This work examines double mutants defective in providing phosphoenolpyruvate (PEP) to the plastids and finds that PEP is essential for multiple steps in plant development, reflecting the central importance of PEP in multiple plant metabolic pathways, including synthesis of aromatic amino acids, lipids, flavonoids, and lignin.
Abstract
Restriction of phosphoenolpyruvate (PEP) supply to plastids causes lethality of female and male gametophytes in Arabidopsis thaliana defective in both a phosphoenolpyruvate/phosphate translocator (PPT) of the inner envelope membrane and the plastid-localized enolase (ENO1) involved in glycolytic PEP provision. Homozygous double mutants of cue1 (defective in PPT1) and eno1 could not be obtained, and homozygous cue1 heterozygous eno1 mutants [cue1/eno1(+/−)] exhibited retarded vegetative growth, disturbed flower development, and up to 80% seed abortion. The phenotypes of diminished oil in seeds, reduced flavonoids and aromatic amino acids in flowers, compromised lignin biosynthesis in stems, and aberrant exine formation in pollen indicate that cue1/eno1(+/−) disrupts multiple pathways. While diminished fatty acid biosynthesis from PEP via plastidial pyruvate kinase appears to affect seed abortion, a restriction in the shikimate pathway affects formation of sporopollonin in the tapetum and lignin in the stem. Vegetative parts of cue1/eno1(+/−) contained increased free amino acids and jasmonic acid but had normal wax biosynthesis. ENO1 overexpression in cue1 rescued the leaf and root phenotypes, restored photosynthetic capacity, and improved seed yield and oil contents. In chloroplasts, ENO1 might be the only enzyme missing for a complete plastidic glycolysis.
INTRODUCTION
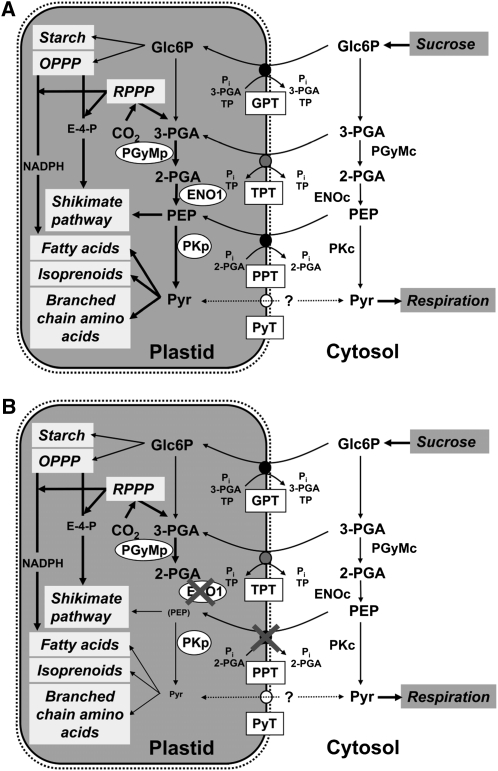
Phosphoenolpyruvate (PEP) plays a central role in plant metabolism. As an intermediate of glycolysis, PEP is indispensable for energy metabolism in the cytosol and delivers ATP and pyruvate by the action of cytosolic pyruvate kinase (PK) (Plaxton, 1996; Givan, 1999). Pyruvate can be fed into the citric acid cycle, yielding NADH for respiratory ATP generation (Fernie et al., 2004). Inside the plastids, PEP may act as a precursor for at least four metabolic pathways (Figure 1A).
Figure 1.
Metabolic Role of PEP in Plastids of Heterotrophic or Mixotrophic Tissues (i.e., Developing Seeds).
In wild-type plants (A), PEP can be imported from the cytosol by PPT, or it may be produced from 3-PGA by the glycolytic sequence involving PGyM and ENO. Both enzymes exist as plastidic and cytosolic forms. In the stroma, PEP together with erythrose 4-phosphate (E-4-P) can enter the shikimate pathway for the biosynthesis of aromatic amino acids and derived compounds, or after conversion to pyruvate by PK, it can be fed into the biosynthesis of fatty acids, isoprenoids, or branched-chain amino acids. Pyruvate may also be imported by a pyruvate transporter (PyT). Other transporters of the phosphate translocator family, such as GPT or the triose phosphate/PT (TPT), may import Glc6P or 3-PGA, respectively. Glc6P can be fed into OPPP and starch biosynthesis. Note that TPT is not likely to be expressed in heterotrophic tissues. The OPPP produces reducing equivalents in the form of NADPH required for anabolic reactions and metabolic intermediates, such as E-4-P. In mixotrophic plastids, 3-PGA and reducing equivalents can be produced by the Calvin cycle (reductive pentose phosphate pathway [RPPP]). By cytosolic glycolysis, imported sucrose can be metabolized to pyruvate, which is subjected to respiration in the mitochondria. In (B), the consequences of a deficiency in both PPT1 and ENO1 are shown. Most likely all metabolic pathways shaded in light gray within the plastids would be negatively affected, which would also feed back on processes taking place in the cytosol.
Together with erythrose 4-phosphate, PEP is fed into the shikimate pathway, which delivers essential aromatic amino acids and a large number of secondary plant products. The initial steps of the shikimate pathway are exclusively localized within the plastid stroma (Herrmann, 1995; Schmid and Amrhein, 1995; Herrmann and Weaver, 1999). Inside the stroma, PEP can also be sequentially metabolized to pyruvate and acetyl-CoA by plastid PK and the pyruvate dehydrogenase complex (Reid et al., 1977; Elias and Givan, 1979; Lernmark and Gardeström, 1994) and thus enter the biosynthesis of fatty acids (Dennis, 1989; Ohlrogge and Jaworski, 1997), which are quantitatively important for triacylglycerol production in oil seeds (e.g., Voelker and Kinney, 2001; Rawsthorne, 2002; Ruuska et al., 2002). Like the shikimate pathway, the de novo biosynthesis of fatty acids for membranes and storage lipids is localized to the plastids (Ohlrogge et al., 1979; Ohlrogge and Jaworski, 1997). Moreover, stromal pyruvate can act as a precursor for the synthesis of branched-chain amino acids (Schulze-Siebert et al., 1984) and together with glyceraldehyde 3-phosphate for the mevalonate-independent way (2-C-methyl-d-erythritol 4-phosphate [MEP] pathway) of isoprenoid biosynthesis (Lichtenthaler, 1999). Unlike plastids from photoautotrophic or mixotrophic tissues, such as green oilseeds (Ruuska et al., 2004; Li et al., 2006; Schwender et al., 2006), in plastids from nongreen tissues the above pathways rely entirely on the provision of carbon skeletons and energy from the cytosol.
In principle, fatty acid biosynthesis in nongreen plastids could be driven by the import of glucose-6-phosphate (Glc6P) or triose phosphates malate or pyruvate (Smith et al., 1992; Kang and Rawsthorne, 1994; Qi et al., 1995; Eastmond and Rawsthorne, 2000; Ruuska et al., 2002; Figure 1A). In Brassicaceae, such as canola (Brassica napus) or Arabidopsis thaliana, PEP is likely to be the predominant precursor for fatty acid biosynthesis in seeds (Schwender and Ohlrogge, 2002; Schwender et al., 2003; Andre et al., 2007, Baud et al., 2007b; Lonien and Schwender, 2009). Hence, a sufficient provision of PEP appears to be essential for lipid biosynthesis and storage (Kubis et al., 2004). In principle, pyruvate generated by cytosolic PK may be imported as precursor for fatty acid biosynthesis, which is supported by feeding experiments with 14C-labeled pyruvate to isolated plastids from B. napus embryos and the subsequent incorporation of 14C into fatty acids (Kang and Rawsthorne, 1994; Eastmond and Rawsthorne, 2000). However, pyruvate deriving from stromal PEP serves as the major precursor for fatty acid biosynthesis in plastids of developing oil seeds (Figure 1A). This notion is supported by the observation that a restriction in plastid-localized PK, which converts PEP to pyruvate, resulted in severely diminished seed oil contents (Andre et al., 2007; Baud et al., 2007b) and by recent 13C feeding experiments of Arabidopsis embryos (Lonien and Schwender, 2009).
PEP can be delivered by the phosphoenolpyruvate/phosphate translocator (PPT) from the cytosol (Fischer et al., 1997) or may be generated inside the plastids by a complete glycolytic pathway (Figure 1A). However, chloroplasts and most nongreen plastids lack the ability to form PEP via glycolysis because the essential enzymes, which convert 3-phosphoglycerate (3-PGA) to PEP (i.e., phosphoglyceromutase [PGyM] and enolase [ENO]) are either absent or, if present, show a low activity (Stitt and Ap Rees, 1979; Schulze-Siebert et al., 1984; Journet and Douce, 1985; Bagge and Larsson, 1986; Van der Straeten et al., 1991; Miernyk and Dennis, 1992; Borchert et al., 1993). By contrast, plastids from lipid storing tissues such as seeds of castor bean (Ricinus communis) or canola have been demonstrated to be capable of catalyzing glycolytic PEP formation (Eastmond and Rawsthorne, 2000).
The import of PEP from the cytosol into the plastid stroma is catalyzed by PPT. The genome of Arabidopsis contains two PPT genes (PPT1, At5g33320; and PPT2, At3g01550), which have been characterized at molecular and functional levels (Knappe et al., 2003). PPT1 is defective in the chlorophyll a/b binding protein underexpressed1 (cue1) mutant (Li et al., 1995; Streatfield et al., 1999), which exhibits a reticulate leaf phenotype with wild-type-like bundle sheath cells but aberrant mesophyll cells and smaller chloroplasts therein (Kinsman and Pyke, 1998). The involvement of a PPT in the delivery of PEP to plastids for the production of aromatic amino acids in certain cell types has already been demonstrated (Streatfield et al., 1999; Voll et al., 2003). The cue1 mutant phenotype could be rescued by feeding aromatic amino acids via the roots (Streatfield et al., 1999) or by the ectopic expression of a C4-type plastid-targeted pyruvate,orthophosphate dikinase (PPDK), capable of producing PEP from pyruvate (Voll et al., 2003), indicating that a shortage of PEP in certain plastids of cue1 is responsible for the mutant phenotype. Moreover, the latter experiment indirectly supports either the presence of a plastid pyruvate transporter in those cell or tissue types where PPT1 is absent, a sufficient rate of pyruvate diffusion across the envelope, or the activity of a plastid-localized malic enzyme, which is capable of producing pyruvate by oxidative decarboxylation of malate (Wheeler et al., 2005). Pyruvate import or generation within the plastid, linked to the activity of PPDK, which is targeted to the cytosol and the plastids (Parsley and Hibberd, 2006), might also be capable of providing PEP for the shikimate pathway in certain tissues. However, the expression level of the Arabidopsis PPDK (At4g15530) based on microarray data is rather poor in most vegetative tissues, but it is enhanced in mature pollen and imbibed seeds (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007). It is hence more likely that, in addition to the import from the cytosol by a PPT, PEP is generated by a complete glycolytic pathway in the plastids.
We have recently identified and functionally characterized the plastid-localized enolase (ENO1) of Arabidopsis, which catalyzes the glycolytic conversion of 2-phosphoglycerate (2-PGA) to PEP (Prabhakar et al., 2009). Most strikingly, the single-copy gene ENO1 is not expressed in photosynthetic tissues but exhibits high transcript abundance in developing siliques. Moreover, both ENO1 and PPT1 are coexpressed during early embryo and seed development (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), suggesting that the task of PEP provision to plastids in these tissues might be shared both by import from the cytosol and by a complete glycolytic pathway within the stroma (Prabhakar et al., 2009).
Here, we demonstrate that Arabidopsis double mutant plants defective in both PPT1 and ENO1 are not viable due to partial gametophytic lethality and impaired sporophyte development, underlining the importance of plastidic PEP (Figure 1B). Ectopic overexpression of ENO1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter could rescue the phenotype of the cue1 mutant. The role of PEP in plastids of generative and vegetative tissues of Arabidopsis is discussed.
