Nitric oxide (NO) regulates many physiological processes in plants. This research demonstrates that NO acts as a redox regulator of a transcription factor network that is a key component of plant defense and systemic acquired resistance. Specifically, NO modifies the NPR1 and TGA1 transcription factors by S-nitrosylating critical amino acids, resulting in enhanced transcription of defense genes.
Abstract
The role of reactive oxygen and nitrogen species in local and systemic defense reactions is well documented. NPR1 and TGA1 are key redox-controlled regulators of systemic acquired resistance in plants. NPR1 monomers interact with the reduced form of TGA1, which targets the activation sequence-1 (as-1) element of the promoter region of defense proteins. Here, we report the effect of the physiological nitric oxide donor S-nitrosoglutathione on the NPR1/TGA1 regulation system in Arabidopsis thaliana. Using the biotin switch method, we demonstrate that both NPR1 and TGA1 are S-nitrosylated after treatment with S-nitrosoglutathione. Mass spectrometry analyses revealed that the Cys residues 260 and 266 of TGA1 are S-nitrosylated and S-glutathionylated even at GSNO concentrations in the low micromolar range. Furthermore, we showed that S-nitrosoglutathione protects TGA1 from oxygen-mediated modifications and enhances the DNA binding activity of TGA1 to the as-1 element in the presence of NPR1. In addition, we observed that the translocation of NPR1 into the nucleus is promoted by nitric oxide. Taken together, our results suggest that nitric oxide is a redox regulator of the NPR1/TGA1 system and that they underline the importance of nitric oxide in the plant defense response.
INTRODUCTION
Nitric oxide (NO) was identified as an important messenger in plant defense signaling against microbial pathogens in the late 1990s (Delledonne et al., 1998; Durner et al., 1998). Subsequently it was shown to be a crucial regulator of many physiological processes in plants, including stomatal closure and plant growth and development (Neill et al., 2002a, 2002b; Pagnussat et al., 2003; Bethke et al., 2004; Zhang et al., 2006; Lee et al., 2008; Seligman et al., 2008). However, less is known about how this redox-active molecule regulates these different events.
NO can regulate physiological processes directly by affecting gene transcription. Transcriptional analyses in response to NO have been done using different techniques, such as cDNA-amplified fragment length polymorphism, microarray analysis, and real-time PCR (Huang et al., 2002; Polverari et al., 2003; Parani et al., 2004). NO-regulated genes are involved in different functional processes, such as signal transduction, defense, and cell death, transport, basic metabolism, and reactive oxygen species production and degradation. Analysis of NO-regulated genes revealed seven families of transcription factor binding sites, including WRKY, GBOX, and OCSE elements; these binding sites are enriched in the promoter region of the NO-regulated genes (Palmieri et al., 2008).
As a readily diffusible free radical, NO reacts with a variety of intracellular and extracellular targets. In this way, NO can act as activator or inhibitor of enzymes, ion channels, or transcription factors and regulate specific processes during abiotic or biotic stress situations in plants (Beltran et al., 2000; Kim et al., 2002; Zhang et al., 2005; Sayed et al., 2007; Asada et al., 2009). In addition to the formation of protein Tyr nitrates (Tedeschi et al., 2005) and metallonitrosyls (Brandish et al., 1998; Russwurm and Koesling, 2004), NO can also form S-nitrosothiols (via S-nitrosylation) (Stamler, 1994; Stamler et al., 2001; Gaston et al., 2003). The majority of all NO-affected proteins seem to be regulated by S-nitrosylation, which occurs either by oxygen-dependent chemical reactions or by the transfer of NO from a nitrosothiol to a protein sulfhydryl group (transnitrosylation). A very important low molecular weight nitrosothiol is S-nitrosoglutathione (GSNO), which is a general physiological transport and storage form of NO in plants and animals (Zhang and Hogg, 2004). The endogenous GSNO concentration is estimated to be in the low micromolar range, but it cannot be exclude that higher concentrations occur locally (Gaston et al., 1993; Kluge et al., 1997). The stability of S-nitrosothiols has been the result of much confusion due to the fact that the presence of traces of contaminating metal ions (especially copper and iron) enhances their degradation. Without impurities, the half-lives for dissolved S-nitrosothiols are in the range of several hours (Hogg, 2000). The decomposition rate is ~5% per hour in water at room temperature. GSNO and S-nitrosothiol levels are controlled by the activity of GSNO reductase (GSNOR), an enzyme that was previously identified as GSH-dependent formaldehyde dehydrogenase (Fliegmann and Sandermann, 1997; Liu et al., 2001). Depending on the conditions, GSNOR metabolizes GSNO to a mixture of products, including GSSG, hydroxylamine, NH3, and GSH sulfinic acid (Jensen et al., 1998), and as a consequence the likelihood of enhanced protein nitrosylation reactions is reduced.
In plants, we are just at the beginning of understanding the regulatory function of protein S-nitrosylation. So far, only a few plant proteins are known to be regulated by S-nitrosylation, including S-adenosylmethionine synthetase (SAMS), metacaspase, peroxiredoxin, and NPR1 (for nonexpresser of pathogenesis-related gene1) (Lindermayr et al., 2006; Belenghi et al., 2007; Romero-Puertas et al., 2007; Tada et al., 2008). SAMS, for example, was shown to be differentially inhibited by NO in Arabidopsis thaliana (Lindermayr et al., 2006). Incubation with GSNO resulted in a blunt, reversible inhibition of SAMS1, whereas SAMS2 and SAMS3 were not significantly affected. Since SAMS catalyzes the synthesis of the ethylene precursor S-adenosylmethionine and NO is known to influence ethylene biosynthesis, this enzyme probably mediates the crosstalk between ethylene and NO signaling.
Furthermore, it was demonstrated that S-nitrosothiols play an important role in plant disease resistance (Feechan et al., 2005). Increased S-nitrosothiol levels disable plant defense responses conferred by distinct resistance gene subclasses, and both basal and nonhost disease resistance are also compromised. Conversely, reduced S-nitrosothiol levels enhance protection against ordinarily virulent microbial pathogens.
It was also shown that GSNOR activity is necessary for the acclimation of plants to high temperature and for normal development and fertility under optimal growth conditions (Lee et al., 2008). The GSNOR mutant shows pleiotropic phenotypes, including failure to grow on nutrient plates, increased numbers of reproductive shoots, and reduced fertility. In wild-type and mutant plants, heat sensitivity is enhanced by NO donors, and the heat sensitivity of GSNOR mutants can be rescued by an NO scavenger. Also, a NO-overproducing mutant is defective in thermotolerance. Although we know that S-nitrosothiol homeostasis is very important for plant growth and development as well as for reactions to abiotic and biotic stress, the exact regulation mechanism is still unclear.
As a possible mechanism, the modification of the DNA binding activity of transcription factors is discussed. The DNA binding affinity of transcription factors can be altered posttranslationally either by phosphorylation or by redox-dependent modifications. The activity of the thiol-containing transcriptional activator OxyR, whose oxidation controls the expression of genes involved in H2O2 detoxification, is modulated by different redox-dependent modifications, including S-nitrosylation (S-NO), S-glutathionylation (S-SG), and the formation of sulfenic acids (S-OH) (Hausladen et al., 1996; Kim et al., 2002). Interestingly, these modified forms of OxyR are transcriptionally active but differ in structure, cooperative properties, DNA binding affinity, and promoter activities. In this way, OxyR can process different redox signals into distinct transcriptional responses (Kim et al., 2002). Furthermore, a group of plant homeodomain transcription factors contains a set of conserved Cys residues that are important for activation of these proteins (Tron et al., 2002). In the oxidized state, the homeodomain transcription factors form intermolecular disulfide bonds, resulting in inefficient DNA binding activity. By contrast, under reducing conditions, the DNA binding activity of these proteins is clearly enhanced.
Another important family of plant transcription factors that is regulated in a redox-dependent manner is the R2R3 MYB family. These proteins have a single Cys residue and this Cys must be reduced to promote DNA binding and transcriptional activity (Heine et al., 2004). Interestingly, NO can also inhibit the DNA binding activity of transcription factors. Posttranslational NO-dependent modification of the Cys residue 53 of Arabidopsis MYB2 results in reduced DNA binding activity of this transcription factor (Serpa et al., 2007). S-Nitrosylation of the Cys residue 53 was detected by biotin switch assay, and as expected for this type of modification, the NO-mediated inhibitory effect was reversed by DTT.
In several cases, transcription factors need to interact with other proteins and bind to the promoter region as multiprotein complexes. NPR1 interacts with members of the TGACG motif binding factor (TGA) family, which bind to elements of the PR1 promoter (Després et al., 2000; Zhou et al., 2000; Fobert and Despres, 2005). NPR1 is a critical component of the salicylic acid (SA)–mediated signal transduction pathway and is a key regulator of systemic acquired resistance (Klessig et al., 2000; Zhou et al., 2000; Durrant and Dong, 2004). The TGA transcription factors belong to the group of bZIP factors. Interestingly, the DNA binding sites for several bZIP factors were enriched in promoter regions of NO-regulated genes (Palmieri et al., 2008). A change in the cellular redox status during the SA-mediated activation of defense leads to reduction of NPR1 to its active monomeric form. Subsequently, the NPR1 monomers are translocated into the nucleus, where they interact with the reduced form of the transcription factor TGA1 (Despres et al., 2003; Pieterse and Van Loon, 2004). This interaction results in an enhanced DNA binding activity of TGA1 to the promoter region of the PR-1 gene and stimulates its expression.
