This work shows that KEG E3 ligase activity is required for regulation of the abundance of the ABA responsive transcription factor ABI5. It shows that KEG undergoes autoubiquitination in response to ABA and is degraded by the 26S proteasome, allowing ABI5 levels to increase and mediate cellular ABA responses.
Abstract
The Arabidopsis thaliana RING-type E3 ligase KEEP ON GOING (KEG) is a negative regulator of abscisic acid (ABA) signaling. Seedlings homozygous for T-DNA insertions in KEG accumulate high levels of the ABA-responsive transcription factor ABSCISIC ACID-INSENSITIVE5 (ABI5). Here, we demonstrate that KEG E3 ligase activity is required for the regulation of ABI5 abundance. KEG ubiquitinates ABI5 in vitro, and a functional KEG RING domain is required to restore the levels of ABI5 in keg-1 to that of the wild type. Overexpression of KEG leads to ABA insensitivity, which correlates with KEG protein levels. In the presence of ABA, ABI5 levels increase drastically via a decrease in ubiquitin-meditated proteasomal degradation. Our results indicate that ABA promotes ABI5 accumulation by inducing the ubiquitination and proteasomal degradation of KEG. A functional RING domain is required for the ABA-induced degradation of KEG, suggesting that the loss is due to self-ubiquitination. Mutations within KEG's kinase domain or treatments with kinase inhibitors prohibit the ABA-induced ubiquitination and degradation of KEG, indicating that phosphorylation, possibly self-phosphorylation, is involved in the ABA regulation of KEG protein levels. We discuss a model for how ABA may negatively regulate KEG protein abundance, leading to accumulation of ABI5 and ABA-dependent cellular responses.
INTRODUCTION
Posttranslational regulation of protein abundance by ubiquitination and subsequent degradation by the 26S proteasome is itself a highly regulated process essential for proper growth and development of all eukaryotes. In the ubiquitination pathway, abnormal and short-lived proteins are modified by the covalent attachment of polymeric ubiquitin chains onto one or more Lys residues. Ubiquitination is catalyzed by the sequential action of three enzymes: E1 (ubiquitin activating), which activates ubiquitin molecules; E2 (ubiquitin conjugating), which accepts the activated ubiquitin from the E1, thus forming an E2-ubiquitin intermediate; and E3 (ubiquitin ligase), which facilitates the transfer of ubiquitin from the E2-ubiquitin intermediate to the target protein. As the substrate recruiting enzymes, E3 ligases confer specificity to the ubiquitination pathway (Vierstra, 2009).
The importance of the ubiquitination pathway is reflected in the abundance of ubiquitination enzymes found in eukaryotic genomes. The majority of ubiquitination enzymes are E3 ligases, a large portion of which are the Really Interesting New Gene (RING) type. The Arabidopsis thaliana genome encodes for ~470 RING-type E3 ligases (Stone et al., 2005). RING-type E3 enzymes have been shown to play important roles in various plant hormone signaling pathways (Hoecker, 2005; Zeng et al., 2006; Dreher and Callis, 2007; Stone and Callis, 2007), including abscisic acid (ABA) signaling, which regulates developmental and physiological processes in plants, including seed dormancy and germination, seedling growth, as well as mediating many abiotic stress responses (Finkelstein et al., 2002). The RING E3 ligase ABI3-INTERACTING PROTEIN2 (AIP2) serves as a negative regulator of ABA signaling by targeting ABSCISIC ACID-INSENSITIVE3 (ABI3) for degradation (Zhang et al., 2005). SALT- AND DROUGHT-INDUCED RING FINGER1 acts upstream of ABI3 and ABI5 in ABA signaling and regulates plant responses to drought and salt stresses (Zhang et al., 2007). RING E3 ligase RING-H2 protein RHA2a regulates ABA-mediated control of seed germination and early seedling development (Bu et al., 2009).
ABI5, a basic domain/leucine zipper (bZIP) transcription factor, has been shown to be essential for the execution of ABA-dependent postgerminative growth arrest (Finkelstein, 1994; Lopez-Molina et al., 2001). The efficiency of the postgerminative ABA-dependent growth arrest is dependent on ABI5 protein accumulation through transcriptional activation and enhanced protein stability (Lopez-Molina et al., 2001; Brocard et al., 2002). KEEP ON GOING (KEG), a multidomain ubiquitin E3 ligase, has been reported to regulate ABI5 levels (Stone et al., 2006). KEG protein consists of a RING and kinase domain followed by a series of ankyrin and HERC2-like repeats, both of which may function as substrate binding modules. Seedlings homozygous for T-DNA insertions in KEG undergo growth arrest immediately after germination, suggestive of increased ABA signaling (Stone et al., 2006). The ability of KEG to interact with ABI5 in vitro, the extremely high levels of ABI5 protein present in keg seedlings, and the ability of abi5-1 mutants to rescue partially the early growth arrest phenotype conferred by keg-1, all support the notion that KEG is required to maintain low levels of ABI5 in the absence of ABA (Stone et al., 2006). However, one of the outstanding questions that remain to be addressed is how ABA signaling regulates KEG activity to promote the accumulation of ABI5.
Although it is well accepted that E3 ligases play an essential role in hormone signal transduction pathways (Stone and Callis, 2007), it is less well understood how these E3s are regulated in response to changing levels of the hormone. Generally, the activity of E3 ligases can be controlled posttranslationally by covalent modifications, such as phosphorylation or conjugation with ubiquitin-like proteins, by noncovalent binding of protein or small-molecule ligands, or by competition among substrates (Deshaies and Joazeiro, 2009). For example, a previously undiscovered mode of E3 regulation by small molecules has been unraveled in plants. The plant hormone auxin fills a cavity in the substrate binding site of its receptor protein, the F-box protein TIR1, thereby creating additional molecular surface to stabilize the binding of TIR1 substrates and increase substrate degradation (Tan et al., 2007). Jasmonoyl-isoleucine, binding to ubiquitin E3 ligase SCFCOI1 (COI1 as the F-box component of Skp/Cullin/F-box E3 complex), promotes SCFCOI1 interaction with JAZ transcriptional repressors, leading to their ubiquitination and degradation by the 26S proteasome (Thines et al., 2007; Staswick, 2008). Another interesting example is FBXL5, an F-box protein that targets the iron regulatory protein IRP2 required for maintaining iron homeostasis for proteasomal degradation. Accumulation of FBXL5 is itself regulated by iron levels (Salahudeen et al., 2009; Vashisht et al., 2009). FBXL5 is degraded upon iron depletion and accumulates when iron binds to its hemerythrin domain (Salahudeen et al., 2009).
A notable feature of RING-type ubiquitin ligase is that the enzymatic activity of the E3 ligase can be monitored through autoubiquitination in vitro (Lorick et al., 1999; Joazeiro and Weissman, 2000), suggesting that the ability to autoubiquitinate is a fundamental feature of a ubiquitin ligase. Whereas many RING E3s have been shown to be ubiquitinated both in vivo and in vitro, often by an autocatalytic process, the physiological consequences are less well characterized. For example, the RING-type E3 ligase Mdm2 regulates its own stability via self-ubiquitination, and this leads to the activation of its target p53 (Fang et al., 2000; Honda and Yasuda, 2000). Autocatalytic ubiquitination could be a simple consequence of E3 activity with no functional impact. Alternatively, it could lead to downregulation of E3 activity owing to degradation by the proteasome and subsequent stabilization of its substrates (Deshaies and Joazeiro, 2009).
In this study, we demonstrate that KEG E3 ligase is posttranslationally modified by ubiquitination and phosphorylation in vivo. In addition, we demonstrate that ABA promotes the autoubiquitination of KEG, leading to KEG protein degradation by the 26S proteasome and ABI5 protein accumulation, which in turn mediate early seedling growth arrest.
