Villins are a class of actin-binding protein with multiple activities. This study uses single filament imaging techniques to evaluate and contrast the functions of Arabidopsis VILLIN1 (VLN1) and VLN3. Both isovariants are shown to bundle actin filaments, whereas VLN3 also severs filaments and can disassemble actin filament bundles in the presence of physiological calcium concentrations.
Abstract
Actin filament bundles are higher-order cytoskeletal structures that are crucial for the maintenance of cellular architecture and cell expansion. They are generated from individual actin filaments by the actions of bundling proteins like fimbrins, LIMs, and villins. However, the molecular mechanisms of dynamic bundle formation and turnover are largely unknown. Villins belong to the villin/gelsolin/fragmin superfamily and comprise at least five isovariants in Arabidopsis thaliana. Different combinations of villin isovariants are coexpressed in various tissues and cells. It is not clear whether these isovariants function together and act redundantly or whether they have unique activities. VILLIN1 (VLN1) is a simple filament-bundling protein and is Ca2+ insensitive. Based on phylogenetic analyses and conservation of Ca2+ binding sites, we predict that VLN3 is a Ca2+-regulated villin capable of severing actin filaments and contributing to bundle turnover. The bundling activity of both isovariants was observed directly with time-lapse imaging and total internal reflection fluorescence (TIRF) microscopy in vitro, and the mechanism mimics the “catch and zipper” action observed in vivo. Using time-lapse TIRF microscopy, we observed and quantified the severing of individual actin filaments by VLN3 at physiological calcium concentrations. Moreover, VLN3 can sever actin filament bundles in the presence of VLN1 when calcium is elevated to micromolar levels. Collectively, these results demonstrate that two villin isovariants have overlapping and distinct activities.
INTRODUCTION
The actin cytoskeleton plays a central role in fundamental physiological processes, including the maintenance of cellular architecture and polarity, cell motility, organelle positioning, vesicle trafficking, cytoplasmic streaming, and cell division. Actin filaments form structural networks comprised of single filaments and prominent higher-order structures made of actin filaments packed closely together. Recent imaging of the actin cytoskeleton in live cells has revealed that it is surprisingly dynamic (Vavylonis et al., 2008; Staiger et al., 2009; Okreglak and Drubin, 2010). In the cortical array of Arabidopsis thaliana epidermal cells, actin filaments are randomly oriented, polymerize rapidly, and are constantly changing position (Staiger et al., 2009). The single filaments are mostly short-lived due to prolific severing activity that fragments them into small pieces, but the stiffer and straighter higher-order structures appear to be rather stable and are long-lived. The latter are often referred to as actin bundles or cables.
Although bundles are a prominent feature of practically all plant cells, their biological function and how they are formed have seldom been studied directly (Higaki et al., 2010). There is one preliminary report of actin bundle formation in vivo (Era et al., 2009), but the mechanism was not well characterized. It is generally accepted, however, that bundles are important for various cellular processes, such as cytoplasmic streaming, as tracks for organelle movement, for positioning of organelles such as chloroplasts and the nucleus, and for opening of stomata (Shimmen et al., 1995; Tominaga et al., 2000; Ketelaar et al., 2002; Shimmen, 2007; Higaki et al., 2009). Bundles are also crucial for the maintenance of cellular architecture and stabilization of transvacuolar strands (Shimmen et al., 1995; Tominaga et al., 2000) as well as for host cell responses to microbial pathogens (Schmidt and Panstruga, 2007; Clément et al., 2009). In addition, studies of hormone signaling and mutant plants suggest that bundles are also involved in cell elongation and cellular morphogenesis (Baluska et al., 2001; Le et al., 2003, 2006; Rahman et al., 2007; Higaki et al., 2010). Several classes of actin binding protein are responsible for the formation of bundles in plants, including fimbrins, two LIM domain–containing (LIM) proteins, certain formins, and villins (Higaki et al., 2007, 2010; Thomas et al., 2009). Whether these proteins cooperate to generate bundles or whether they form different types of bundles that act at different locations to perform specific functions is not clear. Additionally, the bundling proteins comprise multigene families. Functional differences within families are evident from studies on proteins such as the formins (Blanchoin and Staiger, 2010). Thus, there is a need to perform rigorous biochemical characterization of the different isovariants as they might have distinct activities and/or forms of regulation.
Individual actin binding proteins typically have more than one biochemical activity on actin. In addition to bundling filaments, some actin binding proteins can bind to monomers, facilitate polymerization, cap ends, and/or fragment filaments. The villins are one such class of multifunctional protein that is widely expressed in most eukaryotic cells, with the notable exception of Saccharomyces cerevisiae (Bretscher and Weber, 1980; Friederich et al., 1999; Staiger and Hussey, 2004). They belong to the villin/gelsolin/fragmin superfamily, whose members contain gelsolin repeat domains comprised of 125 to 150 amino acids (Friederich et al., 1999; Yin, 1999). Villins comprise a core gelsolin domain, with six gelsolin-homology repeats (G1 to G6), and most are able to sever and cap the barbed ends of actin filaments in the presence of micromolar concentrations of Ca2+ (Glenney et al., 1980; Northrop et al., 1986; Janmey and Matsudaira, 1988; Kumar et al., 2004a). There are exceptions, however, including Drosophila melanogaster Quail and Arabidopsis VILLIN1 (VLN1), which do not exhibit these functions (Matova et al., 1999; Huang et al., 2005). Villins also have a C-terminal headpiece domain (villin headpiece [VHP]) that contributes an additional actin binding site and facilitates the formation of higher-order structures by cross-linking actin filaments into bundles (Friederich et al., 1992, 1999; George et al., 2007).
Plant villin homologs from Lilium longiflorum pollen (135-ABP and 115-ABP) bundle actin filaments in a calcium-calmodulin (Ca2+-CaM)–sensitive manner (Nakayasu et al., 1998; Yokota et al., 1998, 2000, 2003; Vidali et al., 1999; Yokota and Shimmen, 1999). Lily 135-ABP is also able to accelerate polymerization, cap ends, and depolymerize actin filaments in the presence of Ca2+-CaM (Yokota et al., 2005). These plant villins are thought to be involved in the formation of actin filament bundles in pollen tubes (Yokota et al., 1998; Vidali et al., 1999) and root hair cells (Tominaga et al., 2000; Ketelaar et al., 2002).
In addition to bundling, villins are also recognized for the ability to sever or fragment actin filaments like other members of the villin/gelsolin/fragmin superfamily. The ability to sever actin filaments is shared with other unrelated proteins like actin-depolymerizing factor (ADF)/cofilin, twinfilin, and coronin. These proteins enhance disassembly of actin filaments and thus promote the reorganization of the actin cytoskeleton (Ono, 2007). Severing is a prominent feature responsible for the stochastic dynamics of individual actin filaments in live cells (Vavylonis et al., 2008; Staiger et al., 2009; Okreglak and Drubin, 2010), and it is therefore critical to understand the molecular mechanisms that underpin filament breakage.
The Arabidopsis genome encodes five isovariants of villin, VLN1 to VLN5 (Klahre et al., 2000; Huang et al., 2005). Fusion proteins of green fluorescent protein (GFP) with full-length VLN3 and several VHP constructs decorate actin filaments in plant and animal cells, thereby demonstrating their ability to bind actin filaments (Klahre et al., 2000); however, the ability of different villin isovariants to bundle, sever, or nucleate actin filaments in vivo has not been tested. Recent studies demonstrate that VLN1 is a simple bundling protein in vitro that is also capable of protecting actin filaments from ADF1 (Huang et al., 2005). VLN1 does not, however, have capping or severing activities and is not regulated by Ca2+ and CaM. Detailed biochemical analyses described herein were performed to determine whether another isovariant, VLN3, is capable of bundling and/or severing actin filaments in a Ca2+-dependent manner. We demonstrate that VLN3 bundles actin filaments independently of Ca2+ and that it severs filaments in the presence of micromolar [Ca2+].
It is evident from microarray data, RNA gel blots, and promoter:β-glucuronidase (GUS) fusions that multiple villin isovariants coexist in different tissues of the plant during various stages of development (Klahre et al., 2000; Ma et al., 2005; Hruz et al., 2008; Yang et al., 2008). Expression data from single cells such as trichomes (Dai et al., 2010), guard cells and mesophyll cells in leaves (Leonhardt et al., 2004; Yang et al., 2008), and root cells (Birnbaum et al., 2003; Brady et al., 2007; Cartwright et al., 2009) also suggest that more than one isovariant of villin can be present together. Specifically, VLN1 and VLN3 likely coexist in many cell types, although VLN3 is typically more abundant than VLN1. This raises the question of why the two isovariants coexist in cells. Biochemical and in planta functional differences have been demonstrated previously for isovariants of actin and the actin binding proteins, profilin and ADF (Kandasamy et al., 2002, 2007, 2009; Staiger and Blanchoin, 2006). There is also genetic evidence for isovariants exhibiting similar and interchangeable functions, like the vegetative actins and the SCAR isovariants (Zhang et al., 2008; Kandasamy et al., 2009). The villins may therefore have redundant functions, work synergistically, or have unique properties altogether. We hypothesize that these isovariants perform the same function in cells viz bundling actin filaments. At micromolar Ca2+ levels, however, we predict that VLN3 will sever filaments even in the presence of VLN1.
To analyze the functions of VLN1 and VLN3 when present together in vitro, we used time-lapse imaging and evanescent wave microscopy to directly observe the effect of the proteins on single actin filaments in a reconstituted system. We were able to observe bundling by both villins in real time and the mechanism recapitulates a “catch and zipper” activity observed in the cortical actin array of living epidermal cells. Furthermore, in the presence of physiological [Ca2+], VLN3 severed actin filaments in the presence or absence of VLN1 in vitro. Thus, Arabidopsis villin isovariants have overlapping but distinct activities that contribute to the turnover of actin filament bundles. Our study is an important step in understanding how isovariants from the same protein family might work in the complex environment within a cell and how actin binding proteins cooperate to form, maintain, and turn over actin filament bundles.
RESULTS
VLN3 Contains the Most Predicted Ca2+ Binding Sites of Any VLN Isovariant
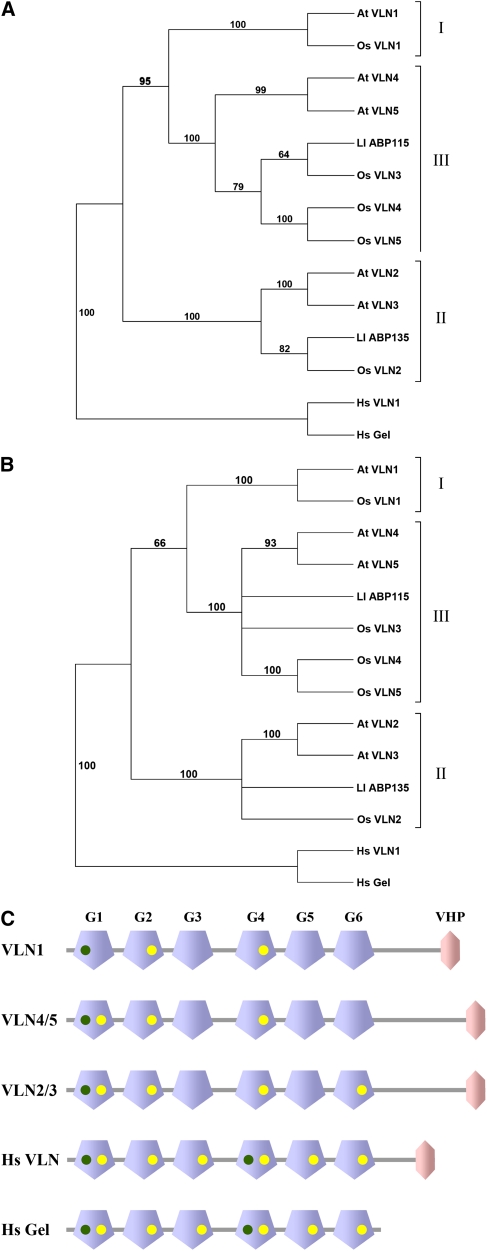
The five isovariants of villin in Arabidopsis (VLN1 to VLN5) are very similar, with any two villins having an amino acid sequence similarity >51%. To analyze the relationship among isovariants, a phylogenetic tree was generated using a sequence alignment of villins from Arabidopsis, rice (Oryza sativa), lily, and human (Homo sapiens) (Figure 1A; see Supplemental Data Set 1 and Supplemental Figure 1 online). Based on a distance-based neighbor-joining analysis, the Arabidopsis villins could be categorized into three distinct groups with well-supported bootstrap values. Specifically, VLN1 fell into a group of its own, designated group I. VLN2 and VLN3 share 88% amino acid sequence similarity and fell into the same group (II), whereas VLN4 and VLN5 comprised group III. The lily villins, 135-ABP and 115-ABP, fell into groups II and III, respectively. We also identified five villin isovariants in rice that had at least six gelsolin-repeat domains and a VHP, with the exception of Os VLN1, which did not have a headpiece. These were also classified into the three groups. The same phylogeny and subgroup classification were obtained with a parsimony-based analysis (Figure 1B). Surprisingly, the clades did not correlate with the expression pattern of the genes. The two lily villins, in spite of being isolated from pollen, did not sort into the same group. Of the five Arabidopsis villin isovariants, VLN1, 2, and 5 are all expressed in pollen, based on microarray data, whereas VLN3 and 4 are not (Pina et al., 2005). VLN1, 2, and 5 did not, however, fall into a single phylogenetic grouping that can be designated as reproductive. This contrasts with other plant cytoskeletal gene families, like profilin or actin (Huang et al., 1996; McDowell et al., 1996; Meagher et al., 1999).
Figure 1.
Plant Villins Can Be Grouped into Three Major Classes by Phylogeny and the Number of Conserved Ca2+ Binding Sites.
(A) and (B) Distance-based (A) and parsimony-based (B) neighbor-joining tree of villins and villin-like proteins from Arabidopsis, rice, lily, and human. The villins or villin-like proteins grouped together with Arabidopsis VLN1 (At VLN1) are designated as group I, those with At VLN2 and 3 as group II, and those with At VLN4 and 5 as group III. Human villin (Hs VLN1) and human gelsolin (Hs Gel) were allocated as an outgroup in the manually rooted tree. Numbers on branches denote percentage bootstrap values (out of 1000 replicates).
(C) Domain structures of Arabidopsis VLNs, human villin, and human gelsolin depicting the Ca2+ binding sites. The modular architecture of the proteins (not drawn to scale) with gelsolin repeat domains in blue (G1 to G6) and the VHP in pink is as produced on the SMART database (http://smart.embl-heidelberg.de/). The Ca2+ binding sites identified by sequence alignment of gelsolin repeat domains of Arabidopsis VLNs and human villin compared with human gelsolin (see Supplemental Figure 2 online) are represented by dots with type 1 in green and type 2 in yellow.
