Comparative analysis of homoeologous regions in soybean (Glycine max) and an orthologous region from common bean (Phaseolus vulgaris) reveal asymmetric evolution between homoeologous regions of soybean as evidenced by structural and gene expression changes. These results shed light on the fate of duplicated genes and chromosome segments after a whole-genome duplication in a paleopolyploid plant.
Abstract
Soybean (Glycine max) has undergone at least two rounds of polyploidization, resulting in a paleopolyploid genome that is a mosaic of homoeologous regions. To determine the structural and functional impact of these duplications, we sequenced two ~1-Mb homoeologous regions of soybean, Gm8 and Gm15, derived from the most recent ~13 million year duplication event and the orthologous region from common bean (Phaseolus vulgaris), Pv5. We observed inversions leading to major structural variation and a bias between the two chromosome segments as Gm15 experienced more gene movement (gene retention rate of 81% in Gm15 versus 91% in Gm8) and a nearly twofold increase in the deletion of long terminal repeat (LTR) retrotransposons via solo LTR formation. Functional analyses of Gm15 and Gm8 revealed decreases in gene expression and synonymous substitution rates for Gm15, for instance, a 38% increase in transcript levels from Gm8 relative to Gm15. Transcriptional divergence of homoeologs was found based on expression patterns among seven tissues and developmental stages. Our results indicate asymmetric evolution between homoeologous regions of soybean as evidenced by structural changes and expression variances of homoeologous genes.
INTRODUCTION
Polyploidy is widespread and recurrent in many plant species as their genomes hold relics of multiple duplication events (Cui et al., 2006). Polyploids are typically grouped into autopolyploids, from spontaneous genome duplication or fusion of unreduced (2n) gametes within a single species; or allopolyploids, from interspecific hybridization of two diverged genomes. In natural populations, allopolyploids are more prevalent than autopolyploids (Jackson and Chen, 2009), and many economically important crop species are polyploids, for instance, potato (Solanum tuberosum; autotetraploid), wheat (Triticum aestivum; allohexaploid), and cotton (Gossypium hirsutum; allotetraploid). Even plants that are cytogenetically diploid have undergone polyploid events during their evolution (paleopolyploid), including maize (Zea mays) and soybean (Glycine max; Shoemaker et al., 1996; Gale and Devos, 1998).
Polyploidization results in homoeologous chromosomes, chromosomal segments, and genes in duplicated genomes. In maize, homoeologous regions have undergone uneven contraction of genic and intergenic regions and expansion by the insertion of retrotransposons (Bruggmann et al., 2006). Collinearity between homoeologous regions of the maize genome is interrupted by inversions and translocations (Wei et al., 2007). In contrast with maize, sorghum (Sorghum bicolor) has experienced uniform expansion by retrotransposon insertions (Messing, 2009; Paterson et al., 2009). In wheat, at the grain hardness locus (Ha), deletions account for a majority of genome rearrangements (Chantret et al., 2005), and at the HMM Glutenin locus, indels (insertions and deletions) are the major contributors to structural differences between the A, B, and D homoeologs (Gu et al., 2006).
Soybean has had at least two putative genome-wide duplications, ~13 and ~59 million years ago (MYA) (Schlueter et al., 2004; Shoemaker et al., 2006; Schmutz et al., 2010). Polyploidy in soybean was seen at different levels: genetic mapping (Shoemaker et al., 1996), cytogenetic mapping (Pagel et al., 2004; Walling et al., 2006), and DNA sequence analyses (Schlueter et al., 2004). The ancestor of genus Glycine was coincident with a polyploidization event 5 to 10 MYA (Doyle and Egan, 2010). Furthermore, observations at molecular and chromosomal levels support the hypothesis that the recent tetraploid event was allopolyploidy and the putative ancestral diploid genomes of soybean are extinct (Gill et al., 2009). Soybean differs from some paleopolyploid genomes in that it has been diploidized (disomic pairing), but the level of genetic collinearity between duplicated segments is much higher than seen in maize, which had a roughly contemporaneous duplication event (Schlueter et al., 2006; Innes et al., 2008; Van et al., 2008).
Several aspects of polyploidization and diploidization are poorly understood. For example, how does diploidization shape a polyploid genome to restore bivalent chromosome behavior and plant fertility, and how does a newly formed polyploid cope with sudden changes in gene dosage across an entire genome? In yeast, it has been shown that diploidization and rescue of dosage may affect certain classes of genes (Scannell et al., 2007), and in plants, fractionation and selective elimination of genes in duplicated segments may play a role in diploidization (Paterson et al., 2006; Thomas et al., 2006).
Soybean is an attractive system for analysis of the effects of genome duplication on chromosome structure and gene fate as there have been two whole-genome duplication events at specific evolutionary time points, and relatives are available (e.g., Phaseolus vulgaris) that do not share the more recent duplication (Choi et al., 2004; Shoemaker et al., 2006). Previous work indicates differing levels of gene conservation between paralogous regions in soybean may be a result of being a product of either the earlier or later duplication event (Schlueter et al., 2006; Innes et al., 2008). Duplicated regions can be assigned to one of the polyploidy events by dating the divergence times of genes within the duplicated segments (Schlueter et al., 2004). Thus, soybean is an excellent system to dissect the effects of multiple polyploid events on the structure and function of a plant genome.
We sequenced two paralogous regions of ~1 Mb each from chromosomes 8 and 15 (hereafter referred to as Gm8 and Gm15). These duplicated segments are derived from the most recent duplication event of ~13 MYA. We also sequenced the orthologous region ~1 Mb from chromosome 5 (referred to as Pv5) from P. vulgaris, which diverged from soybean ~20 MYA (Lavin et al., 2005). Both Glycine and Phaseolus share the 59 million year whole-genome duplication event, but the more recent 13 million year event did not occur in Phaseolus (Choi et al., 2004; Shoemaker et al., 2006). Comparing these three homoeologous/orthologous regions reveals high levels of microcollinearity between the two regions with one region having experienced more extensive structural changes (e.g., lower gene retention rate and higher density of retrotransposons). In addition, we analyzed the transcription of a subset of the paralogous genes in soybean and found transcriptional bias to one homoeologous region and divergent transcription among different tissue types.
RESULTS
Isolation and Sequencing of Homoeologous BACs
We sequenced 20 BACs from two homoeologous loci surrounding a duplicated restriction fragment length polymorphic (RFLP) marker, pA711 (Pagel et al., 2004), to obtain an ~1-Mb window size for each homoeologous region on chromosome 8 (Gm8) and chromosome 15 (Gm15). This was done to determine the level of structural similarity between duplicated loci in the soybean genome and to evaluate the functional fate of duplicated genes in paralogous segments. In addition, we sequenced 10 BACs from P. vulgaris chromosome 5 (Pv5), orthologous to the sequenced soybean regions. All BACs were sequenced to phase II (unfinished sequence containing gaps, in which the order and relative orientation of the sequence contigs are known) or III (finished sequence; one contiguous piece of DNA) (see Supplemental Table 1 online), suitable for comparative analysis (Blakesley et al., 2004). Gm15 had no physical gaps inside the contig. There were two physical gaps in the Gm8 supercontig resulting in three contigs and one physical gap in Pv5 resulting in two contigs.
Inversions in Soybean Homoeologous Regions Are Common in Comparison to Phaseolus
To study the structural variation of these homoeologous/orthologous regions, we first confirmed the order of the contigs on Gm8 and determined the size of the gaps using fluorescence in situ hybridization (FISH) to pachytene chromosomes and DNA fibers of soybean (see Supplemental Figure 1A online). An inversion was observed between Gm8 and Gm15, involving nearly two-thirds of Gm8 and Gm15.
To further explore the structural variations between Gm8 and Gm15, we used Pv5 as an outgroup. FISH to pachytene chromosomes of Phaseolus was done to determine the order of Pv5 contigs (see Supplemental Figure 1B online). DNA sequences in this Pv5 1-Mb window were divided into two segments separated by one physical gap. For Gm15, the orientation of Gm15 segment1 was the same as the corresponding regions from Gm8 and orthologous Pv5. The first few genes on Gm15 segment2, from 168_Gm15 to 173_Gm15, had orthologous genes on Pv5 with the same orientation (Figure 1). On the 3′ end of Gm15, after gene 173, there were two blocks of genes (genes 139 to 149 and 107 to 129) that showed evidence of a complex rearrangement compared with Pv5. In Phasolus, there was an inversion involving both blocks of genes and then genes 107 to 129 were again inverted, relative to Gm15 (Figure 1). For Gm8, genes 107 to 129 have the same orientation as their Pv5 orthologs, but genes 139 to 149 and genes 160 to 173 between Gm8 and Pv5 have been inverted. A translocation also occurred between genes 160 to 173 when comparing Gm8 to Pv5. Thus, using the orthologous region Pv5, it seems that inversions are relatively common to both Gm8 and Gm15.
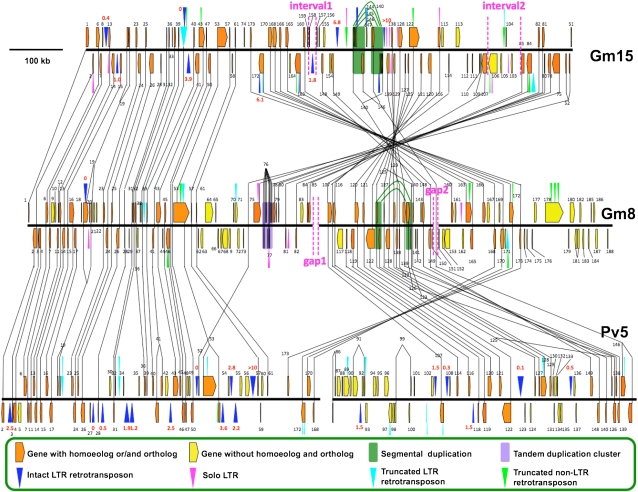
Figure 1.
Annotation Results of Chromosome Segments from Soybean (Gm8 and Gm15) and Phaseolus (Pv5).
The two soybean homoeologous regions, Gm8 and Gm15, and orthologous Phaseolus region, Pv5, are shown. Black bars are sequence contigs. Gm8 has two physical gaps, and Pv5 has one. Pentagons represent genes. Orange genes have homoeologs and/or orthologs (collinear genes), connected by black lines. Yellow genes do not have homoeologs and orthologs (noncollinear genes). Triangles represent transposons: blue triangles are intact LTR retrotranspsons, pink triangles are solo LTRs, cyan triangles are truncated LTR retrotransposons, and green triangles are non-LTR retrotransposons. Shaded boxes show the duplication events: green boxes show segmental duplications. The purple box shows a tandem duplication. Black numbers are the gene number from annotation results. Insertion times of intact LTR retrotransposons are shown in red. Regions between the two pink dotted lines are physical gaps in Gm8 and their corresponding intervals in Gm15.