RESULTS
Rescue of keg's Growth Arrest Phenotype Requires KEG E3 Ligase Activity
KEG contains a functional RING E3 ligase and kinase domain, allowing KEG to posttranslationally modify and regulate protein activity via ubiquitination and phosphorylation, respectively (Stone et al., 2006). KEG loss-of-function seedlings accumulate the ABA-responsive transcription factor ABI5, whose accumulation contributes to the early seedling growth arrest phenotype (Lopez-Molina et al., 2001, 2002; Stone et al., 2006). Therefore, we speculated that the RING domain is essential for KEG function during ABA signaling. To test this hypothesis, complementation experiments were performed using wild-type KEG and KEG harboring mutations within the RING domain that have been previously shown to disrupt E3 ligase activity (Stone et al., 2006). Metal ligand binding residues Cys-29 and His-31 were changed to Ala (KEGAA) (see Supplemental Figure 1A online). We constructed transgenic plants expressing the full-length KEG and KEGAA cDNA fused to the Hemagglutinin (HA)-epitope tag under the control of cauliflower mosaic virus (CaMV) 35S promoter in the keg-1 homozygous background. As the keg-1 homozygous plants do not survive beyond the seedling stage, heterozygous plants, KEG/keg-1, were used for transformations. F1 generation plants were genotyped to obtain plants homozygous for keg-1 with 35S:HA-KEG (keg-1/35S:HA-KEG). F3 generation plants homozygous for both keg-1 and 35S:HA-KEG were identified and used for all analysis. As controls, wild-type Arabidopsis ecotype Columbia-0 (Col-0) plants carrying the empty transformation vector were also produced. At least three independent transgenic lines for each construct were obtained and analyzed for phenotype.
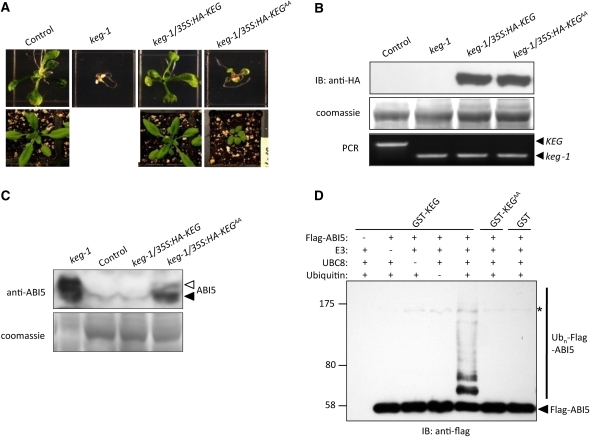
The keg-1 mutant phenotype was fully rescued by the full-length wild-type KEG (Figure 1A). No obvious differences were observed between keg-1/35S:HA-KEG transgenic plants and control plants on Murashige and Skoog (MS) growth medium or in soil (Figure 1A). As shown previously, keg-1 mutants of the same age on MS growth medium displayed a strong postgerminative growth arrest (Figure 1A; Stone et al., 2006). When the F2 population of KEG/keg-1 heterozygous plants harboring the 35S:HA-KEGAA transgene was allowed to grow on MS growth medium for 7 d, we found a new phenotype that was milder than keg-1 (Figure 1A). Compared with keg-1, keg-1/35S:HA-KEGAA plants showed green cotyledons and developed true leaves at 7 d old. When transferred to soil, keg-1/35S:HA-KEGAA plants developed further and produced additional true leaves; however, they were much smaller than control plants and were sterile (Figure 1A). All plants used were confirmed via PCR to be homozygous for the keg-1 mutation, and immunoblotting using HA antibodies was used to detect expression of the transgene, 35S:HA-KEG or 35S:HA-KEGAA (Figure 1B). The results from the phenotypic analysis thus show that full rescue of the keg-1 phenotype requires a functional RING domain.
Figure 1.
Rescue of Growth Arrest and ABI5 Protein Level in the keg-1 Mutants.
(A) Growth of transgenic plants, control (empty transformation vector in Col-0 background), keg-1 (data not shown for the plants in soil), keg-1/35S:HA-KEG (35S:HA-KEG in keg-1 background), and keg-1/35S:HA-KEGAA (RING mutant 35S:HA-KEGAA in keg-1 background) plants on MS medium (7 d old, top panel) and in soil (4 weeks old, bottom panel).
(B) Level of HA-KEG in transgenic plants as determined by immunoblot analysis using anti-HA antibody (top panel). Coomassie blue staining was used to confirm equal loading (middle panel). PCR was used to confirm presence of the T-DNA insertion and homozygosity of the keg-1 mutation (bottom panel). Gene-specific primers were used for the wild-type allele (lane 1) and in combination with T-DNA–specific primers for the mutant allele (lanes 2 to 4).
(C) Levels of ABI5 protein in 7-d-old keg-1, control, keg-1/35S:HA-KEG, and keg-1/35S:HA-KEGAA seedlings as detected by ABI5 antibodies (top panel). Arrowheads indicate the different forms of ABI5. Coomassie blue staining shows levels of loading in each lane (bottom panel).
(D) GST-KEG (E3) is capable of ubiquitinating Flag-ABI5 in vitro in the presence of yeast E1 and Arabidopsis UBC8 (an E2). Omission of GST-KEG, UBC8, or ubiquitin (Ub) from the ubiquitination assay abolishes Flag-ABI5 ubiquitination. KEG with a mutant RING domain, GST-KEGAA, does not ubiquitinate Flag-ABI5. GST protein was use as control. Asterisk indicates a nonspecific band. IB, immunoblot.
Degradation of ABI5 Requires a Functional KEG RING Domain
Postgerminative growth arrest induced by ABA treatment coincides with increases in ABI5 mRNA and protein, implying that ABI5 is the causal agent of this arrest (Lopez-Molina et al., 2001, 2002). keg-1 seedlings undergoing postgerminative growth arrest contain very high levels of ABI5 protein even without ABA treatment. Several lines of evidence indicate that KEG is responsible for the degradation of ABI5 protein at the posttranscriptional level (Stone et al., 2006). We found that the ABI5 protein level in keg-1/35S:HA-KEG transgenic plants was similar to that of the control plants, which was barely detectable without exogenous ABA (Figure 1C). By contrast, the ABI5 protein level in keg-1 mutant plants was extremely high. Compared with keg-1, the level of ABI5 protein detected in keg-1/35S:HA-KEGAA was reduced to some extent but still much higher than in keg-1/35S:HA-KEG and control plants (Figure 1C). The slight decrease of ABI5 protein level observed for keg-1/35S:HA-KEGAA could account for the partial rescue of the keg-1 mutant phenotype by the 35S:HA-KEGAA transgene (Figures 1A and 1C). Multiple forms of ABI5 were observed for keg-1/35S:HA-KEGAA, which is consistent with previous results where two forms of ABI5 were also detected for keg-1 (Figure 1C; Stone et al., 2006). The extremely high levels of ABI5 in keg-1 make clearly resolving the two bands for ABI5 difficult (Figure 1C). Unlike keg-1, in which the two forms of ABI5 are usually similar in abundance, the level of the slower migrating form of ABI5 (open arrow) for keg-1/35S:HA-KEGAA was much lower than that of the faster migrating form of ABI5 (closed arrow; Figure 1C). These results further confirm that KEG E3 ligase activity is required to maintain low levels of ABI5 protein in the absence of ABA.