To understand further the distinction between villin classes, we analyzed their Ca2+ binding motifs. Studies of the x-ray crystal structures of gelsolin reveal two types of Ca2+ binding sites: type 1 sites, where Ca2+ binds at the interface between gelsolin and actin and is important for their interaction; and type 2 sites that are contained within gelsolin, and where Ca2+ binding results in activation of the protein (Choe et al., 2002). Structure-based sequence alignment of the gelsolin-homology domains of Arabidopsis villins and human villin compared with human plasma gelsolin (Figure 1C; see Supplemental Figure 2 online) revealed that all Arabidopsis and lily villins had just one type 1 site (in gelsolin-repeat domain 1 [G1]), whereas human gelsolin and villin have two type 1 sites in G1 and G4, respectively (green dots, Figure 1C; green highlighting, Supplemental Figure 2 online). All six of the type 2 sites present in human villin and gelsolin (yellow dots, Figure 1C; yellow highlighting, Supplemental Figure 2 online) were not conserved in the plant villins (Huang et al., 2005). VLN1 had the least, with only one type 2 site in G2 and another probable site in G4 (Figure 1C). VLN2 and VLN3 had the most, with four type 2 sites in G1, G2, G4, and G6. As VLN1, a Ca2+-insensitive protein that lacks several of the normal villin functions and has the least number of probable Ca2+ binding sites (Huang et al., 2005), it seems likely that the villin isovariants with a greater number of conserved sites will be Ca2+ sensitive. VLN3 is therefore a good candidate to test this hypothesis by studying the biochemical properties of the recombinant protein.
To examine the activities of VLN3 in vitro, the full-length cDNA was obtained from the ABRC and the coding sequence cloned into a T7 vector. The 106-kD nonfusion protein was expressed in Escherichia coli and purified by ion exchange and affinity chromatography (see Supplemental Figure 3A online). The purity of the recombinant protein was >80% and it cross-reacted with an affinity-purified antibody raised against the G1 to G3 domains of VLN1 (Huang et al., 2005; see Supplemental Figure 3B online).
VLN3 Binds to F-Actin in a Ca2+-Insensitive Manner
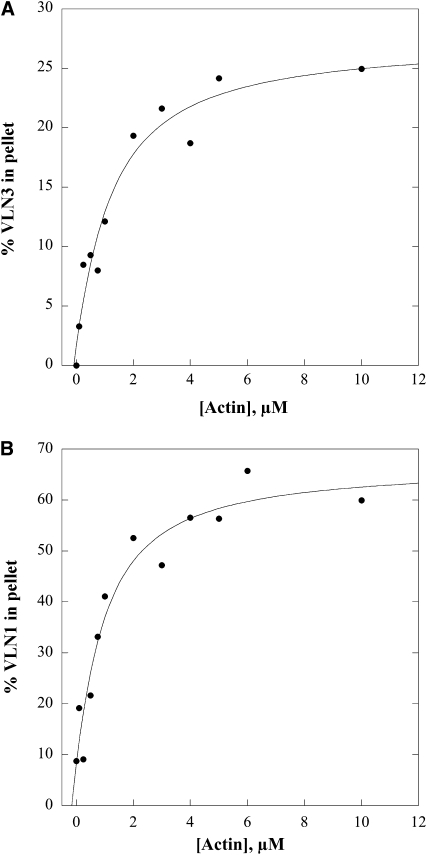
The ability of VLN3 to bind to filamentous actin (F-actin) was analyzed with a variation of the high-speed cosedimentation assay used previously to characterize FIM1 and VLN1 (Kovar et al., 2000; Huang et al., 2005). Apparent equilibrium dissociation constant (Kd) values for VLN3 binding to F-actin were estimated by cosedimentation reactions with 0.5 μM VLN3 or VLN1 and increasing concentrations of phalloidin-stabilized actin. Following centrifugation at high speed, the percentage of villin in the pellet was determined by densitometry. These data were plotted as a function of actin concentration and fitted with a hyperbolic function to estimate apparent dissociation constant values, and representative experiments are shown in Figure 2A (VLN3) and Figure 2B (VLN1). The apparent Kd (±sd) value for VLN3 was 0.9 ± 0.2 μM (n = 6) and was not statistically different from that of VLN1 (0.6 ± 0.4 μM, n = 4; P = 0.3 by Student’s t test). Thus, VLN3 binds to actin filaments with moderate affinity, like the previously characterized isovariant, VLN1.
Figure 2.
VLN3 Binds to F-Actin.
High-speed cosedimentation assays were performed to determine the apparent affinity of villin binding to F-actin. Various amounts of phalloidin-stabilized F-actin were mixed with 0.5 μM VLN3 (A) or VLN1 (B). The final [Ca2+]free, buffered with 1 mM EGTA, was 3 nM. The supernatants and pellets obtained after centrifugation were subjected to SDS-PAGE, and the amount of bound protein in the pellet was determined by densitometry. Percentage of bound VLN was plotted against the concentration of actin and fitted with a hyperbolic curve. The calculated equilibrium dissociation constant (Kd) value for the representative experiment in (A) was 1 μM. The Kd value for the representative experiment in (B) was 0.8 μM.
The binding assays described above were performed at [Ca2+]free <10 nM. To determine whether the binding of VLN3 to F-actin was sensitive to Ca2+, additional experiments were performed in the presence of 100 μM free Ca2+. The average Kd obtained for VLN3 at high Ca2+ was 1.3 ± 0.3 μM (n = 4), which was not statistically different (P = 0.07) from that in low Ca2+ conditions. This indicates that Ca2+ does not affect the affinity of VLN3 for F-actin.
VLN3 Bundles F-Actin in a Ca2+-Independent Manner
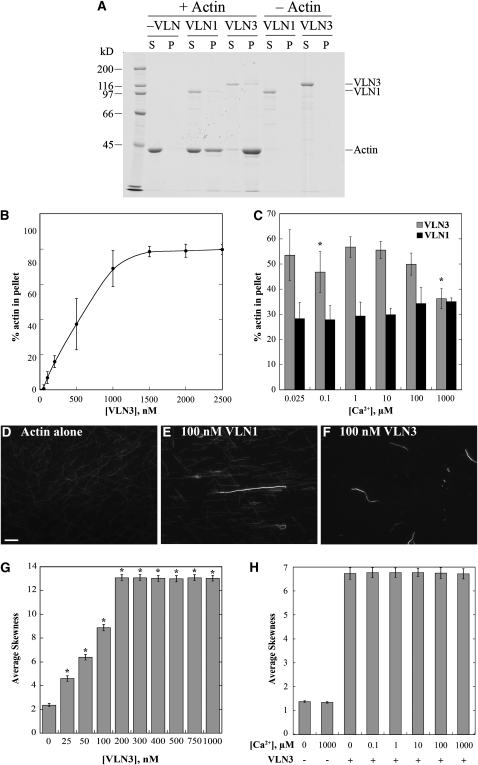
To test whether VLN3 is capable of generating actin filament bundles in vitro, we used a battery of biochemical and fluorescence microscopy assays. Previously, we demonstrated that VLN1 could generate actin filament bundles in a Ca2+-independent manner (Huang et al., 2005). The ability of VLN3 to form higher-order structures with F-actin was initially examined with a low-speed cosedimentation assay (Kovar et al., 2000; Huang et al., 2005). Because individual actin filaments do not sediment at 13,500g, only in the presence of an actin binding protein that is capable of bundling or cross-linking filaments into networks will there be appreciable actin in the pellet. When incubated in the absence of villin, very little polymerized actin sedimented (Figure 3A). On the other hand, in the presence of VLN3 or VLN1, substantial amounts of actin were present in the pellets (Figure 3A). Moreover, the presence of VLN3 resulted in a dose-dependent increase in the percent of actin in the pellet in the absence of Ca2+ (Figure 3B), consistent with bundling or network formation. The low-speed cosedimentation assay was also used to estimate Kd values for VLN3 binding to F-actin. The apparent Kd obtained from similar experiments was 1.1 ± 0.6 μM (n = 3) and was not statistically different (P = 0.7) from that obtained with the high-speed cosedimentation assay.
Figure 3.
VLN3 Bundles F-Actin, and the Activity Is Only Modestly Affected by Ca2+.
(A) The ability of VLN3 to form higher-order structures with F-actin was determined with low-speed cosedimentation assays. Ten micromolar prepolymerized F-actin was incubated with or without VLN1 or VLN3 (25 nM free Ca2+) followed by centrifugation at 13,500g. The supernatants (S) and pellets (P) obtained were subjected to SDS-PAGE and Coomassie stained. The positions of VLN3, VLN1, and actin are marked on the right. Migration of Mr standards is given at the left.
(B) Scatterplot for dose-dependent bundling of actin filaments by VLN3. The low-speed cosedimentation assay was performed by mixing 10 μM prepolymerized F-actin with increasing concentrations of VLN3 in the presence of 25 nM free Ca2+. The supernatants and pellets obtained after centrifugation were subjected to SDS-PAGE and Coomassie stained. Actin bands were analyzed by densitometry, and actin standards on the same gel were used to calculate the percentage of actin in the pellet. Values plotted are means and the error bars represent sd. n = 4.
(C) The effect of Ca2+ on bundling activity of VLN3 was determined by low-speed cosedimentation assays, as in (A), that were performed with 10 μM prepolymerized F-actin and 500 nM VLN3 or VLN1 in the presence of different [Ca2+]free. Percentage of actin in the pellet was determined by densitometry. Values plotted are means, and the error bars represent sd. n = 4. Asterisks represent values that were statistically different (P < 0.05 by t test) from percentage of actin in the pellet at 25 nM Ca2+.
(D) to (F) Micrographs of actin bundles formed in the presence of VLN1 or VLN3. Prepolymerized actin (1 μM) alone (D), with 100 nM VLN1 (E), or 100 nM VLN3 (F) was incubated at room temperature for 15 min and labeled with 1 μM rhodamine-phalloidin. Images taken by wide-field fluorescence microscopy showed filament bundles in the presence of VLN3, similar to VLN1. All exposures were identical for the three representative images and are reflective of those used for skewness analysis (below). Bar in (D) = 10 μm.
(G) VLN3 exhibits dose-dependent bundling based on skewness analysis in vitro. Increasing concentrations of VLN3 have significantly skewed pixel intensities compared with unbundled single actin filament skewness values denoted by asterisks (P < 0.0001 by one-way Student’s t test). The experiment was repeated three times, and mean skewness values (±se) from 150 micrographs per concentration are shown, with 1 μM actin alone controls having a value of 2.4 ± 0.14 and VLN3 saturating at 13 ± 0.3 for 200 nM VLN3.
(H) VLN3 average skewness values are not significantly affected by varying the [Ca2+]free. Conditions include 50 nM VLN3 and 1 μM prepolymerized actin. The experiment was replicated three times, and mean skewness values (±se) for 150 micrographs per Ca2+ concentration from 100 nM to 1 mM are shown. n = 3.
The above experiments were performed at low [Ca2+]free; therefore, we wanted to assess the effects of [Ca2+]free on VLN3 bundling activity. Low-speed cosedimentation assays were performed with a fixed stoichiometry of VLN3 to actin, at [Ca2+]free spanning the physiological range (0.1 to 10 μM). Following incubation of 500 nM VLN3 and 10 μM prepolymerized actin, the mixtures were centrifuged at 13,500g, and the amount of actin in the pellet was determined by densitometry. There was a modest, but statistically significant, decrease in the amount of bundled actin at 100 nM (P = 0.03) and 1 mM Ca2+ (P = 0.04) compared with that at 25 nM Ca2+ (Figure 3C). VLN1, which has been shown to be a Ca2+-independent bundling protein (Huang et al., 2005), exhibited no statistical difference (P > 0.05) in the amount of actin in the pellet in the absence or presence of Ca2+ (Figure 3C). Thus, there was a modest effect of varying the [Ca2+]free on the apparent bundling activity of VLN3 as determined with the low-speed cosedimentation assay. The reduced amount of sedimentable actin at high [Ca2+] could result from changes in bundling, monomer binding or severing activities, or a combination of these properties. Nevertheless, it can be concluded that VLN3 is able to bind and bundle actin filaments efficiently in the absence and presence of Ca2+.
To confirm the formation of higher-order, actin-based structures, fluorescence light microscopy was used to observe the effects of VLN3 on actin filaments at steady state. When 1 μM prepolymerized actin was decorated with rhodamine-phalloidin, predominantly individual actin filaments were visualized (Figure 3D). In comparison, when F-actin was incubated with 100 nM VLN3, in the presence of 100 nM or no free Ca2+, long and thick actin filament bundles were observed (Figure 3F). These were similar to bundles detected when actin and 100 nM VLN1 were incubated together (Figure 3E). To quantitate the generation of higher-order structures from light micrographs, a novel approach was implemented that avoids the drawbacks of hand selecting individual structures and measuring some parameter. Higaki et al. (2009) developed an algorithm to examine the extent of actin bundling in living guard cells of Arabidopsis. The method defines a statistical parameter called skewness (a lack in Gaussian normality) and assumes that a field of single actin filaments will have a Gaussian distribution of pixel intensities, whereas a population with higher-ordered structures shifts or skews the distribution to the left, resulting in higher skewness values. Here, we applied and tested Higaki’s algorithm against reconstituted actin-based structures using phalloidin-decorated actin filaments in the presence or absence of VLN1 or VLN3. Analyses of at least 150 randomly chosen fields of view containing single actin filaments had average skewness values of between 2 and 3 (Figure 3G; see Supplemental Figure 4A online). In the presence of increasing amounts of VLN1 (see Supplemental Figure 4A online) or VLN3 (Figure 3G), average skewness values increased significantly (P < 0.0001, by one-way t test) in a dose-dependent manner and reached saturation at villin concentrations substoichiometric with actin. Moreover, inclusion of 100 μM free Ca2+ did not affect the skewness values over the entire dose series of VLN1 (see Supplemental Figure 4A online). A range of [Ca2+]free from 100 nM to 1 mM also had no significant effect on average skewness values for reactions containing 50 nM VLN3 and 1 μM actin (Figure 3H). Importantly, the average skewness results correlated with the amount of actin in the pellet fraction when the same reactions were subjected to low-speed sedimentation analysis (see Supplemental Figures 4B and 4C online). These results confirm and validate that skewness is an appropriate and robust measure of actin filament higher-order structure formation. Collectively, our findings demonstrate that VLN3 is an actin filament bundling protein and this activity is independent of [Ca2+]free.