In this article, we report the effect of the physiological NO donor GSNO on the NPR1/TGA1 regulation system. We demonstrated that GSNO enhances the DNA binding activity of TGA1 in presence of NPR1 and that both proteins, NPR1 and TGA1, are S-nitrosylated after treatment with the NO donor. Additionally, we observed that the nuclear translocation of NPR1 is promoted by NO. Taken together, these results underline the importance of NO as a redox regulator of transcription in plant defense response.
RESULTS
C172 and C287 of TGA1 Are Involved in the Formation of Intramolecular Structures
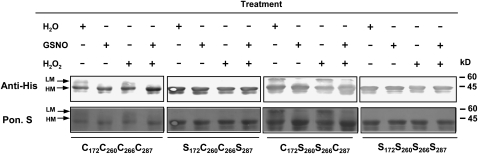
The formation or reduction of intra- or intermolecular disulfide bonds is an important mechanism to regulate the function and activity of proteins. TGA1 has four Cys residues, of which C260 and C266 form an intramolecular bond under oxidative conditions (Despres et al., 2003). We generated several Cys TGA1 mutants (C172S/C287S, C260S/C266S, and C172S/C260S/C266S/C287S) to analyze the effect of GSNO on the formation of intramolecular disulfide bonds. GSNO is a physiological NO donor that is able to S-nitrosylate and S-glutathionylate sulfhydryl groups of Cys residues. TGA1 wild-type proteins and the different Cys mutants were treated with water, GSNO, and/or H2O2 and separated by nonreducing SDS-PAGE. Slight changes of the mobility are diagnostic for the presence of disulfide bonds (Benezra, 1994; Mahoney et al., 1996; Delaunay et al., 2002). In the case of TGA1, low mobility proteins are a sign of formation of disulfide bonds, whereas reduced or modified Cys residues appear as high mobility proteins (Despres et al., 2003). Oxidized forms of wild-type TGA1 proteins occurred in water-treated and H2O2-treated samples (Figure 1). Surprisingly, GSNO-treatment results exclusively in the formation of high mobility proteins. The same tendency can be observed with TGA1-C260S/C266S mutants. By contrast, the TGA1-C172S/C287S double mutants and the TGA1-C172S/C260S/C266S/C287S quadruple mutants do not form low mobility proteins after any treatment. To confirm that the molecular weight difference is due to the redox status of TGA1, recombinant TGA1 and TGA1-C260S/C266S mutants were separated by reducing SDS-PAGE (see Supplemental Figure 1 online). Under these conditions, only high mobility proteins can be observed.
Figure 1.
Analyses of Intramolecular Structures of TGA1.
Recombinant purified proteins of TGA1/WT (left four lanes, CCCC) and three Cys mutants were treated either with 250 mM GSNO, 1 mM H2O2, or both for 30 min, with water treatment as a control. Afterwards, the proteins were separated by nonreducing SDS-PAGE and transferred onto nitrocellulose membrane. Top panel: His-tagged TGA1 proteins were detected with anti-His antibodies. Bottom panel: Ponceau S staining was done to demonstrate equal loading. The position of low mobility (LM) and high mobility (HM) proteins is marked with arrows.
GSNO Promotes DNA Binding Activity of TGA1
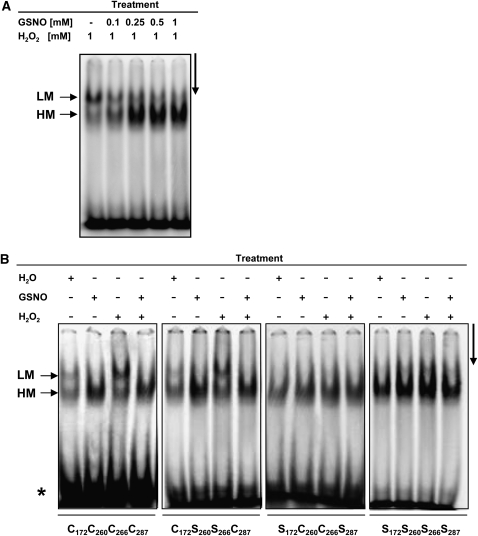
Although it has been described that redox changes do not directly regulate the DNA binding activity of TGA1 (Despres et al., 2003), we analyzed the influence of GSNO on the DNA–TGA1 interaction. The DNA binding ability of the TGA1 proteins was analyzed by electrophoretic mobility shift assays (EMSAs). Recombinant, purified, wild-type TGA1 was incubated with different concentrations of GSNO and 1 mM H2O2 in the presence of the activation sequence-1 (as-1) DNA element as a probe. As shown in Figure 2A, GSNO considerably enhances DNA binding activity of TGA1. Furthermore, wild-type and mutated TGA1 proteins were treated with water, GSNO, H2O2, or a combination of GSNO and H2O2 (Figure 2B). Similar to the formation of structurally different TGA proteins after GSNO treatment (Figure 1), different DNA-TGA complexes are formed under oxidizing conditions (high and low mobility complex). Addition of GSNO, however, resulted exclusively in the formation of high mobility complexes. The same was observed for the C260S/C266S double mutant. Moreover, the TGA1/wild-type (WT) proteins as well as the double mutants C260S/C266S bind the as-1 element with a higher activity after GSNO treatment compared with untreated or H2O2-treated proteins. Interestingly, with the C172S/C287S and the quadruple mutants, mostly high mobility complexes are formed regardless of the treatment. To demonstrate that the observed shifted bands are specific for TGA1–DNA interaction, several control experiments, including super shift and competition analyses, were done (see Supplemental Figure 2 online). Furthermore, S-nitrosylation of TGA1 under the conditions used for EMSAs was shown using the biotin switch assay, which allows specific detection of S-nitrosylated proteins (see Supplemental Figure 3 online).
Figure 2.
DNA Binding Activity of TGA1/WT and TGA1 Mutants.
(A) and (B) EMSAs were done to analyze the DNA binding activity of the treated TGA1 proteins. The asterisks indicate the position of the free probe. LM and HM mark low and high mobility DNA/protein complexes, respectively. The arrows on the right sides indicate the running direction.
(A) Recombinant TGA1/WT was treated with H2O2 and different concentrations of GSNO in the presence of the as-1 DNA element as a probe.
(B) Recombinant TGA1/WT and TGA1 mutants were treated with either H2O, 1 mM GSNO, 1 mM H2O2, or a combination of 1 mM GSNO and 1 mM H2O2.
GSNO Enhances the Binding Activity of TGA1 in the Presence of NPR1
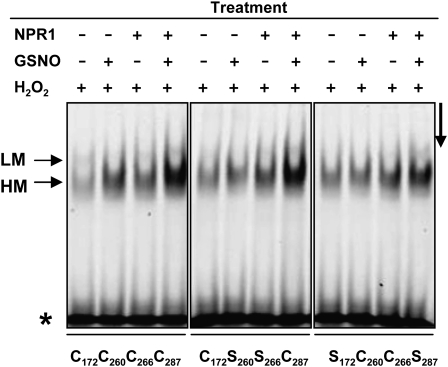
Because NPR1 stimulates the DNA binding activity of TGA1 factors under reducing conditions (Despres et al., 2003), we investigated the effect of GSNO on NPR1’s ability to stimulate TGA1 DNA binding activity. EMSAs demonstrated that under oxidizing conditions, DNA binding activity of TGA1 is very weak (Figure 3, H2O2 treatment). Addition of GSNO or NPR1, however, improves TGA1’s binding activity. More interestingly, the most effective DNA binding activity of TGA1 was observed in the presence of NPR1 and GSNO. This enhanced DNA binding activity could also be observed with both double mutants (C172S/C278S and C260S/C266S).
Figure 3.
EMSA of TGA1/WT and TGA1 Double Mutants under Oxidizing Conditions (1 mM H2O2).
Recombinant proteins were pretreated with water and/or 1 mM GSNO in the presence or absence of NPR1. After incubation with the labeled as-1 element, the samples were separated electrophoretically. The asterisk indicates the position of the free probe. LM and HM mark low and high mobility DNA/protein complexes, respectively. The arrow on the right side indicates the running direction.