RESULTS
A Double Knockout of PPT1 and ENO1 Is Lethal
PPT1 and ENO1 are coexpressed during early embryo and seed development and most likely share the provision of PEP to plastids for anabolic reaction sequences during this developmental stage (Prabhakar et al., 2009). To elucidate the consequences of a restriction in PEP supply to plastids, the eno1-1 mutant allele (Prabhakar et al., 2009) was crossed with the cue1-1 mutant allele (Li et al., 1995), which represents a deletion of the PPT1 gene locus (Streatfield et al., 1999) and likely also at least five expressed genes adjacent to PPT1 (R.E. Häusler and U.-I. Flügge, unpublished data). Among the F2 generation, 300 plants homozygous for cue1-1 were analyzed for the T-DNA insertion in the ENO1 gene. In this screen, no double homozygous mutants and only four eno1 heterozygous plants (i.e., 1.3%) were found (Table 1). This low percentage was far below 50% expected. Moreover, heterozygous eno1-1 mutants in the homozygous cue1-1 background [cue1-1/eno1-1(+/−), ccEe] exhibited stunted shoot growth and retarded flowers and siliques. In the progeny of these ccEe plants (F3 generation), again no double homozygous mutants (ccee) and only four heterozygous mutants (ccEe), all with a retarded growth phenotype, were found among 74 tested plants. This observation suggests that a combined mutation in both genes results in lethality. To confirm this assumption and to further elucidate the observed segregation pattern, crosses between a second mutant allele defective in ENO1 (eno1-2; Prabhakar et al., 2009) and three different cue1 alleles (cue1-1, cue1-3, and cue1-6) were generated. Like cue1-1, cue1-3 is in the background of the ecotype Bensheim (i.e., pOCA106; Li et al., 1995) and is described as a weak allele of cue1, whereas cue1-6 represents a strong allele in the Columbia-0 (Col-0) background (Streatfield et al., 1999). Both cue1-3 and cue1-6 were obtained from ethyl methanesulfonate–mutagenized plants. Analogous to the crosses of eno1-1 with cue1-1, no double homozygous mutants could be isolated in the additional crosses, and the percentage of heterozygous eno1 mutants in the homozygous cue1 backgrounds was again far below expectation (Table 1). These data indicate that lethality of the double homozygous cue1/eno1 mutants is due to a defect in both the PPT1 and ENO1 genes and is not caused by any secondary T-DNA insertions or mutations in the single mutant backgrounds as is evident for cue1-1.
Table 1.
Genetic Analyses of Crosses between cue1 (Male) and eno1 (Female) Mutants
| Mutant Cross | ccEe Plants (%) | Homozygous cue1 Mutants Analyzed in Each Harvest |
| cue1-1 × eno1-1 | 3.79 ± 0.18 | 66 to 71 |
| cue1-1 × eno1-2 | 1.43 ± 0.01 | 68 to 73 |
| cue1-3 × eno1-2 | 16.38 ± 0.22 | 53 to 57 |
| cue1-6 × eno1-2 | 13.08 ± 0.09 | 56 to 65 |
The frequency of heterozygous eno1 mutants in the homozygous cue1 background (ccEe plants) is shown as a percentage of plants analyzed in three independent harvests (mean ± se).
A Heterozygous Mutation of ENO1 in the Background of cue1 Leads to Stunted Shoot Growth and Aberrant Flower Development
Plants heterozygous for eno1 in the homozygous cue1 background (ccEe) exhibited a stunted growth phenotype compared with the wild type or the cue1 single mutants (Figure 2A). Interestingly, growth retardation became evident only after the plants were grown for 4 to 5 weeks on soil (see Supplemental Figure 1A online), whereas plants grown on Murashige and Skoog (MS) agar plates for up to 3 weeks lack any visible growth phenotype of the roots or the shoot (see Supplemental Figure 1B online). Besides the retarded growth of the shoot, the development of the first flowers was severely hampered and resulted in deformed and infertile flowers with underdeveloped stamen (Figures 2B and 2C). At the later stages, flower development was less affected and siliques were formed after pollination (Figures 2D to 2F). However, the size of the siliques was diminished and their length was reduced by >50% compared with wild-type plants or the single mutants (Figure 2G). The number of seeds per silique was severely diminished in all ccEe plants from ~50 seeds in Col-0 or pOCA to <10% in cue1-1/eno1-2(+/−) and between 30 and 40% in cue1-3/eno1-2(+/−) and cue1-6/eno1-2(+/−), respectively (Table 2). There was also a decline in the seed number per silique in the cue1 single mutant alleles, which was less marked for the cue1-3 or cue1-6 allele. According to the gaps in the mature siliques of ccEe plants (Figure 2G), an abortion frequency of seeds of >80% was calculated for the progeny of cue1-1 × eno1-2 crosses and ~45% for both crosses of cue1-3 × eno1-2 and cue1-6 × eno1-2 (Table 2). The cause for the developmental constraints in ccEe plants was further elucidated by expression studies of ENO1. We could show previously that ENO1 is highly expressed in nonroot hair cells of the roots and in the shoot apex but is absent in mature leaves (Prabhakar et al., 2009). As revealed by quantitative RT-PCR (qRT-PCR), ENO1 expression in the roots and the shoot apex of the developing rosettes of cue1-6/eno1-2(+/−) was diminished by 29% ± 4% (n = 3) and 69% ± 6% (n = 3), respectively, compared with the wild type. These data indicate that a heterozygous knockout mutation of ENO1 results in a substantial decrease in ENO1 expression and suggest that the growth retardation in the ccEe lines is based on a gene dosage effect.
Figure 2.
Phenotype of Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe) Compared with Wild-Type and cue1 Plants Grown for 8 Weeks in the Greenhouse.
(A) Comparison of the growth phenotype of Col-0 (1), eno1-2 (2), cue1-1 (3), and cue1-1/eno1-2 (4) plants. The inset shows a detailed view of cue1-1/eno1-2(+/−) (5).
(B) Opened bud of an early (stage 10) wild-type (Col-0) flower.
(C) Opened bud of an early (stage 10) flower of cue1-1/eno1-2(+/−) with degenerated stamens.
(D) Flower and silique development of cue1-6.
(E) Schematic representation of the position of flowers and siliques shown in (D) and (F)
(F) Flower and silique development of cue1-6/eno1-2(+/−) plants.
(G) Destained mature siliques of Col-0 (1), pOCA (2), cue1-1 (3), cue1-6/eno1-2(+/−) (4), and cue1-1/eno1-2(+/−) (5)
Developmental stages of flowers and siliques shown in (D) and (F) are based on the position of the flowers/siliques at the raceme starting from the topmost to lower positions. The numbers in (E) represent the positions of flowers and/or siliques shown in (D) and (F). Bars = 5 mm in (D) and (F).
Table 2.
Frequencies of Seed Abortion, Seed Phenotypes, and Pollen Viability in Wild-Type Arabidopsis (Col-0, Bemsheim [pOCA]), eno1 and cue1 Alleles, and Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe Plants)
| Seed Abortion |
Seed Phenotype (%) |
||||||
| Plant Line | Number of Seeds per Silique | Number of Gaps | Abortion (%) | Class I | Class II | Class III | Nonviable Pollen (%) |
| Col-0 | 50.3 ± 2.2 | 0.25 ± 0.13 | 0.5 (n = 12) | 97.70 | 2.30 | 0.00 | 0.3 ± 0.3 |
| pOCA | 47.9 ± 1.1 | 1.00 ± 0.29 | 2.0 (n = 9) | 95.00 | 5.00 | 0.00 | 0.3 ± 0.3 |
| eno1-1 | 48.9 ± 0.8 | 1.44 ± 0.48d | 2.9 (n = 9) | 80.00 | 17.00 | 3.00 | 6.3 ± 2.0a |
| eno1-2 | 43.8 ± 0.8c | 1.00 ± 0.37 | 2.2 (n = 9) | 76.00 | 20.00 | 4.00 | 8.0 ± 1.0a |
| cue1-1 | 32.9 ± 1.1a | 8.89 ± 0.72a | 21.3 (n = 9) | 70.00 | 25.00 | 5.00 | 0.6 ± 0.3 |
| cue1-3 | 45.3 ± 2.0 | 5.44 ± 0.84a | 10.7 (n = 9) | 91.10 | 8.90 | 0.00 | 0.3 ± 0.3 |
| cue1-6 | 41.0 ± 2.2b | 1.30 ± 0.35c | 3.1 (n = 6) | 91.10 | 8.90 | 0.00 | 0.3 ± 0.3 |
| cue1-1/eno1-2(+/−) | 2.4 ± 0.6a | 12.40 ± 0.65a | 83.6 (n = 9) | 65.40 | 25.00 | 9.60 | 35.0 ± 2.7a |
| cue1-3/eno1-2(+/−) | 20.2 ± 2.3a | 16.50 ± 0.89a | 45.0 (n = 27) | 86.20 | 10.30 | 3.50 | 2.0 ± 0.5 |
| cue1-6/eno1-2(+/−) | 16.2 ± 2.3a | 13.20 ± 1.20a | 44.9 (n = 26) | 64.00 | 26.00 | 10.00 | 29.0 ± 2.0a |
Seeds and seed gaps were counted in 6 to 27 siliques per line (n). The developed seeds were categorized into three classes according to their phenotypic appearance. Class I seeds were wild-type like, class II seeds showed an intermediate reduction in size, and class III seeds a strong reduction in size. Pollen viability was tested by Alexander stain of pollen collected from three plants per line. The data are expressed as mean ± se. Statistical significance of differences between the parameters were assessed by the Welch test with probability values of P < 0.001 (a), P < 0.01 (b), P < 0.02 (c), and P < 0.05 (d).
Female and Male Gametophyte Development and Seed Formation Are Impaired in cue1/eno1(+/−) Plants
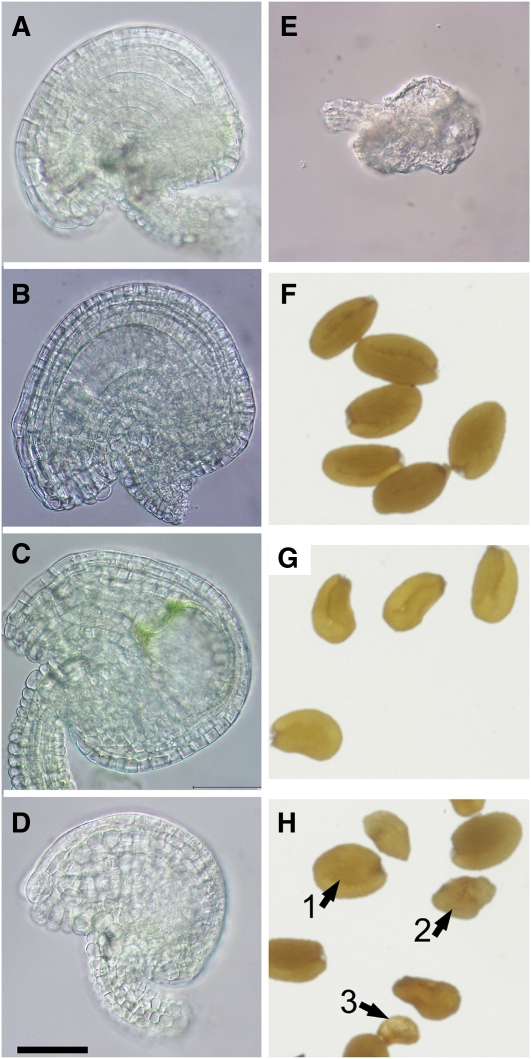
To address the question whether or not gametophyte development was hampered in ccEe, we further analyzed the phenotypes of ovules in flower buds at stage 10 (Smyth et al., 1990) before pollination (Figure 3). Besides ovules with a wild-type-like appearance and a normally developed embryo sac (Figures 3A and 3B), in ~40% of the ovules the embryo sac was diminished in size (Figure 3C) or even absent (Figure 3D). Moreover, there were also ovules completely halted in development (Figure 3E). The above data show that a combined knockout of PPT1 and ENO1 can result in lethality of the female gametophyte.
Figure 3.
Phenotypes of Ovules and Mature Seeds of Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe) Compared with Wild-Type or cue1 Plants.
(A) Ovule of a wild-type (Col-0) plant.
(B) Ovule of cue1-1/eno1-2(+/−) with a wild-type-like appearance.
(C) Ovule of cue1-1/eno1-2(+/−) with a swollen embryo sac.
(D) Ovule of cue1-1/eno1-2(+/−) lacking an embryo sac.
(E) Degenerated ovule of cue1-1/eno1-2(+/−).
(F) Wild-type (pOCA) seeds.
(G) Seeds of cue1-1.
(H) Phenotype of seeds of cue1-1/eno1-2(+/−) showing either a wild-type-like appearance (class I seeds; 1) or that were intermediately and strongly reduced in size (class II, 2; and class III seeds, 3).
Bar = 50 μm in (A) to (E).
[See online article for color version of this figure.]
The remaining developed and matured seeds resulting from unaffected ovules after fertilization could be categorized into three different classes according to their size and phenotypic appearance. Class I seeds were indistinguishable from seeds of cue1 (Figure 3G), which are slightly lighter in color compared with wild-type seeds and hence resemble those from transparent testa mutants (Koornneef, 1990) (Figure 3F). By contrast, class II and class III seeds were intermediately and severely diminished in size, respectively (Figure 3H). Moreover, class III seeds exhibited a wrinkled and shrunken appearance with irregularly shaped testa cells similar to the wrinkled1 (wri1) mutant (Focks and Benning, 1998; Cernac et al., 2006; Baud et al., 2007a). Table 2 shows the distribution of the individual seed classes between the different plant lines. There was also a higher percentage of class II and III seeds in the eno1 single mutant alleles as well as cue1-1 compared with the wild type. A genotypic analysis of all three seed classes revealed that class I seeds were wild-type for ENO1, whereas both class II and class III seeds were heterozygous for eno1. Class I and II seeds were capable of germinating on MS agar plates, whereas class III seeds were not.
