Villin is a major regulator of actin dynamics. This article reports that Arabidopsis thaliana VILLIN5 (VLN5) is a typical member of the villin/gelsolin/framin family and retains a full suite of activities in vitro, including capping, severing, and bundling of actin filaments. Moreover, using reverse genetics, we show that VLN5 is required for the stabilization of actin filaments in the pollen tube.
Abstract
A dynamic actin cytoskeleton is essential for pollen germination and tube growth. However, the molecular mechanisms underlying the organization and turnover of the actin cytoskeleton in pollen remain poorly understood. Villin plays a key role in the formation of higher-order structures from actin filaments and in the regulation of actin dynamics in eukaryotic cells. It belongs to the villin/gelsolin/fragmin superfamily of actin binding proteins and is composed of six gelsolin-homology domains at its core and a villin headpiece domain at its C terminus. Recently, several villin family members from plants have been shown to sever, cap, and bundle actin filaments in vitro. Here, we characterized a villin isovariant, Arabidopsis thaliana VILLIN5 (VLN5), that is highly and preferentially expressed in pollen. VLN5 loss-of-function retarded pollen tube growth and sensitized actin filaments in pollen grains and tubes to latrunculin B. In vitro biochemical analyses revealed that VLN5 is a typical member of the villin family and retains a full suite of activities, including barbed-end capping, filament bundling, and calcium-dependent severing. The severing activity was confirmed with time-lapse evanescent wave microscopy of individual actin filaments in vitro. We propose that VLN5 is a major regulator of actin filament stability and turnover that functions in concert with oscillatory calcium gradients in pollen and therefore plays an integral role in pollen germination and tube growth.
INTRODUCTION
Pollen tubes extend rapidly through the style via tip growth, searching for the ovule to deliver two nonmotile sperm cells to the embryo sac to achieve double fertilization. The growth of pollen tubes is a powerful model system for studying polarized cell expansion or tip growth. This process requires an intact and dynamic actin cytoskeleton (Gibbon et al., 1999; Vidali et al., 2001; Chen et al., 2002; Cole and Fowler, 2006). The actin cytoskeleton assumes distinct distributions in the growing pollen tube. It is less abundant but highly dynamic at the extreme tip of the pollen tube; it forms a dense cortical fringe or collar of actin filaments at the subapex, and it exists as longitudinal actin cables in the shank (Hepler et al., 2001; Lovy-Wheeler et al., 2005; Chen et al., 2009). These spatially separated arrays are believed to perform distinct functions. The apical actin filaments are suggested to regulate exocytic vesicle docking and fusion (Lee and Yang, 2008). Although the function of the actin fringe is not entirely clear, it is proposed to organize vesicles that mediate endocytosis (Lovy-Wheeler et al., 2005; Cardenas et al., 2008), the corresponding region is also the location where cytoplasmic streaming reverses direction. The massive, longitudinal actin cables in the shank of the pollen tube are believed to provide molecular tracks for cytoplasmic streaming and organelle movement. However, how these diverse actin structures are generated and maintained remains poorly understood. Recently, we showed that the Arabidopsis thaliana formin 3 (AFH3) nucleates actin assembly for the generation of actin cables in the pollen tube (Ye et al., 2009). Nevertheless, how AFH3-nucleated actin filaments are subsequently organized into cables or other structures remains to be determined.
The dynamic behavior of the actin cytoskeleton is modulated by the coordinated action of a plethora of actin binding proteins (ABPs) (Staiger and Blanchoin, 2006; Pollard and Cooper, 2009; Staiger et al., 2010). Among these, several classes of ABPs bundle and cross-link actin filaments and are believed to be responsible for the formation and maintenance of higher-order actin filament structures, such as actin cables (Higaki et al., 2007; Thomas et al., 2009). Therefore, we previously proposed that bundling factors (e.g., villins, LIM domain-containing proteins [LIMs], and fimbrins) work in concert with AFH3 to organize actin filaments into actin cables and stabilize them (Ye et al., 2009).
Villin, originally purified from the core actin bundles of intestinal epithelial cell microvilli (Bretscher and Weber, 1979; Matsudaira and Burgess, 1979), is one of the major bundling factors identified in eukaryotic cells. It belongs to a protein superfamily called the villin/gelsolin/fragmin family (Walsh et al., 1984b; Friederich et al., 1989, 1990; McGough et al., 2003; Silacci et al., 2004; Su et al., 2007; Khurana and George, 2008). All of the superfamily members share a core constructed from conserved gelsolin repeats. Gelsolin contains six gelsolin-homology repeats (G1 to G6), whereas severin, fragmin, and CapG contain three gelsolin repeats (G1 to G3), and ABP29 from lily (Lilium longiflorum) pollen contains just two repeats (G1 and G2; Xiang et al., 2007). Due to the presence of an extra headpiece domain at the C terminus that contributes a second actin binding site, villin can bundle actin filaments, in addition to capping and severing actin filaments as gelsolin does. However, not all villins share this full suite of activities; for instance, Arabidopsis VLN1 and Drosophila melanogaster Quail are simple filament bundlers (Matova et al., 1999; Huang et al., 2005). The latter findings emphasize the importance of analyzing the biochemical properties of villins on a case by case basis prior to understanding their function in vivo. Villin is Ca2+ regulated in vitro and its function in cells likely depends on local Ca2+ concentration (Walsh et al., 1984a). Loss of function of villin in mice prevents actin fragmentation induced by Ca2+ and causes destruction of the brush borders, implying that villin is essential for actin rearrangements in response to stimuli (Ferrary et al., 1999). Mutations in Drosophila Quail induce female sterility and cause defects in actin bundle formation in nurse cells (Mahajan-Miklos and Cooley, 1994; Matova et al., 1999).
Villin homologs in plants were originally isolated from lily pollen by biochemical approaches (Nakayasu et al., 1998; Yokota and Shimmen, 1998; Yokota et al., 2003). One lily villin isoform, 135-ABP, binds to and bundles actin filaments in a Ca2+/calmodulin-dependent manner (Yokota et al., 2000). It was shown subsequently to nucleate actin polymerization, to cap the barbed end of actin filaments in a Ca2+/calmodulin-independent manner, and to promote actin disassembly (Yokota et al., 2005). Microinjection of a 135-ABP antibody destroys transvacuolar strands; actin filament bundles in the transvacuolar strand become thinner, confirming that villin plays a major role in stabilizing actin bundles in vivo (Tominaga et al., 2000). Moreover, it was proposed that lily villin is involved not only in bundling actin filaments in the shank of pollen tubes but also in regulating actin dynamics in the apical region in response to oscillatory, tip-high Ca2+ fluxes by capping and disassembling actin filaments (Yokota et al., 2005). Genetic and cytological evidence is urgently needed to support this model. The disassembly activity is probably due to the fragmentation of actin filaments induced by villin. However, there is only limited evidence for actin filament severing by plant villin/gelsolin/fragmin family members, and the threshold of free Ca2+ concentration required for triggering the severing activity has not been determined yet. The Arabidopsis genome contains five villin-like genes (VLN1 to VLN5; Huang et al., 2005), and several green fluorescent protein–villin fusions decorate actin filaments in vivo (Klahre et al., 2000). VLN1 is well characterized biochemically and shown to be a simple Ca2+-insensitive actin bundling protein (Huang et al., 2005). Moreover, VLN1-decorated actin filaments are resistant to depolymerization by ADF1 and latrunculin B (LatB) in vitro, implying a role for VLN1 in stabilizing actin filaments in vivo (Huang et al., 2005). However, the cellular and developmental function of Arabidopsis villins remains to be determined.
In this study, we functionally characterized a villin isovariant, VLN5, that is expressed preferentially in Arabidopsis pollen. VLN5 loss of function destabilizes actin and is associated with growth inhibition of pollen tubes. In vitro biochemical analyses show that VLN5 retains the full suite of activities of villin family members, including filament bundling, barbed-end capping, and Ca2+-dependent severing. A total internal reflection fluorescence microscopy (TIRFM) assay demonstrates the severing activity of VLN5 on individual actin filaments and confirms data from solution-based biochemical assays (Moseley et al., 2006). Moreover, severing is stimulated by physiological Ca2+ concentrations, implying that it is biologically relevant. Thus, we propose that VLN5 is a major actin filament stabilizing factor as well as a regulator of actin dynamics that functions in concert with oscillatory Ca2+ gradients and regulates pollen tube growth.
RESULTS
VLN5 Is Expressed Preferentially in Pollen
There are five villin-like genes in the Arabidopsis genome (Huang et al., 2005), named VLN1 to VLN5. VLN5 initially attracted our attention because it is expressed preferentially in pollen, based on available microarray data (https://www.genevestigator.com/gv/index.jsp; Honys and Twell, 2003; Pina et al., 2005). To analyze the molecular structure and domain composition of VLN5, we aligned its protein sequence with that of the known villin proteins. As shown in Supplemental Figure 1 online, VLN5 retains the overall architecture of a villin family member, including six gelsolin-homology domains (G1 to G6) and a second actin binding motif, the villin headpiece at the C terminus. It shares 32% amino acid sequence identity with human villin and 44% identity with 135-ABP, VLN1, and VLN2. In addition, many important amino acids are conserved in VLN5, implying that VLN5 will maintain the general biochemical activities of villin family members. For example, VLN5 contains the conserved residues for both a site 1 and site 2 Ca2+ regulation site within the G1 domain, whereas VLN1 contains just two conserved residues at site 2 (see Supplemental Figure 1 online; Huang et al., 2005). We used these data to predict that VLN5 has distinct biochemical functions when compared with VLN1, especially regarding regulation by Ca2+.
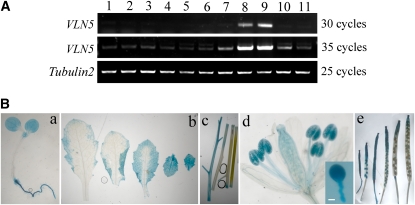
To confirm the expression pattern of VLN5 throughout the Arabidopsis plant, tissue RT-PCR and promoter-β-glucuronidase (GUS) fusions were employed. As shown in Figure 1A, transcript for VLN5 could be detected weakly in all tissues, but it was markedly more abundant in pollen and stamens (Figure 1A, lanes 8 and 9). Our observations are consistent with published microarray expression data (see Supplemental Figure 2 online). Expression patterns were further explored by determining the promoter activity using GUS as a reporter. As shown in Figure 1B, VLN5 promoter activity was detected in various organs, including whole seedling (Figure 1B, a), leaves (Figure 1B, b), stems (Figure 1B, c), flowers (Figure 1B, d), and siliques (Figure 1B, e). However, the promoter activity was highest in pollen, both within anthers and germinated in vitro (Figure 1B, d), which is consistent with the tissue RT-PCR results and the public microarray expression data (see Supplemental Figures 2E and 2F online). Taken together, these data show that VLN5 is preferentially expressed in pollen, which implies that VLN5 may be an important regulator of pollen function.
Figure 1.
VLN5 Is Expressed Preferentially in Pollen.
The expression pattern of VLN5 throughout the Arabidopsis plant was examined by RT-PCR of separated tissues and with a promoter-reporter fusion.
(A) VLN5-specific primers were used to determine tissue expression patterns by RT-PCR. Tubulin2 was used as an internal loading control. The samples are as follows: lane 1, seedling; lane 2, root; lane 3, internode; lane 4, bottom part of stem; lane 5, juvenile leaf; lane 6, adult leaf; lane 7, flower; lane 8, anther; lane 9, pollen; lane 10, silique at early stage; lane 11, silique at late stage.
(B) VLN5 promoter activity was determined using GUS as a reporter. Samples include (a) 7-d-old seedling, (b) leaves, (c) stems, (d) flower, and (e) siliques at different stages. Inset: pollen grain and germinated tube. Bar = 10 μm for the pollen tube.
VLN5 Is Required for Normal Pollen Tube Growth
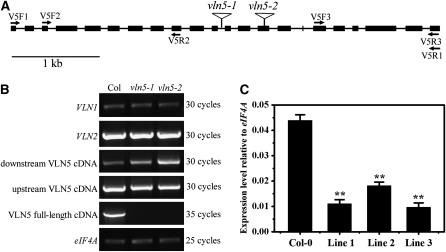
To investigate the in vivo function of VLN5, two T-DNA insertion alleles were analyzed (Figure 2A). No full-length transcript was detected for either allele in homozygous vln5-1 and vln5-2 plants; however, partial transcripts both upstream and downstream of the T-DNA insertion site were identified in both cases (Figure 2B). Based on the lack of a full-length transcript, we assumed that vln5-1 and vln5-2 were knockout lines. In a parallel approach, an RNA interference (RNAi) strategy was also adopted to study the function of VLN5. In transgenic plants carrying a VLN5 RNAi construct driven by the Lat52 promoter, VLN5 transcripts were knocked down significantly (Figure 2C). However, VLN1 and VLN2 transcripts were not reduced (see Supplemental Figure 3 online), indicating that VLN5 transcripts were knocked down specifically in these lines.