We also analyzed the intervals on Gm15 (Gm15 interval 1 and Gm15 interval 2) that correspond to the physical gaps on Gm8 (Gm8 gap 2 and Gm8 gap1) (Figure 1; see Supplemental Figure 1B online). The lengths of Gm15 intervals 1 and 2 were 4.4× and 2.6× shorter than the corresponding Gm8 gaps as determined by fiber-FISH (see Supplemental Table 2 online). Thus, we deduced that there were either deletions in Gm15 or insertions in Gm8. When inspecting the Pv5 corresponding orthologous region of Gm8 gap2, a physical gap on Pv5 was found (see Supplemental Figure 1B online). The length of Gm15 interval1 is shorter compared with the common orthologous structures of physical gaps, Gm8 gap 2 and Pv5 gap (between Pv5 segments 1 and 2). It is likely that a deletion occurred on the ancestral Gm15 interval1, as opposed to two independent insertions on Gm8 and Pv5. A similar situation was found for Gm15 interval2 as there was another physical gap on the end of the Pv5 segment 2, and we were unable to find any overlapping BAC clones to extend the Pv5 segment 2. Thus, we determined that two deletions occurred on Gm15 and that the two intervals are remnants of the putative deletion events.
Gene Fractionation between Gm8 and Gm15
The three genomic regions were manually annotated for genes and repeats (Figure 1; additional details in Supplemental Data Sets 3 to 5 online). Overall, Gm8 had the most genes (130) followed by Pv5 (104) and then Gm15 (88); Gm15 has a lower gene density than Gm8 but both were higher than Pv5 (see Supplemental Figure 2 online).
We determined which soybean homoeologous region had a higher degree of gene fractionation (gene movement) relative to Phaseolus. Because there were three physical gaps in these three regions and in an attempt to be conservative in our approach, only genes flanked by orthologs across all three supercontigs were taken into account. Thus, genes between 2 to 61, 107 to 149, and 168 to 173 were included (Figure 1). We then defined a minimal set of genes present in this region in the ancestral chromosome of Gm8 and Gm15 as defined as genes present in Gm8 and/or Gm15 and also present in Pv5. Genes that have homoeologs without corresponding orthologs may derive from two possibilities: loss from Pv5 after the divergence of Phaseolus and soybean or insertion after the speciation but before the divergence of Gm8 and Gm15. In total, 12 homoeolog pairs belonged to this category, gene1 (on 5′ end of Gm8 and Gm15), between genes 75 to 85 (on 5′ end flanking region of Gm8 gap1), and genes 160 to 166 (on 3′ end flanking region of Gm8 gap2). All these genes are adjacent to physical gaps or the end of the contig; therefore, it was not possible to find an ortholog to anchor to Pv5, and these 12 genes were excluded. In addition, tandemly duplicated genes were considered as a single gene, except for tandem duplications found on at least two of Gm8, Gm15, or Pv5. Based on these criteria, there were 57 orthologs in soybean and Phaseolus of which 41 Pv5 orthologs can be found on both Gm15 and Gm8 and 11 on Gm8, Gm5, and Gm15 (see Supplemental Figure 3 online). Using the gene retention rate to estimate gene fractionation, we found 91% (52/57) and 81% (46/57) gene retention for Gm8-Pv5 and Gm15-Pv5, respectively. Thus, Gm15 had a lower degree of gene retention, or increased gene fractionation, than Gm8.
Since the total gene number in Gm15 was lower than Gm8 (as shown in Supplemental Figure 3 online) and the gene retention rate of Gm15-Pv5 was also lower than Gm8-Pv5, we suspected that there may be other features resulting in this type of organization. Therefore, we looked at Gm15 genes that were (1) noncollinear genes with no homoeolog or ortholog, (2) genes with homoeologs but no ortholog, (3) genes with no homoeolog but with orthologs, or (4) genes with both homoeologs and orthologs. Of the genes on Gm15, 22% were noncollinear genes and 78% were collinear genes (classes b, c, and d together). For Gm8 and Pv5, the percent of noncollinear genes was 25 and 30%, respectively. Thus, the percentages of noncollinear genes in the two soybean homoeologous regions were both lower than Pv5.
Asymmetric Accumulation of Retrotransposons and Solo Long Terminal Repeats
The transposon density in Gm15 was 1.6× higher than Gm8 due to accumulations of retrotransposons, mostly long terminal repeat (LTR) retrotransposons (see Table 1 and Supplemental Figures 2 and 4 online). The density of total LTR retotransposons in Gm15 was ~2× higher than in Gm8 (see Supplemental Figure 5 online), and the density of intact LTR retrotransposons was ~9× higher in Gm15 than in Gm8. Fragmented LTR retrotransposons (i.e., truncated elements and remnants as classified in Supplemental Table 1 online) were also biased to Gm15, ~1.5× higher than in Gm8.
Table 1.
Number of Transposons (LTR Retrotransposon, Non-LTR Retrotransposon, and DNA Transposon) in the Two Soybean Homoeologous Regions, Gm15 and Gm8
| Classes of Transposons | Gm8 | Gm15 |
| LTR retrotransposon | ||
| Intact elements | 1 | 6 |
| Intact elements without TSDs | 0 | 2 |
| Solo LTRs | 5 | 7 |
| Solo LTRs without TSDs | 0 | 2 |
| Truncated element | ||
| Both LTRs partially deleted with TSDs | 2 | 0 |
| Both LTRs partially deleted without TSDs | 3 | 4 |
| One LTR deleted, another partially deleted | 4 | 3 |
| Remnant | ||
| Remnants from insert | 7 | 12 |
| Remnants from LTR | 7 | 11 |
| Total | 29 | 47 |
| Non-LTR retrotransposon LINE | 15 | 17 |
| DNA transposon | 5 | 4 |
Increasing chromosome length from insertions of LTR retrotransposons can be counteracted by deletions of LTR retrotransposons via formation of solo LTRs (Devos et al., 2002). The density of solo LTRs was ~2× higher for Gm15 than Gm8 (see Supplemental Figure 5 online). Thus, similar to the bias seen for LTR retrotransposon density for Gm15, there was also a bias in the density of deletions (formation of solo LTRs) on Gm15. This observation raised the question of whether the higher density of solo LTRs in Gm15 was due to a higher frequency of unequal recombination or whether unequal recombination rates were similar in Gm8 and Gm15, but there were more intact LTR retrotransposons in Gm15 resulting in more solo LTRs. To more conservatively estimate unequal recombination rates, we calculated the number of solo LTRs with target site duplications (TSDs) to LTR retrotransposons with TSDs (Devos et al., 2002; Ma and Bennetzen, 2006). Similar rates of 1.67 and 1.17 were observed (by Fisher’s exact test) for Gm8 and Gm15, respectively; therefore, the bias of solo LTRs density to Gm15 was due to more LTR retrotransposons in Gm15 rather than a higher unequal recombination rate.
Given that the most recent polyploid event in soybean may have been an allopolyploid event (Gill et al., 2009), we hypothesized that the ancestral structures of Gm8 and Gm15 were the same following the divergence of the soybean diploid ancestors. After divergence, two deletions occurred on Gm15 corresponding to the two physical gaps on Gm8. The boundaries of Gm15 intervals 1 and 2 were carefully defined by BLASTn between Gm15 and Gm8 (see Supplemental Table 2 online). These two intervals were remnants of past deletion events. Therefore, we looked for structural features that could be used to infer possible reasons for the deletions. We found that transposon densities were higher for the two intervals on Gm15 than the rest of Gm15. Repeat element densities of both intervals were 2× higher than the Gm15 average (see Supplemental Figure 6 online).
We analyzed the conservation of transposons among the three regions, and only one conserved transposon was found in the two soybean homoeologous regions, between gene40 and gene41, and was not observed in Pv5. Both are polyprotein remnants from a copia-type LTR retrotransposon. Interestingly, the conserved LTR retrotransposon on Gm8 spanning from 175,404 to 187,745 bp has two nested transposon insertions in it, one intact LTR retrotransposon (180,783 to 185,763 bp) and one truncated non-LTR retrotransposon, LINE (186,214 to 187,133 bp). The corresponding conserved LTR retrotransposon on Gm15 has no nested transposons indicating that the nested insertion events on Gm8 occurred after the duplication event.
Timing of LTR Retrotransposon Insertion in Soybean and Phaseolus
The transposon density in Pv5 was similar to Gm15 (see Supplemental Figures 2 and 7 online). However, more intact LTR retrotransposons were found in Pv5, 17 compared with 8 in Gm15, a 1.7× increase in density. This reveals that in these regions, intact LTR retrotransposons were the major force leading to the expansion of Pv5 and resulting in a lower gene density compared with Gm8 and Gm15. The time of insertion for the intact LTR retrotransposons in these three regions were similar, most were <2 million years (61% for Gm8 and Gm15; 70% for Pv5) (see Supplemental Figure 8 online).
Solo LTRs Clusters on Gm15 and Gm8
Solo LTRs can be derived from unequal homologous recombination (Devos et al., 2002). We inspected the distribution of solo LTRs to determine if the occurrence of unequal recombination was even across the entire Gm8 and Gm15. There were nine solo LTRs on Gm15 (see Supplemental Table 1 online). However, there is an enrichment of solo LTRs (seven) in the inversion on Gm15 where the two putative deletions occurred; therefore, we examined the distribution of solo LTRs across this region using a nonoverlapping 20-kb sliding window. A cluster of solo LTRs was found (three solo LTRs within 551 to ~571 kb) (see Supplemental Figure 9 online), indicating that this may be a region with enhanced levels of unequal homologous recombination. Around the cluster, we found a segmental duplication encompassing 64 kb (red box in Supplemental Figure 9 online). Further annotation showed that four genes and one nested LTR retrotransposon structure were involved in this segmental duplication (see Supplemental Figure 10 online). The two nested LTR retrotransposon structures were 100% identical at the DNA sequence level, indicating that this nested structure was formed before the segmental duplication event and excluded the possible origin from a nonreciprocal translocation between Gm8 and Gm15.
Clustering of solo LTRs was also observed in Gm8 where three out of the five total solo LTRs were located within a 70-kb region (see Supplemental Figure 11 online). Structural analyses of this 70-kb segment also revealed tandem duplications resulting in nine copies of gene 76_Gm8, encoding an auxin-responsive protein. Therefore, both Gm8 and Gm15 had duplications/chromosomal rearrangements that were coincident with solo LTRs clusters.
Pseudogenes in Gm8, Gm15, and Pv5
Transposon insertions into genes can result in pseudogenes (Goldberg et al., 1983; Kumar and Bennetzen, 1999) as can other structural mutations. The percentages of pseudogenes derived from transposon insertions to all the genes in the regions were 11 and 17% for Gm8 and Gm15, respectively, and 13% for Pv5 (the same as the average of the two soybean homoeologs (13%). The percentages of pseudogenes to genes in these regions were similar for all three regions, 28 and 27%, for Gm8 and Gm15, respectively, and 29% for Pv5 (see Supplemental Table 3 online).