Filament Bundling Occurs by a Catch and Zipper Mechanism
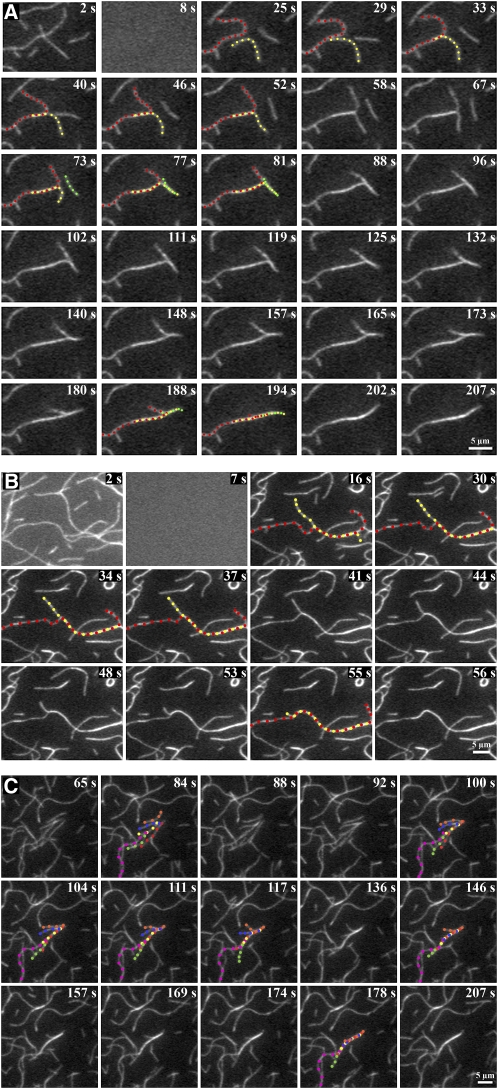
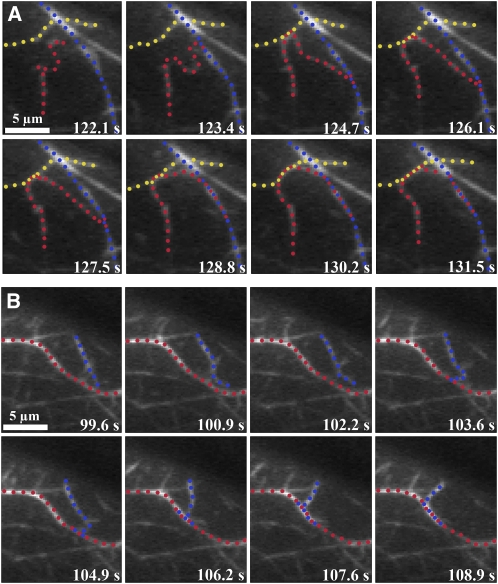
To visualize in real time the formation of villin-induced bundles, we used time-lapse total internal reflection fluorescence (TIRF) microscopy and a reconstituted system. N-ethyl maleimide (NEM)-myosin was used to partially immobilize 25 nM of prepolymerized rhodamine-actin filaments on the cover glass of flow chambers (Amann and Pollard, 2001; Michelot et al., 2005, 2006; Kovar et al., 2006). The actin binding protein was perfused into the chamber and time-lapse images captured to visualize villin-induced bundling. In the absence of villin, most of the actin filaments simply wiggled and very few bundles were observed (see Supplemental Movie 1 online). When VLN3 was perfused through the chamber in the presence of 38.5 nM or 1 μM free Ca2+, some of the individual actin filaments that were positioned close to each other made contact and zippered together to form actin filament bundles (Figures 4A and 4B; see Supplemental Movies 2 and 3 online, respectively). A similar zippering was observed when bundles were formed in the presence of VLN1 (Figure 4C; see Supplemental Movie 4 online) and by the plant formin protein, AFH1 (Michelot et al., 2006). Bundles were also formed by the zippering together of more than two filaments (e.g., Figures 4A and 4C). Thus, in the presence of either VLN3 or VLN1, filament bundles formed by a catch and zipper mechanism.
Figure 4.
Time-Lapse TIRFM Allows Real-Time Visualization of Bundle Formation by Villins in Vitro.
Analysis of bundle formation by time-lapse TIRFM. Prepolymerized rhodamine-actin was weakly adhered to the cover slip of a perfusion chamber with NEM myosin. Twenty-five nanomolar VLN1 (C) or 5 nM VLN3 ([A] and [B]), in the presence of 38.5 nM ([A] and [C]) or 1 μM (B) free Ca2+, was perfused into the chamber and images collected every 1 or 2 s by TIRF microscopy. Two or more filaments marked by different colored dots zipper together to form a bundle represented by alternating dots of the colors marking the individual filaments. Time points on the top right corner of each image indicate the elapsed time from the start of the experiment. See also Supplemental Movies 2 to 4 online. Bars = 5 μm.
Recent advances in fluorescent fusion protein reporters and imaging modalities have allowed actin filament dynamics to be examined with high spatial and temporal resolution in vivo (Era et al., 2009; Staiger et al., 2009; Vidali et al., 2009, 2010). In the cortical array of epidermal cells from dark-grown hypocotyls at least two populations of actin-based structures exist, individual filaments and bundles or cables, and these differ in average length, fluorescence intensity, lifetime, and convolutedness (Staiger et al., 2009). A previous study demonstrates that individual filaments are subject to remarkably dynamic behavior, with rapid elongation rates balanced by prolific severing activity to maintain short filament lifetimes (Staiger et al., 2009). How the more stable bundles form and turn over, however, was not examined. Here, we used time-lapse variable-angle epifluorescence microscopy (VAEM; Konopka and Bednarek, 2008; Staiger et al., 2009) to examine the formation of filament bundles in Arabidopsis epidermal cells expressing the actin reporter GFP-fABD2. As shown in Figure 5 (and Supplemental Movies 5 and 6 online), both individual filaments and short bundles were observed to make contact with an adjacent actin-based structure followed by the zippering together of the two structures along their lengths. In some cases, multiple filaments bundled into one structure, or a single filament participated in the formation of multiple bundles (Figure 5A; see Supplemental Movie 5 online). Examples of this catch and zipper mechanism were observed in every cell imaged. Thus, filament-filament bundling appears to occur in the same manner both in vitro and in vivo.
Figure 5.
Time-Lapse VAEM Allows Real-Time Visualization of Actin Bundle Formation in Vivo.
VAEM was used to image the cortical actin cytoskeleton in dark-grown Arabidopsis hypocotyl epidermal cells 11 d after germination expressing GFP-fABD2, as described previously (Staiger et al., 2009).
(A) A long actin filament (red dots) touches two stationary adjacent filament cables (blue and yellow dots), then bundles by a zippering mechanism (alternating dots). This epidermal cell was imaged at ~1.5-s intervals for 9.4 s. See also Supplemental Movie 5 online. Bar = 5 μm.
(B) Another example of in vivo bundle formation in a hypocotyl epidermal cell expressing GFP-fABD2. Here, a short actin filament (blue dots) makes contact with a stationary filament (red dots), then proceeds to zipper into the stationary filament (red and blue alternating dots). The cortical actin cytoskeleton in this cell was imaged at ~1.5-s intervals for 6.7 s. See also Supplemental Movie 6 online. Bar = 5 μm.
VLN3 Severs Actin Filaments
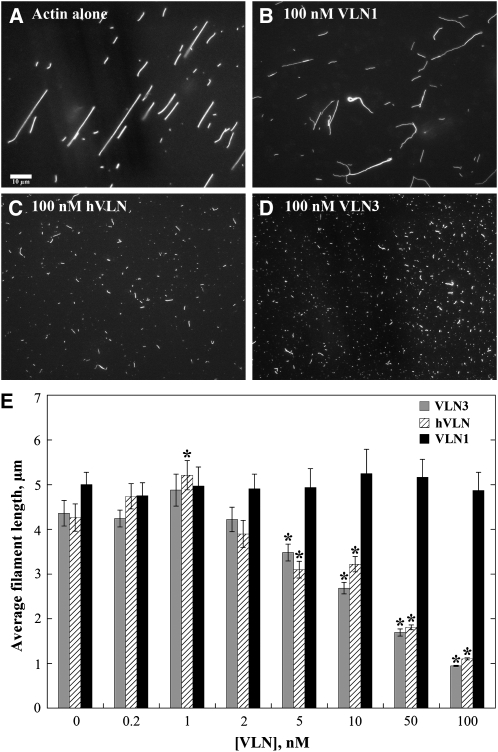
From kinetic analyses of actin assembly and disassembly reactions, it was inferred previously that VLN1 does not sever actin filaments (Huang et al., 2005). To test directly whether VLN3 and VLN1 had severing activity, we examined individual actin filament behavior at steady state and by time-lapse TIRF microscopy. Human villin (hVLN) was used as a positive control (Kumar and Khurana, 2004; Kumar et al., 2004a, 2004b). Experiments were initially performed with wide-field fluorescence light microscopy assays, where prepolymerized actin (1 μM) was incubated with villin in the presence of EGTA or 10 mM free Ca2+ for 15 min followed by the addition of rhodamine-labeled phalloidin to decorate the filaments. When incubated alone or in the presence of various concentrations of VLN1, actin filaments were on average 4 to 5 μm long (Figures 6A, 6B, and 6E). In the presence of 10 mM Ca2+ and 100 nM VLN3 or hVLN, however, the filaments were significantly shorter, ~1 μm in length (Figures 6C to 6E; P > 0.05). This reduction in filament length under high Ca2+ conditions was dose dependent for both VLN3 and hVLN (Figure 6E) and consistent with filament severing.
Figure 6.
The Length of Actin Filaments Is Reduced in a Dose-Dependent Manner by VLN3.
(A) to (D) Micrographs of actin filaments in the absence or presence of villins. One micromolar prepolymerized actin alone or with villin in the presence of 10 mM free Ca2+ was incubated at room temperature for 15 min and labeled with 1 μM rhodamine-phalloidin. Images taken by wide-field fluorescence microscopy showed much shorter filaments in the presence of 100 nM VLN3 (D) or hVLN (C) compared with actin alone (A) or actin in the presence of 100 nM VLN1 (B). Bar = 10 μm.
(E) Average filament lengths as a function of VLN3, hVLN, and VLN1 concentrations were plotted. Values are means and the error bars represent se. n > 200. Asterisks represent the values that were statistically different (P < 0.05 by t test) compared with actin alone. VLN3 and hVLN showed dose-dependent filament shortening, whereas VLN1 had little or no effect on filament length.
The decrease in filament length observed in the presence of VLN3 or hVLN at equilibrium could also be due to monomer sequestration, depolymerization of actin filaments, or inhibition of annealing. To rule these out, and to visualize actin filaments in real time, we used TIRF microscopy. Prepolymerized rhodamine-labeled actin filaments (25 nM) were adhered to the cover glass of a perfusion chamber as described previously. Villin was perfused into the chamber and time-lapse images captured. Analyses with actin filaments alone or actin with VLN1 showed minimal breakage (see Supplemental Movies 1 and 7 online; Figure 7C). In the absence of Ca2+, neither hVLN nor VLN3 had an effect on actin filaments (e.g., for VLN3; see Supplemental Movie 8 online). In the presence of 10 μM free Ca2+, however, hVLN induced breaks along the filaments at concentrations as low as 0.03 nM, demonstrating severing activity (Figures 7A and 7C; see Supplemental Movie 9 online). When perfused in the presence of 10 μM free Ca2+, VLN3 at concentrations between 3 and 10 nM also generated numerous breaks along the filaments (Figures 7B and 7C; see Supplemental Movie 10 online).
Figure 7.
Time-Lapse TIRFM Demonstrates Directly the Severing Activity of Villins and ADF1.
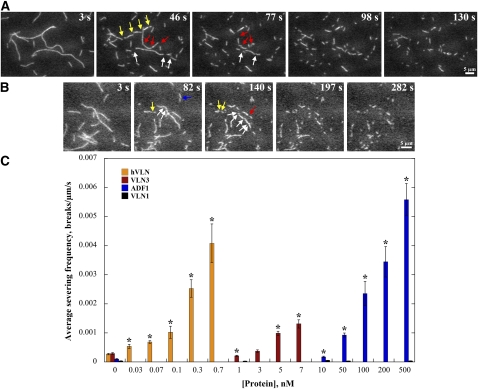
(A) and (B) Rhodamine-actin filaments were adhered to the cover slip of a perfusion chamber, and then 0.5 nM hVLN (A) or 7 nM VLN3 (B), in the presence of 10 μM free Ca2+, was perfused into the chamber and time-lapse images were collected every second. Individual filaments showed breaks (arrows) along their length. The elapsed time in seconds is given in the top right corner of each image. See also Supplemental Movies 9 and 10 online. Bars = 5 μm.
(C) Quantitative analysis of severing activity. Either fluorescence buffer alone or different concentrations of hVLN, VLN3, VLN1, or ADF1 were perfused through chambers containing rhodamine-actin filaments, and time-lapse images were collected at 1- to 5-s intervals. The maximal length of individual filaments was measured, and breaks in each filament were counted during playback of the time-lapse series. Severing frequency was calculated as the number of breaks per unit filament length per unit time. At least three independent experiments in which >20 filaments each were counted were performed for each protein. Error bars represent se. Asterisks represent data points that were statistically different (P < 0.05 by t test) compared with actin alone.
The number of breaks per unit filament length per unit time (breaks/μm/s) was calculated as a quantitative measure for the severing frequency of each protein (Andrianantoandro and Pollard, 2006). Human villin exhibited an average severing frequency (±se) of 0.004 ± 0.0007 breaks/μm/s at 0.7 nM concentration (Figure 7C). The severing was markedly faster at concentrations ≥1 nM, making it impossible to assess quantitatively. Similar problems were faced when collecting time-lapse series of severing in the presence of ≥10 nM VLN3, where filaments tended to stick to glass and/or simply fade away because of rapid severing and loss of fragments. VLN3 did, however, show a significant increase (P < 0.05) in severing frequency at concentrations up to 7 nM, compared with actin alone (Figure 7C). It had an average severing frequency of 0.0013 ± 0.00014 breaks/μm/s at 7 nM. Similar experiments were performed with Arabidopsis ADF1, which was previously suggested not to sever actin filaments (Carlier et al., 1997). We found that ADF1 also severed actin filaments, albeit requiring much higher concentrations than the villins (Figure 7C; see Supplemental Movie 11 online). Thus, VLN3 is a bona fide actin filament severing protein with efficiency between human villin and plant ADF.
VLN3 Severs Actin Filaments in a Ca2+-Dependent Manner
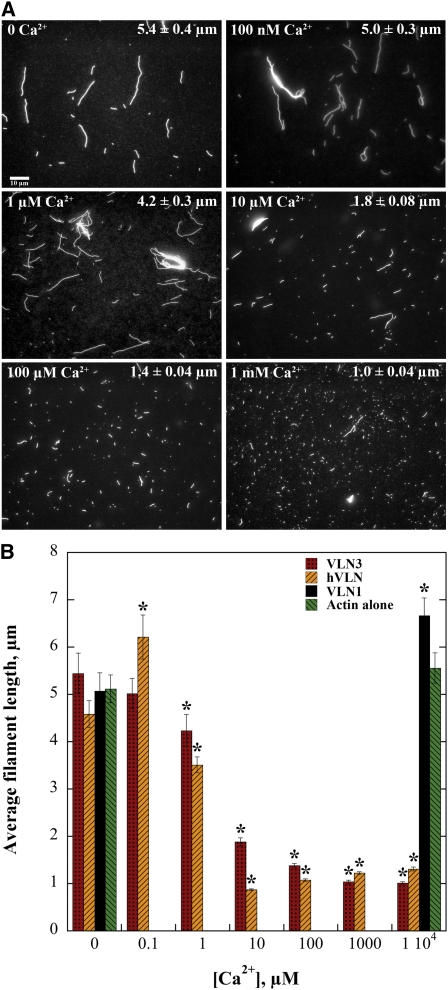
In wide-field fluorescence microscopy experiments, VLN3 showed a reduction in filament length at high [Ca2+]free (Figure 6). We therefore tested whether the activity was affected by varying the [Ca2+]free. At a fixed stoichiometry of actin and VLN3 (1 μM and 50 nM, respectively), filament length decreased as [Ca2+]free increased (Figure 8). There was a statistical difference (P < 0.05 by t test) in the average length of filaments in the presence of 1 μM (mean = 4.2 ± 0.3 μm) to 10 mM (mean = 1.01 ± 0.02 μm) free Ca2+ compared with no Ca2+ (mean = 5.4 ± 0.4 μm; Figures 8A and 8B). There was also a decrease in filament length in the presence of hVLN (Figure 8B), with the difference being significant from 1 μM to 10 mM Ca2+. VLN1, on the other hand, did not reduce filament length in the presence of high [Ca2+]free (Figure 8B). These data suggest that VLN3 shortened the length of actin filaments in a Ca2+-dependent manner, and effective Ca2+ concentrations were in the physiological range of 1 to 10 μM.
