on smooth muscle cells from different tissues. In diseases such as asthma (58) and atherosclerosis (59), smooth muscle cell chemokine receptors are up-regulated. Smooth muscle cells from asthmatics were reported to express a specific group of chemokine receptors with different functions (i.e., CCR2, CCR3, CCR8, and CCR5) (60), as were breast cancer cells (i.e., CXCR4 and CCR7) and melanoma cells (i.e., CCR10, CXCR4, and CCR7) (8). The patterns of chemokine receptor gene expression in breast cancer and melanoma cells apparently differ from those of LAM cells, where CCR10, CXCR2, CCR2, CXCR4, and CX3CR1(8) were found most commonly. Identification of neoplastic cells within a tumor has been based mainly on genetic abnormalities such as point mutations, chromosomal deletions, and instability of genomic regions that could lead to loss of heterozygosity (61, 62). Demonstration of loss of heterozygosity requires a homogeneous or enriched population of cells, as is the case in LAM cells within lung nodule (10). Detection of LOH for TSC2 in cells grown from lung explants is not easy due to growth of multiple types of cells from lung explants. Our finding that cells that migrated in response of CCL2 show LOH for TSC2 suggests that CCL2 could play an important role in LAM cell movement, and thus the potential for metastasis.
Stromal cells might also be involved in LAM cell recruitment to the lung by producing specific growth factors and/or chemokines (5). Selective attraction of LAM cells by CCL2 and CCL2 over-expression by TSC2−/− cells led us to propose a paracrine feedback loop of LAM cell metastasis involving CCL2. Thus, CCL2 and its specific receptors (e.g., CCR2 and CCR10) with potential roles in the pathogenesis of LAM could also represent therapeutic targets.
Acknowledgments
We thank Dr. Martha Vaughan and Dr. Vincent C. Manganiello for helpful discussions and critical review of the manuscript, Michael Spencer for his help with the artwork, and Dr. Angelo M. Taveira-DaSilva and Mario Stylianou for clinical data. We thank The LAM Foundation and the Tuberous Sclerosis Alliance for their assistance in recruiting patients for our studies.
Footnotes
This research was supported by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute. Y.I. was contracted through a Senior Fellowship from the Oak Ridge Institute for Science and Education.
Abbreviations used in this paper: LAM, lymphangioleiomyomatosis; BALF, bronchoalveolar lavage fluid; DLCO, diffusion capacity of carbon monoxide; FEV1, forced expiratory volume in 1 s; LCM, laser capture microdissection; LOH, loss of heterozygosity; PASM, pulmonary artery smooth muscle cells; TSC, tuberous sclerosis complex.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 2.Paget S. The distribution of secondary growths in cancer of the breast. Lancet. 1889;1:571–573. [PubMed] [Google Scholar]
- 3.Hujanen ES, Terranova VP. Migration of tumor cells to organ-derived chemoattractants. Cancer Res. 1985;45:3517–3521. [PubMed] [Google Scholar]
- 4.Moore MA. The role of chemoattraction in cancer metastases. Bioessays. 2001;23:674–676. doi: 10.1002/bies.1095. [DOI] [PubMed] [Google Scholar]
- 5.Liotta LA, Kohn EC. The microenvironment of the tumour-host interface. Nature. 2001;411:375–379. doi: 10.1038/35077241. [DOI] [PubMed] [Google Scholar]
- 6.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cells. 2005;7:211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Singer CA, Salinthone S, Baker KJ, Gerthoffer WT. Synthesis of immune modulators by smooth muscles. Bioessays. 2004;26:646–655. doi: 10.1002/bies.20041. [DOI] [PubMed] [Google Scholar]
- 8.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 9.Steagall WK, Taveira-DaSilva AM, Moss J. Clinical and molecular insights into lymphangioleiomyomatosis. Sarcoidosis Vasc. Diffuse Lung Dis. 2005;22 Suppl. 1:S49–S66. [PubMed] [Google Scholar]