Figure 2.
Identification of Transcript Levels in T-DNA Insertion Mutants and Transgenic RNAi Lines.
(A) Physical structure of the Arabidopsis VLN5 gene. VLN5 contains 22 exons and 21 introns, which are represented by filled boxes and lines, respectively. The position of two independent T-DNA insertion mutants, designated vln5-1 (SAIL_512_F03) and vln5-2 (GABI_225F09), are noted by triangles above the diagram.
(B) Three separate pairs of primers, marked by arrows in (A), were designed to identify the level of VLN5 transcripts. The first pair of primers (V5F1 and V5R1) was designed to amplify VLN5 full-length cDNA, which was not present in vln5-1 and vln5-2 mutant plants. The second and third primer pairs, V5F2/V5R2 and V5F3/V5R3, were used to amplify upstream and downstream transcripts, respectively. Flowers from wild-type Col-0 plants and the two homozygous insertion lines were subjected to RT treatment. eIF4A was used as an internal loading control.
(C) VLN5 transcripts were reduced significantly in VLN5 RNAi flowers. Flowers from wild-type Col-0 and VLN5 RNAi plants were subjected to quantitative real-time PCR analysis. The expression level of the eIF4A gene was used as an internal control. All data represent the mean value of three biological replications. Error bars represent ± sd (n = 3); **P < 0.01.
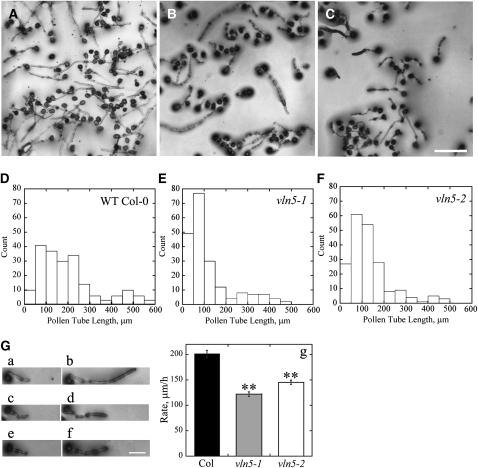
Initially, we tested whether the VLN5 knockout mutants affected pollen germination and pollen tube growth. Surprisingly, the germination rate of pollen from vln5-1 and vln5-2 homozygous plants was not different from that of wild-type Columbia-0 (Col-0) plants (see Supplemental Figure 4 online). We next determined whether vln5 mutants had defects in pollen tube elongation. After 2 h of germination on standard medium in vitro, we found that wild-type Col-0 pollen tubes (Figure 3A) were noticeably longer than vln5 mutant pollen tubes (Figures 3B and 3C). To quantify the differences between wild-type Col-0 and mutants, we measured the length of pollen tubes from numerous images. A histogram of length distributions for pollen tubes showed that the frequency of short pollen tubes increased in vln5-1 and vln5-2 samples compared with the wild type (Figures 3D to 3F). The length of vln5 pollen tubes was significantly shorter than those of wild-type Col-0 (P < 0.01); specifically, the average length (±se) of wild-type Col-0, vln5-1, and vln5-2 pollen tubes was 198.6 ± 9.5 μm, 115.7 ± 7.6 μm, and 133.5 ± 6.7 μm (n = 200), respectively. To measure the growth rate of pollen tubes, we tracked individual growing pollen tubes by light microscopy (Figure 3G, a to f). The average growth rate (±se) of pollen tubes was determined to be 201.7 ± 6.5 μm/h (n = 200), 122.0 ± 4.4 μm/h (n = 200), and 145.3 ± 4.4 μm/h (n = 200) for wild-type Col-0, vln5-1, and vln5-2 pollen tubes, respectively. In addition, pollen tube growth rate was also significantly reduced in three independent vln5 RNAi lines (see Supplemental Figure 5 online). These data suggest that VLN5 is required for normal pollen tube growth. However, no growth phenotype was detected in vln5 root hairs (see Supplemental Figure 6 online), implying that VLN5 is not required for root hair tip growth, which is consistent with its preferential expression in pollen.
Figure 3.
VLN5 Loss-of-Function Mutant Plants Have Retarded Pollen Tube Growth.
(A) to (C) Micrographs of pollen tubes after germination for 2 h in vitro. Pollen was isolated from plants with the following genotypes: wild-type Col-0 (A), homozygous vln5-1 (B), and homozygous vln5-2 (C). Bar in (C) = 100 μm.
(D) to (F) Length distribution of pollen tubes: wild-type Col-0 (D), vln5-1 (E), and vln5-2 (F).
(G) Measurement of growth rates was performed by tracking individual pollen tubes. (a) to (f) Pairs of images from single pollen tubes selected for measurement: (a) wild-type Col-0 pollen tube and (b) the same pollen tube after 30 min of growth; (c) vln5-1 pollen tube and (d) the same pollen tube after 30 min growth; (e) vln5-2 pollen tube and (f) the same pollen tube after 30 min growth. Bar = 50 μm in (f) for (a) to (f). (g) A plot of pollen tube growth rates shows that vln5 mutant pollen tubes had reduced rates compared with the wild type. Wild-type Col-0, black bar; vln5-1, gray bar; vln5-2, white bar. Error bars represent mean values ± se; n = 200. Pollen tube growth rate of vln5 pollen tubes was significantly different from that of wild-type Col-0 pollen tubes as determined by analysis of variance followed by Dunnett post-hoc multiple comparisons; **P < 0.01.
Filamentous Actin Intensity and Organization Do Not Differ between Wild-Type and vln5 Loss-of-Function Pollen Grains and Pollen Tubes
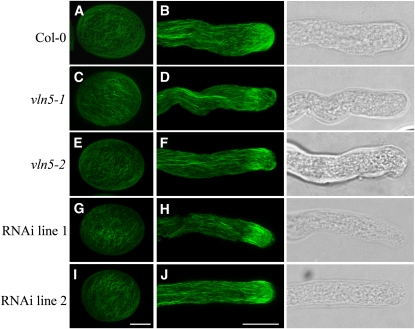
Because villins are key regulators of cytoskeletal organization and function in mammals and flies (Mahajan-Miklos and Cooley, 1994; Ferrary et al., 1999), we wanted to test whether VLN5 loss of function affected the organization of actin in pollen grains or pollen tubes of Arabidopsis. To visualize actin, pollen grains and pollen tubes from wild-type and vln5 homozygous mutant and RNAi plants were subjected to actin staining with Alexa-488 phalloidin. As shown in Figure 4, actin filaments were distributed uniformly throughout pollen grains from wild-type Col-0 (Figure 4A) and vln5 loss-of-function plants (Figures 4C, 4E, 4G, and 4I). Moreover, there was no apparent difference in actin filament intensity and organization when the wild type (Figure 4A) was compared with vln5 loss-of-function pollen (Figures 4C, 4E, 4G, and 4I).
Figure 4.
VLN5 Loss of Function Does Not Affect the Amount or Distribution of Actin Filaments in Ungerminated Pollen Grains and Pollen Tubes.
Pollen grains and pollen tubes from wild-type Col-0 and VLN5 loss-of-function mutants were subjected to actin filament staining with Alexa-488 phalloidin. More than 50 stained pollen tubes were obtained for each genotype, and typical images for each genotype, which represented >85% of the population, are presented. See Supplemental Figure 7 for more examples. The images of actin staining of pollen grains and pollen tubes are projections of confocal optical sections. Bar = 7.5 μm in (I) for pollen grains and 10 μm in (J) for pollen tubes.
(A) Wild-type Col-0 pollen grain.
(B) Wild-type Col-0 pollen tube. On the right is a transmitted light image of the corresponding pollen tube.
(C) vln5-1 pollen grain.
(D) vln5-1 pollen tube and corresponding transmitted light image.
(E) vln5-2 pollen grain.
(F) vln5-2 pollen tube and corresponding transmitted light image.
(G) VLN5 RNAi line 1 pollen grain.
(H) VLN5 RNAi line 1 pollen tube and corresponding transmitted light image.
(I) VLN5 RNAi line 2 pollen grain.
(J) VLN5 RNAi line 2 pollen tube and corresponding transmitted light image.
We also examined the organization of actin in wild-type Col-0 and vln5 loss-of-function pollen tubes. As shown in Figure 4B, normal actin arrays were organized in a distinct pattern in wild-type Col-0 pollen tubes (see Supplemental Figure 7A online). Actin filaments formed longitudinal actin cables in the shank of the pollen tube, they occurred in a dense subapical collar or fringe, and they were less abundant at the extreme apex of the pollen tube (Figure 4B). This pattern is consistent with previous reports of actin organization in pollen tubes from other species (Lovy-Wheeler et al., 2005; Ye et al., 2009). Surprisingly, in vln5 loss-of-function pollen tubes, the actin cytoskeleton maintained a normal distribution (Figures 4D, 4F, 4H, and 4J; see Supplemental Figures 7B to 7E online). In addition, there was no detectable difference in actin filament intensity between wild-type Col-0 and vln5 loss-of-function pollen tubes (see Supplemental Figure 8 online). Therefore, vln5 loss of function does not affect the organization or amount of filamentous actin in pollen grains or tubes.
vln5 Loss of Function Renders Pollen Tube Growth Hypersensitive to LatB
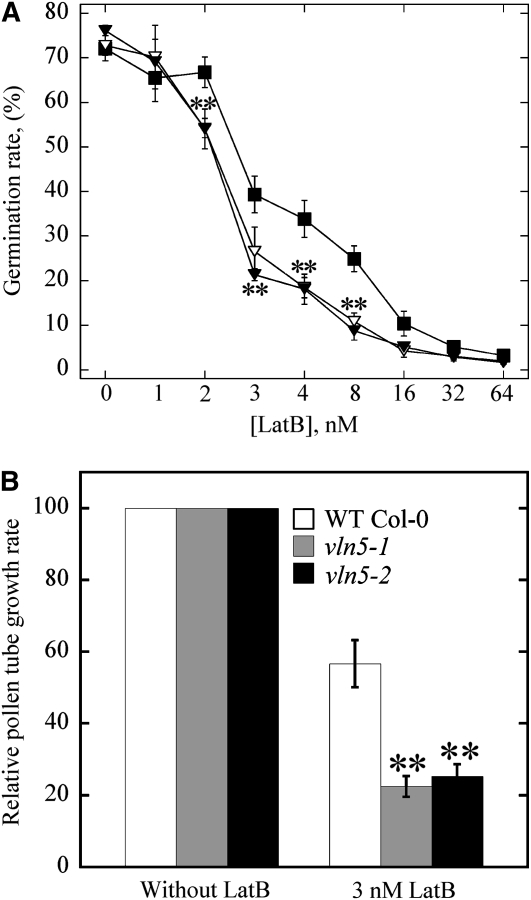
Although actin organization and the levels of filamentous actin were not grossly affected in vln5 loss-of-function pollen, it was still of interest to determine whether the organization and dynamics of actin responded to actin drugs differentially. Initially, we examined the response of pollen germination and pollen tube growth to LatB treatments. LatB prevents actin polymerization by binding with high affinity to monomeric actin (Gibbon et al., 1999). It therefore targets dynamic actin filaments arrays preferentially. To determine whether pollen germination of vln5 mutants responded to LatB differentially, pollen was germinated on medium containing various concentrations of LatB. As shown in Figure 5A, germination of pollen from vln5-1 and vln5-2 mutant plants was significantly more sensitive than pollen from wild-type Col-0 plants at 2, 3, 4, and 8 nM LatB (P < 0.01).
Figure 5.
VLN5 Loss of Function Renders Pollen Germination and Pollen Tube Growth Hypersensitive to LatB.
(A) Germination of vln5 pollen is more sensitive than wild-type pollen to perturbation of the actin cytoskeleton by LatB treatment. Pollen grains from wild-type Col-0 and vln5 plants were germinated on medium containing various concentrations of LatB. Germination was scored after pollen had germinated for 6 h. The average germination rate versus LatB concentration was plotted. Closed boxes, wild-type Col-0; closed triangles, vln5-1; open triangles, vln5-2. Error bars represent se; n ≥ 500. Asterisks denote values that were significantly different from wild-type Col-0 by χ2 test (P < 0.01).