Figure 8.
VLN3 Reduces Filament Length in a Ca2+-Dependent Manner.
(A) Micrographs of actin filaments in the presence of VLN3 at different [Ca2+]free. One micromolar prepolymerized actin was incubated with 50 nM VLN3 in the presence of different [Ca2+]free at room temperature for 15 min and labeled with 1 μM rhodamine-phalloidin. Images were taken by wide-field fluorescence microscopy, and the length of >200 actin filaments was measured from at least three separate images. The [Ca2+]free is indicated in the top left corner of each image, and the average filament length ±se is noted in the top right corner. Bar = 10 μm.
(B) Actin filament length in the presence of villins as a function of [Ca2+]. One micromolar prepolymerized actin was incubated with 50 nM VLN3, VLN1, or hVLN in the presence of different [Ca2+]free. Values plotted are average filament length, and error bars are se. Sample size was at least 200 filaments from at least three separate images for each treatment. Asterisks represent concentrations of Ca2+ with filament lengths that were statistically different (P < 0.05 by t test) compared with the corresponding value in the absence of Ca2+.
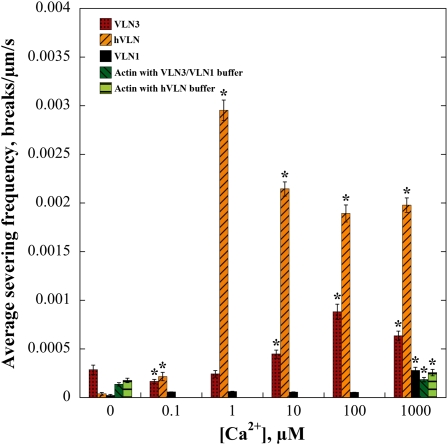
To study the severing frequency of VLN3 in real time at different [Ca2+]free, we returned to time-lapse imaging by TIRF microscopy. At each [Ca2+]free tested, 0.5 nM hVLN, 5 nM VLN1, or 5 nM VLN3 was perfused into chambers containing 25 nM rhodamine-actin filaments, time-lapse images were recorded, and severing frequency determined. When the respective buffers for the recombinant villin proteins, or 5 nM VLN1, were perfused through the chambers in the presence of 1000 μM Ca2+, there was a modest but significant (P < 0.05) increase in the frequency of breaks created compared with no free Ca2+ (Figure 9). This was negligible, however, compared with the severing activity of hVLN, which resulted in high severing frequencies in the presence of [Ca2+]free ≥1 μM (Figure 9). VLN3 also exhibited an increase in severing frequency with increasing [Ca2+]free, up to 100 μM (Figure 9). The magnitude of the severing frequency values for VLN3 was not as high as hVLN. Thus, VLN3 is a Ca2+-dependent severing protein capable of inducing breaks in individual actin filaments.
Figure 9.
Severing Frequency of VLN3 Is Ca2+ Dependent.
Rhodamine-actin filaments were adhered to the cover slip of a TIRF microscopy chamber. Either 0.5 nM hVLN, 5 nM VLN3, or 5 nM VLN1 in the presence of various [Ca2+]free was perfused through the chamber, and time-lapse images were collected every 1 to 5 s. The maximal length of individual filaments was measured, and breaks in each filament counted. A minimum of 20 filaments for each experimental treatment was counted, and the average was plotted against [Ca2+]. Error bars represent se. n is at least 3 for each data set. Asterisks represent data points that were statistically different (P < 0.05 by t test) for severing frequency compared with 0 μM Ca2+.
VLN1 and VLN3 Coexist in Plant Tissues
If VLN1 and VLN3 isovariants are present in the same tissues and cell types, then their distinct activities may have important consequences for actin-based function in those cells. To understand better whether or not these two villins have overlapping expression patterns, we evaluated the previous data and publically available microarray information, as well as performed quantitative real-time PCR (qRT-PCR) on select tissues. Previous RNA gel blot data reveal that VLN1 and VLN3 are ubiquitously expressed throughout the Arabidopsis plant, albeit with VLN1 being less abundant in most tissues and organs (Klahre et al., 2000). A survey of current microarray databases gives a similar impression (Hruz et al., 2008): VLN1 and VLN3 are expressed widely in the plant, but VLN3 is typically more abundant, with the exception of endosperm, pollen, and the shoot apex (see Supplemental Figure 5A online). Promoter:GUS reporter expression studies confirm that VLN1 is present in roots, young leaves, and inflorescences, with strongest VLN1:GUS found in the vasculature of roots, leaves, and filaments of the anthers (Klahre et al., 2000). Overlapping expression of VLN1 and VLN3 in the vasculature of roots is also obvious in the AREX database (Birnbaum et al., 2003; Brady et al., 2007; Cartwright et al., 2009), although VLN3 is present in many more cell types than VLN1, including most notably epidermal cells of the elongation zone and root hairs (see Supplemental Figure 5B online). Strong VLN1:GUS expression was also noted in the guard cells of leaves (Klahre et al., 2000) and is corroborated by microarray analysis of transcripts from guard cell protoplasts where it is coexpressed with VLN3 (Leonhardt et al., 2004; Yang et al., 2008). To evaluate coexpression in inflorescence stems, floral whorls, and mature rosette leaves, we performed qRT-PCR on wild-type plants and compared the results to those obtained from the same tissues isolated from two T-DNA insertion mutants, vln1-4 and vln3-8, respectively (see Supplemental Figure 6 online). Homozygous mutant plants showed little or no expression of the respective transcripts in any tissue tested, indicating that the primer pairs are specific and that the mutants are knockouts for VLN1 and VLN3 (see Supplemental Figure 6 online). VLN1 was expressed at approximately the same level, relative to the glyceraldehyde-3-phosphate dehydrogenase (GAPD) control, in both floral whorls (see Supplemental Figure 6A online) and inflorescence stems (see Supplemental Figure 6B online). VLN3 had threefold lower expression than VLN1 in floral whorls (see Supplemental Figure 6A online), whereas it was twofold higher than VLN1 in inflorescence stems (see Supplemental Figure 6B online). By comparison, in mature rosette leaves, VLN1 levels were quite low and approximately the same as the GAPD control, whereas VLN3 was rather abundant (see Supplemental Figure 6C online). Collectively, these results confirm that VLN1 and VLN3 exhibit spatially and temporally overlapping expression patterns throughout the Arabidopsis plant.
VLN1 Does Not Protect Individual Filaments from Severing by VLN3
To analyze the effects on actin that might result from overlapping and distinct activities of VLN1 and VLN3 proteins, steady state microscopy experiments were performed as before with prepolymerized actin incubated in the presence of villins without Ca2+. For these experiments, we chose a concentration of VLN1 that was saturating for bundle formation, 400 nM (see Supplemental Figures 4A and 4C online), and used 50 nM VLN3. After 15 min, the mix was divided in two. One part was stabilized with rhodamine-labeled phalloidin immediately, whereas the other was treated with Ca2+ before being stabilized. When incubated alone, the actin filaments were qualitatively similar in length before and after the addition of Ca2+ (Figure 10A). In the presence of the villins, filament lengths were similar to actin alone, prior to the addition of Ca2+, and several bundles were observed in the presence of VLN1 or VLN3 (Figure 10A). After the addition of Ca2+, short individual filaments were observed in the presence of VLN3 (Figure 10A), indicating fragmentation. The severing protein was thus activated on the addition of Ca2+ and it markedly shortened single filaments.
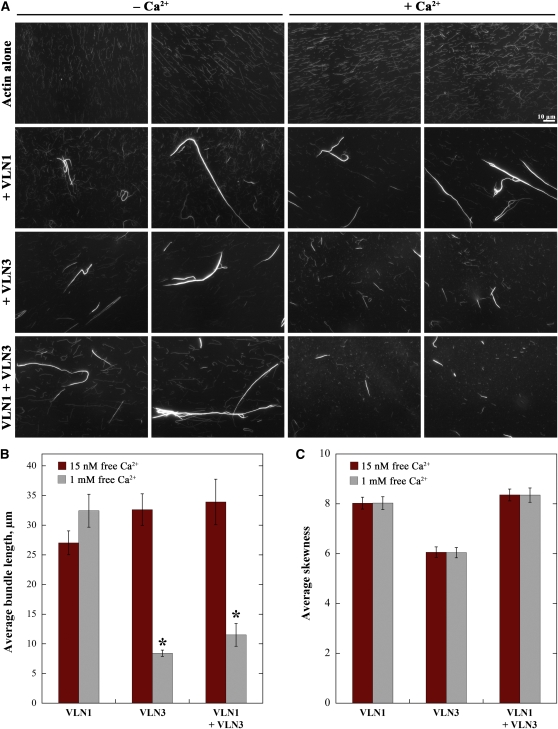
Figure 10.
VLN1 Does Not Protect Filaments or Bundles from Severing by VLN3.
(A) Micrographs of actin filaments in the absence or presence of villins, before and after the addition of Ca2+. One micromolar prepolymerized actin alone or with 400 nM VLN1, 50 nM VLN3, or a combination of VLN1 and VLN3 was incubated at room temperature for 15 min. One millimolar Ca2+ was added to one-half of the mix, which was then incubated for another 15 min. Both were labeled with 1 μM rhodamine-phalloidin, and images were taken by wide-field fluorescence microscopy with identical settings for all treatments. Two representative images for each condition, before (15 nM free Ca2+) and after (1 mM free Ca2+) the addition of Ca2+, are shown. Bar = 10 μm.
(B) and (C) Average length of bundles (B) and average skewness values (C) in the presence of VLN1 and/or VLN3 with or without Ca2+. Experiment was set up as in (A).
(B) The maximal length for >100 bundles was measured from at least 10 images for each data set. Asterisks represent average bundle lengths in the presence of 1 mM free Ca2+ that were statistically different (P < 0.05 by paired t test) from the corresponding controls without Ca2+. The lengths of bundles in the presence of VLN1, VLN3, or both VLN1 and VLN3 without Ca2+ were not statistically different from each other (P > 0.05). In the presence of 1 mM free Ca2+, bundle lengths in the presence of VLN3 with and without VLN1 were not statistically different (P = 0.1).
(C) Mean skewness values were determined from 150 micrographs per treatment with triplicate samples. The addition of 1 mM Ca2+ to samples containing VLN1, VLN3, or the combination of villins did not significantly lower the skewness value compared with the 15 nM Ca2+ controls (P > 0.25). Error bars represent se.
[See online article for color version of this figure.]
Similar results were obtained when actin filaments were coincubated in the presence of both VLN1 and VLN3. Long single filaments and numerous bundles were observed before the addition of Ca2+ (Figure 10A). Following addition of Ca2+, however, all of the individual filaments were short. The presence of a saturating amount of VLN1, a side binding protein, did not affect the shortening of the individual actin filaments. We conclude that VLN1 was unable to protect actin filaments from severing by VLN3.
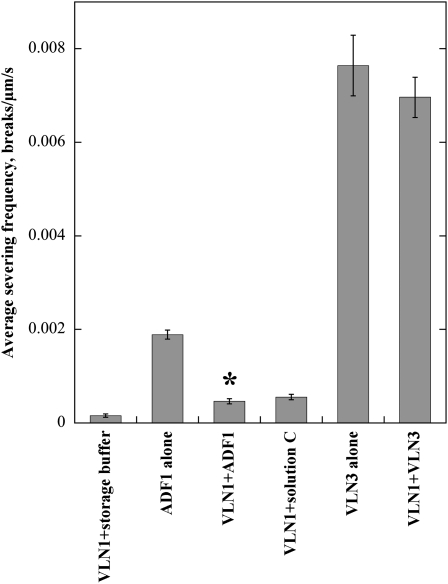
TIRF microscopy experiments were performed to analyze further how the two villin isovariants functioned distinctly at the single filament level. Because previous results had demonstrated that VLN1 protected actin filaments from the effects of ADF1 (Huang et al., 2005), it was of interest to determine whether the presence of VLN1 ameliorated the activities of VLN3. This was evaluated in real time by perfusing 5 nM VLN3 or 200 nM ADF1 into chambers containing 25 nM prepolymerized rhodamine-actin filaments that were previously perfused with 25 nM VLN1. An additional 25 nM VLN1 was added along with the severing protein. Severing by VLN3 was observed in the presence or absence of VLN1 (see Supplemental Movies 12 and 13 online). The average severing frequency of VLN3 in the presence of VLN1 was 0.0069 ± 0.0004 breaks/μm/s and was only 9% less and not statistically different from that of VLN3 alone (0.0076 ± 0.0006 breaks/μm/s; P = 0.4; Figure 11). By contrast, ADF1 severing activity was reduced by >75% in the presence of VLN1 (Figure 11; see Supplemental Movies 14 and 15 online), providing indirect evidence that VLN1 is capable of binding to individual filaments and protecting them from disassembly. Thus, VLN3 severs individual actin filaments in the presence of VLN1.
Figure 11.
VLN3 Severs Individual Actin Filaments in the Presence of VLN1.
Rhodamine-actin filaments were adhered to the cover slip of a TIRF microscopy chamber, and 25 nM VLN1 was perfused into the chamber. Five nanomolar VLN3 in the presence of 10 μM free Ca2+, 200 nM ADF1, or their respective buffers only with an additional 25 nM VLN1 was then perfused into the same chamber, and time-lapse images were collected every 1 to 2 s. VLN3 or ADF1 were also perfused into chambers that had not been pretreated with VLN1. See also Supplemental Movies 12 to 15 online. These experiments were repeated three times, and 20 filaments were measured for each experimental treatment and each replicate. Severing frequency was determined as before. Error bars represent se. Asterisk represents the data point that was statistically different (P < 0.05) for severing frequency from the severing protein alone.