The accumulation of ABI5 protein in keg-1 plants even in the absence of exogenous ABA strongly suggested that KEG is responsible for catalyzing ABI5 ubiquitination. To determine whether KEG could directly ubiquitinate ABI5, in vitro ubiquitination assays were performed using glutathione S-transferase tagged KEG (GST-KEG) and Flag-tagged ABI5 (Flag-ABI5). Due to the large size of the full-length KEG protein (~180 kD), only the RING and kinase domains of KEG was used to produce the recombinant GST-KEG protein. In the presence of yeast E1 and Arabidopsis UBC8 (an E2), GST-KEG can catalyze the ubiquitination of Flag-ABI5 as evident by the higher molecular mass forms of ABI5 detected using Flag antibodies (Figure 1D). The higher molecular mass forms of Flag-ABI5 were not observed when GST-KEG, UBC8, or ubiquitin was omitted from the ubiquitination assays (Figure 1D). Polyubiquitination of ABI5 did not occur when KEG with a nonfunctional RING domain (GST-KEGAA) was used in the assays (Figure 1D), indicating that ABI5 polyubiquitination in vitro is dependent on the integrity of KEG's RING domain.
Overexpression of KEG Renders Plant Insensitive to ABA and Salt
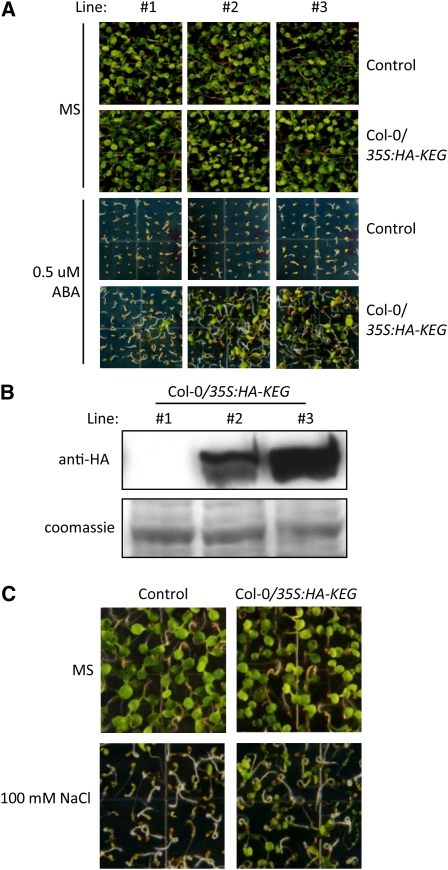
The function of KEG during ABA signaling was further investigated by analysis of KEG-overexpression phenotypes. KEG mutants are hypersensitive to exogenous ABA, which can be attributed to the accumulation of ABI5 (Stone et al., 2006). Overexpression of KEG should have the opposite effect, rendering the plant insensitive to ABA and other ABA-related abiotic stresses, such as high salinity. To determine if this is the case, we constructed transgenic plants expressing the full-length wild-type KEG cDNA fused to the HA-epitope tag under the control of CaMV 35S promoter in wild-type Col-0 background (Col-0/35S:HA-KEG). Wild type Col-0 plants transformed with an empty transformation vector were used as controls. Three independent transgenic lines were used to test ABA sensitivity (Figure 2A). All control plants showed inhibition of cotyledon greening and expansion on MS medium supplemented with 0.5 μM ABA (Figure 2A). By contrast, the KEG-overexpressing lines (Col-0/35S:HA-KEG) showed cotyledon greening and expansion on medium containing 0.5 μM ABA (Figure 2A). The level of ABA insensitivity correlated with the level of HA-KEG protein detected in each of the three lines (Figures 2A and 2B). For example, HA-KEG was barely detectable in line 1, and the ABA sensitivity of this line was very similar to that of the control. By contrast, lines 2 and 3 displayed high levels of HA-KEG and were less sensitive to the inhibitory effects of ABA on cotyledon greening and expansion. Salt treatments result in a rapid accumulation of ABI5 protein, and the abi5 mutant shows salt insensitivity, suggesting that ABI5 mediates transduction of the stress-responsive signal in plants (Lopez-Molina et al., 2001). Therefore, we examined the response of KEG-overexpressing plants to salt stress. Col-0/35S:HA-KEG (line 3), and control transgenic plants were grown on MS growth medium with or without 100 mM sodium chloride (NaCl) for 5 d (Figure 2C). The growth of the control transgenic plants was strongly inhibited; however, KEG-overexpressing transgenic plants of the same age showed cotyledon greening and expansion (Figure 2C). These results are consistent with those from ABI5 mutant plants, which are insensitive to ABA, and further confirm KEG's role in regulating ABA-mediated responses.
Figure 2.
Effects of KEG Overexpression on ABA and Salt Sensitivity.
(A) Three independent lines of Col-0/35S:HA-KEG and control transgenic plants were grown for 7 d on MS medium with or without 0.5 μM ABA.
(B) Levels of HA-KEG fusion protein in Col-0/35S:HA-KEG transgenic lines grown for 7 d on MS medium (top panel). Coomassie blue staining confirms equal loading (bottom panel).
(C) Phenotype of Col-0/35S:HA-KEG transgenic and control plants germinated and grown for 5 d on MS growth medium with (bottom panel) or without (top panel) 100 mM NaCl. Line #3 shown in (A) and (B) were used.
ABA Promotes KEG Degradation via the Ubiquitin-Dependent 26S Proteasome Pathway
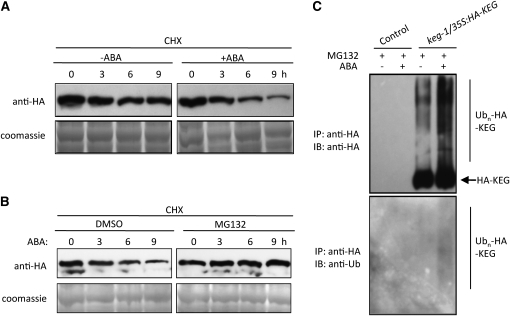
ABA promotes ABI5 protein stabilization and accumulation (Lopez-Molina et al., 2001). Therefore, we presumed that ABA may inhibit KEG activity, allowing ABI5 levels to increase. One way in which ABA may prevent the ubiquitination and degradation of ABI5 is to promote the degradation of KEG. To determine if ABA regulates KEG protein stability, keg-1/35S:HA-KEG transgenic seedlings were treated with ABA and the levels of HA-KEG were observed. Eight-day-old keg-1/35S:HA-KEG seedlings were pretreated with the protein synthesis inhibitor cycloheximide before addition of ABA. KEG protein levels at different time points were detected with HA antibodies. As shown in Figure 3A, the KEG protein level was reduced slightly without ABA treatment and decreased much more rapidly upon ABA treatment. These results suggest that ABA regulates KEG activity through modulating KEG protein abundance.
Figure 3.
ABA Promotes KEG Degradation via the Ubiquitin-Dependent 26S Proteasome Pathway.
(A) Eight-day-old keg-1/35:HA-KEG seedlings were incubated in liquid MS medium supplemented with 500 μM cycloheximide (CHX) followed by treatment with or without 50 μM ABA for the indicated amounts of time. The levels of HA-KEG at each time point were determined by immunoblot with HA antibody (top panel). Coomassie blue staining shows levels of loading in each lane (bottom panel).
(B) Eight-day-old keg-1/35:HA-KEG seedlings were treated with 500 μM CHX and the proteasome inhibitor MG132 (30 μM) or DMSO (control) before treatment with 50 μM ABA for the indicated amounts of time. The levels of HA-KEG protein at indicated time points were determined in the total protein extracts by immunoblot with HA antibody (top panel). Coomassie blue staining shows levels of loading in each lane (bottom panel).
(C) Eight-day-old control and keg-1/35S:HA-KEG seedlings were treated with MG132 followed treatment with (+) or without (−) 50 μM ABA for 9 h. HA-KEG was isolated using anti-HA affinity beads. HA and ubiquitin antibodies were used to detect HA-KEG (top panel) and ubiquitinated HA-KEG (bottom panel), respectively. IP, immunoprecipitation; IB, immunoblot.
