Elimination of Arabidopsis nonspecific phospholipase C (NPC4) results in decreases in diacylglycerol levels, abscisic acid (ABA) sensitivity, and hyperosmotic stress tolerance, whereas overexpression of NPC4 has opposite effects. This indicates that NPC4 promotes ABA response under hyperosmotic stress, whereas NPC-derived diacylglycerol enhances stomatal opening under well-watered conditions.
Abstract
Diacyglycerol (DAG) is an important class of cellular lipid messengers, but its function in plants remains elusive. Here, we show that knockout of the Arabidopsis thaliana nonspecific phospholipase C (NPC4) results in a decrease in DAG levels and compromises plant response to abscisic acid (ABA) and hyperosmotic stresses. NPC4 hydrolyzes various phospholipids in a calcium-independent manner, producing DAG and a phosphorylated head group. NPC4 knockout (KO) plants display decreased ABA sensitivity in seed germination, root elongation, and stomatal movement and had decreased tolerance to high salinity and water deficiency. Overexpression of NPC4 renders plants more sensitive to ABA and more tolerant to hyperosmotic stress than wild-type plants. Addition of a short-chain DAG or a short-chain phosphatidic acid (PA) restores the ABA response of NPC4-KO to that of the wild type, but the addition of DAG together with a DAG kinase inhibitor does not result in a wild-type phenotype. These data suggest that NPC4-produced DAG is converted to PA and that NPC4 and its derived lipids positively modulate ABA response and promote plant tolerance to drought and salt stresses.
INTRODUCTION
Drought and high salinity are two crucial environmental stresses that limit plant growth, productivity, and geographic distribution. Studies in recent years have provided valuable insight into the molecular and cellular mechanisms by which plants respond to and tolerate salinity and drought stresses (Apse et al., 1999; Zhu, 2002; Chinnusamy et al., 2004; Chinnusamy and Zhu, 2009). Both drought and high salinity trigger the production of the phytohormone abscisic acid (ABA), which induces the expression of many genes involved in the stress responses (Finkelstein et al., 2002; Seki et al., 2002; Nambara and Marion-Poll, 2005). Manipulations of key ABA-responsive genes have led to changes in plant tolerance to drought and salt stresses (Kasuga et al., 1999; Umezawa et al., 2006a). Reception of ABA in the cell involves nuclear-localized ABA receptors (Ma et al., 2009; Park et al., 2009) and also membrane-bound G protein–coupled receptor (Pandey et al., 2009). The ABA receptor pyrabactin resistance 1 interacts with the protein phosphatase 2C-like ABA-INSENSITIVE1 (ABI1) (Ma et al., 2009). The function of ABI1 is also regulated by the lipid messenger phosphatidic acid (PA) (Zhang et al., 2004; Mishra et al., 2006).
Increasing evidence indicates that membrane lipids are rich sources for signaling messengers in plant response to hyperosmotic stresses (Testerink and Munnik, 2005; Mishra et al., 2006; Hong et al., 2008). Specifically, PA is produced rapidly in response to hyperosmotic stress, and genetic and pharmacological manipulation of PA levels has resulted in alteration of plant response to ABA, drought, and salinity. Phospholipase D (PLD) hydrolysis of membrane lipids directly produces signaling PA. It has been shown recently that PLDα3, ε, δ, and α1 contribute to hyperosmotic stress–induced PA production and affect plant response to hyperosmotic stress in Arabidopsis thaliana (Hong et al., 2008, 2009; Munnik and Testerink, 2009). Activation of phospholipase C (PLC) is another major route for generating the PA signal. PLC produces diacylglycerol (DAG) that can be phosphorylated by DAG kinase (DGK) to generate PA (den Hartog et al., 2003; Zonia and Munnik, 2004). Plants have two distinctively different families of PLCs. PI-PLC uses primarily phosphoinositides (PIs), whereas nonspecific PLC (NPC) uses common membrane phospholipids, such as phosphatidylcholine (PC) and phosphatidylethanolamine (PE). The signaling function of PI-PLC has been well established in animals. PLC hydrolysis of phosphatidylinositol 4,5-bisphosphate releases two potent second messengers, inositol 1,4,5-trisphosphate and DAG. DAG remains in the membrane and, in animals, activates certain members of the protein kinase C family and other proteins. However, in plants, there is no evidence for the existence of protein kinase C family members, and the target of DAG as a plant signaling molecule remains undefined. Yet, the plant PI-PLC family has been implicated in ABA signaling and salt and drought stress (Hirayama et al., 1995; Sanchez and Chua, 2001; Hunt et al., 2003).
Unlike PI-PLC, NPCs hydrolyze the common membrane lipids PC and PE in a calcium-independent manner (Nakamura et al., 2005). NPCs share sequence similarities with a bacterial PC-PLC that has been characterized in microbial systems (Wang, 2001). PC-PLC isolated from Clostridium perfringens is a potent toxin, with an α-toxin domain (Titball, 1993). This enzyme has hemolytic activity (Saint-Joanis et al., 1989), vascular permeabilization capability (Sugahara et al., 1977), and a platelet-aggregating property (Sugahara et al., 1976). Other nontoxic PC-PLCs are also found in bacteria such as Bacillus cereus and Pseudomonas aeruginosa (Titball, 1991, 1993). Based on sequence homology with bacterial PC-PLC, the Arabidopsis genome is predicted to contain six putative NPC genes designated NPC1-6. These genes encode ~60-kD proteins, consisting of 514– to 538–amino acid residues (Nakamura et al., 2005). NPCs lack the X-Y domains (catalytic domain) and the EF-hand motif (calcium binding domain) of PI-PLC and the calcium-dependent lipid binding C2 domain or PI binding PX and PH domains of PLD. NPC5 is involved in phospholipid degradation and digalactosyldiacylglycerol (DGDG) accumulation in Arabidopsis leaves, but not in roots (Gaude et al., 2008). NPC5 is a cytosolic protein, whereas NPC4 is associated with the plasma membrane (Nakamura et al., 2005; Gaude et al., 2008). Of all the NPC genes, NPC4 is the only one whose expression is greatly induced by phosphate deprivation (Misson et al., 2005), but genetic ablation of NPC4 resulted in a slight decrease of DGDG, whereas the content of major phospholipids did not change during phosphate deprivation (Nakamura et al., 2005). Thus, the metabolic and physiological functions of NPC4 remain elusive. Here, we show that NPC4 plays an important role in plant response to ABA, drought, and high salinity.
RESULTS
Decreased Plant Sensitivity to ABA in NPC4 Knockout Arabidopsis
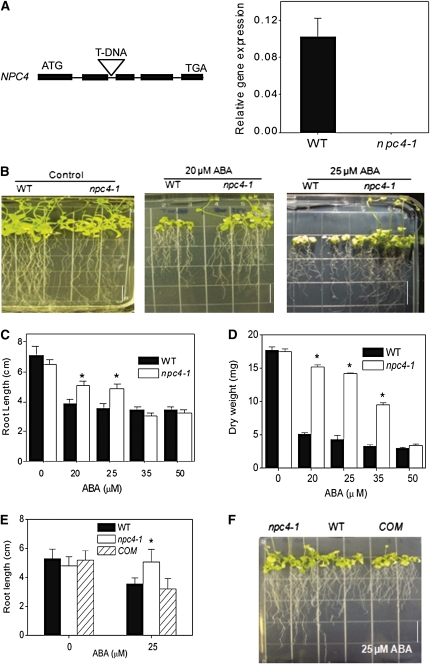
A homozygous T-DNA insertion mutant of NPC4, npc4-1 (SALK_046713), was isolated, and the elimination of NPC4 expression in npc4-1 was verified by quantitative real-time PCR (Figure 1A). We first examined the effect of the npc4-1 mutation on plant response to abiotic stresses and hormones by comparing seedling growth of npc4-1 and the wild type. No overt differences in growth and development were observed when seedlings of the two genotypes were grown on media deficient in phosphate (1 μM), nitrogen (0.6 mM), or potassium (0 μM) or media containing 1 μM indole-3-acetic acid (see Supplemental Figure 1 online). However, a drastic difference in growth occurred between npc4-1 and wild-type seedlings grown on ABA-containing media. The seedlings of npc4-1 had longer primary roots and more lateral roots than wild-type seedlings (Figures 1B and 1C; see Supplemental Figure 2 online). At 25 μM ABA, flowering was inhibited in wild-type plants, but npc4-1 plants were able to produce inflorescences (Figure 1B). The dry weights of npc4-1 rosettes grown on 20 and 25 μM ABA were more than threefold greater than the wild type and at 35 μM ABA, twofold greater than the wild type (Figure 1D). The ABA-resistant root elongation and flowering phenotype of npc4-1 was dosage dependent; at higher concentrations of ABA (35 and 50 μM), root growth of npc4-1 was comparable to the wild type. Genetic complementation of npc4-1 plants with the native NPC4 gene restored the wild-type phenotype to the mutant (Figures 1E and 1F). These results indicate that loss of NPC4 is responsible for reduced sensitivity toward ABA.
Figure 1.
Decreased Sensitivity of NPC4 Knockout Plants to ABA Inhibition of Plant Growth and Development.
(A) T-DNA insertional knockout of NPC4. Left: location of the T-DNA inserted in the NPC4 gene. Solid bars represent exons, and thin lines represent introns. Right: level of NPC4 expression in wild-type and npc4-1 plants as measured by real-time PCR. Relative expression is based on the expression of UBQ10. Three independent experiments were performed; values are means ± sd (n = 3) for one representative experiment.
(B) Seedlings on 0.5× Murashige and Skoog medium containing 0, 20, and 25 μM ABA. Seedlings were germinated on media without ABA for 5 d and then transferred onto media with varied levels of ABA for 2 weeks. Bar = 1.3 cm.
(C) Root length on 0.5× Murashige and Skoog medium supplemented with 0 to 50 μM ABA. Seedlings were germinated on media without ABA for 5 d and then transferred onto media with varied levels of ABA. Primary root length was measured 14 d after transfer. Values are means ± sd (n = 15).
(D) Dry weight of seedlings grown in the presence of 0 to 50 μM ABA. After growing on media with varied levels of ABA for 14 d, seedlings were harvested as measured. Values are means ± sd (n = 3).
(E) Root length of the wild type, npc4-1, and npc4-1 complemented with wild-type NPC4 (COM). Seedlings were grown for 14 d on 0.5× Murashige and Skoog medium in the presence or absence of 25 μM ABA. Values are means ± sd (n = 15).
(F) Seedling phenotypes of the wild type, npc4-1, and npc4-C on 0.5× Murashige and Skoog medium containing 25 μM ABA. Photograph was taken 14 d after transfer to ABA medium. Bar = 1.3 cm.