VLN3 Dismantles Actin Filament Bundles in the Presence of Ca2+ and VLN1
To determine whether VLN3 can sever actin filaments within bundles, the bundles from steady state experiments (Figure 10) were assessed for reductions in length. Moreover, populations of bundles were analyzed for change in skewness values as described above. In the absence of Ca2+, the average length of bundles formed by incubation with a combination of VLN1 and VLN3 was not statistically different than those formed in the presence of VLN1 or VLN3 alone (P > 0.05; Figure 10B, red bars). In the presence of Ca2+, however, the bundled structures generated with VLN3 alone or with VLN3 and VLN1 were ~3 times shorter than their respective controls without Ca2+ (Figure 10B, gray bars). To test whether the shortening of bundles was due to severing and not unbundling, 150 micrographs per treatment were analyzed for skewness (Figure 10C). In the presence of 1 mM Ca2+, average skewness values were not significantly different from 15 nM Ca2+ controls for any combination of villins tested (P > 0.25; Figure 10C). To confirm that skewness values are insensitive to changes in bundle length, VLN1 bundles were mechanically sheared by sonication; values for skewness after 1 and 5 min of sonication were not significantly different from untreated controls, whereas bundle lengths were significantly reduced by sonication (see Supplemental Figures 4D and 4E online). Collectively, these results indicate that VLN3 dismantles actin bundles by severing and shortening rather than depolymerizing or unbundling in the presence of Ca2+. Thus, VLN3 severs actin filament bundles in a Ca2+-dependent manner, regardless of the presence or absence of VLN1.
DISCUSSION
This study provides important insight into the cooperative and antagonist activities of actin binding protein isovariants from the villin/gelsolin/fragmin superfamily. Using state-of-the art biochemical assays and advanced imaging to monitor the behavior of single actin filaments and filament bundles, we characterized two members of the villin gene family with overlapping and distinct functions. The villin family in Arabidopsis has five isovariants, of which VLN1 is unique based on phylogenetic analyses and the number of conserved Ca2+ binding sites (Huang et al., 2005). We predicted that it would also be unique in its failure to sever actin filaments, and this was confirmed by time-lapse TIRF microscopy. Our data further demonstrate unambiguously that another villin isovariant, VLN3, is capable of actin binding and bundling like VLN1 and has Ca2+-dependent severing activity. When present together, as they are likely to occur in trichomes (Dai et al., 2010), guard cells and mesophyll cells in leaves (Leonhardt et al., 2004; Yang et al., 2008) and cells in the root (Birnbaum et al., 2003; Brady et al., 2007; Cartwright et al., 2009), these two proteins can cooperate to form long actin filament bundles. Upon exposure to micromolar [Ca2+], however, the severing function of VLN3 is activated, and this results in the fragmentation of individual filaments as well as bundles irrespective of the presence of VLN1. Taking into account previous findings that VLN1 is able to protect actin filaments from disassembly by ADF1 (Huang et al., 2005), the ability of VLN3 to sever individual filaments and shorten actin bundles containing VLN1 indicates fundamental differences between actin binding protein families in facilitating actin turnover. These observations suggest a revised model for actin filament bundle formation and turnover in cells, based on the divergent activities of villin isovariants.
Actin filament bundles or cables are linked with several important cellular processes in plants (Higaki et al., 2007, 2010; Thomas et al., 2009), but their formation and turnover is poorly understood. Like a recent example from liverwort (Marchantia polymorpha) expressing a Lifeact-Venus reporter (Era et al., 2009), we were able to visualize in real time the formation of actin filament bundles in the cortical array of living Arabidopsis epidermal cells by VAEM imaging. The mechanism, which appears as filament catch and zipper to a neighboring filament or bundle, is mimicked in vitro with a reconstituted system comprising single actin filaments and either recombinant VLN1 or VLN3 imaged by time-lapse TIRF microscopy. The zippering is superficially similar to bundling induced by an unusual plant formin, AFH1 (Michelot et al., 2006). Capture and zippering was also recently described for a novel bundling protein from hair cell stereocilia, TRIOBP, that is implicated in human hereditary deafness (Kitajiri et al., 2010). Our steady state analyses confirmed that when VLN3 and VLN1 are incubated together with actin, numerous bundles are formed. We propose that these isovariants work cooperatively to form large higher-order structures in cells. Several classes of actin-bundling or cross-linking proteins are known to exist in plants. Four families of such proteins are found in Arabidopsis, including the fimbrins, formins, LIM proteins, and villins (Higaki et al., 2007, 2010; Thomas et al., 2009; Staiger et al., 2010). To our knowledge, there have been no studies so far suggesting any cooperation between these families of bundling proteins. The various isovariants of formins and villins are also known to have functions in addition to bundling (Thomas et al., 2009; Blanchoin and Staiger, 2010; Staiger et al., 2010). Different families of bundling proteins in nonplant systems (animals and flies), including Fascin, Forked, espin, fimbrin/plastin, and villins are known to exist together and cooperate or act sequentially during the formation of actin bundles found in specialized cellular structures (Bartles, 2000). For example, massive parallel actin bundles in the neurosensory bristles of Drosophila are formed by sequential action of Forked and Fascin (Tilney et al., 1998, 2000; Wulfkuhle et al., 1998), whereas those in nurse cells require Quail followed by Fascin (Matsudaira et al., 1983; Cant et al., 1994; Mahajan-Miklos and Cooley, 1994). Actin bundles in brush border microvilli of mammalian epidermal cells, on the other hand, are a result of the combined activity of villin, fimbrin/plastin, and small espin (Gerke and Weber, 1983; Matsudaira et al., 1983; Ezzell et al., 1989; Bartles et al., 1998). There have been no reports, however, of cooperativeness between members of the same family of proteins with overlapping expression, like VLN1 and VLN3 (Higaki et al., 2007; Thomas et al., 2009).
There are two fields of thought about the mechanism of bundling by villins. One suggests the formation of dimers of the actin bundling proteins, allowing one actin binding site (e.g., the VHP) on each subunit to interact with separate actin filaments, as implied for lily villin 135-ABP (Yokota et al., 1998, 2005) and demonstrated for human villin (George et al., 2007). Another field suggests the binding of a villin monomer with two actin filaments via its two F-actin binding sites, one in the gelsolin-like core domain and the other in the VHP, thus forming a bundle (Glenney and Weber, 1981; Friederich et al., 1999; Hampton et al., 2008). Our preliminary analytical centrifugation analysis revealed minimal formation of VLN1 or VLN3 dimers. Nevertheless, both proteins bind actin filaments with moderate affinity and generate higher-order structures that resemble filament bundles or cables. Further studies will be required, however, to understand the exact mechanism of bundling by these proteins.
Apart from formation of actin filament cables and networks, dynamic reorganization of the actin cytoskeleton is crucial for the normal functioning of plant and nonplant cells, and filament severing is a significant aspect of this (Ono, 2007; Vavylonis et al., 2008; Staiger et al., 2009; Okreglak and Drubin, 2010). Important insights about cytoskeletal turnover in plant cells was obtained by our recent VAEM analysis of the cortical actin cytoskeleton in epidermal cells of dark-grown hypocotyls of Arabidopsis (Staiger et al., 2009). That study revealed a stochastic behavior of actin filaments dominated by profilic severing, similar to an in vitro biomimetic system of actin turnover and assembly (Michelot et al., 2007). Biochemically, the proteins responsible for severing of the actin were predicted to be either ADF/cofilin or villins (Staiger et al., 2009, 2010). In this study, we performed time-lapse imaging of individual actin filaments in the presence of recombinant VLN3 in vitro, providing direct, quantitative evidence for severing activity by a full-length plant villin. Severing by actin binding proteins is typically inferred in vitro by depolymerization assays with pyrene-labeled actin and fluorimetry assays and/or by imaging of labeled actin filaments in the presence of severing proteins with fluorescence microscopy. Several actin binding proteins from nonplant systems have been shown to sever by these methods, including human plasma gelsolin (Bearer, 1991), human villin (Kumar and Khurana, 2004; Kumar et al., 2004a), mouse formin INF2 (Chhabra and Higgs, 2006), and cofilin from various species (Pavlov et al., 2007). The first plant protein demonstrated to have severing activity by fluorescence microscopy of phalloidin-labeled actin filaments was a gelsolin isolated from poppy (Papaver rhoeas) pollen, Pr ABP80 (Huang et al., 2004). The villins from lily pollen, 135-ABP and 115-ABP, are capable of disassembly, as evidenced by biochemical assays in the presence of Ca2+-CaM (Yokota et al., 2005). Circumstantial evidence for severing of filaments polymerized in vitro, by a shorter homolog of villin (ABP41) that was isolated from pollen of Lilium davidii has been obtained by electron microscopy (Fan et al., 2004). In addition, fluorescence microscopy has been used to observe severing by another low molecular weight protein from lily, ABP29, that is suspected to be a splice variant of 135-ABP (Xiang et al., 2007). Nevertheless, how efficient these villin/gelsolin/fragmin family members are at severing filaments and whether they are functional at physiological Ca2+ levels remain to be determined. A parallel study by Zhang et al. (2010) reveals that recombinant VLN5 also severs actin filaments at micromolar [Ca2+].
TIRF microscopy has recently become an unequivocal way of observing the creation of breaks at the single actin filament level in real time. An evanescent wave from the internally reflected excitation light excites fluorescent filaments in only a narrow region of ~100 nm on the surface of our chamber, thus reducing the background considerably (George, 2004). The clear imaging of breaks permits quantitative analyses of severing as well. The technique has been applied to examine the biochemical activity of proteins like plasma gelsolin (Nag et al., 2009) and budding yeast twinfilin (Moseley et al., 2006). In a landmark study, Andrianantoandro and Pollard (2006) demonstrated direct severing by cofilins from fission yeast, Acanthamoeba, and human. The power of this approach to clarify the molecular mechanism of actin binding protein function is exemplified by plant ADF. Arabidopsis ADF1 was previously suggested to lack the ability to fragment actin filaments and instead argued to depolymerize filaments by monomer binding and increasing the off rate at pointed ends (Carlier et al., 1997; Dong et al., 2001). However, we were able to show severing activity of ADF1 directly by TIRF microscopy. It also allowed us to compare the potency of ADF1 with the villins. The concentrations of ADF1 required to equal the severing frequencies of VLN3 were ~100-fold higher and to equal human villin severing frequency ~1000-fold higher. Thus, plant and human villins sever actin filaments, and this protein family is markedly more potent than plant ADF1 at equivalent concentrations.
Plant actin binding proteins are known to differ in function and regulation from their animal counterparts (Staiger and Blanchoin, 2006). We show here that like human villin, VLN3 is regulated by micromolar [Ca2+]. Both proteins can bundle in the presence of low [Ca2+]. As [Ca2+] increases, they exhibit severing activity. Human villin showed high severing frequency even at 1 μM free Ca2+ in our assays, which was surprisingly lower than the concentration of >200 μM required for severing reported previously (Kumar and Khurana, 2004; Kumar et al., 2004a). The latter was based on a depolymerization assay by fluorimetry, which is indirect and less sensitive than our TIRF assay. VLN3, on the other hand, exhibited a dose-dependent increase in severing frequency with increasing [Ca2+], albeit less than hVLN. There was, however, significant severing by VLN3 in the physiological range of Ca2+ (1 to 10 μM). Thus, VLN3 is likely to be a severing protein in plant cells that is sensitive to fluxes in cytosolic Ca2+ levels.
Several studies on the cytoskeleton and its regulation in plant cells have suggested a role for Ca2+ as a signaling molecule. Pollen tubes and root hairs, two model cell types for actin-mediated tip growth, have an oscillatory, tip-high Ca2+ gradient that is presumed to activate or inactivate Ca2+-dependent actin binding proteins, thereby bringing about changes in actin organization and dynamics (Hepler et al., 2001; Staiger et al., 2010). Furthermore, external availability of Ca2+ or the use of ionophores results in actin reorganization in various types of cells (Yokota et al., 2000; Hepler et al., 2001; Snowman et al., 2002; Cárdenas et al., 2008). Several studies on pollen tubes reveal that increasing the cytosolic [Ca2+] by treating with the Ca2+ ionophore A23187 or mastoparan brings about fragmentation or depolymerization of the actin cytoskeleton (Kohno and Shimmen, 1988; Snowman et al., 2002; Yokota et al., 2005). With evidence of lily villins being extracted from pollen and localized on actin cables in pollen tubes and root hairs (Nakayasu et al., 1998; Yokota et al., 1998, 2000, 2003; Vidali et al., 1999; Yokota and Shimmen, 1999; Tominaga et al., 2000) as well as their being responsible for bundle formation and maintenance in both pollen and root hairs (Shimmen et al., 1995; Tominaga et al., 2000; Ketelaar et al., 2002), the villins have been implicated as Ca2+-regulated proteins involved in actin turnover (Yokota and Shimmen, 2006; Staiger et al., 2010). The lily villins and their splice variants do indeed disrupt actin cables when overexpressed in pollen tubes and stamen hair cells (Tominaga et al., 2000; Yokota et al., 2003; Fan et al., 2004; Xiang et al., 2007; Wang et al., 2008). The biochemical properties of VLN3, although it is an isovariant that is absent from pollen (Pina et al., 2005), point toward the possibility of pollen-expressed VLN2 or VLN5 being responsible for fragmentation of the actin framework in Arabidopsis pollen. In fact, Zhang et al. (2010) verify that VLN5 severs actin in the presence of micromolar [Ca2+] and that it plays an important role in normal pollen tube growth in Arabidopsis. The villin family, with the exception of VLN1, could therefore play an important role in the Ca2+-mediated restructuring of the actin cytoskeleton in other plant cells as well.
As pointed out earlier, different isovariants of the villins have overlapping spatial and temporal expression patterns in Arabidopsis (Klahre et al., 2000; Hruz et al., 2008). Moreover, several different actin binding proteins with similar functions could also have overlapping expression. The possibility of competition or synergies must be considered between such coexisting isovariants or proteins with related function. It has been shown that actin filaments are protected by VLN1 from the disassembly activity of ADF1 in vitro (Huang et al., 2005). Actin filament bundles are in general likely to be resistant to turnover by ADF/cofilin (Michelot et al., 2007), but the basis for this has not been fully explored. We extended the study in a reconstituted system using TIRF microscopy to understand the overlapping or distinct effects of villin isovariants. Individual actin filaments that were incubated with VLN1 before being exposed to VLN3 in the presence of micromolar [Ca2+] are severed to a similar extent whether VLN1 was present or not. This indicated that VLN3 and VLN1 did not compete with each other. Steady state assays to understand the effect of VLN3 on bundles revealed a large number of bright higher-order structures in the presence of VLN1 and/or VLN3 in the absence of Ca2+. The length of these bundles was significantly reduced in the presence of VLN3 and Ca2+, indicating their fragmentation. Moreover, bundle length was still reduced in the presence of VLN1, again suggesting that bundles are not protected from VLN3 activity.
Arabidopsis FIMBRIN1, an actin cross-linking protein, is also known to protect filaments from profilin-mediated disassembly in vivo and in vitro (Kovar et al., 2000). Furthermore, tobacco (Nicotiana tabacum) WLIM1, another actin-bundling protein, stabilizes actin filaments and/or bundles and protects them from latrunculin B both in vitro and in BY2 cells (Thomas et al., 2006). Other examples of accessory proteins that prevent modifications in the actin filament structure include nonplant tropomyosin that interferes with Arp2/3 complex and ADF/cofilin function (Blanchoin et al., 2001). In yeast, however, Aip1 (actin-interacting protein1) and cofilin function together to sever and cap cables decorated by tropomyosin (Okada et al., 2006). This appears to be a mechanism to promote the turnover of actin in order for it to be used for other functions or used directly for assembly in the cell (Okreglak and Drubin, 2010). VLN3 might have a similar function to prune VLN1 bound filaments. This in turn is regulated by Ca2+.