To determine whether KEG protein is degraded via the ubiquitin-proteasome pathway, keg-1/35S:HA-KEG transgenic seedlings were pretreated with the proteasome inhibitor MG132 (Joo et al., 2008) or DMSO (control treatment). Protein synthesis was blocked by treatment with cycloheximide before addition of ABA. As expected, DMSO did not affect KEG protein degradation mediated by ABA. However, MG132 inhibited KEG degradation (Figure 3B), suggesting that the ABA-induced KEG protein degradation is dependent on the 26S proteasome pathway.
To confirm further that the ABA-induced degradation of KEG is mediated by ubiquitin-dependent proteasomal degradation, HA-KEG protein was isolated from control or keg-1/35S:HA-KEG transgenic seedlings treated with ABA in the presence of MG132, and the level of HA-KEG ubiquitination was determined using HA and ubiquitin antibodies (Figure 3C). Without ABA treatment, a low level of ubiquitinated HA-KEG was observed, as evidenced by the higher molecular mass forms of HA-KEG detected using HA antibodies. This correlates with the slow loss of HA-KEG observed in the absence of ABA (Figure 3A). Addition of exogenous ABA greatly increased the level of polyubiquitinated HA-KEG (Figure 3C). The modified form of HA-KEG protein was further confirmed by ubiquitin antibodies, which recognizes only ubiquitinated HA-KEG. As show in the bottom panel of Figure 3C, ubiquitin antibodies detected the higher molecular mass smear of HA-KEG observed following ABA treatment, consistent with the anti-HA blot, but the unmodified HA-KEG could not be detected by ubiquitin antibodies. These results demonstrate that ABA promotes the ubiquitination of KEG and provide further evidence for ABA regulation of KEG protein abundance.
ABA-Induced KEG Degradation Requires a Functional KEG RING Domain
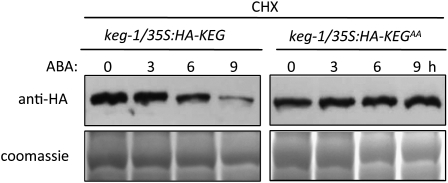
Self-ubiquitination may represent an important mechanism by which E3 ligases regulate their stability. Given that KEG is capable of self-ubiquitination in vitro, which depends upon the presence of a functional RING domain (Stone et al., 2006), we postulate that ABA regulation of KEG abundance may occur via self-ubiquitination. To determine if ABA-induced degradation of KEG is dependent upon KEG's own E3 ligase activity, we compared the degradation of wild-type KEG to that of KEG with a mutated RING domain (KEGAA) in the presence and absence of ABA. keg-1/35S:HA-KEG and keg-1/35S:HA-KEGAA transgenic plants were pretreated with cycloheximide followed by ABA treatment. As previously observed, ABA promoted KEG protein degradation (Figures 3A and 4). However, no obvious change in protein level was observed for HA-KEGAA in the presence of ABA (Figure 4). As the mutations within the RING domain are known to abolish KEG E3 ligase activity, we conclude that the ABA-induced degradation of KEG requires self-ubiquitination activity.
Figure 4.
ABA-Induced KEG Degradation Requires a Functional KEG RING Domain.
Eight-day-old keg-1/35S:HA-KEG and keg-1/35S:HA-KEGAA (RING mutant) transgenic seedlings were incubated with 500 μM cycloheximide (CHX) followed by treatment with 50 μM ABA for the indicated amounts of time. For each time point, equal amounts of protein were analyzed by immunoblot using HA antibody to determine the levels of HA-KEG and HA-KEGAA (top panel). Coomassie blue staining shows loading levels (bottom panel).
ABA-Induced Degradation of KEG Requires the Presence of an Intact Kinase Domain
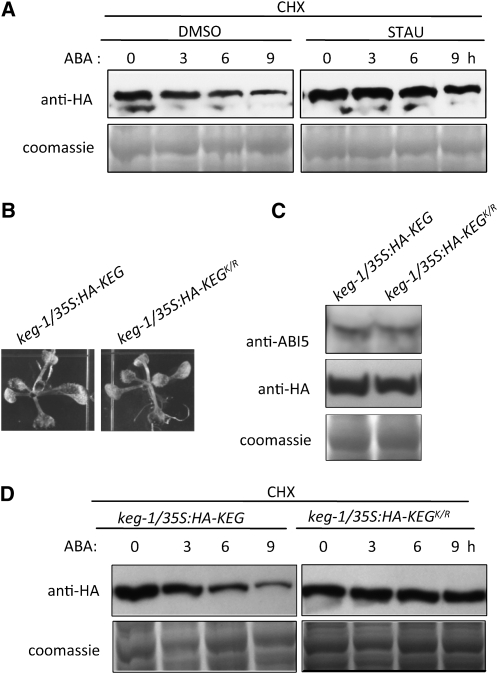
Phosphorylation has been reported to regulate RING E3 ligase autoubiquitination (Dornan et al., 2006; Hunter, 2007). Because KEG contains a kinase domain, we speculated that phosphorylation is involved in KEG ubiquitination and subsequent degradation modulated by ABA. To determine if this is the case, we observed the effects of a general kinase inhibitor staurosporine (STAU) on ABA-induced KEG degradation (Figure 5A). keg-1/35S:HA-KEG transgenic seedlings were pretreated with STAU or DMSO (control) in the presence of cycloheximide after which ABA was added to both treatments, and tissues were collected at the indicated time points. Anti-HA blots show that STAU inhibited KEG degradation compared with the control plants treated with DMSO (Figure 5A). These results indicate that KEG phosphorylation by itself or another kinase plays a role in ABA-induced KEG degradation.
Figure 5.
Phosphorylation Is Required for ABA-Induced Degradation of KEG.
(A) Eight-day-old keg-1/35:HA-KEG transgenic seedlings were treated with the kinase inhibitor STAU (1 μM) or DMSO (control) in the presence of cycloheximide (CHX) followed by 50 μM ABA for the indicated amounts of time. The levels of HA-KEG at each time point were determined by immunoblot analysis using HA antibody (top panel). Coomassie blue staining confirms equal loading (bottom panel).
(B) Phenotype of 7-d-old keg-1/35S:HA-KEG and keg/35S:HA-KEGK/R (kinase mutant) transgenic seedlings grown on MS growth medium.
(C) Levels of ABI5 and HA-KEG as detected by anti-ABI5 and anti-HA for keg-1/35S:HA-KEG and keg/35S:HA-KEGK/R transgenic plants grown for 7 d on MS growth medium without ABA. Coomassie blue staining confirms equal loading.
(D) Eight-day-old keg-1/35S:HA-KEG and keg-1/35S:HA-KEGK/R seedlings were treated with 500 μM CHX followed by 50 μM ABA for the indicated amounts of time. The levels of HA-KEG and HA-KEGK/R at each time point were determined by immunoblot analysis using anti-HA antibody (top panel). Coomassie blue staining shows loading levels (bottom panel).
To explore the role of KEG's kinase domain during ABA-regulated KEG ubiquitination, site-directed mutagenesis was used to mutate the catalytically essential Lys-176 in the kinase ATP binding domain (see Supplemental Figure 1B online) to Arg to produce a kinase mutant (KEGK/R). Transgenic plants were produced expressing the full-length KEG kinase domain mutant fused to the HA-epitope tag under the control of CaMV 35S promoter in the keg-1 background (keg-1/35S:HA-KEGK/R). Similar to keg-1/35S:HA-KEG transgenic plants, keg-1/35S:HA-KEGK/R plants did not show any obvious phenotype in the absence of exogenous ABA, indicating that the mutated KEG can fully rescue the keg-1 phenotype (Figures 1A and 5B). Expression of HA-KEGK/R also rescued the high ABI5 protein level usually observed in the keg-1 mutant to levels observed in keg-1/35S:HA-KEG transgenic plants (Figures 1C and 5C). These results suggest that the kinase domain mutation does not affect KEG's ability to ubiquitinate its substrate.
