The hydrogen peroxide–responsive UDP-glucosyltransferase UGT74E2 from Arabidopsis thaliana is shown to be involved in modulation of plant architecture and water stress response through its activity toward the auxin indole-3-butyric acid (IBA). Evidence is provided that, during water stress, IBA and IBA-glucose levels increase, and auxins help maintain the photosynthetic capacity under stress.
Abstract
Reactive oxygen species and redox signaling undergo synergistic and antagonistic interactions with phytohormones to regulate protective responses of plants against biotic and abiotic stresses. However, molecular insight into the nature of this crosstalk remains scarce. We demonstrate that the hydrogen peroxide–responsive UDP-glucosyltransferase UGT74E2 of Arabidopsis thaliana is involved in the modulation of plant architecture and water stress response through its activity toward the auxin indole-3-butyric acid (IBA). Biochemical characterization of recombinant UGT74E2 demonstrated that it strongly favors IBA as a substrate. Assessment of indole-3-acetic acid (IAA), IBA, and their conjugates in transgenic plants ectopically expressing UGT74E2 indicated that the catalytic specificity was maintained in planta. In these transgenic plants, not only were IBA-Glc concentrations increased, but also free IBA levels were elevated and the conjugated IAA pattern was modified. This perturbed IBA and IAA homeostasis was associated with architectural changes, including increased shoot branching and altered rosette shape, and resulted in significantly improved survival during drought and salt stress treatments. Hence, our results reveal that IBA and IBA-Glc are important regulators of morphological and physiological stress adaptation mechanisms and provide molecular evidence for the interplay between hydrogen peroxide and auxin homeostasis through the action of an IBA UGT.
INTRODUCTION
Reactive oxygen species (ROS) are key signaling molecules that regulate growth and development and coordinate responses to biotic and abiotic stresses in plants (Apel and Hirt, 2004). Under harsh environmental conditions, ROS levels can rise to excessive levels with oxidative damage and cell death as a consequence (Van Breusegem and Dat, 2006). However, the tight regulation of ROS homeostasis by a complex network of ROS-producing and ROS-scavenging enzymes also creates a baseline from which ROS spikes can act as a signal in different cellular processes (Mittler et al., 2004). Hydrogen peroxide (H2O2) was demonstrated to be an effective signaling molecule modulating a diverse set of physiological mechanisms, including senescence (Miao et al., 2004), stomatal closure (Zhang et al., 2001), cell cycle (Potters et al., 2009), and acclimation responses to challenging environmental conditions (Neill et al., 2002). Increased H2O2 levels strongly redirect transcriptional responses, and H2O2-responsive genes are, in addition to a prominent defense response, involved in multiple processes, such as metabolism, energy homeostasis, and protein degradation (Neill et al., 2002; Vandenabeele et al., 2003, 2004; Vanderauwera et al., 2005).
During biotic and abiotic stress events, there is an intimate interplay between ROS and other plant signaling molecules and hormones, such as calcium, salicylic acid (SA), abscisic acid (ABA), jasmonic acid, ethylene, nitric oxide, and gibberellins (Bright et al., 2006; Gudesblat et. al., 2007; Desikan et al., 2008; Galvez-Valdivieso et al., 2009). For example, in guard cells, ABA-dependent H2O2 production is essential for stomatal closure (Desikan et al., 2008) and during root growth; gibberellin signaling contributes to the fine-tuning of ROS levels by inactivating DELLA proteins that transcriptionally regulate ROS-scavenging enzymes and hence modulate biotic and abiotic stress tolerance (Achard et al., 2008). The best known case in which ROS directly influence the action of auxins is a H2O2-dependent mitogen-activated protein kinase cascade that negatively affects auxin sensitivity by downregulation of auxin-inducible gene expression (Nakagami et al., 2006). Interaction between ROS and auxins is also suggested by altered auxin homeostasis through increased H2O2 levels and by observed changes in plant architecture provoked by ROS impingement on auxin signaling (Potters et al., 2007, 2009). Auxin homeostasis could be perturbed by modified auxin redistribution through an effect on PINOID gene expression, which affects polar auxin transport (Pasternak et al., 2005). Similarly, stress-induced changes in the cellular pH gradient will impact chemiosmotically driven auxin uptake, transport, and redistribution (Potters et al., 2007). In addition to the influence of auxin homeostasis through the transcriptional regulation of enzymes involved in its biosynthesis and conjugation (Ljung et al., 2002; Woodward and Bartel, 2005), oxidative degradation of auxins through H2O2-dependent peroxidases occurs as well (Gazarian et al., 1998; Ljung et al., 2002).
Previously, we used catalase-deficient Arabidopsis thaliana plants as a model system to increase in planta H2O2 levels under photorespiration-inducing conditions and monitored the subsequent transcriptional changes through microarray analyses (Vanderauwera et al., 2005). One of the most rapidly and strongly induced transcripts is UGT74E2, which encodes the UDP-glucosyltransferase UGT74E2.
Glycosyltransferases are found in both prokaryotic and eukaryotic species and catalyze the addition of sugars to a wide range of small molecules. Glycosylation of aglycones can alter their activity, solubility, and transport. Therefore, glycosylation is considered an important regulatory mechanism in cellular metabolism (Lim and Bowles, 2004). The range of sugar acceptors in plants is thought to be very diverse, but the exact substrates are known for only a few of the glycosyltransferases, and there is a lack of strict correlation between the structural identity and substrate specificity (Ross et al., 2001). Previously described plant acceptor molecules for glycosylation include hormones (such as auxin, ABA, cytokinin, SA, and brassinosteroids), secondary metabolites (monolignols, anthranilate, caffeic acid, and flavonoids), and xenobiotics (Szerszen et al., 1994; Jackson et al., 2001; Lim et al., 2003; Meßner et al., 2003; Quiel and Bender, 2003; Hou et al., 2004; Poppenberger et al., 2005; Priest et al., 2005; Lanot et al., 2006; Song, 2006; Yonekura-Sakakibara et al., 2007; Dean and Delaney, 2008; Havlová et al., 2008). In the Arabidopsis genome, >100 UDP-glycosyltransferase (UGT) family members are present (Ross et al., 2001).
UGT74E2 is a member of the group L subclass of UGTs (Ross et al., 2001), which includes maize (Zea mays) iaglu and Arabidopsis UGT84B1, both described as auxin glycosyltransferases, and UGT74F2, which has been recognized previously as SA and anthranilate (a Trp precursor) glycosyltransferase (Szerszen et al., 1994; Jackson et al., 2001; Quiel and Bender, 2003; Song, 2006). Here, we demonstrate that UGT74E2 is an early H2O2-responsive gene coding for an UGT preferentially glycosylating indole-3-butyric acid (IBA). Ectopic overexpression of UGT74E2 in Arabidopsis, in addition to perturbing IBA and the general auxin homeostasis, alters plant architecture and improves stress tolerance. Our results provide insight into the regulation of auxin homeostasis in plants and give molecular evidence for the interplay between H2O2 and auxin signaling through the action of an IBA UGT.
RESULTS
H2O2-Inducible UGT74E2 Is Rapidly Induced by Abiotic Stresses
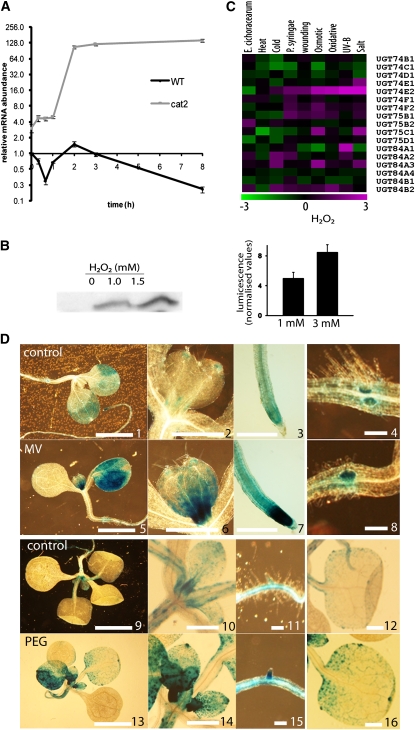
Previously, using catalase-deficient plants as a model system, we identified a comprehensive set of H2O2-responsive genes (Vandenabeele et al., 2004; Vanderauwera et al., 2005). When these catalase-deficient plants are exposed to photorespiratory-inducing conditions, such as high light intensities, cellular levels of H2O2 increase, triggering significant changes in the transcriptome. One of the most H2O2-responsive transcripts, UGT74E2, encodes a UGT, showing a >100-fold induction of transcript levels within 2 h of high-light treatment (Figure 1A). At transcript and protein levels, UGT74E2 was also induced in ProUGT74E2:LUC and wild-type seedlings in response to exogenous H2O2 (Figure 1B). The strong responsiveness of the UGT74E2 transcript and protein levels was a first indication for a role of UGT74E2 in H2O2-mediated stress signal transduction. As UGT74E2 belongs to the UGTs of the L group (Ross et al., 2001), we assessed the abiotic and biotic stress responsiveness of 17 UGTs in available microarray experiments (Figure 1C) (Zimmermann et al., 2004). Of all UGTs, expression of UGT74E2 was most strongly induced by oxidative stress and was the most responsive to a wide variety of different stresses (Figure 1C). These microarray-based expression data were confirmed by RT-PCR analysis of plants exposed to salt, polyethylene glycol (PEG), and dehydration stress. In all cases, UGT74E2 transcripts were induced early and strongly (see Supplemental Figure 1 online). Spatial expression patterns of UGT74E2 were studied using ProUGT74E2:GUS (β-glucuronidase) promoter reporter gene fusion constructs. Under unstressed conditions, GUS expression was visible in developed leaves and at the bases and tips of young leaf primordia (Figure 1D). In developing leaves, the UGT74E2 promoter activity was most prominent in the hydathodes and on the margins of petioles and bases (Figure 1D). As leaves expanded progressively with age, staining largely disappeared in these organs (Figure 1D). In roots, the activity of the UGT74E2 promoter was the strongest in the root tip. The same pattern was observed in lateral roots, in which GUS activity could be observed very early in the first-formed lateral root primordia, in the hypocotyl-root junction, and afterward in lateral root primordia (pericycle cells). Root primordia also remained strongly stained after protrusion (Figure 1D), but later GUS staining became restricted to the root tips. In general, induction was strong and rapid in leaves and even more pronounced in young leaves under stress conditions. Oxidative stress imposed by the herbicide methyl viologen (MV), a ROS propagator, induced GUS activity in expanded leaves and especially in young leaves (Figure 1D). A similar expression pattern was observed under osmotic stress imposed by PEG (Figure 1D). Under these treatments, induced GUS staining in roots was again confined to the tips but was also observed in the root vasculature (Figure 1D). These findings indicate that UGT74E2 expression is stress inducible via a H2O2-dependent mechanism.
Figure 1.
Induction of UGT74E2 by H2O2 and Environmental Stresses.
(A) Rapid induction of the UGT74E2 transcript by high-light treatment in catalase-deficient (cat2) and wild-type background (Vanderauwera et al., 2005) as determined by quantitative PCR. Error bars are se (n = 3).
(B) Left side: Induction of UGT74E2 protein levels in 14-d-old wild-type seedlings exposed for 5 d to increased concentrations of exogenous H2O2 as determined by protein immunoblots. Right side: UGT74E2 transcript induction was quantified in pUGT74E2:LUC seedlings treated with 1 and 3 mM H2O2 for 5 d. Error bars are se (n = 3).
(C) Stress-related expression patterns of the Arabidopsis group L glycosyltransferases. Log2-transformed relative expression values of the group L glycosyltransferases in publicly available microarray data on biotic and abiotic stress treatments (Genevestigator) (Zimmermann et al., 2004). Magenta, green, and black indicate upregulation, downregulation, and no change versus control experiments, respectively.
(D) Histochemical GUS staining of pUGT74E2:GUS seedlings under control conditions (1 to 4 and 9 to 12) or exposed for 5 h to 50 μM MV (5 to 8) or 500 mM PEG (13 to 16). On 12-d-old seedlings (1 to 8), pUGT74E2:GUS expression is visible in the distal tip and base of leaf primordia (1 and 2), mesophyll of cotyledons (1), first formed lateral root primordia in the hypocotyl-root junction (4), and root tip (1 and 3). On 16-d-old seedlings (9 to 16), GUS staining is prominent in young leaf primordia and developing leaf tips and stipules (9 and 10), in hydathodes of developed leaves and on margins of petioles and bases (12), and in roots tips and lateral root primordia (11). MV or PEG stress increased pUGT74E2:GUS expression in these tissues (5 to 8 and 13 to 16). Bars = 2 mm in panels 1, 2, 5, 6, 9, and 13, 0.5 mm in panels 3, 7, 10, 12, 14, and 16, and 0.1 mm in panels 4, 8, 11, and 15.