- 10.Carsillo T, Astrinidis A, Henske EP. Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. Proc. Natl. Acad. Sci. USA. 2000;97:6085–6090. doi: 10.1073/pnas.97.11.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N. Engl. J. Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- 12.Strizheva GD, Carsillo T, Kruger WD, Sullivan EJ, Ryu JH, Henske EP. The spectrum of mutations in TSC1 and TSC2 in women with tuberous sclerosis and lymphangiomyomatosis. Am. J. Respir. Crit. Care Med. 2001;163:253–258. doi: 10.1164/ajrccm.163.1.2005004. [DOI] [PubMed] [Google Scholar]
- 13.Sato T, Seyama K, Kumasaka T, Fujii H, Setoguchi Y, Shirai T, Tomino Y, Hino O, Fukuchi Y. A patient with TSC1 germline mutation whose clinical phenotype was limited to lymphangioleiomyomatosis. J. Intern. Med. 2004;256:166–173. doi: 10.1111/j.1365-2796.2004.01356.x. [DOI] [PubMed] [Google Scholar]
- 14.Bittmann I, Rolf B, Amann G, Lohrs U. Recurrence of lymphangioleiomyomatosis after single lung transplantation: new insights into pathogenesis. Hum. Pathol. 2003;34:95–98. doi: 10.1053/hupa.2003.50. [DOI] [PubMed] [Google Scholar]
- 15.Karbowniczek M, Astrinidis A, Balsara BR, Testa JR, Lium JH, Colby TV, McCormack FX, Henske EP. Recurrent lymphangiomyomatosis after transplantation: genetic analyses reveal a metastatic mechanism. Am. J. Respir. Crit. Care Med. 2003;167:976–982. doi: 10.1164/rccm.200208-969OC. [DOI] [PubMed] [Google Scholar]
- 16.Crooks DM, Pacheco-Rodriguez G, DeCastro RM, McCoy JP, Jr, Wang JA, Kumaki F, Darling T, Moss J. Molecular and genetic analysis of disseminated neoplastic cells in lymphangioleiomyomatosis. Proc. Natl. Acad. Sci. USA. 2004;101:17462–17467. doi: 10.1073/pnas.0407971101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonetti F, Pea M, Martignoni G, Zamboni G, Iuzzolino P. Cellular heterogeneity in lymphangiomyomatosis of the lung. Hum. Pathol. 1991;22:727–728. doi: 10.1016/0046-8177(91)90298-4. [DOI] [PubMed] [Google Scholar]
- 18.Hoon V, Thung SN, Kaneko M, Unger PD. HMB-45 reactivity in renal angiomyolipoma and lymphangioleiomyomatosis. Arch. Pathol. Lab. Med. 1994;118:732–734. [PubMed] [Google Scholar]
- 19.Guinee DG, Jr, Feuerstein I, Koss MN, Travis WD. Pulmonary lymphangioleiomyomatosis: diagnosis based on results of transbronchial biopsy and immunohistochemical studies and correlation with high-resolution computed tomography findings. Arch. Pathol. Lab. Med. 1994;118:846–849. [PubMed] [Google Scholar]
- 20.Adema GJ, de Boer AJ, Vogel AM, Loenen WA, Figdor CG. Molecular characterization of the melanocyte lineage-specific antigen gp100. J. Biol. Chem. 1994;269:20126–20133. [PubMed] [Google Scholar]
- 21.Matsumoto Y, Horiba K, Usuki J, Chu SC, Ferrans VJ, Moss J. Markers of cell proliferation and expression of melanosomal antigen in lymphangioleiomyomatosis. Am. J. Respir. Cell Mol. Biol. 1999;21:327–336. doi: 10.1165/ajrcmb.21.3.3693. [DOI] [PubMed] [Google Scholar]
- 22.Goncharova EA, Goncharov DA, Eszterhas A, Hunter DS, Glassberg MK, Yeung RS, Walker CL, Noonan D, Kwiatkowski DJ, Chou MM, et al. Tuberin regulates p70 S6 kinase activation and ribosomal protein S6 phosphorylation: a role for the TSC2 tumor suppressor gene in pulmonary lymphangioleiomyomatosis (LAM) J. Biol. Chem. 2002;277:30958–30967. doi: 10.1074/jbc.M202678200. [DOI] [PubMed] [Google Scholar]
- 23.Pacheco-Rodriguez G, Steagall WK, Crooks DM, Stevens LA, Hashimoto H, Li S, Wang JA, Darling TN, Moss J. TSC2 loss in lymphangioleiomyomatosis cells correlated with expression of CD44v6, a molecular determinant of metastasis. Cancer Res. 2007;67:10573–10581. doi: 10.1158/0008-5472.CAN-07-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 25.Bacon K, Baggiolini M, Broxmeyer H, Horuk R, Lindley I, Mantovani A, Maysushima K, Murphy P, Nomiyama H, Oppenheim J, et al. Chemokine/chemokine receptor nomenclature. J. Interferon Cytokine Res. 2002;22:1067–1068. doi: 10.1089/107999002760624305. [DOI] [PubMed] [Google Scholar]
- 26.IUIS/WHO Subcommittee on Chemokine Nomenclature. Chemokine/chemokine receptor nomenclature. Cytokine. 2003;21:48–49. [Google Scholar]