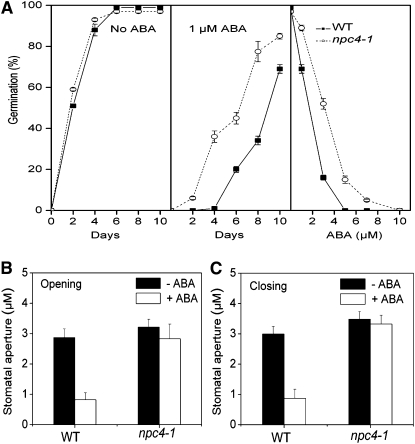
One of the best known effects of ABA is to induce seed dormancy and inhibit seed germination. In the absence of ABA, nearly 100% of seeds of the wild type and npc4-1 germinated within 4 d (Figure 2A, left panel). In the presence of ABA, npc4-1 seeds broke dormancy earlier than the wild type and were more resistant than the wild type to ABA inhibition of seed germination. In the presence of 1 μM ABA for 4 d, 36% of npc4-1 seeds germinated, whereas no wild-type seeds germinated (Figure 2A, middle panel). After 10 d at 1 μM ABA, npc4-1 seeds showed 33% more germination than wild-type seeds. At 5 μM ABA, the germination of wild-type seeds was completely inhibited, but 10% npc4-1 seeds still germinated (Figure 2A, right panel). Another well-documented function of ABA is to promote the stomatal closure. To test whether npc4-1 was altered in ABA-mediated stomatal movement, stomatal peels of wild-type and npc4-1 plants were incubated with 50 μM ABA. ABA promoted stomatal closure and inhibited stomatal opening in the wild type but not in npc4-1 leaf peels (Figure 2B).
Figure 2.
Decreased ABA Sensitivity of NPC4 Knockout in Seed Germination and Stomatal Movement.
(A) Seed germination rates in the absence of ABA (left), the presence of 1 μM ABA (middle), or different concentrations of ABA (right). Fully desiccated seeds were germinated on 0.5× Murashige and Skoog with or without ABA. Germination was scored at 10 d after transferring seeds from 4°C. One hundred seeds per genotype were measured in each experiment. Values are means ± sd (n = 3).
(B) Opening of closed stomata as affected by ABA. Wild-type and mutant epidermal peels were incubated in the dark without ABA to close stomata. The peels were then placed under light for 3 h in the presence of 50 μM ABA. Stomata aperture was imaged using a light microscope. The images obtained were measured using Image Pro software. Values are means ± se (n = 15).
(C) Closure of open stomata in the presence of ABA. Epidermal peels were placed under light for 3 h to fully open stomata of both mutant and the wild type. The peels were treated with 50 μM ABA for 3 h. Values are means ± sd (n = 40).
We also identified T-DNA insertional knockout mutants for the other five NPCs and examined the effect of these mutations on plant response to ABA. At 25 μM ABA, all other NPC-KOs, npc1-1, npc2-1, npc3-1, npc5-1, and npc6-1, displayed ABA-inhibited growth similar to wild-type seedlings and markedly different from npc4-1 seedlings, which displayed ABA resistance (see Supplemental Figure 2 online). Lateral root number was higher in npc4-1 than the wild type, whereas lateral root number was similar among the wild type, npc1-1, npc2-1, npc3-1, and npc6-1 but lower in npc5-1 in response to ABA (see Supplemental Figure 2 online). The results indicate that the decreased ABA sensitivity was specific to NPC4.
Different Expression Patterns of the NPC Family
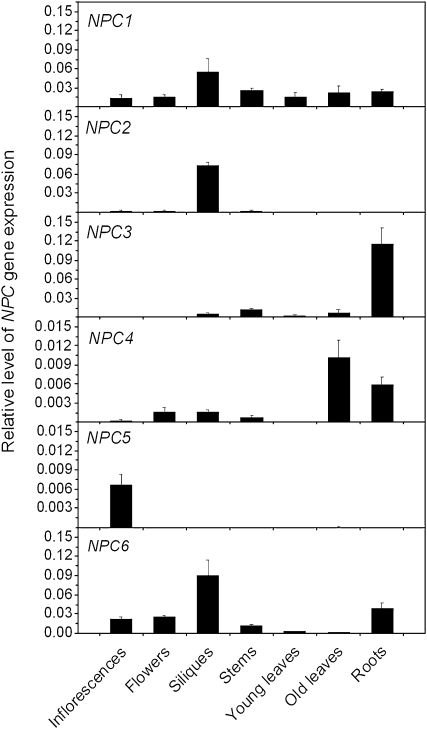
The expression patterns of the NPC gene family were determined by quantitative real-time PCR using RNA from different tissues of 6-week-old Arabidopsis plants. The highest level of NPC4 expression was detected in mature leaves and then roots, and its expression in young leaves and reproductive tissues was low (Figure 3). By comparison, NPC5 was expressed mostly in inflorescences. The highest level of expression for NPC3 was in roots, whereas for NPC2 and NPC6, expression was highest in siliques. NPC1 was expressed in similar levels all the tissues examined (Figure 3). Analysis of available transcriptomic data (http://bar.utoronto.ca) indicates that the expression pattern of the NPCs varies in root cell types (see Supplemental Figure 3A online). The database data failed to distinguish NPC4 and NPC5 due to high sequence similarity. However, our analysis using gene-specific primers showed that NPC5 was mostly expressed in inflorescences, and its level of expression in leaves and roots was much lower than that of NPC4 (Figure 3). Thus, the combined NPC4/5 transcript level in leaves and roots may result mostly from the NPC4 expression. The level of NPC4 was higher in root stele and endodermis cells than cortex and lateral root caps. In guard cells, the transcript level of NPC4 was induced threefold by ABA (Yang et al., 2008; http://bar.utoronto.ca). The expression of other NPCs was detected in guard cells, but ABA treatments did not increase expression of NPC1, 2, or 6 (see Supplemental Figure 3B online). Some increase in NPC3 occurred, but its level of expression was lower than that of NPC4. These data indicate that NPC gene family members have overlapping but mostly distinguishable patterns of expression in Arabidopsis tissues and cells. ABA induction of NPC4 expression was detected in leaves; the increase was transient, with the highest level of expression at 12 h after ABA treatment (Figure 4).
Figure 3.
Expression Pattern of the NPC Gene Family Members in Arabidopsis Tissues.
Total RNA was isolated from 6-week-old soil-grown Arabidopsis plants. The level of gene expression was normalized to that of UBQ10. Values are means ± sd; n = 3 biological replicates.
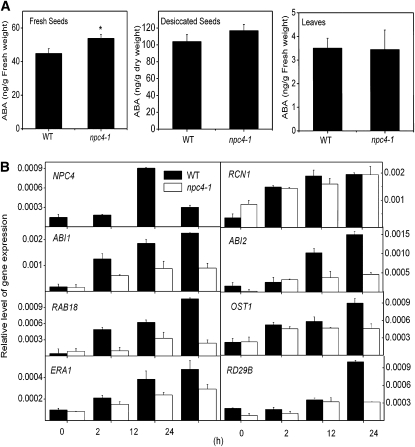
Figure 4.
ABA Content and the Expression of NPC4 and ABA-Responsive Genes in the Wild Type and npc4-1.
(A) ABA content of fresh seeds, desiccated seeds, and leaves of 4-week-old Arabidopsis plants. ABA was extracted and analyzed by the use of liquid chromatography-MS/MS. Values are means ± sd (n = 3). *Significant at P < 0.05 compared with the wild type based on Student’s t test.
(B) Levels of gene expression in response to ABA as measured by quantitative real-time PCR. Four-week-old Arabidopsis plants were treated with 50 μM ABA, and samples were harvested at indicated time intervals for RNA extraction. Relative expression is based on the expression of UBQ10. Values are means ± sd (n = 3).
ABA Content and the Expression of ABA-Responsive Genes
To investigate whether altered responses to ABA were due to changes in ABA levels, ABA content was measured in freshly picked and desiccated seeds and leaf tissue of npc4-1 and the wild type. The level of ABA in freshly picked npc4-1 seeds was significantly higher (20%) than wild-type seeds (Figure 4A), and the difference became smaller in desiccated seeds (Figure 4A). However, the ABA content in rosette leaves was similar in npc4-1 and the wild type (Figure 4A). These results indicate that the decreased sensitivity of npc4-1 toward ABA is not attributable to lower ABA content than wild-type tissue, and NPC4 likely is involved in plant response to ABA.
We then tested whether npc4-1 had altered expression of ABA-responsive genes (Figure 4). The basal level of expression of ABI1, ENHANCED RESPONSE TO ABA1 (ERA1), Response to ABA (RAB18), and Open Stomata1 (OST1) were similar in wild-type and npc4-1 leaves. The basal levels of Response to Desiccation 29B (RD29B) and ABI2 were lower in npc4-1 than the wild type, whereas Root Curls in NPA1 (RCN1) was higher in npc4-1 (Figure 4B). When the plants were treated with ABA, the expression of the genes tested was induced in wild-type and npc4-1 leaves. However, the level of induction of ABI1, ABI2, RAB18, OST1, RD29B, and ERA1 was lower in npc4-1 than in the wild type. The level of RCN1 expression in response to ABA in the wild type and npc4-1 was similar, but the magnitude of induction in npc4-1 was lower than the wild type (Figure 4B). These results are consistent with the phenotypic changes indicating that npc4-1 plants have decreased, but not abolished, sensitivity to ABA.
Decreased DAG Content in the NPC4 Knockout
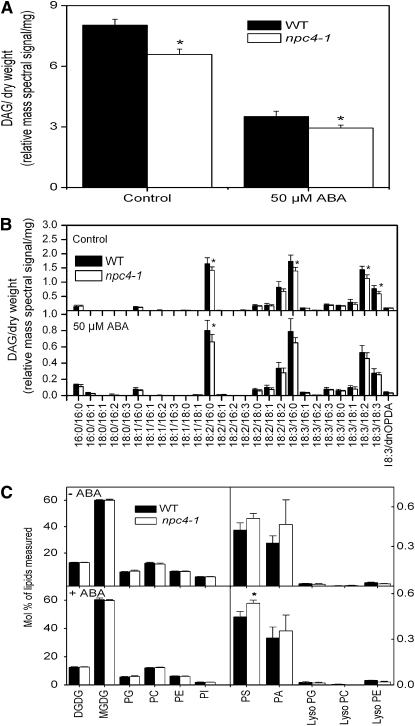
To determine the effect of the NPC4 loss on DAG level, we analyzed the relative levels of DAG molecular species in the wild type and npc4-1 using electrospray ionization tandem mass spectrometry (ESI-MS/MS). Under normal growth conditions, the DAG level in 4-week-old wild-type leaves was ~4% of total glycerophospholipids and galactolipids, and the DAG content in npc4-1 leaves was ~80% of the wild type (Figures 5A and 5B). Treatment of leaves with 50 μM ABA resulted in a >50% decrease in DAG level in both the wild type and npc4-1, and the leaves of npc4-1 still had a lower level of DAG than the wild type. The molecular species of DAG in Arabidopsis leaves that produced the largest mass spectral signals were 16:0/18:2, 16:0/18:3, and 18:2/18:3 combinations, followed by 18:2/18:2 and 18:3/18:3 DAG. The levels of all the major DAG species were decreased in npc4-1 leaves (Figure 5B). Under normal and ABA-treated conditions, DGDG, monogalactosyldiacylglycerol (MGDG), PC, PE, PA, phosphatidylglycerol (PG), and phosphatidylinositol (PI) were similar in wild-type and npc4-1 plants. However, phosphatidylserine (PS) was elevated in npc4-1 compared with wild-type plants, suggesting that NPC4 may use this lipid as a substrate (Figure 5C).