(B) The growth of vln5-1 and vln5-2 pollen tubes was more sensitive than wild-type pollen to LatB treatment. To measure the growth rate of pollen tubes, pollen from wild-type Col-0 and vln5 mutant plants was initially germinated for 2 h in standard germination medium. Individual pollen tubes were subsequently tracked to measure the growth rate of pollen tubes. To determine the effect of LatB on growth rates, 3 nM LatB was added to the germination medium. The growth rate of pollen tubes from wild-type Col-0, vln5-1, and vln5-2 in standard germination medium was normalized to 100%, and the relative growth rate of pollen tubes in 3 nM LatB was plotted. Wild-type Col-0 grew significantly better than did vln5 mutant pollen in the presence of LatB. Error bars represent mean ± se (n > 72); **P < 0.01 (Student’s t test).
To determine whether the growth of vln5 pollen tubes was also hypersensitive to LatB treatment, pollen tubes were grown in standard germination medium in the presence or absence of LatB. Considering that half-maximal inhibition of pollen tube growth occurs at LatB concentration of 2 to 7 nM, as reported previously for lily, Tradescantia, and maize (Zea mays) pollen (Gibbon et al., 1999; Vidali et al., 2001), 3 nM LatB was used in this assay. Indeed, it was shown previously that 3 nM LatB treatment reduced pollen tube growth substantially for wild-type Arabidopsis pollen (Ye et al., 2009). As shown in Figure 5B, the presence of 3 nM LatB reduced the growth rate of vln5-1 and vln5-2 pollen tubes to 22.5 and 25.3% of that in standard germination medium, respectively. By comparison, 3 nM LatB reduced the growth rate of wild-type Col-0 pollen tubes to 56.7% of that in standard germination medium. Loss of function of VLN5 therefore increased the sensitivity of pollen tube growth to LatB treatment significantly (P < 0.01). The growth of VLN5 RNAi pollen tubes was also more sensitive to LatB treatment than that of wild-type Col-0 pollen tubes (see Supplemental Figure 9 online). Collectively, these data demonstrate that both pollen germination and pollen tube growth are significantly more prone to perturbation by LatB treatment in vln5 mutants, which implies that VLN5 may be involved in these processes via stabilizing actin filaments.
The Actin Cytoskeleton in vln5 Mutants and VLN5 RNAi Pollen Tubes Is Sensitive to Perturbation with LatB
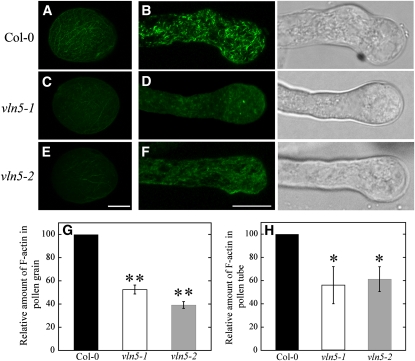
We next sought to visualize the response of the actin cytoskeleton to LatB treatment directly. To do so, actin filaments were stained with Alexa-488 phalloidin after treatment with 100 nM LatB for 30 min. As shown in Figure 6, actin filaments became markedly shorter and less abundant in both wild-type Col-0 pollen grains (Figure 6A) and wild-type Col-0 pollen tubes (Figure 6B), consistent with net depolymerization as reported quantitatively for maize (Gibbon et al., 1999) and poppy (Papaver rhoeas; Snowman et al., 2002) pollen tubes. However, actin filaments disappeared almost completely in vln5 mutant pollen grains (Figures 6C and 6E) and pollen tubes (Figure 6D and 6F) after treatment with 100 nM LatB for 30 min. Changes in average pixel intensity from fluorescence micrographs were used to assess the relative levels of actin in filamentous form in both pollen grains and pollen tubes. The amount of F-actin in both wild-type Col-0 pollen grains and wild-type Col-0 pollen tubes was normalized to 100%. The relative amount (average ± sd; n = 10) of F-actin was reduced to 52.5% ± 3.8% and 39.2% ± 2.9% of that in treated wild-type Col-0 pollen grains for vln5-1 and vln5-2 pollen grains, respectively. This suggests that wild-type Col-0 has significantly better resistance to LatB treatment than vln5-1 and vln5-2 pollen grains (P < 0.01). In addition, the relative amount (average ± sd; n ≥ 27) of F-actin was reduced to 56.1% ± 16.0% and 61.3% ± 10.7% of that in treated wild-type Col-0 pollen tubes for vln5-1 and vln5-2 pollen tubes, respectively, suggesting that VLN5 loss-of-function reduced the resistance of pollen tubes to LatB treatment significantly (P < 0.01). These results indicate that VLN5 is required for stabilizing actin filaments in both pollen grains and pollen tubes.
Figure 6.
Pollen Grains and Pollen Tubes with Reduced VLN5 Levels Have Unstable Actin Filaments.
Ungerminated pollen grains and pollen tubes treated with 100 nM LatB for 30 min were subjected to actin staining with Alexa-488 phalloidin. Transmitted light images of each pollen tube are shown to the right of the fluorescence microscope image.
(A) Wild-type Col-0 pollen grain.
(B) Wild-type Col-0 pollen tube.
(C) vln5-1 pollen grain.
(D) vln5-1 pollen tube.
(E) vln5-2 pollen grain.
(F) vln5-2 pollen tube. Bar = 7.5 μm in (E) for pollen grains and 10 μm in (F) for pollen tubes.
(G) Quantification of relative F-actin level in pollen grains treated with 100 nM LatB. The amount of F-actin in treated wild-type Col-0 pollen grains was normalized to 100%. Error bars represent ± sd (n = 10); **P < 0.01 (Student’s t test).
(H) Quantification of relative F-actin level in pollen tubes treated with 100 nM LatB. The amount of F-actin in treated wild-type Col-0 pollen tubes was normalized to 100%. Error bars represent ± sd (n > 27); *P < 0.05 (Student’s t test).
vln5 Loss of Function Renders Pollen Tube Growth Resistant to Cytochalasin D Treatment
We next sought to determine whether the growth of vln5 pollen tubes responds to other actin inhibitors differentially. Cytochalasin D (CD) alters actin dynamics by capping the barbed end of actin filaments (Cooper, 1987). Considering that pollen germination and pollen tube growth have similar sensitivity to CD treatment and that the half-maximal inhibition of pollen tube growth occurs at ~200 and 500 nM CD, as reported previously for tobacco (Nicotiana tabacum) and maize pollen (Geitmann et al., 1996; Gibbon et al., 1999), respectively, these concentrations were selected for our assays. The growth rate of pollen tubes was normalized to 100% in standard germination medium. Surprisingly, the growth of vln5 pollen tubes was more resistant to CD treatments compared with wild-type Col-0 pollen tubes. As shown in Supplemental Figure 10 online, the presence of 200 nM CD reduced the growth rate of vln5-1 and vln5-2 pollen tubes to just 91.2 and 86.6% of that in standard germination medium, respectively, and the presence of 500 nM reduced the growth rate of vln5-1 and vln5-2 pollen tubes to 49.0 and 46.3% of that in standard germination medium, respectively. By comparison, the presence of 200 and 500 nM CD reduced the pollen tube growth rate of wild-type Col-0 pollen tubes to 58.7 and 14.5% of that in standard germination medium, respectively. In summary, VLN5 loss of function leads to pollen tube growth that is resistant to CD treatments. In other words, vln5 loss of function is partially rescued by treatment with a filament capping agent.
Recombinant VLN5 Binds to and Bundles Actin Filaments in Vitro
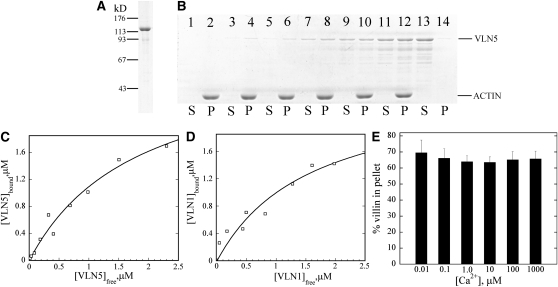
Given that VLN5 appears to be required for the stability of actin filaments in pollen and for normal pollen tube growth, we investigated the molecular basis for VLN5 regulation of actin dynamics in vitro. Recombinant VLN5 was expressed in Escherichia coli as a nonfusion protein and purified to near homogeneity using a combination of ammonium sulfate precipitation and several chromatographic fractionation steps (Figure 7A). The ability of purified VLN5 to bind actin filaments was determined with a high-speed cosedimentation assay. As shown in Figure 7B, VLN5 bound to actin filaments in a dose-dependent manner. To determine the equilibrium dissociation constant (Kd) value for this interaction, VLN5 in the pellet ([VLN5]bound) was plotted as a function of VLN5 in the supernatant ([VLN5]free) and the resulting data fitted with a hyperbolic function. In a representative experiment shown in Figure 7C, the Kd was determined to be 0.55 μM. A mean Kd value (±sd) of 0.6 ± 0.1 μM was determined for the binding of VLN5 to actin filaments from three independent experiments. By comparison, the Kd value was determined to be 0.7 ± 0.3 μM (n = 3) for the well-characterized VLN1 (Figure 7D), which is close to the published value (Huang et al., 2005). The Kd for actin filament side binding is also very close to that of human villin, which has a Kd value of 4.4 μM for the wild-type form and 0.6 μM for the phosphorylated form (Zhai et al., 2001).
Figure 7.
VLN5 Binds to Actin Filaments with High Affinity.
(A) Recombinant VLN5 expressed in bacteria was purified to homogeneity with several chromatography steps. Three micrograms of purified protein separated on an SDS-PAGE gel and stained with Coomassie blue is shown.
(B) A high-speed cosedimentation assay was employed to determine the affinity of VLN5 for actin filaments. Three micromolar polymerized actin was incubated with various concentrations of VLN5 and the mixtures were centrifuged at 200,000g for 1 h to separate bound versus unbound VLN5. After centrifugation, equal amounts of pellets (P) and supernatants (S) were separated by SDS-PAGE and stained with Coomassie blue. Samples loaded in lanes 1 to 12 contained actin plus 0 (lanes 1 and 2), 0.1 (lanes 3 and 4), 0.2 (lanes 5 and 6), 0.5 (lanes 7 and 8), 1 (lanes 9 and 10), and 2 μM (lanes 11 and 12) VLN5, whereas lanes 13 and 14 contained 2 μM VLN5 only.
(C) Actin filaments were sedimented in the presence of various concentrations of VLN5, as shown in (B). The amount of VLN5 in pellets and supernatants from the various samples was determined with Image J. The concentration of VLN5 in the pellet ([VLN5]bound) as a function of the concentration of VLN5 in the supernatant ([VLN5]free) was plotted, and the data were fitted with a hyperbolic function to calculate the equilibrium dissociation constant (Kd) value. In this representative experiment, the calculated Kd value was 0.55 μM.
(D) The Kd value for VLN1, used as a positive control for binding experiments, was determined to be 0.63 μM in this representative experiment.
(E) To determine the effects of Ca2+ on VLN5 binding to actin filaments, 1 μM VLN5 was incubated with 3 μM F-actin in the presence of various [Ca2+]free. The percentage of VLN5 that sedimented with actin filaments was plotted as a function of [Ca2+] free. Error bars represent ±sd (n = 3).
Like VLN1 (Huang et al., 2005) and VLN3 (see companion paper, Khurana et al., 2010), VLN5 binding to actin filaments was calcium insensitive over 5 orders of magnitude in free Ca2+ concentration (Figure 7E). Given that the binding of 135-ABP to actin filaments was shown to be regulated by Ca2+/calmodulin (Yokota et al., 2000), we sought to determine whether the binding of VLN5 to actin filaments is regulated by Ca2+/calmodulin as well. Indeed, the amount of VLN5 in the pellet was modestly but significantly reduced in the presence of 50 μM calmodulin and 1 mM free Ca2+ (see Supplemental Figure 11 online, lane 6; P < 0.05), suggesting that the binding of VLN5 to actin filaments is regulated by Ca2+/calmodulin.