- 27.Balkwill F. Cancer and the chemokine network. Nat. Rev. Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Takeuchi F, Wang JA, Fuller C, Pacheco-Rodriguez G, Moss J, Darling TN. MCP-1 overexpressed in tuberous sclerosis lesions acts as a paracrine factor for tumor development. J. Exp. Med. 2005;202:617–624. doi: 10.1084/jem.20042469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper JA. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schliwa M. Action of cytochalasin D on cytoskeletal networks. J. Cell Biol. 1982;92:79–91. doi: 10.1083/jcb.92.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, Withoff S, Verma IM. Inflammation-associated cancer: NF-κB is the lynchpin. Trends Immunol. 2005;26:318–325. doi: 10.1016/j.it.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Ferrans VJ, Yu ZX, Nelson WK, Valencia JC, Tatsuguchi A, Avila NA, Riemenschn W, Matsui K, Travis WD, Moss J. Lymphangioleiomyomatosis (LAM): a review of clinical and morphological features. J. Nippon Med. Sch. 2000;67:311–329. doi: 10.1272/jnms.67.311. [DOI] [PubMed] [Google Scholar]
- 34.Ding M, Cui S, Li C, Jothy S, Haase V, Steer BM, Marsden PA, Pippin J, Shankland S, Rastaldi MP, et al. Loss of the tumor suppressor Vhlh leads to upregulation of Cxcr4 and rapidly progressive glomerulonephritis in mice. Nat. Med. 2006;12:1081–1087. doi: 10.1038/nm1460. [DOI] [PubMed] [Google Scholar]
- 35.Ceradini DJ, Gurtner GC. Homing to hypoxia: HIF-1 as a mediator of progenitor cell recruitment to injured tissue. Trends Cardiovasc. Med. 2005;15:57–63. doi: 10.1016/j.tcm.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Taveira-DaSilva AM, Stylianou MP, Hedin CJ, Kristof AS, Avila NA, Rabel A, Travis WD, Moss J. Maximal oxygen uptake and severity of disease in lymphangioleiomyomatosis. Am. J. Respir. Crit. Care Med. 2003;168:1427–1431. doi: 10.1164/rccm.200206-593OC. [DOI] [PubMed] [Google Scholar]
- 37.Semenza GL. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 38.Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG., Jr TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003;4:147–158. doi: 10.1016/s1535-6108(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 39.Pan J, Mestas J, Burdick MD, Phillips RJ, Thomas GV, Reckamp K, Belperio JA, Strieter RM. Stromal derived factor-1 (SDF-1/CXCL12) and CXCR4 in renal cell carcinoma metastasis. Mol. Cancer. 2006;5:56. doi: 10.1186/1476-4598-5-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceradini DJ, Kulkarni AR, Callaghan MJ, Tepper OM, Bastidas N, Kleinman ME, Capla JM, Galiano RD, Levine JP, Gurtner GC. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat. Med. 2004;10:858–864. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- 41.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 42.LaVoie HA. The role of GATA in mammalian reproduction. Exp. Biol. Med. 2003;228:1282–1290. doi: 10.1177/153537020322801107. [DOI] [PubMed] [Google Scholar]
- 43.Kruger B, Schroppel B, Ashkan R, Marder B, Zulke C, Murphy B, Kramer BK, Fischereder M. A monocyte chemoattractant protein-1 (MCP-1) polymorphism and outcome after renal transplantation. J. Am. Soc. Nephrol. 2002;13:2585–2589. doi: 10.1097/01.asn.0000031701.53792.54. [DOI] [PubMed] [Google Scholar]
- 44.Rovin BH, Lu L, Saxena R. A novel polymorphism in the MCP-1 gene regulatory region that influences MCP-1 expression. Biochem. Biophys. Res. Commun. 1999;259:344–348. doi: 10.1006/bbrc.1999.0796. [DOI] [PubMed] [Google Scholar]
- 45.Simeoni E, Hoffmann MM, Winkelmann BR, Ruiz J, Fleury S, Boehm BO, Marz W, Vassalli G. Association between the A-2518G polymorphism in the monocyte chemoattractant protein-1 gene and insulin resistance and type 2 diabetes mellitus. Diabetologia. 2004;47:1574–1580. doi: 10.1007/s00125-004-1494-4. [DOI] [PubMed] [Google Scholar]
- 46.Keszei M, Nagy A, Kozma GT, Radosits K, Tolgyesi G, Falus A, Szalai C. Pediatric asthmatic patients have low serum levels of monocyte chemoattractant protein-1. J. Asthma. 2006;43:399–404. doi: 10.1080/02770900600710433. [DOI] [PubMed] [Google Scholar]
