Abstract
The future bioinformatics needs of the Arabidopsis community as well as those of other scientific communities that depend on Arabidopsis resources were discussed at a pair of recent meetings held by the Multinational Arabidopsis Steering Committee and the North American Arabidopsis Steering Committee. There are extensive tools and resources for information storage, curation, and retrieval of Arabidopsis data that have been developed over recent years primarily through the activities of The Arabidopsis Information Resource, the Nottingham Arabidopsis Stock Centre, and the Arabidopsis Biological Resource Center, among others. However, the rapid expansion in many data types, the international basis of the Arabidopsis community, and changing priorities of the funding agencies all suggest the need for changes in the way informatics infrastructure is developed and maintained. We propose that there is a need for a single core resource that is integrated into a larger international consortium of investigators. We envision this to consist of a distributed system of data, tools, and resources, accessed via a single information portal and funded by a variety of sources, under shared international management of an International Arabidopsis Informatics Consortium (IAIC). This article outlines the proposal for the development, management, operations, and continued funding for the IAIC.
The Multinational Arabidopsis Steering Committee (MASC) and the North American Arabidopsis Steering Committee (NAASC) hosted workshops in Nottingham, UK (April 15 to 16, 2010) and Washington DC (May 10 to 11, 2010) to consider the future bioinformatics needs of the Arabidopsis community as well as other science communities that depend vitally on Arabidopsis resources. The outcomes of both workshops were presented and discussed at the International Conference on Arabidopsis Research (ICAR) in Yokohama, Japan. The focus of the workshops was on Arabidopsis because of its unique and essential role as a reference organism for all seed plant species. The development of the highly annotated “gold standard” Arabidopsis genome sequence has been an invaluable resource for plant and crop sciences. This platform provides important information and working practices for other species and for comparative genomic and evolutionary studies. Arabidopsis tools and resources for information storage, curation, and retrieval have been developed over recent years primarily through the activities of The Arabidopsis Information Resource (TAIR), the Nottingham Arabidopsis Stock Centre (NASC), and the Arabidopsis Biological Resource Center, among others. However, the Arabidopsis community and funding agencies recognize the need for a single data management infrastructure. The key challenge is to develop and fund this resource in a sustainable and transparent manner.
Global challenges surrounding food and energy security require intelligent plant breeding strategies that will be dependent on a central Arabidopsis information resource to aid our understanding of gene function and associated phenotype in many different environments. The knowledge accrued in Arabidopsis informs our understanding of the genetic basis of plant processes and crop traits. To date, this has accumulated primarily through analysis of single genes. However, gene products do not act alone but rather in complex interacting networks. Thus, the challenge for the Arabidopsis community is to understand this higher level of complexity, to a significant extent through the application of new high volume, quantitative experimental techniques. The goals of these efforts are to develop gene/protein/metabolite networks that will enable systems-level modeling of plant processes and ultimately to translate these findings to crop plants. To achieve these goals, we must develop novel approaches to data management, integration, and access.
The UK workshop addressed three principal issues: the types of data generated by the Arabidopsis community, the types of data used by the community, and future needs of the community. The objective was to produce recommendations for the type of infrastructure necessary to address the challenges and opportunities associated with the application of new technologies and recommendations for a sustainable funding model to support this infrastructure. These recommendations were considered and expanded upon at the US workshop with the ultimate goal of generating solutions to the issues discussed in the first meeting. It was recognized that cohesive, cooperative, and long-term international collaboration will be critical to successfully maintain an Arabidopsis database infrastructure that is essential for plant biology research worldwide.
The workshop participants concluded that there is a continued need for a central Arabidopsis information resource, based on the productivity of the Arabidopsis community and the critical importance of the findings generated by this community. For example, ∼3000 Arabidopsis publications are currently published in peer-reviewed journals each year, a nearly 10-fold increase since the early 1990s; and in 2009, TAIR was accessed by 335,692 unique visitors and had nearly 20 million page views. Furthermore, the importance of a current, well-organized, and carefully curated Arabidopsis genome to researchers studying other plants, including crops, cannot be overstated. In the future, this resource should be part of a larger infrastructure that would be dynamic and responsive to new directions in plant biology research.