S-Nitrosylation of NPR1 and TGA1
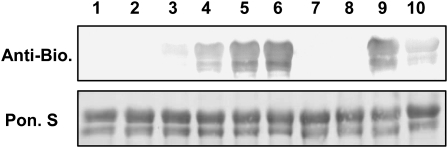
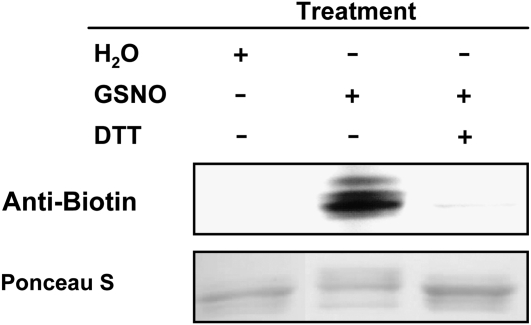
As demonstrated, GSNO enhances the DNA binding activity of TGA1 alone and also in the presence of NPR1. This raises the question as to which kind of modifications resulted from the GSNO treatment of TGA1 and NPR1. GSNO is able to S-nitrosylate and S-glutathionylate thiol groups of Cys residues. For detection of S-nitrosylated thiol groups, we used the biotin switch method, which was developed by Jaffrey et al. (2001), and specifically detects S-nitrosylated proteins. As shown in Figure 4 and in Supplemental Figure 4 online, TGA1 wild type and both double mutants (CSSC and SCCS) are S-nitrosylated after GSNO treatment, whereas treatment with GSH did not give a signal in the biotin switch assay. Furthermore, S-nitrosylation of TGA1 and the double mutants can be abolished by adding a reducing agent such as DTT to the S-nitrosylated proteins. No S-nitrosylation can be demonstrated for the quadruple mutant because these proteins no longer have any Cys residues. Additionally, recombinant and purified NPR1 were also subjected to the biotin switch assay, and S-nitrosylation could be detected only after GSNO treatment, while exposure to GSH gave no signal. Addition of DTT abolished S-nitrosylation of NPR1 (Figure 5).
Figure 4.
S-Nitrosylation of Recombinant TGA1.
Ten micrograms of purified recombinant TGA1 was treated with increased concentrations of GSNO (0.05, 0.1, 0.25, and 0.5 mM; lanes 3 to 6) and underwent the biotin switch method. Additionally, TGA1 was S-nitrosylated with 0.5 mM GSNO and reduced again with 10 mM DTT before and after biotinylation (lanes 7 and 10, respectively). Furthermore, GSNO-treated TGA1 underwent the biotin switch method without biotin-HPDP (lane 8) or without ascorbate (lane 9). Control treatments were done with water (lane 1) and 0.5 mM GSH (lane 2). Proteins were separated under nonreducing conditions by SDS-PAGE and blotted onto nitrocellulose membrane. Biotinylated proteins were detected using antibiotin antibodies. Ponceau S staining demonstrated equal loading.
Figure 5.
S-Nitrosylation of Recombinant NPR1.
Ten micrograms of purified recombinant NPR1 was treated with 0.1 mM GSNO, and S-nitrosylation was analyzed using the biotin switch assay (lane 2). Furthermore, NPR1 was treated with 0.1 mM GSNO and reduced with 10 mM DTT before undergoing the biotin switch method (lane 3). Control treatment was done with water (lane 1). Proteins were separated by nonreducing SDS-PAGE and blotted onto nitrocellulose membrane. Biotinylated proteins were detected using antibiotin antibodies. Ponceau S staining demonstrated equal loading.
Mass Spectrometry Analyses of GSNO-Treated TGA1
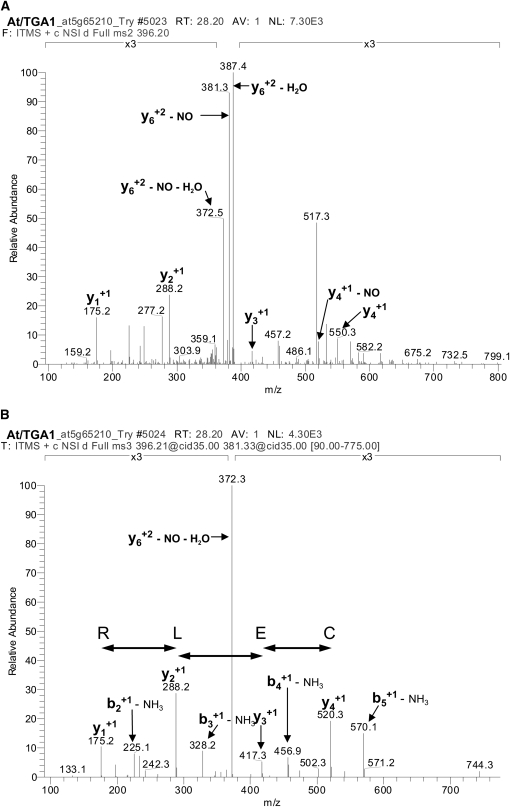
In parallel with the biotin switch assay, we used mass spectrometric analyses to detect GSNO-mediated modifications of the Cys residues of TGA1, as this method also allows the detection of S-glutathionylation. We treated recombinant purified TGA1 with 10, 100, and 500 μM GSNO for 10 min, digested the proteins with trypsin, and analyzed the Cys-containing peptides for their modifications (Table 1). C260 and C266 were found to be S-nitrosylated and S-glutathionylated after all treatments. By contrast, C287 was S-nitrosylated and S-glutathionylated just after treatment with 500 μM GSNO, and C172 was S-glutathionylated after treatment with 10, 100, and 500 μM GSNO. At the highest GSNO concentration, C172 was also found to be S-nitrosylated. The mass spectrometric analysis of the peptide 170QICNOELR175, containing an S-nitrosylated Cys residue, is shown in Figure 6. MS2 analysis of the S-nitrosylated peptide (396.2) results in loss of the NO group (381.3) (Figure 6A). The amino acid sequence was verified with the following MS3 analysis (Figure 6B). In Table 2, expected and observed mass-to-charge (m/z) values of the analyzed peptides containing unmodified, S-nitrosylated, or S-glutathionylated Cys residues are summarized. In some cases, we observed a mass difference of ±0.98, which is due to modification during trypic digestion. An increase or loss of 0.98 mass units results from an N-terminal amidation and deamidation, respectively.
Table 1.
Determination of Cys Modifications of TGA1
| 10 μM GSNO |
0.1 mM GSNO |
0.5 mM GSNO |
||||
| S-NO | S-SG | S-NO | S-SG | S-NO | S-SG | |
| C172 | − | + | − | + | + | + |
| C260 | + | + | + | + | + | + |
| C266 | + | + | + | + | + | + |
| C287 | − | − | − | − | + | + |
Purified recombinant TGA1 was treated with different concentrations of GSNO for 10 min. Residual GSNO was removed by gel filtration. After tryptic digestion, the Cys-containing peptides were analyzed for their modification by nano-liquid chromatography tandem mass spectrometry.
Figure 6.
Mass Spectrometry Analysis of S-Nitrosylated 170QICNOELR175.
(A) and (B) Purified, recombinant TGA1 was treated with 500 μM GSNO for 10 min at room temperature. After removal of excess GSNO by gel filtration and tryptic digestion for 1 h at 37°C, the peptides were analyzed by mass spectrometry.
(A) IT MS2 CID spectra of nitrosylated TGA1 peptide QICNOELR.
(B) IT MS3 CID spectrum of neutral loss fragment (−NO) from (A).
Table 2.
Determination of Modified Cys Residues of TGA1 after Treatment with GSNO
| Sequence | Unmodified (M+H)+: Expected/Observed m/z | +NO (M+H)+: Expected/Observed m/z | +GS (M+H)+: Expected/Observed m/z |
| 170QICELR175 | 761.40/761.39 | 791.39/792.40 | 1066.48/1066.48 |
| 242VLLPHFDVLTDQQLLDVCNLK263 | 2423.30/2423.30 | 2453.29/2453.30 | 2728.38/2728.38 |
| 264QSCQQAEDALTQGMEK279 | 1766.77/1766.77 | 1796.76/1796.77 | 2071.85/2071.85 |
| 280LQHTLADCVAAGQLGEGSYIPQVNSAMDR308 | 3044.46/3044.46 | 3074.45/3074.45 | 3349.54/3349.54 |
Purified recombinant TAG1 was treated with 500 μM GSNO for 10 min at room temperature. After tryptic digestion (1 h, 37°C) Cys-containing peptides were analyzed for their modifications (S-nitrosylation and S-glutathionylation) by mass spectrometry. For each peptide, expected and observed m/z values are shown.
NO-Dependent Transport of NPR1-GFP into the Nucleus
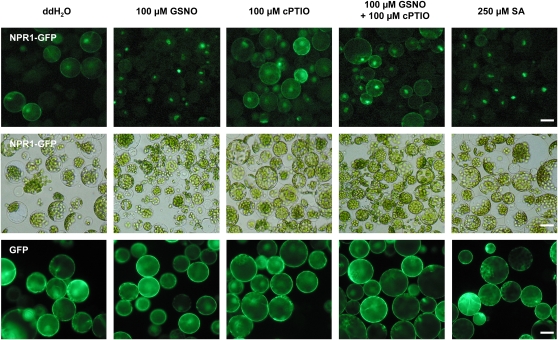
As already mentioned, NPR1 is retained in the cytosol in its inactive oligomeric form. To interact with TGA1, NPR1 monomers must be translocated into the nucleus. For this reason, we investigated whether GSNO causes the accumulation of NPR1 in the nucleus. We treated Arabidopsis mesophyll protoplasts harboring a 35S:NPR1-GFP (for green fluorescent protein) construct with GSNO and analyzed the distribution of NPR1-GFP within the cells by fluorescent microscopy. As shown in Figure 7, we could observe the translocation of NPR1 into the nucleus, while in the control treatment with H2O, the NPR1-GFP fusion protein is distributed in the cytosol. Scavenging of NO with 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) prevented the translocation into the nucleus. Treatment with SA was used as positive control. Addition of the NO scavenger cPTIO alone did not induce nuclear transloction of NPR1-GFP. Taken together, these results indicate that a NO-dependent mechanism is responsible for the translocation into the nucleus.