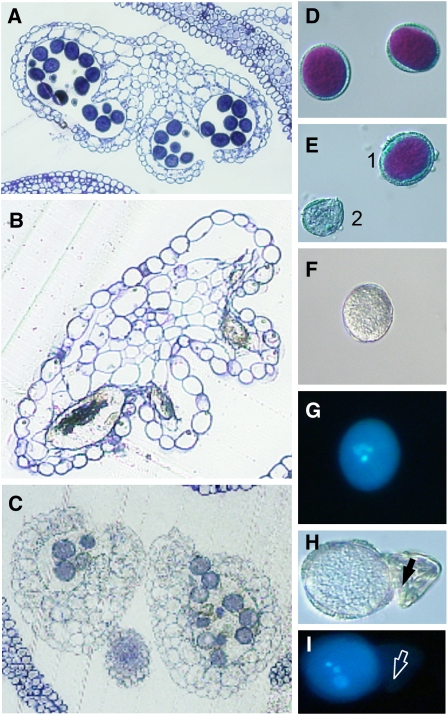
We further analyzed the effect of a double knockout of PPT1 and ENO1 on the development of the male gametophyte. Cross sections of anthers of ccEe plants revealed a reduced number of pollen compared with the wild type (Figures 4A to 4C). Moreover, there was a large portion of underdeveloped pollen in anthers of cue1-1/eno1-2(+/−) and cue1-6/eno1-2(+/−) plants (Figures 4B and 4C). Vitality staining showed that the underdeveloped pollen grains of the ccEe plants were dead (Figures 4D and 4E). Likewise the cellular structure of the underdeveloped pollen was disrupted and the DNA of the nuclei in the three cellular stage was not detectable by 4',6-diamidino-2-phenylindole (DAPI) staining (Figures 4F to 4I). In the cue1-1/eno1-2(+/−) and cue1-6/eno1-2(+/−) double mutants, the lethality frequency of the pollen was up to 35 and 29%, respectively, compared with <1% in wild-type plants and the single mutants. Interestingly, ccEe plants obtained from crosses of eno1-2 with the weak cue1-3 allele showed only 2% dead pollen, suggesting that at least pollen viability can be maintained if PPT1 is present, albeit functionally impaired (Table 2).
Figure 4.
Cross Sections of Pollen Sacs as well as Vitality (Alexander) and DAPI Staining of Pollen from Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe) Compared with the Wild Type.
(A) Cross section of a wild-type (Col-0) pollen sac.
(B) Cross section of a pollen sac of cue1-1/eno1-2(+/−).
(C) Cross section of a pollen sac from cue1-6/eno1-2(+/−).
(D) Vitality staining of wild-type pollen (pOCA).
(E) Vitality staining of pollen from cue1-1/eno1-2(+/−) with an example of a viable (1) and an aborted (2) pollen.
(F) Bright-field image of wild-type pollen (pOCA).
(G) DAPI staining of the wild-type pollen shown in (F).
(H) Bright-field image of wild-type-like pollen and degenerated pollen (arrow) of cue1-1/eno1-2(+/−).
(I) DAPI staining of the pollen grains shown in (H). The degenerated pollen is marked by an arrow.
[See online article for color version of this figure.]
The Pollen Exine Structure Is Impaired and Phenolic Compounds Are Less Abundant in Anthers and Mature Pollen of ccEe Plants
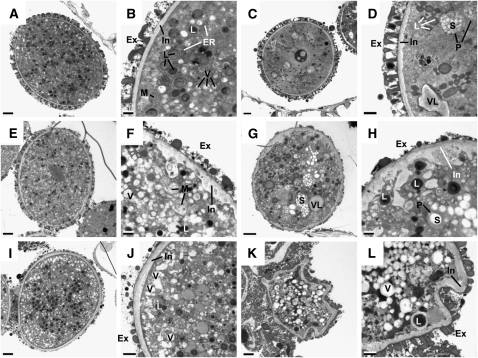
To further elucidate the reason for the high lethality rate of ccEc pollen, we analyzed the ultrastructure of developing pollen grains of wild-type and ccEe plants. As shown in Figure 5, pollen grains from ccEe plants exhibit a variety of phenotypes different from wild-type pollen (Figures 5A and 5B). The most prominent features of ccEe compared with wild-type pollen grains were an irregularly shaped intine (Figures 5D, 5F, 5H, and 5L) and a strongly diminished exine structure (Figures 5F to 5J). In individual cases, pollen development was completely disrupted (Figures 5K and 5L). Moreover, in some of the mutant pollen, massive starch accumulation was found in the plastids (Figures 5D, 5G, and 5H), whereas starch granules in wild-type pollen were less abundant (Figures 5A and 5B). Interestingly, lipid bodies and vacuoles were found at nearly the same density in ccEe pollen grains (Figures 5C to 5J) compared with the wild type (Figures 5A and 5B), suggesting that fatty acid biosynthesis appears not to be a major metabolic restriction during pollen maturation. Apart from dead pollen (Figures 5K and 5L), deposition of exine material was severely diminished in pollen of ccEe plants (Figures 5C to 5L), suggesting that sporopollonin deposition was restricted. Sporopollonin represents the major constituent of the exine and consists of both long-chain fatty acids and phenolic compounds derived from the phenylpropanoid metabolism (Guilford et al., 1988; Wiermann et al., 2001; Dobritsa et al., 2009). Moreover, a comparison of autofluorescence emission from developing anthers captured by confocal laser scanning microscopy revealed that phenolic compounds were less abundant both in the anthers and in the pollen exine of ccEe plants compared with the wild type (see Supplemental Figure 2 online). Likewise, diphenylborate-2-aminoethyl–stained mature pollen grains of ccEe plants exhibited considerably diminished yellow/green fluorescence both from the wall and the cytoplasm of the pollen grains compared with the wild type (see Supplemental Figure 3 online).
Figure 5.
Cross Sections of Differently Affected Pollen Grains of Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe) Analyzed by Transmission Electron Microscopy in the Tricellular Stage in Comparison to Wild-Type Col-0.
(A) Cross section of a wild-type (Col-0) pollen grain.
(B) Close-up of the wild-type pollen grain shown in (A).
(C) to (L) Phenotypic changes in the ultrastructure of pollen grains observed in pollen sacs of ccEe plants.
(C) Pollen grain of cue1-1/eno1-2(+/−) with a wild-type-like appearance.
(D) Close-up of the pollen grain shown in (C) with increased numbers of starch granules in the plastids and a slightly deformed and swollen intine.
(E) Pollen grain of cue1-1/eno1-2(+/−) with a wild-type-like size but affected exine and intine structures.
(F) Close-up of the pollen grain shown in (E) with a focus on the underdeveloped exine structure and the strongly deformed intine.
(G) Pollen grain of cue1-1/eno1-2(+/−) with a deformed pollen wall, high numbers of starch granules, and large vacuole-like structures.
(H) Close-up of the pollen grain shown in (G) with a focus on the impaired exine and intine structures.
(I) Pollen grain of cue1-6/eno1-2(+/−) with a wild-type-like appearance but an impaired exine structure.
(J) Close-up of the pollen grain shown in (I) with a focus on the underdeveloped exine structure.
(K) Strongly deformed pollen grain of cue1-6/eno1-2(+/−).
(L) Close-up of the pollen grain shown in (K).
Ex, exine; In, intine; L, lipid body; ER, endoplasmatic reticulum; V, vacuoles; VL, vacuole-like bodies; M, mitochondria; P, plastids; S, starch granules. Bars = 2 and 0.5 μm for the overviews and close-ups, respectively.
In Vitro Germination of Pollen Is Severely Diminished in ccEe Plants
The developmental constrains of pollen from ccEe plants combined with their aberrant ultrastructure and diminished contents of phenolic compounds are reflected in a severely diminished in vitro germination rate. As is shown in Supplemental Figure 4 online, <20% of the pollen grains were capable of germinating in the lines cue1-1/eno1-2(+/−) and cue1-6/eno1-2(+/−), whereas almost 80% of pollen from wild-type plants germinated on agar plates.
Male and Female Transmission Efficiencies of the cue1 and eno1 Mutation Were Diminished in Segregating ccEe Plants
Having established that a high lethality frequency of both the female and male gametophytes in ccEe plants was involved in the failure to establish double homozygous cue1/eno1 mutant plants, we further analyzed the segregation pattern of the individual genotypes in the F2 generation of crosses between cue1-1 or cue1-6 and eno1-2. To determine female and male transmission efficiencies (TEs), reciprocal crosses between eno1-2 and cue1-1 were performed. A detailed analysis of the segregation patterns is contained in Supplemental Table 1 online. The major outcome of this analysis can be summarized as follows. For the mutation in the PPT1 gene, both female and male TEs were similar (30%) but lower than the expected value of 50% for each gametophyte. By contrast, the female TE for the mutation in the ENO1 gene was about half (16.7%) compared with the male TE (31.3%), suggesting that a lesion of ENO1 in the background of the homozygous cue1 mutant has a much stronger effect on female gametophyte development (in particular on embryo sac development) than a lesion in the PPT1 gene in the homozygous eno1 background.
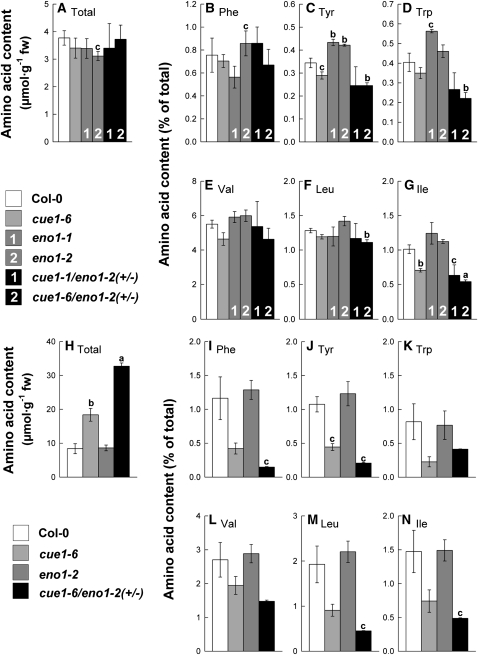
Contents of Aromatic and Branched-Chain Amino Acids Are Diminished in Flowers and Rosette Leaves of cue1/eno1(+/−) Double Mutants
The proposed function of the PPT in vegetative tissues is the provision of PEP for the shikimate pathway (Fischer et al., 1997), and it has previously been shown that a mutation in the PPT1 gene in cue1 mutants leads to lower contents of Phe in leaves (Voll et al., 2003). Moreover, branched-chain amino acid synthesis commences from plastidic pyruvate via acetolactate synthase (Schulze-Siebert et al., 1984). To gain information on the steady state contents of amino acids in those tissues where both PPT1 and ENO1 are expressed compared with tissues where ENO1 is absent, we analyzed the amino acid composition in mature flowers (stage 13-14; Smyth et al., 1990) and rosette leaves of ccEe plants and the single mutants compared with wild-type plants (Figure 6; see Supplemental Figure 5 online).
Figure 6.
Contents of Selected Amino Acids Extracted from Flower Buds or Rosette Leaves of the Wild Type (Col-0), cue1-6 and eno1 Single Mutant, and the Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe).
Flower buds ([A] to [G]) and rosette leaves ([H] to [N]). The data represent the mean ± se of n = 5 ([A] to [G]) or n = 3 ([H] to [N]) independent experiments. Statistical significance of differences between the parameters were assessed by the Welch test with probability values of P < 0.001 (a), P < 0.01 (b), and P < 0.05 (c) indicated above the respective bars. Contents of total soluble amino acid ([A] and [H]) were estimated from the sum of all recognized proteinogenic amino acid after separation by HPLC. The relative contents of the aromatic amino acids Phe, Tyr, Trp ([B] to [D] and [I] to [K]) and the branched-chain amino acids Val, Leu, and Ile ([E] to [G] and [L] to [N]) were expressed as a percentage fraction of the total amino acid content in flower buds (A) and rosette leaves (H), respectively.
In flowers, the content of total free amino acids (i.e., the sum of all detected amino acids after HPLC separation) ranged between 3 and 4 μmol·g−1 fresh weight (FW) and was not significantly affected in the individual mutant lines compared with wild-type plants (Figure 6A). Of the aromatic amino acids, Phe contents varied between the lines but did not show a significant change in either of the mutants compared with wild-type plants (Figure 6B). By contrast, Tyr content was significantly increased in both alleles of the eno1 mutant but only slightly diminished in cue1-6 as well as in the ccEe plants (Figure 6C). Similarly, the Trp content was increased in both eno1 mutant alleles but decreased in the ccEe plants (Figure 6D). Of the branched-chain amino acids, the contents of Val and Leu were not significantly altered between the lines (Figures 6E and 6F), whereas Ile content was diminished in cue1-6 and the ccEe plants (Figure 6G). In Supplemental Figure 3 online, the contents of a broader range of proteogenic amino acids are shown for the mutant compared with wild-type plants. Of the major amino acids, Gln content was significantly diminished by ~50% in both eno1 mutant alleles and the ccEe plants (see Supplemental Figure 5B online), whereas Glu content was increased in cue1-6 but not consistently affected in both ccEe alleles (see Supplemental Figure 5A online). Thr and Gly contents showed a trend of an increase in both eno1 alleles but were diminished in cue1-6 (see Supplemental Figures 5E and 5F online), whereas Ser content remained unaffected (see Supplemental Figure 5D online). The content of Ala was increased in all mutant lines compared with the wild-type plants (see Supplemental Figure 5G online). His content was significantly diminished by 30% in the ccEe plants (see Supplemental Figure 5H online). It has recently been shown that His exerts control on developmental processes (Mo et al., 2006). Interestingly, both Arg and Lys contents were significantly increased in cue1-6 but exhibited wild-type-like levels in both eno1 alleles and the ccEe plants (see Supplemental Figures 5I and 5J online). An increased Arg content in cue1 has previously been reported by Streatfield et al. (1999) and He et al. (2004). For the latter report, a new cue1 mutant allele (nos1) has been isolated in a screen for nitric oxide (NO) overproducers.