DATA TYPES AND USES: NOW AND IN THE FUTURE
The kinds of data currently generated by Arabidopsis researchers are diverse and in a variety of formats (Table 1). They vary in volume and complexity, and although some of these data types are common among plant species, many have become available first in Arabidopsis, a pattern that is likely to be repeated for future technologies. Overall, the volume of data is dramatically increasing, particularly due to the exponential growth of next-generation sequencing of genomes, chromatin, and RNA and, on a smaller scale, expanding proteome and metabolome data sets. The quantity of assembled data will require novel storage and display capabilities. In the future, we must deal with sophisticated new data sets including, but not limited to, high-resolution microarray data, image data, cell-type-specific or time series expression profiles, protein localization data, protein activation and relocation data, protein–protein interaction data, and promoter structure and transcription factor binding sites (both positional and temporal). All these data sets will be used to generate systems-level models that must also be stored in an accessible way. Because Arabidopsis has become the most important reference plant, with unmatched tools and resources, it likely will be the plant system in which traditional and novel forms and quantities of data will first become available.
Table 1.
Types of Data Deposited by Arabidopsis Researchers
| Published Literature |
| Genomes |
| Metabolome, catalog of metabolites |
| Proteome |
| Protein sequence and structure |
| Protein subcellular localization |
| Protein modifications |
| Interactome |
| cDNA sequence |
| Gene expression data |
| Genetic variation and accession genomes, single nucleotide polymorpisms and indels |
| Quantitative trait loci |
| Expression quantitative trait loci |
| Alternative splicing |
| Phenomics and phenotypic data |
| Epigenetic data |
| Exogenous small molecules |
Integration of these different data types will therefore be a key issue, both vertical integration, in which all available Arabidopsis information is accessible, and horizontal integration, whereby it is possible to move easily between different species. This horizontal integration process will naturally begin with genome/ortholog alignment with plant/crop genomes and extend to other data sets as the depth and complexity of the data from other plant species becomes sufficiently rich. As annotation and curation is increasingly inferred from several types of data, users will demand clear audit trails that indicate the provenance of the data pertaining to genes and their products. Currently, TAIR plays a key role in providing an authoritative stamp for community-approved annotation (for example, defining a working complete set of gene models); the need for this is dramatically increased, not made redundant, in the face of a data explosion.
It also is important that data are readily available in convenient formats and via tools that are accessible to a range of users. Development of software based on an open source model should be a fundamental principle, as this approach most efficiently leverages expertise and capacity across the fields of genomics and systems biology and has been shown by experience to produce the most trusted and adaptable software tools. Most of the challenges in data growth and diversity faced by the Arabidopsis community are not unique; cooperative tool development with researchers working on other species will ensure that useful software is developed in a cost-effective manner.
The highly curated and characterized gene/protein/metabolite networks developed in Arabidopsis will prove invaluable for systems biology approaches that seek to construct and constrain a range of models, which in turn will provide a framework for interpretation of a variety of complex results. The high standard of curation and data annotation in the Arabidopsis community makes these resources important to researchers in other communities seeking to gain valuable functional insights into their own data. Examples include crop scientists as well as those studying model organisms and other less-well-studied plant species. These wider applications underpin efforts to understand the molecular basis of plant growth and development and, ultimately, crop yield.
The high volumes of data now generated in biological research increases the importance of efficient and flexible tools for data analysis, inspection, and visualization. At present, the community's ability to access and analyze data is limited by the highly heterogeneous and often complicated (sometimes out of necessity) nature of many bioinformatics tools. Traditionally, genome browsers have provided a basic framework through which additional annotation can be visualized. However, new data types are pushing the limits of visualization. For example, data on genomic variation, such as that generated by the 1001 Arabidopsis genomes project, will help to link genotype to plant phenotype; however, the resources and tools needed to access and analyze these data are still in the early stages of development. Thus, we anticipate an ongoing need for the development of production-level web software with easy-to-use interfaces, integrated analysis tools, and uniform access to multiple data types.