Figure 7.
NO-Dependent Transport of NPR1-GFP into the Nucleus.
Protoplasts of plants stably transformed with a 35S:NPR1-GFP construct were isolated and treated with double-distilled water, 100 μM GSNO, 100 μM cPTIO, 100 μM GSNO + 100 μM cPTIO, or 250 μM SA for 20 h. Localization of NPR1-GFP fusion protein was analyzed with a fluorescence microscope (top row, ×200). Center row shows the same cells in a bright-field micrograph. Protoplasts of plants stably transformed with a 35S:GFP construct were used as control to demonstrate that the different treatments have no effect of GFP (bottom row, ×200). Bars = 10 μm.
TGA1 Cys Mutants Exhibit Altered PR-1, PR-2, and PR-5 Expression
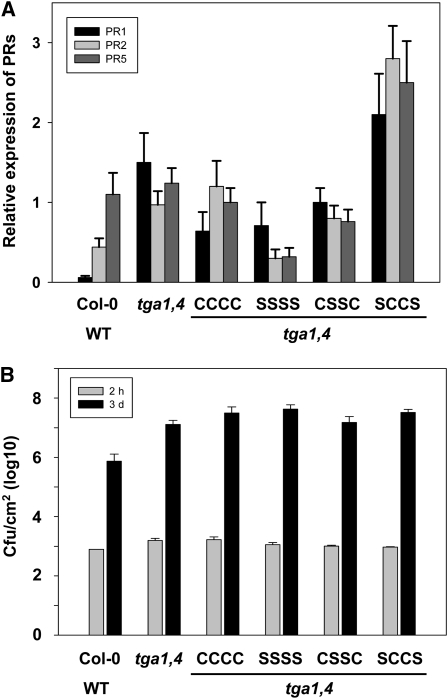
Arabidopsis tga1 and tga4 double knockout plants are impaired in PR-1, PR-2, and PR-5 expression and are a optimal tools to demonstrate the effect of TGA1 Cys mutants on expression of these SA marker genes. Therefore, double and quadruple TGA1 Cys mutants were transformed into tga1/tga4 double knockout plants under control of a 35S promoter. Expression of the introduced TGA1 variants has been proofed by PCR using TGA1-specific primers (see Supplemental Figure 5 online). Transcription of the SA marker genes PR-1, PR-2, and PR-5 has been analyzed by quantitative RT-PCR (Figure 8A). We found, that tga1/tga4 double knockout plants transformed with TGA1-WT, TGA1-C172S/C260S/C266S/C287S, or TGA1-C260S/C266S having similar relative expression levels for the tested SA marker genes as Columbia-0 (Col-0)/WT and tga1/tga4 double knockout plants (between 0.2 and 1.5 relative expression). Interestingly, double knockout plants transformed with TGA1-C172S/C287S have significant higher transcript levels of PR1, PR2, and PR5 in comparison to Col-0/WT and tga1/tga4 double knockout plants.
Figure 8.
Analyses of PR-1, PR-2, and PR-5 Expression and Basal Resistance in tga1/tga4 Double Knockout Plants Complemented with Different TGA1 Variants.
All experiments were done with Col-0 wild type, tga1/tga4 double knockout (tga1,4) plants, and tga1,4 complemented with TGA1-WT (CCCC), TGA1-C172/260/266/287S (SSSS), TGA1-C260/266S (CSSC), and TGA1-C172/287S (SCCS).
(A) Quantitative RT-PCR analysis of the amount of PR-1, PR-2, and PR-5 transcripts. Pools of three plants were used for each analysis. Experiments were repeated three times with up to three independent lines per construct. Data represent the average of all experiments per lines. The error bars represent se.
(B) For infection experiments, leaves of 4-week-old plants were infiltrated with virulent P. syringae DC3000 (105 colony-forming units/mL). Samples were collected 2 h (gray) and 3 d (black) postinfection. The experiment was repeated three times with similar results. Data represent values of one experiment averaged from nine leaf samples per genotype. Error bars represent se.
Since tga1/tga4 double knockout plants display enhanced disease susceptibility, we performed bacterial infection experiments to see whether the different TGA1 constructs can complement the enhanced disease susceptibility phenotype in the double knockout. As shown is Figure 8B, tga1/tga4 double knockout plants supported significantly higher pathogen growth than Col-0/WT plants and none of the different TGA1 variants could complement susceptibility of the double knockout.
DISCUSSION
In recent years, intracellular redox changes have come into focus as major regulators of key cellular functions in plant physiology and pathophysiology. Redox signaling is a process wherein free radicals, such as reactive oxygen and nitrogen species, act as messengers in biological systems mainly through modification of Cys residues. Transient and reversible redox-mediated modification of functional protein Cys residues, such as those located in the catalytic sites of enzymes or in the DNA binding domains of transcription factors, and the consequent formation of either intramolecular disulfide bonds or mixed protein glutathione disulfides, are major redox-related signaling mechanisms. Reactive oxygen species and reactive nitrogen species have been shown to serve as diffusible intra- and intercellular signals for activation of various physiological reactions in plants (Durner and Klessig, 1999; Neill et al., 2002a, 2002b; Feechan et al., 2005; Kotchoni and Gachomo, 2006). For example, NO activates a number of defense genes and has emerged as one of the pivotal mediators of disease resistance in plants (Delledonne et al., 1998; Durner et al., 1998; Huang et al., 2002). Furthermore, it has been observed that NO production occurs within the same time frame with that of H2O2, and a critical balance between the two redox molecules regulates cellular outcomes, such as sensitivity or resistance to a given stress situation (Delledonne et al., 2001).
NPR1 and TGA1 are well-described redox-regulated signaling compounds (Despres et al., 2003). Both proteins interact in their reduced state, which results in enhanced DNA binding of TGA1 and activation of PR gene expression (Despres et al., 2003; Pieterse and Van Loon, 2004). We demonstrated that TGA1 is structurally modified by GSNO and H2O2 (Figure 1). Under oxidizing conditions (H2O2) or conditions without protection from oxidation (H2O), TGA1 forms disulfide bonds leading to an inactive conformation. Diagnostic for the formation of an intramolecular disulfide bond is the lower mobility during electrophoretic separation (Delaunay et al., 2002). After GSNO treatment, only high mobility proteins can be detected, demonstrating that GSNO protects TGA1 from oxygen-mediated modifications. Different electrophoretic mobility of oxidized and reduced TGA1 was also demonstrated by Després et al. (2003). They showed that in vitro–translated TGA1 treated with diamide forms an intramolecular disulfide bond, whereas the C260N mutant has the same electrophoretic mobility under reducing and oxidizing conditions. Furthermore, a C260N+C266S mutant and the treatment of wild-type plants with SA, which leads to the reduction of TGA1 Cys residues, enables the TGA1/NPR1 interaction in yeast and Arabidopsis. This suggests the presence of a disulfide bond between C260 and C266 under oxidizing conditions.
Surprisingly, in the TGA1 double mutant C172S/C287S, we could not observe a disulfide bond formation between C260 and C266. Probably the mutation of the two Cys residues results in enormous steric alterations that prohibit the formation of an intramolecular disulfide bond between C260 and C266. In the TGA1 C260S/C266S mutant, however, low mobility proteins could be observed under oxidizing conditions, demonstrating that disulfide bond formation also occurs between C172 and C287. Interestingly, tga1 tga4 knockout plants transformed with the TGA1-C172S/C287S mutant showed hyperexpression of the defense-related genes PR-2 and PR-5 (Figure 8). These results demonstrate that reduction of these Cys residues is important for TGA1 activity, since the mutations mimic their reduced status.
Under oxidizing conditions, TGA1 forms a high and a low mobility complex in the EMSA, whereas only high mobility complexes appeared after treatment with GSNO. Obviously, the binding activity of TGA1 is altered under oxidizing conditions, resulting in the binding of more than one TGA1 protein per as-1 fragment. Addition of GSNO probably alters the conformation of TGA1. This results in a different DNA binding behavior, and the formation of low mobility complexes is not possible any more. This would suggest that different structural conformations of TGA1 resulting from the different redox conditions have different functions. Different redox modifications of a protein can have unique functional effects as it is described for the bacterial transcription factor OxyR (Kim et al., 2002). In this case, exposure of OxyR to GSNO or S-nitrosylated Cys and H2O2 results in formation of disulfide bonds, S-OH, S-NO, and S-SG. Interestingly, these alternatively modified proteins have different activities. S-hydroxylation, S-nitrosylation, and S-glutathionylation produce unique conformational changes in OxyR, which result in different DNA binding activities in the following order: OxyR-SSG > OxyR-SOH > OxyR-SNO. Furthermore, the fact that different multiple active forms of the transcription activator produce distinct alterations in the same DNA structure suggests that varied motifs or binding sites for OxyR forms with different affinities exist and have different consequences for activation of alternative genes.