UGT74E2 Is an IBA Glucosyltransferase
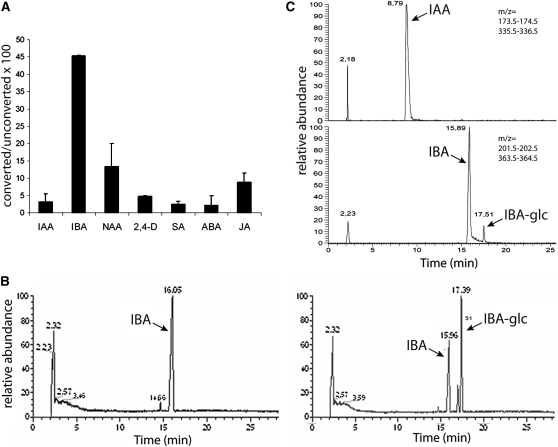
UGT74E2 encodes a putative UGT, but its activity and substrate have not been previously demonstrated. As two other members of the Arabidopsis group L glycosyltransferases had been reported to glucosylate auxin and SA (Jackson et al., 2001; Song, 2006), we examined whether the purified recombinant protein 6xHIS-UGT74E2 could glucosylate different plant hormones (indole-3-acetic acid [IAA], IBA, the synthetic auxin analogs naphthaleneacetic acid [NAA] and 2,4-D, SA, ABA, and jasmonic acid) in an in vitro assay. In this assay, 2 μg of the recombinant protein was incubated for 3 h with the different hormones, together with a saturating concentration (5 mM) of uridine-5′-diphosphoglucose as the Glc donor. Relative conversion rates of the aglycones to the glucosylated forms were calculated by liquid chromatography–coupled mass spectrometry (LC-MS). Whereas only low levels of the glucosylated forms were detected for the other hormones, the glycosylation activity was prominent toward IBA (Figure 2A), and formation of IBA-Glc was undetectable in the absence of UGT74E2 (Figure 2B). Because of the remarkable specificity toward IBA, we compared IAA and IBA in replicated short-term assays (3 and 5 min) and a range of substrate concentrations (from 5 pM to 5 μM): again, no glucosylated IAA was found, whereas IBA-Glc was clearly observed when IBA was supplied at a concentration of 1 and 5 μM (Figure 2C). Therefore, a Lineweaver-Burk plot at substrate concentrations of 1 and 5 μM revealed significant intercept and slope values (P < 0.05) only for a reaction time of 5 min, yielding Km and Vmax values of 1.39 μM and 0.9 × 10−3 μM/s, respectively, although similar values had been computed for a 3-min reaction (Km 1.47 μM and Vmax 1.6 × 10−3 μM/s). These data indicate that UGT74E2 is an auxin glucosyltransferase that preferentially uses IBA as substrate.
Figure 2.
Identification of UGT74E2 as an IBA Glucosyltransferase.
(A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin IBA is the preferred substrate. Error bars are se (n = 2). JA, jasmonic acid.
(B) Separation by liquid chromatography of buffered samples containing UDP-Glc and IBA without (left) and with (right) addition of recombinant UGT74E2. Peak identity was determined by mass spectrometry. A significant proportion of IBA was glucosylated only in the presence of UGT74E2.
(C) Separation by liquid chromatography of buffered samples containing UDP-Glc and IAA (top) or IBA (bottom) in short-term incubation assays (10 min). No IAA-Glc could be detected, whereas a significant proportion of IBA was glucosylated.
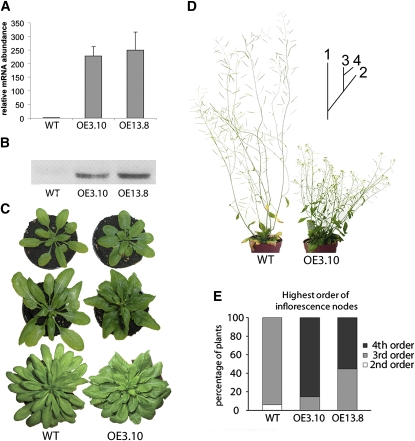
To examine the effect of increased UGT74E2 levels in vivo, Arabidopsis transgenic lines containing p35S:UGT74E2 (see Methods) were constructed (UGT74E2OE plants). For further studies, we selected two independent overexpressing (OE) lines, OE3.10 and OE13.8, based on their high UGT74E2 expression levels as determined by real-time RT-PCR (Figure 3A) and protein gel blot analysis (Figure 3B). Because IBA and IAA can be interconverted (Bartel et al., 2001), overexpression of an IBA glucosyltransferase could deregulate the general auxin homeostasis. To confirm this assumption, IAA, IBA, and Glc conjugate levels (IAA-Glc and IBA-Glc) were measured in wild-type and UGT74E2OE whole seedlings at 1.04 growth stage, corresponding approximately to 16-d-old plants (Boyes et al., 2001). Analysis by liquid chromatography indicated that IBA and IBA-Glc levels were significantly higher in UGT74E2OE lines than those of the wild-type plants (Table 1). These data are in accordance with the catalytic specificity of the recombinant enzyme in vitro. IAA-Glc decreased by ~30% in UGT74E2OE seedlings, while the free IAA levels were comparable between transgenic and wild-type plants. The levels of the inactive amide conjugate IAA-Glu and oxidized IAA (oxIAA) had increased by on average 48 and 27%, respectively. IAA-Ala, IAA-Asp, and methylated IAA levels (metIAA) were not affected (Table 1, experiment 1). Elevated IBA-Glc (P = 5 × 10−4) and reduced IAA-Glc (P = 9 × 10−6) levels in the transgenic lines were consolidated in an independent experiment that included a water stress treatment (see below). Taken together, these results suggest that the transgenic plants respond to the overproduction of an IBA-UGT by repressing the IAA-ester conjugation pathway, possibly due to competition for UDP-Glc with the IBA conjugation pathway, and by inducing the IAA oxidative pathways, without altering free IAA levels.
Figure 3.
Modulation of Arabidopsis Architecture by Overexpression of UGT74E2.
(A) UGT74E2 mRNA levels in wild-type and two UGT74E2OE plants assessed by real-time RT-PCR. Error bars are se (n = 3).
(B) UGT74E2 protein levels in wild-type and UGT74E2OE plants assessed by protein gel blot with leaves from 1-month-old plants. For each lane, 200 μg of soluble protein was loaded.
(C) Rosette shapes of 3-, 4-, and 5-week-old (top to bottom) wild-type and UGT74E2OE plants growing under long-day (top, middle) or short-day (bottom) regimes, respectively. Note the intense green color of UGT74E2OE plants and the compressed rosette structure.
(D) Mature UGT74E2OE plants that were shorter and displayed a higher level of shoot branching than the wild type. Diagram illustrates the concept of inflorescence node order, in which 1 is the primary inflorescence and 2 to 4 are the subsequent orders of axillary nodes.
(E) Higher order of inflorescence branching of UGT74E2OE plants compared with wild-type plants (P < 0.001, n = 9 to 16).
Table 1.
Disturbed Auxin Levels in UGT74E2-Overexpressing Plants
| Auxins | Wild Type | UGTOE3.10 | UGTOE13.8 |
| Experiment 1: Control | |||
| IBAa | 0.48 ± 0.14 | 1.03 ± 0.27 | 1.37 ± 0.33 |
| IBA-Glca | 1,980 ± 330 | 3,287 ± 462 | 5,196 ± 768 |
| IAA | 88.98 ± 18.07 | 82.75 ± 8.76 | 88.60 ± 5.73 |
| IAA-Glc | 304.59 ± 29.45 | 193.51 ± 30.04 | 224.24 ± 6.27 |
| oxIAA | 305.29 ± 27.91 | 370.39 ± 52.50 | 407.48 ± 18.79 |
| IAA-Glu | 13.94 ± 3.00 | 19.65 ± 4.51 | 21.68 ± 4.48 |
| IAA-Asp | 6.22 ± 1.32 | 6.83 ± 1.40 | 7.89 ± 1.24 |
| IAA-Ala | 5.01 ± 1.61 | 4.06 ± 0.74 | 5.21 ± 0.57 |
| metIAA | 85.79 ± 12.48 | 82.97 ± 8.65 | 99.52 ± 4.99 |
| Experiment 2: PEG | |||
| IBA | 5.67 ± 0.24 | 5.95 ± 2.05 | 7.33 ± 1.89 |
| IBA-Glca | 21,002.09 ± 4,319.61 | 29,964.16 ± 15,223.65 | 31,770.65 ± 4,721.77 |
| IAA | 58.21 ± 4.03 | 97.68 ± 2.69 | 111.47 ± 7.97 |
| IAA-Glca | 2,081.62 ± 358.24 | 1,233.26 ± 114.94 | 1,003.56 ± 71.46 |
Measurements of endogenous auxin content (pmol g−1 tissue fresh weight) under control or PEG-induced stress conditions. Five replicates were tested statistically by ANOVA. Data are means ± se. Statistically significant differences are indicated in bold (n = 5, P < 0.05).
For experiment 1, IBA and IBA-Glc measurements (n = 9 to 10) were P = 0.06 for IBA and P = 0.004 for IBA-Glc. Hormone levels for wild-type and transgenic lines grown under control conditions for experiment 2 were of similar magnitude as observed in experiment 1; P = 5 × 10−4 for IBA-Glc and P = 9 × 10−6 for IAA-Glc.
Under control growth conditions, no defects in root gravitropism were observed in the transgenic lines, while root length was slightly reduced in transgenic plants grown on Murashige and Skoog (MS) media without sucrose (see Supplemental Table 1 online); root growth rate was similar to that of wild-type plants on MS media with 0.5% sucrose (see Supplemental Figure 1B online). To investigate the effect of increased IBA and IBA-Glc levels on the auxin response, root elongation assays were performed. While root growth can be stimulated by low concentrations of externally applied auxin, excess amounts inhibit primary root elongation and promote secondary root initiation (Ludwig-Müller et al., 1993). UGT74E2OE seedlings transferred to plates supplemented with auxins exhibited a wild-type sensitivity to IAA, IBA, NAA, and 2,4-D in primary root elongation inhibition assays. Exogenous IBA-Glc did not show inhibitory effects on root elongation in wild-type and transgenic lines (see Supplemental Figure 1B online). The addition of the auxin transport inhibitors naphthylphthalamic acid (NPA) and 2,3,5-triiodobenzoic acid did not differentially inhibit root length in either wild-type or transgenic plants (see Supplemental Figure 1B online). Also, lateral root induction by both IAA and IBA was similar in all the lines, while IBA-Glc had no effect at all (see Supplemental Figure 1C online). To explore the role of the IBA transport or its conversion into IAA during lateral root development, lateral root induction was monitored on plates supplemented with IAA, IBA, and IBA-Glc in the absence or presence of NPA. As reported before (Casimiro et al., 2001), NPA decreased IAA lateral root induction in all the lines. Surprisingly, NPA further increased the induction of lateral roots caused by IBA (see Supplemental Figure 1C online). For all the lines, no lateral root development could be observed in the presence of 5 μM NPA or NPA in combination with IBA-Glc.
UGT74E2 Overexpression Affects Arabidopsis Architecture
The UGT74E2OE plants displayed phenotypes that differed from those of the wild-type plants: they had a more compact rosette structure with visibly shorter petioles, although the total biomass (fresh weight) was largely unaffected (Supplemental Table 1 online). This compressed appearance was even more pronounced under short-day growth conditions (Figure 3C). UGT74E2OE leaves had a dark-green color with an increased chlorophyll concentration of 10 to 15% (see Supplemental Table 1 online). After inflorescence emergence, UGT74E2OE lines developed a clear shoot branching phenotype. Mature UGT74E2OE plants were also shorter in stature than wild-type plants (Supplemental Table 1 online). Interestingly, the primary inflorescence, which first arises from the rosette, is usually outgrown by its side branches in UGT74E2OE plants, in contrast with wild-type plants, in which the primary apex dominates its side branches. Furthermore, in UGT74E2OE plants, the shoot branching was of a higher order than that of wild-type plants (Figures 3D and 3E). Loss of apical dominance is a phenotype often linked to altered auxin homeostasis (Estelle and Somerville, 1987; Bak and Feyereisen, 2001), supporting the biochemical data that UGT74E2 is an auxin glucosyltransferase (Figure 2). UGT74E2OE plants showed a mild delay in flowering of up to 1 week (see Supplemental Figure 2B online). To assess whether the changes in overall leaf morphology were underpinned by changes at the cellular level, fully expanded third leaves of 3-week-old plants were analyzed by scanning electron microscopy. Epidermal cell density and stomatal density and length were not significantly affected by UGT74E2 overexpression under short-day or long-day growth conditions (see Supplemental Figure 3 online).
To explore the effects of IBA and IBA-Glc on plant architecture and development, wild-type seedlings were germinated and grown on medium supplemented with auxins. Both IBA and IBA-Glc reduced the leaf area (see Supplemental Figure 2A online), but the compact rosette phenotype of UGT74E2OE plants could not be mimicked by external IBA or IBA-Glc applications. Growth on 5 μM of IBA or IBA-Glc significantly delayed bolting in wild-type plants. Whereas nontreated plants started to bolt after 20 d, it took on average 70 d in the presence of exogenous IBA or IBA-Glc. Similarly, UGT74E2OE plants only started bolting after 27 d (see Supplemental Figure 2B online). These results suggest that besides IAA (Frankowski et al., 2009), IBA and IBA-Glc also could inhibit flower induction, which might be the reason for the delayed flowering phenotype displayed by the UGT74E2OE plants.
Two mutants with a T-DNA insertion into the second exon of the Arabidopsis UGT74E2 gene, ugt74e2-01, and the promoter region, ugt74e2-09, were examined. The T-DNA insertions impaired the UGT74E2 full-length transcript accumulation (see Supplemental Figure 4 online). Homozygous knockout plants had a wild-type phenotype, including the rosette phenotype and flowering time (see Supplemental Figure 5A online) and root length and root inhibition responses to IAA, IBA, IBA-Glc, and NAA (see Supplemental Figure 5B online).
Transcriptional Impact of Elevated UGT74E2 Expression
The downstream effects of elevated UGT74E2 levels on the Arabidopsis transcriptome were determined by microarray analysis. Three pools per genotype (Columbia-0 [Col-0] and UGT74E2OE) of in vitro–grown seedlings were harvested at growth stage 1.04 and total RNA was prepared. RNA of each pool was hybridized to a full-genome microarray (Affymetrix GeneChip ATH1). After processing, normalization, and multiple testing corrections, in total 31 probe sets with a P value <0.05 and a 1.5-fold change were retained as differential, of which 13 were upregulated and 18 were downregulated (see Supplemental Table 2 online). In addition to four transcription factors, functional categories that were represented included glycosylhydrolases (three upregulated and one downregulated) and genes involved in hormone signaling/response (two upregulated and four downregulated).