A different picture emerged from the amino acid spectrum of mature rosette leaves (i.e., where ENO1 is not expressed). The total amino acid content was doubled in cue1-6 and increased even threefold in cue1-6/eno1-2(+/−) compared with the wild type and the eno1-2 mutant (Figure 6H). This increase in the total amino acid content reflects specifically higher contents of Glu, Gln, Asp, Asn, Ser, and Thr. When expressed as a percentage of the total free amino acids, the content of the major amino acids remained nearly unaltered between the plants (see Supplemental Figures 5K to 5O online). By contrast, the relative content of Gly was diminished in cue1-6 and cue1-6/eno1-2(+/−) (see Supplemental Figure 5P online), whereas Ala was significantly increased in both these lines (see Supplemental Figure 5Q online). The relative contents of minor amino acids, such as the aromatic amino acids Phe, Tyr, and Trp (Figures 6I to 6K), as well as the branched chain amino acids Val, Leu, and Ile (Figures 6L to 6N) were significantly decreased on the expense of the major amino acids in cue1 and in particular in ccEe plants. However, on an absolute scale, only Phe was lowered in cue1-6/eno1-2(+/−), whereas Tyr content remained unaltered and Trp content was even slightly enhanced. In contrast with flowers, relative and absolute contents of His and Arg were significantly increased in rosette leaves of cue1-6/eno1-2(+/−) (see Supplemental Figures 5R and 5S online).
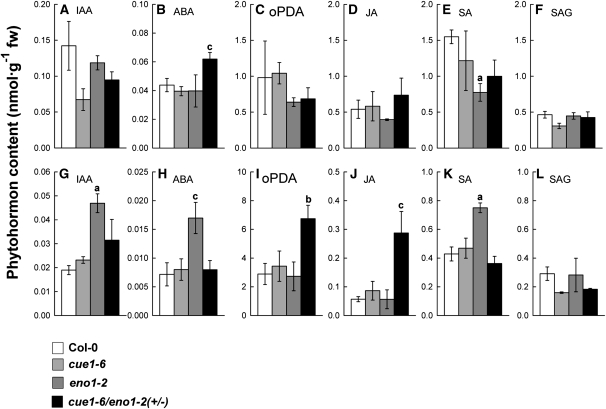
Phytohormone Levels Are Altered in Flowers and Rosette Leaves of ccEe Plants
To gain additional information for the underlying reason of the aberrant growth and developmental phenotype in ccEe plants, we analyzed the levels of phytohormones and signal molecules in the wild type, the cue1-6 and eno1-2 single mutants, as well as in cue1-6/eno1-2(+/−) (Figure 7). The growth regulator indole-3-acetic acid (IAA) has been of particular interest, as it can derive from the aromatic amino acid Trp (Bartel, 1997). In flowers, the IAA content seemed to be decreased in cue1-6 and cue1-6/eno1-2(+/−) (Figure 7A), and in rosette leaves, it was significantly increased (i.e., doubled) only in eno1-2 (Figure 7G). Cytokinin levels were below detection limits with the method applied. However, with the aid of an ultra performance liquid chromatography/time-of-flight mass spectrometry (TOF-MS) analysis of relative amounts of cis/trans zeatin could be detected, and a twofold increase in rosette leaves of cue1-6 and cue1-6/eno1-2(+/−) was found. The content of abscisic acid (ABA) was significantly increased in flowers but not in rosette leaves of ccEe plants (Figures 7B and 7H). Interestingly, in rosette leaves, the ABA level was significantly increased in leaves of the eno1-2 single mutant (Figure 7H). Strikingly, levels of jasmonic acid (JA) and its precursor 12-oxo-phytodienoic acid (oPDA) were significantly increased in rosette leaves of ccEe plants (Figures 7I and 7J) but remained unaltered in flowers (Figures 7C and 7D). High levels of JA have been shown to inhibit mitosis and thereby plant growth (Zhang and Turner, 2008). Salicylic acid (SA) responded with a decline in flowers and an increase in rosette leaves only in the eno1-2 single mutant (Figures 7E and 7K). The content of the SA glucoside (SAG) remained unaltered in all lines both in flowers and in rosette leaves (Figures 7F and 7L).
Figure 7.
Contents of Phytohormones in Flowers or Rosette Leaves of Col-0 Wild Type, cue1-6 and eno1-2 Single Mutants, and the Heterozygous eno1-2 Mutant in the Homozygous cue1-6 Background (cue1-6/eno1-2[+/−]).
Flowers ([A] to [F]) and rosette leaves ([G] to [L]). Phytohormones identified by liquid chromatography–mass spectrometry separation were IAA ([A] and [G]), ABA ([B] and [H]), oPDA ([C] and [I]), JA ([D] and [J]), SA ([E] and [K]), and SAG ([F] and [L]). The data represent the mean ± se of n = 3 independent experiments. Statistical significance of differences between the parameters was assessed by the Welch test with probability values of P < 0.01 (a), P < 0.02 (b), and P < 0.05 (c) indicated above the respective bars.
Secondary Plant Products Are Diminished in ccEe Plants
A variety of secondary plant products derive from phenylpropanoid metabolism, such as flavonoids (Dinkova-Kostova, 2008), anthocyanidin (Chalker-Scott, 1999), and lignin (Boerjan et al., 2003). Phenylpropanoids are synthesized from the aromatic amino acid Phe as one of the end products of the shikimate pathway. Flavonoids play a key role in UV protection (Dinkova-Kostova, 2008) or as putative signal molecules (e.g., Buer et al., 2007). In some species, flavonoids are also constituents of the cuticle or of suberin layers (Holloway, 1983; Mintz-Oron et al., 2008). It is conceivable that a decreased flux into aromatic amino acids and derived compounds (such as flavonoids) leads to disturbed development of the flowers as was observed for the ccEe plants (see Figure 2).
Flavonoid contents in mature flowers were quite variable between the lines and ranged between 6.51 and 9.64 nmol·g−1 FW (referred to naringinin as a flavonoid standard) in the control plant pOCA and in Col-0 wild type, respectively. There was no clear trend of diminished flavonoid content in the cue1 or eno1 single mutants compared with the wild type (see Supplemental Table 2 online). By contrast, flavonoid contents in cue1-1 appeared to be increased compared with the control plant. The flavonoid content in the ccEe plant [cue1-6/eno1-2(+/−)] of 5.56 nmol·g−1 FW was significantly diminished by 42% compared with the wild type but only 10% compared with the cue1-6 single mutant. Contents of anthocyanins were close to the detection limit in the flower buds of all lines investigated and therefore are not shown.
Lignin, as a constituent of cell walls (e.g., of the xylem elements), also derives from the phenylpropanoid metabolism. Staining with ACF (astrablue, chrysoidin, neofuchsin) revealed that lignification of specific tissues in the inflorescence stem of ccEe plants appeared to be diminished compared with the wild type or the cue1 single mutants (see Supplemental Figure 6 online; Turner and Sieburth, 2003). Interestingly, the interfascicular sclerenchyma cells in stems of Col-0 (see Supplemental Figure 6A online) and cue1-6 (see Supplemental Figure 6B online) showed a strong lignification, whereas the xylem elements were only faintly stained. By contrast, there was almost no detectable lignification of the sclerenchyma cells in stems of cue1-6/eno1-2(+/−) (see Supplemental Figure 6C online) or cue1-1/eno1-2(+/−) (see Supplemental Figure 6D online). In both lines, the only lignified cells appeared to be those of the xylem elements. It is so far unknown which factors control the degree of lignification in the individual cell types.
Is the Growth Phenotype of the ccEe Plants Caused by Impaired Cuticle Wax Synthesis?
In a recent report, Beaudoin et al. (2009) observed alterations in shoots and flower development in the kcr1 mutant, which is defective in β-ketoacyl-CoA reductase and, thus, disturbed in fatty acid elongation. These alterations were very similar to those observed for the ccEe plants. A complete knock out of this gene leads to embryo lethality. Interestingly, trichomes of kcr1 exhibited a distorted phenotype similar to those of the eno1 single mutant alleles (Prabhakar et al., 2009), which is also evident for the ccEe plants. It has been proposed that KCR1 is involved in the synthesis of cuticle waxes and the composition of sphingolipids. Toluidine blue (TB) staining of the kcr1 mutant revealed a loss of cuticle integrity.
We also addressed the question of cuticle integrity by TB staining (see Supplemental Figure 7 online). Interestingly, ccEe plants exhibited a positive staining with TB when compared with wild-type, cue1, and eno1 plants, suggesting a lack of cuticle integrity when the expression of ENO1 is reduced in the cue1 background.
Further insights into structural differences of the cuticle between the individual lines were obtained by scanning electron microscopy of inflorescence stem surfaces (see Supplemental Figure 8 online). Interestingly, the density of wax crystals was largely diminished on stems of the ccEc plants compared with the wild type and the cue1 and eno1 single mutants, suggesting diminished cuticle wax deposition.
A thorough cuticle wax analysis revealed that the total wax content was significantly increased rather than decreased in cue1 and ccEc plants (see Supplemental Figure 9A online). Moreover, there were some changes in the relative content of certain wax components, such as C29 aldehyde, C27 alkane, and C26 and C29 alcohol, which were significantly increased in ccEe (see Supplemental Figures 9C to 9E, 9H, and 9J online) or decreased, such as C29 ketone (see Supplemental Figure 9M online). It is likely that such compositional changes of cuticle wax components increase the wettability of the leaf and stem surface and, hence, the accessibility for TB.
Chlorophyll leaching experiments, which were performed as a further test for cuticle integrity, did not show any significant differences between the individual lines (see Supplemental Figure 10 online). In addition, light microscopy analysis of the adaxial leaf surface suggests a higher stomatal density in ccEe compared with the wild-type and both single mutant plants (see Supplemental Figure 11 online). Moreover, stomata of ccEe appear smaller and round rather than oval shaped (see Supplemental Figures 11G and 11H online) compared with the wild type and the single mutants (see Supplemental Figures 11A to 11F online). Most relevant for TB staining, stomatal pores of ccEe plants were widely open.
Finally, the expression of genes involved in cuticle wax biosynthesis, such as CER1, CER10, WAX2, BDG, KCR1, and KCR2 was not significantly affected (see Supplemental Figure 12 online). In summary, the phenotype of ccEe plants is not caused by an impaired cuticle wax biosynthesis, hence ruling out a shortage of lipid supply for wax biosynthesis due to the combined lesions in PPT1 and ENO1.
Ectopic Overexpression of ENO1 Rescues the cue1 Phenotype
In a previous report, we could demonstrate that ectopic overexpression of a C4-type PPDK, driven by the CaMV 35S promoter could rescue the cue1 leaf phenotype (Voll et al., 2003). PPDK is capable of producing PEP from pyruvate and thus replenishes PEP in plastids of those tissues where PPT1 is missing (e.g., in cells of the vasculature of the leaf) (Knappe et al., 2003). As ENO1 is not expressed in mature leaves (Prabhakar et al., 2009), PEP supply relies entirely on the import by the PPT and not on plastid glycolysis. Hence, we asked the question whether ectopic overexpression of ENO1 in the cue1-6 mutant background is capable of rescuing the cue1 phenotype. Of several transformed lines tested for ENO1 expression, cue1-6 ENO1 (4) and (5) showed the highest transcript abundance of ENO1 (Figure 8B). As a control, ENO1 was also overexpressed in wild-type Col-0, yielding two lines [Col-0 ENO1 (A) and (C)] with an increased transcript level of ENO1 (Figure 8B). As shown in Figures 8C and 8D, ENO1 overexpressors in the cue1-6 background had indeed a wild-type-like appearance (i.e., the reticulate leaf phenotype was completely rescued) (Figure 8D). However, cue1-6 transformants with a low transcript abundance of ENO1 [e.g., cue1-6 ENO1 (1)] still showed the cue1 phenotype (Figure 8C, 4), indicating that a threshold level of ENO1 transcripts had to be exceeded to exert a rescuing effect. A similar observation has been reported for PPDK-overexpressing lines in the cue1-6 background (Voll et al., 2003). Moreover, overexpression of ENO1 could also rescue the retarded growth phenotype of cue1-6 roots (Figure 8E) but had virtually no effect on the phenotypic appearance of roots and shoots when overexpressed in wild-type plants (Figure 8F). These data show that the only missing enzyme for a complete plastidic glycolytic pathway appears to be ENO1 and not plastidic PGyM, at least in those tissues where PPT1 is expressed in wild-type plants.