We demonstrated that GSNO treatment has an effect on NPR1 and TGA1 activity (Figures 2, 3, and 7) and that NPR1 and TGA1 are both S-nitrosylated by GSNO (Figures 4 and 5). In the presence of GSNO, TGA1 DNA binding activity is considerably enhanced. This is quite surprising, since DNA binding of TGA1 is described to be not redox regulated (Despres et al., 2003). Apparently, the GSNO-dependent modifications results in conformational changes of TGA1, which allow a better DNA binding ability. However, since GSNO is able to both S-nitrosylate and S-glutathionylate Cys residues, we cannot say exactly which type of modification is responsible for the increased DNA binding activity of TGA1. The covalently attached NO or glutathione have various different features, such as size and hydrophobicity, which influence their environment in different ways and altered the chemical features of the modified protein. Different effects on protein function after S-glutathionylation or S-nitrosylation of the same Cys residue are described for several proteins (Aracena et al., 2003, 2005; Martinez-Ruiz and Lamas, 2007).
Systemic acquired resistance involves the production of SA. SA induces NO production through a NOS-dependent route in Arabidopsis, concluding that NO is a downstream signal in the SA-induced plant defense response (Zottini et al., 2007). A potential target for the produced NO is the transcription factor TGA1. As mentioned before, DNA binding activity of TGA1 is improved in the presence of the NO donor GSNO, probably due to S-nitrosylation of critical Cys residues. Alteration of the redox conditions influences the DNA binding activity of several transcription factors (Tron et al., 2002; Heine et al., 2004; Toledano et al., 2004). However, the ability of TGA1 to bind its cognate promoter element in vitro was unchanged even in the presence of a molar excess of several redox-regulating compounds (Despres et al., 2003). Instead, reducing conditions are required for interaction of TGA1 with NPR1, which acts as a cofactor to stimulate TGA1’s DNA binding activity.
Redox-dependent interaction with NPR1 is only described for TGA1 and TGA4, while TGA2, TGA3, TGA5, TGA6, and TGA7 interact with NPR1 independently of the cellular redox status (Zhang et al., 1999; Zhou et al., 2000; Despres et al., 2003). The Cys residues C260 and C266 of TGA1 form a disulfide bond under oxidizing conditions, which precludes interaction with NPR1. These two redox-sensitive Cys residues are also conserved in TGA4, but not in the other TGA isoforms. Site-directed mutagenesis of both Cys residues enables the interaction with NPR1, concluding that both residues are controlling the interaction (Despres et al., 2003). Furthermore, the redox status of C260 and C266 of TGA1 and TGA4 is shifted considerably after SA treatment to become predominantly reduced. However, tga1/tga4 knockout plants transformed with the TGA1-C172S/C287S mutant showed hyperexpression of the defense-related genes PR-1, PR-2, and PR-5 (Figure 8A), suggesting that reduction of these Cys residues is also important for TGA1 activity, since the mutations mimic their reduced status.
Surprisingly, hyperexpression of these defense-related genes could not complement enhanced disease susceptibility phenotype in these plants (Figure 8B). It is likely that the expression level of these defense genes, and consequently the amount of synthesized PR-1, PR-2, and PR-5 proteins, is just not high enough to increase basal resistance in the double knockouts. As shown in Figure 8A, the expression levels of the analyzed defense-related genes are just about twofold higher in the tga1/tga4 plants complemented with TGA1-C172S/C287S than in the double knockout plants. Furthermore, mutation of these two Cys residues might negatively influences the stability of the protein in planta. Taken together, we could demonstrate that the redox status of C172 and C287 is important for the intramoleculare structure of TGA1 and that opening of the disulfide bond and GSNO-dependent modification of the Cys residues positively affect DNA binding activity of this transcription factor.
Interestingly, NPR1 enhances not only the DNA binding activity of the reduced TGA1 (Despres et al., 2003) but also the DNA binding activity of the GSNO-treated TGA1 (Figure 3). Apparently, the GSNO-dependent modifications result in conformational changes of TGA1 and/or NPR1, which allow a more effective TGA1–NPR1 interaction and as consequence a more effective DNA binding of TGA1. Furthermore, it is possible that the GSNO-dependent modifications just protect TGA1 from formation of disulfide bonds, which would result in lower DNA binding activity of TGA1.
As already mentioned, the SA-dependent SAR signaling pathway involves NPR1 as master regulator, which interacts with TGA1 and further improves its DNA binding activity. NO promotes the ability of NPR1 to enhance DNA binding activity of TGA1 (Figure 3). Furthermore, it was shown that treatment of Arabidopsis wild-type and transgenic 35S:NPR1-GFP plants with SA induces S-nitrosylation of endogenous NPR1 and the NPR1-GFP proteins and accumulation of GFP-labeled NPR1 in the nucleus (Tada et al., 2008). The NO-mediated enhanced activity of NPR1 is quite surprising since S-nitrosylation of NPR1 facilitates its oligomerization, which keeps it in the cytosol and is essential for NPR1 homeostasis upon SA induction (Tada et al., 2008). The monomerization of NPR1 is catalyzed by thioredoxins, which reduce NPR1 and allow the transloction into the nucleus. This nuclear translocation of NPR1 is required for interaction of NPR1 with TGA transcription factors and for PR gene expression. Interestingly, the nuclear translocation of NPR1 is also induced by GSNO/NO (Figure 7). However, the S-nitrosylation–mediated oligomerization is not seen as an inhibitory effect of NPR1 signaling but rather as a step prior to monomer accumulation. From this point of view, the observed NO-mediated nuclear transloction of NPR1 is not contradictory to the results described by Tada et al. (2008). It is conceivable that NPR1 monomers are just the transport form of this protein. Since we demonstrated a positive effect of GSNO/NO on the NPR1-mediated DNA binding activity of TGA1, it is possible that NPR1 is S-nitrosylated again in the nucleus. Interestingly, it is also described that NO can induce SA production (Durner et al., 1998; Huang et al., 2004). As a consequence, the observed nuclear translocation of NPR1 after GSNO treatment would be just a result of the SA-induced redox changes.
In addition to pathogen-mediated SAR, a nonpathogen-induced systemic resistance (ISR) has been described. Interestingly, the signaling pathway of ISR involves also NPR1, but it is independent of SA (Pieterse et al., 1996, 1998). Furthermore, many of the known defense-related genes, such as SA-inducible PR-1, PR-2, and PR-5 and the ethylene- and jasmonate-inducible genes HEL, CHIB, PDF1.2, ATVSP, LOX1, LOX2, and PAL1, were not found to be upregulated in Arabidopsis inoculated with the nonpathogenic Pseudomonas fluorescens WCS417r (Wang et al., 2005a). More interestingly, NO seems to be also involved in manifesting the ISR in plants (Heil, 2001; Wang et al., 2005b). However, neither the signaling mechanism of NO nor the function of NPR1 in this pathway is known. Probably NPR1 is also S-nitrosylated like in SAR, but in the case of ISR, it is not interacting with TGA1, but probably with another TGA isoform, another transcription factor, or just another signaling partner. Taken together, NPR1-dependent signaling seems to be much more complex than assumed so far, and the participation of NPR1 in two different signaling pathways makes it quite difficult to analyze the function of NPR1 for both resistance reactions separately. This complexity could also be an explanation for the contradictory observations that NO induces on one hand nuclear translocation of NPR1 (reported in this article) and on the other hand NPR1 oligomerization, which keeps NPR1 in the cytosol (Tada et al., 2008).
In sum, we could demonstrate that both proteins, TGA1 and NPR1, are redox modified by GSNO and that these modifications enhance the DNA binding activity of TGA1. More interestingly, we observed that the translocation of NPR1 into the nucleus is promoted by NO. These results suggest a regulatory role of NO for the NPR1/TGA1 system and add an important aspect to the described redox control of plant defense response (Despres et al., 2003).