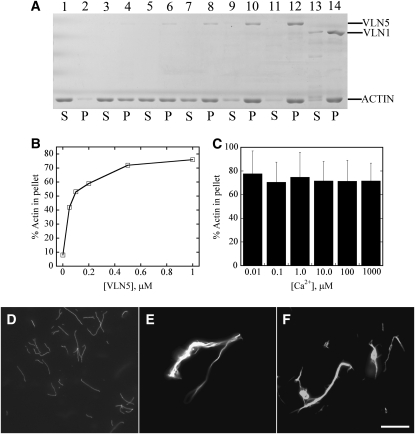
Most villins characterized to date exhibit actin filament bundling activity, with the exception of a prototype villin (Hofmann et al., 1992). Indeed, bundling is the only villin-like property maintained by Arabidopsis VLN1 (Huang et al., 2005). Therefore, it is reasonable to expect that VLN5 will bundle actin filaments in vitro. To test this, a low-speed cosedimentation assay was employed. As shown in Figure 8, the presence of VLN5 increased the amount of sedimentable actin in a dose-dependent manner (Figures 8A and 8B). Because actin alone does not sediment appreciably under these conditions (Figure 8A, lane 2), this suggests that VLN5 has bundling activity, and the effect was similar to that of VLN1 (Figure 8A, lane 14; Huang et al., 2005). Moreover, the bundling activity of VLN5 was Ca2+ insensitive over 5 orders of magnitude in free Ca2+ concentration (Figure 8C). To confirm the bundling activity of VLN5, actin filaments in the presence and absence of villin were visualized directly by fluorescence light microscopy after staining with rhodamine-phalloidin. Actin filaments appeared to be individual structures in the absence of villin (Figure 8D), whereas in the presence of 1 μM VLN5 they were organized into massive bundles (Figure 8E). This effect was similar to that observed for VLN1 (Figure 8F; Huang et al., 2005). These results demonstrate that VLN5 bundles actin filaments in a Ca2+-insensitive manner.
Figure 8.
VLN5 Bundles Actin Filaments in a Calcium-Insensitive Manner.
A low-speed cosedimentation assay and fluorescence microscopy were employed to determine the bundling activity of VLN5.
(A) VLN5 bundles actin filaments in a dose-dependent manner. Reactions containing actin alone, actin plus various concentrations of VLN5, or actin plus 1 μM VLN1 were incubated for 30 min and then sedimented at 13,600g for 30 min. Samples for the supernatant (S) and pellet (P) were separated by SDS-PAGE; lanes 1, 3, 5, 7, 9, 11, and 13 represent supernatant of actin alone, actin plus 50 nM VLN5, actin plus 100 nM VLN5, actin plus 200 nM VLN5, actin plus 500 nM VLN5, actin plus 1 μM VLN5, and actin plus 1 μM VLN1, respectively. Samples in lanes 2, 4, 6, 8, 10, 12, and 14 represent pellet of actin alone, actin plus 50 nM VLN5, actin plus 100 nM VLN5, actin plus 200 nM VLN5, actin plus 500 nM VLN5, actin plus 1 μM VLN5, and actin plus 1 μM VLN1, respectively.
(B) Percentage of actin sedimented in the low-speed assay as a function of the concentration of VLN5.
(C) VLN5 bundles actin filaments in a calcium-insensitive manner. To determine whether the bundling activity of VLN5 is regulated by calcium, actin filaments were incubated with 1 μM VLN5 in the presence of various [Ca2+]free. The amounts of actin in the pellet were determined in the presence of different [Ca2+]free. The percentage of actin sedimented at 10 nM Ca2+ was 78%, at 100 nM Ca2+ was 71%, at 1 μM Ca2+ was 75%, at 10 μM Ca2+ was 72%, at 100 μM Ca2+ was 71%, and at 1 mM Ca2+ was 72%. Three independent experiments were conducted; error bars represent ±sd.
(D) to (F) Micrographs of actin filaments in the presence or absence of villin stained with rhodamine phalloidin.
(D) Individual actin filaments in the absence of villin. The image was captured at a 500-ms exposure time.
(E) Actin filament bundles formed in the presence of 1 μM VLN5. The image was captured at a 150-ms exposure time.
(F) Actin bundles formed in the presence of 1 μM VLN1. The image was captured at a 150-ms exposure time. Bar = 10 μm.
It was reported previously that the bundling activity of 135-ABP is reduced or inhibited by Ca2+/calmodulin (Yokota et al., 2000); therefore, we sought to determine whether this type of regulation is conserved for VLN5. Initially, the low-speed cosedimentation assay was employed. The results showed that the presence of 50 μM CaM and 1 mM free Ca2+ significantly decreased (P < 0.05) the amount of actin in the pellet (see Supplemental Figure 12A, lane 6, and Supplemental Figure 12B online) compared with that for VLN5 alone (see Supplemental Figure 12A, lane 4, and Supplemental Figure 11B online), which suggests that Ca2+/CaM inhibits the bundling activity of VLN5. The effect of Ca2+/CaM on the bundling activity of VLN5 was confirmed by direct visualization of actin filaments under a fluorescence light microscope. Actin filaments behaved as individual filaments in the absence of VLN5 (see Supplemental Figure 12C online), whereas actin filaments were organized into numerous, prominent bundles in the presence of 500 nM VLN5 (see Supplemental Figure 12D online), consistent with the previous observations (Figure 8E). However, the actin bundle structures were smaller and less abundant in the presence of Ca2+/CaM, and more individual actin filaments appeared in each field (see Supplemental Figure 12E online). Collectively, these results suggest that regulation of the bundling activity of VLN5 by Ca2+/CaM is conserved for VLN5.
VLN5 Caps the Barbed End of Actin Filaments
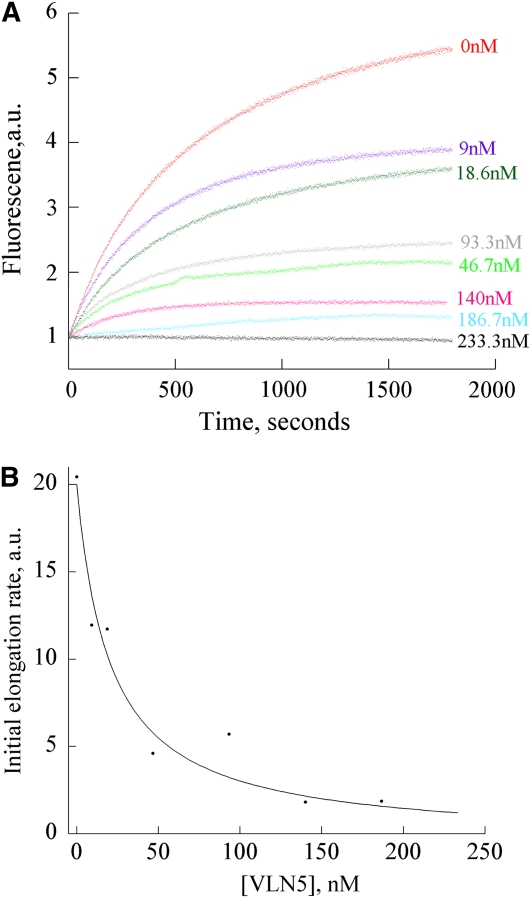
Most villins bind to the barbed end of actin filaments with high affinity and prevent subunit addition and loss (Glenney et al., 1981; Walsh et al., 1984b); this is known as capping activity. To determine whether VLN5 caps the barbed end of actin filaments, a seeded actin elongation assay was performed (Huang et al., 2003). In these assays, profilin-actin is used to suppress nucleation and the addition of actin to the pointed ends of filaments but permits addition at barbed ends unless they are capped. As shown in Figure 9A, VLN5 decreased the initial rate of actin elongation in a dose-dependent manner, confirming its barbed end capping activity. A representative Kd value of 20.4 nM was estimated by fitting the data to Equation 1 (Figure 9B; see Methods). From three independent experiments, a mean Kd value (±sd) of 15.8 ± 5.2 nM was calculated for VLN5 capping. To determine whether the capping activity of VLN5 is regulated by Ca2+, we performed the elongation assay in the presence of 10 μM free Ca2+. A mean Kd value (±sd) of 5.5 ± 1.6 nM was calculated for VLN5 capping, which suggested that VLN5 has a higher affinity for the barbed end of actin filaments in the presence of 10 μM Ca2+. However, there is no order of magnitude difference in the Kd value compared with that at 10 nM Ca2+. The affinity for the capping activity of VLN5 is low compared with that of human villin, which has a Kd value of 0.19 nM (Walsh et al., 1984b), but in the same range as Arabidopsis heterodimeric capping protein (Huang et al., 2003). This suggests that VLN5 binds to the barbed end of actin filaments with moderate affinity and is different from VLN1, which lacks capping activity altogether (Huang et al., 2005).
Figure 9.
VLN5 Caps the Barbed Ends of Actin Filaments.
(A) Preformed F-actin seeds (0.8 μM) were incubated with different concentrations of VLN5, and 1 μM G-actin, saturated with 4 μM human profilin I, was added to initiate actin elongation at the barbed end. Polymerization was monitored by tracking the increase in pyrene-actin fluorescence upon assembly. Concentrations of VLN5, from top to bottom, were 0, 9, 18.6, 93.3, 46.7, 140, 186.7, and 233 nM. A single representative experiment (n = 3) is shown.
(B) Initial rates of elongation as a function of VLN5 concentration were plotted for the representative experiment shown in (A). The data were fit with Equation 1 (see Methods) to determine an apparent Kd value of 20.4 nM for VLN5 binding to filament barbed ends.
VLN5 Stabilizes Actin Filaments at Low [Ca2+] but Depolymerizes Actin Filaments in the Presence of Physiological [Ca2+]
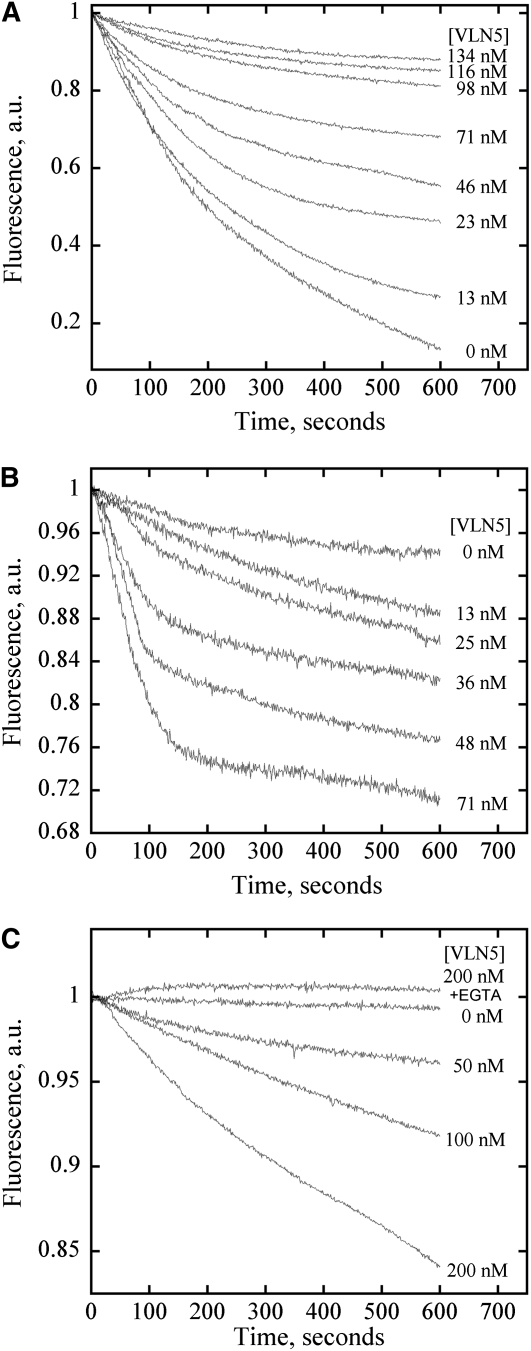
Given that VLN5 binds to and bundles actin filaments and caps the barbed end of actin filaments, we sought to determine whether VLN5 stabilizes actin filaments against depolymerization in vitro. To test this, a dilution-mediated actin depolymerization assay was performed. In this assay, actin filaments were diluted in low ionic strength solution and actin depolymerization could occur from both pointed and barbed ends of actin filaments. As shown in Figure 10A, VLN5 inhibited actin depolymerization in a dose-dependent fashion in the presence of 10 nM free Ca2+, suggesting that VLN5 protects actin filaments from depolymerization. This could be due to the side binding and/or capping activity. Considering that most villin family proteins are reported to be calcium responsive, we decided to determine whether VLN5 exerts different effects on actin filaments under high [Ca2+]. In this assay, actin filaments were diluted into polymerization buffer. As shown in Figure 10B, VLN5 enhanced dilution-mediated actin depolymerization in a dose-dependent manner in the presence of 10 μM free Ca2+, suggesting that VLN5 acts on actin filaments differently under physiological versus low Ca2+ conditions. The difference here could be due to the activation of severing activity. Therefore, the interpretation of the results above could be that VLN5 severed actin filaments to generate more pointed ends, consequently enhancing actin depolymerization. Indeed, this assay was used to quantify the severing activity between different villin-related proteins (Revenu et al., 2007). It has been reported that actin is buffered with equimolar profilin in pollen tubes (Vidali and Hepler, 1997; Gibbon et al., 1999; Snowman et al., 2002). Therefore, to mimic the situation in vivo, we tested the effect of VLN5 on profilin-mediated actin depolymerization. As shown in Figure 10C, VLN5 prevented actin depolymerization in the presence of 10 nM free Ca2+, whereas it enhanced actin depolymerization in a dose-dependent manner in the presence of 10 μM free Ca2+. Taken together, these data suggest that VLN5 stabilizes actin filaments in the presence of 10 nM free Ca2+ but promotes actin depolymerization in the presence of 10 μM free Ca2+.