Figure 8.
Ectopic Overexpression of ENO1 Rescues the cue1 Phenotype.
(A) RT-PCR with RNA extracted from leaves of wild-type (Col-0) or transformants overexpressing ENO1 in the Col-0 background.
(B) RT-PCR with RNA extracted from leaves of cue1-6 or transformants overexpressing ENO1 in the cue1-6 background.
(C) Rosette phenotypes of the wild type (1), an ENO1-overexpressing line [cue1-6 ENO1 (4)] with a high transcript abundance of ENO1 (2), the cue1-6 mutant (3), and an ENO1-overexpressing line [cue1-6 ENO1 (1)] with a low transcript abundance of ENO1 (4).
(D) Growth phenotype of flowering Col-0 wild-type (1), an ENO1-overexpressing line in the cue1-6 background [cue1-6 ENO1 (4)] with a high transcript abundance of ENO1 (2), and cue1-6 (3) plants. The inset shows the rescue of the leaf phenotype of cue1-6 overexpressing ENO1 [line cue1-6 ENO1 (4)] in comparison with cue1-6 (5).
(E) Rescue of the retarded root phenotype of the cue1-6 mutant by overexpression of ENO1 in the cue1-6 background. Three cue1-6 mutant plants are compared with one ENO1-overexpressing line [i.e., cue1-6 ENO1 (4)].
(F) Phenotypic comparison of wild-type Col-0 with Col-0 plants overexpressing ENO1.
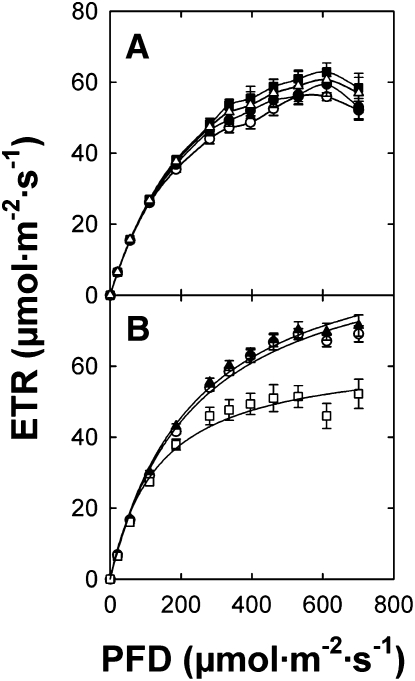
Ectopic Overexpression of ENO1 Restores Photosynthetic Electron Transport in cue1 but Has No Effect on Photosynthetic Performance of Wild-Type Plants
It has been shown previously that the rate of photosynthetic electron transport (ETR) is impaired in different alleles of the cue1 mutant (Streatfield et al., 1999; Voll et al., 2003), most likely based on decreased density of mesophyll cells characteristic of the leaf phenotype. Hence, the rescue of the cue1 mutant phenotype by overepressing ENO1 might restore ETR. We addressed this issue by measuring ETR with the aid of pulse amplitude modulation fluorometry. As shown in Figure 9, ETR in cue1-6 plants overexpressing ENO1 recovered to wild-type levels (Figure 9B). Moreover, there was no effect on ETR in wild-type plants overexpressing ENO1 (Figure 9A), indicating that a complete glycolytic pathway in chloroplasts does not interfere with ETR.
Figure 9.
Light Saturation Curves of Photosynthetic ETRs Determined by Imaging Pulse Amplitude Modulation Fluorometry.
(A) Comparison of light saturation curves between Col-0 (open circles), eno1-2 (triangles), Col-0 ENO1 (A) (closed circles), and Col-0 ENO1 (C) (squares).
(B) Comparison of light saturation curves between Col-0 (cirlces), cue1-6 (squares), and cue1-6 ENO1 (4) (triangles).
The data represent the mean ± se of n = 9 measurements on individual plants per line.
Overexpression of ENO1 Can Partially Rescue the Diminished Silique and Seed Production of cue1
As ENO1 together with PPT1 is most likely involved in the provision of PEP as precursor for fatty acid biosynthesis and for the shikimate pathway in plastids of seeds, we determined seed yield of the mutant plants and ENO1-overexpressing lines in the cue1 or Col-0 background (Table 3). For the cue1-6 mutant allele, silique number, seed weight, and number of seeds per plant were reduced significantly compared with wild-type plants. The diminished harvest of cue1 is most likely a consequence of decreased maximum photosynthesis rates and retarded growth of the shoots. Interestingly, the number of siliques and seeds was increased in both alleles of the eno1 mutant. However, the seed weight per plant was reduced, thus leading to an overall decline in the specific seed weight by 20% compared with the wild type. Two lines overexpressing ENO1 in the Col-0 background lacked any trend of a changed yield of siliques or seeds. Line Col-0 ENO1 (C) yielded similar numbers of siliques and specific seed weights as the wild type but exhibited lower numbers of seeds per plant, whereas Col-0 ENO1 (A) had similar seed numbers as the wild type but a slightly decreased specific seed weight (Table 3). The decreased number of siliques relative to the wild type was similar in cue1-6 plants overexpressing ENO1 [i.e., the lines cue1-6 ENO (4) and (5)] compared with the cue1-6 mutant plants, despite a complete rescue of the cue1 leaf and root phenotype (Table 3). Moreover, the number of seeds was considerably lower in both cue1-6 ENO1 lines than in wild-type plants but was increased compared with cue1-6, whereas the specific seed weight in cue1-6 ENO1 was similar to wild-type plants (Table 3). These data indicate that ectopic overexpression of ENO1 in the wild-type background had no effect on harvest parameters, whereas overexpression of ENO1 in the cue1 background could partially rescue the diminished silique and seed production characteristic for the cue1 mutant.
Table 3.
Harvest Parameters of Wild-Type Arabidopsis (Col-0), cue1-6, and ENO1-Overexpressing Lines in the Col-0 or cue1-6 Background
| Plant Line | Siliques per Plant | Seed Weight per Plant (mg) | Specific Seed Weight (μg·seed−1) | Number of Seeds per Plant |
| Col-0 | 208.5 ± 10.2 (100) | 104.2 ± 5.9 (100) | 14.4 ± 0.7 (100) | 7239 |
| cue 1-6 | 188.7 ± 6.7 (91) | 49.0 ± 2.4 (47)a | 12.4 ± 0.5 (86) | 3941 |
| Col-0 ENO1 (A) | 201.7 ± 7.6 (97) | 79.5 ± 5.9 (76)b | 14.5 ± 1.1 (101) | 5466 |
| Col-0 ENO1 (C) | 199.4 ± 5.4 (96) | 89.8 ± 7.7 (79) | 12.4 ± 0.6 (86)d | 7266 |
| cue 1-6 ENO1 (4) | 180.5 ± 9.0 (87)d | 79.1 ± 4.1 (76)b,a | 13.2 ± 0.2 (92) | 5989 |
| cue 1-6 ENO1 (5) | 186.2 ± 12.4 (88) | 82.1 ± 7.0 (86)c,a | 14.0 ± 0.6 (97)d | 5875 |
Plants were grown in a temperature-controlled greenhouse during Febuary and March. The data are expressed as mean ± se. The specific seed weight was estimated from 100 to 200 seeds counted. The data represent the mean value ± se of 15 individual plants per line. Statistical significance of differences between the parameters was assessed by the Welch test with probability values of P < 0.001 (a), P < 0.01 (b), P < 0.02 (c), and P < 0.05 (d). The bold letters in italics refer to cue1-6 as a control.
Seed Oil Content and Other Storage Compounds Are Severely Diminished in ccEe Seeds
In a next step, we analyzed the storage compounds in seeds (fatty acid content and composition as well as protein and carbohydrate contents) of all lines used in this study (Tables 4 and 5). Total fatty acids were quantified as a measure for seed oil content. The total lipid content in the wild-type plants Col-0 and Bensheim (pOCA) was 8.13 μg seed−1 and 8.45 μg seed−1, respectively (Table 4). It was slightly reduced in the cue1 and eno1 mutant alleles. This decrease was significant according to Welch test for cue1-6 and eno1-1 but less marked in cue1-1 and eno1-2. Of the ccEe plants, class I seeds showed a high, wild-type-like oil content of 7 to 10 μg·seed−1, while in class II seeds, the oil content was reduced to ~2.2 to 4.3 μg·seed−1, and class III seeds contained strongly reduced amounts of oil (1.9 to 2.5 μg·seed−1). The strong reduction in oil content in class II and class III seeds thus correlated with the seed size and was accompanied by an increase in the contents of saturated fatty acids (i.e., 16:0, 18:0, and 22:0), while the amounts of unsaturated fatty acids (i.e., 18:3 and 20:1) decreased (Table 4). The strong decrease in 20:1 content is indicative of a reduction in the amount of storage lipids versus membrane lipids in the seeds because 20:1 is restricted to the triacylglycerol pool. These changes are also reflected in a strong decrease in the desaturation index (ID), which indicates the number of double bonds in all unsaturated fatty acid classes divided by the number of all saturated fatty acid classes. Furthermore, the increase in 16:0 and the decrease in 18:3 affected the ratio of C16 to C18 fatty acids (for a complete comparison of fatty acid composition, see Supplemental Figure 13 online). Overexpression of ENO1 in the Col-0 background had no severe effect on lipid content and composition. The average lipid content was slightly less in Col-0 ENO1. This was supported by an increase in the C16/C18 ratio and a decrease in ID suggesting a diminished rather than an enhanced production of storage lipids.
Table 4.
Lipid Contents and Fatty Acid Composition in Seeds of Wild-Type Arabidopsis (Col-0 and pOCA), eno1 and cue1 Alleles, and ENO1-Overexpressing Lines in the Col-0 or cue1-6 Background and Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe Plants)
| Plant Line | Total Lipid (μg·Seed−1) | C16/C18 Ratio | C20:1 (Mol %) | ID | n |
| pOCA | 8.45 ± 0.29 | 0.123 | 13.60 ± 0.28 | 11.15 | 5 |
| cue1-1 | 6.63 ± 0.73 | 0.112 | 13.08 ± 0.47 | 12.91 | 5 |
| cue1-3 | 6.77 ± 0.44c | 0.109 | 13.96 ± 0.47 | 13.35 | 5 |
| Col-0 | 8.13 ± 0.27 | 0.104 | 13.56 ± 0.23 | 10.71 | 10 |
| cue1-6 | 5.70 ± 0.35a | 0.112 | 12.85 ± 0.32 | 12.41 | 5 |
| eno1-1 | 5.53 ± 0.29a | 0.142 | 12.98 ± 0.27 | 10.02 | 10 |
| eno1-2 | 7.77 ± 0.49 | 0.132 | 13.26 ± 0.18 | 10.28 | 10 |
| Col-0 ENO1 | 7.17 ± 0.55 | 0.154 | 12.63 ± 0.27d | 9.37 | 10 |
| cue1-6 ENO1 | 7.21 ± 0.30d,b | 0.137 | 13.49 ± 0.29 | 9.88 | 10 |
| cue1-1/eno1-1(+/−) (class III) | 2.51 ± 0.41b | 0.208 | 8.92 ± 0.72b | 5.56 | 5 |
| cue1-1/eno1-1(+/−) (class II) | 3.98 ± 0.44d | 0.141 | 10.83 ± 0.66 | 8.54 | 5 |
| cue1-1/eno1-1(+/−) (class I) | 10.40 ± 0.27b | 0.099 | 14.03 ± 0.29 | 13.02 | 5 |
| cue1-1/eno1-2(+/−) (class III) | 2.10 ± 0.20b | 0.278 | 4.24 ± 0.52a | 3.45 | 5 |
| cue1-1/eno1-2(+/−) (class II) | 2.25 ± 0.32b | 0.284 | 5.49 ± 0.55a | 3.26 | 5 |
| cue1-1/eno1-2(+/−) (class I) | 6.50 ± 0.13 | 0.121 | 11.67 ± 0.25 | 8.75 | 5 |
| cue1-3/eno1-2(+/−) (class III) | 2.18 ± 0.18a | 0.249 | 7.51 ± 0.61a | 3.98 | 5 |
| cue1-3/eno1-2(+/−) (class II) | 4.26 ± 0.37b | 0.162 | 10.35 ± 0.74c | 6.52 | 5 |
| cue1-3/eno1-2(+/−) (class I) | 7.00 ± 0.23 | 0.119 | 12.53 ± 0.20d | 9.76 | 5 |
| cue1-6/eno1-2(+/−) (class III) | 1.91 ± 0.19a | 0.323 | 6.01 ± 0.95b | 2.80 | 5 |
| cue1-6/eno1-2(+/−) (class II) | 2.62 ± 0.15a | 0.287 | 8.53 ± 0.73c | 3.90 | 5 |
| cue1-6/eno1-2(+/−) (class I) | 7.07 ± 0.26d | 0.132 | 11.62 ± 0.05c | 8.76 | 5 |
Lipid contents and fatty acid composition determined on individual seeds (n = 5 to 10) and C16/C18 ratios as well as the desaturation index (ID) were calculated from the mol % of individual fatty acids shown in Supplemental Figure 13 online. The data represent the mean value ± se. Statistical significance of differences between the parameters was assessed by the Welch test with probability values of P < 0.001 (a), P < 0.01 (b), P 0.02 (c), and P < 0.05 (d). Bold letters in italics refer to cue1-6 as a control.