Figure 5.
DAG, Phospholipid, and Galactolipid Content and Effect of ABA on the Wild Type and npc4-1.
(A) Total DAG content and its changes in response to ABA. Four-week-old soil-grown Arabidopsis plants were treated with 50 μM ABA for 1 h. Total lipids were extracted from leaves and profiled using ESI-MS/MS. Asterisk denotes that the difference between the wild type and mutant is significant with P < 0.05 as determined by Student’s t test. Values are means ± se (n = 5).
(B) DAG molecular species with and without ABA treatment. Molecular species are presented as a:b/c:d, where a and c represent the number of carbons in the two fatty acids of DAG, and c and d represent the number of double bonds in the two fatty acids, respectively. dnOPDA, dinor-oxophytodienoic acid. Asterisk denotes that the difference between the wild type and mutant is significant with P < 0.05 as determined by Student’s t test. Values are means ± se (n = 5).
(C) Total phospholipid and galactolipid content in leaves and its changes in response to ABA. Four-week-old soil-grown Arabidopsis plants were treated with ABA for 1 h. Asterisk denotes that the difference between the wild type and mutant is significant with P < 0.05 as determined by Student’s t test. Values are means ± se (n = 5).
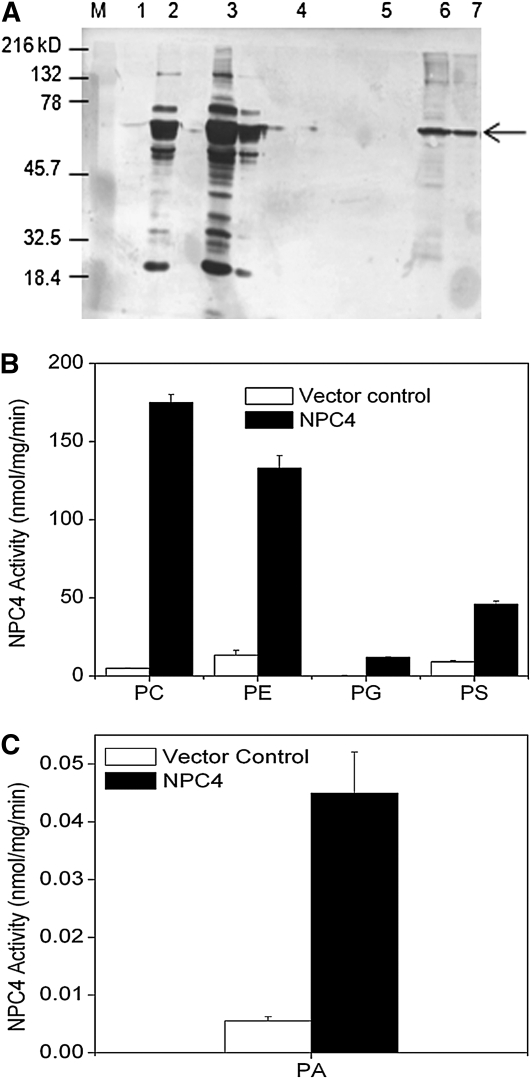
To test whether NPC4 used PS and PA as substrates, we expressed NPC4 in Escherichia coli and measured its activity toward different phospholipids (Figure 6A). NPC4 displayed a high activity toward PC and PE (Figures 6B and 6C). In addition, NPC4 used PG and PS as substrates, but these activities were lower than the hydrolytic activity with PC or PE (Figure 6B). Furthermore, using NPC4 purified from E. coli, NPC4 displayed reproducible hydrolytic activity on 16:0/16:0 PA, although the activity on this PA was much lower than its activity on PC or PE (Figure 6C). However, no NPC4 activity was detected when 18:1/18:1 PA was used as substrate. Previously, NPC4 was reported to hydrolyze PC and PE; some activity toward phosphoinositol-4,5-bisphosphate was noted, but this activity was ~10-fold less than that on PC (Nakamura et al., 2005).
Figure 6.
NPC4 Activity toward Different Phospholipids.
(A) Immunoblotting of NPC4 protein produced in E. coli. NPC4 from total E. coli lysate was purified and separated on 10% SDS-PAGE and transferred onto a filter and blotted with anti-His antibody. M, marker; 1, total lysate from empty vector; 2 and 3, total lysate harboring NPC4; 4, empty vector extract bound to His-agarose beads; 5, bound protein eluted from empty vector extract; 6, NPC4 extract bound to His-agarose beads; 7, NPC4 eluted from beads. Arrow indicates the 60-kD NPC4 protein.
(B) Lipid hydrolysis activity assayed in the presence of different phospholipids using crude E. coli lysate. DAG was quantified indirectly by measuring the release of the phosphorylated head group using a colorimetric phosphate assay. Values are means ± sd (n = 3). Vector control refers to the lysate from E. coli harboring the empty vector. Lipid abbreviations are described in the legend to Figure 5.
(C) NPC4 activity on PA using purified NPC4. DAG was quantified using gas chromatography (GC) analysis of fatty acid methyl esters derived from the DAG. Vector control refers to the lysate from E. coli harboring the empty vector that was purified in the same manner as NPC4. Values are means ± sd (n = 3).
Increased ABA Sensitivity in NPC4-Overexpressing Plants
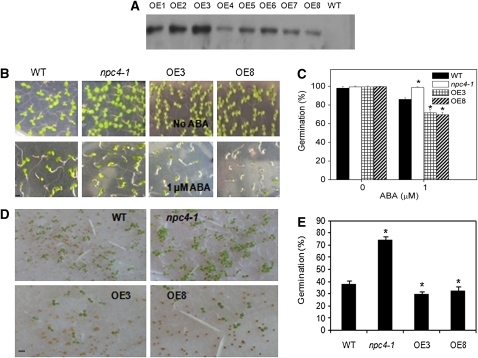
To further investigate the function of NPC4, we generated transgenic Arabidopsis lines overexpressing STREP-tagged NPC4 (NPC4-OE) under the control of the cauliflower mosaic virus 35S promoter. Production of NPC4-STREP in plants was confirmed by immunoblotting using anti-STREP antibodies (Figure 7A). Several independent lines were tested for their stress responses, and two representative transgenic lines were analyzed further. NPC4-OE seeds were more sensitive to ABA inhibition of germination than wild-type and npc4-1 seeds, and some of the seedlings did not develop green cotyledons but exhibited an etiolated phenotype after germination (Figure 7B). At 1 μM ABA, unlike npc4-1 seeds that displayed a higher germination rate than wild-type seeds, the germination of NPC4-OE seeds was 20% lower than the wild type (Figures 7B and 7C).
Figure 7.
Increased Sensitivity of NPC4-Overexpressing Plants to ABA Inhibition of Seed Germination.
(A) Production of Strep-tagged NPC4 in Arabidopsis wild-type plants. Proteins were extracted from the leaves of NPC4-STREP transgenic plants and separated by 10% SDS-PAGE and transferred to a polyvinylidene difluoride membrane. Ten micrograms of total protein was loaded onto each lane. The membrane was blotted with anti-STREP antibody conjugated with horseradish peroxidase. Lanes OE1 through OE8 represent different transgenic lines harboring the NPC4-STREP overexpression construct.
(B) Germination on 0.5× Murashige and Skoog medium in the presence of 1 μM ABA. Photographs were taken at 7 d after seeds were sown. Bar = 2 mm.
(C) Percentage germination of seeds of the wild type, npc4-1, OE3, and OE8 on 0.5× Murashige and Skoog medium with 0 or 1 μM ABA. One hundred seeds per genotype were measured. Values are means ± sd, n = 3, and experiments were repeated three times to ensure reproducibility.
(D) and (E) Precocious germination. Freshly picked seeds were germinated on Whatman filter paper (D), and the germination percentage was quantified after 7 d (E). One hundred and fifty seeds per genotype were measured. Values are means ± sd; n = 3. *Significant at P < 0.05 compared with the wild type based on Student’s t test. Bar = 3 mm.
ABA maintains dormancy in seeds and prevents precocious germination. Genetic studies in Arabidopsis revealed that the first ABA peak occurs maternally right before the maturation phase of the offspring; the second ABA peak occurs in the embryo itself and is essential for the induction of dormancy (Karssen et al., 1983). To test precocious germination in these genotypes, freshly harvested wild-type, npc4-1, and OE seeds were germinated on filter paper. Fresh npc4-1 seeds germinated at a much greater rate than wild-type seeds (80% versus 40%), whereas only ~30% of NPC4-OE seeds germinated (Figures 7D and 7E). These results suggest that NPC4 promotes ABA action and seed dormancy.
Opposite Responses to Hyperosmotic Stresses by NPC4 Knockout and Overexpression
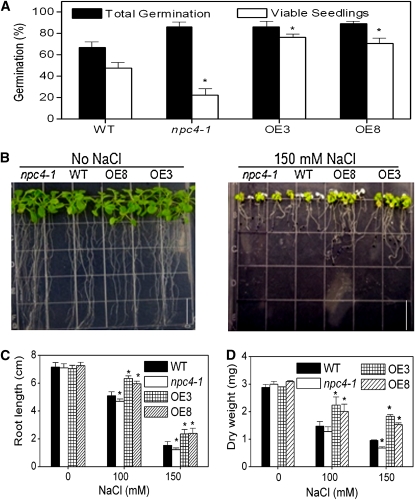
ABA plays an important role in sensing and adaptation to abiotic stresses, such as drought and salinity (Yamaguchi-Shinozaki and Shinozaki, 1994). Since npc4-1 plants exhibited reduced ABA sensitivity in both germination and vegetative growth, we investigated the effect of NPC4 alterations on plant tolerance to salinity and water deficits. Wild-type and npc4-1 seeds were sown on 200 mM NaCl, and the percentage of plants in which emergence of the radicle was observed was scored as total germination; seedlings with green cotyledons were scored as viable seedlings. Seedlings of npc4-1 were hypersensitive to NaCl; wild-type seeds produced more than twice as many viable seedlings as npc4-1 (Figure 8A). By contrast, the number of viable OE (OE3 and OE8) seedlings was 60% higher than the wild type, and almost all germinated OE seedlings were viable. The total germination was lower in the wild type than npc4-1 and OE seeds under high salinity.