Figure 10.
VLN5 Stabilizes Actin Filaments in the Presence of 10 nM Free Ca2+ but Enhances Actin Depolymerization in the Presence of 10 μM Ca2+.
(A) VLN5 protects actin filaments from dilution-mediated depolymerization in the presence of 10 nM free Ca2+. In this assay, the amount of F-actin is proportional to pyrene fluorescence. Five micromolar F-actin (100% pyrene labeled) was incubated with various concentrations of VLN5 for 5 min at room temperature and diluted 25-fold into low-ionic strength Buffer G in the presence of 10 nM Ca2+. Under these conditions, actin depolymerization could occur at both pointed and barbed ends of actin filaments. However, the loss of subunits from barbed ends of actin filaments contributes mainly to depolymerization, which was blocked by capping of VLN5. Therefore, VLN5 prevents actin depolymerization in a dose-dependent manner.
(B) VLN5 enhances actin depolymerization following dilution in the presence of 10 μM Ca2+. Five micromolar F-actin (50% pyrene labeled) was incubated with various concentrations of VLN5 for 5 min at room temperature and diluted 50-fold in polymerization solution (1× KMEI) in the presence of 10 μM Ca2+. Given that actin depolymerization was induced in polymerization solution and that the final actin concentration drops to the value close to the critical concentration at the barbed end of actin filaments, actin depolymerization mainly occurs at the pointed end and is slow in the absence of VLN5. However, the addition of VLN5 may induce the severing of actin filaments, which increases the number of actin filaments. Although the newly generated barbed ends of actin filaments were capped by VLN5, the increased number of pointed ends would enhance actin depolymerization.
(C) VLN5 stabilizes actin filaments from profilin-mediated actin depolymerization in the presence of 2 mM EGTA but enhances profilin-mediated actin depolymerization in the presence of 10 μM free Ca2+.
VLN5 Severs Actin Filaments
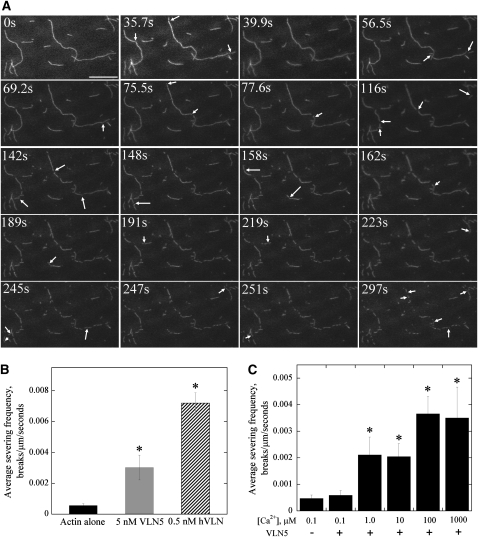
Most villins, with the exception of Arabidopsis VLN1 and Drosophila Quail, are able to sever actin filaments. To determine whether VLN5 severs actin filaments, a TIRFM assay was employed to track the behavior of individual actin filaments over time. As shown in Figure 11, VLN5 (5 nM) resulted in the shortening of initially long single actin filaments in the presence of 1 μM free calcium. Careful observations established that this was due to fragmentation or severing of actin filaments and that the number of breaks in a field increased over time (see Supplemental Movies 1 and 2 online). In the absence of VLN5, few breaks were observed (see Supplemental Movie 3 online), demonstrating that the experimental system or photodamage had minimal effects on the actin filaments. To quantify the severing activity of villins, the average severing frequency was calculated as the number of breaks, per unit length of actin filament, per second (i.e., breaks/μm/s; Andrianantoandro and Pollard, 2006). As shown in Figure 11B, the average severing frequency of VLN5 at 5 nM was 0.003 ± 0.0008 breaks/μm/s, which is significantly higher than that for actin alone (0.0006 ± 0.0001 breaks/μm/s). However, it was less potent than that of human villin at 0.5 nM, which resulted in an average severing frequency of 0.0072 ± 0.0007 breaks/μm/s (Figure 11B; see Supplemental Figure 13 and Supplemental Movie 4 online). To determine whether the severing frequency of VLN5 is dependent on calcium, VLN5 at 5 nM was incubated with actin filaments in the presence of various concentrations of free calcium. No severing was observed in the presence of 100 nM free calcium and 5 nM VLN5 (see Supplemental Figure 13B and Supplemental Movie 5 online), but actin filaments were severed efficiently in the presence of 10 μM, 100 μM, and 1 mM free calcium (see Supplemental Figures 13C to 13E and Supplemental Movies 6 to 8 online). To compare the severing frequency of VLN5 in the presence of various concentrations of free calcium, the average severing frequency of VLN5 was quantified. As shown in Figure 11C, the severing activity of VLN5 was only triggered after free calcium was elevated to 1 μM. These results demonstrated that VLN5 can sever F-actin in vitro in a calcium-dependent manner.
Figure 11.
Direct Visualization of VLN5 Severing Activity by Time-Lapse TIRFM.
(A) Prepolymerized, 25 nM rhodamine-labeled actin filaments were introduced into a perfusion chamber, where they were attached to the coverglass by NEM-myosin. Five nanomolar VLN5 was injected into the perfusion chamber in the presence of 1 μM free Ca2+. Individual actin filaments showed an increasing number of breaks (arrows) as time elapsed. Bar = 20 μm. See Supplemental Movie 1 online for the entire series.
(B) The severing activity of VLN5 is less potent than that of human villin (hVLN). Either 5 nM VLN5 or 0.5 nM hVLN was perfused into a chamber containing 25 nM actin filaments in the presence of 1 μM free Ca2+. The average severing frequency was plotted in the presence or absence of villins. Error bars represent ± se (n = 15). Asterisks represent values that were significantly different from that of the no villin control (*P < 0.05 by a Student’s t test).
(C) The severing activity of VLN5 is Ca2+ dependent. Five nanomolar VLN5 in the presence of various concentrations of Ca2+ was perfused into the perfusion chamber and time-lapse images were collected. At least 15 filaments for each experimental treatment were selected to calculate the average severing frequency, and the average severing frequency was plotted against [Ca2+]. Error bars represent ± se; n = 3 for each data set. Asterisks represent values that were significantly different from that of the no villin control (*P < 0.05 by a Student’s t test).
DISCUSSION
How distinct actin filament structures in pollen tubes are generated, maintained, and turned over remains an open question. To this end, we functionally characterized a new Arabidopsis villin isovariant, VLN5, that is preferentially expressed in pollen. In vitro biochemical analyses indicate that VLN5 is a typical villin family member; it retains the full suite of activities on actin, including barbed-end capping, filament bundling, Ca2+-dependent severing, and Ca2+/CaM inhibition of bundling. Loss of function of VLN5 correlates with the instability of actin filaments in pollen tubes and grains, confirming its role as a stabilizer of actin filaments in vivo. Free Ca2+ concentrations that are within the range of the tip-focused oscillatory calcium gradient (Holdaway-Clarke et al., 1997; Messerli et al., 2000) are sufficient to trigger VLN5 severing activity in vitro, implying that VLN5 could be involved in regulating actin dynamics in the apical region of pollen tubes by severing actin filaments.
VLN5 Retains a Full Suite of Villin Family Activities
Although the villins from Arabidopsis are reasonably well conserved in terms of overall domain organization and amino acid sequence composition, the activity of each isovariant can vary greatly, as we showed previously for VLN1 (Huang et al., 2005) and as demonstrated in the companion paper for VLN3 (Khurana et al., 2010). These results urge us to analyze the full activities of each protein isovariant in detail, before assuming their potential functions in vivo. A combination of low-speed cosedimentation assays and direct fluorescence light microscopy visualization of actin filament arrays was employed to determine whether VLN5 bundles actin filaments. The results demonstrated that VLN5 binds to and bundles actin filaments (Figures 7 and 8). This implies that VLN5 may be involved in organizing actin filaments into longitudinal bundles and stabilizing actin filaments in the pollen tube. Indeed, VLN5 was shown to stabilize actin filaments from dilution- and profilin-mediated actin depolymerization under low [Ca2+] conditions (Figures 10A and 10C), confirming that VLN5 does indeed stabilize actin filaments. It was shown recently that dimer formation is necessary for the bundling activity of human villin (George et al., 2007), which provides a different view of the requirement for the bundling activity of villin and gives insight into the molecular mechanisms underlying the bundling activity of villin. Therefore, it is important to determine whether this is the case for VLN5 or other plant villin-related proteins in general. Interestingly, VLN1 and VLN3 do not form dimers in vitro, yet they bind and bundle actin filaments with high affinity (Khurana et al., 2010), suggesting that the mechanisms underlying bundling and stabilization may not be universal. In addition, the binding and bundling activity of VLN5 is regulated by Ca2+/CaM (see Supplemental Figures 11 and 12 online), which is consistent with a previous report that the activity of ABP-135 is regulated by Ca2+/CaM (Yokota et al., 2000). This property further distinguishes VLN5 from VLN1 (Huang et al., 2005).
Capping of the filament barbed ends by certain ABPs, like the heterodimeric capping protein Arabidopsis CP (Huang et al., 2003, 2006), stabilizes actin filaments by preventing subunit loss or addition at that end. To determine whether VLN5 caps actin filaments, a seeded actin elongation assay was employed. The results demonstrate that VLN5 binds to the barbed end of actin filaments and prevents the addition of actin and profilin-actin complex, confirming its capping activity (Figure 9). The substoichiometric amounts of VLN5 sufficient to achieve this effect are consistent with high-affinity binding and capping of filament barbed ends. Indeed, an apparent Kd for barbed ends was calculated to be 16 nM for VLN5. This value is higher than that for human villin (Walsh et al., 1984b) but similar to that reported for Arabidopsis CP and Papaver rhoeas ABP80 (Huang et al., 2003, 2004, 2006). This feature of VLN5 will allow it to prevent subunit addition and loss at the barbed ends of actin filaments in vivo, thereby allowing the maintenance of a large actin-profilin pool and stabilization of the existing actin filaments in the pollen tube. Indeed, VLN5 stabilizes actin filaments from profilin-induced actin depolymerization in the presence of 10 nM free calcium (Figure 10C). Additionally, growth of vln5 pollen tubes is less sensitive to the actin capping reagent, CD, than is that of wild-type Col-0 pollen tubes (see Supplemental Figure 10 online), which implies that the capping activity of VLN5 could be biologically relevant.
Several pieces of evidence indicate that VLN5 can sever actin filaments. In solution-based biochemical assays, VLN5 enhances dilution- and profilin-mediated actin depolymerization in the presence of 10 μM free Ca2+ (Figures 10B and 10C). This effect could result from monomer binding, severing, or a combination of these activities. The most convincing and direct evidence for severing comes from time-lapse TIRFM movies of the behavior of individual filaments in the presence of VLN5 (Figure 11; see Supplemental Movies 3 to 7 online). After incubation with VLN5 in the presence of 1 μM Ca2+, numerous breaks developed along rhodamine-actin filaments. This provides evidence for the severing activity of a plant villin. Similar data for VLN3 show that it is also capable of severing actin filaments in a Ca2+-dependent manner, albeit VLN3 is quantitatively less potent than human villin (Khurana et al., 2010). Indeed, VLN5 is also quantitatively less potent than human villin (Figure 11B), but its activity is similar to that of VLN3 (Khurana et al., 2010).