- 47.Hwang SY, Cho ML, Park B, Kim JY, Kim YH, Min DJ, Min JK, Kim HY. Allelic frequency of the MCP-1 promoter -2518 polymorphism in the Korean population and in Korean patients with rheumatoid arthritis, systemic lupus erythematosus and adult-onset Still’s disease. Eur. J. Immunogenet. 2002;29:413–416. doi: 10.1046/j.1365-2370.2002.00346.x. [DOI] [PubMed] [Google Scholar]
- 48.Govindarajan B, Brat DJ, Csete M, Martin WD, Murad E, Litani K, Cohen C, Cerimele F, Nunnelley M, Lefkove B, et al. Transgenic expression of dominant negative tuberin through a strong constitutive promoter results in a tissue-specific tuberous sclerosis phenotype in the skin and brain. J. Biol. Chem. 2005;280:5870–5874. doi: 10.1074/jbc.M411768200. [DOI] [PubMed] [Google Scholar]
- 49.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang G, Rosen DG, Zhang Z, Bast RC, Jr, Mills GB, Colacino JA, Mercado-Uribe I, Liu J. The chemokine growth-regulated oncogene 1 (Gro-1) links RAS signaling to the senescence of stromal fibroblasts and ovarian tumorigenesis. Proc. Natl. Acad. Sci. USA. 2006;103:16472–16477. doi: 10.1073/pnas.0605752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsui K, Riemenschneider K, Hilbert WSL, Yu ZX, Takeda K, Travis WD, Moss J, Ferrans VJ. Hyperplasia of type II pneumocytes in pulmonary lymphangioleiomyomatosis. Arch. Pathol. Lab. Med. 2000;124:1642–1648. doi: 10.5858/2000-124-1642-HOTIPI. [DOI] [PubMed] [Google Scholar]
- 52.Taveira-DaSilva AM, Hedin C, Stylianou MP, Travis WD, Matsui K, Ferrans VJ, Moss J. Reversible airflow obstruction, proliferation of abnormal smooth muscle cells, and impairment of gas exchange as predictors of outcome in lymphangioleiomyomatosis. Am. J. Respir. Crit. Care Med. 2001;164:1072–1076. doi: 10.1164/ajrccm.164.6.2102125. [DOI] [PubMed] [Google Scholar]
- 53.Ryu JH, Myers JL, Swensen SJ. Bronchiolar disorders. Am. J. Respir. Crit. Care Med. 2003;168:1277–1292. doi: 10.1164/rccm.200301-053SO. [DOI] [PubMed] [Google Scholar]
- 54.Henske EP. Metastasis of benign tumor cells in tuberous sclerosis complex. Genes Chromosomes Cancer. 2003;38:376–381. doi: 10.1002/gcc.10252. [DOI] [PubMed] [Google Scholar]
- 55.Murakami T, Cardones AR, Finkelstein SE, Restifo NP, Klaunberg BA, Nestle FO, Castillo SS, Dennis PA, Hwang ST. Immune evasion by murine melanoma mediated through CC chemokine receptor-10. J. Exp. Med. 2003;198:1337–1347. doi: 10.1084/jem.20030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murakami T, Cardones AR, Hwang ST. Chemokine receptors and melanoma metastasis. J. Dermatol. Sci. 2004;36:71–78. doi: 10.1016/j.jdermsci.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 57.Schecter AD, Calderon TM, Berman AB, McManus CM, Fallon JT, Rossikhina M, Zhao W, Christ G, Berman JW, Taubman MB. Human vascular smooth muscle cells possess functional CCR5. J. Biol. Chem. 2000;275:5466–5471. doi: 10.1074/jbc.275.8.5466. [DOI] [PubMed] [Google Scholar]
- 58.Schecter AD, Berman AB, Yi L, Mosoian A, McManus CM, Berman JW, Klotman ME, Taubman MB. HIV envelope gp120 activates human arterial smooth muscle cells. Proc. Natl. Acad. Sci. USA. 2001;98:10142–10147. doi: 10.1073/pnas.181328798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lucas AD, Bursill C, Guzik TJ, Sadowski J, Channon KM, Greaves DR. Smooth muscle cells in human atherosclerotic plaques express the fractalkine receptor CX3CR1 and undergo chemotaxis to the CX3C chemokine fractalkine (CX3CL1) Circulation. 2003;108:2498–2504. doi: 10.1161/01.CIR.0000097119.57756.EF. [DOI] [PubMed] [Google Scholar]
- 60.Murray LA, Syed F, Li L, Griswold DE, Das AM. Role of chemokines in severe asthma. Curr. Drug Targets. 2006;7:579–588. doi: 10.2174/138945006776818674. [DOI] [PubMed] [Google Scholar]
- 61.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 62.Devilee P, Cleton-Jansen AM, Cornelisse CJ. Ever since Knudson. Trends Genet. 2001;17:569–573. doi: 10.1016/s0168-9525(01)02416-7. [DOI] [PubMed] [Google Scholar]