VLN3 (this study) and VLN5 (Zhang et al., 2010), but not VLN1, exhibit Ca2+-regulated severing activity. It is likely that this is important for actin filament turnover in response to changes in Ca2+ levels in plant cells, such as in tip-growing pollen tubes (Staiger et al., 2010). Similar to pollen tubes, the actin cytoskeleton undergoes dramatic rearrangements in root hairs as well (Era et al., 2009). Injection of an anti-lily villin antibody into growing root hairs of Arabidopsis causes disruption of bundles and changes in nuclear localization (Ketelaar et al., 2002). Microarray data reveal the presence of all five villin isovariants in Arabidopsis roots, with VLN3 having the highest expression levels (Ma et al., 2005). More specifically, VLN3 along with VLN2 and VLN4 are expressed in root hairs (Birnbaum et al., 2003; Brady et al., 2007; Cartwright et al., 2009). It would not be surprising that VLN3 and the other isovariants play a key role in the growth of, and organelle positioning in, root hairs. In addition, the villins might be important players in the regulation of the actin cytoskeleton in guard cells. Actin bundles are known to form transiently during stomatal movements (Higaki et al., 2009), and the villins, specifically VLN3 and VLN1, are expressed in guard cells (Leonhardt et al., 2004; Yang et al., 2008). The predicted role of the villins in the dynamic reorganization of actin in the epidermal cells of the hypocotyl (Staiger et al., 2009) and their specific expression in this tissue (Ma et al., 2005) implies the importance of these proteins in cell expansion. Reverse-genetic methods will provide vital evidence to support these predictions.
METHODS
Phylogenetic Analysis
Protein sequences for the villins in Arabidopsis thaliana were obtained from the National Center for Biotechnology Information Entrez database (http://www.ncbi.nlm.nih.gov/; Klahre et al., 2000) and compared with sequences available in The Arabidopsis Information Resource database (http://www.Arabidopsis.org). Villin-like protein sequences in Oryza sativa ssp japonica were obtained from the Venter Institute’s rice database (http://rice.plantbiology.msu.edu/; International Rice Genome Sequencing Project, 2005; Ohyanagi et al., 2006) and the TIGR/MSU Rice Genome Annotation Resource (http://rice.plantbiology.msu.edu/; Yuan et al., 2005; Ouyang et al., 2007). Five sequences for putative villins were identified and named according to their proximity to the Arabidopsis villins in the phylogenetic tree generated as mentioned below. Multiple sequence alignments were performed with ClustalW (MacVector version 7.2.3; Accelrys Software). The alignment was imported into a Phylogenetic Analysis Using Parsimony (PAUP) program (Swofford, 1998), and distance- and parsimony-based neighbor-joining trees were generated with manual rooting keeping human villin and gelsolin as the outgroup, tree-bisection-reconnection branch swapping, and 1000 replicates to generate bootstrap values.
Protein Production
Full-length cDNA for VLN3 was obtained from the ABRC (Ohio State University; EST clone H2G4T7). The coding sequence was amplified by PCR, cloned into pCR2.1 (Invitrogen), and confirmed by sequencing. It was subcloned into the vector pET23d (Novagen) and transformed into Escherichia coli BL21 (DE3) Codon Plus to be expressed as a nonfusion protein. Bacterial cultures were grown overnight at 37°C, subcultured into fresh Luria-Bertani media, and grown at 37°C for 2 h. This was followed by overnight induction at 15°C after addition of 0.4 mM isopropyl β-d-thiogalactopyranoside. Cells were collected by centrifugation and resuspended in Solution A (50 mM Tris-HCl, pH 8.0, 10 mM KCl, 1 mM DTT, and 0.01% sodium azide), supplemented with a 1:200 dilution of a stock solution of protease inhibitors (Ren et al., 1997). The cell suspension was sonicated and then clarified by centrifugation. The supernatant was applied to a DEAE-Sepharose column (Amersham Biosciences) preequilibrated with Solution A. Proteins were eluted with a linear gradient of KCl (10 to 500 mM). Fractions were analyzed by SDS-PAGE, and villin-containing fractions were pooled and dialyzed against Solution B (50 mM Tris-HCl, pH 8.0, 0.2 mM EGTA, 1 mM DTT, 0.01% NaN3, and 1:200 protease inhibitors). Dialyzed villin was applied to a Cibacron Blue 3G-A column (Sigma-Aldrich) preequilibrated with Solution B. Proteins were eluted with a step gradient of increasing KCl concentration (0.25, 0.35, 0.5, and 1 M) and 0.1 M ATP. Villin-containing fractions were identified by SDS-PAGE, pooled, and dialyzed against Solution C (10 mM Tris-HCl, pH 8.0, 140 mM KCl, and 1 mM DTT). Purified villin was concentrated with a 100-kD cutoff Centricon (Millipore), frozen in liquid nitrogen, and stored at −80°C. The concentration of VLN3 was determined by Bradford assay (Bio-Rad Protein Assay) using BSA as a standard.
Arabidopsis VLN1 and ADF1 were purified as described previously (Carlier et al., 1997; Huang et al., 2005). Human VLN plasmid was kindly provided by S. Khurana (University of Tennessee Health Science Center, Memphis, TN), and the recombinant protein was purified roughly as described by Panebra et al. (2001) as a glutathione S-transferase fusion protein, followed by removal of the glutathione S-transferase tag with thrombin (Kovar et al., 2000).
Actin was isolated from rabbit skeletal muscle acetone powder (Spudich and Watt, 1971), and monomeric Ca-ATP-actin was purified by gel filtration chromatography on Sephacryl S-300 (Pollard, 1984) in Buffer G (5 mM Tris-HCl, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM DTT, and 0.1% NaN3).
High- and Low-Speed Cosedimentation Assays
The ability of the purified recombinant villin to bind to F-actin was examined with a high-speed cosedimentation assay (Kovar et al., 2000; Huang et al., 2005). Actin was prepolymerized in the presence of equimolar phalloidin. In a 200-μL reaction volume, 1 μM of the putative actin binding protein was incubated with 5 μM phalloidin-stabilized F-actin in the presence of 1× F-buffer (10× stock: 50 mM Tris-HCl, pH 7.5, 5 mM DTT, 5 mM ATP, 1 M KCl, and 50 mM MgCl2) with 1 mM EGTA for 30 min at 23°C. The free Ca2+ concentration in the reaction was 3 nM as determined with EqCal software (version 1.1; Biosoft). Following incubation, 80 μL of the reaction mix (total sample) was transferred to another tube with 20 μL of 5× SDS-PAGE sample buffer (1× comprises 40 mM Tris, pH 6.8, 100 mM DTT, 1% [w/v] SDS, and 10% [v/v] glycerol) The remaining mix was centrifuged for 30 min at 186,000g at 4°C. Supernatant (80 μL) was removed to a separate tube and 20 μL of 5× sample buffer was added to it. After discarding the remaining supernatant, 150 μL of 2× sample buffer was added to the pellet. The samples were then separated by 10% SDS-PAGE and stained with Coomassie Brilliant Blue R (Sigma-Aldrich).
The apparent affinity of VLN3 for F-actin was determined by sedimentation of reactions with a fixed amount of VLN3 and a range of concentrations of phalloidin-stabilized F-actin. Increasing amounts of actin (0.1, 0.25, 0.5, 0.75, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, and 10.0 μM) were incubated with 0.5 μM villin for 30 min at 23°C followed by a 30-min spin at 186,000g. The free Ca2+ concentration was <10 nM in these reactions. To determine Kd values, equal volumes of the pellet were separated by SDS-PAGE, gels were scanned, and the intensity of VLN3 was measured with LabWorks software (version 4.0.0.8; UVP BioImaging Systems). Standard curves were generated from the intensity of known amounts of purified VLN3 loaded on the same gel, and these were used to quantify the amount of bound protein. The percentage of bound protein as a function of actin concentration was plotted and fitted to a hyperbolic function using Kaleidagraph (version 4.0.1; Synergy Software). The effect of Ca2+ on the binding was also analyzed by performing the assay in the presence of 1 mM EGTA and 111 μM Ca2+ to give a final free [Ca2+] of 100 μM.
Bundling activity of VLN3 was analyzed with a low-speed cosedimentation assay (Kovar et al., 2000; Huang et al., 2005). For the dose-dependent bundling assay, increasing amounts of VLN3 were incubated with 10 μM prepolymerized actin in the presence of 1× KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0) in a 100-μL reaction. Following a 45-min incubation at 23°C, the mixture was centrifuged at 13,500g at 4°C for 40 min. Similar to the high-speed assay, 20 μL of 5× sample buffer was added to 80 μL of the supernatant, and 50 μL 2× sample buffer was added to the pellet. The samples were examined by 10% SDS-PAGE followed by staining with Coomassie Brilliant Blue R. The intensity of the bands was measured, and the percentage of actin in the pellet was calculated from a standard curve for known amounts of actin loaded on the same gel.
The effect of Ca2+ on bundling activity was also examined by incubating 500 nM VLN3 with 10 μM F-actin in the presence of 1× KMEI and various concentrations of free Ca2+ (0.1, 1, 10, and 100 μM and 1 mM).
Wide-Field Fluorescence Microscopy
Fluorescence microscopy was used to observe individual actin filaments alone or in the presence of villin at steady state (Blanchoin et al., 2000; Huang et al., 2003). For analyzing the dose-dependent reduction in filament length, 1 μM polymerized actin was incubated at 23°C with various amounts of villins in 1× KMEI and 10 mM free Ca2+. After 15 min, 1 μM rhodamine-phalloidin (Sigma-Aldrich) was added to the tube. Following another 15-min incubation, the mixture was diluted to ~5 nM F-actin with fluorescence buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 100 mM DTT, 100 μg/mL glucose oxidase, 15 mg/mL glucose, 20 μg/mL catalase, and 0.5% methylcellulose). In the assays where Ca2+ was added later, the mixture, comprising 1 μM F-actin alone or with 400 nM VLN1, 50 nM VLN3, or a combination of VLN1 and VLN3, was divided in two following the 15-min incubation. One half was labeled with rhodamine-phalloidin, diluted to 5 nM final F-actin concentration, and imaged. To the other half, 1 mM Ca2+ was added. This was incubated for an additional 15 min before imaging. The filaments were observed by epifluorescence illumination under a Nikon Microphot SA microscope equipped with a ×60, 1.4–numerical aperture Planapo objective, and digital images were captured with an ORCA-ER 12-bit CCD camera (Hamamatsu Photonics) using MetaMorph software (version 6.0; Universal Imaging). MetaMorph was also used to measure the lengths of all actin filaments in at least four different images from the same slide. The intensity and width of individual actin filaments were recorded, and structures with higher intensity (above an average intensity of ~3500 arbitrary units) and/or width (>4 pixels in an 800% enlarged image) than normal were considered to be bundles and were not included in the counts of filament length. In assays of bundle turnover, filament bundles were defined as any structure with a region that had an intensity above 1000 arbitrary units (exposure time, 400 ms; e-gain, 200) and their maximal lengths measured with MetaMorph. A minimum of 100 bundles were traced in at least 10 different images from three independent experiments for each condition.
Determination of Skewness
Skewness was previously used by Higaki et al. (2009) to monitor actin filament bundling in vivo. Here, we used their algorithm to compare single actin filaments and reconstituted actin filament bundles generated with VLN1 and VLN3 in vitro. All proteins were clarified at 186,000g for 45 min prior to each experiment. Each 103-μL reaction comprised 1× KMEI and 1 μM prepolymerized actin filaments, plus or minus recombinant villins. Reactions were supplemented with 1 μM rhodamine phalloidin and then imaged (or first incubated with various [Ca2+] for 15 min and then supplemented with rhodamine phalloidin) as described previously. For experiments testing the synergy or competition between VLN1 and VLN3, 103-μL reactions were incubated with 400 nM VLN1 and then supplemented with 50 nM VLN3 and an additional 400 nM VLN1. To establish the efficacy of skewness on severed actin bundles versus intact actin bundles, 1 μM VLN1 was used to prepare bundles, and half the reaction was sheared by sonication for 1 or 5 min in a water bath sonicator. All reactions were diluted at a ratio of 3 μL in 40 μL of fluorescence buffer, and filaments were visualized using polylysine-coated cover slips (Huang et al., 2003). A fixed exposure time and gain setting were selected such that both single filaments and higher-order structures could be visualized without saturation of pixels. Micrographs were analyzed in ImageJ using Higaki et al. (2009) parameters (i.e., minor window level and threshold adjustments of precisely the same extent per image per concentration, followed by Higaki’s skewness macro available at http://hasezawa.ib.k.u-tokyo.ac.jp/zp/Kbi/HigStomata). To correlate skewness with another independent measure of actin bundle formation, after samples were removed for microscopy, the remainder of each reaction was subjected to cosedimentation analysis as described above. Skewness experiments and the corresponding low-speed cosedimentation assays were performed in triplicate with independent batches of VLN1 and VLN3, and average values (±se) between replicate experiments were plotted.
TIRF Microscopy
Individual severing events along actin filaments as well as bundling were imaged by time-lapse TIRF microscopy. Rhodamine-labeled muscle actin (Cytoskeleton) in Buffer G was polymerized in the presence of 1× KMEI for at least 1 h at room temperature. A mixture of rhodamine-actin and unlabeled actin in a 1:1 ratio was used for experiments with ADF1. Glass flow chambers, with a capacity of ~40 μL, were prepared by separating a 24 × 60-mm cover slip from a standard microscope slide using strips of Parafilm (Amann and Pollard, 2001; Kovar and Pollard, 2004). Chambers were perfused with 5 nM NEM-treated myosin (rabbit skeletal muscle myosin; Cytoskeleton) in myosin buffer (0.5 M KCl, 10 mM MgCl2, and 10 mM imidazole, pH 7.0). NEM-myosin was left in the chamber for ~15 s, followed by a myosin buffer wash. BSA (1% [w/v]) was flowed into the chamber and incubated for at least 2 min, followed by a fluorescence buffer wash. A final concentration of 25 nM of filamentous rhodamine-actin in fluorescence buffer was perfused and allowed to settle for ~5 min. Fluorescence buffer alone, or a mixture of actin binding proteins in the presence of 0.5 mM thesit (polidocanol; Sigma-Aldrich; Harris and Higgs, 2006), with or without Ca2+ diluted in fluorescence buffer, was introduced after placing the chamber on the microscope stage. In the competition assays, 25 nM VLN1 was perfused into the chamber first. After 2 to 3 min, 5 nM VLN3 with 10 μM Ca2+, or the appropriate buffer along with an additional 25 nM VLN1, was flowed into the same chamber. Chambers that had not previously been subjected to VLN1 were also perfused with VLN3 or ADF1.
Actin filaments were observed by TIRF illumination on an Olympus IX-71 inverted microscope equipped with a ×60, 1.45–numerical aperture TIRFM Planapo objective using a 543-nm HeNe laser line (Melles Griot Photonics). The time course of bundling or severing was acquired at 1- to 5-s intervals with an ORCA-EM-CCD camera (model C9100-12; Hamamatsu Photonics) using IPLab software (version 3.9; Scanalytics). Typical exposure times were 650 ms (or 1100 ms for 1:1 rhodamine-unlabeled actin) with the electronic gain set at 230. A data table with elapsed times between individual frames was recorded for each experiment.