METHODS
Production of TGA1 and NPR1 in Escherichia coli and Purification of Recombinant Protein
E. coli strain BL21 DE3 pLysS (Invitrogen) harboring the plasmids pDEST-42 (Invitrogen) was grown in Luria-Bertani medium supplemented with 100 μg/mL ampicillin until A600 ~0.5 was reached. Production of recombinant 6xHis-tagged proteins was induced with 1 mM isopropyl-β-d-thiogalactopyranoside. After overnight incubation at 10°C (NPR1) or 20°C (TGA1), bacterial cells were harvested by centrifugation. For protein isolation, the cells were resuspended in an appropriate volume of extraction buffer (50 mM NaH2PO4, pH 7.5, 2 mM EDTA, 20% [v/v] glycerol, 20 mM β-mercaptoethanol, 1 mM DTT, and 1 mg/mL lysozyme) and incubated on ice for 30 min and disrupted by sonication. Cellular debris was removed by centrifugation (20,000g, 20 min, 4°C). The recombinant 6xHis-tagged proteins were purified by immobilized metal chelate affinity chromatography using the Ni-NTA metal affinity matrix (Qiagen) according to the instructions of the manufacturer. Adsorbed proteins were washed with 20 volumes of washing buffer (50 mM NaH2PO4, pH 7.5, 300 mM NaCl, 20 mM imidazole, 0.5% [v/v] Triton X-100, 20 mM β-mercaptoethanol, and 1 mM DTT) and eluted from the affinity matrix with extraction buffer (50 mM NaH2PO4, pH 7.5, 300 mM NaCl, and 250 mM imidazol)
EMSAs
The EMSAs were performed as described previously (Ausubel et al., 1989; Carey, 1991). The as-1 element [consensus sequence 5′-TGACG(N7)TGACG-3′] represents an important DNA binding site within the promoter regions of various stress-related genes, such as the PR-1 or the GST gene (Garreton et al., 2002; Despres et al., 2003). In addition, it is a well-studied target sequence for the TGA1 transcription factor, and for this reason, a DNA sequence representing as-1 was used as probe. The oligonucleotides as-1F (5′-GGTCGAGCTGACGTAAGGGATGACGCACG-3′) and as-1R (5′-GGCGTGCGTCATCCCTTACGTCAGCTCGA-3′) were combined at a final concentration of 100 μM, denatured at 95°C for 5 min, and annealed at 20°C for 30 min. One hundred nanograms of double-stranded DNA were radioactively labeled with [α-32P]dCTP by Klenow exo−. The addition of 20 mM EDTA stopped the fill-in reaction. Finally, the radioactively labeled DNA fragments were separated from nonincorporated radionucleotides by filtration through a Micro Biospin P6 column (Bio-Rad). Approximately 1.5 × 108 cpm/μg double-stranded DNA resulted as specific activity. The recombinant purified TGA1 and NPR1 were reduced in the presence of 10 mM DTT on ice for 30 min before the DTT was removed by gel filtration. The following procedures were performed at dark conditions. Freshly reduced proteins were pretreated with 1 mM GSNO or left untreated at room temperature for 30 min. Subsequently, 250 ng TGA1 and 500 ng NPR1 were incubated with 120 pg radioactively labeled as-1 fragment (2 × 104 cpm). These binding assays were performed in 15 μL of binding buffer [20 mM HEPES-KOH, pH 7.9, 100 mM KCl, 20% (v/v) glycerol, and 0.05 μg/μL poly(dI-dC)] in the absence or presence of 1 mM H2O2 for a further 30 min. After the addition of 2 μL loading dye (50 mM EDTA, pH 8.0, 50 mM Tris/HCl, pH 8.0, 1.25 mg/mL bromphenolblue, and 1.25 mg/mL xylencyanol), the reactions were separated on a native polyacrylamide gel (4%) in 1× Tris/glycine/EDTA at 4°C. The gel was dried under vacuum and autoradiographed between intensifying screens at −80°C.
Gel Electrophoresis and Immunoblot Analysis
Proteins were separated by SDS-PAGE on 12% polyacrylamide gels (Laemmli, 1970), transferred onto polyvinylidene fluoride membranes, and blocked with 1% nonfat milk powder and 1% BSA. The blots were incubated with antibiotin mouse monoclonal antibody conjugated with alkaline phosphatase at a dilution of 1:10,000 for 1 h. Cross-reacting protein bands were visualized using 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium as substrates.
Biotin Switch Method
The in vitro S-nitrosylation and subsequent biotinylation of S-nitrosylated proteins were done as described by Jaffrey et al. (2001) with minor modifications. Recombinant and purified NPR1 and TGA1 were treated with GSNO for 20 min at room temperature. Blocking of non-nitrosylated free Cys residues was done by incubation with 20 mM methyl methanethionsulfonate and 2.5% SDS at 50°C for 20 min with frequently vortexing. Residual methyl methanethionsulfonate was removed by precipitation with 5 volumes of −20°C acetone, and the proteins were resuspended in 0.1 mL HENS buffer (HEN buffer containing 1% SDS) per mg protein. Biotinylation was achieved by adding 2 mM N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (biotin-HPDP) and 1 mM ascorbate and incubating at room temperature for 1 h.
Biotin-labeled proteins were separated by nonreducing SDS-PAGE on 12% polyacrylamide gels (Laemmli, 1970), transferred onto polyvinylidene fluoride membranes, and blocked with 1% nonfat milk powder and 1% BSA. The blots were incubated with antibiotin mouse monoclonal antibody conjugated with alkaline phosphatase (Sigma-Aldrich) at a dilution of 1:10,000 for 1 h. Cross-reacting protein bands were visualized using 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium as substrates.
Nano-HPLC-MS2/3 and Data Analysis
For mass spectrometry analyses, proteins were treated with 500 μM GSNO for 10 min at room temperature. After removal of excess GSNO using Micro Bio-Spin 6 columns (Bio-Rad), proteins were digested with trypsin at 37°C for 1 h in 50 mM NH4HCO3, pH 5.5. The reaction was performed in darkness to avoid light-dependent decomposition of the modifications. The trypsin:protein ratio was 1:20.
All nano-HPLC-MS2/3 experiments were performed on a Flux Rheos 2200 nanoflow system connected to a linear ion trap-Fourier transform mass spectrometer (LTQ-Orbitrap; Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Proxeon Biosystems).
Protein digests were analyzed by online nano-liquid chromatography-MS/MS. The samples were separated on an in-house made 10-cm reversed phase capillary emitter column (inner diameter 75 μm, 5 μm ProntoSIL 120-5-C18 ace-EPS; Bischoff) using 120-min linear gradients from 0 to 40% acetonitrile/0.1% formic acid at a flow rate of 270 nL/min.
The mass spectrometer was operated in the data-dependent mode to automatically switch between MS, MS2, and MS3 acquisition. Survey full-scan MS spectra (m/z 350 to 1800) were acquired in the Orbitrap with resolution R = 7500 at m/z 400. The six most intense ions were then sequentially fragmented in the linear ion trap using collisionally induced dissociation at a normalized collision energy of 35 V. In the case of a resulting neutral loss of 9.7 and 14.5 m/z, respectively, in the MS2 spectra of the three most abundant peaks (indicating the loss of NO), these fragments were selected for further fragmentation (MS3). Former target ions selected for MS2 were dynamically excluded for 30 s. Total cycle time was ~3 s. The general mass spectrometry conditions were as follows: spray voltage, 1.4 kV; no sheath and auxiliary gas flow; and ion transfer tube temperature, 200°C. Ion selection thresholds were 500 counts for MS2 and 500 counts for MS3. An activation q = 0.25 and activation time of 30 ms were applied in both MS2 and MS3 acquisitions.
Peptides and proteins were identified via automated database searching (Bioworks 3.3.1, SP1) of all MS2 and MS3against an in-house curated database containing the protein entries from The Arabidopsis Information Resource proteome database (TAIR, version 7), E. coli, porcine trypsin, and the human keratins (36,361 protein sequences). Spectra were searched with a mass tolerance of 1.5 atomic mass unit for the parent mass and 1 atomic mass unit for the MS2 and MS3 fragment masses with semitryptic specificity allowing two miscleavages. All modifications were set to be variable: oxidation of Met and nitrosylation and glutathionylation of Cys.
Preparation of Arabidopsis thaliana Protoplasts
Arabidopsis protoplasts were isolated as described by Abel and Theologis (1994) with some modifications. In brief, leaves of 4-week-old Arabidopsis plants were cut in 1-mm strips and incubated in a cellulase/macerozyme solution (1.25% cellulase [Serva], 0.3% macerozyme [Serva], 0.4 M mannitol, 20 mM KCl, and 20 mM MES, pH 5.7). The protoplast solution was vacuum infiltrated to reduce the protoplasting time and to increase the viability of the protoplasts. After infiltration, the leaf tissues were gently shaken (40 rpm) for 100 min at room temperature, and the protoplasts were harvested by filtration through nylon membrane (71 μm). After centrifugation (100g, 2 min, 4°C), the protoplasts were washed two times with 4°C cold W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES, pH 5.7). The quality of the isolated protoplasts was analyzed by microscopy, and the concentration was adjusted to ~50 protoplasts per μL.
Site-Directed Mutagenesis
The modification of single nucleotide residues was performed as previously described (Lindermayr et al., 2003). Briefly, for mutation, a pair of oligonucleotides was synthesized harboring the desired alterations. The size of the primers was adjusted to yield a melting temperature of 68°C using the following formula: Tm = 81.5 + 0.41 x GC (%) – 675/number of bases – sequence deviation (%). For amplification, 20 ng plasmid DNA was used in a total volume of 15 μL, including 1 μM each primer, 200 μM deoxynucleotide triphosphate, and 1 unit of PfuTurbo DNA polymerase (Stratagene). After denaturation (2 min at 94°C), 18 cycles were conducted, consisting of 45 s at 94°C, 30 s at 55°C, and 15 min at 72°C, followed by a final extension step at 72°C for 10 min. Subsequently, the parental and hemiparental template DNA was digested with DpnI, and the amplified plasmids were transformed into E. coli DH5α. The mutation was verified by sequencing.