UGT74E2 Mediates Abiotic Stress Responses
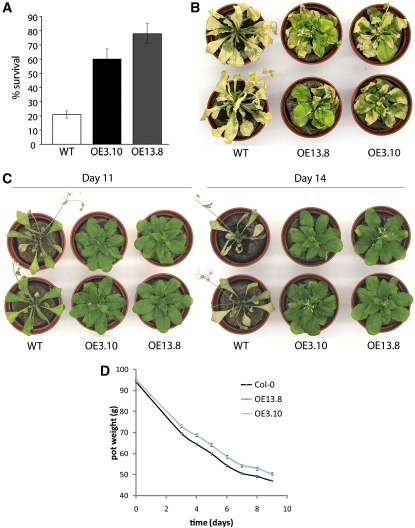
We assessed the potential impact of the UGT74E2 overexpression on stress tolerance. One-month-old transgenic and wild-type seedlings grown on MS agar plates were transferred to plates supplemented with 150 mM NaCl for 8 d. Survival rates of the transgenic seedlings were markedly higher than those of wild-type seedlings (Figure 4A). In soil, 3.5-week-old plants growing under a controlled watering regime were irrigated regularly with 500 mM NaCl. After 2 weeks, wild-type plants showed severe foliar chlorosis and necrosis, while transgenic plants remained green and appeared viable. After 3 weeks of salinity stress, transgenic plants started bolting in contrast with the wild-type plants (Figure 4B).
Figure 4.
Increased Tolerance of UGT74E2OE Plants against Osmotic Stress.
(A) One-month-old wild-type and overexpressing transgenic seedlings grown on MS agar plates and transferred to plates supplemented with 150 mM NaCl. Survival was assessed 8 d after transfer. Overexpressing lines showed significantly increased survival compared with wild-type plants (P < 0.05, Student’s t test). Measurements were made on two independent replicates of 20 seedlings each. Error bars indicate se.
(B) Plants of wild-type Col-0 and two independent UGT74E2OE lines grown under a controlled watering regime for 3.5 weeks followed by watering for 3 weeks with 500 mM NaCl.
(C) Plants of wild-type Col-0 and two independent UGT74E2OE lines grown under a controlled watering regime for 3.5 weeks and deprived from further watering for 12 d. On day 13, plants were rewatered and observed for recovery on day 14.
(D) Plants were grown with 1.5 g water/g dry soil for 3.5 weeks, after which both groups were not watered for 9 d. The total weight (pot + soil + plant) was recorded at different times. During the dehydration period, the water economy of the UGT74E2OE plants was significantly better (P < 0.05, Student’s t test) than that of the wild-type plants already after 3 d. Error bars indicate se (n = 6).
For drought stress experiments, wild-type and UGT74E2OE plants were grown under water-sufficient conditions. After 2 weeks, plants were watered in a controlled way by bringing the total weight of each pot to a target weight (see Methods) for 3.5 weeks; thereafter, plants were separated into a control (further watering) and drought-treated (no watering) group. After 10 d without watering, the first signs of wilting were already visible in 60% of the wild-type plants, while all UGT74E2OE plants had a healthy appearance. After 11 d, all wild-type plants suffered clearly from water loss, in contrast with the UGT74E2OE plants that still looked healthy (Figure 4C). After 12 d of drought, all wild-type plants looked severely dehydrated and the UGT74E2OE plants showed the first signs of wilting. On day 13 of the drought treatment, all plants were rewatered to allow recovery. After 24 h of rewatering, all UGT74E2OE plants had regained turgor, while none of the wild-type plants presented signs of recuperation (Figure 4C). In agreement with the increased tolerance, high UGT74E2 expression resulted already within 3 d in significantly (P < 0.05) reduced water loss per pot under drought stress condition (Figure 4D). These data indicate that, in accordance with the upregulation of UGT74E2 by osmotic and salt stress treatments (Figure 1), transgenic UGT74E2OE plants exhibited increased tolerance to salinity and drought stress. Comparison of the drought tolerance of the wild type, UGT74E2OE, and ugt74e2 mutants revealed that the knockout mutants and the wild-type plants did not differ obviously (see Supplemental Figure 5C online).
Additionally, the effect of osmotic stress on auxin homeostasis in wild-type and transgenic lines was examined by growing seedlings on medium with or without PEG (for average hormone levels in seedlings grown on PEG-containing medium, see Table 1, experiment 2). In both lines, the levels of IAA-Glc and IBA-Glc had increased 6- (P < 2 × 10−16) and 10-fold (P < 2 × 10−16), respectively, whereas only a slight increase was observed for IBA (P = 3 × 10−3) and no change at all for IAA (P = 0.9) (Table 1, experiment 2). These data are in accordance with the induction of the UGT74E2 transcript and protein under abiotic stresses (Figure 1; see Supplemental Figure 1 online).
We assessed whether the observed water stress tolerance was associated with improved photosynthesis. Under normal conditions, in the transgenic plants, the maximal electron transport rate (ETRmax), the maximum photochemical efficiency of photosystem II (PSII) in the dark-adapted state (ΦPSIImax), the photochemical quenching (qP; Table 2), the maximal CO2 assimilation rates, and the stomatal conductance did not change significantly (see Supplemental Table 1 online). However, in detached transgenic leaves, the ETRmax and qP were higher than those of the wild-type plants. ΦPSIImax decreased by dehydration stress equally for all the lines (Table 2). The significantly increased photosynthetic performance and associated energetic advantage specifically under dehydration conditions are in agreement with the increased drought and salt stress tolerance of the transgenic plants.
Table 2.
Photosynthetic Parameters under Well-Watered and Dehydrated Conditions
| Parameters | Wild type | UGTOE3-10 | UGTOE13-8 |
| Well watered | |||
| ETRmax | 94.90 ± 0.06 | 114.10 ± 0.06 | 108.80 ± 0.12 |
| ΦPSIImax | 0.817 ± 0.003 | 0.810 ± 0.002 | 0.812 ± 0.002 |
| qP | 0.48 ± 0.02 | 0.45 ± 0.03 | 0.50 ± 0.01 |
| Dehydration | |||
| ETRmax | 17.7 ± 1.5 | 33.1 ± 3.4 | 31.3 ± 3.5 |
| ΦPSIImax | 0.69 ± 0.01 | 0.74 ± 0.01 | 0.72 ± 0.01 |
| qP | 0.28 ± 0.03 | 0.46 ± 0 .02 | 0.43 ± 0.03 |
Photosynthesis was measured at 500 μmol m−2 s−1 as described in Methods. Well-watered wild-type and transgenic plants were grown in soil for 51 d under short-day conditions. Data are means ± se (n = 3). Dehydration treatments were applied for 1.5 h on detached fully expanded leaves from 1-month-old plants growing under long-day regime. Statistically significant differences are indicated in bold (n = 10, P < 0.05). ETRmax, maximal electron transfer rate; ΦPSIImax, maximal photochemical yield of photosystem II; qP, photochemical quenching.
Next, we investigated whether exogenously applied IBA or IBA-Glc could lead to increased water stress tolerance. Two-week-old seedlings were exposed to osmotic stress (54.7 g/L mannitol). Plants were either germinated and grown in the presence of IBA or IBA-Glc before the supply of both mannitol (M) and auxin (M+IBA-IBA or M+IBA-Glc-IBA-Glc treated), or the auxins were added only when the plants were transferred to the mannitol-containing plates (M+IBA and M+IBA-Glc). The chlorophyll fluorescent variable ETRmax was recorded after 8 d of stress exposure. In wild-type and UGT74E2OE seedlings, mannitol treatments resulted in a 25% and only 9% decrease in ETRmax, respectively (see Supplemental Table 3 online). The addition of IBA or IBA-Glc from germination or only during the water stress period could not mimic the photosynthetic response observed in the transgenic lines. On the contrary, at the assayed concentrations, accumulation of IBA or IBA-Glc had a negative effect on the ETRmax.
Finally, we examined whether the stress response in UGT74E2OE plants depended on increased IAA levels induced by stress. After 8 d of water stress treatment (400 g/L PEG), wild-type seedlings grown with additional IAA supply (wild-type PEG+IAA-IAA treated) mimicked the response of UGT74E2OE plants. However, addition of IAA at the beginning of the stress treatment had no effect (wild-type PEG+IAA; see Supplemental Table 3 online). These results suggest that IBA, IBA-Glc, and IAA affect mechanisms that control photosynthesis and that, under stress, photosynthesis can be protected by increased IAA levels.
Perturbation of IBA Metabolism Impinges on the ABA Response
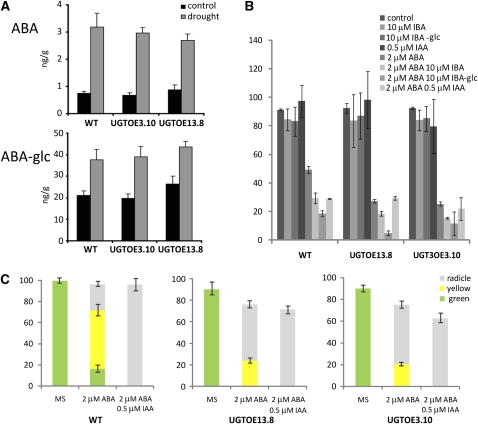
Because the phytohormone ABA plays a pivotal role in plant growth functions, such as stomatal opening, flowering time, and adjustment to abiotic stress conditions (Finkelstein et al., 2002; Quesada et al., 2003; Mishra et al., 2006; Shen et al., 2006), both the observed differences in water use efficiency (Figures 4D) and delayed flowering of UGT74E2OE plants prompted us to study the effect of perturbed IBA homeostasis on ABA responses. Like auxin, ABA levels are regulated by the relative rates of biosynthesis, catabolism, conjugation, and transport. The main conjugation pathway occurs through ABA glucosylation that forms ABA-Glc esters (Cutler and Krochko, 1999; Nambara and Marion-Poll, 2005) that have been suggested to be a storage and/or transport form of ABA or a signal molecule (Sauter et al., 2002; Lee et al., 2006). To explore the possibility that IBA-dependent changes in ABA homeostasis account for the increased water stress tolerance displayed by UGT74E2OE plants, we quantified ABA and ABA-Glc levels per fresh weight by LC-MS in both transgenic and wild-type seedlings (developmental growth stage 1.04) under control and dehydration stress conditions. In nonstressed seedlings of wild-type and UGT74E2OE lines, the levels of ABA and ABA-Glc were similar (Figure 5A). Also under dehydration conditions, levels of free ABA and ABA-Glc increased similarly in all the lines, confirming that UGT74E2 is not an ABA glucosyltransferase, as previously demonstrated in in vitro assays (Figure 2A), and that IBA deregulation does not significantly affect free ABA levels.
Figure 5.
ABA Homeostasis and Response in Transgenic Lines.
(A) Relative abundance of ABA and ABA-Glc per unit fresh weight of wild-type and UGT74E2OE plants quantified by HPLC-MS under control and dehydrated conditions. Data are based on three biological replicates (means ± se).
(B) Percentage of emerged radicles for wild-type and UGT74E2OE lines scored 3 d after seed imbibition in the absence (control) or presence of 2 μM ABA, 10 μM IBA, 10 μM IBA-Glc, 0.5 μM IAA, or a combination of each auxin with ABA at the same concentration as mentioned before. Data are means ± se and are based on three biological replicates of 50 seeds each and P < 0.05.
(C) Percentage of emerged green cotyledons 8 d after imbibition in wild-type and transgenic lines grown as described in (B). Data are means ± se and are based on three replicates of 50 seeds each and P < 0.05.
We analyzed the effect of disturbed IBA homeostasis on ABA signaling by assessing the ABA sensitivity of transgenic lines during germination and seedling establishment. ABA inhibition of seed germination was determined by scoring the percentage of seeds with radicle emergence in the presence of 2 μM ABA. Under normal conditions, no differences in germination were observed, but in the presence of ABA, the germination of UGT74E2OE lines was clearly delayed and more inhibited than that of wild-type seeds (Figures 5B and 5C). ABA inhibition of early seedling development was measured by scoring greening of cotyledons 8 d after germination. The seedlings were subclassed into three development stages: seedlings at stage 0.5 (root), cotyledons without chloroplast development (yellow cotyledons), and seedlings at stage 1.0 (true leaves >1 mm). UGT74E2OE plants were hypersensitive to ABA, showing higher inhibition of cotyledon emergence, greening, and development of true leaves than the wild-type plants. Transgenic seedlings were developmentally arrested because none of the cotyledons could green and no seedlings with visible true leaves could be detected after 8 d (Figure 5C). The enhanced ABA sensitivity displayed by transgenic plants suggests that IBA homeostasis affects the regulation of ABA signaling by increasing the sensitivity to high ABA levels during germination and seedling development.
To confirm that the increased ABA sensitivity in UGT74E2OE plants resulted from the increased levels of IBA or IBA-Glc, we tested whether the combination of IBA, IBA-Glc, or IAA and ABA synergistically repressed germination and seedling establishment. Germination was slightly affected on medium supplemented with 10 μM IBA or IBA-Glc, but not at 0.5 μM IAA in wild-type or transgenic lines (Figure 5B). After 8 d, no developmental arrest could be observed by auxin treatment (Figure 5C). However, in combination, IBA, IBA-Glc, or IAA and ABA markedly inhibited germination after 3 d in all the lines and, more pronouncedly, in the transgenic lines (Figure 5B). While IBA and IBA-Glc in combination with ABA stopped further germination and axis elongation of radicles in all the lines, a small proportion of wild-type seedlings could green or further germinate after 8 d of growth in the presence of IAA together with ABA. Transgenic seedlings were more sensitive to the synergic effect of IAA and ABA on germination that also blocked seedling establishment (Figure 5C). Taken together, these observations strengthen the notion that auxins enhance ABA inhibition of germination as well as seedling development. Hence, IBA deregulation in UGT74E2OE plants magnifies the crosstalk between auxin and ABA during early seedling growth.