Figure 8.
Opposite Effects of npc4-1 and NPC4-OE on Tolerance to Salt Stress.
(A) Quantification of total germination and viable growth of wild-type, npc4-1, and NPC4-OE seedlings on 0.5× Murashige and Skoog medium plus 200 mM NaCl for 2 weeks. Total germination represents the percentage of radicle emergence, and viable seedlings refer to seedlings not bleached and remaining green. One hundred seeds per genotype were germinated. Values are means ± sd; n = 3. *Significant at P < 0.05 compared with the wild type based on Student’s t test.
(B) Seedling phenotypes under salt stress. Five-day-old seedlings were transferred to 0.5× Murashige and Skoog medium with and without NaCl. Photographs were taken 10 d after transfer. Bar = 1.3 cm.
(C) Root length of wild-type, npc4-1, and OE seedling grown on 0.5× Murashige and Skoog medium plus 0, 100, or 150 mM NaCl for 10 d. Values are means ± sd (n = 15).
(D) Biomass production under salt stress. The rosettes from the wild type, KO, and OE were harvested on day 10 from plants subjected to salt stress as described in (C) and oven dried for 2 d at 60°C. Values are means ± sd (n = 15). Three independent experiments were performed with similar results, and the values of one experiment are presented.
To test the effect of NPC4 alterations on seedling growth in response to NaCl, 5-d-old seedlings grown on 0.5× Murashige and Skoog medium were transferred to NaCl-containing plates. Ten days after growth on 100 mM NaCl, npc4-1 primary roots were 8% shorter than the wild type, whereas OE roots were 20% longer than wild-type roots. On plates containing 150 mM NaCl, the primary root length of npc4-1 was 80% of the wild-type root length; OE roots were 55% longer than the wild type (Figures 8B and 8C). Biomass accumulation in npc4-1, NPC4-OE, and wild-type seedlings was similar in the absence of NaCl (Figure 8D). However, in the presence of 100 mM NaCl, the dry weight of npc4-1 seedlings was 85% of wild-type seedlings, whereas NPC4-OE seedlings weighed 150% as much as wild-type seedlings (Figure 8D). At 150 mM NaCl, npc4-1 seedlings accumulated ~70% of the biomass of wild-type seedlings, whereas OE accumulated twofold more biomass than the wild type (Figure 8D).
To determine whether the response was specific to salt stress, npc4-1, NPC4-OE, and wild-type seedlings were tested for their responses to water deficits. Murashige and Skoog plates were infused with polyethylene glycol (PEG) to a water potential of −0.25 or −0.45 MPa (Verslues and Bray, 2004). The length of npc4-1 primary roots was ~90 and 80% of wild-type seedlings on −0.25 and −0.45 MPa, respectively (Figures 9A and 9B). The primary roots of NPC4-OE seedlings were ~20 and 15% longer than those of wild-type seedlings on −0.25 and −0.45 MPa, respectively. Compared with the wild type, npc4-1 seedlings accumulated ~18 and 5% less biomass on −0.25 and −0.45 MPa, respectively (Figure 9C). By contrast, NPC4-OE seedlings accumulated ~35 and 30% more biomass than wild-type seedlings on −0.25 and −0.45 MPa, respectively (Figure 9C).
Figure 9.
Opposite Effects of npc4-1 and NPC4-OE on Tolerance to Water Deficiency.
(A) Seedling morphology of npc4-1 and NPC4-OE plants germinated on PEG-infused plates. Four-day-old seedlings were transferred to 0.5× Murashige and Skoog medium and 1.5% agar plates infused with overlay solution containing 0 and 170 g/L−1 PEG that corresponded to −0.25 and −0.45 MPa. Photographs were taken 10 d after seedlings were transferred. Control represents seedlings that were grown on 0.5× Murashige and Skoog 1% agar plates. Bar = 1.3 cm.
(B) Root length of wild-type, npc4-1, and OE seedlings grown on 0.5× Murashige and Skoog medium, −0.25 or −0.45 MPa PEG for 10 d. Values are means ± sd (n = 15). *Significant at P < 0.05 compared with the wild type based on Student’s t test.
(C) Biomass production under drought stress. The rosettes from wild-type, npc4-1, and OE plants in (B) were harvested on day 10 from plants subjected to drought stress and oven dried for 2 d at 60°C. Dry weights were measured, and values are means ± sd (n = 15). Three independent experiments were performed with similar results, and the values represent one experiment.
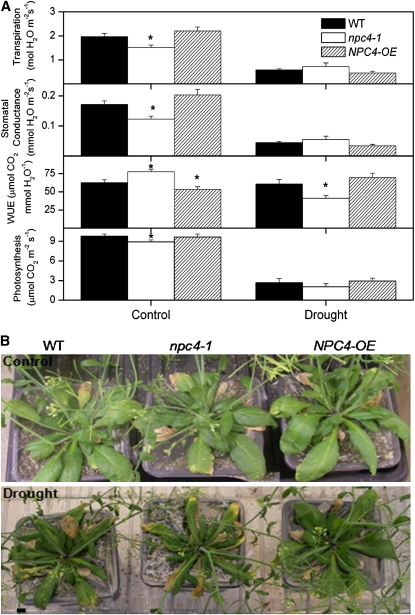
The effect of npc4-1 and OE was investigated in plants grown in soil under drought conditions. Drought conditions were simulated by reducing and maintaining the soil water capacity to 35% and controls were maintained at 100%. Under well-watered control conditions, transpiration rate and stomatal conductance were lower in npc4-1 than the wild type and higher in OE compared with the wild type. However, under drought conditions, transpiration rate and stomatal conductance were higher in npc4-1 compared with the wild type and lower in OE compared with the wild type (Figure 10A). Lower levels of photosynthesis and water use efficiency were observed in npc4-1 than wild-type plants under drought stress, whereas OE plants had higher water use efficiency than the wild type (Figure 10A). Under control growth conditions, no differences in phenotype were observed among the genotypes. Under drought conditions, a visible reduction in leaf turgor and size was observed in npc4-1 plants compared with the wild type. By contrast, OE plants were more turgid than wild-type plants (Figure 10B). These results support the conclusion that NPC4 promotes tolerance to hyperosmotic stresses.
Figure 10.
Effect of npc4-1 and NPC4-OE on Tolerance to Water Deficiency in Soil-Grown Plants.
(A) Transpiration rate, stomatal conductance, water use efficiency (WUE) (A/gs; i.e., photosynthetic rate/stomatal conductance), and photosynthetic rate of wild-type, npc4-1, and OE plants under control and water deficit conditions. Seven-day-old seedlings were transferred to pots containing water saturated soil. Control conditions were maintained at 100% field capacity, and water deficit conditions were maintained at 35% for a period of 10 d. Drought parameters were measured and recorded using a LI-6400 portable photosynthesis system (LI-COR) over a 10-d period. Six plants were measured per genotype. Experiments were conducted three independent times with similar results. Values are means ± se (n = 24) of one experiment. *Significant at P < 0.05 compared with the wild type based on Student’s t test.
(B) Phenotypes of wild-type, npc4-1, and OE plants under control and water deficit conditions. Photographs were taken at the end of the experiments, and plants were 6 weeks old. Three independent experiments were performed with similar results, and the photos represent one experiment. Bar = 6 mm.
Restoring npc4-1 to Wild Type–Like ABA Response by DAG and PA
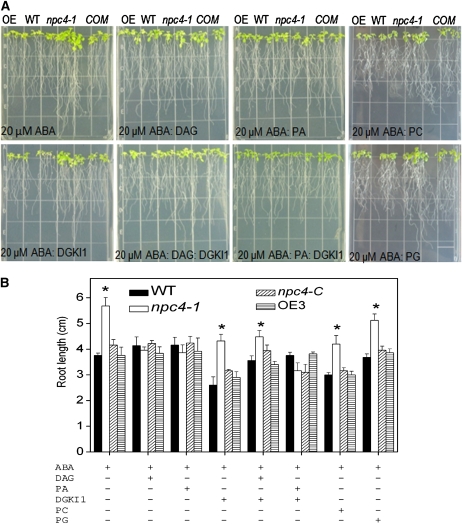
To determine how NPC4 functions in plant response to stress, we tested whether the NPC product DAG restores the npc4-1 phenotype to the wild type in the presence of ABA. Short-chain DAGs and short-chain phospholipids (8:0/8:0 PA, 8:0/8:0 PC, and 8:0/8:0 PG) were used in this experiment. The short chains were chosen for addition to the growth media because these compounds are more water soluble than 18C- or 16C-acyl chain lipids. Short-chain analogs of DAG and phospholipids are also known to more readily undergo transbilayer movement (Horman and Pownall, 1988). The presence of 8:0/8:0 DAG restored npc4-1 root growth so that in the presence of ABA, npc4-1 roots were the same length as those of wild-type seedlings (Figure 11A). The addition of 8:0/8:0 PA also restored npc4-1 root growth to the wild-type phenotype in the presence of ABA. Short-chain PC or PG had no such effect (Figure 11A). This result suggested that NPC4-produced DAG might be phosphorylated to PA by DGK. We then added the DAG kinase inhibitor 1 (DGKI1) to inhibit the conversion of DAG to PA. When DGKI1 was added together with DAG in the presence of ABA, DAG was no longer able to fully restore the npc4-1 root growth to the wild-type phenotype. Instead, the root growth of npc4-1 remained less sensitive to ABA inhibition than the wild type in the presence of 100 μM DGKI1 (Figure 11B). On the other hand, when PA and DGKI1were added in the presence of ABA, PA was able to increase the ABA sensitivity of npc4-1 to the level of the wild type. DGKI1 by itself at this concentration displayed no significant effect on root growth. These results indicate that the DAG produced by NPC4 is converted to PA, and the NPC4/DGK pathway plays a role in ABA signaling events.
Figure 11.
Addition of 8:0/8:0 DAG or 8:0/8:0 PA Restored npc4-1 Seedlings’ ABA Sensitivity to the Wild Type.
(A) Seedling morphology and root growth of npc4-1, OE, COM, and the wild type in the presence of 20 μM ABA without or with 8:0/8:0 DAG, 8:0/8:0 PA, and/or DGKI1. Four-day-old seedlings were transferred to 0.5× Murashige and Skoog medium with or without 20 μM 8:0/8:0 DAG, 8:0/8:0 PA, and DGKI1 and grown for 10 d. COM, the wild type complemented with NPC4. Bar = 1.3 cm.