Table 5.
Protein and Carbohydrate Contents in Seeds of Wild-Type Arabidopsis (Col-0 and pOCA), eno1 and cue1 Alleles, and ENO1-Overexpressing Lines in the Col-0 or cue1-6 Background and Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe Plants)
| Plant Line | Total Protein (μg·Seed−1) | Lipid/Protein Ratio | Sucrose | Glucose (ng·Seed−1) | Fructose | Seed Weight (μg·Seed−1) |
| pOCA | 10.28 ± 0.23 | 0.82 | 308 ± 31 | 5.10 ± 0.66 | 4.23 ± 0.44 | 33.33 ± 3.43 |
| cue1-1 | 10.60 ± 0.96 | 0.63 | 277 ± 10 | 7.61 ± 1.34 | 7.02 ± 1.01d | 15.72 ± 1.67d |
| cue1-3 | 10.98 ± 0.22 | 0.62 | 309 ± 4 | 4.99 ± 0.94 | 3.92 ± 1.72 | 26.67 ± 1.91 |
| Col-0 | 10.56 ± 0.31 | 0.77 | 249 ± 13 | 2.34 ± 0.60 | 2.39 ± 0.51 | 30.00 ± 1.17 |
| cue1-6 | 7.74 ± 0.18b | 0.74 | 374 ± 24c | 9.24 ± 4.14 | 7.58 ± 2.20 | 24.76 ± 1.91 |
| eno1-1 | 10.95 ± 0.12 | 0.51 | 387 ± 24b | 9.42 ± 1.98 | 7.02 ± 2.30 | 36.19 ± 0.95c |
| eno1-2 | 10.42 ± 0.29 | 0.75 | 317 ± 25 | 12.76 ± 1.07b | 8.23 ± 0.16b | 22.86 ± 2.86 |
| Col-0 ENO1 | 10.74 ± 0.44 | 0.67 | 341 ± 33d | 4.70 ± 0.60d | 3.91 ± 0.87 | 28.57 ± 3.76 |
| cue1-6 ENO1 | 9.86 ± 1.07 | 0.73 | 307 ± 14c | 5.65 ± 0.91d | 4.21 ± 0.60d | 28.10 ± 3.79 |
| cue1-1/eno1-2(+/−) (class III) | 1.50 ± 0.35a | 1.67 | 22 ± 1b | 6.88 ± 1.79 | 10.51 ± 1.70d | 7.62 ± 0.95a |
| cue1-1/eno1-2(+/−) (class II) | 4.83 ± 0.55b | 0.82 | 177 ± 8d | 2.45 ± 0.64d | 3.17 ± 0.74 | 13.33 ± 1.91b |
| cue1-1/eno1-2(+/−) (class I) | 12.14 ± 0.38c | 0.86 | 293 ± 10 | 6.59 ± 1.10 | 5.21 ± 1.21 | 27.62 ± 5.04 |
The contents of protein and carbohydrates (sucrose, glucose, and fructose) were referred to individual seeds of mutant and wild-type plants. The individual seed weight was determined in three to six batches of 35 seeds per line. The data represent the mean value ± se. Statistical significance of differences between the parameters were assessed by the Welch test with probability values of P < 0.001 (a), P < 0.01 (b), P < 0.02 (c), and P < 0.05 (d).
Mature Arabidopsis seeds store similar amounts of protein and lipids when as a percentage of the dry weight (Chen et al., 2009). As shown in Table 5, total protein content was similar in the single cue1 and eno1 mutants compared with the respective wild-type or control plants, leading to a decline in the lipid/protein ratio by 10 to 20% in the cue1 alleles and up to 30% in the eno1 alleles (Table 5). These data indicate that oil content rather than protein content responds more strongly when PPT1 and ENO1 are impaired. Similar to the seed oil content, protein content was severely diminished in class III seeds of the segregating ccEe plants, whereas class II seeds showed an intermediate decline in protein content.
Seeds of Arabidopsis contain sucrose as major carbohydrate. In mature seeds, starch was below the detection limit of the coupled enzymatic assay applied. Total carbohydrate contents were not appreciably affected in the single mutants compared with the respective wild-type plants (Table 5). Similar to seed oil and protein, sucrose content was severely diminished in class III and intermediately reduced in class II seeds of the ccEe plants. Interestingly the levels of both hexoses (glucose and fructose) were appreciably increased in the strong cue1 alleles as well as in both eno1 mutant alleles relative to the respective wild-type plants. Likewise, contents of both hexoses were enhanced in class III seeds of the ccEe plants, whereas class II seeds contained diminished hexose contents relative to the wild-type-like class I seeds. It is conceivable that a block in oil production leads to a diminished sucrose consumption by glycolysis (Lonien and Schwender, 2009) and results in enhanced hydrolytic cleavage of sucrose by invertase activities. Again, overexpression of ENO1 in the Col-0 or cue1-6 backgrounds had no strong effect on seed protein and carbohydrate contents. Increased carbohydrate contents have been observed in seeds of the wri1 mutant, defective in seed carbohydrate use (Focks and Benning, 1998; Lonien and Schwender, 2009).
DISCUSSION
In this report, we analyzed the central role of PEP in plant metabolism with the aid of Arabidopsis mutants impaired in PEP provision to the plastids. For this purpose, we crossed three different alleles of the cue1 mutant (defective in PPT1; Li et al., 1995; Streatfield et al., 1999; Knappe et al., 2003; Voll et al., 2003) with two alleles of the eno1 mutant (Prabhakar et al., 2009). Interestingly, plants lacking both the PPT1 and ENO1 genes could not be obtained as double homozygous lines and even heterozygous eno1 mutants in the homozygous cue1 background exhibited a severe dwarfish phenotype combined with aberrant flower, silique, and seed development. This result was surprising as Arabidopsis contains a second functional PPT gene (i.e., PPT2), which is expressed, apart from the roots, in most vegetative and generative tissues (see Supplemental Table 3 and Supplemental Figure 14 online). Moreover, Arabidopsis contains a PPDK gene, which could provide PEP from pyruvate in plastids. However, neither PPT2 nor PPDK is capable of compensating for the deficiencies in PPT1 and ENO1; thus, normal development of Arabidopsis plants depends on the presence of these two genes. Furthermore, it is surprising that a heterozygous defect in the ENO1 gene in the cue1 background has such a strong impact on whole-plant development, in particular as ENO1 is not expressed, for instance, in mature leaves. Hence, it is likely that fully functional PPT1 and ENO1 activities are required in early plant development (i.e., in the apical and leaf meristems, where both genes are highly expressed) (Prabhakar et al., 2009). A detailed analysis of the spatial and temporal expression pattern of genes involved in PEP and pyruvate metabolism in plastids is contained in Supplemental Table 3 and Supplemental Figure 14 online based on publicly available microarray data and the aid of the efp browser of the University of Toronto (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi; Winter et al., 2007).
In addition, we could rescue the cue1 phenotype by constitutive overexpression of ENO1, indicating that (1) a full complement of plastidial glycolytic enzymes can provide a sufficient flux into PEP to compensate for a deficiency in its import by PPT1 and (2) that ENO, but not PGyM, is the only missing enzyme for a complete glycolysis in chloroplasts and root plastids. However, overexpression of ENO1 could not improve seed yield and seed oil contents in wild-type plants.
The Segregation of cue1 and eno1 Revealed Constraints in Gametophyte and Sporophyte Development
The offspring of crosses between cue1 and eno1, using three alleles of the former and two alleles of the latter, did not yield double homozygotes, indicating that lethality of the ccee plants is independent of the ecotypes (i.e., Col-0 or Bensheim) or secondary mutations. Moreover, even the percentage of heterozygous eno1 mutants in the homozygous cue1 background (ccEe plants) was far below expectations (Table 1). Likewise, the percentage of heterozygous cue1 mutants in the homozygous eno1 (Ccee plants) background was less than half the expected number (see Supplemental Table 1B online). Strikingly, ccEe plants exhibited a pronounced phenotype in the sporophyte, including growth retardation, deformed flowers, and shortened siliques (Figure 2), whereas the sporophyte of Ccee plants was not noticeably compromised during vegetative development. As ENO1 expression was decreased in those vegetative tissues where ENO1 transcripts were highly abundant in wild-type plants (i.e., in roots and the shoot apex), the constraints in vegetative development in ccEe plants were most likely due to a diminished function of the ENO1 protein. Such sporophytic constraints were also reflected in the male and female TE (see Supplemental Table 1C online) calculated from the genotype distribution in the offsprings of reciprocal crosses between cue1-1 (male) × eno1-2 (female) and eno1-1 (male) × cue1-1 (female). For the cue1-1 mutation, the male and female TEs of ~30% were very similar, whereas for the eno1-2 mutation, the female TE was decreased to ~15% compared with the male TE, which again was close to 30%. Hence, the additional decline in the female TE for the eno1-2 mutation suggests sporophytic rather than gametophytic constraints.
Restrictions in the Shikimate Pathway and in Fatty Acid Biosynthesis Are the Main Reasons for Impaired Gametophyte Development
The underlying reasons for developmental constraints of female (Figure 3) and male gametophytes (Figure 4) in ccEe plants are based most likely on a restriction of metabolism starting from plastidial PEP.
While PEP imported into nongreen plastids by the PPT has to be generated by cytosolic glycolysis and/or gluconeogenesis, its formation inside the plastid ought to commence from imported Glc6P or 3-PGA and the subsequent conversion to PEP by plastid glycolysis involving PGyM and ENO1 (Figure 1A). It has been been shown previously that Glc6P can enter the plastid via one of the two Glc6P/phosphate translocators (GPTs) of Arabidopsis (Kammerer et al., 1998). Arabidopsis mutants defective in GPT1 could not be obtained as homozygous lines (Niewiadomski et al., 2005). In the heterozygous gpt1 mutant, both female and male gametophytes show high rates of lethality comparable to those of the ccEe plants. For developing pollen of the heterozygous gpt1 mutant, it could be shown that, besides the absence of starch, oil production seemed to be also diminished in phenotypically modified pollen. As Glc6P is required for the OPPP (Kruger and von Schaewen, 2003), by which reducing equivalents in form of NADPH are provided for anabolic reaction sequences, such as fatty acid biosynthesis, the absence of GPT1 in the haploid gametophytes was proposed to lead to a deficiency in membrane formation and eventually death (Niewiadomski et al., 2005). Hence, NADPH supply by the OPPP for fatty acid biosynthesis appears to be crucial for early gametophyte development.
Here, we could show that, in turn, a restriction in PEP supply to plastids leads to a high lethality rate of gametophytes in ccEe plants. It is tempting to speculate that, at least in the female gametophytes PEP, after conversion to pyruvate, serves as the major substrate for fatty acid biosynthesis. Hence, diminished fatty acid provision may lead to a halt in ovule development in ccEe plants (Figure 3) similar to the heterozygous gpt1 mutant (Niewiadomski et al., 2005).
However, diminished fatty acid supply cannot explain the high lethality rate of male gametophytes in ccEe plants. Ultrastructural analysis revealed that pollen of the ccEe plants contained at least similar numbers of lipid bodies and vacuoles compared with wild-type pollen (Figure 5), indicating that fatty acid biosynthesis is not likely to be restricted during male gametophyte development in ccEe plants. It is hence conceivable that lethality of pollen in the ccEe plants is rather due to a restriction in the shikimate pathway. This notion is supported by autofluorescence imaging of phenolic compounds in pollen sacs (see Supplemental Figure 2 online) and individual pollen of ccEe plants compared with the wild type (see Supplemental Figure 3 online). In both cases, phenolic compounds were severely diminished. Moreover, flowers of ccEe plants contained significantly decreased levels of flavonoids (see Supplemental Table 2 online), again indicating a restriction of the shikimate pathway in floral organs.
The exine structure, which is strongly defective in pollen of ccEe plants, consists mainly of sporopollonin, which is formed by the diploid cells of the pollen sac secretory tapetum (Ariizumi et al., 2004). Sporopollonin is an extremely rigid substance containing both long-chain fatty acids and phenolic compounds derived from the phenylpropanoid metabolism (Guilford et al., 1988; Wiermann et al., 2001). Hence, the impaired exine formation observed in the majority of the pollen (80%) in ccEe plants is most likely due to a diminished gene dosage of ENO1 in the absence of PPT1 in the tapetum cells rather than an absence of both proteins in the microspores. The heterozygous knockout of ENO1 in the cue1 background might thus hamper sporopollonin production by the tapetum cells due to diminished PEP provision for the shikimate pathway in the plastids therein.