To determine if the substitution in the kinase domain affects the ability of ABA to increase KEG degradation, keg-1/35S:HA-KEGK/R and keg-1/35S:HA-KEG seedlings were treated as before to compare the degradation of HA-KEGK/R to that of HA-KEG in the presence of ABA. Anti-HA blots show that the levels of HA-KEGK/R decreased more slowly than did HA-KEG levels in the presence of ABA (Figure 5D). These results indicate that an intact kinase domain is required for ABA to increase the degradation of KEG. Another possible explanation for the stabilization of KEG upon substitution of the invariant Lys is that the Lys may serve as a site for ubiquitination. To determine if this was the case, we compared efficiency of His-Flag-KEG and His-Flag-KEGK/R self-ubiquitination in vitro. If the Lys does serve as a target for ubiquitin attachment, then the Lys-to-Arg substitution should decrease the level of His-Flag-KEGK/R self-ubiquitination. However, the level and pattern of His-Flag-KEGK/R self-ubiquitination was comparable to that of His-Flag-KEG (see Supplemental Figure 2 online), suggesting that the Lys is not a ubiquitin attachment site, at least in vitro.
Phosphorylation Is a Prerequisite for ABA-Induced Ubiquitination of KEG
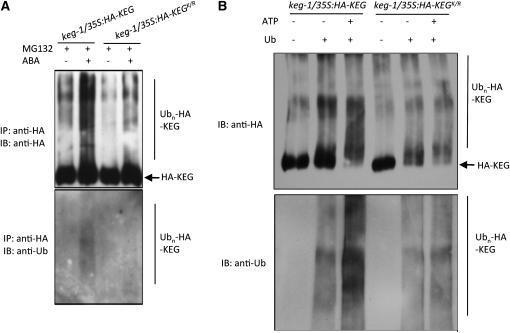
As shown above, ABA promotes the ubiquitination and degradation of KEG. To determine if KEG's kinase domain plays a role in the ABA-mediated increase in KEG ubiquitination, we compared the increase in the level of ubiquitinated HA-KEGK/R in the presence of ABA to that of HA-KEG. KEG proteins were isolated from keg-1/35S:HA-KEG and keg-1/35S:HA-KEGK/R transgenic seedlings treated with or without ABA in the presence of MG132 and subjected to blotting with HA and ubiquitin antibodies. Compared with HA-KEG, less ubiquitinated HA-KEGK/R was observed in the presence of ABA (Figure 6A). These results suggest that an intact kinase domain is required for ABA-induced KEG ubiquitination and subsequent degradation.
Figure 6.
Phosphorylation Promotes KEG Self-Ubiquitination.
(A) Eight-day-old keg-1/35S:HA-KEG and keg-1/35S:HA-KEGK/R transgenic seedlings were treated with MG132 for 16 h followed by treatments with (+) or without (−) 50 μM ABA for 9 h. HA-KEG and HA-KEGK/R proteins were isolated from total protein extracts using anti-HA affinity beads and subjected to immunoblotting using HA and ubiquitin (Ub) antibodies to detect HA-KEG/KEGK/R (top panel) and ubiquitinated HA-KEG/HA-KEGK/R (bottom panel), respectively. IP, immunoprecipitation; IB, immunoblot.
(B) HA-KEG and HA-KEGK/R proteins were immunoprecipitated from 8-d-old keg-1/35S:HA-KEG and keg-1/35S:HA-KEGK/R transgenic seedlings, respectively, using anti-HA affinity beads. Immunoprecipitated proteins were used in vitro kinase assays with (+) or without (−; control) ATP followed by ubiquitination assays. Ubiquitin (Ub) was omitted from assays as a control. Anti-HA and anti-Ub were used to detect HA-KEG/KEGK/R (top panel) and ubiquitinated HA-KEG/KEGK/R (bottom panel), respectively. IP, immunoprecipitation; IB, immunoblot.
ABA may regulate KEG ubiquitination by modulating the levels of KEG self-phosphorylation or phosphorylation by another kinase. ProQ Diamond staining, which specifically detects phosphorylated proteins, showed that KEG is phosphorylated in vivo in the presence and absence of ABA and ABA does not increase the level of KEG phosphorylation (see Supplemental Figure 3 online). These results suggest that ABA may instead modulate the site of phosphorylation and not necessarily the extent of KEG phosphorylation. To explore further the role of phosphorylation in regulating KEG autoubiquitination, HA-KEG and HA-KEGK/R were immunoprecipitated from keg-1/35S:HA-KEG and keg-1/35S:HA-KEGK/R transgenic plants, respectively, with anti-HA affinity beads and used in an in vitro phosphorylation assay followed by a ubiquitination assay. As shown in Figure 6B, compared with nonprephosphorylated HA-KEG (−ATP), prephosphorylated HA-KEG (+ATP) is more efficiently ubiquitinated. The increase in ubiquitination observed for prephosphorylated HA-KEG was not observed for prephosphorylated HA-KEGK/R. These results suggest phosphorylation increases KEG's ability to self-ubiquitinate.
DISCUSSION
The Ub/26S proteasome pathway has been directly or indirectly implicated in the action of all major plant hormones, including auxin, gibberellins, ABA, jasmonic acid, and ethylene (Hellmann and Estelle, 2002; Smalle and Vierstra, 2004; Dreher and Callis, 2007; Stone and Callis, 2007). Hormone signaling often leads to a secondary modification of targets that enhances either their degradation or stability. Many of these target proteins are transcriptional activators or repressors and affecting their half-lives is an efficient control point in hormone signaling. For example, the ABA-responsive bZIP transcription activator ABI5 has been reported to be one potential target for the ubiquitin ligase activity of KEG during postgerminative development (Stone et al., 2006). In keg mutants, ABI5 levels are significantly higher than in wild-type Arabidopsis plants (Stone et al., 2006). Here, we provide further evidence demonstrating that KEG directly targets ABI5 for ubiquitination, leading to its subsequent degradation. First, overexpression of HA-KEG in keg-1 mutants can fully rescue the initial growth arrest phenotype, as well as reduce the high ABI5 protein level associated with the phenotype. Furthermore, KEG is capable of ubiquitinating ABI5 protein in vitro. A functional RING domain is required to rescue fully the keg-1 mutant phenotype and to restore wild-type levels of ABI5. The kinase domain also plays a role in KEG-regulated postgerminative development since keg-1 transgenic plants expressing the KEG protein with a nonfunctional RING domain can grow much better than the keg-1 mutants, even though the levels of ABI5 were still higher than those observed for the wild type.
Plant transcriptional regulators are targets of E3 ligases in a variety of hormone responses, including rice (Oryza sativa) SLENDER RICE1 targeted by SCFGID2 (Sasaki et al., 2003) and Arabidopsis RGA targeted by SCFSLY1 (Dill et al., 2004) in gibberellin responses, ETHYLENE INSENSITIVE3 targeted by SCFEBF1/EBF2 in ethylene signaling (Guo and Ecker, 2003; Potuschak et al., 2003), JASMONATE-ZIM-DOMAIN (JAZ) transcriptional repressors targeted by SCFCOI1 in jasmonate signaling (Chini et al., 2007; Thines et al., 2007), auxin/indole-3-acetic acid transcriptional repressors targeted by the SCFTIR1/AFB in auxin signaling (Gray et al., 2001; Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005; Tan et al., 2007), and ABI3 and ABI5 targeted by RING-type E3 ligase AIP2 and KEG, respectively, in ABA signaling (Zhang et al., 2005; Stone et al., 2006). Hormone-dependent transcriptional activation has to be tightly regulated to avoid inappropriate cellular responses. The experimental evidence for how hormones regulate these E3 ligases to modulate hormonal responses is limited. For the SCF-type E3 ligase, jasmonoyl-isoleucine and auxin have been reported to have a similar way of regulating E3 activity. Both hormones promote the interaction of transcriptional repressors (JAZs or auxin/indole-3-acetic acid transcriptional repressors) with F-box proteins (COI1 or TIR1/AFBs) in the SCF complex, leading to the proteasomal degradation of the repressors and release of the transcriptional activator (MYC2 or ARFs, respectively) and gene transcription (Chico et al., 2008).