(B) Root length of NPC4-KO, OE, COM, and the wild type in response to 20 μM ABA in the presence of 8:0/8:0 DAG, 8:0/8:0 PA, 8:0/8:0 PG, 8:0/8:0 PC, and/or DGKI1. Values are means ± sd (n = 10). *Significant at P < 0.05 compared with the wild type based on Student’s t test. Experiments were performed at least three times with similar results, and the values presented represent one experiment. COM, the wild type complemented with NPC4. Lipid abbreviations are described in the legend to Figure 5. DAGKI1, DAG kinase inhibitor 1.
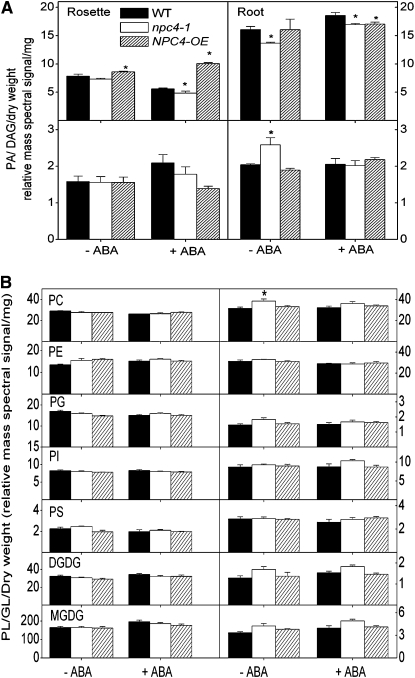
We then measured the level of DAG and PA, as well as other glycerolipids, in rosettes and roots of wild-type, npc4-1, and NPC4-OE seedlings. Under the control condition, DAG content in npc4-1 rosette was lower than the wild type, whereas OE was higher. After treatment with ABA, a decrease in DAG content was observed in wild-type and npc4-1 rosettes compared with the control, but an increase in DAG occurred in NPC4-OE rosettes (Figure 12A, left panel). The DAG content in npc4-1 rosettes was ~85% that of the wild type, whereas DAG in NPC4-OE was 40% higher than the wild type after ABA treatment (Figure 12A, right panel). In roots without ABA treatment, the DAG content of NPC4-OE and the wild type was similar, but npc4-1 contained ~20% less DAG than the wild type. ABA treatment resulted in an increase of DAG content in wild-type and npc4-1 roots, but the DAG increase was subtle in NPC4-OE roots (Figure 12A, right panel). These results indicate that NPC4 alterations directly impact DAG content both in roots and rosettes with or without ABA, but the effect differs between rosettes and roots.
Figure 12.
Glycerolipid Contents in Roots and Rosettes of Wild-Type, npc4-1, and OE Seedlings in Response to ABA.
(A) DAG and PA changes in the wild type, npc4-1, and OE in response to ABA. Five-day-old seedlings were transferred to 0.5× Murashige and Skoog vertical plates and grown for 10 d. Seedlings treated with ABA and infused with 50 μM ABA, and controls were infused with the carrier solution for 1 h. Total lipids were extracted and profiled using tandem ESI-MS/MS. Values are means ± se (n = 3 to 5). *Significant at P < 0.05 compared with the wild type based on Student’s t test.
(B) Phospholipid and galactolipid changes in response to ABA in the wild type, npc4-1, and OE. Plant growth conditions and treatment are same as described in (A). Values are means ± se (n = 4 or 5). *Significant at P < 0.05 compared with the wild type based on Student’s t test. Lipid abbreviations are described in the legend to Figure 5.
Without ABA treatment, PA contents of the wild type, npc4-1, and NPC4-OE were similar in rosettes, but npc4-1 roots had 20% more PA than wild-type and OE roots. After ABA treatment for 1 h, the PA content in the wild type remained unchanged, and PA in npc4-1 decreased, whereas PA in NPC4-OE increased compared with corresponding roots without ABA treatment (Figure 12A, bottom right panel). In rosettes treated with ABA, the PA content in the wild type and npc4-1 increased, whereas the PA content in NPC4-OE decreased. In rosettes without ABA treatment, the contents of other glycerolipids were comparable among the wild type, npc4-1, and NPC4-OE, except that npc4-1 rosettes had more PS than NPC4-OE and npc4-1 and NPC4-OE had a higher level of PE than the wild type. ABA treatment for 1 h did not result in major changes, but there was a trend toward an increase in galactolipids MGDG and DGDG with a concomitant decrease in phospholipids PC, PG, and PS (Figure 12B, left panel). The trend of opposite changes in galactolipids versus phospholipids also occurred when lipids from ABA-treated roots were compared those without ABA, but PE was the phospholipid displaying a decrease (Figure 12B, left panel). In the absence of ABA treatment, the PC level in npc4-1 roots was higher than the wild type and NPC4-OE. Under control and ABA-supplemented growth conditions, the levels of PE, PI, PG, and PS were similar in wild-type and NPC4-OE rosettes and roots. The levels of MGDG and DGDG tended to be higher in npc4-1 roots than the wild type and NPC4-OE.
DISCUSSION
NPC4 Positively Modulates Plant Response to Hyperosmotic Stresses and ABA
Nonspecific PLC is a new family of phospholipases recently described in plants (Wang, 2001; Nakamura et al., 2005). The study of their function in plants has so far been limited to their role in phosphate deficiency (Nakamura et al., 2005; Gaude et al., 2008). Results from this work provide insight regarding the metabolic and physiological functions of NPC4 and its derived DAG. Our results show that manipulations of NPC4 alter plant response to high salinity and drought; npc4-1 displayed hypersensitivity to salt and water deficits, while OE plants were more tolerant to hyperosmotic stress and produced more biomass than did the wild type. In addition, the study indicates that the reduced tolerance of npc4-1 plants to abiotic stress results from a positive role of NPC4 in mediating ABA response. ABA is known to be involved in plant tolerance to abiotic stress, such as drought and salinity (Finkelstein et al., 2002). NPC4-KO plants showed reduced sensitivity to ABA in seed germination, seed dormancy, and root growth, whereas NPC4-OE was increased in ABA sensitivity. ABA levels in npc4-1 were higher than in the wild type in seeds but similar in leaves, indicating that NPC4 is most likely involved in ABA responses other than ABA accumulation. Additionally, stress-responsive and ABA-inducible genes, such as RD29B, ABI1, ABI2, RAB18, ERA1, and OST1, were downregulated in npc4-1. The opposite effects of NPC4 ablation and overexpression suggest that NPC4 acts as a positive regulator of ABA and hyperosmotic stress responses in Arabidopsis.
NPC4 Is Involved in the Production of Basal and Stress-Induced Production of DAG
Ablation of NPC4 decreases DAG in both leaves and roots, and the decrease occurred in plants grown two different conditions, in leaves of soil-grown plants and in 0.5× Murashige and Skoog plate-cultured rosettes under normal, unstressed growth conditions. The results suggest that NPC4 is involved in the production of a basal level of DAG in both tissues. Under stressed conditions, such as drought (see Supplemental Figure 4 online), and ABA treatments (Figure 5A), the steady level of DAG decreased in rosettes. The decrease may result from a decrease in the activity of de novo lipid biosynthesis, as much of the steady state DAG may be produced via de novo biosynthesis and used for the biosynthesis of PC, PE, and other glycerolipids. Separate pools may exist for DAG used for signaling and for neutral and polar lipid biosynthesis. In addition, the decrease of DAG could result from an increase in DGKs. On the other hand, DAG in roots increased in both the wild type and npc4-1 in response to ABA, indicating that the production and regulation of DAG differs in roots and rosettes. Consistent with this notion, overexpression of NPC4 increases the DAG level in leaves but not roots. This might be due to the higher level of expression of NPC4 in leaves than roots and/or quick conversion of DAG to PA in NPC4-OE roots.
It is worth noting that very little is known about the steady state cellular level of DAG and its molecular species in plants. This analysis by ESI-MS/MS allows for measurement of DAG species and comparative analysis of DAG in different plant samples. DAG levels are expressed as relative mass spectral signal where a value of 1 equals the signal for 1 nmol of a synthetic 15:0/15:0 DAG added as an internal standard. This mass spectral signal comparison is necessary because variation in ionization and fragmentation efficiency among different acyl glycerol species makes it impossible to use the MS responses of the plant and internal standard DAG species to determine DAG amount without knowing the mass spectral response factors for each DAG molecular species, and these response factors have not been determined for plant DAGs (Han and Gross, 2001).
NPC4 and DAG Promote Stomatal Opening under Well-Watered Conditions
The results suggest that the production of DAG is one route by which NPC4 mediates ABA and hyperosmotic stress response. DAG may function directly as a mediator and/or phosphorylated by DGK to PA. DAG in itself is a potent messenger in animal cells, but its role in signaling in plants is unknown. A previous study using Vicia faba and synthetic DAG indicated that DAG promoted stomatal opening by activating an ion pump in the guard cell plasma membrane and inhibits stomatal closure by possibly inhibiting K+ efflux (Lee and Assmann, 1991). This study shows that under well-watered control conditions, npc4-1 plants had lower stomatal conductance and transpiration rates than the wild type, whereas NPC4-OE tended to have higher stomatal conductance than the wild type. These differences are associated with the opposite changes in DAG levels in the NPC4-KO and OE leaves. The stomata density of the npc4-1 leaves was not altered, and detached leaves from well-watered npc4-1 plants also lost less water than the wild type (see Supplemental Figure 5A online). Adding DAG to the closed stomata promoted stomatal opening more for npc4-1 than the wild type (see Supplemental Figure 5B online). The results suggest that npc4-1 leaves have a smaller stomatal aperture than those of the wild type and provide genetic evidence that NPC4-produced DAG is involved in promoting stomatal opening under well-watered conditions (Figure 13).
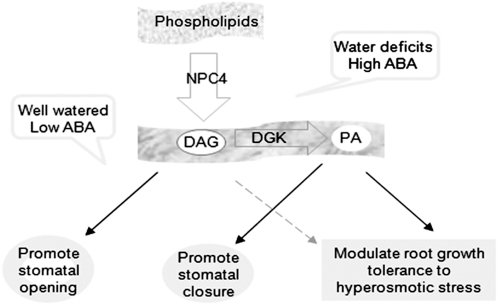
Figure 13.
A Working Model for the Function of NPC4 and Derived DAG in Mediating Plant Response to ABA and Hyperosmotic Stress.
In well-watered plants, NPC4 contributes to the basal production of DAG that promotes stomatal opening. Water deficits increase the level of ABA, which stimulates NPC4 to produce DAG. The stress-induced DAG is phosphorylated by DGK to PA that promotes ABA-promoted stomatal closure and root response to ABA and hyperosmotic stress.