In summary, our biochemical results suggest that VLN5 retains a full suite of activities on actin, similar to those reported previously for human villin (Walsh et al., 1984b), but markedly different from those of VLN1 (Huang et al., 2005). This could be due to the difference of amino acids at Ca2+ regulation sites. Indeed, compared with VLN1, VLN5 retains all conserved residues for site 1 and site 2 Ca2+ regulation sites within the G1 domain (see Supplemental Figure 1 online; Huang et al., 2005). The significance of these putative Ca2+ regulation sites could be verified by mutagenesis in the future.
VLN5 Is a Major Player in Stabilizing Actin Filaments in the Pollen Tube
Pollen tube growth is one of the best-characterized actin-based cell elongation systems. Actin filaments form distinct arrays that have a polarized distribution throughout the pollen tube (Hepler et al., 2001; Cheung and Wu, 2008; Chen et al., 2009). How these actin arrays are generated, maintained, and turned over remains an open question. Considering that VLN5 bundles and caps actin filaments, it is reasonable to speculate that VLN5 participates in stabilizing actin filaments and maintaining actin arrays in the pollen tube. Indeed, compared with wild-type Col-0 pollen tubes, actin filaments in the shank of vln5 pollen tubes become unstable after being subjected to LatB treatment, indicating that VLN5 is indeed required for stabilizing actin filaments in the pollen tube. A deficiency in VLN5 levels may increase the action of other depolymerizing factors, for instance, ADF. Indeed, it was shown that VLN1-decorated actin filaments are resistant to the depolymerizing effect of ADF1 (Huang et al., 2005). In a companion study, VLN1 also protected actin filaments against the severing activity of ADF1 (Khurana et al., 2010). Therefore, it is reasonable to speculate that decoration of actin filaments with VLN5 will prevent the action of ADF in the shank of the pollen tube. However, we could not detect any major perturbations in actin distribution when vln5 and VLN5 RNAi pollen tubes (Figures 4D, 4F, 4H, and 4J) were compared with wild-type pollen. One possibility is that VLN5 is functionally redundant with other villin isoforms expressed in pollen. The likely candidates are VLN2 and VLN1, since they are both expressed in pollen, according to the microarray data (https://www.genevestigator.com/gv/index.jsp; Honys and Twell, 2003; Pina et al., 2005), although their expression is less abundant than VLN5.
We also cannot rule out the possibility that VLN5 functions overlap with other bundling factors, for instance, fimbrins, LIMs and formins. Indeed, Arabidopsis FIMBRIN1 was shown to be a major cross-linker and stabilized actin from profilin-induced depolymerization in vitro and in vivo (Kovar et al., 2000). Therefore, the pollen-expressed fimbrins (i.e., FIM5, FIM4, and FIM3; Pina et al., 2005) may functionally overlap with VLN5. Recombinant WLIM1 was also shown to bind to and bundle actin filaments and stabilize actin filaments against depolymerization (Thomas et al., 2006), and overexpression of WLIM1 induces massive actin cable formation in Nicotiana benthamiana leaves (Thomas et al., 2007). Recently, L. longiflorum LIM1, which is enriched in pollen, was also shown to bind to and bundle actin filaments and stabilize actin filaments against LatB-mediated actin depolymerization in vitro (Wang et al., 2008). Therefore, pollen-expressed LIM domain–containing proteins may function similarly in Arabidopsis. Finally, a specific Arabidopsis formin, AFH1, was shown to bundle actin filaments directly (Michelot et al., 2005, 2006). Therefore, some pollen-expressed formin with bundling activity may have functions that overlap with VLN5. Genetic analyses and direct biochemical measurements will be needed to test this hypothesis. In addition, actin dynamics (e.g., actin filament severing frequency) might be functionally altered, although no defects in overall organization were detected in vln5 pollen tubes. Indeed, loss of function of VLN5 renders pollen tube growth resistant to CD treatment, supporting this speculation (see Supplemental Figure 10 online). However, measuring dynamic parameters of the actin cytoskeleton directly in vln5 pollen tubes will provide insight into this in the future. This should be possible with live-cell actin markers (e.g., GFP-ABD2; Sheahan et al., 2004; Vidali et al., 2009). Indeed, Staiger et al. (2009) visualized the behavior of cortical actin in epidermal cells directly and found that severing activity dominates the dynamic behavior of individual actin filaments. It will be of substantial interest to test whether villin isoforms are responsible for the severing events of actin filaments under calcium stimulating conditions.
Based on the biochemical properties of recombinant VLN5, and considering the existence of oscillatory calcium gradients in the pollen tube, it is plausible to predict that VLN5 binds to and stabilizes actin cables in the shank of pollen tubes by bundling and capping actin filaments and regulates actin dynamics in the apical region in response to tip-high Ca2+ fluxes by capping and severing actin filaments, as previously predicted for the potential function of 135-ABP (Yokota et al., 2005). Certainly, more cytological evidence is needed to support this model.
In summary, we show here that VLN5 is a versatile molecule, with activities including capping, bundling, and severing of actin filaments. VLN5 loss-of-function destabilizes actin filaments and correlates with the inhibition of pollen tube growth. We propose that VLN5 is a major regulator of actin stability as well as dynamics and functions in concert with oscillatory calcium gradients in the pollen tubes. However, further cytological and genetic analyses are certainly needed to gain insight into the function of VLN5.
METHODS
RT-PCR Analysis
Total RNA was isolated from inflorescence tissues of wild-type, VLN5 T-DNA insertion mutants (SAIL_512_F03 and GABI_225F09), and VLN5 RNAi lines with TRIzol reagent (Invitrogen). For cDNA synthesis, 2 μg of total RNA from different tissues was used for reverse transcription with MMLV reverse transcriptase (Promega) according to the manufacturer’s recommendations. To confirm the expression level of VLN5, 1 μL reaction product was used as template for PCR analysis to amplify cDNA fragments of VLN5 with specific primers (see Supplemental Table 1 online). eIF4A was used as an internal control with specific primers (see Supplemental Table 1 online) to amplify an ~0.5-kb DNA fragment. PCR products were checked by 1% agarose gel electrophoresis.
To perform quantitative real-time PCR, cDNA was synthesized using the M-MLV reverse transcription reagents with oligo-d(T)18 according to the manufacturer’s instructions (Promega). Quantitative real-time PCR was performed with specific primers qVLN5F and qVLN5R (see Supplemental Table 1 online) using the Rotor Gene3000 sequence detection system (Corbett Research) according to the manufacturer’s instructions in a 20-μL reaction volume containing SYBR Green dye chemistry (TaKaRa). The amplicon size was 140 bp. The level of eIF4A transcript amplified with primers qeIF4AF and qeIF4AR was used as an internal control. The relative transcript of the VLN5 gene was calculated using the 2ΔCt method, in which ΔCt = Ct (eIF4A) – Ct (VLN5).
GUS Staining
The promoter of VLN5 (1091 bp upstream from the ATG) was amplified with primers shown in Supplemental Table 1 online and was fused with GUS and transformed into wild-type plants. T3 plants were subjected to GUS staining, which was performed as described by Jefferson et al. (1987); the samples were cleared as described by Malamy and Benfey (1997). To analyze the expression pattern of VLN5, five plants each from nine independent T3 lines were selected. The images of GUS staining were taken with a Leica DM4000 microscope equipped with Leica Application Suite software.
Pollen Germination and Pollen Tube Growth Measurement
In vitro pollen germination was performed essentially according to Ye et al. (2009). Pollen was harvested from newly opened flowers and placed onto pollen germination medium consisting of 1 mM CaCl2, 1 mM Ca(NO3)2, 1 mM MgSO4, 0.01% (w/v) H3BO3, and 18% (w/v) sucrose solidified with 0.8% (w/v) agar, pH 7.0. The plates were cultured at 28°C under high humidity. To determine the effects of LatB (Calbiochem) on pollen germination, various concentrations of LatB were included in the germination medium. To determine the effects of LatB on pollen tube growth, liquid germination medium with or without LatB was added after the average length of pollen tubes grew up to ~150 μm in standard germination medium. Micrographs of pollen grains and pollen tubes were collected with an IX71 microscope equipped with a ×10 objective, and digital images were acquired with a Retiga EXi Fast 1394 CCD camera using Image-Pro Express 6.3 software. At least 500 pollen grains were counted to calculate the average germination rate for each genotype or treatment. To determine the growth rate of pollen tubes, >70 pollen tubes were measured for each condition.
Actin Staining and Quantification in Pollen Grains and Pollen Tubes
To visualize the actin cytoskeleton in pollen grains and pollen tubes, pollen was fixed with 300 μM N-(maleimidobenzoyloxy)-succinimide in germination medium for 1 h. The fixative was subsequently washed out with TBS-T (50 mM Tris, 200 mM NaCl, and 0.05% Nonidet P-40, pH 7.4), and the actin cytoskeleton was stained with 200 nM Alexa-488 phalloidin (Molecular Probes) in the same buffer. Actin filaments were visualized with a confocal laser scanning microscope (Leica) equipped with a ×100 objective (1.46-NA HC PLAN). The fluorescent phalloidin was excited with the 488-nm line of an argon laser, and step size was set at 0.5 μm to collect the optical section. Images were prepared by generating projections of the optical sections through an individual pollen grain or pollen tube. To determine the effect of LatB on the organization of actin in the pollen grains and tubes, pollen grains and tubes were treated with 100 nM LatB and were then subjected to actin staining as described above. To measure the amount of actin filaments in pollen grains and pollen tubes after treatment with 100 nM LatB for 30 min, the images were collected under the same conditions. The amount of F-actin was analyzed by measuring the pixel intensity of individual pollen grains and tubes. The images were subsequently processed and analyzed with Image J software (http://rsbweb.nih.gov/ij/; version 1.38).
Statistical Analysis
Several statistical analysis methods were performed to characterize the phenotypic difference between wild-type and VLN5 loss-of-function mutants. The statistical analysis of pollen germination rate data was conducted by the χ2 test with SPSS 13.0. For those experiments that meet the assumptions of analysis of variance, they were analyzed in SPSS 13.0 followed by Dunnett post-hoc multiple comparisons. Other statistical analyses were performed using Student’s t test.
Protein Purification
The coding region of VLN5 was amplified by RT-PCR from floral tissue RNA using the forward primer V5F1 and the reverse primer V5R1 (see Supplemental Table 1 online). After verifying the authenticity of the sequence, the sequence was cloned into the pET23d expression vector (Novagen). The VLN5-pET23d plasmid was transformed into Escherichia coli BL21 (DE3) strain, and protein expression was induced by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside overnight at 16°C. E. coli cells were collected by centrifugation at 6000 rpm (Beckman JA-10), and after washing with 50 mM Tris-HCl, pH 8.0, the pellet was resuspended in the same buffer and cells were broken by sonication. The sonicate was clarified at 43,667g for 30 min, and the supernatant was precipitated with 45% ammonium sulfate. The pellet was resuspended in Cibacron Blue Buffer B containing 500 mM KCl, 0.2 mM EGTA, 0.01% NaN3, and 10 mM Tris-HCl, pH 8.0, supplemented with a 1:100 dilution of protease inhibitor cocktail (Ren et al., 1997), and then clarified at 43,667g for 20 min. The supernatant was applied to a Cibacron Blue 3GA column (Sigma-Aldrich) preequilibrated with Cibacron Blue Buffer B and eluted with Cibacron Blue Buffer C containing 1 M KCl, 0.2 mM EGTA, 0.01% NaN3, and 10 mM Tris-HCl, pH 8.0. Protein fractions containing VLN5 were pooled and dialyzed against HA Buffer A (10 mM KH2PO4 and 500 mM KCl, pH 7.0) and were then applied to a HA column preequilibrated with HA Buffer A. Bound proteins were eluted with buffer containing 40 mM KH2PO4 and 500 mM KCl, pH 7.0. The pooled protein fractions were dialyzed against buffer containing 250 mM KCl and 10 mM Tris-HCl, pH 8.0, for 40 min and were then applied to a Q-Sepharose column (Amersham Pharmacia Biotech) preequilibrated with the same buffer. Bound proteins were eluted with buffer containing 350 mM KCl and 10 mM Tris-HCl, pH 8.0. Purified VLN5 was dialyzed against 10 mM Tris-HCl, pH 8.0, aliquoted, and flash frozen in liquid nitrogen and stored at −80°C. The concentration of VLN5 was determined by the Bradford assay (Bio-Rad) using BSA as a standard.