The stack of 100 frames from each time-lapse series was used to observe and count breaks along single filaments in all images following the addition of protein. The data table was used to calculate the elapsed time following perfusion. Filament lengths were also measured using the length tool of IPLab and a conversion factor of 1 pixel = 0.267 μm for the ×60 objective. Severing frequency for each actin filament was calculated as the number of breaks, per unit filament length, per unit time (i.e., breaks/μm/s). The severing frequency for a particular protein in a certain condition was estimated by calculating the average severing frequency from at least 20 filaments that were longer than 10 μm. At least three time-lapse series were captured for each experimental condition. Montages from time-lapse experiments were created in MetaMorph by selecting specific frames and adjusting contrast and brightness levels uniformly across all images. Figures for publication were generated in Adobe Photoshop CS5 (version 12.0; Adobe Systems).
Live-Cell Imaging with VAEM
The cortical actin cytoskeleton in epidermal cells from Arabidopsis hypocotyl cells expressing GFP-fABD2 was examined by time-lapse VAEM (Konopka and Bednarek, 2008) according to methods described previously (Staiger et al., 2009). Briefly, etiolated seedlings 11 d after germination were mounted in water between a slide and cover glass and positioned on the inverted TIRF microscope platform described above. Fluorescence from GFP-fABD2 was excited with a 100-mW Ar laser (Melles Griot Photonics) attenuated to 20% power with neutral density filters. Emitted light passed through a triple-wavelength dichroic and emission filter (448-543-633TBDR; Omega Optical), and images were captured at 1.5-s intervals with a Hamamatsu ORCA-EM 512 × 512 CCD camera operated by IPLab software. Filament bundling was examined upon playback of the time-lapse image stack in IPLab or ImageJ, and montages of still images were prepared with Photoshop CS5.
Quantitative Real-Time PCR
Inflorescence stems, floral whorls, and mature rosette leaves from wild-type Arabidopsis Columbia-0, vln1-4 (SALK_133579), and vln3-8 (SAIL_110_E04) homozygous mutant plants, grown under standard long-day conditions, were harvested at 44 d after germination. Plant material was flash frozen in liquid nitrogen upon harvest and ground to a fine powder with mortar and pestle. RNA isolation was performed using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Two-step qRT-PCR was performed as follows: Isolated RNA was measured for concentration and relative quality via spectrophotometry, and then 2 μg RNA was treated with RQ1 DNase (Promega) to remove contaminating DNA. Random primers (NEB) and MMLV reverse transcriptase (Promega) were used to synthesize cDNA in accordance with product protocols. qRT-PCR was performed using 2× SYBR Green master mix (Applied Biosystems) using a method adapted from the Applied Biosystems protocol and Udvardi et al. 2008 and a StepOne Plus real-time PCR system (Applied Biosystems). Gene-specific VLN1 (5′-GGCATCTAAATCTAATGCTTTGTCT-3′; 5′-ATGTCTAGGCTAAGTAAAGACATT-3′) and VLN3 (5′-CTATCAAGCACGCAAAAGAGG-3′; 5′-TAATGTCGTGTGTTCTGGCAG-3′) primers were used, and relative expression levels were compared after normalization to GAPD transcript levels (5′-CACTTGAAGGGTGGTGCCAAG-3′; 5′-CCTGTTGTCGCCAACGAAGTC-3′). Three biological and technical replicates were performed per treatment. Results were analyzed using comparative ΔCT with GAPD controls (Rieu and Powers, 2009). Mean results (±se) were plotted.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At VLN1, NP_029567 (At2g29890); At VLN2, NP_565958 (At2g41740); At VLN3, NP_567048 (At3g57410); At VLN4, NP_194745 (At4g30160); At VLN5, NP_200542 (At5g57320); Ll ABP135, AAD54660; Ll ABP115, BAC77209; Hs VLN, NP_009058; Hs Gel, CAA28000; Os VLN1, Os05g06110; Os VLN2, Os03g24220; Os VLN3, Os06g44890; Os VLN4, Os04g51440; and Os VLN5, Os08g14230.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of Villins and Villin-Like Proteins.
Supplemental Figure 2. Most Ca2+ Binding Sites Are Conserved in VLN3.
Supplemental Figure 3. Purification of Recombinant VLN3.
Supplemental Figure 4. Validation of Skewness as a Measure of Actin-Filament Bundling in Vitro with VLN1.
Supplemental Figure 5. VLN1 and VLN3 Coexist in Plant Tissues and Cells.
Supplemental Figure 6. Quantitative Real-Time PCR Analysis of VLN1 and VLN3 Expression in Wild-Type and Mutant Floral Whorls, Inflorescence Stems, and Mature Rosette Leaves.
Supplemental Data Set 1. Nexus File of Multiple Sequence Alignment for Villin and Villin-Like Proteins.
Supplemental Movie 1. Time-Lapse TIRFM Series of 25 nM Rhodamine-Actin Filaments Treated with Buffer Only.
Supplemental Movie 2. Time-Lapse TIRFM Series of Filament Bundle Formation in the Presence of 5 nM VLN3.
Supplemental Movie 3. Another Example of Bundle Formation in the Presence of 5 nM VLN3.
Supplemental Movie 4. Time-Lapse Series of Filament Bundle Formation in the Presence of 25 nM VLN1.
Supplemental Movie 5. Time-Lapse VAEM Visualization of Actin Bundle Formation in Vivo.
Supplemental Movie 6. Time-Lapse VAEM Visualization of Actin Bundle Formation in Vivo.
Supplemental Movie 7. Actin Filaments in the Presence of 25 nM VLN1 Show Minimal Breakage.
Supplemental Movie 8. VLN3 Has No Effect on Actin Filaments in the Absence of Ca2+.
Supplemental Movie 9. Actin Filaments Are Severed by 0.5 nM Human Villin in the Presence of Ca2+.
Supplemental Movie 10. Actin Filaments Are Severed by 7 nM VLN3 in the Presence of Ca2+.
Supplemental Movie 11. Actin Filaments Are Severed by 200 nM ADF1.
Supplemental Movie 12. Time-Lapse TIRFM Series of Actin Filaments Exposed to 5 nM VLN3 Alone.
Supplemental Movie 13. Time-Lapse TIRFM Series of Actin Filaments Precoated with 25 nM VLN1, Followed by Addition of 5 nM VLN3 and 25 nM VLN1.
Supplemental Movie 14. Time-Lapse TIRFM Series of Actin Filaments Exposed to 200 nM ADF1 Alone.
Supplemental Movie 15. Time-Lapse TIRFM Series of Actin Filaments Precoated with 25 nM VLN1, Followed by Addition of 200 nM ADF1 and 25 nM VLN1.
Supplemental Movie Legends.
Supplementary Material
Acknowledgments
We thank colleagues in the Staiger lab and the Purdue Cytoskeletal Group for their continuous help and advice. This work was supported by grants from the U.S. National Science Foundation (MCB-0130576), by the Physical Biosciences Program of the Office of Basic Energy Sciences, U.S. Department of Energy, under Contract DE-FG02-04ER15526, and by a Department of Energy–sponsored Center for Direct Catalytic Conversion of Biomass to Biofuels (C3Bio), an Energy Frontier Research Center (DE-SC0000997). The TIRF microscope facility was supported in part with funds from the Bindley Bioscience Center at Purdue University. We thank the ABRC for the cDNA stock of VLN3 as well as T-DNA insertion lines and S. Khurana (University of Tennessee Health Science Center, Memphis, TN) for the human villin plasmid.
References
- Amann K.J., Pollard T.D. (2001). Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. USA 98: 15009–15013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T.D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24: 13–23 [DOI] [PubMed] [Google Scholar]
- Baluska F., Busti E., Dolfini S., Gavazzi G., Volkmann D. (2001). Lilliputian mutant of maize lacks cell elongation and shows defects in organization of actin cytoskeleton. Dev. Biol. 236: 478–491 [DOI] [PubMed] [Google Scholar]
- Bartles J.R. (2000). Parallel actin bundles and their multiple actin-bundling proteins. Curr. Opin. Cell Biol. 12: 72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartles J.R., Zheng L., Li A., Wierda A., Chen B. (1998). Small espin: A third actin-bundling protein and potential forked protein ortholog in brush border microvilli. J. Cell Biol. 143: 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearer E.L. (1991). Direct observation of actin filament severing by gelsolin and binding by gCap39 and CapZ. J. Cell Biol. 115: 1629–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Amann K.J., Higgs H.N., Marchand J.B., Kaiser D.A., Pollard T.D. (2000). Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404: 1007–1011 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T.D., Hitchcock-DeGregori S.E. (2001). Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr. Biol. 11: 1300–1304 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Staiger C.J. (2010). Plant formins: Diverse isoforms and unique molecular mechanism. Biochim. Biophys. Acta 1803: 201–206 [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.-Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Bretscher A., Weber K. (1980). Villin is a major protein of the microvillus cytoskeleton which binds both G and F actin in a calcium-dependent manner. Cell 20: 839–847 [DOI] [PubMed] [Google Scholar]
- Cant K., Knowles B.A., Mooseker M.S., Cooley L. (1994). Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J. Cell Biol. 125: 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas L., Lovy-Wheeler A., Kunkel J.G., Hepler P.K. (2008). Pollen tube growth oscillations and intracellular calcium levels are reversibly modulated by actin polymerization. Plant Physiol. 146: 1611–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.-F., Laurent V., Santolini J., Melki R., Didry D., Xia G.-X., Hong Y., Chua N.-H., Pantaloni D. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J. Cell Biol. 136: 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright D.A., Brady S.M., Orlando D.A., Sturmfels B., Benfey P.N. (2009). Reconstructing spatiotemporal gene expression data from partial observations. Bioinformatics 25: 2581–2587 [DOI] [PubMed] [Google Scholar]
- Chhabra E.S., Higgs H.N. (2006). INF2 is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem. 281: 26754–26767 [DOI] [PubMed] [Google Scholar]
- Choe H., Burtnick L.D., Mejillano M., Yin H.L., Robinson R.C., Choe S. (2002). The calcium activation of gelsolin: Insights from the 3Å structure of the G4-G6/actin complex. J. Mol. Biol. 324: 691–702 [DOI] [PubMed] [Google Scholar]
- Clément M., Ketelaar T., Rodiuc N., Banora M.Y., Smertenko A., Engler G., Abad P., Hussey P.J., de Almeida Engler J. (2009). Actin-depolymerizing factor2-mediated actin dynamics are essential for root-knot nematode infection of Arabidopsis. Plant Cell 21: 2963–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Wang G., Yang D.S., Tang Y., Broun P., Marks M.D., Sumner L.W., Dixon R.A., Zhao P.X. (2010). TrichOME: A comparative omics database for plant trichomes. Plant Physiol. 152: 44–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.-H., Xia G.-X., Hong Y., Ramachandran S., Kost B., Chua N.-H. (2001). ADF proteins are involved in the control of flowering and regulate F-actin organization, cell expansion, and organ growth in Arabidopsis. Plant Cell 13: 1333–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Era A., Tominaga M., Ebine K., Awai C., Saito C., Ishizaki K., Yamato K.T., Kohchi T., Nakano A., Ueda T. (2009). Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant Cell Physiol. 50: 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzell R.M., Chafel M.M., Matsudaira P.T. (1989). Differential localization of villin and fimbrin during development of the mouse visceral endoderm and intestinal epithelium. Development 106: 407–419 [DOI] [PubMed] [Google Scholar]
- Fan X., Hou J., Chen X., Chaudhry F., Staiger C.J., Ren H. (2004). Identification and characterization of a Ca2+-dependent actin filament-severing protein from lily pollen. Plant Physiol. 136: 3979–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederich E., Vancompernolle K., Huet C., Goethals M., Finidori J., Vandekerckhove J., Louvard D. (1992). An actin-binding site containing a conserved motif of charged amino acid residues is essential for the morphogenic effect of villin. Cell 70: 81–92 [DOI] [PubMed] [Google Scholar]
- Friederich E., Vancompernolle K., Louvard D., Vandekerckhove J. (1999). Villin function in the organization of the actin cytoskeleton: Correlation of in vivo effects to its biochemical activities in vitro. J. Biol. Chem. 274: 26751–26760 [DOI] [PubMed] [Google Scholar]
- George N. (2004). TIRF microscopy: The evanescent wave of the future. Am. Lab. 36: 26–28 [Google Scholar]
- George S.P., Wang Y., Mathew S., Kamalakkannan S., Khurana S. (2007). Dimerization and actin-bundling properties of villin and its role in the assembly of epithelial cell brush borders. J. Biol. Chem. 282: 26528–26541 [DOI] [PubMed] [Google Scholar]
- Gerke V., Weber K. (1983). Isolation and characterization of mammalian villin and fimbrin, the two bundling proteins of the intestinal microvilli. Eur. J. Cell Biol. 31: 249–255 [PubMed] [Google Scholar]
- Glenney J.R., Jr., Bretscher A., Weber K. (1980). Calcium control of the intestinal microvillus cytoskeleton: Its implications for the regulation of microfilament organizations. Proc. Natl. Acad. Sci. USA 77: 6458–6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenney J.R., Jr., Weber K. (1981). Calcium control of microfilaments: uncoupling of the F-actin-severing and -bundling activity of villin by limited proteolysis in vitro. Proc. Natl. Acad. Sci. USA 78: 2810–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton C.M., Liu J., Taylor D.W., DeRosier D.J., Taylor K.A. (2008). The 3D structure of villin as an unusual F-actin crosslinker. Structure 16: 1882–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.S., Higgs H.N. (2006). Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 406: 190–214 [DOI] [PubMed] [Google Scholar]
- Hepler P.K., Vidali L., Cheung A.Y. (2001). Polarized cell growth in higher plants. Annu. Rev. Cell Dev. Biol. 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Higaki T., Kojo K.H., Hasezawa S. (2010). Critical role of actin bundling in plant cell morphogenesis. Plant Signal. Behav. 5: 484–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T., Kutsuna N., Sano T., Kondo N., Hasezawa S. (2009). Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 61: 156–165 [DOI] [PubMed] [Google Scholar]
- Higaki T., Sano T., Hasezawa S. (2007). Actin microfilament dynamics and actin side-binding proteins in plants. Curr. Opin. Plant Biol. 10: 549–556 [DOI] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Chaudhry F., Franklin-Tong V.E., Staiger C.J. (2004). A gelsolin-like protein from Papaver rhoeas pollen (PrABP80) stimulates calcium-regulated severing and depolymerization of actin filaments. J. Biol. Chem. 279: 23364–23375 [DOI] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Kovar D.R., Staiger C.J. (2003). Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278: 44832–44842 [DOI] [PubMed] [Google Scholar]
- Huang S., McDowell J.M., Weise M.J., Meagher R.B. (1996). The Arabidopsis profilin gene family. Evidence for an ancient split between constitutive and pollen-specific profilin genes. Plant Physiol. 111: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Robinson R.C., Gao L.Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C.J. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Janmey P.A., Matsudaira P.T. (1988). Functional comparison of villin and gelsolin. Effects of Ca2+, KCl, and polyphosphoinositides. J. Biol. Chem. 263: 16738–16743 [PubMed] [Google Scholar]