Under drought stress, however, npc4-1 plants tended to lose more water and had lower water use efficiency than the wild type. The ratios of stomatal conductance in the well-watered plant to those under drought conditions were 3.3 for the wild type, 2.0 for npc4-1, and 4.5 for NPC4-OE (Figure 10A). Thus, in response to drought, the OE and wild-type plants displayed greater decreases than npc4-1 in stomatal conductance to reduce transpirational water loss. This difference indicates that stomata in NPC4-ablated leaves are less responsive to water deficits than those in the wild type. Drought is known to increase ABA production, and the stomatal behavior in npc4-1 epidermal peels suggests that NPC4 is involved in stomatal movements in response to ABA. These results support the conclusion that NPC4 renders plants more responsive to ABA and water availability in plants coping with hyperosmotic stress. Under water deficits and high levels of ABA, NPC4-derived DAG may be converted to PA that has been shown to promote stomatal closure (Zhang et al., 2004; Figure 13).
DAG Is Converted to PA That Mediates the NPC4 Effect on Hyperosmotic Response
The results from root growth indicate that NPC4-produced DAG is phosphorylated to PA that mediates the NPC4 effect on plant response to ABA and hyperosmotic stresses. Evidence for this proposition is that both DAG and PA restore the npc4-1 response to ABA to the wild-type response, whereas addition of the DGK inhibitor that blocks the DAG to PA conversion prevents response restoration by DAG. In plants, DAG has been shown to be phosphorylated to PA under hyperosmotic stress conditions (Arisz et al., 2009; Darwish et al., 2009; Munnik and Testerink, 2009). PI-PLC has been thought to be the enzyme activated to produce DAG, but the enzymatic step for DAG production was not genetically established. NPC4 is associated with the plasma membrane (Nakamura et al., 2005), and these analyses show that its expression and activity are induced by ABA and hyperosmotic stresses (http://bar.utoronto.ca). The results suggest that NPC4 contributes to the production of DAG used for PA formation. The PA content in well-watered npc4-1 roots was higher than in wild-type roots. NPC4 had PA-hydrolyzing activity, and the ablation of NPC4 might attenuate PA degradation in the knockout leaves. However, the reason for the increase requires further investigation because the PA-hydrolyzing activity of NPC4 was much lower than that of PC hydrolysis under these assay conditions. In addition, the steady state level of PA in plants can be influenced by various enzymes, including multiple forms of PLDs, DGKs, PA phosphatases, and other NPCs, as well as enzymes in de novo PA biosynthesis (Wang et al., 2006). The PA-hydrolyzing activity of NPC4 was not detected using NPC4 expressed in E. coli previously (Nakamura et al., 2005). Our study shows that using purified NPC4 is needed for the detection of NPC4 PA hydrolyzing activity since crude E. coli extracts harbor phosphatidylglycerolphosphate phosphatase activity that dephosphorylates PA (Icho and Raetz, 1983; Icho, 1988).
In summary, opposite physiological performances exhibited by NPC4-KO and OE plants indicate that NPC4 is a positive mediator in the plant response to ABA and hyperosmotic stress. Moreover, the results provide new insight into the roles of NPC4 and derived DAG in regulating stomatal movement and water loss in plants with varied water status. We propose that under well-watered conditions, the NPC4 and its derived DAG are involved in promoting stomatal opening (Figure 13). Under stress, however, DAG produced by NPC4 is converted to PA, and the NPC4/DGK pathway promotes root and plant growth in response to ABA and water deficit. These results provide an impetus for further investigation to determine the function of the new PLC family and the signaling function of DAG in plants.
METHODS
Plant Growth and Treatments
Seeds of NPC4-KO and the wild type of Arabidopsis thaliana (Columbia ecotype) were sown in potting soil (Fafard; superfine germinating mix containing Canadian sphagnum peat moss, perlite, vermiculite, wetting agent, starter nutrients, and dolomitic limestone) and kept at 4°C for 2 d. Plants were grown in a growth chamber with cool white fluorescent light of 100 μmol m−1 s−1 under 12-h-light/12-h-dark and 23°C/22°C cycles. Four- to six-week-old plants were used to prepare epidermal peels. Seed germination was performed on 0.5× Murashige and Skoog medium supplemented with 1.5% sucrose. Seeds were sterilized in 70% ethanol for 5 min followed by 20% bleach for 3 min and then rinsed three times with sterile, distilled water. Sterilized seeds were sown on medium containing 0 to 10 μM ABA and stratified at 4°C for 48 h. Germination was scored at 2-d intervals for 10 d. For precocious germination, fresh seeds of the wild type, NPC4-KO, and NPC4-OE were collected directly from yellow siliques on plants at the same time from the plants grown under the same conditions. Seeds were placed on moistened Whatman filter paper for germination, and germination percentage was calculated after 7 d of plating the seeds. For root elongation assays, 5-d-old seedlings were transferred to 0 to 50 μM ABA and grown under cool white fluorescent light of 100 μmol m−1 s−1 under 16-h-light/8-h-dark and 23°C/22°C cycles for 2 weeks. For rescue experiments, 8:0/8:0 DAG, PC, PG, and PA (Avanti Polar Lipids) were prepared by drying the chloroform solvent under a stream of nitrogen gas and vortexing the dried lipids in water followed by sonication. DGKI1 (R59022, 6-[2-[4-[(4-fluorophenyl) phenylmethylene]-1-piperidinyl] ethyl]-7-methyl-5H-thiazolo-[3,2-a]-pyrimidin-5-one) (Calbiochem) was dissolved in DMSO as described by Gomez-Merino et al. (2005). These solutions were added to the surface of 20 μM ABA agar plates (200 μL of 100 μM). For NaCl (0 to 150 mM) and PEG (−0.25 and −0.45 MPa) experiments, 5-d-old seedlings were transferred and grown under cool white fluorescent light of 100 ~mol m−1 s−1 under 12-h-light/12-h-dark and 23°C/22°C cycles for 10 d. PEG plates were prepared as described by Verslues and Bray (2004). Briefly, 1.5% agar plates containing 0.5× Murashige and Skoog medium were solidified and then were overlayed with a solution containing 6 mM MES with 0 or 170 g/L PEG (molecular weight, 8000; Sigma-Aldrich), which gave rise to an MPa of −0.25 and −0.45, respectively. The solution was allowed to sit for 24 h, and excess solution was removed from the plates. Control plates were 1% agar plates containing 0.5× Murashige and Skoog medium.
A gravimetric approach was adapted to create water limitation to study the effect of NPC4 alteration on stomatal regulation (Sheshshayee et al., 2005). Arabidopsis seeds were sown in potting soil and stratified for 2 d and then grown for 10 d under 12 h light/12 h dark. The seedlings were transplanted to pots containing soil saturated with a known weight of water (soil saturation was achieved by adding 5 times the weight of the soil in water). The pots were covered with thick polyethylene sheets to prevent evaporation. The first set of plants was maintained at 100% field water capacity (FC), and the second set was gradually brought down to 35% FC (drought stress). The pots were weighed daily during the experimental period, and the difference in weight in subsequent days was corrected by adding water to maintain the required FC. Stomatal conductance (gs) and other gas exchange-related parameters were measured on fully expanded leaf using a portable gas exchange system (LICOR6400-XT; LICOR Biosciences). Measurements were taken on the first 4 d following the onset of drought stress.
Knockout Mutant Isolation and Complementation
T-DNA insertion mutants in NPC1, NPC2, NPC3, NPC4, NPC5, and NPC6, designated npc1-1-npc6-1, were identified from the Salk T-DNA knockout collection. Seeds were obtained from the ABRC. The NPC-KO homozygous T-DNA insertion mutants were isolated by PCR screening using NPC gene-specific primers (see Supplemental Table 1 online) and a T-DNA left border primer: 5′-TGGTTCACGTAGTGGCCATC-3′. A single T-DNA insert was verified by cosegregation of the mutant allele with kanamycin resistance in a 3:1 ratio. The loss of transcription of NPC4 was confirmed using NPC4-specific primers 5′-AGCATCAAATGCTGCTGCTCAACC-3′ (forward) and 5′-TCCACCCACACACAAGAGAAGTGA-3′ (reverse) by real-time PCR as described below. The real-time PCR cycling conditions comprised 90-s denaturation at 95°C and 50 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with the final cycle terminated at 72°C for an additional 10 min. For complementation of the NPC4-KO mutant, the native NPC4 gene including its native promoter (1085 bp upstream of the start codon) was amplified from wild-type Arabidopsis plants and cloned into Asc1 sites of PEC291 vector. The primers used for amplification were 5′-ATGGCGCGCCGACAGGGTTTGTGATGCTACAATG-3′ (forward) and 5′-ATGGCGCGCCGGCGCGACTTTGTATTCCTATTC-3′ (reverse). The construct was introduced into the JM109 strain to test for positive clones. The resulting positive clone was transformed in Agrobacterium tumefaciens C58 and was used to transform npc4-1 plants by floral dip (Clough and Bent, 1998). Hygromycin-resistant T3 transgenic plants were screened for homozygous lines. Homozygous lines did not segregate when placed on hygromycin.
Real-Time PCR
Total RNA was isolated using a rapid cetyl-trimethyl-ammonium bromide method (Stewart and Via, 1993), and RNA was precipitated using 2 M LiCl at 4°C. RNA integrity was checked on 1% (w/v) agarose gel. Eight micrograms of total RNA was digested with DNaseI according to the manufacturer’s instructions (Ambion). Real-time PCR was performed as described by Li et al. (2006). Three independent experiments were performed; values are means ± sd (n = 3) for one representative experiment. Briefly, RNA was reverse transcribed using an iScript cDNA synthesis kit (Bio-Rad). The efficiency of cDNA synthesis was analyzed by real-time PCR amplification of UBQ10 (At4g05320). The level of gene expression was normalized to that of UBQ10. PCR was performed with the MyiQ sequence detection system (Bio-Rad) using SYBR Green to monitor double-stranded DNA synthesis. Reaction mixes contained 7.5 μL 2xSYBR Green Master Mix reagent kit (Bio-Rad), 1.0 ng cDNA, and 200 nM of each gene-specific primer in a final volume of 15 μL. The primers for different genes were listed in Supplemental Table 1 online. The PCR cycling conditions were as follows: one cycle of 95°C for 90 s to facilitate denaturation and 35 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with the final cycle terminated at 72°C for an additional 10 min.
Measurement of Stomatal Aperture
Stomatal aperture was measured using a procedure described by Zhang et al. (2004). Briefly, epidermal peels from 6-week-old plants were floated in a solution of 5 mM MES-KOH, pH 6.15, 30 mM KCl, and 1 mM CaCl2. After incubation for 3 h in the dark to ensure the closure of stomata, 50 μM ABA was added and incubated under cool white light (150 μmol m−1 s−l) at room temperature for 3 h to induce stomatal opening. To test the effect of ABA on stomatal closure, epidermal peelings were incubated in 5 mM MES-KOH, pH 6.15, 30 mM KCl, and 1 mM CaCl2 for 3 h under cool white light (150 μmol m−1 s−l) at room temperature to ensure the opening of stomata. After incubation, 50 μM ABA was added and the peels incubated under cool white light (150 μmol m−1 s−l) at room temperature for 3 h. Epidermal peels were observed under a microscope; stomatal aperture was recorded using a digital camera and analyzed with IMAGEPRO software (Media Cybernetics).