A CONTINUED NEED FOR AN ARABIDOPSIS COMMUNITY PORTAL
The value of an Arabidopsis information portal should be measured primarily through its ability to facilitate and stimulate high-quality science. There is strong justification for such a resource that provides a vital service to what is a large and vibrant scientific community. This community comprises not only those working directly on Arabidopsis but also researchers working on other plants and animals. In particular, scientists working on all the major crop plants look to Arabidopsis data to inform their research. Arabidopsis is likely to continue to play a nodal role due to its well-annotated genome and its wealth of genetic and genomic resources, which make it unique among plant species in being well suited to systems biology research.
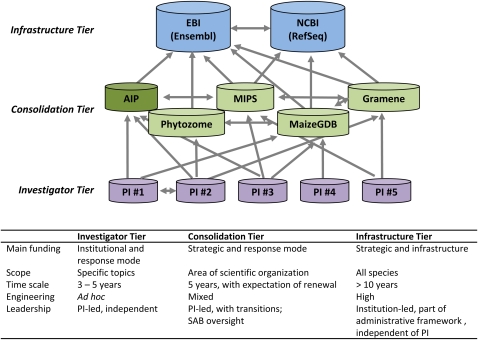
Clearly there is a need to define a manageable scope for any information resource. One division is between archives and interpreted resources. Archives (for relatively unprocessed data) often can be very broad in scope, and for many data types, a specific Arabidopsis repository may not be needed. Another set of resources can then provide interpreted views of the archived data for specific purposes. One can think of such interpreted resources as existing in three tiers (Figure 1) (Parkhill et al., 2010). The first tier consists of local databases that feature novel or highly specialized data resources run mainly by individual researchers focused on a narrow biological question. In the second tier, data are consolidated into forms that are more readily useable by a larger community (an Arabidopsis information portal belongs in this level). In effect, a community trusts a resource of this type with custodianship of its data. As different data types are brought into the community portal, the challenge will be to set priorities as to what should be consolidated and how data can and should be integrated. Input from the community both directly and through scientific advisory boards will be critical in setting the priorities, scope, and standards for quality control. The third tier enables cross-species comparisons of data sets, by integrating the outputs of differently focused resources; currently, this is mainly feasible for genomes and gene expression. Work should be directed to developing common data formats and tools for interrogation, to facilitate exchange between databases for different species, and to ensure that Arabidopsis information can be fully exploited by bioinformatics resources being developed to serve communities for which Arabidopsis is a key model organism (e.g., crop science).
Figure 1.
Three Tiers of Data Resources.
Proposed scope for an information resource to house interpreted biological resources (modified from Parkhill et al., 2010). The lowest and most fundamental tier consists of local databases of specialized data resources run mainly by individual investigators. The second tier provides a layer of consolidation into more durable and useable forms for a larger defined community; an Arabidopsis community portal belongs in this level and is indicated as AIP. The third tier enables cross-species comparisons; this requires an integrated set of diverse resources.
AN INTERNATIONAL ARABIDOPSIS INFORMATICS CONSORTIUM
The Arabidopsis community has a strong tradition of international cooperation (e.g., multinational sequencing initiative, multinational steering committee, international stock centers, annual international meeting etc.). The development of a new international Arabidopsis informatics initiative is a logical next step to manage the increasing amounts and types of data and will allow the leveraging of resources, knowledge, and collaborations. In our view, there is a strong justification and incentive to expand the current informatics structure into an international organization, the International Arabidopsis Informatics Consortium (IAIC). The consortium will need to be dynamic and represent the evolving needs and capacities of the community while reflecting the funding interests of the respective countries.
We propose that the IAIC be made up of a distributed system of data, tools, and resources that would be funded by a variety of sources under an international management and scientific advisory board. Participants at these workshops emphasized the importance of a unified front-end interface. We therefore envisage that the core of the IAIC will be the Arabidopsis Information Portal (AIP) that will interact with and link to resources across the globe, including Arabidopsis data sets generated in individual laboratories, information from other species, and other biological data sets. We propose that all data be accessed via the AIP and that the AIP combine outputs into a single user-friendly interface. The AIP will enable optimized use of data, tools, and resources to maximize the return on public research investment for the wider scientific community.