In ABA signaling, ABI5 is a positive regulator of ABA signaling. ABI5 protein levels dramatically increase when seedlings are treated with ABA or are exposed to stress, which increases intracellular ABA levels. The increase in ABI5 protein levels is accomplished via reduced proteolysis (Lopez-Molina et al., 2001). SUMO modification has been found to protect the inactive form of the ABI5 from proteolytic degradation (Miura et al., 2009). Previous evidence (Stone et al., 2006) and evidence from this study support the conclusion that KEG is an E3 ligase for ABI5 in vivo. Therefore, the decrease in KEG E3 activity toward ABI5 in the presence of ABA is a prerequisite for the accumulation of ABI5. Here, our evidence demonstrates that KEG protein degradation is much more rapid with ABA treatment compared with without ABA. The ABA-induced reduction in KEG protein levels was efficiently blocked by the 26S proteasome inhibitor MG132. Furthermore, ubiquitination of KEG proteins increased following ABA treatment. Based on this evidence, we concluded that ABA regulates KEG protein level by inducing ubiquitination and subsequent degradation through the 26S proteasome pathway. Mutations within KEG's RING domain stabilize the protein and abolish the ABA-induced degradation, suggesting that KEG's ubiquitin ligase function is essential for its own degradation, and KEG is therefore regulated by autoubiquitination.
A few RING E3 ligases have been reported to be subjected to autoubiquitinaton and this tends to inhibit their E3 ligase activity (Deshaies and Joazeiro, 2009). One example is the mammalian RING-type E3 ligase Mdm2, which is reported to regulate its own stability via self-ubiquitination (Fang et al., 2000; Honda and Yasuda, 2000). In response to stimuli such as DNA damage, Mdm2 ubiquitinates and targets itself for proteasomal degradation. This leads to the activation of its ubiquitination target, the transcription factor p53, which then induces cell cycle arrest or apoptosis. Similar to Mdm2, the levels of KEG itself may regulate ABI5 degradation, and KEG's abundance is regulated by ABA levels. High levels of ABA induce KEG autoubiquitination and degradation, leading to the accumulation of ABI5.
Together, these observations raise the question as to how the switch from ABI5 ubiquitination to KEG autoubiquitination is influenced by ABA. KEG contains a kinase domain; therefore, phosphorylation maybe another posttranslational modification that regulates KEG's E3 ligase activity toward substrate- or self-ubiquitination. Many RING E3 ligases have been shown to be regulated positively or negatively by cycles of phosphorylaion and dephosphorylation (Hunter, 2007; Deshaies and Joazeiro, 2009). For instance, mammalian COP1 autoubiquitination is stimulated by ATM phosphorylation, which produces an allosteric alteration (Dornan et al., 2006), and the activity of Mdm2 toward p53 is increased by Akt/PKB phosphorylation through reduced autoubiquitination (Feng et al., 2004). As for KEG, phosphorylation (probably self-phosphorylation) also could be one method to regulate positively its autoubiquitination. Pretreatment of seedlings with a general kinase inhibitor staurosporine impedes ABA-induced degradation of KEG, indicating that phosphorylation is involved in this process. The KEG kinase mutant, which is much more stable than KEG in the presence of ABA, is not ubiquitinated in response to ABA. In addition, prephosphorylation of KEG results in increased self-ubiquitination in vitro. Together, these results suggest that KEG's phosphorylation is required for ABA-induced self-ubiquitination and subsequent degradation. However, the number of phosphate groups per KEG molecule seems to be similar in the presence and absence of ABA. The interpretation could be that in response to ABA there is a phosphorylation rearrangement in KEG. The outcome of the rearrangement dictates whether KEG ubiquitinates itself or its substrate. The possibility that another kinase may phosphorylate KEG in response to ABA still remains.
Several reports have indicated that ABA signaling is tightly controlled and that ABI5 plays a central role in the postgerminative growth regulation of ABA responses (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001). However, ABI5 accumulation is not sufficient to result in postgerminative growth arrest. Transgenic plants overexpressing ABI5 show increased ABI5 protein but grow normally in the absence of ABA (Lopez-Molina et al., 2001; Stone et al., 2006). Comparing the keg-1 mutant with ABI5-overexpressing transgenic plants, we observed that the keg-1 mutant accumulates additional higher molecular mass forms of ABI5 that are not observed in ABI5-overexpressing plants (Stone et al., 2006). Previous studies have shown that ABA activates SnRK2s (SnRK2.2, SnRK2.3, and SnRK2.6), resulting in phosphorylation of ABI5 and ABI5-like bZIP transcription factors (Kobayashi et al., 2005; Fujii et al., 2007; Fujii and Zhu, 2009; Nakashima et al., 2009), indicating that ABI5 is phosphorylated in response to ABA and phosphorylation activates the transcription factor. The severe postgerminative growth phenotype of keg-1 suggests that the additional forms of ABI5 may be the phosphorylated active ABI5, which is responsible for the growth arrest. Therefore, keg-1knockout results in two effects on ABI5 in the absence of or in low ABA. First, loss of the KEG E3 activity leads to an increase in ABI5. Second, the absence of KEG somehow activates the kinases, probably SnRK2s, which phosphorylate and activate ABI5. This hypothesis correlates with the reduced levels of the higher molecular mass form of ABI5 observed in keg-1/35S:HA-KEGAA seedlings, which may represent the activated form of ABI5. The reduced higher molecular mass form of ABI5 may explain why the phenotype of keg-1/35S:HA-KEGAA was not as severe as keg-1. KEG loss-of-function mutants actually simulate the situation where plants are exposed to high levels of ABA. Consequently, it is plausible that plants remove the abundant KEG protein to allow ABI5 to accumulate and be activated in the presence of ABA.
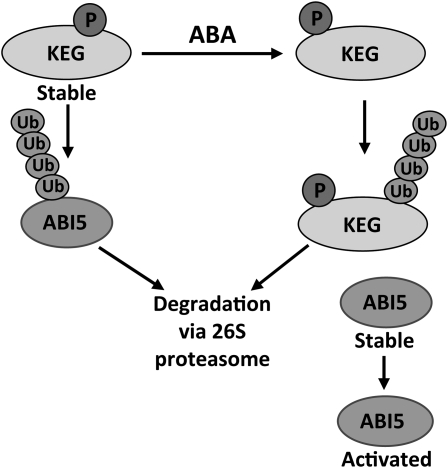
A tentative model for regulation of KEG and ABI5 activity by ABA is shown in Figure 7. Under normal growth conditions, KEG is stable and continually ubiquitinates and targets ABI5 for degradation, thus keeping ABI5 levels low and allowing seedling growth. In the presence of ABA, KEG is ubiquitinated and degraded by the 26S proteasome. ABA-induced KEG degradation may involve de novo phosphorylation by KEG itself or probably by another kinase. It is possible that phosphorylation could induce a conformational change in KEG that may efficiently expose the ubiquitin conjugating site, which would result in KEG autoubiquitination and subsequent degradation. Consequently, ABI5 becomes stable and activates the downstream response genes that are responsible for the postgerminative growth arrest. Meanwhile, ABA seems to induce slightly KEG mRNA (Wawrzynska et al., 2008), possibly as part of a feedback loop, enabling the creation of a reserve of KEG protein that can rid the seedling of high levels of ABI5 to turn off the ABA signaling pathway. Our model presents an example in plants where a hormone promotes E3 ligase autoubiquitination to activate a signaling pathway. However, it would be interesting to explore the detailed mechanism of how the ABA signal is perceived and transduced to KEG and how phosphorylation directly affects KEG autoubiquitination.