We next examined pseudogenization of genes with duplicates (homoeologs and locally duplicated genes) and genes without duplicates (single copy genes). Our hypothesis was that genes with homoeologs would be more likely to be pseudogenized as the other copy could complement the loss. We used genes 2 to 61, 75 to 85, 107 to 149, and 168 to 173 (Figure 1) for analysis of pseudogenization (Figure 1). The percentage of pseudogenes with duplicates to total pseudogenes was 56% for Gm8 and 61% for Gm15. Pseudogenes without duplicates to total pseudogenes was 44 and 39% for Gm8 and Gm15, respectively. However, in Pv5, the composition was quite different, 10% for pseudogenes with duplicates versus 90% for pseudogenes without duplicates. The pseudogenization rates for genes with duplicates (looking at only duplicated genes) were 21 and 18% for Gm8 and Gm15, respectively, similar to 20% in Pv5. Contrary to our hypothesis, however, duplicated genes were less likely to be pseudogenized than single copy genes for which the pseudogenization rate was 34 and 47% for Gm8 and Gm15, respectively, and 25% for Pv5.
We next examined the location of transposon insertions into genes (i.e., 5′ untranslated region [UTR], exon, intron, or 3′ UTR). For Gm8 and Gm15, ~67 and ~65% of the transposon insertions in genes were into introns, respectively, and ~33 and ~29% into exons for Gm8 and Gm15, respectively (see Table 2). Approximately 6% of insertions into Gm15 genes were in 3′ UTRs. In Pv5, the most frequent insertion site was also introns (~64% in introns, ~27% in exons, and ~9% in 3′ UTRs). No insertions into 5′ UTR were found in any of the three regions.
Table 2.
Analyses of Genes with Transposon Insertions in Gm8, Gm15, and Pv5
| Features of Genes with Transposon Insertions | Gm8 | Gm15 | Pv5 |
| Number of genes with transposon insertions | 13 | 11 | 10 |
| Percentage of genes with transposon insertions | 10.0% | 12.5% | 9.6% |
| Number of transposons inserted into a gene | |||
| Genes with one transposon | 9 | 5 | 9 |
| Genes with two transposons | 3 | 6 | 1 |
| Genes with three transposons | 1 | 0 | 0 |
| Classes of inserted transposons | |||
| LTR retrotransposons | 8 | 9 | 9 |
| Solo LTRs | 2 | 3 | 0 |
| Non-LTR retrotransposons | 8 | 5 | 2 |
| Insertion site in gene | |||
| 5′ End | 0 | 1 | 0 |
| Exon | 6 | 6 | 3 |
| Intron | 12 | 10 | 7 |
| 3′ End | 0 | 0 | 1 |
| Insertions into genes with homoeologs | |||
| One homoeolog with transposon insertion | 6 | 7 | NAa |
| Both homoeologs with transposon insertions | 2 | 2 | NAa |
| Insertions into genes with orthologs | |||
| Number of genes with orthologs that have insertions | 5 | 5 | 3 |
| One homoeolog and ortholog with insertions | 2 | 0 | 2 |
| Both homoeologs and ortholog with insertions | 1 | 1 | 1 |
Not applicable.
Among the three major categories of transposons (DNA transposons and non-LTR and LTR retrotransposons), ~63% of transposon insertions into genes were LTR retrotransposons for Gm8 and Gm15 combined compared with ~82% for Pv5. Interestingly, 14 solo LTRs were found in the two soybean homoeologs, of which six were found inside genes (two in exons and three in introns). Multiple retrotransposon insertions into individual genes were also observed. In the soybean homoeologs, ~58% were multiple insertions into a single gene. However, in Pv5, among the nine genes with transposon insertions, only one gene had more than one transposon insertion.
Synonymous Substitution Rates Are Biased to Gm8
Synonymous (Ks) and nonsynonymous (Ka) substitution rates were calculated to estimate evolutionary pressures on genes in these regions. After filtering out truncated genes, 47 pairwise comparisons for homoeologs from the two soybean regions were used to calculate Ks. The mean Ks was 0.1586 ± 0.0788 (red boxes in Figure 2, zones 1 and 4; see Supplemental Data Set 6 online for detail), and the Ks distribution between Gm8-Gm15 ranged from 0.04 to 0.38 (see Supplemental Figure 12 online). Previous studies have shown that a recent genome duplication in soybean occurring ~13 MYA was correlated with the distribution of Ks ranging from 0.03 to ~0.39 (Schlueter et al., 2004; Shoemaker et al., 2006; Schmutz et al., 2010). Because the distribution of Ks between Gm8 and Gm15 in this study located within the range of the distribution of Ks from the recent genome duplication in soybean, we concluded that the divergence time for Gm8 and Gm15, 13 million years, precedes the polyploidization event ~5 to 10 MYA (Doyle and Egan, 2010; Gill et al., 2009).
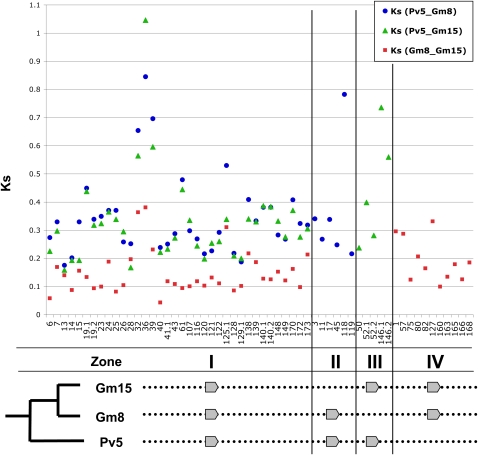
Figure 2.
Distribution of Ks Values of Genes in Both Soybean Homoeologs and Phaseolus.
Blue circles are Ks values from Pv5-Gm8; green triangles are Ks values from Pv5-Gm15; red boxes are Ks values from Gm8-Gm15. Different zones show the Ks from different homoeologous and orthologous counterparts. Zone I shows the Ks from the pairwise comparison of the two soybean homoeologs and one Phaseolus ortholog. Zone II shows the Ks from the sequence comparisons of Pv5 orthologs and Gm8 homoeologs. Zone III shows the Ks from Pv5 orthologs and Gm15 homoeolog comparisons. Zone IV shows the Ks from homoeologs between Gm8 and Gm15. Numbers below the plot are the annotated gene numbers. Ks value of gene114 (Pv5-Gm15) is ~2.8 due to extensive variation outside the conserved protein domain, and it is not shown here.
Orthologs from Pv5 were used as outgroups to compare the Ks of orthologous pairs between Gm8-Pv5 and Gm15-Pv5. After filtering truncated genes, 36 genes were available for this analysis (Figure 2, zone 1; see Supplemental Data Set 6 online for detail), and the mean Ks between Gm8-Pv5 and Gm15-Pv5 was 0.3471 ± 0.1446 and 0.3260 ± 0.1581, respectively. Comparing the two groups of Ks values, 75% of Ks values in Gm8-Pv5 were larger than their counterpart ones in Gm15-Pv5. A two-pair sample t test for these two groups of Ks values indicated that the Ks of genes between Gm8-Pv5 were significantly higher than between Gm15-Pv5 (P < 0.05).
Using the Ka/Ks ratio to evaluate evolutionary pressure (neutral versus positive versus negative) revealed that almost all genes have Ka/Ks < 1 (0.2327 ± 0.1106 for Gm8-Pv5 versus 0.2501 ± 0.1662 for Gm15-Pv5). This suggests that almost all the genes in the two soybean homoeologs were under purifying selection. The one exception was gene40 (1.026 for Gm15-Pv5). No statistically significant differences were found for either Ka or for Ka/Ks ratios across the two homoeologous regions.
Genes on Gm8 Are More Highly Expressed Than Their Gm15 Homoeologs
We assessed functional diversification of duplicated genes by analyzing transcriptional differences between soybean homoeologous genes by developing Sequenom MassArray assays for 105 exon-derived single nucleotide polymorphisms (SNPs). Following data quality control filtering, 71 assays representing 29 homoeologous gene pairs (see Supplemental Data Set 2 online) were of sufficient quality for downstream analyses. These assays were used to quantify the proportion of transcripts derived from the Gm8 and Gm15 gene copies for each gene pair.
The proportion of Gm8 and Gm15 transcript was quantified for the 71 assays over seven different soybean tissue types (large pod, small pod, flower, leaf, cotyledon, hypocotyls, and root). Multiple assays were analyzed for 18 of the 29 homoeologous gene pairs. The gene profiles of these assays were similar within each gene pair (see Supplemental Figure 13 online). Supplemental Figure 14 online compares the Gm8 transcript proportions among assays compared with the average Gm8 proportion for each gene. These data indicate that the assays cross-validate one another and give reliable data across tissue types. The only obvious example where the assays did not cross-validate was the gene23 leaf tissue. In this case, the three assays each detected vastly different proportions of the Gm8-Gm15 transcript (see Supplemental Figure 13 online). These differences may be the result of differential splicing between the homoeologous transcripts.
To identify the presence of transcriptional bias between homoeologous genes, the Gm8-Gm15 transcript proportions from cDNA templates were compared by t test with the Gm8-Gm15 proportions from DNA controls. Only 10 out of the 71 assays showed no significant homoeologous transcriptional bias for all seven tissue types. For the 29 gene pairs studied, the transcriptional biases tended to favor the Gm8 rather than Gm15 (Figure 3). The mean Gm8 transcript proportion was 0.582, and this average bias was similar across tissue types (maximum was small pod at 0.601 and minimum was leaf at 0.545).
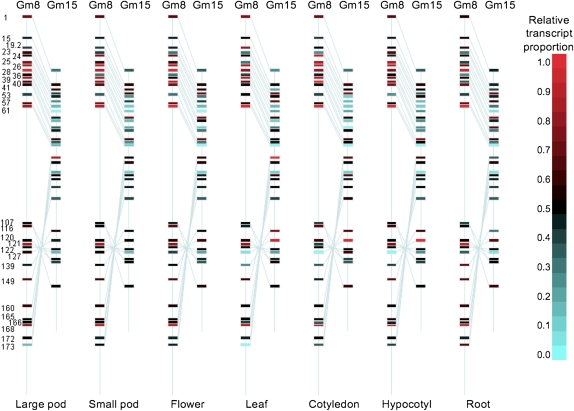
Figure 3.
Heat Maps of Relative Transcript Abundance of Homoeologous Genes on Gm8 and Gm15 for Seven Different Tissue Types.
The relative positions of 29 tested genes (top to bottom) are shown as colored rectangles along the Gm8 and Gm15 contigs. The numbers on the left are gene annotation results of the 29 tested genes. The transcription level of each gene is indicated as high (red) or low (blue) relative to the respective homoeologous counterpart. Homoeologs with approximately equal transcription levels are shown as black. The scale on the right indicates the coloration used to depict relative transcript proportions between homoeologous copies.