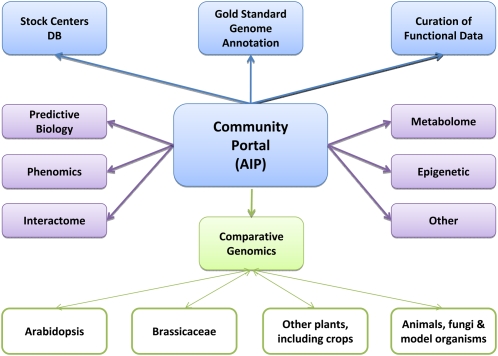
To ensure that the IAIC is built on strong foundations, we propose that the IAIC has a core consisting of four parts: (1) the AIP as outlined above; (2) a Gold Standard Genome Annotation (i.e., a finished genome [no gaps], annotated with protein and nonprotein coding genes, including some level of experimental support behind the functional predictions) and gene models that are revised by curation or targeting programming based on feedback and new data; (3) genome/sequence curation that provides functional information on each gene, its product(s), and associated regulatory landscape in a genomic context; and (4) stocks and resources database(s).
Using the core as the basis for the IAIC, additional noncore modules can then easily be added to form the IAIC, as illustrated in Figure 2. Indeed, such a model as proposed here with a clearly defined set of standards allows for any data, resource, or tools generated across the globe to become part of the IAIC. In this approach, the user does not face a dispersed landscape of data; instead, resources are federated giving the user the impression of a seamless whole. Furthermore, a distributed model allows the workload, human expertise, innovation, and costs to be shared across many sites that are internationally located. The proposed model for the IAIC produces additional resilience and flexibility by providing opportunities to bring together creativity and energy from many places. A federated approach also has the advantage of specialization with each module being able to focus on a particular area of expertise. Examples of such a distributed informatics model exist for other organisms, such as WormBase for Caenorhabditis species and FlyBase for Drosophila species.
Figure 2.
The Structure of the IAIC.
IAIC consists of core of four components in blue: (1) the AIP, which is the central hub of the consortium, provides a single user interface to access to all the constituent parts of the consortium, sets standards, and provides training; (2) gold standard genome annotation; (3) curation of functional data; and (4) stock center database(s) to enable rapid access to resources. Noncore modules are illustrated in purple; those listed in the figure are just examples and are not meant to be an exhaustive list. The comparative genomics module (in green) provides one example of how the IAIC will link out to other plant species.
The proposed modular structure provides an ideal opportunity for the IAIC to link out and interact with other plant species. In fact, workshop participants noted that an essential function of the IAIC would be to ensure that the distributed set of resources that make up the IAIC could easily be leveraged to benefit other plant communities. We propose that the most effective way to achieve this would be to develop a noncore module in comparative genomics that would allow integration of data from other species as it reaches sufficient depth and quality. The module could then grow at varying rates depending on the data sets available, ease of integration, and interoperability. We envisage that such a module could consist of four layers: (1) Arabidopsis, natural variation and genome evolution; (2) other Brassicaceae, nearest relatives enabling wider genome associations; orthology, natural variation, evolution, and crop traits; (3) crop genomes, evolution, orthology, and crop traits; and (4) other species. Such a module would not only allow other plant and crop researchers to access Arabidopsis information but would also enable Arabidopsis researchers to link out to appropriate orthologs and associated data in other plant species. To ensure that there is interoperability between data and resources generated in other communities, it will be essential for the IAIC to establish strong links with other plant data providers, to allow exchange of information, best practice, and to help build a common framework.