Figure 7.
Model for ABA Regulation of KEG Activity to Determine ABI5 Abundance.
In the absence of ABA, KEG is stable and continually ubiquitinates ABI5. Ubiquitinated ABI5 is degraded by the 26S proteasome. ABA promotes KEG autoubiquitination and subsequent degradation by the 26S proteasome. As KEG is phosphorylated in the presence and absence of ABA, ABA-induced KEG degradation may involve a rearrangement in phosphorylation status. This may be accomplished by KEG phosphorylating itself or another kinase. Reduction in KEG abundance leads to the accumulation of ABI5 and subsequent growth arrest.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana seeds, ecotype Col-0 wild type, mutants, and transgenic seeds were surface-sterilized with 50% (v/v) bleach and 0.1% Triton X-100. After cold treatment at 4°C for 2 d, seeds were germinated and grown on 0.5× MS medium containing 0.8% agar and 1% sucrose plates under continuous light at 22°C. For plants grown in soil, 7-d-old seedlings were transferred from MS plates to soil and grown under photoperiodic cycles of 16 h light and 8 h dark at 22°C in a growth chamber. For salt or ABA treatment, the MS medium was supplemented with indicated amounts of ABA or NaCl. keg-1 (Salk_049542) seeds obtained from the ABRC (Alonso et al., 2003; Stone et al., 2006) were a gift from the Callis laboratory (University of California, Davis).
Plasmid Construction and Site-Directed Mutagenesis
Total RNA was extracted from Col-0 tissues using Trizol (Sigma-Aldrich) following the manufacturer's instructions. Reverse transcription (RT) was performed using total RNA extracted from seedlings and oligo(dT)16 primer with SuperScript II reverse transcriptase (Invitrogen). RT product was used in PCRs to amplify the predicted KEG open reading frame using Phusion polymerase (Finnzymes). The full-length KEG cDNA (4878 bp) and partial cDNA regions of KEG encoding the RING and kinase domain (1383 bp) were cloned into the gateway entry vector pDONR 201 (Invitrogen). Point mutations were introduced with Phusion site-directed mutagenesis kit (Finnzymes) to generate the RING domain mutation (C-29-A, H-31-A; KEGAA) and kinase domain mutation (K-176-R; KEGK/R). For plant transformation, full-length and mutated KEG cDNAs were introduced into the pEarleyGate 201 plant transformation vector (Earley et al., 2006) via the Gateway method to produce in-frame fusions with the HA tag under the control of the CaMV 35S promoter. For control plant transformation vectors, the sequence between attB sites of pEarleyGate 201 was deleted. For bacterial expression, wild-type and mutated KEG cDNAs were recombined into the pDEST 565 or modified pDEST 527 vectors (Addgene plasmids 11520 and 11518, respectively; donated by Dominic Esposito, National Cancer Institute, Frederick) to obtain fusion proteins with His-GST tag or Flag-His tag, respectively. To produce the modified pDEST 527, a Flag tag was introduced N-terminal to the HIS tag via an NdeI restriction site. The ABI5 open reading frame was amplified from pUNI 51 obtained from ABRC and introduced into pDONR 201 and then recombined into the modified pDEST 527 to produce a Flag-His fusion protein. UBC8 expression plasmid was constructed previously (gift from Judy Callis, University of California, Davis; Kraft et al., 2005). All the clones were confirmed by sequencing. Primers used are listed in Supplemental Table 1 online.
Plant Transformation and Transgenic Plants Selection
pEarleyGate 201 with KEG full-length wild type and mutated cDNAs were introduced into Agrobacterium tumefaciens strain GV3101. The transgenic Arabidopsis plants were generated using the flower dipping method (Clough and Bent, 1998). For the complementation of keg-1, keg T-DNA insertion (Salk_049542) heterozygous plants were used for the transformation. Transgenic plants were selected by growing on half-strength MS salts plus 0.8% agar and 50 μM dl-phosphinothricin (Sigma-Aldrich). Transformants were transferred to soil and allowed to set seeds. For keg-1 T-DNA mutant background, T-DNA homozygous plants were confirmed by PCR. Primers used for genotyping plants are listed in Supplemental Table 1 online. F3 generations of transgenic plants were used for all experiments.
Protein Extraction, Immunoprecipitation, and Immunoblot Analysis
Unless indicated otherwise, 7-d-old seedlings were used for all experiments. keg-1/35S:HA-KEG transgenic plants were germinated and grown on MS plates for 3 d and then transferred to liquid MS medium for additional 4 d of growth. Cycloheximide (500 μM) was added 2 h before ABA treatment. ABA was added at a final concentration of 50 μM. Ethanol (the solvent for ABA) was used as a negative control. For proteasome inhibition, 30 μM MG132 was added 16 h before ABA treatment. DMSO (the solvent for MG132) was used as a negative control. For the kinase inhibitor treatments, 1 μM staurosporine was added 2 h before ABA treatment. DMSO (solvent for staurosporine) was used as a negative control. Seedlings were collected at the indicated time points after treatment, frozen in liquid nitrogen, and stored at −80°C until use. At least three independent repetitions were performed for each experiment.
Total protein was extracted from seedling tissues using 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 20 mM NaF, 10 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 5% glycerol, and protease inhibitor cocktail tablets (Roche Diagnostics). The concentration of protein was determined using the Bradford reagent (Sigma-Aldrich) with BSA as a standard.
For immunoblot analysis, 15 μg of total protein per lane was separated on 5% SDS-PAGE. After electrophoresis, proteins were electrotransferred to polyvinylidene fluoride membrane. After blocking for 1 h in TBST (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween 20) with 5% nonfat dry milk at room temperature, membranes were incubated with mouse anti-HA (Sigma-Aldrich) at 1:10,000 dilution for 2 h or rabbit anti-ABI5 (gift from Richard Vierstra and Lisa Farmer, University of Wisconsin; Stone, et al., 2006) at 1:1000 dilution overnight. Following three washings with TBST, membranes were incubated with horseradish peroxidase–conjugated goat anti-mouse or anti-rabbit IgG (Sigma-Aldrich). After three washings with TBST, the membranes were visualized using an enhanced Lumi-Light Western Blotting Substrate kit (Thermo Scientific) following the manufacturer's instructions. Coomassie Brilliant Blue staining was used to show protein loading levels.
For immunoprecipitation, HA-KEG, HA-KEGAA (RING mutant), or HA-KEGK/R (kinase mutant) was isolated from total protein extracted from keg-1/35S:HA-KEG, keg-1/35S:HA-KEGAA, and keg-1/35S:HA-KEGK/R transgenic plants, respectively, by incubation with anti-HA affinity beads (Sigma-Aldrich) at 4°C overnight, washed with 20 mM Tris-HCl and 150 mM NaCl three times, and eluted with 50 μL 1× SDS loading buffer. After boiling for 5 min, 5 μL of supernatant was separated by 5% SDS-PAGE. Immunoprecipitated proteins were detected by immunoblot analysis on polyvinylidene fluoride membranes with anti-HA (Sigma-Aldrich) at 1:10,000 dilutions or antiubiquitin (BostonBiochem) at 1:1250 as described above. For phosphoprotein analysis, immunoprecipitated proteins were separated on 5% SDS-PAGE and stained with ProQ Diamond or Sypro Ruby according to the manufacturer's instructions (Molecular Probes).