However, the amplitude of Gm8 transcript bias had regional differences along the Gm8 and Gm15 contigs (Figure 3). Gm8 transcript biases were considerably higher in the noninverted segment (first 15 gene pairs tested, average 0.633) than in the inverted segment (last 14 tested gene pairs, average 0.528). Interestingly, regional differences were variable across tissue types. Cotyledons exhibited the strongest regional differences. The average cotyledon Gm8 transcript proportion was 0.674 for the first 15 gene pairs but was reduced to 0.464 in the 14 gene pairs located within the inverted segment, whereas roots essentially had no regional bias.
Transcriptional Divergence of Homoeologous Genes and Correlation with Nucleotide Divergence
Homoeologous transcript proportions of Gm8 and Gm15 genes exhibited a range of similarities and differences across tissue types. Among all the sampled tissues, homoeolog transcript proportions were significantly correlated with one another (P < 0.05; see Supplemental Figure 15 online), except for root versus cotyledon (R2 = 0.116; P = 0.071), which exhibited a wider range of variation. Several genes showed substantial differences in homoeolog bias across different tissue types (see Supplemental Figure 13 online). The data have been condensed in Figure 4 and reordered from left to right according to genes exhibiting low to high rates of statistically significant variation across tissue types. Several of the genes showed evidence for consistently high transcriptional variation across tissue types over multiple assays, for instance, genes 139 and 24. We observed a wide range of divergences among the gene pairs on Gm8-Gm15 in terms of nucleotide sequence and transcription patterns. We postulated that gene pairs with high levels of nucleotide divergence (based on Ks, Ka, and Ka/Ks) may also exhibit increased levels of transcriptional divergence among tissues types.
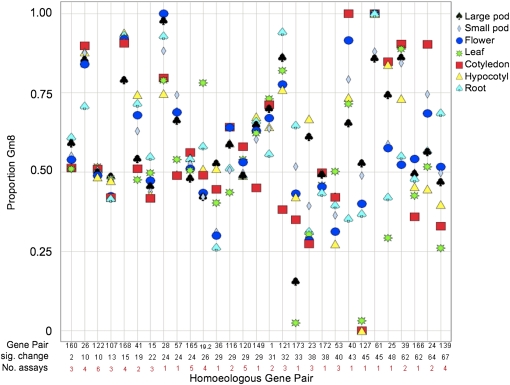
Figure 4.
Assessment of Tissue-Specific Transcriptional Divergence between Homoeolog Pairs.
The relative transcriptional proportions of the Gm8 homoeologs are shown for 29 tested gene pairs. The mean Gm8 proportion across two biological replicates is shown in the plot for each gene pair across seven tissue types. The percentage of statistically significant (P < 0.05) tissue × tissue differences detected in a factorial pairwise comparison of the homoeolog proportions among seven tissue types is shown below each gene pair number. There was a wide range (2 to 67%) of transcription variations among homoeolog pairs. The genes are ordered from lowest to highest percentage of tissue × tissue differences among the seven tissue types. Therefore, the gene pairs on the left infrequently exhibited significant tissue-specific transcriptional variation, and the genes on the right frequently exhibited significant tissue-specific transcriptional variation. The red number shown below each percentage indicates the number of informative assays for each homoeolog pair.
To address this question, we defined the level of transcriptional divergence between each homoeologous pair as the standard error of the Gm8-Gm15 transcript proportions among the seven tissue types. We also defined the degree of absolute expression bias between each homoeologous pair as the average of the absolute values of transcript proportions among tissues that have been subtracted by 0.5. Using these metrics, we found a moderate correlation between nonsynonymous divergence (Ka) and homoeolog transcriptional divergence among tissue types (R = 0.427; P = 0.023) (see Supplemental Figure 16 online). A moderate correlation was also observed between the ratio of nonsynonymous divergence to synonymous divergence (Ka/Ks value) and transcriptional divergence (R = 0.474; P = 0.011). For nonsynonymous divergence and absolute transcript bias, the correlation was weak (R = 0.386; P = 0.042). These data indicate that homoeolog pairs with increasing rates of nonsynonymous sequence divergence tend to exhibit more expression bias and less consistent relative expression abundances across tissue types.
Finally, we examined the top and bottom four genes (15% tails) with the least and most divergent transcription patterns based on ranking by transcriptional divergence using the standard error of the Gm8-Gm15 transcript proportions among all the seven tissues. The top 15% of genes with the most transcriptional variation included a tetratricopeptide repeat-containing protein, a Pro-rich family protein, a DNA binding bromodomain-containing protein, and an RNA binding protein. The four genes with the least divergent transcription included a VQ motif-containing protein, a type II homeodomain-leucine zipper protein, a protease lipid transfer protein family protein, and a protein of unknown function. None of the eight genes were colocalized on the two homoeologous regions nor was there an obvious association with predicted gene function.
DISCUSSION
The Origin of the Variation between the Two Soybean Homoeologous Regions
By comparing related species of different ploidies, genome variation between subgenomes in one organism can be traced back to specific evolutionary events (e.g., genome divergence or polyploidization). For instance, variation at the grain hardness locus (Ha) from the A, B, or D genome of hexaploid wheat was compared with their diploid and tetraploid progenitors (Chantret et al., 2005). However, all modern diploid Glycines (both annuals and perennials) are at 2n = 40 (Doyle and Egan, 2010), and the ancestral diploid genomes of soybean are hypothesized to be extinct (Gill et al., 2009; Doyle and Egan, 2010). As a result, it is impossible to trace the differences between Gm8 and Gm15 that were derived from the divergence of the ancestral genome versus those from the polyploidization/diploidization event.
The putative evolution of soybean includes the divergence of ancestral soybean genomes ~13 MYA and an allopolyploidization event ~5 to 10 MYA (Gill et al., 2009; Doyle and Egan, 2010; Schmutz et al., 2010). Therefore, the structural variation between Gm8 and Gm15 (inversions and deletions) could be derived either from the divergence event (~13 MYA), the polyploidization (5 to 10 MYA), or ensuing diploidization/fractionation. That is, the variation between the two soybean homoeologus regions might occur within the time frame of 13 MYA to the time point of forming the modern soybean genome. Likewise, the functional variation (transcriptional bias and Ks bias) between Gm8 and Gm15 may be due to divergence of the ancestral genomes, similar to the expression bias to certain subgenomes in cotton (Chaudhary et al., 2009). Our observations further support that soybean could be an allopolyploid, consistent with the hypothesis inferred from centromere analysis (Gill et al., 2009).
Genomic Variation at the Structural Level and Gene Level in Gm8, Gm15, and Pv5
When comparing the two soybean homoeologous regions with each other, inversions and deletions were observed. In addition, using the Phaseolus orthologous region to investigate genome variation in soybean revealed that inversions account for the majority of genome rearrangements and interruptions to gene collinearity. The role of inversions in causing genome rearrangement in legumes is consistent with maize, another paleopolyploid crop, where comparisons between maize and rice (Oryza sativa) showed that most genome rearrangements were caused by inversions (~74% of genome rearrangements) (Wei et al., 2007).
Gm15 has a lower gene retention rate (increased gene fractionation) than Gm8, but the percentage of the noncollinear gene is similar to Gm8. We speculate that the noncollinear genes on Gm15 may be derived from gene insertions, which seems more likely than losing both the homoeolog and ortholog independently from Gm8 and Pv5. Thus, considering both the gene retention rate (estimated from the minimal ancestral gene set) and gene gain (percentage of noncollinear genes), both Gm8 and Gm15 have similar rates of gene gain, but Gm15 has a lower gene retention rate. Combining these results with structural analyses, we conclude that there has been asymmetric evolution of Gm8 and Gm15, as Gm15 has a lower gene retention rate and undergone two deletion events.
The gene retention rate for homoeologs in soybean in Gm8 and Gm15, 91 and 81%, respectively, is higher than the 77% reported in another soybean homoeologous region (Innes et al., 2008) with a similar duplication time. Previous reports in maize, another paleopolyploid plant, found 32% collinearity of genes between homoeologous blocks that diverged ~5 MYA (Lai et al., 2004) and 20 to ~35% collinearity between blocks that diverged ~10 MYA (Bruggmann et al., 2006), both lower than Gm8 and Gm15, 75 and 78%, respectively. Therefore, at the gene level, homoeologous regions in soybean appear to be much more stable than the roughly contemporaneous duplicates in maize.
LTR Retrotransposon Activity in Gm8, Gm15, and Pv5
We observed only one conserved transposon between Gm8 and Gm15, indicating that conserved transposons are rare in the soybean homoeologous regions from the 13 million year event. There were no detectable conserved transposons in the orthologous regions of soybean and Phaseolus. One reason for this is that after the divergence from the common ancestor of soybean and Phaseolus, the common transposons derived from the common ancestor decayed due to mutation and/or multiple rounds of nested transposon insertions. These observations are consistent with a previous study of collinear regions between four cereals: rice, sorghum, barley (Hordeum vulgare), and wheat (Ramakrishna et al., 2002). Wheat and barley have similar divergence time frame of 10 to 14 MYA (Wolfe et al., 1989) as the two soybean homoeologous regions. In these comparisons, there were no detectable conserved transposons.
The density of intact LTR retrotransposons in Gm8 and Gm15, both euchromatic regions, was 0.94 and 8.8 LTR retrotransposons/Mb, respectively. For other soybean homoeologous regions, excluding pericentromeric regions, it was 7.8 LTR retrotransposons/Mb (Wawrzynski et al., 2008). In addition, Gm15 had higher densities of other classes of LTR retrotransposons than Gm8, for instance, fragmented LTR retrotransposons (truncated and remnants of LTR retrotransposons) and total LTR retrotransposons from all classes. Thus, variation of LTR retrotransposon density in soybean euchromatic regions is large and varies between homoeologous regions. We note that Gm15 had more extensive structural changes (deletions and lower gene retention rate) than Gm8, which suggests a correlation between structural changes and repeat DNA accumulation. It is possible that asymmetry of repeats and rearrangements might be more broadly found between duplicated segments in the soybean genome. Most of the intact LTR retrotransposons were inserted in the last 3 MYA in both species. This could indicate that LTR retrotransposons were more active 2 to 3 MYA or, alternatively, that intact LTR retrotransposons older than 3 million years have decayed. In the Phaseolus orthologous region, the intact LTR retrotransposon density was higher, 14.7 insertions/Mb. This indicates that within the same time frame, LTR retrotransposons may have been more active in Phaseolus than soybean or that LTR retrotransposons older than 3 million years have decayed more rapidly in soybean.
Solo LTRs clusters accompanied a segmental duplication on Gm15 and nine tandemly duplicated genes on Gm8. Thus, regions containing irregular structures shared one common feature, a higher density of solo LTRs. Solo LTRs are derived from unequal homologous recombination between LTR retrotransposons (Devos et al., 2002). The solo LTR clusters here reveal that many LTR retrotransposons were removed by unequal recombination events. Several models for segmental duplications involve transposable elements (Fiston-Lavier et al., 2007). Our data suggest that LTR retrotransposons are coincident with increased structural complexity and may have played a role in several rearrangements via some mechanisms, for instance, unequal recombination, that resulted in a higher density of solo LTRs and segmental/tandem duplications.