ENSURING THE SUSTAINABILITY OF AN INTERNATIONAL ARABIDOPSIS INFORMATICS CONSORTIUM
Management and Operations
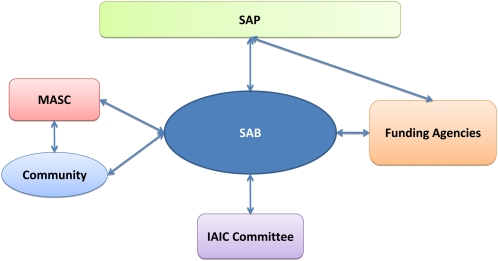
To ensure the IAIC fulfills the objectives outlined above, we propose the establishment of an International Scientific Advisory Board (SAB), an IAIC Committee, and a Scientific Advisory Panel (SAP). The role of the SAB would be to (1) direct future activities of the IAIC, both core components and noncore modules; (2) help to encourage compliance with the standards set out by the AIP; (3) liaise with funding agencies in the respective countries involved in the IAIC; (4) act as a point of contact for principal investigators (PIs)/groups wishing to contribute to the IAIC; and (5) liaise with the community to ensure that the IAIC continues to anticipate and serve the needs of the community. The SAB will be formed by a minimum of one scientist from each of the countries involved in supporting the IAIC. The SAB will be selected in consultation with MASC and the funding agencies supporting the IAIC. It will be essential for SAB members to have the appropriate expertise in technical implementation and community needs. Members of the funding agencies supporting the IAIC would be invited to be observers at SAB meetings. The IAIC Committee would consist of the PIs leading the core component and noncore modules of the IAIC. To ensure that membership of the IAIC Committee does not cross over with membership of the SAB, SAB members should not lead core components or modules of the IAIC. We recommend that a chairperson that is not involved in any part of the IAIC be appointed by the SAB and oversee the IAIC Committee. The committee would report to and interact with the SAB. We propose that the committee meet twice a year, once at ICAR and one virtual meeting. A SAP will also be formed to review the progress of the IAIC. SAP members will be selected from the Arabidopsis and wider research communities and consist of a set of advisors that are distinct from the SAB and IAIC committee. The SAP could assist with midterm review and end-of-grant reviews. The managerial structure of the IAIC is outlined in Figure 3.
Figure 3.
Management Structure of the IAIC.
The management of the IAIC is split into three levels. (1) IAIC Committee consisting of the PIs leading the core components and noncore modules of the IAIC. This committee would report to and interact with the SAB. (2) SAB, consisting of a minimum of one scientist from each of the countries involved in the IAIC. The SAB would oversee the development of the IAIC and interact with the funding agencies, MASC, and the community. (3) SAP, which would review the progress of the IAIC.
Since the funding streams supporting both core components and noncore modules are expected to come from different international funding sources, efficient operation of the IAIC will require careful planning. We therefore propose that the establishment of the IAIC is divided into two phases: (1) development of the IAIC and (2) operation of the IAIC. In phase 1, we recommend that the SAB is appointed and begins liaising with funding agencies to determine possible mechanisms for setting up the core components and noncore modules. In some cases, this might require the establishment of specific calls for proposals, while in other cases existing funding schemes may already be in place. Irrespective of the mechanisms that funding agencies are able to provide, we strongly recommend that funding for the core components be secured in advance of noncore modules. During the first phase of the IAIC, the SAB will also develop a suggested list of noncore modules and appoint the IAIC Committee Chair. There are likely to be many examples of projects that currently exist that could easily be adapted to become part of the IAIC. The SAB would help identify and liaise with such projects and provide information regarding the funding mechanisms available to adapt or establish these modules to become a part of the IAIC. PIs will be encouraged to apply for funds in specific countries to adapt or establish components of the AIP.
While there may appear to be an overlap of functions between the SAP (reporting to the funding agencies) and the SAB (liaising with the funding agencies and reporting to the SAP), experience in other areas has shown that these two boards can fulfill very different roles. In particular, the SAB can have a more private and direct interaction with the scientists and PIs overseeing work within the consortium; thus, the SAB has the opportunity to be more constructively critical of these scientists and the project.
Funding
During the workshops there were wide ranging discussions of the current and future funding mechanisms for informatics and cyberinfrastructure, and it was concluded, for the reasons that are clearly articulated by Chandras et al. (2009), that commercial, semicommercial, and cross-subsidy models are not feasible approaches for funding the IAIC. Instead, since the use, development, and contribution of data, tools, and resources are international, a transnational funding structure appears to be the most common sense mechanism for providing support for the IAIC, providing good value for money for scientists and funders alike. Coordinated, international support for the IAIC would increase the number of financial stakeholders and spread the burden of long-term funding, and because the whole will be greater than the sum of its parts, we envisage that a distributed model that is internationally funded would encourage a variety of funding bodies to become involved and support this endeavor.
Given the critical nature of the core components to the success of the project, a greater stability and, therefore, financial commitment from the funding agencies involved is required for the core of the IAIC in contrast with noncore modules of the IAIC. While there may be some turnover of the noncore components, driven either scientifically or financially, a stable core means that the resource remains sustainable over time.