ESI-MS/MS Analysis of DAG and PA
Lipid extraction was performed as described by Welti et al. (2002). Briefly, rosettes and roots were excised from Arabidopsis plants and immersed immediately in 3 mL of hot isopropanol containing 0.01% butylated hydroxytoluene at 75°C. Samples were kept at 75°C for 15 min, and 1.5 mL of chloroform and 0.6 mL of water were added. Samples were placed on a shaker for 1 h, and the solvent was transferred to a new tube. The samples were reextracted using chloroform:methanol (2:1) at least four times for 30 min each time on a shaker. The lipid extracts were combined and washed with 1 M KCl followed by washing with 1 mL of water. The solvent was evaporated under a stream of nitrogen gas and the remaining tissue was oven dried at 100°C and weighed.
An automated ESI-MS/MS approach was used for DAG and phospholipid analysis. The samples were dissolved in 1 mL of chloroform. An aliquot of 5 to 300 μL of extract in chloroform was used. Precise amounts of internal standards, obtained and quantified as previously described (Welti et al., 2002), were added. Phospholipid and galactolipid amounts were as described by Xiao et al. (2010), and 0.93 nmol 15:0/15:0 DAG was added. The sample and internal standard mixture were combined with solvents, such that the ratio of chloroform/methanol/300 mM ammonium acetate in water was 300/665/35, and the final volume was 1.4 mL.
Unfractionated lipid extracts were introduced by continuous infusion into the ESI source on a triple quadrupole mass spectrometer (4000 Q-TRAP; Applied Biosystems). Samples were introduced using an autosampler (LC Mini PAL; CTC Analytics) fitted with the required injection loop for the acquisition time and presented to the ESI needle at 30 μL/min. Phospholipid and galactolipid scans and scan parameters were described by Xiao et al. (2010). DAG was detected by a series of neutral loss scans that detected DAG species as [M + NH4]+ ions. The scans targeted losses of various fatty acids as neutral ammoniated fragments: NL 259.2 (15:0, for the internal standard); NL 273.2 (16:0); NL 271.2 (16:1); NL 269.2 (16:2); NL 267.2 (16:3); NL 301.2 (18:0); NL 299.2 (18:1); NL 297.2 (18:2); NL 295.2 (18:3); NL 309.2 (18:4-O or oxophytodienoic acid); and NL 281.2 (16:4-O or dinor-oxophytodienoic acid). For DAG scans, the scan speed was 60 μ per second. The collision energy, with nitrogen in the collision cell, was +35 V, declustering potential was +100 V, entrance potential was +15 V, and exit potential was +12 V. Sixty of each DAG continuum scan were averaged in MCA mode.
For all analyses the collision gas pressure was set on low, and the mass analyzers were adjusted to a resolution of 0.7 μm full width at half height. The source temperature (heated nebulizer) was 100°C, the interface heater was on, +5.5 kV was applied to the electrospray capillary, the curtain gas was set at 20 (arbitrary units), and the two ion source gases were set at 45 (arbitrary units). For all analyses, the background of each spectrum was subtracted, the data were smoothed, and peak areas integrated using a custom script and Applied Biosystems Analyst software.
Phospholipids and galactolipids were determined by comparison of the peak for each lipid species to those of two internal standards of the same class (Welti et al., 2002; Xiao et al., 2010). Data were corrected for chemical or instrumental noise in the samples (Jao et al., 2009). The molar amount of each phospholipid species in the sample was calculated and normalized to the sample dry weights to produce data in the units nmol/mg.
For DAG analysis, data from the neutral loss scans were corrected for overlap of isotopic variants (A + 2 peaks) in higher mass lipids were applied. Both corrections for overlap within spectra, due to (A + 2 peaks) of [M + NH4]+ minus the neutral loss fragment, and overlap across spectra, due to the (A + 2 peaks) of the neutral loss fragments, were performed. The corrected signals corresponding to each DAG molecular species were combined, and DAG species were quantified in relation to the internal standard. Because there is variation in ionization efficiency among acyl glycerol species with different fatty acyl groups (Han and Gross, 2001), the ratio of the MS response of the plant DAG species to the MS response of the DAG internal standard does not provide a value that is directly proportional to the DAG content of each species. DAG amounts are thus expressed as relative mass spectral signal/mg dry weight, where a signal of 1.0 represents a signal equal to the signal of 1 nmol 15:0/15:0 DAG (the internal standard).
ABA Content
Seeds or leaf tissues were ground in liquid nitrogen and 0.5 mL of 1-propanol:H2O:HCl (2:1:0.002 v/v) was added to the homogenate and vortexed well. One milliliter of dichloromethane and ABA internal standards were added. The samples were vortexed and centrifuged at 11,300g for 1 min. The lower phase was collected, and ABA was quantified by MS as described by Pan et al. (2008).
Enzyme Activity Assay of NPC4 Expressed in Escherichia coli
The NPC4 cDNA was amplified from 5′RACE pollen cDNA library using Phusion high fidelity Taq polymerase (New England Biolabs). The PCR product was cloned into pCR 2.1-TOPO (Invitrogen) vector to screen for a positive clone. A positive clone was selected, purified, and cloned into pET28a(+) expression vector with a His tag under the control of the T7 promoter and transformed into E. coli Rosetta (DE3). To induce the expression of NPC4, 0.1 mM isopropyl β-d-thiogalactopyranoside was added when the culture had reached an A600 of ~0.4. The culture was then incubated for 36 h at 12°C. The cells were harvested by centrifugation (15,000g, 2 min) and resuspended in extraction buffer containing 50 mM Tris-HCl, pH 7.3, 50 mM NaCl, 5% glycerol, 1 mM DTT, and 0.5 mM PMSF. Cells were lysed by sonication and centrifuged (1500g, 10 min) to remove cell debris. The supernatant was assayed for PLC activity as described below.
Protein concentration in the supernatant was quantified using Bradford reagent (Bio-Rad) with BSA as a standard, according to the manufacturer’s instructions. The PLC assay was performed using previously defined conditions (Nakamura et al., 2005). Briefly, the reaction mixture contained 100 μL of enzyme, 100 μL of substrate, and 300 μL of assay buffer (50 mM Tris-HCl, pH 7.3, 50 mM NaCl, and 5% glycerol). The substrate was prepared as follows: 100 μL of 10 μg/μL 18:1/18:1 PC (Avanti Polar Lipids), PE made by transphosphatidylation of egg PC (Avanti Polar Lipids), soybean (Glycine max) PS (Avanti Polar Lipids), 16:0/16:0 PA (Matreya), and 18:1/18:1 PG (Avanti Polar Lipids) were dried under a stream of nitrogen gas and suspended in 1 mL of 250 mM Tris-HCl, pH 7.3, and 0.25% deoxycholate. Complete resuspension of the lipids was verified before addition to each reaction. The samples were incubated for 1 h at 37°C. The reaction was stopped by the addition of 1 mL of chloroform:methanol (1:2) or 0.75 mL ethyl acetate followed by vigorous vortexing and addition 0.75 mL of 0.45% NaCl. In reactions that were terminated with ethyl acetate, DAG was quantified by thin layer chromatography (TLC) followed by GC analysis. To measure phosphate released from the head group, EDTA was added to the aqueous phase removed from the reactions terminated using chloroform/methanol, to a final volume and concentration of 1 mL and 50 mM, respectively. Two units of calf alkaline phosphatase was added to each tube and incubated for 120 min at 37°C. Total phosphate released was quantified colorimetrically following a method described by Chen et al. (1956). For TLC, the solvent from the organic, upper phase of the ethyl acetate extract of the reaction was evaporated under a stream of nitrogen gas, and the residue was dissolved in 20 μL of chloroform/methanol (2:1, v/v) and separated on a TLC plate. The plate was developed in petroleum ether/ethyl ether/acetic acid (50:50:1, v/v/v). The DAG band was scraped from the TLC plate, 5 μL of 5.4 μM/mL 17:0 TAG was added to the samples as an internal standard, and the mixture was transmethylated in methanol containing 1% H2SO4 and 0.05% butylated hydroxytoluene at 90°C for 1 h. One milliliter of hexane and 1 mL of water were added, and the upper phase was removed for GC analysis. The fatty acid methyl esters were separated on a Shimadzu GC-17A gas chromatograph supplied with a hydrogen flame ionization detector and a capillary column DB-5MS (30 m; 0.25 mm i.d.) with helium carrier at 11 mL/min. The oven temperature was maintained at 170°C for 3 min and then increased linearly to 210°C (5°C min−1). Fatty acid methyl esters were identified by comparison of their retention times with known standards (37-component FAME mix; Supelco 47885-U).
Immunoblotting, Detection, and Purification of NPC4-His
Bacterial cells were harvested by centrifugation and resuspended in an extraction buffer (50 mM Tris-HCl, pH 7.3, 50 mM NaCl, 5% glycerol, 1 mM DTT, and 0.5 mM PMSF). Cells were lysed by sonication and centrifuged at 1500g for 5 min. The supernatant and total cell lysates were separated by 10% SDS-PAGE. After electrophoresis, proteins were transferred to a polyvinylidene difluoride membrane. The membrane was blotted with anti-His antibody (1:10,000) conjugated with alkaline phosphatase for 2 h. The protein bands were visualized by alkaline phosphatase reaction. For purification of NPC4-His, Ni-NTA agarose (Qiagen) was used according to the manufacturer’s instructions. Briefly, 3 mL supernatant containing NPC4-His or empty vector control were added to 0.5 mL of Ni-NTA agarose and incubated for 1 h with gentle agitation at 4°C. Samples were washed four times, and Ni-NTA agarose was resuspened in 1 mL assay buffer (50 mM Tris-HCl, pH 7.3, 50 mM NaCl, and 5% glycerol). Samples were used for immunoblotting and PLC activity.
Immunoblotting and Detection of NPC4-STREP
Total proteins were extracted from 4-week-old Arabidopsis leaves using buffer A (50 mM Tris-HCl, pH 7.5, 10 mM KCl, 1 mM EDTA, 1 mM DTT, and 0.5 mM PMSF). After centrifugation at 15,000g for 5 min, 10 μg of supernatant proteins were separated by 10% SDS-PAGE. The proteins (10 μg) were transferred to a polyvinylidene difluoride membrane, and the membrane was blotted with anti-STREP antibody (1:10,000) conjugated with horseradish peroxidase for 1 h. The membrane was washed four times for 10 min with PBS/T buffer (1× phosphate saline buffer and 0.05% Tween 20). The membrane was incubated with LumiGLO substrate for 1 min and then exposed to x-ray film.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: NPC1, Atg07230; NPC2, At2g26870; NPC3, At3g03520; NPC4, At3g03530; NPC5, At3g03540; NPC6, At3g48610; RD29B, At5g52300; RAB18, AT5g66400; UBQ10, At4g05320; ABI1, At4g26080; ABI2, At5g57050; OST1, At4g33950; ERA1, At5g40280; and RCN1, At1g25490. Identification numbers for T-DNA mutants are as follows: npc1-1, SALK 027871; npc2-1, SALK 018011; npc3-1, SALK 065482; npc4-1, SALK_046713; npc5-1, SALK 045037; and npc6-1, SALK 077041.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Root Length of npc4-1 and Wild-Type Seedlings Grown in Media Deficient in Phosphorous, Potassium, or Nitrogen or Containing IAA.