Heterogeneous transposon density was observed not only between homoeologous regions but also within individual homoeologous regions. More structural changes (deletions) were found in the two Gm15 intervals, where there was higher transposon density, suggesting a correlation between structural changes and transposable elements. It is known that transposons can induce major genome rearrangements, including deletions, duplications, inversions, macrotransposition, and reciprocal translocations (Gray, 2000; Caceres et al., 2001; Lonnig and Saedler, 2002; Huang and Dooner, 2008; Lee et al., 2008; Zhang et al., 2009). We hypothesize that Gm15, with a higher transposon density, underwent more extensive structural changes (i.e., higher density of LTR retrotransposon, solo LTRs, and lower gene retention rate) and that transposable elements were likely an active force in this remodeling of a homoeologous chromosome in soybean (see Supplemental Figure 17 online).
Pseudogenization Event in Gm8, Gm15, and Pv5
Pseudogenization rates can vary among species. Combining the pseudogene data from intergenic regions (genomic regions between annotated genes) and annotated genes (Benovoy and Drouin, 2006; Thibaud-Nissen et al., 2009; Zou et al., 2009), the percentage of pseudogenes in Arabidopsis thaliana and O. sativa ssp japonica was 16 and 42%, respectively. We observed similar rates of 28% in the two soybean homoeologs and 29% for Phaseolus, intermediate to Arabidopsis and rice. The extent of pseudogenization in these two Phaseoloid legume genomes was similar in these regions. Due to polyploidy, though, pseudogenes may be complemented by a paralog. We found that the percentage of pseudogenes with paralogs to all pseudogenes was 56 and 61% for Gm8 and Gm15, respectively. In rice, two studies focusing on either intergenic regions (Zou et al., 2009) or on annotated genes only (Thibaud-Nissen et al., 2009) found that the percentage of pseudogenes with paralogs was 28 and 75%, respectively. In Arabidopsis, the percentage of pseudogenes with paralogs was 47% (Zou et al., 2009). Therefore, our results from these two soybean homoeologous regions are concordant with previous studies. Pseudogenization rates for duplicated genes in the two soybean homoeologs and in Phaseolus were similar, ~20%, but pseudogenization rates of single copy genes were higher, ~38 and 25% for soybean and Phaseolus, respectively. Explanations for this observation may include that the two homoeologous counterparts have transcriptionally diverged temporally and/or spatially, resulting in complementary expression patterns. Thus, pseudogenization of either of the homoeologs could be deleterious, leading to selection against pseudogenization of these transcriptionally complementary homoeologs. This is concordant with our observation of transcriptional variation across tissues. Another possibility is that the duplicated copies of pseudogenized homoeologs have been degraded beyond recognition or that retrotransposition resulting in processed pseudogenes (PΨgs) may be a common mechanism for generating pseudogenized single-copy genes in soybean.
Transposon insertions into introns was ~2× higher than into exons, similar to results from the adh1 locus (Alcohol Dehydrogenase 1) region of rice, maize, and sorghum (Tikhonov et al., 1999; Ammiraju et al., 2008). The lack of insertions into coding regions supports the model of deleterious insertions: mutations resulting from insertions into exons or regulatory sequences are deleterious to an individual (Montgomery et al., 1987). However, transposon insertions into introns could still be deleterious as not every intron is without function. Introns can encode microRNAs (Ying and Lin, 2009), contain RNA splicing regulatory motifs (Ponthier et al., 2006), or promote intron-mediated enhancement (Mascarenhas et al., 1990; Rose et al., 2008). Introns made larger by transposon insertions could affect pre-mRNA splicing and impact plant fitness, as intron size is critical for the splicing efficiency of pre-mRNA (Klinz and Gallwitz, 1985). Another possibility is that different retotransposon families have insertion sites biases for exons or introns (Goldberg et al., 1983; Kumar and Bennetzen, 1999). The observed bias of more transposons in introns than exons could be due to the activity of specific retrotransposon families favoring intron insertion sites.
Asymmetric Evolution at Synonymous Sites between Soybean Homoeologus Regions
In addition to asymmetric gene retention/loss, synonymous divergence (Ks) analyses also revealed asymmetric sequence divergence between paralogs from Gm8 and Gm15. This is congruent with previous observations between duplicated segments in rice (Wang et al., 2005). However, in our study, the size of duplicated blocks and the degree of synteny are greater than the study in rice and the duplication more recent.
Based on neutral theory, the mutation rate is equal to Ks (Kimura, 1968; Chamary et al., 2006; Duret, 2009). Ks values in Gm8 were significantly larger than Gm15, indicating a higher mutation rate for Gm8. The structural features of these two soybean homoeologous regions are very different. For instance, Gm15 has undergone more extensive structural changes and has a higher transposon density. These two factors are known to correlate with mutation rates (Chiaromonte et al., 2001; Hardison et al., 2003) and may have contributed to the asymmetric evolution between Gm8 and Gm15. Nucleotide divergence and repeat elements were correlated in noncoding regions of mammalian genomes, suggesting that some regions are more susceptible to mutation and repeat element insertions (Chiaromonte et al., 2001; Hardison et al., 2003). By contrast, in soybean, Gm15 with more repeat DNA insertions accumulated fewer mutations at synonymous sites in genic regions. Thus, there may be different mechanisms that determine the effect of repeat elements on substitution rates in soybean genic regions.
In addition to DNA rearrangement and transposon density, many factors may contribute to variation in mutation rates across a genome (Ellegren et al., 2003; Wolfe and Li, 2003; Baer et al., 2007). These factors include local recombination rate, gene density, functionally related genes, transcriptional coupled repair (TCR), pattern of gene expression, base composition (GC content and CpG dinucleotide), distance to telomere, and chromatin structure (Hardison et al., 2003; Navarro and Barton, 2003; Chuang and Li, 2004; Arndt et al., 2005; Hellmann et al., 2005; Duret and Arndt, 2008; Tian et al., 2008; Berglund et al., 2009; Duret, 2009). For soybean, a very notable cause is ancestral polymorphisms that may have been present in the ancestral genomes contributing Gm15 and Gm8. The present-day soybean genome was likely a merging of two previously diverged genomes via an allopolyploid event (Gill et al., 2009). Thus, the higher synonymous divergence found in Gm8 may be traced to ancestral polymorphisms. Therefore, the observed asymmetric evolution could be a combination of both ancestral polymorphisms and structural changes following the divergence event.
Asymmetric Evolution of Individual Homoeologous Pairs
The two soybean homoeologous regions were generally under negative selection (Ka/Ks <1), except for gene 40_Gm15 (VQ motif-containing protein). Interestingly, its homoeolog, 40_Gm8, was still under negative selection. The Ks values were similar between 40_Gm8-Pv5 and 40_Gm15-Pv5, but the Ka values were different, resulting in a higher Ka/Ks in 40_Gm8-Pv5, 1.026 versus 0.5 in 40_Gm15-Pv5. For nonsynonymous sites, mutation rate and selection constraints contribute to substitution rate. Based on neutral theory that the mutation rate is equal to Ks (Kimura, 1968; Chamary et al., 2006; Duret, 2009), mutation rates for 40_Gm8 and 40_Gm15 were similar. However, they experienced different evolutionary constraints, resulting in different nonsynonymous substitution rates (Ka) and, therefore, different Ka/Ks values. Based on these observations, a small percentage of soybean homoeologs may be under different evolutionary constraints.
Gene expression as measured by transcript abundance was biased toward the Gm8 homoeolog (40_Gm8), 70% for Gm8 versus 30% for Gm15. The variation of expression patterns among the seven tissues was large (e.g., 40_Gm15 was silenced in the cotyledon, suggesting highly transcriptional divergence between Gm8_40 and Gm15_40). Surprisingly, although their evolutionary fates appear different, their divergent value of 0.043 (synonymous substitution rate, Ks) was one of the smallest among all the homoeologous genes in these regions. In contrast with homoeolog pair gene40, some homoeolog pairs showed a relatively slow rate of transcriptional divergence within the same time frame. For instance, homoeolog pair gene122 had little transcriptional divergence and no strong transcript bias or variation of expression patterns among the seven tissues. Thus, within the same evolution time frame of ~13 million years, a few homoeolog pairs diverged rapidly.
Expression Biases Are Correlated with Sequence Divergence
Our results indicate that expression divergence increases over time after duplication, similar to studies on human and yeast (Gu et al., 2002; Li et al., 2005). When comparing both homoeologous regions, nonsynonymous changes were correlated with biased expression and homoeolog transcriptional divergence; that is, more amino acid changes were coupled with a higher degree of transcriptional bias to one homoeolog and a higher degree of homoeolog transcriptional divergence among different tissue types and developmental stages.
Asymmetric expression pattern was also observed between paralogs in Arabidopsis and rice (Blanc and Wolfe, 2004; Ganko et al., 2007; Throude et al., 2009) and between homoeologs in cotton (Chaudhary et al., 2009). Our expression results showed evidence for transcriptional divergence among different tissue types of some homoeolog pairs, consistent with findings in other plant species. Furthermore, our results show evidence of regional transcriptional bias between clusters of genes on one homoeologous region versus the other.
Several factors can affect gene expression, including gene GC content of the gene (Kudla et al., 2006), intron size (Ren et al., 2006), intron motifs (Rose et al., 2008), methylated transposon distribution (Hollister and Gaut, 2009), and chromosomal rearrangements (Marques-Bonet et al., 2004). Most notable for our study is the negative correlation between transcript level and density of recent transposon insertions that may be methylated (Hollister and Gaut, 2009). Gm15 had a greater total transposon density and more recent LTR retrotransposon insertions (more intact LTR retrotransposons) than Gm8, and we observed that Gm8 homoeologs were more highly expressed. Thus, transposons may contribute to the observed transcriptional variation between the two soybean homoeologous regions.
It is possible that the differences in homoeolog expression levels are influenced by homoeolog ancestry. The soybean genome, putatively an allopolyploid (Gill et al., 2009), may be the combination of two diverged ancestral genomes. As a result, the expression differences may be partially attributable to the differential transcription states of the donor parents. Thus, it is possible that the biased gene expression observed in the present-day soybean genome results from a combination of factors: inherited differences from previously diverged ancestral genomes and/or postdivergence structural changes between homoeologs (i.e., retrotransposon insertions, deletions, and inversions).