Plant Transformation
Transformation of Arabidopsis plants was done by the floral dip technique (Clough and Bent, 1998). Inflorescences of Arabidopsis plants were dipped into Agrobacterium tumefaciens solution (OD600 ~0.8) with 5% sucrose and 0.05% Silvett L-77. To increase the infection efficiency, plants were covered with a plastic wrap for 2 d to guarantee high humidity.
Real-Time Quantitative RT-PCR
Total RNA extractions were performed from 200 mg leaf tissue using the TRIzol reagent according to the supplier’s instructions (Invitrogen). Each mRNA sample was reverse transcribed using Superscript II reverse transcriptase (Invitrogen) according to the protocol of the supplier. Inactivation of the reverse transcriptase was done by incubating the mixture at 70°C for 15 min, and the cDNA solution was stored at −20°C.
For real-time quantitative RT-PCR, the following gene-specific primer pairs were designed for PR-1 (PR1-f, 5′-GTGCCAAAGTGAGGTGTAACAA-3′; PR1-r, 5′-CGTGTGTATGCATGATCACATC-3′), PR-2 (PR2-f, 5′-GTCTGAATCAAGGAGCTTAGCC-3′; PR2-r, 5′-GATGGACTTGGCAAGGTATCG-3′), and PR-5 (PR5-f, 5′-ATGTGAGCCTCGTAGATGGTTAC-3′; PR5-r, 5′-GATCCATGACCTTAAGCATGTCG-3′). All primer pairs were checked for amplification specificity and an efficiency superior to 80% using a serial cDNA dilution. Real-time quantification was performed using a 7500 real-time PCR system (Applied Biosystems). Individual PCR reaction mixtures contained 4 μL of diluted cDNA (1:15), 10 μL of Sybr Green Mastermix (Thermo Fisher Scientific), and 250 μM of each primer in a final volume of 20 μL. In all experiments, three biological replicates of each sample and two technical (PCR) replicates were performed. Normalization of real-time quantitative RT-PCR data was done by geometric averaging of multiple internal control genes (Ubiquitin 9 [At5g18380] and S16 [At5g18380 and At2g09990]) (Vandesompele et al., 2002). The stability of the reference genes was tested and normalization was performed using GeNorm (Vandesompele et al., 2002).
Pathogen Infection
Virulent Pseudomonas syringae DC3000 was grown on King’s B medium and infiltrated with a syringe into leaves of 4-week-old plants at a concentration of 105 colony-forming units mL−1 in 10 mM MgCl2. Mock inoculation was performed using 10 mM MgCl2. In planta bacterial titers were determined by shaking leaf discs from infected leaves in 10 mM MgCl2 supplemented with 0.01% Silwet-L77 at 28°C for 1 h. The resulting bacterial suspensions were serially diluted, and spots of 20 μL per dilution were grown on King’s B medium and counted.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries or the Arabidopsis Genome Initiative database under the following accession numbers: NM_105102 (At1g64280), NM_125919 ( At5g65210), and NM_121041 (At5g10030). The tga1-1 and tga4-1 mutants were identified in the T-DNA insertion collection from the SALK Institute Genomic Laboratory (http://signal.salk.edu) (SALK_082821 and SALK_127923 for tga1-1 and tga4-1, respectively). tga1/tga4 double knockouts were provided by X. Dong (Duke University, Durham, NC) (Kesarwani et al., 2007).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Molecular Weight Difference of TGA1 Is Due to Its Redox Status.
Supplemental Figure 2. TGA1 Binds Specifically to the as-1 Element.
Supplemental Figure 3. S-Nitrosylation of TGA1 under Conditions Used for EMSA Aassays.
Supplemental Figure 4. S-Nitrosylation of Recombinant TGA1/WT and Cys Mutants.
Supplemental Figure 5. Analysis of Expression of Introduced TGA1 Variants.
Acknowledgments
We thank Elke Mattes, Lucia Gößl, Birgit Geist, Marion Wenig, Claudia Knappe, Gaby Römling, and Rosina Ludwig for excellent technical assistance. We also thank Corina Vlot, Xinnian Dong, and Yasuomi Tada for fruitful discussion.
References
- Abel S., Theologis A. (1994). Transient transformation of Arabidopsis leaf protoplasts: A versatile experimental system to study gene expression. Plant J. 5: 421–427 [DOI] [PubMed] [Google Scholar]
- Aracena P., Sanchez G., Donoso P., Hamilton S.L., Hidalgo C. (2003). S-glutathionylation decreases Mg2+ inhibition and S-nitrosylation enhances Ca2+ activation of RyR1 channels. J. Biol. Chem. 278: 42927–42935 [DOI] [PubMed] [Google Scholar]
- Aracena P., Tang W., Hamilton S.L., Hidalgo C. (2005). Effects of S-glutathionylation and S-nitrosylation on calmodulin binding to triads and FKBP12 binding to type 1 calcium release channels. Antioxid. Redox Signal. 7: 870–881 [DOI] [PubMed] [Google Scholar]
- Asada K., Kurokawa J., Furukawa T. (2009). Redox- and calmodulin-dependent S-nitrosylation of the KCNQ1 channel. J. Biol. Chem. 284: 6014–6020 [DOI] [PubMed] [Google Scholar]
- Ausubel F.M., Brent R, Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. (1989). Current Protocols in Molecular Biology. (New York: Greene Publishing Associates and Wiley-Interscience; ). [Google Scholar]
- Belenghi B., Romero-Puertas M.C., Vercammen D., Brackenier A., Inze D., Delledonne M., Van Breusegem F. (2007). Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J. Biol. Chem. 282: 1352–1358 [DOI] [PubMed] [Google Scholar]
- Beltran B., Orsi A., Clementi E., Moncada S. (2000). Oxidative stress and S-nitrosylation of proteins in cells. Br. J. Pharmacol. 129: 953–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R. (1994). An intermolecular disulfide bond stabilizes E2A homodimers and is required for DNA binding at physiological temperatures. Cell 79: 1057–1067 [DOI] [PubMed] [Google Scholar]
- Bethke P.C., Gubler F., Jacobsen J.V., Jones R.L. (2004). Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta 219: 847–855 [DOI] [PubMed] [Google Scholar]
- Brandish P.E., Buechler W., Marletta M.A. (1998). Regeneration of the ferrous heme of soluble guanylate cyclase from the nitric oxide complex: Acceleration by thiols and oxyhemoglobin. Biochemistry 37: 16898–16907 [DOI] [PubMed] [Google Scholar]
- Carey J. (1991). Gel retardation. Methods Enzymol. 208: 103–117 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Delaunay A., Pflieger D., Barrault M.B., Vinh J., Toledano M.B. (2002). A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111: 471–481 [DOI] [PubMed] [Google Scholar]
- Delledonne M., Xia Y., Dixon R.A., Lamb C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Delledonne M., Zeier J., Marocco A., Lamb C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., Fobert P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15: 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C., DeLong C., Glaze S., Liu E., Fobert P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290 [PMC free article] [PubMed] [Google Scholar]
- Durner J., Klessig D.F. (1999). Nitric oxide as a signal in plants. Curr. Opin. Plant Biol. 2: 369–372 [DOI] [PubMed] [Google Scholar]
- Durner J., Wendehenne D., Klessig D.F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant W.E., Dong X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42: 185–209 [DOI] [PubMed] [Google Scholar]
- Feechan A., Kwon E., Yun B.W., Wang Y., Pallas J.A., Loake G.J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. USA 102: 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegmann J., Sandermann H., Jr. (1997). Maize glutathione-dependent formaldehyde dehydrogenase cDNA: A novel plant gene of detoxification. Plant Mol. Biol. 34: 843–854 [DOI] [PubMed] [Google Scholar]
- Fobert P.R., Despres C. (2005). Redox control of systemic acquired resistance. Curr. Opin. Plant Biol. 8: 378–382 [DOI] [PubMed] [Google Scholar]
- Garreton V., Carpinelli J., Jordana X., Holuigue L. (2002). The as-1 promoter element is an oxidative stress-responsive element and salicylic acid activates it via oxidative species. Plant Physiol. 130: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston B., et al. (1993). Endogenous nitrogen oxides and bronchodilator S-nitrosothiols in human airways. Proc. Natl. Acad. Sci. USA 90: 10957–10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston B.M., Carver J., Doctor A., Palmer L.A. (2003). S-nitrosylation signaling in cell biology. Mol. Interv. 3: 253–263 [DOI] [PubMed] [Google Scholar]
- Hausladen A., Privalle C.T., Keng T., DeAngelo J., Stamler J.S. (1996). Nitrosative stress: Activation of the transcription factor OxyR. Cell 86: 719–729 [DOI] [PubMed] [Google Scholar]
- Heil M. (2001). Induced systemic resistance (ISR) against pathogens - A promising field for ecological research. Perspect. Plant Ecol. Evol. Syst. 4: 65–79 [Google Scholar]
- Heine G.F., Hernandez J.M., Grotewold E. (2004). Two cysteines in plant R2R3 MYB domains participate in REDOX-dependent DNA binding. J. Biol. Chem. 279: 37878–37885 [DOI] [PubMed] [Google Scholar]