Supplemental Figure 2. Phenotype of Six NPC Knockout Mutants and Wild-Type Seedlings in Response to ABA.
Supplemental Figure 3. Gene Expression of the NPCs in Root Cells and Guard Cells and Their Response to ABA.
Supplemental Figure 4. Drought Effect on DAG Content in the Wild Type and npc4-1.
Supplemental Figure 5. Water Loss from Detached Leaves and DAG and PG Effect on Stomatal Opening.
Supplemental Table 1. Primers for Real-Time PCR.
Supplemental References.
Supplementary Material
Acknowledgments
We thank Xiangqing Pan for hormone analysis, Sung Chul Bahn for help with gene cloning, Tara Wood for help on knockout mutant isolation, and Shivakumar Devaiah for help on water loss experiments, ABA-mediated stomatal movement, and knockout mutant isolation. The work was supported by grants from the USDA (2007-35318-18393) and the National Science Foundation (IOS-0818740) to X.W. Instrument acquisition and method development at the Kansas Lipidomics Research Center was supported by the National Science Foundation (EPS 0236913, MCB 0455318, and DBI 0521587), Kansas Technology Enterprise Corporation, K-IDeA Networks of Biomedical Research Excellence (INBRE) of the National Institutes of Health (P20RR16475), and Kansas State University.
References
- Apse M.P., Aharon G.S., Snedden W.A., Blumwald E. (1999). Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Arisz S.A., Testerink C., Munnik T. (2009). Plant PA signaling via diacylglycerol kinase. Biochim. Biophys. Acta 1791: 869–875 [DOI] [PubMed] [Google Scholar]
- Chen P.S., Toribara T.Y., Warner H. (1956). Microdetermination of phosphorus. Anal. Chem. 28: 1756–1758 [Google Scholar]
- Chinnusamy V., Schumaker K., Zhu J.K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J. Exp. Bot. 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J.K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12: 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Darwish E., Testerink C., Khalil M., El-Shihy O., Munnik T. (2009). Phospholipid signaling responses in salt stressed rice leaves. Plant Cell Physiol. 50: 986–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hartog M., Verhoef N., Munnik T. (2003). Nod factor and elicitiors activate different phospholipids signaling pathways in suspension-cultured alfalfa cells. Plant Physiol. 132: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R.R., Gampala S.S., Rock C.D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14 (suppl.): S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude N., Nakamura Y., Scheible W.R., Ohta H., Dormann P. (2008). Phospholipase C5 (NPC5) is involved in galactolipids accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 56: 28–39 [DOI] [PubMed] [Google Scholar]
- Gomez-Merino F.C., Arana-Ceballos F.A., Trejo-Tellez L.I., Skirycz A., Bearly C.A., Dormann P., Muller-Roeber B. (2005). Arabidopsis AtDGK7, the smallest member of plant diacylglycerol kinases (DGKs), displays unique biochemical features and saturates at low substrate concentration: The DGK inhibitor differentially affects AtDGK2 and AtDGK7 activity in vitro and alters plant growth and development. J. Biol. Chem. 280: 34888–34899 [DOI] [PubMed] [Google Scholar]
- Han X., Gross R. (2001). Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 295: 88–100 [DOI] [PubMed] [Google Scholar]
- Hirayama T., Ohto C., Mizoguchi T., Shinozaki K. (1995). A gene encoding a phosphatidylinositol-specific phospholipase C is induced by dehydration and salt stress in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 92: 3903–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Devaiah S.P., Bahn S.C., Thamasandra B.N., Li M., Welti R., Wang X. (2009). Phospholipase Dε and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J. 58: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Pan X., Welti R., Wang X. (2008). Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horman R., Pownall H.J. (1988). Transbilayer diffusion of phospholipids: Dependence on headgroup structure and acyl chain length. Biochim. Biophys. Acta 938: 155–166 [DOI] [PubMed] [Google Scholar]
- Hunt L., Mills L.N., Pical C., Leckie C.P., Aitken F.L., Kopa J., Muller-Roeber B., McAinsh M.R., Hetherington A.M., Gray J.E. (2003). Phospholipase C is required for the control of stomatal aperture by ABA. Plant J. 34: 47–55 [DOI] [PubMed] [Google Scholar]
- Icho T. (1988). Membrane-bound phosphatases in Escherichia coli: Sequence of the pgpA gene. J. Bacteriol. 170: 5110–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icho T., Raetz C.R. (1983). Multiple genes for membrane-bound phosphatases in Escherichia coli and their action on phospholipid precursors. J. Bacteriol. 153: 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao C.Y., Roth M., Welti R., Salic A. (2009). Metabolic labeling and direct imaging of choline phospholipids in vivo. Proc. Natl. Acad. Sci. USA 106: 15332–15337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen C.M., Brinkhorst-van der Swan D.L.C., Breekland A.E., Koornneef M. (1983). Induction of dormancy during seed development by endogenous abscisic acid: Studies on abscisic acid deficient genotype of Arabidopsis thaliana (L.) Heynh. Planta 157: 158–165 [DOI] [PubMed] [Google Scholar]
- Kasuga M., Liu Q., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1999). Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Lee Y., Assmann S.M. (1991). Diacylglycerols induce both ion pumping in patch-clamped guard-cell protoplasts and opening of intact stomata. Proc. Natl. Acad. Sci. USA 88: 2127–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Qin C., Welti R., Wang X. (2006). Double knockouts of phospholipases Dζ1 and Dζ2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol. 140: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Mishra G., Zhang W., Deng F., Zhao J., Wang X. (2006). A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312: 264–266 [DOI] [PubMed] [Google Scholar]
- Misson J., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T., Testerink C. (2009). Plant phospholipids signaling: In a nutshell. J. Lipid Res. 50: S260–S265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Awai K., Masuda T., Yoshioka Y., Takamiya K., Ohta H. (2005). A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 280: 7469–7476 [DOI] [PubMed] [Google Scholar]
- Nambara E., Marion-Poll A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Pan X., Welti R., Wang X. (2008). Simultaneous quantification of major phytohormones and related compounds in crude plant extracts by liquid chromatography-electrospray tandem mass spectrometry. Phytochemistry 69: 1773–1781 [DOI] [PubMed] [Google Scholar]
- Pandey S., Nelson D.C., Assmann S.M. (2009). Two novel GPCR-Type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of start proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Joanis B., Garnier T., Cole S. (1989). Gene cloning shows the alpha-toxin of Clostridium perfringens to contain both sphingomyelinase and lecithinase activities. Mol. Gen. Genet. 219: 453–460 [DOI] [PubMed] [Google Scholar]
- Sanchez J.P., Chua N.H. (2001). Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13: 1143–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Monitoring the expression of around 7,000 Arabidopsis genes under ABA treatments using a full length cDNA microarray. Funct. Integr. Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Sheshshayee M.S., Bindumadhava H., Ramesh R., Prasad T.G., Lakshminarayana M.R., Udayakumar M. (2005). Oxygen isotope enrichment (Δ18O) as a measure of time-averaged transpiration rate. J. Exp. Bot. 56: 3033–3039 [DOI] [PubMed] [Google Scholar]
- Stewart C.N., Jr, Via L.E. (1993). A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14: 748–758 [PubMed] [Google Scholar]
- Sugahara T., Takahashi T., Yamaya S., Ohsaka A. (1976). In vitro aggregation of platelets induced by alpha-toxin (phospholipaseC) of Clostridium perfringens. Jpn. J. Med. Sci. Biol. 29: 255–263 [PubMed] [Google Scholar]
- Sugahara T., Takahashi T., Yamaya S., Ohsaka A. (1977). Vascular permeability increase by α-toxin (phospholipase C) of Clostridium perfringens. Toxicon 15: 81–87 [DOI] [PubMed] [Google Scholar]
- Testerink C., Munnik T. (2005). Phosphatidic acid: A multifunctional stress signaling lipid in plants. Trends Plant Sci. 10: 368–375 [DOI] [PubMed] [Google Scholar]
- Titball R.W. (1993). Bacterial phospholipases C. Microbiol. Rev. 57: 347–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titball R.W., Leslie D.L., Harvey S., Kelly D.C. (1991). Haemolytic and sphingomyelinase activities of Clostidium perfringens alpha-toxin are dependent on a domain homologous to that of an enzyme from the human arachidonic acid pathway. Infect. Immun. 59: 1872–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T., Fujita M., Fujita Y., Yamaguchi-Shinozaki K., Shinozaki K. (2006a). Engineering drought tolerance in plants: discovering and tailoring genes unlock the future. Curr. Opin. Biotechnol. 17: 113–122 [DOI] [PubMed] [Google Scholar]
- Verslues P.E., Bray E.A. (2004). LWR1 and LWR2 are required for osmoregulation and osmotic adjustment in Arabidopsis. Plant Physiol. 136: 2831–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. (2001). Plant phospholipases. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 211–231 [DOI] [PubMed] [Google Scholar]
- Wang X., Devaiah S.P., Zhang W., Welti R. (2006). Signaling functions of phosphatidic acid. Prog. Lipid Research 45: 250–278 [DOI] [PubMed] [Google Scholar]
- Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H., Rajashekar C.B., Williams T.D., Wang X. (2002). Profiling membrane lipids in plant stress responses: role of phospholipase Dα in freezing- induced lipid changes in Arabidopsis. J. Biol. Chem. 277: 31994–32002 [DOI] [PubMed] [Google Scholar]
- Xiao S., Gao W., Chen Q.F., Chan S.W., Zheng S.X., Ma J., Wang M., Welti R., Chye M.L. (2010). Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell 22: 1463–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low- temperature, or high salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Costa A., Leonhardt N., Siegel R.S., Schroeder J.I. (2008). Isolation of a strong Arabidopsis guard cell promoter and its potential as a research tool. Plant Methods 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Qin C., Zhao J., Wang X. (2004). Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc. Natl. Acad. Sci. USA 101: 9508–9513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L., Munnik T. (2004). Osmotically induced cell swelling versus cell shrinking elicits specific changes in phospholipids signals in tobacco pollen tubes. Plant Physiol. 134: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.