DISCUSSION
UGT74E2 Perturbs Auxin Homeostasis
Auxins are critical regulators of transcriptional changes that drive essential developmental processes, including directional growth responses (tropisms), control of plant architecture, abiotic and biotic stress responses, and flower and embryo development (Robert and Friml, 2009). Despite the wide range of auxin types in plants, most research conducted on endogenous auxin has been focused on the primary free auxin, IAA. The other abundant, naturally occurring auxin IBA has received scant attention, and in vivo studies on its function are rather limited, although concentrations in several plant species approach those of IAA and in Arabidopsis seedlings constitute ~25 to 30% of the total free auxin pool (Ludwig-Müller et al., 1993; Bartel et al., 2001; Rashotte et al., 2003; Poupart et al., 2005; Strader et al., 2008). Like IAA, the activity of IBA affects lateral root induction and elongation of roots, shoots, and hypocotyls (Zolman et al., 2000; Rashotte et al., 2003), as well as adventitious root initiation (Ludwig-Müller and Hilgenberg, 1995), callus regeneration (Márton and Browse, 1991), and induction of auxin-responsive reporter genes (Oono et al., 1998; Ulmasov et al., 1999).
Different lines of evidence have suggested that IBA and IAA can be interconverted: IBA is synthesized from IAA and, vice versa, can be converted to IAA in a process paralleling fatty acid β-oxidation (Ludwig-Müller and Epstein, 1994; Bartel et al., 2001; Zolman et al., 2001a, 2001b, 2007, 2008) and act as a precursor to IAA. Many Arabidopsis mutants with reduced root growth sensitivity to IBA, but normal sensitivity to IAA, have defective β-oxidation (Bartel et al., 2001; Zolman et al., 2001a, 2001b, 2007, 2008). However, reports on several IBA-resistant, IAA-sensitive mutants without defects in β-oxidation suggest that IBA has direct auxin effects independently of its conversion to IAA. This partial independence of both auxins is further supported by accounted differences in IAA and IBA polar transport (Zolman et al., 2000; Rashotte et al., 2003; Poupart et al., 2005).
Plants use various mechanisms to spatially and temporally regulate IAA concentration and gradients (auxin homeostasis). Besides de novo biosynthesis, degradation, and transport, two main groups of IAA conjugates are synthesized and hydrolyzed (Ljung et al., 2002; Woodward and Bartel, 2005): ester-type conjugates, in which the carboxyl group of IAA is linked to sugars (for example, Glc) or cyclic polyols (such as inositol), and amide-type conjugates in which the carboxyl group forms an amide bond with amino acids or polypeptides (Jackson et al., 2002; Ljung et al., 2002; Staswick et al., 2005; Park et al., 2007).
Unlike IAA, the pool of IBA is largely ester conjugated (Ludwig-Müller and Epstein, 1993). Besides IBA synthetase activities in maize and Arabidopsis, IBA glucosylation activity in Arabidopsis (Ludwig-Müller and Epstein, 1993; Ludwig-Müller, 2007) and IBA-Ala hydrolase in wheat (Triticum aestivum) (Campanella et al., 2004), the identity of the plant genes involved in IBA biosynthesis, conjugation, and/or hydrolysis remained unknown (Ludwig-Müller and Epstein, 1993). Here, we demonstrate that UGT74E2 is an IBA glucosyltransferase with a high and preferred activity toward IBA in in vitro assays and that its overexpression resulted in increased IBA-Glc levels in planta (Figure 2, Table 1).
The ectopic expression of UGT74E2 also affected the general auxin homeostasis in planta with increased free IBA and IBA-Glc levels (Table 1), suggesting that the IBA synthesis is induced to compensate for the increased IBA-glucosyltransferase activity. Moreover, in the UGT74E2 transgenic lines, three glycosylhydrolases are transcriptionally upregulated (see Supplemental Table 2 online). Assuming a positive feedback due to elevated IBA-Glc levels, this response could reflect the upregulation of IBA-glycosylhydrolases, making these genes good candidates to encode currently unidentified IBA-Glc hydrolases.
For IAA, the Glc-esterified pool decreased in UGT74E2OE seedlings, while IAA-Glu and oxIAA levels increased (Table 1). Although it cannot be excluded that transgenic plants have a reduced free IAA content in specific organs or cell types, total levels of free IAA were not affected. A similar increase in enzymatic substrate and product was observed in transgenic plants overexpressing the IAA-glucosyltransferase UGT84B1. Possibly, the plants overcompensate for overexpression of UGT74E2 by the synthesis or release of the free hormones, triggered by their increased conjugation rate, to maintain cellular homeostasis. Interestingly, the effect on IAA-Glu and oxIAA accumulation by UGT84B1 overexpression was opposite (Jackson et al., 2002), underlining the complex homeostasis of auxin and its conjugates.
Modification of IBA Homeostasis Impinges on Plant Root and Shoot Development
Ectopic expression of the group L UGTs, UGT84B1 (Jackson et al., 2002) and UGT74E2 (this study), led to similar morphological changes, such as a rounded shape of the rosette leaves, short petioles, dwarfed stature, and loss of apical dominance (Figure 3; see Supplemental Table 1 online). Additionally, UGT74E2OE plants showed increased chlorophyll accumulation and delayed flowering (see Supplemental Table 1 and Supplemental Figure 2B online). Exogenously applied IBA or IBA-Glc could not mimic the architectural changes that are apparent in the gain-of-function lines, but decreased the leaf area and had an inhibitory effect on the flowering time (see Supplemental Figures 1 and 2B online). Therefore, the increased levels of IBA and IBA-Glc could explain the delayed flowering phenotype displayed by UGT74E2OE plants. Despite the many reports concerning the influence of IAA on floral induction, its role in this process has not been elucidated yet (Frankowski et al., 2009). Our study shows that the exogenous application of other auxins, such as IBA and IBA-Glc, can postpone the flowering time. Moreover, the observation that free IBA, but not IAA, levels increase in UGT74E2OE plants indicates that increased IBA levels probably affect the shoot morphogenesis directly rather than through a conversion to IAA.
The reason for the inability to mimic all transgenic phenotypes through exogenous application might be that in these types of feeding experiments, endogenous auxin homeostasis mechanisms (catabolism, transport, and distribution) impair a similar IBA/IBA-Glc long-term perturbation as provoked by ectopic overexpression of UGT74E2. An unresolved question remains regarding the cellular function of IBA-Glc. Whereas, in contrast with IBA, IBA-Glc is not an active auxin in root growth assays (see Supplemental Figure 1 online), its in vivo accumulation correlates with leaf and shoot growth alterations (see Supplemental Figure 2 online). This apparent discrepancy between root and shoot phenotypes could be explained by the possibility that IBA-Glc acts as an IBA storage form in roots that can be exported to green tissues in response to internal or external stimuli. In leaves, IBA-Glc could become biologically active or be used as a slow-release form of IBA. In the overexpressing plants, morphological changes might result from a perturbed spatial auxin distribution and transport. Overaccumulation of IBA in UGT74E2OE plants could inhibit the IAA transport by competing for the same transporters. Alternatively, altered IBA/IBA-Glc ratios induced by ectopic UGT74E2 expression might affect the IBA sensitivity, steering alterations in plant morphology.
UGT74E2 Is Involved in Stress-Induced Morphological Adaptations
Commonly observed symptoms of stressed plants include growth retardation and reduced metabolism that might be caused by reallocation of metabolic resources between different physiological pathways to maximize plant survival under stress conditions. Because a wide variety of biotic and abiotic stresses can induce the formation of ROS, they might play a role in controlling developmental processes and modulate auxin sensitivity (Potters et al., 2007, 2009). Recent studies have shown that oxidative stress induces a broad spectrum of auxin-like effects in Arabidopsis seedlings, such as elongation inhibition of roots, cotyledons, and leaves, effects consistent with alterations in auxin levels and/or distribution. The similar effects of a wide variety of stresses on growth, referred to as stress-induced morphological responses, hint at a common mechanism for the control of IAA levels in cells and/or tissues (Leyser, 2005; Pasternak et al., 2005; Potters et al., 2009).
UGT74E2 overexpression not only affected the plant architecture but also allowed transgenic plants to survive prolonged periods of drought and high salinity (Figure 4), suggesting a relationship between architectural changes and environmental stress resistance. Profuse shoot branching had already been observed in Arabidopsis plants treated with medium UV-B radiation and strong wind exposure (Brodführer, 1955). Also, stunted petioles and, hence, compressed rosette structure might provide a stress avoidance strategy by preventing direct soil water evaporation through expansion of ground coverage. Based on the phenotype and stress tolerance of UGT74E2OE plants and the stress inducibility of the gene, we can hypothesize that UGT74E2 is one component of the ROS signaling pathway that alters the auxin responsiveness, leading to stress-induced reorientation of growth, directly relevant for plant adaptation. In this sense, the tissue-specific production of UGT74E2 (Figure 1) reveals a clear role of these IBA-modifying proteins in the auxin distribution. UGT74E2 is expressed at sites of auxin synthesis and where the hormone accumulates: lateral root primordia, root tips, primordial leaf tips, and hydathodes. This asymmetric distribution of auxin is required for correct specification of cell fates (Leyser, 2005). The auxin gradient pattern is maintained by polar auxin transport (Ikeda et al., 2009) and is crucial for the morphogenesis of organs, such as embryonic cotyledons and roots, leaf and flower primordia in shoot meristems, primordia of lateral roots, as well as leaf vascular pattern and shape (Zgurski et al., 2005; Scarpella et al., 2010). The absence of substantial upregulation of auxin-responsive genes (see Supplemental Table 2 online) suggests that the transgenic plants are adapted to the constitutive enhancement of IBA and IBA-Glc levels. Induction of UGT74E2 expression under stress (Figure 1; see Supplemental Figure 1 online) reflects its involvement in the alteration of free IBA levels and the consequent changes in auxin distribution that will affect auxin-dependent stress responses. Hence, we can postulate that UGT74E2 has a function in auxin gradient disruption under stress conditions. Following this idea, ectopic expression of UGT74E2 could affect auxin gradient, thus mimicking the effect of stress on auxin homeostasis. The permanent auxin deregulation in the transgenic plants might, for example, improve osmotic adjustment or decrease water consumption and also shape the morphology of rosettes as part of an adaptative stress response. The wild-type phenotype observed in ugt74e2 knockout lines might result from redundant activities of one of the more than 70 UGTs with activity toward IBA, such as UGT84B1 (Jackson et al., 2001) or its closest homolog UGT74E1.
Auxins Might Assist in Safeguarding the Photosynthetic Capacity during Water Stress
In maize, both free and conjugated IBA increased under water-limiting conditions (Ludwig-Müller and Hilgenberg, 1995). Our observations that IBA and IBA-Glc are strongly induced by osmotic stress and that constitutively increased levels of free and glucosylated IBA (Table 1) correlate with water stress tolerance imply a significant role of IBA in adaptation to water stress. Similarly, overproduction of WES1, a stress-inducible IAA-Asp–conjugating GH3 enzyme, reduced growth and increased biotic and abiotic stress tolerance in Arabidopsis plants by directing IAA into the IAA-Asp catabolic pathway (Park et al., 2007). In this context, UGT74E2 could represent another stress-responsive component in the auxin modulation through a direct effect on IBA homeostasis and, thereby, indirectly on IAA homeostasis by steering IAA into conjugative and oxidative pathways.
Although we could mimic the photosynthetic response of UGT74E2OE plants during water stress by exogenously supplied IAA using PEG-stressed wild-type seedlings, IBA and IBA-Glc accumulation failed to simulate the effects of stress on the photosynthetic parameters (see Supplemental Table 3 online). These results suggest that the protective auxin-dependent response relies on an increase in IAA levels that is triggered by water stress signaling (see Supplemental Table 3B online). The IAA treatment was effective only when seedlings were germinated and grown on medium supplemented with IAA but not when IAA was supplied at the beginning of the stress period. In other words, establishment of an appropriate IAA gradient is necessary to provide positional cues for the response. As UGT74E2OE plants accumulate high levels of IBA and IBA-Glc, the IBA-IBA-Glc conversion to IAA is important for the proper effect on photosynthesis displayed by the transgenic plants under stress, but the IAA levels were not affected in wild-type or transgenic plants under PEG-induced water stress. Nevertheless, it cannot be excluded that in the transgenic plants the free IAA content increases under stress in specific organs or cell types, raising the possibility that a localized increase in IAA, maybe in chloroplasts, leads to a protective effect on photosynthesis and, thus, on energy metabolism. As a consequence, plants would cope better with the stress by having the increased cellular energy to expend on homeostasis and osmotic balance.
Another possibility might be that IBA-IBA-Glc homeostasis in UGT74E2OE plants positively affects the IAA transport in photosynthetic tissues under water stress, allowing IAA to act favorably on photosynthesis. Thus, photosynthesis, like so many other physiological processes, is apparently modulated by auxins, allowing plants to adjust or adapt their photosynthesis to environmental cues. Interestingly, a stress-responsive chlorophyll binding protein in the light-harvesting complex of PSII (ELIP2) was upregulated in UGT74E2OE seedlings (see Supplemental Table 2 online). ELIP2 has been reported to protect chloroplasts against photooxidative damage and to be required also for normal chlorophyll accumulation in deetiolated seedlings (Rossini et al., 2006). Furthermore, in pea (Pisum sativum), ELIP is strongly induced during UV-B and salt stress (Sävenstrand et al., 2004), and in Arabidopsis, ELIP2 is triggered by diverse abiotic stresses (Tzvetkova-Chevolleau et al., 2007), possibly to protect photosynthetic components against stress-induced damage. However, further studies will be needed to clarify the role of IBA and IBA-Glc on photosynthetic processes under stress.