In Vitro Ubiquitination and Phosphorylation Assays
For ubiquitination assays, a portion of wild-type KEG or RING mutant KEG (KEGAA), consisting of only the RING and kinase domain, were expressed as His-GST fusions, ABI5 was expressed as a Flag-His tagged fusion, and Arabidopsis UBC8 was expressed as a His-tagged fusion protein. All proteins were expressed in Escherichia coli strain Rosetta (DE3) and purified using nickle-charged resin (Bio-Rad) according to the manufacturer's protocols. Ubiquitination assays were performed as described previously (Stone et al., 2006). Briefly, reactions (30 μL) containing 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 0.05 mM ZnCl2, 1 mM ATP (Sigma-Aldrich), 0.2 mM DTT, 10 mM phosphocreatine, 0.1 unit of creatine kinase (Sigma-Aldrich), 50 ng of yeast E1 (BostonBiochem), 150 ng of purified His-AtUBC8 (E2), 2 μg of ubiquitin (BostonBiochem), 300 ng His-GST-KEG or His-GST-KEGAA (RING mutant), and/or 300 ng Flag-His-ABI5 were incubated at 30°C for 2 h. Reactions were stopped by adding SDS sample buffer (125 mM Tris-HCl, pH 6.8, 20% [v/v] glycerin, 4% [w/v] SDS, and 10% [v/v] β-mercaptoethanol) and analyzed by SDS-PAGE followed by immunoblots using anti-GST (Sigma-Aldrich) or anti-Flag antibodies (Sigma-Aldrich).
For kinase assays, HA-KEG and HA-KEGK/R was immunoprecipitated from total protein extracted from keg-1/35S:HA-KEG and keg-1/35S:HA-KEGK/R transgenic plants, respectively, with anti-HA affinity beads as described above. Bead-bound HA-tagged proteins were incubated at 30°C for 30 min in 45 μL kinase assay buffer (20 mM Tris-HCl, pH 7.5, 10 mM MnCl2, and 10 μM ATP). Beads were then washed three times with 50 mM Tris-HCl, pH 7.5, and 150 mM NaCl. Ubiquitination assays were then performed by adding 45 μL ubiquitination assay buffer as described above to the beads, incubated at 30°C for 2 h, and analyzed by SDS-PAGE followed by immunoblots using HA or ubiquitin antibodies.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: KEG, 4515105706; ABI5, 2049427; and UBC8, 1005720488.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment Showing the Conserved Mutation Sites in KEG RING and Kinase Domains.
Supplemental Figure 2. Time Course for the Detection of KEG and KEGK/R Self-Ubiquitination.
Supplemental Figure 3. ProQ Diamond Staining Showing the Phosphorylation of KEG in the Presence and Absence of ABA.
Supplemental Table 1. List of Primers.
Acknowledgments
We thank Judy Callis (University of California, Davis) for KEG mutant seeds and At-UBC8 cDNA and for comments and discussions on the manuscript. We also thank Richard Vierstra and Lisa Farmer (University of Wisconsin, Madison) for the gift of ABI5 antibody. Research in the Stone laboratory is supported by grants from the Human Frontier Science Program Organization and the Natural Sciences and Engineering Research Council of Canada.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Brocard I.M., Lynch T.J., Finkelstein R.R. (2002). Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129: 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q., Li H., Zhao Q., Jiang H., Zhai Q., Zhang J., Wu X., Sun J., Xie Q., Wang D., Li C. (2009). The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol. 150: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico J.M., Chini A., Fonseca S., Solano R. (2008). JAZ repressors set the rhythm in jasmonate signaling. Curr. Opin. Plant Biol. 11: 486–494 [DOI] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Deshaies R.J., Joazeiro C.A. (2009). Ring domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005a). The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jürgens G., Estelle M. (2005b). Plant development is regulated by a family of auxin receptor F-box proteins. Dev. Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan D., Shimizu H., Mah A., Dudhela T., Eby M., O'Rourke K., Seshagiri S., Dixit V.M. (2006). ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 313: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Dreher K.A., Callis J. (2007). Ubiquitin, hormones and biotic stress in plants. Ann. Bot. (Lond.) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Fang S., Jensen J.P., Ludwig R.L., Vousden K.H., Weissman A.M. (2000). Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J. Biol. Chem. 275: 8945–8951 [DOI] [PubMed] [Google Scholar]
- Feng J., Tamaskovic R., Yang Z., Brazil D.P., Merlo A., Hess D., Hemmings B.A. (2004). Stabilization of Mdm2 via decreased ubiquitination is mediated by protein kinase B/Akt-dependent phosphorylation. J. Biol. Chem. 279: 35510–35517 [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5: 765–771 [Google Scholar]
- Finkelstein R.R., Gampala S.S., Rock C.D. (2002). Abscisic acid signalling in seeds and seedling. Plant Cell 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Lynch T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Zhu J. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Kepinski S., Rouse D., Leyser O., Estelle M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Hellmann H., Estelle M. (2002). Plant development: Regulation by protein degradation. Science 297: 793–797 [DOI] [PubMed] [Google Scholar]
- Hoecker U. (2005). Regulated proteolysis in light signalling. Curr. Opin. Plant Biol. 8: 469–476 [DOI] [PubMed] [Google Scholar]
- Honda R., Yasuda H. (2000). Activity of MDM2, a ubiquitin ligase, toward or itself is dependent on the RING finger domain of the ligase. Oncogene 19: 1473–1476 [DOI] [PubMed] [Google Scholar]
- Hunter T. (2007). The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol. Cell 28: 730–738 [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A., Weissman A.M. (2000). RING finger proteins: Mediators of ubiquitin ligase activity. Cell 102: 549–552 [DOI] [PubMed] [Google Scholar]
- Joo S., Liu Y., Lueth A., Zhang S. (2008). MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54: 129–140 [DOI] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., Yamamoto A., Hattori T. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44: 939–949 [DOI] [PubMed] [Google Scholar]
- Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.S., Deng X.W., Callis J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitylation enzymes of Arabidopsis. Plant Physiol. 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Chua N.H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41: 541–547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA dependent growth arrest during germination. Plant J. 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Lorick K.L., Jensen J.P., Fang S., Ong A.M., Hatakeyama S., Weissman A.M. (1999). RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96: 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Lee J., Jin J.B., Yoo C.Y., Miura T., Hasegawa P.M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F-box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Salahudeen A.A., Thompson J.W., Ruiz J.C., Ma H.W., Kinch L.N., Li Q.M., Grishin N.V., Bruick R.K. (2009). An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326: 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A., Itoh H., Gomi K., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Jeong D.H., An G., Kitano H., Ashikari M., Matsuoka M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Smalle J., Vierstra R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Staswick P.E. (2008). JAZing up jasmonate signalling. Trends Plant Sci. 13: 66–71 [DOI] [PubMed] [Google Scholar]
- Stone S.L., Callis J. (2007). Ubiquitin ligases mediate growth and development by promoting protein death. Curr. Opin. Plant Biol. 10: 624–632 [DOI] [PubMed] [Google Scholar]
- Stone S.L., Hauksdottir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Williams L.A., Farmer L.M., Vierstra R.D., Callis J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Calderon-Villalobos L.I., Sharon M., Zheng C., Robinson C.V., Estelle M., Zheng N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Vashisht A.A., et al. (2009). Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326: 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra R.D. (2009). The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wawrzynska A., Christiansen K.M., Lan Y., Rodibaugh N.L., Innes R.W. (2008). Powdery mildew resistance conferred by loss of the ENHANCED DISEASE RESISTANCE1 protein kinase is suppressed by a missense mutation in KEEP ON GOING, a regulator of abscisic acid signaling. Plant Physiol. 148: 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.R., Vega-Sanchez M.E., Zhu T., Wang G.L. (2006). Ubiquitination-mediated protein degradation and modification: An emerging theme in plant-microbe interactions. Cell Res. 16: 413–426 [DOI] [PubMed] [Google Scholar]
- Zhang X., Garreton V., Chua N.H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yang C., Li Y., Zheng N., Chen H., Zhao Q., Gao T., Guo H., Xie Q. (2007). SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]