Conclusions
The soybean genome is a mosaic structure consisting of homoeologous segments (Shoemaker et al., 1996; Walling et al., 2006). Our observations of asymmetric evolution between Gm8 and Gm15 may be representative of other homoeologous segments of soybean genome. We hypothesize that after genome divergence ~13 MYA, the progenitor genomes merged to form the ancestral tetraploid soybean genome ~5 to 10 MYA and that transposon activity, fractionation, and other structural processes shuffled the genome to form the present-day soybean genome. Meanwhile, because of gradual accumulation of local structural difference around one copy relative to its homoeologous counterpart, expression patterns between homoeologous pairs changed, resulting in a subset of genes that show transcriptional bias or divergence among different tissue types. It remains to be seen what role these processes played in the domestication and breeding of soybean into the crop plant that it is today.
METHODS
BAC Selection
Two BACs (Gm_UMb001_24D13 and Gm_UMb001_05F05) derived from soybean (Glycine max) homoeologous regions centered around the duplicated RFLP locus, pA711 (AQ842034), were sequenced in a previous study (Schlueter et al., 2007). These BACs were blasted (Altschul et al., 1990) against the Williams 82 BAC end sequence database to begin chromosome walking in the reference genotype. As there were physical gaps in the homoeologous region located on Gm8, multiple approaches were used to screen libraries and choose overlapping BACs from two BAC libraries, GmWBb and GmWBc. We took advantage of sequence resources (ESTs and BAC end sequences), genetic mapping (sequence tag site derived from SNPs), and hybridization-based markers (overgos) (http://www.soymap.org/) to extend BAC contigs. Information from soybean paralogs and Phaseolus orthologs information derived from sequenced BACs were also applied to extend the contig (see Supplemental Data Set 1 online). Positive clones were scored using a custom program, ComboScreenJ (http://pbr.agry.purdue.edu/coMboscreenj_doc/).
For Phaseolus vulgaris, RFLP marker A711 (pA711) (AQ842034) was used to screen the PVGBa BAC library to find BACs orthologous to the soybean regions. We used similar approaches (above) to extend the orthologous contig (see Supplemental Data Set 1 online).
BAC Sequencing
DNA from selected BACs was extracted using the Qiagen Large Construct kit and sheared with a Hydroshear (Genemachines) to average size of 5 to 9 kb. Sheared fragments were blunt-ended with mung bean (Vigna radiata) nuclease (New England Biolabs), dephosphorylated with shrimp alkaline phosphatase (USB), and “A” tails added by incubation with Taq DNA polymerase in the presence of deoxynucleotide triphosphates. These fragments were inserted into the vector pCR4TOPO using the TA cloning kit (Invitrogen) following the manufacturer’s instructions. The resulting DNA was electroporated into DH10B electroMAX cells (Invitrogen). Clones were picked using a Qpix colony picker (Genetix) into 384-well culture trays (Genetix) filled with 60 μL terrific broth culture medium plus 8% glycerol. After overnight growth (16 h), cultures were frozen at −80°C until needed. R.E.A.L. Prep 96 Plasmid kits (Qiagen) were used to prepare DNA minipreps from 1.3-mL cultures grown in deep 96-well plates for 16 h at 37°C shaking at 300 rpm. DNA was resuspended in 50 μL water, with 4 μL used for sequencing reactions. Clones were sequenced from both directions using Big Dye Terminator chemistry (Applied Biosystem) and run on an ABI3730 capillary sequencer after terminator cleanup using Squeeky-Clean (Bio-Rad) 96-well column plates. Base calling and quality assessment were done using PHRED (Ewing et al., 1998), assembled by PHRAP, and edited with CONSED (Gordon et al., 1998). Some gaps were filled by a combination of primer walking and shotgun sequencing of subclones using the EZ-Tn5 <TET-1> insertion kit (Epicentre Biotechnologies) with extremes at both sides of the sequencing gaps. Final error rate was estimated using CONSED.
FISH
FISH mapping of BACs on pachytene chromosomes and extended DNA fibers was performed as described previously (Walling et al., 2006). For soybean pachytene FISH (see Supplemental Figure 1A online), BACs were labeled with digoxigenin-UTP (DIG), Biotin-UTP (Roche), or fluorescein isothiocyanate (FITC; Molecular Probes). Labeling system I is as follows: GmWBb46B19 and GmWBb71E11 were labeled with DIG (shown as red), GmWBb77J08 and GmWBc63M16 were labeled with biotin (shown as blue), and GmWBc29O09 was labeled with FITC (shown as green). Labeling system II is as follows: GmWBb46B19 and GmWBb71E11 were labeled with digoxigenin (red), GmWBb77J08 and GmWBc63M16 with FITC (green), and GmWBc29O09 with digoxigenin (red). For P. vulgaris, PVGBa84A11, PVGBa34D21, PVGBa90L08, and PVGBa110H03 were used to determine the direction and orientation of the clones relative to soybean (see Supplemental Figure 1B online). Again, two labeling systems were used (labeling system I, PVGBa34D21 was labeled with digoxigenin, PVGBa90L08 with biotin, and PVGBa110H03 with FITC; for labeling system II, PVGBa84A11 was labeled with biotin, PVGBa34D21 with digoxigenin, and PVGBa110H03 with FITC).
Comparative Sequence Analysis
The two large sequence contigs from soybean were compared using BLASTn with the E-value 1e-4. The Artemis Comparison Tool (Carver et al., 2005) was used to visualize conserved genomic regions and identify structural homoeology.
Transposon Sequence Analysis
To identify LTR retrotransposons, we used de novo and similarity searches. For de novo searches, assembled contigs were screened for putative LTR retrotransposons using LTR_STRUC (McCarthy and McDonald, 2003). Candidate LTR retrotransposons were manually checked for the following features: (1) terminal TG/CA inverted repeat in the LTRs, (2) primer binding site, (3) polypurine tract, (4) and short TSDs. LTR retrotransposons with all these features were defined as intact retrotransposons. To find LTR transposons missed by LTR-STRUC, the contigs were searched manually for the above characteristics.
For similarity searches, an intact LTR retrotransposon database was constructed from all the intact LTR retrotransposons found. To make a more comprehensive database, additional intact LTR retrotransposons were added using the same methodology to search other sequenced BACs sequenced from the SoyMap Project (http://www.soymap.org). A combination of nucleotide and protein similarity searches was used to annotate sequence contigs. For protein similarity, BLASTx searches of the contigs against the National Center for Biotechnology Information (NCBI) protein nonredundant database with cutoff value of 1e-10 were used. For nucleotide similarity, cross_match (http://www.phrap.org) was used to screen the genomic regions with the library described above. Solo LTRs were defined as genomic fragments that aligned to the 5′ and 3′ends of one LTR but did not extend to the internal region of the intact LTR retrotransposon. Sequences flanking solo LTRs were analyzed for TSDs. To identify non-LTR retrotransposons and DNA transposons, BLASTx was used to compare the contigs against the NCBI protein nonredundant database with a cutoff value of 1e-10.
Insertion times of the LTR retrotransposons were estimated by aligning the 5′ and 3′ LTR sequences with MUSCLE (Edgar, 2004) followed by manual inspection, and sequence divergence (estimated number of substitutions per site between sequences) was calculated with MEGA4.0 (Tamura et al., 2007) using the Kimura 2 parameter model. The time of LTR retrotransposon insertion was estimated using the methodology and synonymous substitution rate (1.3 × 10−8) reported earlier (Ma and Bennetzen, 2004).
Gene Annotation and Sequence Divergence Analysis
Three gene prediction methodologies were used for gene prediction. First, ab initio gene prediction methods included FGENESH (Salamov and Solovyev, 2000), GENSCAN (Burge and Karlin, 1997), and GeneMARK (Lomsadze et al., 2005) with dicot (Medicago trunculata and Arabidopsis thaliana) models. Second, gene prediction methods with phylogenic evidence included FGENESH-2 (http://linux1.softberry.com/berry.phtml) using the homoeologous regions from soybean. Finally, gene prediction with mRNA evidence was done using GenomeThreader (Gremme et al., 2005) with cDNA from plantGDB (http://www.plantgdb.org/) and ESTs from GenBank as the mRNA resource. In addition to gene prediction programs, homology searches using BLASTx against the nonredundant protein database in NCBI were also used for gene prediction. We used Apollo (Lewis et al., 2002) to integrate all prediction methods into a visualization interface. Repeatmasker (http://www.repeatmasker.org/) was used to mask transposons (cutoff 250) using the custom database described above and uploaded into Apollo. These data were manually inspected and integrated to provide a gene and repeat annotation.
Coding sequences of predicted genes were confirmed by ORF finder (http://www.geneinfinity.org/sms_orffinder.html), and amino acid sequences were aligned with MUSCLE (Edgar, 2004) followed by manual inspection. Sequence divergence was calculated with the codeml program in PAML (Yang, 1997) implemented in PAL2NAL (http://www.bork.eMbl.de/pal2nal/) (Suyama et al., 2006).
Mutations can result in disrupted ORFs, including premature stop codons and frame shifts (by single base mutation or larger indels), and can lead to nonfunctional proteins or pseudogenization (Zheng and Gerstein, 2007). Pseudogenes were classified into two categories in this study: pseudogenes with transposon insertions (transposon-induced pseudogenes) and pseudogenes without transposon insertions (nontransposon-induced pseudogenes). If a gene had transposon insertions into exons or the 5′/3′ UTRs, but not into introns, it was classified as a pseudogene with transposon insertions. The deduced protein sequences of the remaining genes were then used to query the NCBI nonredundant protein database using BLASTp, and the alignments were manually inspected. Furthermore, protein sequences from genes having paralogs/orthologs were aligned using ClustalW2 (Larkin et al., 2007). If the alignment was disrupted, the genomic DNA sequence around the disrupted region was inspected to determine if it was derived from either a protein truncation or a missing predicted exon(s). If the truncation covered more than 15% of the gene, it was classified as a pseudogene without transposon insertion.
Plant Growth Conditions and Tissue Sampling for Gene Expression Analyses
The G. max cultivar “Williams 82” was grown in a greenhouse on Metromix soil with 16 h light/8 h dark, daytime temperature ~30°C, and night temperature ~22°C with daily watering. Seven tissue types (large pod, small pod, flower, leaf, cotyledon, hypocotyl, and root) were collected for total RNA extraction. Cotyledons and hypocotyls were collected 3 d after emergence (DAE) and leaves, roots, and flowers were collected 60 DAE. Large pod (longer than 4 cm) and small pod (smaller than 2 cm) were collected 81 DAE. Samples were taken from at least six plants and flash-frozen in liquid nitrogen followed by the total RNA extraction.
For total RNA isolation, samples from four individual plants were combined and ground in liquid nitrogen with Tri reagent (Molecular Research Center). To avoid DNA contamination, extracted total RNA was treated with DNaseI (Promega) and confirmed by RT-PCR using tubulin-specific primers. Two biological replicates of each tissue type were sampled. Two independent cDNA synthesis technical replicates were generated for each biological replicate, and cDNA was synthesized from RNA using Superscript (Invitrogen) according to the manufacturer's instructions. Additionally, DNA from leaves was isolated from Williams 82 from two leaves and two biological replicates using a standard CTAB extraction protocol. Thus, four biological replicates of DNA templates were analyzed.