IBA Is Involved in Water Stress Responses, Independently of ABA
The water stress–tolerant phenotype of UGT74E2OE seems be a direct consequence of increased IBA and IBA-Glc levels rather than derived from an altered ABA homeostasis. First, ABA levels were similar in wild-type and transgenic plants, both under control or dehydration conditions (Figure 5A). Although the Arabidopsis ABA glycosylhydrolase BGL1 transcripts are downregulated (see Supplemental Table 2 online), the levels of ABA-Glc are identical in transgenic and wild-type plants (Figure 5A). Moreover, real-time quantitative RT-PCR analysis revealed that the expression levels of the transcripts for the UGT71B6 (ABA-specific UGT) or 9-cis-epoxycarotenoid dioxygenase, a key enzyme in ABA biosynthesis, did not differ in the wild-type and transgenic lines. Together, these findings indicate that perturbations in the IBA levels do not affect the ABA biosynthesis. Second, the microarray analysis did not imply a genuine ABA-dependent transcriptional response (see Supplemental Table 2 online). Third, the stomatal conductance of UGT74E2OE plants was similar to that of wild-type plants (see Supplemental Table 1 online). Thus, high IBA-IBA-Glc levels in the transgenic plants do seemingly not affect the ABA-dependent stomatal regulation (Acharya and Assmann, 2009), suggesting that the improved water stress tolerance of UGT74E2OE plants (Figure 4) might be associated with the drought adaptation process rather than with the ABA response adjustments (Park, 2007). In this view, the modified expression of an osmotic stress–inducible vacuolar calcium binding protein in the UGT72E4 plants (see Supplemental Table 2 online) might provide an interesting lead as a component involved in the IBA-specific signaling in response to osmotic stress.
Auxins Amplify ABA-Induced Inhibition of Seed Germination
The precise mechanism on how perturbed IBA homeostasis affects water use efficiency and water tolerance without influencing the ABA responses remains speculative because altered interactions between auxin- and ABA-dependent responses cannot be excluded during early seedling development in the transgenic plants. Both germination and seedling growth were more sensitive to ABA in UGT74E2OE plants (Figures 5B and 5C). At these developmental stages, crosstalk between ABA- and auxin-dependent responses has been reported to take place, in which either ABA-dependent repression of growth is potentiated by auxin (Liu et al., 2007) or auxin repression of embryonic axis elongation is enhanced by ABA (Belin et al., 2009). Here, we demonstrate that high levels of IBA, IBA-Glc, or IAA enhance the ABA inhibition of germination as well as of seedling development (Figure 5). Moreover, accumulation of IBA and IBA-Glc in combination with ABA prevented further postgermination growth. The increased sensitivity of UGT74E2OE plants to ABA, or ABA simultaneously with auxin, during germination and early seedling growth (Figure 5) highlights the role of IBA and IBA-Glc in these processes.
Conclusion
In summary, plant morphogenesis, including shoot branching, leaf area, and sprouting of axillary buds, is affected by IBA and/or IBA-Glc homeostasis. Moreover, IBA-Glc can act as a physiologically active form of auxin on processes, such as rosette shape and flowering. Because of its spatial restriction to younger tissues, UGT74E2 might play a prominent role in stress-induced protective architectural changes in plants. The mechanisms that steer the IBA/IBA-Glc–dependent water stress–tolerant phenotype are less clear. Several lines of evidence hint at an action through improved osmotic adjustment or decreased water consumption rather than through a direct modulation of ABA-dependent protective responses. One attractive, but highly speculative, hypothesis is that, similar to the ABA-dependent protective stomatal closure as a drought stress response in fully developed leaves, the correlation between the stress-inducible UGT74E2 expression and the enhanced auxin levels in newly formed tissues reflects a previously undescribed, auxin-dependent mechanism to protect these tissues during water stress events. Clearly, manipulation of IBA homeostasis might be a new avenue in crop protection for water stress tolerance. In future research, the analysis of crosses between UGT74E2 overexpressors and mutants blocked in IBA biosynthesis, transport, and oxidation to IAA would allow us to investigate the timing and importance of IBA and IBA-to-IAA conversion in plant development, in addition to their role during water stress.
METHODS
Cloning of UGT74E2
A BX815725 full-length GSLT cDNA template provided by Genoscope (Centre National de Séquençage, Evry, France) was used as template for PCR reactions with PLATINUM Pfx DNA polymerase (Invitrogen) with primers 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCACCATGAGAGAAGGATCTCATCTT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAACAAAACATAGAAACAAACT-3′. The PCR product was cloned by recombination into pDONR221 (Invitrogen) to generate the entry vector.
Plant Material, Growth Conditions, and Transformation
All Arabidopsis thaliana lines used were in the Col-0 ecotype, unless otherwise specified. Seeds were sterilized by overnight incubation with chlorine gas (100 mL 12% NaOCl and 3 mL 37% HClO). Plants were grown on 4.3 g/L MS medium (Duchefa), 0.5 g/L 2-(N-morpholino)ethanesulfonic acid, 0.1 g/L myo-inositol, 5 g/L sucrose, 7 g/L plant tissue culture agar (LabM), 0.5 mg/L nicotinic acid, 0.5 mg/L pyridoxin, and 1 mg/L thiamin at 22°C and were given 65 μE m–2 s–1 radiation in a 16-h-light/8-h-dark photoperiod. Arabidopsis plants were transformed via the Agrobacterium tumefaciens–mediated floral dip (Clough and Bent, 1998). Kanamycin-resistant plants were selected on MS medium with 35 mg/L kanamycin (Sigma-Aldrich). For auxin treatments, seedlings were grown under yellow-filtered light.
Soil-grown plants were cultivated in controlled culture rooms either under an 8-h photoperiod of 300 μmol E m–2 s–1 or under a 16-h photoperiod of 85 μE m–2 s–1. Day/night temperature and relative humidity were 22/18°C and 60/50%, respectively.
Generation of Arabidopsis Col-0 Overexpression Lines
UGT74E2 was cloned by recombination from the Gateway-compatible pDONR221 (Invitrogen) to the p35S overexpression vector pK7WG2 (Karimi et al., 2002). The construct was transformed into Arabidopsis Col-0 by Agrobacterium-mediated floral dip. Overproduction of UGT74E2 was confirmed by real-time RT-PCR. Homozygous lines with a single T-DNA locus were selected by segregation and protein gel blot analysis.
Identification of Homozygous Insertion, Mutants at the UGT74E2 Locus
T-DNA insertional mutant lines containing a single T-DNA insertion in the At UGT74E2 gene (SALK_016116) or promoter region (SALK_091130) were selected from the SALK T-DNA collection. To identify mutants homozygous for the T-DNA insertion, genomic DNA of seedlings was subjected to PCR genotyping with the insertion primer LBb1-3 (5′-ATTTTGCCGATTTCGGAAC-3′) and either the forward flanking primers, 16LP (5′-ATCAAATGTTGGAACTCGCTG-3′) and 9LP (5′-CGTTTTTCCGTTAAATATTTTAAATG-3′), or the reverse ones, 16RP (5′-AGAAAGCTGGGTCACAAAACATAGAAACAAACTCAT-3′) and 9RP (5′-AAGACCTTTTGAGGCTAAGCG-3′). The cycling parameters were one cycle at 94°C for 3 min followed by 35 cycles (at 94°C for 45 s, at 53°C for 30 s, and at 72°C for 1 min) and a 10-min extension at 72°C.
For RT-PCR analysis, total RNA was isolated with the TRIzol reagent (Invitrogen), and cDNA was synthesized from 2 μg total RNA with the SuperScript II RNase H-reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Each cDNA sample was diluted 1:10, of which 2 μL was used for PCR amplification with At UGT74E2-specific primers (UGT-F, 5′-TAACTTCTTCCACACTTCTCATAATCT-3′, and UGT-R, 5′-ACAACAAAAACTAGAGTCAGTAACAAC-3′). PCR amplification of the Arabidopsis ACTIN2 gene with gene-specific primers (forward primer, 5′-TCGGTGGTTCCATTCTTGCT-3′, and reverse primer, 5′-GCTTTTTAAGCCTTTGATCTTGAGAG-3′) was done as a loading control. Cycling parameters were one cycle (at 94°C for 2 min) followed by 30 cycles (at 94°C for 30 s, at 50°C for 30 s, and at 72°C for 2 min) and a 5-min extension at 72°C. PCR products were detected by SYBR Green-saved stained (Invitrogen) 2% agarose gels. All reactions were done in triplicate with two independent RNA samples.
IBA-Glc Synthesis
1H and 13C NMR spectra were recorded on an AV600 spectrometer (Bruker) (600 MHz for 1H and 150 MHz for 13C) (chemical shifts quoted relative to CDCl3 or CD3OD). All synthesized compounds used in the enzyme assays were analyzed at the Australian National University Microanalytical Facility (Perth, Australia). Flash chromatography was done on silica gels (BDH) with the specified solvents. Thin layer chromatography was performed on silica gel 60 F254 (Merck) aluminum-backed plates that were stained by heating (>200°C) with 5% sulfuric acid in ethanol. Percentage yields for chemical reactions as described are quoted only for those compounds that were purified by column chromatography. Purity was assessed by thin layer chromatography or 1H NMR spectroscopy.
For 2,3,4,6-tetra-O-benzyl-1-O-(4-[1H-indol-3-yl]butanoyl)-β-d-glucose, IBA (390 mg, 1.9 mmol) was added to a solution of tetra-O-acetyl-glucosyl trichloroacetimidate (Schmidt and Michel, 1985) (1.3 g, 1.9 mmol) in CH2Cl2 (20 mL) and the solution stirred (at room temperature) for 8 h. Concentration of the mixture and flash chromatography of the residue (EtOAc/hexanes, 1:3) yielded the ester 2,3,4,6-tetra-O-benzyl-1-O-(4[1H-indol-3-yl]butanoyl)--d-glucose as a colorless oil (1.1 g, 83%). The physical properties of the compound are: Rf 0.40 (EtOAc/hexanes, 3:7); 1H NMR (600 MHz, CDCl3) δ 7.94 (br s, 1H), 7.58 (d, J = 8 Hz, 1H), 7.35 to 7.22 (m, 20H), 7.19 to 7.10 (m, 3H), 6.90 (s, 1H), 5.64 (d, J = 8.1 Hz, 1H), 4.89 (A part of ABq, J = 10.9 Hz, 1H), 4.84 to 4.81 (m, 2H), 4.75 (A part of ABq, J = 11.5 Hz, 1H), 4.73 (A part of ABq, J = 11.4 Hz, 1H), 4.61 (A part of ABq, J = 12.1 Hz, 1H), 4.54 (A part of ABq, J = 10.8 Hz, 1H), 4.48 (A part of ABq, J = 12.2 Hz, 1H), 3.75 to 3.71 (m, 4H), 3.60 to 3.56 (m, 2H), 2.79 (dt, J = 2.6, 7.3 Hz, 1H), 2.42 to 2.39 (m,1H), 2.36 to 2.31 (m, 1H), 2.11 to 2.08 (m, 2H); 13C NMR (150 MHz, CDCl3) δ 172.0, 138.3, 138.0, 137.9, 137.8, 136.3, 128.4-127.3, 121.9, 121.6, 119.2, 118.8, 115.2, 111.1, 93.9, 84.7, 81.0, 77.1, 75.7, 75.4, 75.0, 74.9, 73.4, 68.1, 33.6, 24.8, 24.2; and high-resolution MS mass-to-charge ratio (m/z) 726.3443 ([M + H]+ C46H47NO7).
For 1-O-(4-[1H-indol-3-yl]butanoyl)-β-d-glucopyranose, Palladium-on-charcoal (10%, 200 mg) was added to a solution of the ester 2,3,4,6-tetra-O-benzyl-1-O-(4[1H-indol-3-yl]butanoyl)--d-glucose (0.65 g, 0.90 mmol) in methanol:EtOAc:CH3COOH (3:2:2, 35 mL) and the solution stirred under one atmosphere of hydrogen gas (1 atm., 30 min). The mixture was filtered through a Celite pad and concentration of the mixture followed by flash chromatography of the residue (methanol/EtOAc, 1:9) yielded the ester X as a colorless oil (0.18 g, 56%). The physical properties of the compound are: Rf 0.28 (methanol/EtOAc, 1:9); 1H NMR (600 MHz, CD3OD) δ 7.52 (d, J = 8 Hz, 1H), 7.32 (d, J = 8.2 Hz, 1H), 7.06 (t, J = 8 Hz, 1H), 7.02 (s, 1H), 6.98 (t, J = 8 Hz, 1H), 5.49 (d, J = 8.1 Hz, 1H), 3.82 (dd, J = 1.9, 12.1 Hz, 1H), 3.67 (dd, J = 4.7, 11.9 Hz, 1H), 3.41 to 3.32 (m, 4H), 2.79 (t, J = 7.3 Hz, 1H), 2.46 to 2.38 (m,2H), 2.04 to 1.98 (m, 2H); 13C NMR (150 MHz, CDCl3) δ 174.2, 138.2, 128.7, 123.1, 12.2, 119.4, 119.3, 115.4, 112.1, 95.6, 78.8, 78.0, 73.9, 71.1, 62.3, 34.5, 26.5, 25.3; and high-resolution MS m/z 366.1567 ([M + H]+ of C18H23NO7).
Stress Treatments
For the high-light induction assays, 6-week-old catalase-deficient (CAT2HP1) and control (PTHW) plants were exposed to continuous high-light irradiation (Vanderauwera et al., 2005). Middle-aged leaves of 20 to 30 plants per line were sampled 0, 3, and 8 h after the onset of the high-light stress and pooled for RNA analysis.