We therefore propose that the core components of the IAIC should be stably funded on a 5-year rolling basis with the appropriate review and renewal at time points consistent with the funding body/bodies supporting the core components. We suggest several options for core funding. Option 1 would be unitary funding for all core components from a single national funding agency. For option 2, all core components are funded by a consortium of national agencies or a consortium of international agencies. For option 3, core components are funded separately by national or international agencies. For option 4, options 1, 2, or 3 are combined with an institutional commitment from the core host(s), a university or research institute, to house one or all core components. The latter could be a commitment in cash or in kind. And for option 5, funding for thematically related data-generating projects might be top-sliced or taxed as a way of funding the core of the IAIC and allowing immediate dissemination of data from these projects through inclusion in the IAIC. The success of this depends on the number of related projects supported by a funder and the degree to which sufficient funding could be raised. This supports the now usual institutional policy for data sharing and has the advantage of adjusting the funding to the core on the basis of national need. However, it is possible that fluctuation of the data-generating projects with time might compromise long-term planning for the core of the IAIC.
We envisage that each of the noncore modules will be funded nationally or through consortia of national/international funding agencies with shared policy priorities. An internationally distributed funding model for the IAIC provides plurality of funding, spreads the costs and the risks, and generates added value for both core components and noncore modules investment. The separation of funding priorities between the core components and noncore modules allows financial sustainability to be prioritized and distributed between these activities, thus providing greater stability for the core. This separation also provides considerably more flexibility in the spectrum of models, which might be adopted simultaneously across the IAIC.
Technology and Standards
The technological sustainability of the IAIC will depend on several features, including openness, standards, intelligent new web-based solutions, widely applicable tools, and a centralized body to enforce standards. Openness, in the context of data, means that none are proprietary or subject to use restrictions and that raw data are easily downloadable. Openness in the context of database tools means that the underlying code for these is developed following an open source and collaborative model.
In using a distributed model for the IAIC, whereby data from geographically dispersed sites are accessed and linked through one portal (AIP), the development of clear standards to allow archiving, exchange, and mining of data will be critical. For the data contained in the AIP to be easily accessed and used, adherence to community standards for metadata will also become increasingly important. Examples of such standards for microarray expression data (MIAME) and for proteomics data (MIAPE) already exist, while others such as those for metabolomics data still need to be developed. In order for dispersed sites to feed data to the AIP on the fly and to ensure machine readability of AIP resources by other databases and software tools, intelligent web-based solutions, such as web services, should be employed. Again, standards will need to play a role to make sure that the most current data are available via the AIP and also to ensure that there is interoperability across the IAIC.
To meet these challenges, the AIP will help develop and establish standards for existing data, tools, and resources. These would assist current projects to be adapted to become part of IAIC and ensure interoperability between all parts of the IAIC. The AIP would also ensure that future resources conform to the necessary standards if they wish to become a noncore module of the IAIC. To be effective, the IAIC will need to interact and learn from the wealth of research communities that are also tackling the challenges of archiving, exchanging, and mining data to ensure that the IAIC is part of a common technological framework whereby the information in IAIC can be brought to other communities and vice versa. It is particularly important for the AIP to set the requirements for interoperability that the noncore resources (components of the IAIC) would need to meet; the AIP should make it relatively easy for these contributed resources to meet the standards through good engineering, documentation, training, etc.
It will also be essential for the AIP to provide training for researchers wishing to access data in the IAIC as well as for those generating data, tools, and resources and wishing to interact with/become part of the IAIC.
CONCLUSIONS
This is a critical moment for Arabidopsis informatics; the current model for the curation and delivery of Arabidopsis data is being challenged in the very near term, while the amount of data is accumulating at a rapidly increasing rate. This presents a challenge to the community to review its needs and priorities. These should be articulated clearly and appropriately to national funding agencies that support major users and generators of Arabidopsis data. There is now an opportunity for plant biologists to develop a new international approach to informatics and cyberinfrastructure that will meet new needs for data integration, access, and analysis. The workshop participants concluded that the development and maintenance of plant data, tools, and resources, including those of Arabidopsis, would require significant support by funding agencies. However, the IAIC would leverage funding from a variety of sources, develop richer tools than a single group, and help to establish and set standards for informatics resources. As proposed, a federated, international model could facilitate inclusion of data and resources developed by, and for, other plant communities. Our recommendations are not without risks, and other model organisms face similar issues in sustaining their informatics resources and may well come to different conclusions about the best path forward. In the context of Arabidopsis and the tightly knit, yet global, group of researchers that study it, a well-executed implementation of these recommendations should establish a sustainable informatics platform to serve the broad range of needs and applications that we, and scientists studying other species, have for Arabidopsis data.