Gene Expression Analyses
The Sequenom MassArray system has been previously used for high-throughput quantification of transcript ratios between maize (Zea mays) alleles (Springer and Stupar, 2007) and cotton (Gossypium hirsutum) homoeologous genes (Chaudhary et al., 2009). In this study, we adopted this technology to quantify the transcript ratios of homoeologous soybean genes residing on contigs Gm8 and Gm15.
SNPs were identified between the coding regions of Gm8 and Gm15 homoeologs using phytozome and BLAST2 sequence alignments (http://www.phytozome.net/). These SNPs were used to distinguish the Gm8 and Gm15 homoeologous gene copies in downstream transcriptional analyses. BLAST searches of the soybean genome were performed using the phytozome soybean database to identify SNP sequences specific to the Gm8 and Gm15 regions, such that additional gene copies elsewhere in the genome would not interfere with transcriptional analyses of the homoeologous copies. Six hundred and nineteen SNPs were submitted for Sequenom MassArray assay design at the University of Minnesota BioMedical Genomics Center. Sequenom MassArray assays for 375 SNPs were identified, and 105 SNP assays were selected for transcriptional analyses.
To quantify homoeologous transcript ratios, PCR and extension PCR reactions on the cDNA and DNA control templates were performed according to the manufacturer's specifications (Sequenom). All templates (including each pair of cDNA technical replicates) were run through the Sequenom PCR process twice, resulting in four technical replicates for each cDNA biological replicate. Mass spectrometry quantification of homoeologous gene ratios was performed at the University of Minnesota Genotyping Facility. The resulting data were run through a quality control pipeline to remove unusable data. Assays identified as “Bad Spectra,” having a frequency of uncertainty >0.2, or an unused extension primer frequency >0.5 were removed (Chaudhary et al., 2009). Monomorphic data points (values showing 1:0 transcript ratios between the Gm8 and Gm15 gene copies) were removed when in obvious disagreement with the other technical replicates. Assays with high rates of questionable data points based on these filtering criteria were removed from downstream analyses. The data from the DNA control templates were then used to identify the most quantitatively unbiased assays. Assays with discarded data for two or more of the four DNA biological replicates were removed from further analyses. The technical replicate values were averaged for each DNA template biological replicate to estimate the assay biases. A perfectly unbiased assay should generate proportions of 0.5 for both Gm8 and Gm15 from the DNA control templates. Therefore, assays with mean Gm8 and Gm15 DNA proportions >0.6 or <0.4 were removed from further analyses. Following these data filtration steps, 71 assays representing 29 homoeologous gene pairs remained in the analysis.
Two-tailed homoscedastic t tests between the DNA control and cDNA data determined the statistical significance of transcriptional biases between the homoeologous genes for each of the 497 assay × tissue type combinations (see Supplemental Data Set 2 online). Significance thresholds were set at P value < 0.05. We also applied two-tailed homoscedastic t tests (P < 0.05) to identify significantly altered homoeolog transcript proportions between tissue types. Factorial pairwise comparisons of the seven tissue types result in a total of 21 tissue × tissue comparisons per assay. Additionally, most gene pairs are represented by more than one assay; therefore, the number of tissue × tissue comparisons varies between 21 and 126 per gene pair.
For graphical and data display purposes, the Gm8-Gm15 proportions generated from the DNA controls were used to standardize the cDNA transcript proportions for each assay. Each Gm8 and Gm15 homoeolog should be present at equal proportions (0.5) in the DNA control templates. To correct for assay biases, 0.5 was subtracted from the measured DNA proportions to generate a correction factor for the Gm8 and Gm15 homoeologs in each assay. This correction factor was subsequently subtracted from each cDNA proportion measurement, thus correcting for biases inherent to the assay. The correction factor was not applied to monoallelic transcript measurements (proportions 0.0 or 1.0); corrected measurements that fell below 0.0 or exceeded 1.0 were set to the respective monoallelic levels. The corrected cDNA technical replicate values were averaged for each biological replicate. The mean transcript proportions and standard deviations among biological replicates were computed for each assay (see Supplemental Data Set 2 online).
For graphing and summary purposes, transcript data were averaged among assays for gene pairs with multiple assays. Microsoft Excel, PowerPoint, and Spotfire DecisionSite 9.1.1 software applications were used to generate figures and tables of the homoeologous transcription data.
Accession Numbers
Sequence data for the sequenced BACs from this article can be found in the GenBank/EMBL databases under the accession numbers in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. FISH Mapping to Confirm the Orientation of Sequence Contigs.
Supplemental Figure 2. Gene Density (Genes/Mb) and Transposon Density (Transposons/Mb) in the Two Soybean Homoeologous Regions and the Orthologous Phaseolus Region.
Supplemental Figure 3. Number of Collinear/Noncollinear Genes within Regions Anchored by Orthologs across Gm8, Gm15, and Pv5.
Supplemental Figure 4. Number of Different Categories of Transposons in the Two Soybean Homoeologous Regions.
Supplemental Figure 5. Insertion Density (Transposons/Mb) of Different Categories of Transposons in the Two Soybean Homoeologs.
Supplemental Figure 6. Transposon Density in Gm15 Intervals 1 and 2.
Supplemental Figure 7. Number and Density of Transposons in the Orthologous Phaseolus Region.
Supplemental Figure 8. Insertion Time (MYA) of Intact LTR Retrotransposons in the Two Soybean Homoeologous Regions and the Orthologous Phaseolus Region.
Supplemental Figure 9. Distribution of Transposon and Unequal Recombination Rate in Gm15 43l to ~749-kb Region.
Supplemental Figure 10. Segmental Duplication in Gm15 (Red Box in Supplemental Figure 9).
Supplemental Figure 11. Successive Tandem Duplication Event Observed in Gm8.
Supplemental Figure 12. The Distribution of Divergence Rates of Homoeologous Genes in Gm8 and Gm15.
Supplemental Figure 13. Gm8-Gm15 Transcriptional Proportion Profiles for 29 Homoeologous Gene Pairs across Seven Different Tissue Types.
Supplemental Figure 14. Validation of the Assays Used for Gm8 and Gm15 Transcription Comparisons.
Supplemental Figure 15. Homoeolog Expression Differences among Tissue Types.
Supplemental Figure 16. Correlation between Sequence Divergence and Transcript Divergence.
Supplemental Figure 17. A Hypothesis for the Evolutionary History of Soybean Homoeologous Regions.
Supplemental Table 1. The Sequenced BAC Information in Gm8, Gm15, and Pv5.
Supplemental Table 2. Defining Homoeologous Segments on the Two Soybean Homoeologous Regions.
Supplemental Table 3. Pseudogene Features.
Supplemental Data Set 1. PCR Probes and DNA Sequences for Screening BAC Libraries and Databases to Extend Gm8, Gm15, and Pv5.
Supplemental Data Set 2. Homoeolog-Specific Expression Assay Information and Data.
Supplemental Data Set 3. Annotation of Genes in Gm8, Gm15, and Pv5.
Supplemental Data Set 4. Coordinates and Directions of Genes in Gm8, Gm15, and Pv5.
Supplemental Data Set 5. Features of Transposons in Gm8, Gm15, and Pv5.
Supplemental Data Set 6. Ks Values among Homoeologs and Orthologs from Soybean Gm8/Gm15 and Phaseolus Pv5.
Acknowledgments
We thank Dinesha Walek and William Haun for providing assistance in generating the homoeologous transcription data. We thank the National Science Foundation (DBI 0501877 and IOS 0822258) for funding to S.A.J. to support this work.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Ammiraju J.S., et al. (2008). Dynamic evolution of oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell 20: 3191–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt P.F., Hwa T., Petrov D.A. (2005). Substantial regional variation in substitution rates in the human genome: Importance of GC content, gene density, and telomere-specific effects. J. Mol. Evol. 60: 748–763 [DOI] [PubMed] [Google Scholar]
- Baer C.F., Miyamoto M.M., Denver D.R. (2007). Mutation rate variation in multicellular eukaryotes: Causes and consequences. Nat. Rev. Genet. 8: 619–631 [DOI] [PubMed] [Google Scholar]
- Benovoy D., Drouin G. (2006). Processed pseudogenes, processed genes, and spontaneous mutations in the Arabidopsis genome. J. Mol. Evol. 62: 511–522 [DOI] [PubMed] [Google Scholar]
- Berglund J., Pollard K.S., Webster M.T. (2009). Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 7: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakesley R.W., et al. (2004). An intermediate grade of finished genomic sequence suitable for comparative analyses. Genome Res. 14: 2235–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggmann R., et al. (2006). Uneven chromosome contraction and expansion in the maize genome. Genome Res. 16: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C., Karlin S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Caceres M., Puig M., Ruiz A. (2001). Molecular characterization of two natural hotspots in the Drosophila buzzatii genome induced by transposon insertions. Genome Res. 11: 1353–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T.J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J. (2005). ACT: The Artemis Comparison Tool. Bioinformatics 21: 3422–3423 [DOI] [PubMed] [Google Scholar]
- Chamary J.V., Parmley J.L., Hurst L.D. (2006). Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 7: 98–108 [DOI] [PubMed] [Google Scholar]
- Chantret N., et al. (2005). Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 17: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary B., Flagel L., Stupar R.M., Udall J.A., Verma N., Springer N.M., Wendel J.F. (2009). Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 182: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaromonte F., Yang S., Elnitski L., Yap V.B., Miller W., Hardison R.C. (2001). Association between divergence and interspersed repeats in mammalian noncoding genomic DNA. Proc. Natl. Acad. Sci. USA 98: 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.K., et al. (2004). A sequence-based genetic map of Medicago truncatula and comparison of marker colinearity with M. sativa. Genetics 166: 1463–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J.H., Li H. (2004). Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS Biol. 2: E29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., et al. (2006). Widespread genome duplications throughout the history of flowering plants. Genome Res. 16: 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos K.M., Brown J.K., Bennetzen J.L. (2002). Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12: 1075–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Egan A.N. (2010). Dating the origins of polyploidy events. New Phytol. 186: 73–85 [DOI] [PubMed] [Google Scholar]
- Duret L. (2009). Mutation patterns in the human genome: More variable than expected. PLoS Biol. 7: e1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L., Arndt P.F. (2008). The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 4: e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., Smith N.G., Webster M.T. (2003). Mutation rate variation in the mammalian genome. Curr. Opin. Genet. Dev. 13: 562–568 [DOI] [PubMed] [Google Scholar]
- Ewing B., Hillier L., Wendl M.C., Green P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Fiston-Lavier A.S., Anxolabehere D., Quesneville H. (2007). A model of segmental duplication formation in Drosophila melanogaster. Genome Res. 17: 1458–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M.D., Devos K.M. (1998). Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95: 1971–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganko E.W., Meyers B.C., Vision T.J. (2007). Divergence in expression between duplicated genes in Arabidopsis. Mol. Biol. Evol. 24: 2298–2309 [DOI] [PubMed] [Google Scholar]