For in vitro salt stress survival assays, 1-month-old seedlings grown on MS agar plates at 22°C and 100 μE m–2 s–1 were transferred to new MS plates supplemented with 150 mM NaCl. Survival rate was scored after 8 d.
H2O2 stress was imposed by transferring 14-d-old pUGT74E2:LUC or wild-type seedlings to new MS 0.5% sucrose plates supplemented with 1 or 3 mM H2O2 for 5 d.
Drought tolerance was investigated by two experimental settings. First, seeds from wild-type Col-0 and two independent UGT74E2OE lines (grown on the same tray under optimal growth conditions) were used. All plants were grown in separate pots on Jiffy-7 soil pellets (Jiffy Products) in a controlled growth chamber under a 16-h light regime at 21°C and 70% relative humidity for 2 weeks under normal watering. After 2 weeks, pots were irrigated in a controlled manner by bringing the total weight of each pot (plastic container, soil, and plant) to 63 g with water two to three times weekly to ensure that all plants received a similar watering regime. After 3.5 weeks, all pots were brought to the target weight a last time and separated into two groups, one with further controlled watering and one without watering (five plants per line). After 13 d without watering, the drought-treated plants were rewatered, and recovery was checked after 24 h. For the in-soil salt stress assay, a similar procedure was followed, but instead of a watering stop, watering was continued with water containing 500 mM NaCl.
In a second, semiautomated drought stress experiment, plants (six plants per line) were germinated in cylindrical polypropylene pots (200 mL, 53-mm diameter, 88-mm height; VWR International) filled with ~90 g of soil (Saniflor professional potting compost containing 20% organics; white peat, garden peat, fertilizer based on calcium and magnesium; pH 5.0 to 6.5; electric conductivity of 450 μS/n) at 1.75 g water/g dry soil water concentrations until growth stage 1.04 (Boyes et al., 2001), corresponding to a plant age of ~14 d. Wild-type and transgenic plants were further grown for 3.5 weeks under 1.50 g water/g dry soil water regime until bolting. During this period, changes in pot plus plant weight were corrected by adding water on a daily basis. After bolting, watering was stopped for 13 d, and the total weight of each individual pot (pot, soil, and plant) was recorded regularly.
For treatments with PEG, seedlings at developmental stage 1.04 were carefully removed from the MS 0.5% sucrose agar plates and immersed in MS liquid medium supplemented with 10% PEG6000 and incubated at 22°C under continuous light for 2 d. Seedlings were used for hormone profiling.
For salt stress transcript analysis, sterilized Arabidopsis Col-0 seeds were sown in growth boxes on conatin ingrafts (LifeRaftR) supported by floats (Raft Float). The boxes were closed with ventilated lids (Osmotek; kindly provided by Julia Knutova). Nine seeds were placed per raft and transferred to solid medium in the growth boxes (Kilian et al., 2007). The boxes were stratified for 2 d at 4°C and incubated for 11 d in an 8-h-light/16-h-dark regime. Membrane rafts were transferred to liquid medium, supported by the floats. On day 18 after sowing, plants were mock treated or salt stressed by adding NaCl up to a concentration of 150 mM in the liquid medium. After 3 h, the roots were harvested and snap frozen.
For dehydration stress, plants grown on MS plates until stage 1.04 were exposed to 30% relative humidity for 10 h as described previously (Lee et al., 2006). Seedlings were harvested for ABA measurement.
ABA Inhibition of Seed Germination and Early Seedling Development Assays
Wild-type and transgenic seeds were sown on MS 0.5% sucrose agar plates with or without 2 μM ABA, 0.5 mM IAA, 10 mM IBA, 10 mM IBA-Glc, or a combination of 2 mM ABA with the above-mentioned concentration of IAA, IBA, or IBA-Glc. The percentage of seed germination was scored 3 d after imbibition and examined under a dissecting microscope. Germination is defined as an obvious protrusion of the radicle through the seed coat. For the ABA inhibition of early seedling development, the percentage of seedlings with green, yellow, or radicle was scored 9 d after imbibition. Each experiment was done in triplicate of 100 seeds each.
Measurement of Root Length and Lateral Root Response
For auxin-responsive root elongation assays, seedlings were grown vertically for 4 d on MS 0.5% sucrose agar medium before transfer to medium supplemented with the indicated auxin or auxin transport inhibitor concentrations. The lengths of primary roots were measured after 8 or 10 d. Digital images of seedlings were captured and the root length quantified with the ImageJ program (http://rsb.info.nih.gov/ij/).
For lateral root assays, 4-d-old seedlings grown vertically on MS 0.5% sucrose agar medium were transferred to new plates supplemented with either different concentrations of the indicated auxin or auxin plus the transport inhibitor NPA and grown for eight additional days. Primordia emerging from the primary root, as seen under a dissecting microscope, were counted as lateral roots.
Quantitative Real-Time PCR
Total RNA and cDNA were prepared with TRIzol reagent (Invitrogen) and with Superscript II RNase H-reverse transcriptase (Invitrogen), respectively, according to the manufacturer's instructions. The cDNA was synthesized from 2 μg total RNA, and each cDNA sample was diluted 1:5, of which 2 μL was used for UGT74E2 transcript abundance measurement by means of the real-time PCR kit (Invitrogen) with Universal ProbeLibrary probe #138 (Roche Diagnostics) and primers 5′-GAATCGTCCTCATACCCGAAT-3′ and 5′-GCTTTGGACCCATTTCAACA-3′. The gene coding for ACTIN-RELATED PROTEIN7 was used for normalization with Universal ProbeLibrary probe #147 (Roche Diagnostics) and primers 5′-ACTCTTCCTGATGGACAGGTG-3′ and 5′-CTCAACGATTCCATGCTCCT-3′. Amplification was monitored in real time with the iCycler iQ (Bio-Rad) sequence detection system. For each reaction, the crossing point was determined with the Fit Point Method of the iCycler Software 3.1. PCR reactions were done in triplicate with three independent RNA samples.
Promoter-GUS and LUC Analysis
Genomic DNA was isolated from Arabidopsis (Col-0) with the DNeasy plant kit (Qiagen) according to the manufacturer's instructions. The 1500-bp region upstream of the UGT74E2 start codon was amplified by PCR with the Platinum Taq High Fidelity DNA polymerase (Invitrogen) and the forward and reverse primers. The PCR product was cloned into pDONR221 and by recombination into pBGWFS7 (Karimi et al., 2002) or into pKGWL7, generating transcriptional pUGT74E2:GFP:GUS or pUGT74E2:LUC fusions, respectively. The construct was transformed into Arabidopsis Col-0 by Agrobacterium-mediated floral dip. Multiple transformants with a single insertion locus were selected by segregation analysis. For the treatments with PEG and MV, seedlings at developmental stage 1.02 were carefully removed from MS agar plates, transferred to 96-well plates, and immersed in MS or MS supplemented with 50 mM PEG6000 or 50 mM MV solutions for 5 h. For mannitol treatment, seeds were sown on half-strength MS and 1% sucrose agar plates with or without 25 mM mannitol and grown until developmental stage 1.04.
GUS assays were done according to Beeckman and Engler (1994). Samples were mounted in Tris-saline buffer or lactic acid and photographed with a stereomicroscope (Stemi SV11; Zeiss) or with a differential interference contrast microscope (Leica). Luciferase activity was measured essentially as described (Chinnusamy et al., 2002).
Purification of Recombinant UGT74E2
The UGT74E2-coding sequence was transferred by recombination from pDONR221 to the 6xHIS pDEST17 vector (Invitrogen). The expression vector was transformed into Escherichia coli BL21 DE3 pLys. A culture of 100 mL Luria-Bertani broth with 100 μg/mL ampicillin was inoculated and grown overnight at 37°C and 220 rpm. This culture was diluted to 500 mL with an optical density (OD600) of 0.125, grown for 2 h at room temperature and 220 rpm, and induced with 0.2 mM isopropyl-1-thio-β-d-galactopyranoside. After 2 h of induction at room temperature and 220 rpm, cells were harvested by centrifugation at 4000g for 5 min and resuspended in 2 mL of buffer (750 mM sucrose and 200 mM Tris-HCl, pH 8.0). Lysozyme (1 mg) and 14 mL of half-strength buffer were added immediately. After incubation at 4°C for 45 min, the cells were harvested by centrifugation and osmotically shocked in 25 mL of one-third strength PBS containing the protease inhibitor cocktail Complete without EDTA (Roche Diagnostics). Cell debris was removed by centrifugation at 100,000g for 15 min. One milliliter of 50% Talon resin (BD Biosciences) was added and incubated for 3 h under gentle shaking. Talon beads were washed and eluted according to the manufacturer's description. Fractions of 0.5 mL were collected and analyzed by SDS-PAGE (15% [w/v] gel) and protein hybridization with an anti-HIS antibody (Qiagen). Protein concentration of the eluted fractions was determined with Bio-Rad Protein Assay DC with BSA as reference.
Glucosyltransferase Assay
The reaction mix in a volume of 0.1 mL contained 50 mM HEPES, pH 7.6, 2.5 mM MgSO4, 10 mM KCl, 5 mM UDP-Glc, and 14.4 mM 2-mercaptoethanol. Samples were prepared with 1 mM hormone, with or without 2 μg purified UGT74E2, and incubated for 3 h at 30°C. Reactions were terminated by adding 10 μL trifluoroacetic acid. Of each sample, 10 μL was injected by means of a SpectraSystem AS1000 autosampler (Thermo Separation Products), cooled at 4°C, onto a reversed phase Luna C18(2) column (2.1 × 150 mm, 3 μm; Phenomenex). A gradient separation (SpectraSystem P1000XR HPLC pump; Thermo Separation Products) was run from 0.1% aqueous triethylammonium acetate (solvent A, pH 5) to acetonitrile (0.1% triethylammonium acetate; solvent B) under the following conditions: column oven 40°C; flow 0.25 mL min–1; time 0 min, 5% solvent B; time 36 min, 100% solvent B. UV/visible (UV/Vis) absorption between 200 and 450 nm was measured with a SpectraSystem UV6000LP detector (Thermo Separation Products) at a scan rate of 5 Hz. Full mass spectrometry scans (m/z 100 to 700) with an electrospray ionization source, operated in the negative mode, coupled to an LCQ Classic (Thermo Quest) mass spectrometer, were taken under the following conditions: spray voltage, 4.5 kV; sheath gas, 63 (arbitrary); and capillary temperature, 265°C. Similar results were obtained in three independent experiments.
To evaluate the kinetics of IBA-Glc production by UGT74E2, the reaction time was first tested by supplying 5 and 10 μM of IBA for 5, 10, 15, 20, and 30 min. No increased levels of IBA-Glc were observed when 10 μM substrate instead of 5 μM was added or when the reaction times were under 10 min. Therefore, the enzymatic reactions were done for 3 and 5 min with a range of substrate concentrations (5, 50, and 500 pM; 5, 25, 50, and 500 nM; and 1 and 5 μM). Each reaction was done in triplicate. Instead of the SpectraSystem HPLC system, an Acquity UPLC (Waters) consisting of a binary solvent manager, a sample manager, and a 2996 PDA detector was coupled to the LCQ Classic (same parameter values as mentioned above). A 15-min gradient of 95% solvent A (solvent A, 0.1% aqueous acetic acid; solvent B, acetonitrile, 0.1% acetic acid) to 10% solvent A at 400 μL/min was run on a reverse-phase Acquity UPLC BEH C18 column (150 × 2.1 mm, 1.7 μm; Waters) operated at 55°C. UV/Vis absorption spectra were collected between 200 and 450 nm at a rate of 20 Hz. The integration of the peak area was based on the UV/Vis absorption at 225 nm. To determine the Km and Vmax values, a Lineweaver-Burk plot was constructed by regressing the reciprocal of the product concentration (1/μM) versus the reciprocal of the reaction rate (s/μM).
Plant Protein Extraction and Protein Gel Blot Analysis of UGT74E2
Approximately 200 mg of leaf tissue was ground in liquid nitrogen, mixed with 400 μL buffer (10 mM HEPES/KOH, pH 7.9, containing 400 mM NaCl, 0.5 mM DTT, 0.1 mM EDTA, 5% glycerol, and 0.5 mM phenylmethylsulfonylfluoride), then clarified by centrifugation (12,000g for 20 min). Samples corresponding to 175 μg (cat2 and wild-type lines) or 30 μg (UGT74E2OE lines) of protein were resolved by SDS-PAGE on 12% polyacrylamide gels and analyzed by immunoblotting with rabbit antisera raised against UGT74E2 and antirabbit IgG antibody conjugated with horseradish peroxidase (GE Healthcare). Antibody-protein complexes were visualized with an ECL kit (GE Healthcare) and Kodak X-OMAT film (Eastman Kodak). UGT74E2 polyclonal antibodies were produced against two specific peptides that were also used for one further affinity purification of the antibodies (Eurogentec).
Microarray Analysis of UGT74E2OE Lines
Arabidopsis Col-0 and homozygous UGT74E2OE seeds were sown on half-strength MS plant medium in triplicate. Plantlets per triplicate were harvested and pooled at developmental stage 1.04. Total RNA was isolated with TRIzol reagent (Invitrogen). The concentration of total RNA was determined with a Nanodrop ND-1000 spectrophotometer (Agilent Technologies), and the quality was examined with the RNA 6000 Nano Assay (Agilent Technologies). Each triplicate of Arabidopsis Col-0 and homozygous AT PHB4OE plants was hybridized to one Affymetrix chip (Genechip Arabidopsis ATH1 Genome Array). For each hybridization, 5 μg of total RNA was used. Affymetrix chips were analyzed at the VIB Microarray Facility (Leuven, Belgium) under the manufacturer's conditions for reverse transcription, labeling, hybridization, and scanning (https://www.affymetrix.com). Raw data were processed with the Bioconductor software package (Gentleman et al., 2004) and the statistical algorithm RMA (Irizarry et al., 2003). The expression values were analyzed with the limma bioconductor package (Smyth, 2004), and a false discovery rate correction for multiple testing was applied (Benjamini and Hochberg, 1995). Probe sets with a differential regulation of at least 1.5-fold and a P value <0.05 were selected.