Actin was purified from rabbit muscle acetone powder according to previously published methods (Spudich and Watt, 1971) and further purified with Sephacryl S-300 chromatography (Pollard, 1984). For fluorimetry experiments, actin was labeled on Cys-374 with pyrene iodoacetamide (Pollard, 1984). VLN1 was purified essentially according to Huang et al. (2005), and human profilin was purified according to methods described previously (Fedorov et al., 1994). Arabidopsis thaliana CaM2 was purified by phenyl-Sepharose affinity chromatography described as previously (Safadi et al., 2000).
Cosedimentation Assays
To determine the actin binding and bundling properties of VLN5, low- and high-speed cosedimentation assays were performed roughly according to Kovar et al. (2000). Briefly, proteins were preclarified at 140,000g before starting the experiment. Various concentrations of VLN5 or VLN1 were incubated with 3 μM F-actin in 1× KMEI (10× stock: 500 mM KCl, 10 mM MgCl2, 10 mM EGTA, and 100 mM imidazole-HCl, pH 7.0) in the presence of 10 nM free Ca2+ (calculated with the EQCAL program; Biosoft). After incubation at 25°C for 30 min, the mixtures were centrifuged at either 200,000g for 1 h in an Optima MAX-XP ultracentrifuge (Beckman) at 4°C or at 13,600g for 30 min in a microcentrifuge at 4°C. Equal amounts of pellet and supernatant samples were separated by 12.5% SDS-PAGE and stained with Coomassie Brilliant Blue R. To determine apparent equilibrium dissociation constant (Kd) values, the amount of VLN5 or VLN1 in the pellet and supernatant was analyzed by Image J software (http://rsbweb.nih.gov/ij/; version 1.38). A Kd value for VLN5 and VLN1 bound to actin filaments was calculated by fitting a hyperbolic function to the data from plots of protein in the pellet as a function of protein in the supernatant using Kaleidagraph v3.6 software (Synergy Software).
High- and low-speed cosedimentation assays were also used to determine whether the actin binding and bundling activities of VLN5 are regulated by calcium or calcium/CaM. Three micromolar F-actin was incubated with 1 μM VLN5 in the presence of various concentrations of free Ca2+ (calculated with the EQCAL program; Biosoft). The amount of VLN5 or actin in the supernatant and pellet was determined in the high- or low-speed cosedimentation assay to establish whether the binding of VLN5 to F-actin or the bundling activity of VLN5 was regulated by calcium, respectively. To determine whether the binding of VLN5 to actin filaments is regulated by Ca2+/calmodulin, three micromolar actin filaments were incubated with 500 nM VLN5 with or without calmodulin (50 μM) in the presence of 1 mM free Ca2+. The amount of VLN5 or actin in the supernatant and pellet was determined to see whether the binding or bundling activity of VLN5 is regulated by calcium/calmodulin.
Barbed-End Capping Assay
To determine whether VLN5 binds and caps the barbed-end of actin filaments, a seeded elongation assay was performed according to methods described previously (Huang et al., 2003). Actin filaments at a final concentration of 0.8 μM were incubated with various concentrations of VLN5 for 5 min at room temperature. Elongation was monitored by tracking the increase of pyrene fluorescence after the addition of 1 μM 5% pyrene-labeled actin monomers, saturated with 4 μM human profilin, to the actin filament mixture. The time course of actin elongation was tracked by monitoring the increase of pyrene fluorescence with a fluorimeter (QuantaMaster Luminescence QM 3 PH fluorimeter; Photon Technology International) with the excitation set at 365 nm and emission at 407 nm. The final [Ca2+]free was 10 nM or 10 μM. The initial elongation rate was plotted versus VLN5 concentration, and the equilibrium binding constant of VLN5 for the barbed end of actin filaments was determined by fitting the data with Equation 1:
where Vi is the observed rate of elongation, Vif is the rate of elongation when all the barbed ends are free, Vib is the rate of elongation when all the barbed ends are capped, [ends] is the concentration of barbed ends, and [VLN5] is the concentration of VLN5.
The data were modeled with Kaleidagraph software.
Actin Depolymerization Assays
Dilution-mediated actin depolymerization was performed roughly according to published methods (Huang et al., 2003). To test the effect of VLN5 on stabilizing actin filaments, the following procedure was used. Briefly, preassembled 5 μM F-actin (100% pyrene labeled) was incubated with various concentrations of VLN5 in the presence of 10 nM free Ca2+ for 5 min at room temperature. Depolymerization was initiated by diluting the mixtures 25-fold into low-ionic strength Buffer G in the presence of 10 nM free Ca2+. To test the effect of VLN5 on enhancing actin depolymerization, the depolymerization assay was performed roughly according to Revenu et al. (2007), with minor modifications. Briefly, preassembled 5 μM F-actin (50% pyrene labeled) was incubated with various concentrations of VLN5 in the presence of 10 μM free Ca2+ for 5 min at room temperature and diluted 50-fold into polymerization solution (1× KMEI) with 10 μM free Ca2+.
Profilin-mediated actin depolymerization was performed as described previously (Huang et al., 2004). Actin depolymerization was initiated by adding equimolar amounts of human profilin to 2.5 μM F-actin (50% pyrene labeled) in the presence of various concentrations of VLN5. Depolymerization was induced in the presence of 10 nM or 10 μM free Ca2+ and monitored by determining the decrease in pyrene fluorescence.
Fluorescence Microscopy of Actin Filaments
To visualize individual actin filaments, they were labeled with equal molar rhodamine-phalloidin (Sigma-Aldrich) and imaged by epifluorescence microscopy as described previously (Blanchoin et al., 2000). Four micromolar actin was incubated in 1× KMEI with various concentrations of VLN5 for 30 min at room temperature. The reaction mixture was then diluted to 10 nM in fluorescence buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 100 mM DTT, 100 mg/mL glucose oxidase, 15 mg/mL glucose, 20 mg/mL catalase, and 0.5% methylcellulose) before imaging. Actin filaments were observed under an IX71 microscope (Olympus) equipped with a ×60, 1.42–numerical aperture oil objective, and digital images were collected with a Retiga EXi Fast 1394 CCD camera (QImaging) using Image-Pro Express 6.3 software. Filament length was measured using Image J (http://rsbweb.nih.gov/ij/; version 1.38).
Direct Visualization of Actin Filament Severing by TIRFM
Visualization of actin filaments with TIRF illumination was roughly according to Amann and Pollard (2001) and Michelot et al. (2005). Glass flow cells were coated with a mixture of 10 nM NEM-myosin for 1 min and washed with 1% BSA in fluorescence buffer. Actin filaments (25 nM) in fluorescence buffer were perfused into the flow cells, allowed to settle, and placed on the microscope for TIRF illumination. After finding a field of interest, 5 nM VLN5 in fluorescence buffer at different [Ca2+]free was perfused into the flow cells. Actin filaments were visualized under an Olympus IX-71 microscope equipped with a ×60, 1.45–numerical aperture Planapo objective (Olympus) by TIRF illumination. Images were captured as a time-lapse series at 2-s intervals using a Hamamatsu ORCA-EM-CCD camera (model C9100-12) controlled by IPLab software (v3.9; Scanalytics). To quantify the severing activity of VLN5 or human villin, >15 filaments with lengths >10 μm were selected for calculating the average severing frequency as the numbers of breaks, per unit filament length, per unit time (i.e., breaks/μm/s).
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL or Arabidopsis Genome Initiative database under the following accession numbers: human villin (HV; NP_009058), lily 135-ABP (AAD54660), Arabidopsis VLN1 (NP_029567; At2g29890), VLN2 (NP_565958; At2g41740), and VLN5 (NP_200542; At5g57320).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. VLN5 Retains Conserved Residues at Both Type 1 and Type 2 Ca2+ Binding Sites in the G1 Domain.
Supplemental Figure 2. VLN5 Is Expressed Preferentially in Mature Pollen and at Higher Levels Than Other Arabidopsis Villins.
Supplemental Figure 3. The Level of VLN1 and VLN2 Transcripts Was Not Reduced in VLN5 RNAi Flowers.
Supplemental Figure 4. VLN5 Loss of Function Does Not Affect Pollen Germination Rate.
Supplemental Figure 5. Pollen Tube Growth Rate Was Reduced in VLN5 RNAi Lines.
Supplemental Figure 6. Root Hair Length of vln5 Homozygous Mutant Plants Is Not Significantly Different from That of Wild-Type Col-0.
Supplemental Figure 7. Actin Distribution in Wild-Type Col-0 and VLN5 Loss-of-Function Pollen Tubes.
Supplemental Figure 8. VLN5 Loss of Function Does Not Alter the Level of Actin Polymer in Pollen Tubes.
Supplemental Figure 9. Pollen Tube Growth of VLN5 RNAi Plants Is Hypersensitive to LatB Treatment.
Supplemental Figure 10. VLN5 Loss of Function Renders the Growth of Pollen Tubes Resistant to Cytochalasin D.
Supplemental Figure 11. The Amount of VLN5 Sedimented Was Decreased in the Presence of Ca2+/Calmodulin.
Supplemental Figure 12. Ca2+/CaM Inhibits the Bundling Activity of VLN5.
Supplemental Figure 13. Direct Visualization of VLN Severing Activity by Time-Lapse TIRFM.
Supplemental Table 1. List of Primers Used in Genotyping, Plasmid Construction, and RT-PCR.
Supplemental Movie 1. Time-Lapse Evanescent Wave Microscopy of Actin Filament Severing by 5 nM VLN5 in the Presence of 1 μM Free Ca2+.
Supplemental Movie 2. This Movie Shows a Time-Lapse Series of Actin Filament Severing after the Addition of 5 nM VLN5 in the Presence of 1 μM Free Ca2+.
Supplemental Movie 3. This Movie Shows a Time-Lapse Series of Actin Filament Behavior in the Absence of VLN5.
Supplemental Movie 4. This Movie Shows a Time-Lapse Series of Actin Filament Severing after the Addition of 0.5 nM Human Villin in the Presence of 1 μM Free Ca2+.
Supplemental Movie 5. This Movie Shows a Time-Lapse Series of Actin Filament Severing after the Addition of 5 nM VLN5 in the Presence of 100 nM Free Ca2+.
Supplemental Movie 6. This Movie Shows a Time-Lapse Series of Actin Filament Severing after the Addition of 5 nM VLN5 in the Presence of 10 μM Free Ca2+.
Supplemental Movie 7. This Movie Shows a Time-Lapse Series of Actin Filament Severing after the Addition of 5 nM VLN5 in the Presence of 100 μM Free Ca2+.
Supplemental Movie 8. This Movie Shows a Time-Lapse Series of Actin Filament Severing after the Addition of 5 nM VLN5 in the Presence of 1 mM Free Ca2+.
Supplemental References.
Supplemental Movie Legends.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology (2007CB947600 and 2006CB910606), the National Natural Science Foundation of China (31071179, 30771088, 30821007, and 30900721), and the Chinese Academy of Sciences (hundred talents program and KJCX2-YW-L08). This work was also partially supported by the CAS/SAFEA International Partnership Program for Creative Research Teams, the Knowledge Innovation Program of the Chinese Academy of Sciences (110500Q002), and the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry. Research in the laboratory of C.J.S. was supported by the Physical Biosciences Program of the Office of Basic Energy Sciences, U.S. Department of Energy, under Contract DE-FG02-04ER15526. The TIRF facility at Purdue was established in part with a grant from the Bindley Bioscience Center. We thank the ABRC and the Nottingham Arabidopsis Stock Centre for providing T-DNA insertion lines. We also thank Zhenbiao Yang (University of California, Riverside) for help with immunostaining and Ying Sun (Hebei Normal University) for providing the AtCaM2 prokaryotic expression plasmid and S. Khurana (University of Tennessee Health Science Center, Memphis, TN) for the human villin plasmid.