- Hogg N. (2000). Biological chemistry and clinical potential of S-nitrosothiols. Free Radic. Biol. Med. 28: 1478–1486 [DOI] [PubMed] [Google Scholar]
- Huang X., Kiefer E., von Rad U., Ernst D., Foissner I., Durner J. (2002). Nitric oxide burst and nitric oxide-dependent gene induction in plants. Plant Physiol. Biochem. 40: 625–631 [Google Scholar]
- Huang X., Stettmaier K., Michel C., Hutzler P., Mueller M.J., Durner J. (2004). Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 218: 938–946 [DOI] [PubMed] [Google Scholar]
- Jaffrey S.R., Erdjument-Bromage H., Ferris C.D., Tempst P., Snyder S.H. (2001). Protein S-nitrosylation: A physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3: 193–197 [DOI] [PubMed] [Google Scholar]
- Jensen D.E., Belka G.K., Du Bois G.C. (1998). S-Nitrosoglutathione is a substrate for rat alcohol dehydrogenase class III isoenzyme. Biochem. J. 331: 659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesarwani M., Yoo J., Dong X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 144: 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.O., Merchant K., Nudelman R., Beyer W.F., Jr., Keng T., DeAngelo J., Hausladen A., Stamler J.S. (2002). OxyR: A molecular code for redox-related signaling. Cell 109: 383–396 [DOI] [PubMed] [Google Scholar]
- Klessig D.F., et al. (2000). Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA 97: 8849–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge I., Gutteck-Amsler U., Zollinger M., Do K.Q. (1997). S-nitrosoglutathione in rat cerebellum: Identification and quantification by liquid chromatography-mass spectrometry. J. Neurochem. 69: 2599–2607 [DOI] [PubMed] [Google Scholar]
- Kotchoni S.O., Gachomo E.W. (2006). The reactive oxygen species network pathways:an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J. Biosci. 31: 389–404 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lee U., Wie C., Fernandez B.O., Feelisch M., Vierling E. (2008). Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20: 786–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Fliegmann J., Ebel J. (2003). Deletion of a single amino acid residue from different 4-coumarate:CoA ligases from soybean results in the generation of new substrate specificities. J. Biol. Chem. 278: 2781–2786 [DOI] [PubMed] [Google Scholar]
- Lindermayr C., Saalbach G., Bahnweg G., Durner J. (2006). Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J. Biol. Chem. 281: 4285–4291 [DOI] [PubMed] [Google Scholar]
- Liu L., Hausladen A., Zeng M., Que L., Heitman J., Stamler J.S. (2001). A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410: 490–494 [DOI] [PubMed] [Google Scholar]
- Mahoney C.W., Pak J.H., Huang K.P. (1996). Nitric oxide modification of rat brain neurogranin. Identification of the cysteine residues involved in intramolecular disulfide bridge formation using site-directed mutagenesis. J. Biol. Chem. 271: 28798–28804 [DOI] [PubMed] [Google Scholar]
- Martinez-Ruiz A., Lamas S. (2007). Signalling by NO-induced protein S-nitrosylation and S-glutathionylation: convergences and divergences. Cardiovasc. Res. 75: 220–228 [DOI] [PubMed] [Google Scholar]
- Neill S.J., Desikan R., Clarke A., Hancock J.T. (2002a). Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol. 128: 13–16 [PMC free article] [PubMed] [Google Scholar]
- Neill S.J., Desikan R., Clarke A., Hurst R.D., Hancock J.T. (2002b). Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 53: 1237–1247 [PubMed] [Google Scholar]
- Pagnussat G.C., Lanteri M.L., Lamattina L. (2003). Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiol. 132: 1241–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M.C., Sell S., Huang X., Scherf M., Werner T., Durner J., Lindermayr C. (2008). Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: A bioinformatics approach. J. Exp. Bot. 59: 177–186 [DOI] [PubMed] [Google Scholar]
- Parani M., Rudrabhatla S., Myers R., Weirich H., Smith B., Leaman D.W., Goldman S.L. (2004). Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol. J. 2: 359–366 [DOI] [PubMed] [Google Scholar]
- Pieterse C.M., Van Loon L.C. (2004). NPR1: The spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 7: 456–464 [DOI] [PubMed] [Google Scholar]
- Pieterse C.M., van Wees S.C., Hoffland E., van Pelt J.A., van Loon L.C. (1996). Systemic resistance in Arabidopsis induced by biocontrol bateria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8: 1773–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse C.M., van Wees S.C., van Pelt J.A., Knoester M., Laan R., Gerrits H., Weisbeek P.J., van Loon L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10: 1571–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polverari A., Molesini B., Pezzotti M., Buonaurio R., Marte M., Delledonne M. (2003). Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol. Plant Microbe Interact. 16: 1094–1105 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas M.C., Laxa M., Matte A., Zaninotto F., Finkemeier I., Jones A.M., Perazzolli M., Vandelle E., Dietz K.J., Delledonne M. (2007). S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell 19: 4120–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russwurm M., Koesling D. (2004). NO activation of guanylyl cyclase. EMBO J. 23: 4443–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed N., Baskaran P., Ma X., van den Akker F., Beuve A. (2007). Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proc. Natl. Acad. Sci. USA 104: 12312–12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligman K., Saviani E.E., Oliveira H.C., Pinto-Maglio C.A., Salgado I. (2008). Floral transition and nitric oxide emission during flower development in Arabidopsis thaliana is affected in nitrate reductase-deficient plants. Plant Cell Physiol. 49: 1112–1121 [DOI] [PubMed] [Google Scholar]
- Serpa V., Vernal J., Lamattina L., Grotewold E., Cassia R., Terenzi H. (2007). Inhibition of AtMYB2 DNA-binding by nitric oxide involves cysteine S-nitrosylation. Biochem. Biophys. Res. Commun. 361: 1048–1053 [DOI] [PubMed] [Google Scholar]
- Stamler J.S. (1994). Redox signaling: Nitrosylation and related target interactions of nitric oxide. Cell 78: 931–936 [DOI] [PubMed] [Google Scholar]
- Stamler J.S., Lamas S., Fang F.C. (2001). Nitrosylation. the prototypic redox-based signaling mechanism. Cell 106: 675–683 [DOI] [PubMed] [Google Scholar]
- Tada Y., Spoel S.H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., Zuo J., Dong X. (2008). Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedeschi G., Cappelletti G., Negri A., Pagliato L., Maggioni M.G., Maci R., Ronchi S. (2005). Characterization of nitroproteome in neuron-like PC12 cells differentiated with nerve growth factor: Identification of two nitration sites in alpha-tubulin. Proteomics 5: 2422–2432 [DOI] [PubMed] [Google Scholar]
- Toledano M.B., Delaunay A., Monceau L., Tacnet F. (2004). Microbial H2O2 sensors as archetypical redox signaling modules. Trends Biochem. Sci. 29: 351–357 [DOI] [PubMed] [Google Scholar]
- Tron A.E., Bertoncini C.W., Chan R.L., Gonzalez D.H. (2002). Redox regulation of plant homeodomain transcription factors. J. Biol. Chem. 277: 34800–34807 [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ohara Y., Nakayashiki H., Tosa Y., Mayama S. (2005a). Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol. Plant Microbe Interact. 18: 385–396 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yang Q., Tosa Y., Nakayashiki H., Mayama S. (2005b). Nitric oxide-overproducing transformants of Pseudomonas fluorescens with enhanced biocontrol of tomato bacterial wilt. J. Gen. Plant Pathol. 71: 33–38 [Google Scholar]
- Zhang J., Jin B., Li L., Block E.R., Patel J.M. (2005). Nitric oxide-induced persistent inhibition and nitrosylation of active site cysteine residues of mitochondrial cytochrome-c oxidase in lung endothelial cells. Am. J. Physiol. Cell Physiol. 288: C840–C849 [DOI] [PubMed] [Google Scholar]
- Zhang L., Wang Y., Zhao L., Shi S., Zhang L. (2006). Involvement of nitric oxide in light-mediated greening of barley seedlings. J. Plant Physiol. 163: 818–826 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fan W., Kinkema M., Li X., Dong X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96: 6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Hogg N. (2004). The mechanism of transmembrane S-nitrosothiol transport. Proc. Natl. Acad. Sci. USA 101: 7891–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.M., Trifa Y., Silva H., Pontier D., Lam E., Shah J., Klessig D.F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13: 191–202 [DOI] [PubMed] [Google Scholar]
- Zottini M., Costa A., De Michele R., Ruzzene M., Carimi F., Lo Schiavo F. (2007). Salicylic acid activates nitric oxide synthesis in Arabidopsis. J. Exp. Bot. 58: 1397–1405 [DOI] [PubMed] [Google Scholar]