Purification of IAA, IAA Conjugates, IBA, and IBA Metabolites
Plant material (0.04 to 0.1 g fresh weight) was homogenized in liquid nitrogen and extracted overnight in 80% methanol. To the extracts, 300 pmol phenyl-13C6-IAA (Cambridge Isotope Laboratory) was added. After overnight extraction at −18°C in the dark, cell debris was removed by centrifugation (15 min at 16,100g; Eppendorf Centrifuge 5415D). The supernatant was passed over a RP-C18 solid-phase extraction cartridge (500 mg; Varian) as described (Prinsen et al., 2000). The pigments were retained on the cartridge, whereas the effluent contained all compounds of interest. At this stage, the effluent was divided into three equal aliquots. One aliquot was analyzed for IBA and IBA metabolites by means of LC-MS/MS under scan, parent scan, and multiple reaction monitoring (MRM) conditions. A second aliquot was further purified for free IAA, whereas the third aliquot was processed for IAA conjugates after alkaline hydrolysis in 7 n NaOH for 2 h at 100°C under nitrogen-saturated atmospheric conditions. After hydrolysis, the samples were titrated to pH 4.0 and desalted with a RP-C18 cartridge. The aliquot used for the analysis of free IAA as well as the desalted hydrolyzed fraction containing IAA and hydrolyzed IAA conjugates were both separately passed over a DEAE-Sephadex cartridge (2 mL). An RP-C18 cartridge was coupled underneath and the DEAE cartridge was eluted by 6% formic acid. The IAA, concentrated on the RP-C18, was eluted with 2× 1 mL diethylether and further evaporated under a nitrogen stream. Before analysis on LC-MS/MS, the IAA samples were resolved in 100 μL of acidified methanol and methylated with ethereal diazomethane, dried under a nitrogen stream (TurboVap, LV evaporator; Zymark), and resolved in 100 μL methanol to transfer samples to inserts. Samples were dried under nitrogen and resolved in 20% methanol prior to quantification.
Quantification of IAA and IAA Conjugates by Micro LC-(ES+)-MS/MS
IAA samples were analyzed with a micro LC-electrospray (ES+)-MS/MS (Waters). The HPLC system consisted of a 522 solvent delivery pump and a 465 autosampler (Kontron Instruments). Samples (25 μL) were injected on a RP-C18 Prodigy ODS (3), 5 μm, 100 Å, 100 × 1.00 mm column (Phenomenex) and eluted with a methanol 0.01 M NH4OAc solvent gradient (1 min at 20% methanol; linear gradient from 20% methanol to 90% methanol in 2 min; 3.5 min at 90% methanol; 5 min at 20% methanol). A constant flow rate of 60 μL/min was used. The effluent was introduced into the ES source, Z-spray probe (source temperature, 80°C; capillary voltage, +3.5kV; cone voltage, 20 V), delivered to the VG Quattro II tandem mass spectrometer for analysis, and quantified by MRM of [MH]+ combined with the m/z of 190>130 and 196>136 of methylated IAA and 13C6-IAA, respectively. The chromatograms obtained were processed by means of Masslynx 3.4 (Waters). Concentrations were calculated following the principles of isotope dilution and were expressed in pmol g−1 fresh weight.
Identification and Quantification of IBA and IBA Metabolites by Micro LC-(ES+)-MS/MS
IBA was quantified by LC-(ESI+)-MS/MS MRM of [MH]+ combined with the m/z of 204>186 of IBA together with a full production scan. IBA conjugates were identified by a parent scan of 204, identifying all metabolites of IBA. IBA glucosides were quantified with MRM of [MH]+ combined with the m/z of 366>204 identified with the parent scan together with a purified IBA-Glc reference solution. LC and MS conditions were as described for the IAA analysis.
Measurement of ABA and ABA-Glc Levels
Sampling for hormone analysis was performed as described above. Samples were immediately frozen in liquid nitrogen and stored at −80°C. Frozen samples were lyophilized for 48 h, and the plant hormones and metabolites were extracted and fractionated by HPLC under conditions described previously (Chiwocha et al., 2003, 2005). d4-ABA (obtained from Sue Abrams, NRC-PBI, Saskatoon, Canada) was used as the internal standard for quantification of ABA. ABA levels were analyzed by MRM mode essentially as described (Chiwocha et al., 2003), except that an Agilent 1100 capillary LC system linked to a 6330 LC/MSD Trap XCT Ultra system was used. Endogenous ABA was quantified with the QuantAnalysis software (Agilent Technologies).
Statistical Analysis
Weighted analysis of variance (ANOVA) was done to compare the hormone levels in extracts of the transgenic lines with those in wild-type plants with the lm function in R version 2.6.1 (Venables and Ripley, 2002). Whenever necessary, data were Box-Cox transformed to obtain homoscedasticity. All significance thresholds were set at 0.05. In a first experiment, the levels of IBA, IBA-Glc, IAA, IAA esters, oxIAA, IAA-Glc, IAA-Asp, IAA-Ala, and metIAA were analyzed by a one-way ANOVA with five replicates for each line, except for IBA and IBA-Glc, for which nine replicates were available. In the case of a significant model, differences in mean value between the lines were evaluated with the summary function. Significant line differences in the levels of IAA-Glc and IBA-Glc were verified in a second experiment. A final experiment considered the effect of stress on the hormone levels (IBA, IBA-Glc, IAA, IAA-Glc, and metIAA) by growing seedlings on control medium as well as on medium containing PEG. After two-way ANOVA, none of the analyses yielded a significant (α = 0.05) interaction term. The same statistical approach was also applied to look for significant differences in ETR value between the transgenic lines and the wild type, when a stress condition (growth on PEG or mannitol) was applied, or between the transgenic lines and the hormone-treated wild type. A significant interaction was observed between the line effect and the stress effect; thus, one-way ANOVAs were done. Because of the number of tests, a more stringent significance threshold of 0.01 was adopted.
Auxin Effect on Plant Morphology and Stress Tolerance
Wild-type and transgenic seedlings were germinated and grown on MS 0.5% sucrose plates or magenta boxes or supplemented with 0.5 μM IAA, 5 and 10 μM IBA, or 5 and 10 μM IBA-Glc, Leaf areas were scored at the indicated time and calculated with the ImageJ (http://rsb.info.nih.gov/ij/). The effect of auxin on bolting and flower development was scored after 2 months. Data were based on three replicates of 25 seeds each.
Wild-type plants and transformants were grown on MS 0.5% sucrose agar plates for 2 weeks and transferred to new plates supplemented with 400 g/L PEG8000 (−0.7 MPa), 54.7 g/L mannitol (−0.75 MPa), or both PEG and auxin (PEG+IAA, PEG+IBA, PEG+IBA-Glc, M+IAA, M+IBA, and M+IBA-Glc). Seedlings were also germinated on plates supplemented with IBA, IBA-Glc, or IAA and after 2 weeks transferred to new PEG or M plates supplemented with IBA (PEG+IBA-IBA treated), IBA-Glc (M+IBA-Glc-IBA-Glc treated), or IAA (PEG+IAA-IAA treated). Chlorophyll a fluorescence was measured after 8 or 6 d of stress treatment. In all cases, the concentration of auxin was 0.5 μM IAA, 5 μM IBA, and 5 μM IBA-Glc.
Photosynthetic Measurements
Chlorophyll a fluorescence, net CO2 assimilation (A), stomatal conductance, and transpiration rates (E) were determined on fully expanded attached leaves of 51-d-old plants growing under short-day growth conditions (two leaves from three plants of each line) with a portable gas exchange system (LI-6400; Li-Cor) equipped with a chamber fluorometer (Li-6400-40). A photosynthetic photon flux density of 500 μmol m−2 s−1 was supplied by red (630 nm) and blue (470 nm) LEDs, and the fraction of blue light was 10%. The CO2 concentration of the air entering the leaf chamber and the temperature were adjusted to 360 ppm and 25°C, respectively. The gas exchange parameters were calculated automatically using the software of the photosynthesis system.
The maximal chlorophyll fluorescence level (Fm and Fm’) were measured under a saturating pulse of 0.8 s (8000 μmol photon m−2 s−1). The maximal photochemical yield of PSII (ΦPSIImax), the effective ΦPSII in the light, and the photochemical quenching (qP) were determined as (Fm-Fo)/Fm, (Fm'–Fs)/Fm', and (Fm'-Fs)/(Fm'-Fo'), respectively (Genty et al., 1989), where Fm and Fm’ and Fo and Fo’ are maximal and minimal fluorescence in dark-adapted and light-adapted leaves, respectively, and Fs the steady state fluorescence level.
For the data in Table 2 (dehydration growth conditions) and Supplemental Table 3 online, the chlorophyll a fluorescence was measured with an Imaging-PAM M-Series chlorophyll fluorescence System (Heinz Walz).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries or the Arabidopsis Genome Initiative database under the following accession numbers: UGT74E2 (At1g05680), Zea mays iaglc (L34847), UGT84B1 (At2g23260), UGT74F2 (At2g43829), WES1 (At4g27260), ELIP2 (At4g14690), BGL1 (At1g52400), ACTIN2 (At3g18780), ACTIN-RELATED PROTEIN7 (At3g60830), DIN2 BETA-GLUCOSIDASE30 (At3g60140), POLYGALACTURONASE/PECTINASE (At5g14650), and GLUCOSYL HYDROLASE (At4g02290). The GenBank accession numbers for knockout lines mentioned in this article are ugt74e2-01 (SALK_016116) and ugt74e2-09 (SALK_091130).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. UGT74E2 Stress Induction and Auxin Effect on Root Development.
Supplemental Figure 2. Auxin Effects on Plant Morphology.
Supplemental Figure 3. Analysis of Epidermal Cells in UGT74EOE Plants.
Supplemental Figure 4. Identification of Homozygous Insertion Mutants at the UGT74E2 Locus.
Supplemental Figure 5. Phenotype and Auxin Response of UGT74E2 Knockout Mutants.
Supplemental Table 1. Phenotypic Comparison of Wild-Type and UGT74E2OE Plants.
Supplemental Table 2. Differential Genes in UGT74E2OE Plants as Determined by Full-Genome Microarray Analysis.
Supplemental Table 3. Auxin Effects on Photosynthesis.
Supplementary Material
Acknowledgments
We thank Mariusz Kowalczyk and Karin Ljung for the in planta quantification of IAA and IAA conjugate levels (Department of Forest Genetics and Plant Physiology, Umeå Plant Science Centre, Swedish University of Agricultural Sciences, Umeå, Sweden). This work was supported by grants from the Research Fund of the Ghent University (“Geconcerteerde onderzoeksacties” No. 12051403 and “Personele Ondersteuning Zware Onderzoeksinfrastructuur I/00003/02). V.B.T. is the recipient of a Marie Curie Intra-European Fellowship for Career Development (PIEF-GA-2008-221427). O.V.A. and I.D.C. are grateful to the Agency for Innovation by Science and Technology in Flanders for predoctoral fellowships.