Supplementary Material
Acknowledgments
The UK meeting and participation by UK representatives at the US meeting were supported by Biotechnology and Biological Science Research Council Awards ISIS 2079 and International Workshop 2068. The US meeting and participation of US representatives at the UK meeting were supported by National Science Foundation Award MCB-1037009.
IAIC WORKSHOP PARTIPANTS AND AFFILIATIONS
Primary Contributors (workshop organizers and significant contributors to the final report):
Ruth Bastow, Genomic Arabidopsis Resource Network, UK; Jim Beynon, University of Warwick, UK; Mark Estelle, University of California, San Diego; Joanna Friesner, NAASC; Erich Grotewold, Arabidopsis Biological Resource Center and Ohio State University; Irene Lavagi, MASC, UK; Keith Lindsey, Durham University, UK; Blake Meyers, University of Delaware; Nicholas Provart, University of Toronto, Canada.
Secondary Contributors (workshop attendees and report section contributors):
Philip Benfey, Duke University; Ewan Birney, European Bioinformatics Institute, UK; Pascal Braun, Dana-Farber Cancer Institute and Harvard Medical School; Volker Brendel, PlantGDB and Iowa State University; Robin Buell, Michigan State University; Mario Caccamo, Biotechnology and Biological Science Research Council Genome Analysis Centre, UK; Jim Carrington, Oregon State University; Mike Cherry, Saccharomyces DB and Stanford University; Joseph Ecker, Salk Institute; Janan Eppig, The Mouse Genome Database and the Jackson Laboratory; Mark Forster, Syngenta; Rodrigo Gutiérrez, Pontificia Universidad Católica de Chile; Pierre Hilson, VIB Ghent, Belgium; Eva Huala, TAIR, Carnegie Institution for Science; Manpreet Katari, New York University; Paul Kersey, European Bioinformatics Institute, UK; Joerg Kudla, Muenster University, Germany; Hong Ma, Fudan University, China, and Pennsylvania State University; Minami Matsui, RIKEN, Japan; Kathy Matthews, FlyBase and Indiana University; Sean May, MASC, NASC, and University of Nottingham, UK; Klaus Mayer, Munich Information Center for Protein Sequences, Germany; Andrew Millar, University of Edinburgh, UK; Harvey Millar, University of Western Australia, Australia; Eric Mjolsness, University of California, Irvine; Todd Mockler, Oregon State University; Basil Nikolau, Iowa State University; Magnus Nordborg, Gregor Mendel Institute, Austria; Chris Rawlings, Rothamsted Research, UK; Paul Schofield, University of Cambridge, UK; Heiko Schoof, Eu-SOL and Max Planck Institute, Germany; Julian I. Schroeder, University of California, San Diego; Taner Z. Sen, MaizeGDB and U.S. Department of Agriculture–Agricultural Research Service, Iowa State University; Dan Stanzione, iPlant Collaborative and University of Texas at Austin; Chris Town, Craig Venter Institute; Tetsuro Toyoda, RIKEN, Japan; Todd Vision, University of North Carolina at Chapel Hill and NESCent; Sean Walsh, ETH Zurich, Switzerland; Xiujie Wang, Chinese Academy of Sciences, China; Doreen Ware, Gramene and U.S. Department of Agriculture–Agricultural Research Service, Cold Spring Harbor Laboratory; Wolfram Weckwerth, University of Vienna, Austria; Weicai Yang, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China.
References
- Chandras C., Weaver T., Zouberakis M., Smedley D., Schughart K., Rosenthal N., Hancock J.M., Kollias G., Schofield P.N., Aidinis V. (2009). Models for financial sustainability of biological databases and resources. Database (Oxford) 2009: bap017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Birney E., Kersey P. (2010). Genomic information infrastructure after the deluge. Genome Biol. 11: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.