- Gill N., Findley S., Walling J.G., Hans C., Ma J., Doyle J., Stacey G., Jackson S.A. (2009). Molecular and chromosomal evidence for allopolyploidy in soybean, Glycine max (L.) Merr. Plant Physiol. 151: 1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R.B., Hoschek G., Vodkin L.O. (1983). An insertion sequence blocks the expression of a soybean lectin gene. Cell 33: 465–475 [DOI] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P. (1998). Consed: A graphical tool for sequence finishing. Genome Res. 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Gray Y.H. (2000). It takes two transposons to tango: Transposable-element-mediated chromosomal rearrangements. Trends Genet. 16: 461–468 [DOI] [PubMed] [Google Scholar]
- Gremme G., Brendel V., Sparks M.E., Kurtz S. (2005). Engineering a software tool for gene structure prediction in higher organisms. Inf. Softw. Technol. 47: 965–978 [Google Scholar]
- Gu Y.Q., et al. (2006). Types and rates of sequence evolution at the high-molecular-weight glutenin locus in hexaploid wheat and its ancestral genomes. Genetics 174: 1493–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Nicolae D., Lu H.H., Li W.H. (2002). Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 18: 609–613 [DOI] [PubMed] [Google Scholar]
- Hardison R.C., et al. (2003). Covariation in frequencies of substitution, deletion, transposition, and recombination during eutherian evolution. Genome Res. 13: 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann I., Prufer K., Ji H., Zody M.C., Paabo S., Ptak S.E. (2005). Why do human diversity levels vary at a megabase scale? Genome Res. 15: 1222–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister J.D., Gaut B.S. (2009). Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 19: 1419–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.T., Dooner H.K. (2008). Macrotransposition and other complex chromosomal restructuring in maize by closely linked transposons in direct orientation. Plant Cell 20: 2019–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes R.W., et al. (2008). Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol. 148: 1740–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S., Chen Z.J. (2009). Genomic and expression plasticity of polyploidy. Curr. Opin. Plant Biol. 13: 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1968). Evolutionary rate at the molecular level. Nature 217: 624–626 [DOI] [PubMed] [Google Scholar]
- Klinz F.J., Gallwitz D. (1985). Size and position of intervening sequences are critical for the splicing efficiency of pre-mRNA in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 13: 3791–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., Lipinski L., Caffin F., Helwak A., Zylicz M. (2006). High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 4: e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Bennetzen J.L. (1999). Plant retrotransposons. Annu. Rev. Genet. 33: 479–532 [DOI] [PubMed] [Google Scholar]
- Lai J., Ma J., Swigonova Z., Ramakrishna W., Linton E., Llaca V., Tanyolac B., Park Y.J., Jeong O.Y., Bennetzen J.L., Messing J. (2004). Gene loss and movement in the maize genome. Genome Res. 14: 1924–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lavin M., Herendeen P.S., Wojciechowski M.F. (2005). Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 54: 575–594 [DOI] [PubMed] [Google Scholar]
- Lee J., Han K., Meyer T.J., Kim H.S., Batzer M.A. (2008). Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons. PLoS ONE 3: e4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.E., et al. (2002). Apollo: A sequence annotation editor. Genome Biol. 3: RESEARCH0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.H., Yang J., Gu X. (2005). Expression divergence between duplicate genes. Trends Genet. 21: 602–607 [DOI] [PubMed] [Google Scholar]
- Lomsadze A., Ter-Hovhannisyan V., Chernoff Y.O., Borodovsky M. (2005). Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 33: 6494–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonnig W.E., Saedler H. (2002). Chromosome rearrangements and transposable elements. Annu. Rev. Genet. 36: 389–410 [DOI] [PubMed] [Google Scholar]
- Ma J., Bennetzen J.L. (2004). Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. USA 101: 12404–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Bennetzen J.L. (2006). Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc. Natl. Acad. Sci. USA 103: 383–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T., Caceres M., Bertranpetit J., Preuss T.M., Thomas J.W., Navarro A. (2004). Chromosomal rearrangements and the genomic distribution of gene-expression divergence in humans and chimpanzees. Trends Genet. 20: 524–529 [DOI] [PubMed] [Google Scholar]
- Mascarenhas D., Mettler I.J., Pierce D.A., Lowe H.W. (1990). Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol. Biol. 15: 913–920 [DOI] [PubMed] [Google Scholar]
- McCarthy E.M., McDonald J.F. (2003). LTR_STRUC: A novel search and identification program for LTR retrotransposons. Bioinformatics 19: 362–367 [DOI] [PubMed] [Google Scholar]
- Messing J. (2009). Synergy of two reference genomes for the grass family. Plant Physiol. 149: 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery E., Charlesworth B., Langley C.H. (1987). A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet. Res. 49: 31–41 [DOI] [PubMed] [Google Scholar]
- Navarro A., Barton N.H. (2003). Chromosomal speciation and molecular divergence–accelerated evolution in rearranged chromosomes. Science 300: 321–324 [DOI] [PubMed] [Google Scholar]
- Pagel J., Walling J.G., Young N.D., Shoemaker R.C., Jackson S.A. (2004). Segmental duplications within the Glycine max genome revealed by fluorescence in situ hybridization of bacterial artificial chromosomes. Genome 47: 764–768 [DOI] [PubMed] [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Paterson A.H., Chapman B.A., Kissinger J.C., Bowers J.E., Feltus F.A., Estill J.C. (2006). Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces, and Tetraodon. Trends Genet. 22: 597–602 [DOI] [PubMed] [Google Scholar]
- Ponthier J.L., Schluepen C., Chen W., Lersch R.A., Gee S.L., Hou V.C., Lo A.J., Short S.A., Chasis J.A., Winkelmann J.C., Conboy J.G. (2006). Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J. Biol. Chem. 281: 12468–12474 [DOI] [PubMed] [Google Scholar]
- Ramakrishna W., Dubcovsky J., Park Y.J., Busso C., Emberton J., SanMiguel P., Bennetzen J.L. (2002). Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162: 1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X.Y., Vorst O., Fiers M.W., Stiekema W.J., Nap J.P. (2006). In plants, highly expressed genes are the least compact. Trends Genet. 22: 528–532 [DOI] [PubMed] [Google Scholar]
- Rose A.B., Elfersi T., Parra G., Korf I. (2008). Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 20: 543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamov A.A., Solovyev V.V. (2000). Ab initio gene finding in Drosophila genomic DNA. Genome Res. 10: 516–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D.R., Butler G., Wolfe K.H. (2007). Yeast genome evolution–The origin of the species. Yeast 24: 929–942 [DOI] [PubMed] [Google Scholar]
- Schlueter J.A., Dixon P., Granger C., Grant D., Clark L., Doyle J.J., Shoemaker R.C. (2004). Mining EST databases to resolve evolutionary events in major crop species. Genome 47: 868–876 [DOI] [PubMed] [Google Scholar]
- Schlueter J.A., Lin J.Y., Schlueter S.D., Vasylenko-Sanders I.F., Deshpande S., Yi J., O'Bleness M., Roe B.A., Nelson R.T., Scheffler B.E., Jackson S.A., Shoemaker R.C. (2007). Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics 8: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlueter J.A., Scheffler B.E., Schlueter S.D., Shoemaker R.C. (2006). Sequence conservation of homeologous bacterial artificial chromosomes and transcription of homeologous genes in soybean (Glycine max L. Merr.). Genetics 174: 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Shoemaker R.C., Polzin K., Labate J., Specht J., Brummer E.C., Olson T., Young N., Concibido V., Wilcox J., Tamulonis J.P., Kochert G., Boerma H.R. (1996). Genome duplication in soybean (Glycine subgenus soja). Genetics 144: 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R.C., Schlueter J., Doyle J.J. (2006). Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 9: 104–109 [DOI] [PubMed] [Google Scholar]
- Springer N.M., Stupar R.M. (2007). Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell 19: 2391–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. (2006). PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34: W609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thibaud-Nissen F., Ouyang S., Buell C.R. (2009). Identification and characterization of pseudogenes in the rice gene complement. BMC Genomics 10: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throude M., et al. (2009). Structure and expression analysis of rice paleo duplications. Nucleic Acids Res. 37: 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Wang Q., Zhang P., Araki H., Yang S., Kreitman M., Nagylaki T., Hudson R., Bergelson J., Chen J.Q. (2008). Single-nucleotide mutation rate increases close to insertions/deletions in eukaryotes. Nature 455: 105–108 [DOI] [PubMed] [Google Scholar]
- Tikhonov A.P., SanMiguel P.J., Nakajima Y., Gorenstein N.M., Bennetzen J.L., Avramova Z. (1999). Colinearity and its exceptions in orthologous adh regions of maize and sorghum. Proc. Natl. Acad. Sci. USA 96: 7409–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van K., Kim D.H., Cai C.M., Kim M.Y., Shin J.H., Graham M.A., Shoemaker R.C., Choi B.S., Yang T.J., Lee S.H. (2008). Sequence level analysis of recently duplicated regions in soybean [Glycine max (L.) Merr.] genome. DNA Res. 15: 93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling J.G., Shoemaker R., Young N., Mudge J., Jackson S. (2006). Chromosome-level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics 172: 1893–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yu L., Lai F., Liu L., Wang J. (2005). Molecular evidence for asymmetric evolution of sister duplicated blocks after cereal polyploidy. Plant Mol. Biol. 59: 63–74 [DOI] [PubMed] [Google Scholar]
- Wawrzynski A., et al. (2008). Replication of nonautonomous retroelements in soybean appears to be both recent and common. Plant Physiol. 148: 1760–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F., et al. (2007). Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 3: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H., Gouy M., Yang Y.W., Sharp P.M., Li W.H. (1989). Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 86: 6201–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H., Li W.H. (2003). Molecular evolution meets the genomics revolution. Nat. Genet. 33(Suppl): 255–265 [DOI] [PubMed] [Google Scholar]
- Yang Z. (1997). PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556 [DOI] [PubMed] [Google Scholar]
- Ying S.Y., Lin S.L. (2009). Intron-mediated RNA interference and microRNA biogenesis. Methods Mol. Biol. 487: 387–413 [DOI] [PubMed] [Google Scholar]
- Zhang J., Yu C., Pulletikurti V., Lamb J., Danilova T., Weber D.F., Birchler J., Peterson T. (2009). Alternative Ac/Ds transposition induces major chromosomal rearrangements in maize. Genes Dev. 23: 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D., Gerstein M.B. (2007). The ambiguous boundary between genes and pseudogenes: The dead rise up, or do they? Trends Genet. 23: 219–224 [DOI] [PubMed] [Google Scholar]
- Zou C., Lehti-Shiu M.D., Thibaud-Nissen F., Prakash T., Buell C.R., Shiu S.H. (2009). Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol. 151: 3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]