References
- Achard P., Renou J.-P., Berthomé R., Harberd N.P., Genschik P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18: 656–660 [DOI] [PubMed] [Google Scholar]
- Acharya B.R., Assmann S.M. (2009). Hormone interactions in stomatal function. Plant Mol. Biol. 69: 451–462 [DOI] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Bak S., Feyereisen R. (2001). The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol. 127: 108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B., LeClere S., Magidin M., Zolman B.K. (2001). Inputs to the active indole-3-acetic acid pool: De novo synthesis, conjugate hydrolysis, and indole-3-butyric acid β-oxidation. J. Plant Growth Regul. 20: 198–216 [Google Scholar]
- Beeckman T., Engler G. (1994). An easy technique for the clearing of histochemically stained plant tissue. Plant Mol. Biol. Rep. 12: 37–42 [Google Scholar]
- Belin C., Megies C., Hauserová E., Lopez-Molina L. (2009). Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21: 2253–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57: 289–300 [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright J., Desikan R., Hancock J.T., Weir I.S., Neill S.J. (2006). ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 45: 113–122 [DOI] [PubMed] [Google Scholar]
- Brodführer U. (1955). Der Einfluss einer abgestuften Dosierung von ultravioletter Sonnenstrahlung auf das Wachstum der Pflanzen. Planta 45: 1–56 [Google Scholar]
- Campanella J.J., Olajide A.F., Magnus V., Ludwig-Müller J. (2004). A novel auxin conjugate hydrolase from wheat with substrate specificity for longer side-chain auxin amide conjugates. Plant Physiol. 135: 2230–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I., Marchant A., Bhalerao R.P., Beeckman T., Dhooge S., Swarup R., Graham N., Inzé D., Sandberg G., Casero P.J., Bennett M. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Stevenson B., Lee B.-h., Zhu J.-K. (2002). Screening for gene regulation mutants by bioluminescence imaging. Sci. STKE 2002: pl10. [DOI] [PubMed] [Google Scholar]
- Chiwocha S.D.S., Abrams S.R., Ambrose S.J., Cutler A.J., Loewen M., Ross A.R.S., Kermode A.R. (2003). A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: An analysis of hormone regulation of thermodormancy in lettuce (Lactuca sativa L.) seeds. Plant J. 35: 405–417 [DOI] [PubMed] [Google Scholar]
- Chiwocha S.D.S., Cutler A.J., Abrams S.R., Ambrose S.J., Yang J., Ross A.R.S., Kermode A.R. (2005). The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 42: 35–48 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cutler A.J., Krochko J.E. (1999). Formation and breakdown of ABA. Trends Plant Sci. 4: 472–478 [DOI] [PubMed] [Google Scholar]
- Dean J.V., Delaney S.P. (2008). Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 132: 417–425 [DOI] [PubMed] [Google Scholar]
- Desikan R., et al. (2008). The histidine kinase AHK5 integrates endogenous and environmental signals in Arabidopsis guard cells. PLoS ONE 3: e2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle M.A., Somerville C. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206: 200–206 [Google Scholar]
- Finkelstein R.R., Gampala S.S.L., Rock C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankowski K., Kęsy J., Wojciechowski W., Kopcewicz J. (2009). Light- and IAA-regulated ACC synthase gene (PnACS) from Pharbitis nil and its possible role in IAA-mediated flower inhibition. J. Plant Physiol. 166: 192–202 [DOI] [PubMed] [Google Scholar]
- Galvez-Valdivieso G., Fryer M.J., Lawson T., Slattery K., Truman W., Smirnoff N., Asami T., Davies W.J., Jones A.M., Baker N.R., Mullineaux P.M. (2009). The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21: 2143–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazarian I.G., Lagrimini L.M., Mellon F.A., Naldrett M.J., Ashby G.A., Thorneley R.N.F. (1998). Identification of skatolyl hydroperoxide and its role in the peroxidase-catalysed oxidation of indol-3-yl acetic acid. Biochem. J. 333: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B., Briantais J.-M., Baker N.R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990: 87–92 [Google Scholar]
- Gudesblat G.E., Iusem N.D., Morris P.C. (2007). Guard cell-specific inhibition of Arabidopsis MPK3 expression causes abnormal stomatal responses to abscisic acid and hydrogen peroxide. New Phytol. 173: 713–721 [DOI] [PubMed] [Google Scholar]
- Havlová M., Dobrev P.I., Motyka V., Štorchová H., Libus J., Dobrá J., Malbeck J., Gaudinová A., Vanková R. (2008). The role of cytokinins in responses to water deficit in tobacco plants over-expressing trans-zeatin O-glucosyltransferase gene under 35S or SAG12 promoters. Plant Cell Environ. 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Hou B., Lim E.-K., Higgins G.S., Bowles D.J. (2004). N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 279: 47822–47832 [DOI] [PubMed] [Google Scholar]
- Ikeda Y., Men S., Fischer U., Stepanova A.N., Alonso J.M., Ljung K., Grebe M. (2009). Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat. Cell Biol. 11: 731–738 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Ooi S.L., Wu Z., Boeke J.D. (2003). Use of mixture models in a microarray-based screening procedure for detecting differentially represented yeast mutants. Stat. Appl. Genet. Mol. Biol. 2: Article 1 [DOI] [PubMed] [Google Scholar]
- Jackson R.G., Kowalczyk M., Li Y., Higgins G., Ross J., Sandberg G., Bowles D.J. (2002). Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: Phenotypic characterisation of transgenic lines. Plant J. 32: 573–583 [DOI] [PubMed] [Google Scholar]
- Jackson R.G., Lim E.-K., Li Y., Kowalczyk M., Sandberg G., Hoggett J., Ashford D.A., Bowles D.J. (2001). Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 276: 4350–4356 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D'Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Lanot A., Hodge D., Jackson R.G., George G.L., Elias L., Lim E.-K., Vaistij F.E., Bowles D.J. (2006). The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. Plant J. 48: 286–295 [DOI] [PubMed] [Google Scholar]
- Lee K.H., Piao H.L., Kim H.-Y., Choi S.M., Jiang F., Hartung W., Hwang I., Kwak J.M., Lee I.-J., Hwang I. (2006). Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Leyser O. (2005). The fall and rise of apical dominance. Curr. Opin. Genet. Dev. 15: 468–471 [DOI] [PubMed] [Google Scholar]
- Lim E.-K., Bowles D.J. (2004). A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 23: 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E.-K., Higgins G.S., Li Y., Bowles D.J. (2003). Regioselectivity of glucosylation of caffeic acid by a UDP-glucose:glucosyltransferase is maintained in planta. Biochem. J. 373: 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.-P., Montgomery T.A., Fahlgren N., Kasschau K.D., Nonogaki H., Carrington J.C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52: 133–146 [DOI] [PubMed] [Google Scholar]
- Ljung K., Hull A.K., Kowalczyk M., Marchant A., Celenza J., Cohen J.D., Sandberg G. (2002). Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 49: 249–272 [PubMed] [Google Scholar]
- Ludwig-Müller J. (2007). Indole-3-butyric acid synthesis in ecotypes and mutants of Arabidopsis thaliana under different growth conditions. J. Plant Physiol. 164: 47–59 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J., Epstein E. (1993). Indole-3-butyric acid in Arabidopsis thaliana. II. In vivo metabolism. J. Plant Growth Regul. 13: 189–195 [Google Scholar]
- Ludwig-Müller J., Epstein E. (1994). Indole-3-butyric acid in Arabidopsis thaliana. III. In vivo biosynthesis. Plant Growth Regul. 14: 7–14 [Google Scholar]
- Ludwig-Müller J., Hilgenberg W. (1995). Characterization and partial purification of indole-3-butyric acid synthetase from maize (Zea mays). Physiol. Plant. 94: 651–660 [Google Scholar]
- Ludwig-Müller J., Sass S., Sutter E.G., Wodner M., Epstein E. (1993). Indole-3-butyric acid in Arabidopsis thaliana. I. Identification and quantification. Plant Growth Regul. 13: 179–187 [Google Scholar]
- Márton L., Browse J. (1991). Facile transformation of Arabidopsis. Plant Cell Rep. 10: 235–239 [DOI] [PubMed] [Google Scholar]
- Meßner B., Thulke O., Schäffner A.R. (2003). Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta 217: 138–146 [DOI] [PubMed] [Google Scholar]
- Miao Y., Laun T., Zimmermann P., Zentgraf U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Mishra G., Zhang W., Deng F., Zhao J., Wang X. (2006). A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312: 264–266 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004). The reactive oxygen gene network in plants. Trends Plant Sci. 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nakagami H., Soukupová H., Schikora A., Žárský V., Hirt H. (2006). A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 281: 38697–38704 [DOI] [PubMed] [Google Scholar]
- Neill S., Desikan R., Hancock J. (2002). Hydrogen peroxide signalling. Curr. Opin. Plant Biol. 5: 388–395 [DOI] [PubMed] [Google Scholar]
- Oono Y., Chen Q.G., Overvoorde P.J., Köhler C., Theologis A. (1998). age Mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10: 1649–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.-M. (2007). Auxin homeostasis in plant stress adaptation response. Plant Signal. Behav. 2: 306–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-E., Park J.-Y., Kim Y.-S., Staswick P.E., Jeon J., Yun J., Kim S.-Y., Kim J., Lee Y.-H., Park C.-M. (2007). GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 282: 10036–10046 [DOI] [PubMed] [Google Scholar]
- Pasternak T., Potters G., Caubergs R., Jansen M.A.K. (2005). Complementary interactions between oxidative stress and auxin control plant growth responses at plant, organ, and cellular level. J. Exp. Bot. 56: 1991–2001 [DOI] [PubMed] [Google Scholar]
- Poppenberger B., Fujioka S., Soeno K., George G.L., Vaistij F.E., Hiranuma S., Seto H., Takatsuto S., Adam G., Yoshida S., Bowles D. (2005). The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 102: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potters G., Pasternak T.P., Guisez Y., Jansen M.A.K. (2009). Different stresses, similar morphogenic responses: Integrating a plethora of pathways. Plant Cell Environ. 32: 158–169 [DOI] [PubMed] [Google Scholar]
- Potters G., Pasternak T.P., Guisez Y., Palme K.J., Jansen M.A.K. (2007). Stress-induced morphogenic responses: Growing out of trouble? Trends Plant Sci. 12: 98–105 [DOI] [PubMed] [Google Scholar]
- Poupart J., Rashotte A.M., Muday G.K., Waddell C.S. (2005). The rib1 mutant of Arabidopsis has alterations in indole-3-butyric acid transport, hypocotyl elongation, and root architecture. Plant Physiol. 139: 1460–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest D.M., Jackson R.G., Ashford D.A., Abrams S.R., Bowles D.J. (2005). The use of abscisic acid analogues to analyse the substrate selectivity of UGT71B6, a UDP-glycosyltransferase of Arabidopsis thaliana. FEBS Lett. 579: 4454–4458 [DOI] [PubMed] [Google Scholar]
- Prinsen E., Van Laer S., Öden S., Van Onckelen H. (2000). Auxin analysis. Plant Hormone Protocols: Methods in Molecular Biology, Vol. 141, Tucker G.A., Roberts J.A., (Totowa, NJ: Humana Press; ), pp. 49–65 [DOI] [PubMed] [Google Scholar]
- Quesada V., Macknight R., Dean C., Simpson G.G. (2003). Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 22: 3142–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiel J.A., Bender J. (2003). Glucose conjugation of anthranilate by the Arabidopsis UGT74F2 glucosyltransferase is required for tryptophan mutant blue fluorescence. J. Biol. Chem. 278: 6275–6281 [DOI] [PubMed] [Google Scholar]
- Rashotte A.M., Poupart J., Waddell C.S., Muday G.K. (2003). Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 133: 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H.S., Friml J. (2009). Auxin and other signals on the move in plants. Nat. Chem. Biol. 5: 325–332 [DOI] [PubMed] [Google Scholar]
- Ross J., Li Y., Lim E.-K., Bowles D.J. (2001). Higher plant glycosyltransferases. Genome Biol. 2: reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini S., Casazza A.P., Engelmann E.C.M., Havaux M., Jennings R.C., Soave C. (2006). Suppression of both ELIP1 and ELIP2 in Arabidopsis does not affect tolerance to photoinhibition and photooxidative stress. Plant Physiol. 141: 1264–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter A., Dietz K.-J., Hartung W. (2002). A possible stress physiological role of abscisic acid conjugates in root-to-shoot signalling. Plant Cell Environ. 25: 223–228 [DOI] [PubMed] [Google Scholar]
- Sävenstrand H., Olofsson M., Samuelsson M., Strid Å. (2004). Induction of early light-inducible protein gene expression in Pisum sativum after exposure to low levels of UV-B irradiation and other environmental stresses. Plant Cell Rep. 22: 532–536 [DOI] [PubMed] [Google Scholar]
- Scarpella E., Barkoulas M., Tsiantis M. (2010). Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. 2: a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R.R., Michel J. (1985). O-(α-d-glucopyranosyl)trichloroacetimidate as a glucosyl donor. J. Carbohydr. Chem. 4: 141–169 [Google Scholar]
- Shen Y.-Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826 [DOI] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article 3 [DOI] [PubMed] [Google Scholar]
- Song J.T. (2006). Induction of a salicylic acid glucosyltransferase, AtSGT1, is an early disease response in Arabidopsis thaliana. Mol. Cells 22: 233–238 [PubMed] [Google Scholar]
- Staswick P.E., Serban B., Rowe M., Tiryaki I., Maldonado M.T., Maldonado M.C., Suza W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L.C., Monroe-Augustus M., Rogers K.C., Lin G.L., Bartel B. (2008). Arabidopsis iba response5 suppressors separate responses to various hormones. Genetics 180: 2019–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szerszen J.B., Szczyglowski K., Bandurski R.S. (1994). iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265: 1699–1701 [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T., Franck F., Alawady A.E., Dall'Osto L., Carrière F., Bassi R., Grimm B., Nussaume L., Havaux M. (2007). The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant J. 50: 795–809 [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F., Dat J.F. (2006). Reactive oxygen species in plant cell death. Plant Physiol. 141: 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S., Vanderauwera S., Vuylsteke M., Rombauts S., Langebartels C., Seidlitz H.K., Zabeau M., Van Montagu M., Inzé D., Van Breusegem F. (2004). Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39: 45–58 [DOI] [PubMed] [Google Scholar]
- Vandenabeele S., Van Der Kelen K., Dat J., Gadjev I., Boonefaes T., Morsa S., Rottiers P., Slooten L., Van Montagu M., Zabeau M., Inzé D., Van Breusegem F. (2003). A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc. Natl. Acad. Sci. USA 100: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S., Zimmermann P., Rombauts S., Vandenabeele S., Langebartels C., Gruissem W., Inzé D., Van Breusegem F. (2005). Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W.N., Ripley B.D. (2002). Linear statistical models. Modern Applied Statistics with S, 4th ed., Statistics and Computing Series, Venables W.N., Ripley B.D., (Heidelberg, Germany: Springer; ), pp. 139–181 [Google Scholar]
- Woodward A.W., Bartel B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Niida R., Saito K. (2007). Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282: 14932–14941 [DOI] [PubMed] [Google Scholar]
- Zgurski J.M., Sharma R., Bolokoski D.A., Schultz E.A. (2005). Asymmetric auxin response precedes asymmetric growth and differentiation of asymmetric leaf1 and asymmetric leaf2 Arabidopsis leaves. Plant Cell 17: 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang L., Dong F., Gao J., Galbraith D.W., Song C.-P. (2001). Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 126: 1438–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Martinez N., Millius A., Adham A.R., Bartel B. (2008). Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180: 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Monroe-Augustus M., Thompson B., Hawes J.W., Krukenberg K.A., Matsuda S.P.T., Bartel B. (2001b). chy1, an Arabidopsis mutant with impaired β-oxidation, is defective in a peroxisomal β-hydroxyisobutyryl-CoA hydrolase. J. Biol. Chem. 276: 31037–31046 [DOI] [PubMed] [Google Scholar]
- Zolman B.K., Nyberg M., Bartel B. (2007). IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 64: 59–72 [DOI] [PubMed] [Google Scholar]
- Zolman B.K., Silva I.D., Bartel B. (2001a). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
- Zolman B.K., Yoder A., Bartel B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156: 1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.