However, an exine phenotype similar to that observed for pollen from ccEe plants has recently been reported for an Arabidopsis mutant defective in CYP704, a cytochrome P450–dependent long-chain fatty acid ω-hydroxylase involved in sporopollonin biosynthesis (Dobritsa et al., 2009), indicating that in the case of the cyp704 mutant, an impaired provision of long-chain fatty acids by the pollen sac tapetum compromises exine formation.
Seed Development in ccEe Plants Is Impaired Due to a Restriction in Fatty Acid Biosynthesis
Unlike plastids of nongreen tissues, plastids of developing oil seeds are mixotrophic and obtain substantial parts of their energy, reducing power and 3-PGA by photosynthesis (Ruuska et al., 2004; Li et al., 2006). In knockdown mutants or antisense plants of plastid-localized PK, seed development was severely compromised (Andre et al., 2007; Baud et al., 2007b), indicating that pyruvate as precursor for fatty acid biosynthesis is provided from PEP inside the plastids rather than by import from the cytosol. Moreover, the wri1 mutant, defective in a transcription factor (Cernac and Benning, 2004) that regulates the expression of genes involved in carbohydrate metabolism (e.g., glycolysis), showed a similar phenotype (Focks and Benning, 1998; Baud and Graham, 2006; Baud et al., 2007a). This aspect of embryo development has recently been further analyzed by elegant 13C flux studies (Lonien and Schwender, 2009) with mutant plants compromised in PKp (Baud et al., 2007b) or with wri1 (Focks and Benning, 1998; Ruuska et al., 2002; Baud et al., 2007a). Both pkp1 pkp2 double mutants and wri1 exhibit a severe decrease in seed oil contents. Hence, in Col-0 wild-type plants, the majority (89%) of plastidic pyruvate used for fatty acid biosynthesis in seeds was derived from PKp, and the residual might be shared by pyruvate import and oxidative decarboxylation by plastidic ME4. In our approach, seed oil content was severely diminished in class II and III seeds of the segregating ccEe plants, which were heterozygously mutated in the ENO1 gene in the background of the homozygous cue1 mutants (cf. Table 4), thus supporting the significance of PEP supply to plastids for seed oil production.
The Phenotype of ccEe Plants Is Not Connected to a Restriction in Cuticle Wax Biosynthesis
At a first glance, the sporophytic and gametophytic phenotype of ccEe plants resembled in many aspects mutant plants impaired in fatty acid elongation, such as kcr1, defective in a β-ketoacyl-CoA reductase (Beaudoin et al., 2009). Homozygous kcr1 mutants were embryo lethal, and siliques of heterozygous kcr1 mutants contained 25% aborted seeds. The remaining seeds appeared white and transparent. To analyze the function of KCR1 during vegetative development, Beaudoin et al. (2009) created RNA interference (RNAi) plants, which exhibited a variety of phenotypes, including deformed flowers, stunted growth of rosette leaves, as well as shortened and crooked siliques. These are very similar to the phenotypes of ccEe plants. Interestingly KCR1 RNAi plants also showed deformed trichomes similar to those of the eno1 single mutants (Prabhakar et al., 2009) and ccEe plants. Staining with TB and additional experimental approaches revealed that the phenotype of the KCR1 RNAi plants was most likely due to disturbed cuticle wax biosynthesis (Beaudoin et al., 2009).
Here, we observed a similar leakiness of the cuticle in leaves and stems of ccEe plants (see Supplemental Figure 7 online), suggesting impaired cuticle wax biosynthesis. In parallel to the enhanced TB staining, we found decreased density in epicuticular wax of stems (see Supplemental Figure 8 online). Surprisingly, wax amounts were not altered or even increased in ccEe plants (see Supplemental Figure 9 online). However, comparable observations have been made with other cuticle mutants in the past. Increased chlorophyll leaching from leaves and enhanced TB staining of leaves in mutants compared with the wild type were accompanied by significant increases in wax amounts (Aharoni et al., 2004; Schnurr et al., 2004; Kurdyukov et al., 2006) and not by decreases as could intuitively be expected. This leads to the conclusion that it must be ultrastructural changes in wax arrangement or in wax deposition to the cutin polymer probably combined with an increased stomatal aperture (see Supplemental Figure 11 online) leading to the observed effects of enhanced cuticular permeability and not necessarily decreases in wax amounts. These data also suggest that a restriction in fatty acid biosynthesis in the apical meristem, which would lead inevitably to a restriction in cuticle wax biosynthesis starting from stored oleosomes (Panikashvili and Aharoni, 2008), is not the main reason for the developmental phenotype of ccEe plants.
The Retarded Growth Phenotype of ccEe Plants Might Originate from an Increased Jasmonate Content and Disturbed Amino Acid Metabolism
To further elucidate the underlying reasons for the retarded growth phenotype of ccEe plants, the spectrum of proteinogenic free amino acids and phytohormones was determined in rosette leaves and flowers of wild-type, single mutant, and ccEe plants. Interestingly, leaves of both cue1 and ccEe plants contained elevated levels of total free amino acids, due to increased amounts of the major amino acids. By contrast, contents of minor amino acids, which in principle derive from plastidial PEP, such as aromatic and branched-chain amino acids, were severely diminished on a relative but not on an absolute scale. Similar changes in the amino acid spectrum have recently been reported for the Phe insensitive growth (pig) mutant of Arabidopsis (Voll et al., 2004). However, the underlying molecular reason for the disturbed amino acid composition in the pig mutant has not been further resolved. Strinkingly Ala, His, and Arg were dramatically increased in leaves of ccEe plants both on a relative and absolute basis. Arg has been proposed to act as a precursor for the synthesis of NO in plants (Guo et al., 2003), which exerts multiple effects on plant growth and development (del Rio et al., 2004).
Unlike in rosette leaves, in flowers, total free amino acid remained unchanged in the single mutants and the ccEe plants compared with the wild type. Of the aromatic amino acids, only Trp was significantly decreased in cue1-6/eno1-2(+/−). Since Trp can serve as a precursor for auxin biosynthesis (Bartel, 1997), the stunted growth of the shoot and constraints in early flower development could hence also be linked to modified auxin availability (Pagnussat et al., 2009; Vanneste and Friml, 2009). However, the auxin IAA decreased in flowers of cue1-6 and ccEe plants, whereas its content remained nearly unchanged in leaves of the same lines (Figure 7). Hence, diminished auxin contents can be ruled out as an underlying reason for the developmental constraints in ccEe plants. Interestingly, the eno1-2 single mutant exhibited a significant increase in IAA, ABA, and SA contents. However, increased levels of these compounds did not result in any obvious growth phenotype of the eno1 mutant. ABA levels were only slightly but significantly enhanced in flowers of ccEe plants compared with the wild type and the single mutants. However, it appears unlikely that the moderate increase in ABA levels is the major reason for constraints in flower development.
In rosette leaves, but not in flowers, contents of JA and its precursor oPDA exhibited a significant fourfold increase in ccEe compared to wild-type plants. It has been demonstrated that an increase in jasmonate levels causes growth retardation (e.g., in the cev1 mutant defective in the cellulose synthase CeSA3) (Ellis et al., 2002) or in the fatty acid oxygenation upregulated8 mutant (Rodríguez et al., 2010). Wound-induced increased JA levels inhibit mitosis and thus result in stunted growth or a bonsai phenotype of the shoot (Zhang and Turner, 2008). It is hence likely that increased JA levels are the major reason for growth retardation of ccEe plants. However, as JA derives from fatty acid metabolism, an increase in its content, as observed here, cannot be directly linked to the lesion in PPT1 and a decline in ENO1.
Cytokinin levels were increased in rosette leaves of both cue1 and ccEe plants. Pyruvate delivered from plastidic PEP by the action of PKp can enter the biosynthesis of isoprenoids via the MEP pathway. The MEP pathway has been shown to provide the prenyl group of the cytokinins trans-zeatin and isopentenyl adenine via plastid-localized isopentenyltransferases (Kasahara et al., 2004). However, an impairment of the MEP pathway in ccEe plants would most likely lead to diminished rather than increased cytokinin levels.
Considering the expression profile of ENO1 and PPT1 (see Supplemental Table 3 and Supplemental Figure 14 online), it is hard to conceive as to why a heterozygous lesion in ENO1 in the background of the cue1 mutant has such a dramatic effect on shoot development in Arabidopsis. Unlike PPT1, ENO1 is not expressed in mature leaves (see Prabhakar et al., 2009). However, both genes share expression in the apical shoot meristem and in the meristem of young leaves (Prabhakar et al., 2009). Hence, we conclude that a disturbance of the developmental program rather than in metabolism is the major cause for the retarded growth phenotype in ccEe plants. In leaves, PPT1 expression is found in the vasculature, including the bundle sheath. In particular, PPT1 is highly expressed in xylem parenchyma cells (Knappe et al., 2003), which led to the assumption that certain metabolic signals deriving from the phenylpropanoid pathway are generated in these cell types and may control mesophyll development (Voll et al., 2003). The link of the deficiency of PPT1 in vegetative tissues and the occurrence of the reticulate leaf phenotype of the cue1 mutant remains obscure and is currently the subject of intense investigations.
Constitutive ENO1 Overexpression Rescues the cue1 Phenotype
It has previously been shown that constitutive overexpression of the C4-type PPDK of Flaveria trinervia targeted to the plastids rescued the reticulate leaf phenotype of the cue1 mutant (Voll et al., 2003). Thus, a limitation of PEP import into the stroma via PPT1 can be compensated by PEP generation from pyruvate within the plastids. As ENO1 is not expressed in mature leaves (see Supplemental Table 3 and Supplemental Figure 14 online; Prabhakar et al., 2009), PEP provision by plastid-localized glycolysis can be excluded. Here, we could demonstrate that constitutive overexpression of ENO1 could rescue both the leaf and root phenotypes of cue1. Thus, our data demonstrate (1) that ENO1 is the only missing enzyme for a complete glycolytic pathway within plastids, at least in those cells where PPT1 is expressed, and (2) that sufficient flux through plastid glycolysis exists to compensate the restriction in PEP import into the stroma by PPT1. It is likely that in chloroplasts and in plastids from heterotrophic tissues, which lack ENO1 expression, glycolysis ceases at the step of 3-PGA to 2-PGA conversion (i.e., the reaction catalyzed by plastid-localized PGyM).
According to the ARAMEMNON database (http://aramemnon.botanik.uni-koeln.de/; Schwacke et al., 2004), the genome of Arabidopsis contains 24 putative PGyM genes. Of the 24 PGyM genes, there are six genes encoding proteins with a strongly predicted N-terminal transit peptide for plastid targeting (At1g22170, At1g58280, At1g78050, At3g52155, At5g22620, and At5g62840). Whereas At1g22170 and At1g78050 are not expressed in photosynthetic active tissues, the transcripts of of the other four are also abundant in leaves, suggesting that 2-PGA can be formed also in chloroplasts. It has been demonstrated that both PPTs of Arabidopsis are capable of a 2-PGA/PEP counterexchange (Knappe et al., 2003). It is hence conceivable that the PPT (PPT1 and/or PPT2) controls to certain extent the flux of photoassimilates into secondary metabolism (i.e., the shikimate pathway) by sensing the metabolic status within the mesophyll. Under conditions that favor the accumulation of 3-PGA and 2-PGA (catalyzed by PGyM) in the chloroplast stroma (e.g., as a consequence of sink limitation in the cytosol), 2-PGA/PEP counterexchange by the PPT would increase and hence open the way in the direction of the production of aromatic amino acids and follow-up products. As has recently been shown for tobacco (Nicotiana tabacum) plants with an antisense inhibition of a cytosolic ENO, a limitation in cytosolic PEP production results in a reticulate leaf phenotype similar to that of cue1, suggesting that in wild-type plants, cytosolic PEP supply is sufficient (Voll et al., 2009).
Constitutive overexpression of ENO1 in wild-type plants had neither an obvious effect on the final size of the transformants and leaf morphology nor on the photosynthetic capacity. However, a more detailed analysis is required to address the question as to whether a complete glycolytic pathway within the chloroplast stroma fine-tunes primary and secondary metabolism. In chloroplasts, the glycolytic sequence involving PGyM and ENO, for instance, would withdraw 3-PGA fixed by ribulose-1,5-bisphosphate carboxylase/oxygenase in the light and could hence result in a depletion of Calvin cycle intermediates combined with a decline in the production of photoassimilates.
Concluding Remarks
Our data clearly show that both PPT1 and ENO1 are indispensable for survival of Arabidopsis plants and demonstrate that plastidial PEP plays a crucial role for normal plant development. In the future, it will be challenging to gain more information on the fine-tuning of plastid-to-cytosol communication involving plastidial transport proteins, such as the pyruvate transporter and PPT2. For both transporters, there is lack of information on their functional involvement in plant development and metabolism because (1) the pyruvate transporter has not yet been identified and (2) there are no PPT2 knockout mutants available to date.