References
- Amann K.J., Pollard T.D. (2001). Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. USA 98: 15009–15013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T.D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24: 13–23 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Amann K.J., Higgs H.N., Marchand J.B., Kaiser D.A., Pollard T.D. (2000). Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404: 1007–1011 [DOI] [PubMed] [Google Scholar]
- Bretscher A., Weber K. (1979). Villin: The major microfilament-associated protein of the intestinal microvillus. Proc. Natl. Acad. Sci. USA 76: 2321–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas L., Lovy-Wheeler A., Kunkel J.G., Hepler P.K. (2008). Pollen tube growth oscillations and intracellular calcium levels are reversibly modulated by actin polymerization. Plant Physiol. 146: 1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Wong E.I., Vidali L., Estavillo A., Hepler P.K., Wu H.M., Cheung A.Y. (2002). The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Qu X., Wu Y., Huang S. (2009). Regulation of actin dynamics in pollen tubes: Control of actin polymer level. J. Integr. Plant Biol. 51: 740–750 [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2008). Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59: 547–572 [DOI] [PubMed] [Google Scholar]
- Cole R.A., Fowler J.E. (2006). Polarized growth: Maintaining focus on the tip. Curr. Opin. Plant Biol. 9: 579–588 [DOI] [PubMed] [Google Scholar]
- Cooper J.A. (1987). Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 105: 1473–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A.A., Pollard T.D., Almo S.C. (1994). Purification, characterization and crystallization of human platelet profilin expressed in Escherichia coli. J. Mol. Biol. 241: 480–482 [DOI] [PubMed] [Google Scholar]
- Ferrary E., et al. (1999). In vivo, villin is required for Ca(2+)-dependent F-actin disruption in intestinal brush borders. J. Cell Biol. 146: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederich E., Huet C., Arpin M., Louvard D. (1989). Villin induces microvilli growth and actin redistribution in transfected fibroblasts. Cell 59: 461–475 [DOI] [PubMed] [Google Scholar]
- Friederich E., Pringault E., Arpin M., Louvard D. (1990). From the structure to the function of villin, an actin-binding protein of the brush border. Bioessays 12: 403–408 [DOI] [PubMed] [Google Scholar]
- Geitmann A., Wojciechowicz K., Cresti M. (1996). Inhibition of intracellular pectin transport in pollen tubes by monensin, brefeldin A and cytochalasin D. Bot. Acta 109: 373–381 [Google Scholar]
- George S.P., Wang Y., Mathew S., Srinivasan K., Khurana S. (2007). Dimerization and actin-bundling properties of villin and its role in the assembly of epithelial cell brush borders. J. Biol. Chem. 282: 26528–26541 [DOI] [PubMed] [Google Scholar]
- Gibbon B.C., Kovar D.R., Staiger C.J. (1999). Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11: 2349–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenney J.R., Kaulfus P., Weber K. (1981). F-actin assembly modulated by villin - Ca++-dependent nucleation and capping of the barbed end. Cell 24: 471–480 [DOI] [PubMed] [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Higaki T., Sano T., Hasezawa S. (2007). Actin microfilament dynamics and actin side-binding proteins in plants. Curr. Opin. Plant Biol. 10: 549–556 [DOI] [PubMed] [Google Scholar]
- Hofmann A., Eichinger L., Andre E., Rieger D., Schleicher M. (1992). Cap100, a novel phosphatidylinositol 4,5-bisphosphate-regulated protein that caps actin filaments but does not nucleate actin assembly. Cell Motil. Cytoskeleton 23: 133–144 [DOI] [PubMed] [Google Scholar]
- Holdaway-Clarke T.L., Feijo J.A., Hackett G.R., Kunkel J.G., Hepler P.K. (1997). Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell 9: 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D., Twell D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Chaudhry F., Franklin-Tong V.E., Staiger C.J. (2004). A gelsolin-like protein from Papaver rhoeas pollen (PrABP80) stimulates calcium-regulated severing and depolymerization of actin filaments. J. Biol. Chem. 279: 23364–23375 [DOI] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Kovar D.R., Staiger C.J. (2003). Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278: 44832–44842 [DOI] [PubMed] [Google Scholar]
- Huang S., Gao L., Blanchoin L., Staiger C.J. (2006). Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol. Biol. Cell 17: 1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Robinson R.C., Gao L.Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C.J. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S., George S.P. (2008). Regulation of cell structure and function by actin-binding proteins: Villin's perspective. FEBS Lett. 582: 2128–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana P., Henty J.L., Huang S., Staiger A.M., Blanchoin L., Staiger C.J. (2010). Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22: 2727–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Friederich E., Kost B., Louvard D., Chua N.H. (2000). Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiol. 122: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Staiger C.J., Weaver E.A., McCurdy D.W. (2000). AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J. 24: 625–636 [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Yang Z. (2008). Tip growth: Signaling in the apical dome. Curr. Opin. Plant Biol. 11: 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovy-Wheeler A., Wilsen K.L., Baskin T.I., Hepler P.K. (2005). Enhanced fixation reveals the apical cortical fringe of actin filaments as a consistent feature of the pollen tube. Planta 221: 95–104 [DOI] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Cooley L. (1994). The villin-like protein encoded by the Drosophila quail gene is required for actin bundle assembly during oogenesis. Cell 78: 291–301 [DOI] [PubMed] [Google Scholar]
- Malamy J.E., Benfey P.N. (1997). Analysis of SCARECROW expression using a rapid system for assessing transgene expression in Arabidopsis roots. Plant J. 12: 957–963 [DOI] [PubMed] [Google Scholar]
- Matova N., Mahajan-Miklos S., Mooseker M.S., Cooley L. (1999). Drosophila quail, a villin-related protein, bundles actin filaments in apoptotic nurse cells. Development 126: 5645–5657 [DOI] [PubMed] [Google Scholar]
- Matsudaira P.T., Burgess D.R. (1979). Identification and organization of the components in the isolated microvillus cytoskeleton. J. Cell Biol. 83: 667–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A.M., Staiger C.J., Min J.K., Simonetti K.D. (2003). The gelsolin family of actin regulatory proteins: Modular structures, versatile functions. FEBS Lett. 552: 75–81 [DOI] [PubMed] [Google Scholar]
- Messerli M.A., Creton R., Jaffe L.F., Robinson K.R. (2000). Periodic increases in elongation rate precede increases in cytosolic Ca2+ during pollen tube growth. Dev. Biol. 222: 84–98 [DOI] [PubMed] [Google Scholar]
- Michelot A., Derivery E., Paterski-Boujemaa R., Guerin C., Huang S., Parcy F., Staiger C.J., Blanchoin L. (2006). A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr. Biol. 16: 1924–1930 [DOI] [PubMed] [Google Scholar]
- Michelot A., Guerin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C.J., Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17: 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.B., Maiti S., Goode B.L. (2006). Formin proteins: Purification and measurement of effects on actin assembly. Methods Enzymol. 406: 215–234 [DOI] [PubMed] [Google Scholar]
- Nakayasu T., Yokota E., Shimmen T. (1998). Purification of an actin-binding protein composed of 115-kDa polypeptide from pollen tubes of lily. Biochem. Biophys. Res. Commun. 249: 61–65 [DOI] [PubMed] [Google Scholar]
- Pina C., Pinto F., Feijo J.A., Becker J.D. (2005). Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. (1984). Polymerization of ADP-actin. J. Cell Biol. 99: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. (2009). Actin, a central player in cell shape and movement. Science 326: 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H., Gibbon B.C., Ashworth S.L., Sherman D.M., Yuan M., Staiger C.J. (1997). Actin purified from maize pollen functions in living plant cells. Plant Cell 9: 1445–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenu C., Courtois M., Michelot A., Sykes C., Louvard D., Robine S. (2007). Villin severing activity enhances actin-based motility in vivo. Mol. Biol. Cell 18: 827–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safadi F., Reddy V.S., Reddy A.S. (2000). A pollen-specific novel calmodulin-binding protein with tetratricopeptide repeats. J. Biol. Chem. 275: 35457–35470 [DOI] [PubMed] [Google Scholar]
- Sheahan M.B., Staiger C.J., Rose R.J., McCurdy D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136: 3968–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silacci P., Mazzolai L., Gauci C., Stergiopulos N., Yin H.L., Hayoz D. (2004). Gelsolin superfamily proteins: Key regulators of cellular functions. Cell. Mol. Life Sci. 61: 2614–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowman B.N., Kovar D.R., Shevchenko G., Franklin-Tong V.E., Staiger C.J. (2002). Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14: 2613–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246: 4866–4871 [PubMed] [Google Scholar]
- Staiger C.J., Blanchoin L. (2006). Actin dynamics: old friends with new stories. Curr. Opin. Plant Biol. 9: 554–562 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Poulter N.S., Henty J.L., Franklin-Tong V.E., Blanchoin L. (2010). Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 6: 1969–1986 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Sheahan M.B., Khurana P., Wang X., McCurdy D.W., Blanchoin L. (2009). Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J Cell Biol 184: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Wang T., Dong H., Ren H. (2007). The villin/gelsolin/fragmin superfamily proteins in plants. J Integr. Plant Biol. 49: 1183–1191 [Google Scholar]
- Thomas C., Hoffmann C., Dieterle M., Van Troys M., Ampe C., Steinmetz A. (2006). Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 18: 2194–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Moreau F., Dieterle M., Hoffmann C., Gatti S., Hofmann C., Van Troys M., Ampe C., Steinmetz A. (2007). The LIM domains of WLIM1 define a new class of actin bundling modules. J. Biol. Chem. 282: 33599–33608 [DOI] [PubMed] [Google Scholar]
- Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., Steinmetz A. (2009). Actin bundling in plants. Cell Motil. Cytoskeleton 66: 940–957 [DOI] [PubMed] [Google Scholar]
- Tominaga M., Yokota E., Vidali L., Sonobe S., Hepler P.K., Shimmen T. (2000). The role of plant villin in the organization of the actin cytoskeleton, cytoplasmic streaming and the architecture of the transvacuolar strand in root hair cells of Hydrocharis. Planta 210: 836–843 [DOI] [PubMed] [Google Scholar]
- Vidali L., Hepler P.K. (1997). Characterization and localization of profilin in pollen grains and tubes of Lilium longiflorum. Cell Motil. Cytoskeleton 36: 323–338 [DOI] [PubMed] [Google Scholar]
- Vidali L., McKenna S.T., Hepler P.K. (2001). Actin polymerization is essential for pollen tube growth. Mol. Biol. Cell 12: 2534–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., Bezanilla M. (2009). Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T.P., Weber A., Davis K., Bonder E., Mooseker M. (1984a). Calcium dependence of villin-induced actin depolymerization. Biochemistry 23: 6099–6102 [DOI] [PubMed] [Google Scholar]
- Walsh T.P., Weber A., Higgins J., Bonder E.M., Mooseker M.S. (1984b). Effect of villin on the kinetics of actin polymerization. Biochemistry 23: 2613–2621 [DOI] [PubMed] [Google Scholar]
- Wang H.J., Wan A.R., Jauh G.Y. (2008). An actin-binding protein, LlLIM1, mediates calcium and hydrogen regulation of actin dynamics in pollen tubes. Plant Physiol. 147: 1619–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Huang X., Wang T., Zhang Y., Liu Q., Hussey P.J., Ren H. (2007). ACTIN BINDING PROTEIN 29 from Lilium pollen plays an important role in dynamic actin remodeling. Plant Cell 19: 1930–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Zheng Y., Yan A., Chen N., Wang Z., Huang S., Yang Z. (2009). Arabidopsis formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 21: 3868–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Muto S., Shimmen T. (2000). Calcium-calmodulin suppresses the filamentous actin-binding activity of a 135-kilodalton actin-bundling protein isolated from lily pollen tubes. Plant Physiol. 123: 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Shimmen T. (1998). Actin-bundling protein isolated from pollen tubes of lily. Biochemical and immunocytochemical characterization. Plant Physiol. 116: 1421–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Tominaga M., Mabuchi I., Tsuji Y., Staiger C.J., Oiwa K., Shimmen T. (2005). Plant villin, lily P-135-ABP, possesses G-actin binding activity and accelerates the polymerization and depolymerization of actin in a Ca2+-sensitive manner. Plant Cell Physiol. 46: 1690–1703 [DOI] [PubMed] [Google Scholar]
- Yokota E., Vidali L., Tominaga M., Tahara H., Orii H., Morizane Y., Hepler P.K., Shimmen T. (2003). Plant 115-kDa actin-filament bundling protein, P-115-ABP, is a homologue of plant villin and is widely distributed in cells. Plant Cell Physiol. 44: 1088–1099 [DOI] [PubMed] [Google Scholar]
- Zhai L., Zhao P., Panebra A., Guerrerio A.L., Khurana S. (2001). Tyrosine phosphorylation of villin regulates the organization of the actin cytoskeleton. J. Biol. Chem. 276: 36163–36167 [DOI] [PubMed] [Google Scholar]