- Kandasamy M.K., Burgos-Rivera B., McKinney E.C., Ruzicka D.R., Meagher R.B. (2007). Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell 19: 3111–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M.K., McKinney E.C., Meagher R.B. (2002). Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 13: 251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M.K., McKinney E.C., Meagher R.B. (2009). A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 21: 701–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T., Faivre-Moskalenko C., Esseling J.J., de Ruijter N.C.A., Grierson C.S., Dogterom M., Emons A.M.C. (2002). Positioning of nuclei in Arabidopsis root hairs: An actin-regulated process of tip growth. Plant Cell 14: 2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajiri S.-i., et al. (2010). Actin-bundling protein TRIOBP forms resilient rootlets of hair cell stereocilia essential for hearing. Cell 141: 786–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Friederich E., Kost B., Louvard D., Chua N.H. (2000). Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiol. 122: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno T., Shimmen T. (1988). Accelerated sliding of pollen tube organelles along Characeae actin bundles regulated by Ca2+. J. Cell Biol. 106: 1539–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Bednarek S.Y. (2008). Variable-angle epifluorescence microscopy: A new way to look at protein dynamics in the plant cell cortex. Plant J. 53: 186–196 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Harris E.S., Mahaffy R., Higgs H.N., Pollard T.D. (2006). Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124: 423–435 [DOI] [PubMed] [Google Scholar]
- Kovar D.R., Pollard T.D. (2004). Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA 101: 14725–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D.R., Staiger C.J., Weaver E.A., McCurdy D.W. (2000). AtFim1 is an actin filament crosslinking protein from Arabidopsis thaliana. Plant J. 24: 625–636 [DOI] [PubMed] [Google Scholar]
- Kumar N., Khurana S. (2004). Identification of a functional switch for actin severing by cytoskeletal proteins. J. Biol. Chem. 279: 24915–24918 [DOI] [PubMed] [Google Scholar]
- Kumar N., Tomar A., Parrill A.L., Khurana S. (2004a). Functional dissection and molecular characterization of calcium-sensitive actin-capping and actin-depolymerizing sites in villin. J. Biol. Chem. 279: 45036–45046 [DOI] [PubMed] [Google Scholar]
- Kumar N., Zhao P., Tomar A., Galea C.A., Khurana S. (2004b). Association of villin with phosphatidylinositol 4,5-bisphosphate regulates the actin cytoskeleton. J. Biol. Chem. 279: 3096–3110 [DOI] [PubMed] [Google Scholar]
- Le J., El-Assal S.E.-D., Basu D., Saad M.E., Szymanski D.B. (2003). Requirements for Arabidopsis ATARP2 and ATARP3 during epidermal development. Curr. Biol. 13: 1341–1347 [DOI] [PubMed] [Google Scholar]
- Le J., Mallery E.L., Zhang C., Brankle S., Szymanski D.B. (2006). Arabidopsis BRICK1/HSPC300 is an essential WAVE-complex subunit that selectively stabilizes the Arp2/3 activator SCAR2. Curr. Biol. 16: 895–901 [DOI] [PubMed] [Google Scholar]
- Leonhardt N., Kwak J.M., Robert N., Waner D., Leonhardt G., Schroeder J.I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Sun N., Liu X., Jiao Y., Zhao H., Deng X.W. (2005). Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 138: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan-Miklos S., Cooley L. (1994). The villin-like protein encoded by the Drosophila quail gene is required for actin bundle assembly during oogenesis. Cell 78: 291–301 [DOI] [PubMed] [Google Scholar]
- Matova N., Mahajan-Miklos S., Mooseker M.S., Cooley L. (1999). Drosophila Quail, a villin-related protein, bundles actin filaments in apoptotic nurse cells. Development 126: 5645–5657 [DOI] [PubMed] [Google Scholar]
- Matsudaira P., Mandelkow E., Renner W., Hesterberg L.K., Weber K. (1983). Role of fimbrin and villin in determining the interfilament distances of actin bundles. Nature 301: 209–214 [DOI] [PubMed] [Google Scholar]
- McDowell J.M., Huang S., McKinney E.C., An Y.-Q., Meagher R.B. (1996). Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142: 587–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher R.B., McKinney E.C., Kandasamy M.K. (1999). Isovariant dynamics expand and buffer the responses of complex systems: The diverse plant actin gene family. Plant Cell 11: 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A., Berro J., Guérin C., Boujemaa-Paterski R., Staiger C.J., Martiel J.-L., Blanchoin L. (2007). Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 17: 825–833 [DOI] [PubMed] [Google Scholar]
- Michelot A., Derivery E., Paterski-Boujemaa R., Guérin C., Huang S., Parcy F., Staiger C.J., Blanchoin L. (2006). A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Curr. Biol. 16: 1924–1930 [DOI] [PubMed] [Google Scholar]
- Michelot A., Guérin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C.J., Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17: 2296–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.B., Okada K., Balcer H.I., Kovar D.R., Pollard T.D., Goode B.L. (2006). Twinfilin is an actin-filament-severing protein and promotes rapid turnover of actin structures in vivo. J. Cell Sci. 119: 1547–1557 [DOI] [PubMed] [Google Scholar]
- Nag S., Ma Q., Wang H., Chumnarnsilpa S., Lee W.L., Larsson M., Kannan B., Hernandez-Valladares M., Burtnick L.D., Robinson R.C. (2009). Ca2+ binding by domain 2 plays a critical role in the activation and stabilization of gelsolin. Proc. Natl. Acad. Sci. USA 106: 13713–13718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu T., Yokota E., Shimmen T. (1998). Purification of an actin-binding protein composed of 115-kDa polypeptide from pollen tubes of lily. Biochem. Biophys. Res. Commun. 249: 61–65 [DOI] [PubMed] [Google Scholar]
- Northrop J., Weber A., Mooseker M.S., Franzini-Armstrong C., Bishop M.F., Dubyak G.R., Tucker M., Walsh T.P. (1986). Different calcium dependence of the capping and cutting activities of villin. J. Biol. Chem. 261: 9274–9281 [PubMed] [Google Scholar]
- Ohyanagi H., et al. (2006). The Rice Annotation Project Database (RAP-DB): Hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 34: D741–D744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ravi H., Smith E.M., Goode B.L. (2006). Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: A coordinated mechanism for severing and capping filaments. Mol. Biol. Cell 17: 2855–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V., Drubin D.G. (2010). Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J. Cell Biol. 188: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. (2007). Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol. 258: 1–82 [DOI] [PubMed] [Google Scholar]
- Ouyang S., et al. (2007). The TIGR Rice Genome Annotation Resource: Improvements and new features. Nucleic Acids Res. 35: D883–D887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panebra A., Ma S.X., Zhai L.W., Wang X.T., Rhee S.G., Khurana S. (2001). Regulation of phospholipase C-gamma(1) by the actin-regulatory protein villin. Am. J. Physiol. Cell Physiol. 281: C1046–C1058 [DOI] [PubMed] [Google Scholar]
- Pavlov D., Muhlrad A., Cooper J., Wear M., Reisler E. (2007). Actin filament severing by cofilin. J. Mol. Biol. 365: 1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pina C., Pinto F., Feijó J.A., Becker J.D. (2005). Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 138: 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. (1984). Polymerization of ADP-actin. J. Cell Biol. 99: 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A., Bannigan A., Sulaman W., Pechter P., Blancaflor E.B., Baskin T.I. (2007). Auxin, actin and growth of the Arabidopsis thaliana primary root. Plant J. 50: 514–528 [DOI] [PubMed] [Google Scholar]
- Ren H., Gibbon B.C., Ashworth S.L., Sherman D.M., Yuan M., Staiger C.J. (1997). Actin purified from maize pollen functions in living plant cells. Plant Cell 9: 1445–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I., Powers S.J. (2009). Real-time quantitative RT-PCR: Design, calculations and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S.M., Panstruga R. (2007). Cytoskeletal functions in plant-microbe interactions. Physiol. Mol. Plant Pathol. 71: 4–6 [Google Scholar]
- Shimmen T. (2007). The sliding theory of cytoplasmic streaming: fifty years of progress. J. Plant Res. 120: 31–43 [DOI] [PubMed] [Google Scholar]
- Shimmen T., Hamatani M., Saito S., Yokota E., Mimura T., Fusetani N., Karaki H. (1995). Roles of actin filaments in cytoplasmic streaming and organization of transvacuolar strands in root hair cells of Hydrocharis. Protoplasma 185: 188–193 [Google Scholar]
- Snowman B.N., Kovar D.R., Shevchenko G., Franklin-Tong V.E., Staiger C.J. (2002). Signal-mediated depolymerization of actin in pollen during the self-incompatibility response. Plant Cell 14: 2613–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. (1971). The regulation of rabbit skeletal muscle contraction: I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246: 4866–4871 [PubMed] [Google Scholar]
- Staiger C.J., Blanchoin L. (2006). Actin dynamics: Old friends with new stories. Curr. Opin. Plant Biol. 9: 554–562 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Hussey P.J. (2004). Actin and actin-modulating proteins. The Plant Cytoskeleton in Cell Differentiation and Development, Hussey P.J., (Oxford, UK: Blackwell Publishers; ), pp. 32–80 [Google Scholar]
- Staiger C.J., Poulter N.S., Henty J.L., Franklin-Tong V.E., Blanchoin L. (2010). Regulation of actin dynamics by actin-binding proteins in pollen. J. Exp. Bot. 61: 1969–1986 [DOI] [PubMed] [Google Scholar]
- Staiger C.J., Sheahan M.B., Khurana P., Wang X., McCurdy D.W., Blanchoin L. (2009). Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J. Cell Biol. 184: 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. (1998). PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4. (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Thomas C., Hoffmann C., Dieterle M., Van Troys M., Ampe C., Steinmetz A. (2006). Tobacco WLIM1 is a novel F-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 18: 2194–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., Steinmetz A. (2009). Actin bundling in plants. Cell Motil. Cytoskeleton 66: 940–957 [DOI] [PubMed] [Google Scholar]
- Tilney L.G., Connelly P.S., Vranich K.A., Shaw M.K., Guild G.M. (1998). Why are two different cross-linkers necessary for actin bundle formation in vivo and what does each cross-link contribute? J. Cell Biol. 143: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L.G., Connelly P.S., Vranich K.A., Shaw M.K., Guild G.M. (2000). Regulation of actin filament cross-linking and bundle shape in Drosophila bristles. J. Cell Biol. 148: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M., Yokota E., Vidali L., Sonobe S., Hepler P.K., Shimmen T. (2000). The role of plant villin in the organization of the actin cytoskeleton, cytoplasmic streaming and the architecture of the transvacuolar strand in root hair cells of Hydrocharis. Planta 210: 836–843 [DOI] [PubMed] [Google Scholar]
- Udvardi M.K., Czechowski T., Scheible W.-R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J.-Q., Hao S., O'Shaughnessy B., Pollard T.D. (2008). Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science 319: 97–100 [DOI] [PubMed] [Google Scholar]
- Vidali L., Burkart G.M., Augustine R.C., Kerdavid E., Tüzel E., Bezanilla M. (2010). Myosin XI is essential for tip growth in Physcomitrella patens. Plant Cell 22: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Rounds C.M., Hepler P.K., Bezanilla M. (2009). Lifeact-mEGFP reveals a dynamics apical F-actin network in tip growing plant cells. PLoS ONE 4: e5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., Yokota E., Cheung A.Y., Shimmen T., Hepler P.K. (1999). The 135 kDa actin-bundling protein from Lilium longiflorum pollen is the plant homologue of villin. Protoplasma 209: 283–291 [Google Scholar]
- Wang T., Xiang Y., Hou J., Ren H.-Y. (2008). ABP41 is involved in the pollen tube development via fragmenting actin filaments. Mol. Plant 1: 1048–1055 [DOI] [PubMed] [Google Scholar]
- Wulfkuhle J.D., Petersen N.S., Otto J.J. (1998). Changes in the F-actin cytoskeleton during neurosensory bristle development in Drosophila: The role of singed and forked proteins. Cell Motil. Cytoskeleton 40: 119–132 [DOI] [PubMed] [Google Scholar]
- Xiang Y., Huang X., Wang T., Zhang Y., Liu Q., Hussey P.J., Ren H. (2007). ACTIN BINDING PROTEIN 29 from Lilium pollen plays an important role in dynamic actin remodeling. Plant Cell 19: 1930–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Costa A., Leonhardt N., Siegel R.S., Schroeder J.I. (2008). Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H.L. (1999). Gelsolin. Guidebook to the Cytoskeletal and Motor Proteins, Kreis T., Vale R., (New York: Oxford University Press; ), pp. 99–102 [Google Scholar]
- Yokota E., Muto S., Shimmen T. (2000). Calcium-calmodulin suppresses the filamentous actin-binding activity of a 135-kilodalton actin-bundling protein isolated from lily pollen tubes. Plant Physiol. 123: 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Shimmen T. (1999). The 135-kDa actin-bundling protein from lily pollen tubes arranges F-actin into bundles with uniform polarity. Planta 209: 264–266 [DOI] [PubMed] [Google Scholar]
- Yokota E., Shimmen T. (2006). The actin cytoskeleton in pollen tubes: Actin and actin binding proteins. The Pollen Tube, Malho R., (Berlin/Heidelberg, Germany: Springer; ), pp. 139–155 [Google Scholar]
- Yokota E., Takahara K.-i., Shimmen T. (1998). Actin-bundling protein isolated from pollen tubes of lily. Plant Physiol. 116: 1421–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Tominaga M., Mabuchi I., Tsuji Y., Staiger C.J., Oiwa K., Shimmen T. (2005). Plant villin, lily P-135-ABP, possesses G-actin binding activity and accelerates the polymerization and depolymerization of actin in a Ca2+-sensitive manner. Plant Cell Physiol. 46: 1690–1703 [DOI] [PubMed] [Google Scholar]
- Yokota E., Vidali L., Tominaga M., Tahara H., Orii H., Morizane Y., Hepler P.K., Shimmen T. (2003). Plant 115-kDa actin-filament bundling protein, P-115-ABP, is a homologue of plant villin and is widely distributed in cells. Plant Cell Physiol. 44: 1088–1099 [DOI] [PubMed] [Google Scholar]
- Yuan Q., Ouyang S., Wang A., Zhu W., Maiti R., Lin H., Hamilton J., Haas B., Sultana R., Cheung F., Wortman J., Buell C.R. (2005). The Institute for Genomic Research Osa1 rice genome annotation database. Plant Physiol. 138: 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Mallery E.L., Schlueter J., Huang S., Fan Y., Brankle S., Staiger C.J., Szymanski D.B. (2008). Arabidopsis SCARs function interchangeably to meet actin related protein 2/3 activation thresholds during morphogenesis. Plant Cell 20: 995–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Qu X., Bao C., Khurana P., Wang Q., Xie Y., Zheng Y., Chen N., Blanchoin L., Staiger C.J., Huang S. (2010). Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22: 2749–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.