METHODS
Plant Material and Growth Conditions
Wild-type and mutant Arabidopsis thaliana plants, ecotype Col-0 or the transgenic control plant pOCA106, ecotype Bensheim (Li et al., 1995), were used in all experiments. The cue1 alleles (cue1-1, cue1-3, and cue1-6) were isolated in a genetic screen based on the aberrant expression of CAB3 (Li et al., 1995; Streatfield et al., 1999). The eno1 mutant alleles (eno1-1, N521328; and eno1-2, N421734) were provided by the Nottingham Arabidopsis Stock Centre (http://Arabidopsis.info/). Arabidopsis plants were germinated and grown on soil for 4 to 5 weeks in a temperature-controlled greenhouse at a 17-h-light/7-h-dark cycle. The average photon flux density on plant level was 70 to 100 μmol·m2·s1. For growth of plants in sterile culture, seeds were surface sterilized and germinated on agar supplemented with half-strength MS medium containing 2% sucrose (Duchefa). Plantlets were grown in a Percival growth cabinet at day/night temperatures of 21°C/18°C and a 17-h-light/7-h-dark cycle at an average photon flux density of 50 to 80 μmol·m2·s1.
Generation of cue1 eno1 Double Mutants and Segregation Analysis
Immature flowers of homozygous single mutants were emasculated and manually cross-pollinated. The crosses of cue1-1 × eno1-1, cue1-1 × eno1-2, cue1-3 × eno1-2, and cue1-6 × eno1-2 were performed using the eno1-1 or eno1-2 plants as the female parent. In the reciprocal cross of eno1-2 × cue1-1, the cue1-1 plant was used as the female parent. The F1 plants heterozygous for both alleles were selfed, and the heterozygous double mutants were identified by genotyping in the F2 or higher populations.
Genotyping of eno1 Mutants by PCR
Wild-type and mutant alleles of ENO1 were identified by PCR on genomic DNA extracted according to Edwards et al. (1991). The insertion site of the T-DNA from the GABI-Kat collection (Rosso et al., 2003) was identified using the T-DNA–specific primer GABI LB pAC161 08409 (5′-ATATTGACCATCATACTCATTGC-3′) in combination with the appropriate gene-specific primers, for example, the primer pairs ENO1-1 (sense) 5′-CAGGATGATTGGAGCTCATGG-3′ and ENO1-1 (antisense) 5′-CGACATATCTCTGAGCATCTG-3′ for the eno1-1 mutant allele and ENO1-2 (sense) 5′-ATCATGTCTGAGATTCCGTCG-3′ and ENO1-2 (antisense) 5′-CTGACCTTCACTTCCCATCTG-3′ for the eno1-2 mutant allele. For genotyping of the seed classes I, II, and III of the ccEc plants, 50 seeds of each class was selected and DNA extracted by the Extract-N-Amp-Seed kit (Sigma-Aldrich).
Genotyping of cue1 Mutants in the T3 Generation
The mutant alleles cue1-6 and cue1-1 were obtained from ethyl methanesulfonate–mutagenized seeds or after γ-ray treatment (deletion of the PPT1 locus), respectively. The hetero- or homozygous T-DNA insertion in the ENO1 gene was analyzed by PCR on genomic DNA, whereas heterozygous mutations in the PPT1 gene were tested in the progeny of the F2 plants after self-fertilization. Those plants that were heterozygous for cue1 in the F2 generation again segregated in the F3 generation and exhibited homozygous cue1 plants with its characteristic phenotype. Moreover, as mutant plants homozygous for the mutation in both genes could not be isolated, the occurrence of a heterozygous mutation in the PPT1 gene in the homozygous eno1 mutant background was analyzed by pollen viability tests in the F3 generation after self-fertilization of the F2 plants. Only those plants showing a pollen abortion of >10% in the F3 generation were considered to have been heterozygous for cue1 in the F2 generation.
TE
The TE of reciprocal crosses of cue1-1 and eno1-2 was calculated according to Blanvillain et al. (2008) and is defined as the number of mutated alleles divided by the number of total alleles times 100.
Expression Analysis
For RT-PCR analysis, total RNA was isolated from the individual lines using the RNA plant mini kit (Qiagen) according to the manufacturer’s instruction. Oligo(dT)-primed cDNA from 2 μg of total RNA (DNase treated) was synthesized using the Bioscript reverse transcriptase system (Bioline). ENO1 was amplified with the primer pair ENO1 (sense) 5′-CACCATGGCTTTGACTACAAAACC-3′ and ENO1 (antisense) 5′-TCATGGTGATCGGAAAGCTTCACC-3′. Actin2 served as a control with the primer pair Actin (sense) 5′-TAACTCTCCCGCTATGTATGT-3′ and Actin (antisense) R 5′-CCACTGAGCACAATGTTACCGTAC-3′. The amplifications was performed with the following settings, 94°C for 5 min followed by 35 cycles at 94, 55, and 72°C, 30 s each, and a final extension at 72°C for 7 min.
For qRT-PCR analyses, total RNA was isolated using the RNAeasy Plant mini kit (Qiagen) according to the manufacturer’s protocol, and cDNA was synthesized from 2 μg of total RNA as described above. The qRT-PCR reactions were conducted in the presence of the Power SYBR Green master mix (ABI Prism) according to the manufacturer’s instructions in a 7300 Real-Time PCR system (Applied Biosystems) equipped with Sequence Detection Software v1.4. Expression levels of candidate genes were normalized to Actin2 expression levels as a control. The qRT-PCR experiments were done on three independent biological samples and three technical replicates. The primer sequences of genes used in qRT-PCR experiments are listed in Supplemental Table 4 online, including those involved in cuticle wax biosynthesis, such as CER1 (Aarts et al., 1995), WAX2 (Chen et al., 2003), CER10/ECR (Zheng et al., 2005), BDG (Kurdyukov et al., 2006), KCR1, and KCR2 (Beaudoin et al., 2009).
Overexpression of ENO1
Plants constitutively overexpress ENO1 were generated by fusing the ENO1 cDNA to the CaMV 35S promoter and transforming the construct into Arabidopsis. PCR primers were designed for the full-length coding sequence of ENO1 (At1g74030). A sense primer ENO1-GW-F (5′-CACCATGGCTTTGACTACAAAACC-3′) containing the start codon, and an antisense primer containing the stop codon ENO1-2GW-R (5′-TCATGGTGATCGGAAAGCTTCACC-3′) was designed for cloning into the Gateway entry vector pENTR D-TOPO. The PCRs were run at 95°C for 2 min followed by 35 cycles (30 s at 95°C, 30 s at 59°C, and 1 min at 72°C) and a final extension at 72°C for 7 min. Full-length ENO1 was recombined by LR reaction into the Gateway destination vector pGWB2 and transformed into DH10B competent cells. Transgenic plants were generated by vacuum infiltration of Arabidopsis flowers using Agrobacterium tumefaciens cultures containing the appropriate construct (Clough and Bent, 1998). The primary transformants were allowed to flower and produce seeds. Transformants were selected with kanamycin and verified by PCR analysis.
Microscopy and Histochemical Analyses
For all histochemical analyses, the Eclipse E800 microscope (Nikon) equipped with differential interference contrast and fluorescence optics was used. Images were captured using a 1-CCD color video camera (KY-F1030; JVC) operated by the DISKUS software package (Technisches Buero Hilgers).
Pollen viability was assessed using Alexander staining (Alexander, 1969). Pollen grains released from the anthers were directly transferred to the staining solution and further analyzed by light microscopy. Male gametophyte development was analyzed by DAPI staining (Park et al., 1998) on mature, dehisced, and indehiscent pollen.
Phenolic compounds of mature pollen grains were visualized by fluorescence microscopy in the presence of a fluorescence enhancer. Pollen grains were incubated for 15 to 30 min in 0.25% diphenylborate-2-aminoethyl and 0.005% Triton X-100 and the fluorescence subsequently visualized with an excitation of 330 nm < λ < 380 nm and an emission filter λ > 420 nm.
Lignification of cell walls was analyzed in cross-sections of inflorescence stems 1 mm above the rosette with ACF (i.e. equal portions of astrablue, chrysoidin and neofuchsin solution at a concentration of 1 mg·mL−1 each; Etzold, 2002). In the presence of ACF, lignin stains red, suberin and cutin yellow, and cellulose cell walls are contrasted blue. The cross-sections were immersed in the ACF staining solution and immediately analyzed by light microscopy.
Developing ovules were analyzed by differential interference contrast light microscopy. Ovules were removed from the pistil or the silique and immersed in a droplet of water on a slide and covered with a cover slip. The whole-mount ovules were examined.
TB staining of rosette leaves and stems was essentially done as described by Tanaka et al. (2004).
Confocal Laser Scanning Microscopy
Phenolic compounds in anthers were analyzed by confocal laser scanning microscopy (excitation, 488 nm; emission, 500 to 550 nm) with a Leica SP2 microscope. Images were captured by the LCS software and further processed using Adobe Photoshop 6.
Scanning Electron Microscopy
Cutical wax crystals on the epidermis of inflorescence stems were visualized by scanning electron microscopy with a Leica Electron Optics 430i using the image analysis software SIS version 2.11. The stem samples were placed untreated (i.e., not sputtered) directly into the vacuum of the scanning electron microscopy device.
Ultrastructural Analysis
For transmission electron microscopy of pollen, stamens were taken from different flower stages and fixed with 2% glutaraldehyde in 50 mM phosphate buffer, pH 7.2, overnight at 4°C. Samples were postfixed in 1% osmium tetroxide for 8 h on ice, dehydrated in a graduated acetone series, including a step with 1% uranylacetate (in 50% acetone, 2 h), embedded in Spurr’s resin, and polymerized at 60°C for 72 h. Ultrathin sections (60 to 70 nm) were cut with a diamond knife (Diatome) on a Leica EM UC6 ultramicrotome (Leica Microsystems) and mounted on pioloform-coated copper grids. The sections were stained with lead citrate and uranyl acetate (Reynolds, 1963) and viewed with a JEOL JEM-2100 transmission electron microscope operated at 80 kV. Micrographs were taken using a 4k × 4k Gatan UltraScan 4000 CCD camera. The sections were also stained using TB stain (a 4:1 mixture of 1% TB, 1% borax, and 1% pyromin A) for light microscopy viewing.
In Vitro Germination of Pollen
The germination efficiency of pollen grains was assessed in vitro according to Fan et al. (2001). For each experiment, freshly anther-dehisced flowers were randomly collected from five different plants and were dipped on agar plates to transfer the pollen grains to the germination medium (5 mM MES/Tris, pH 5.8, 1 mM KCl, 10 mM CaCl2, 0.8 mM MgSO4, 1.5 mM boric acid, 1% [w/v] agar, 16.6% [w/v] sucrose, 3.65% [w/v] sorbitol, and 10 μg mL−1 myo-inositol). For the germination assay, plates were incubated for 20 h in a climate-controlled chamber at 22°C with 100% humidity in low light (30 μmol m−2 s−1) supplied by fluorescent tubes. For each agar plate, 100 pollen were counted and the germination percentage calculated.
Chlorophyll Leaching Assay
Chlorophyll leaching from rosette leaves was determined as described by Aharoni et al. (2004). Whole rosettes of 3-week-old plants were rinsed with tap water, weighed, and transferred to tubes containing 30 mL of 80% ethanol. The rosettes were incubated under gently agitation in the dark. For chlorophyll determination, 1 mL of the ethanol was removed from each sample every 10 min during the first hour and finally after 90 and 120 min. Chlorophyll contents were measured at 664 and 647 nm.
Amino Acid Analysis
For the analysis of free amino acid contents, the tissue (100 to 200 mg) was ground to a fine powder under liquid nitrogen and subsequently extracted with 50 μL of 1 M perchloric followed by 50 μL of 0.1 M perchloric acid. The pH of the extracts was adjusted to 7.0 with 9.3 μL of 5 M potassium carbonate. The mixture was centrifuged for 30 min at 20,800g at 4°C. The supernatants were transferred to fresh tubes for HPLC analysis. The samples were precolumn derivatized with o-phthalaldehyde (Lindroth and Mopper, 1979) and separated on a Hewlett Packard HP1100 HPLC system, using a Nucleodur 100-5 C18 ec column (Macherey and Nagel). An optimized stepwise gradient ranging from 7% buffer A (50% methanol and 50% acetonitrile) and 93% buffer B (40 mM sodium acetate, pH 6.5) to 80% buffer A was used for elution at 35°C for 33 min at a flow rate of 1 mL·min−1 after injection of 5-μL sample. Derivatives were detected by excitation at 230 nm and their fluorescence at 450 nm. Alternatively freshly ground frozen plant tissue was extracted for 20 min at 4°C sequentially with 900 μL 80% (v/v), 300 μL 50% (v/v), and 300 μL 80% (v/v) aqueous methanol (buffered with 2.5 mM HEPES/KOH, pH 7.5). Upon precolumn derivatization with orthophtalaledehyde, 5 μL of the derivates were subjected to HPLC analysis using a Hyperclone C18 BDS column (Phenomenex) connected to an Ultimate 3000 HPLC system coupled with the RF2000 fluorescence detector (Dionex). Fluorescent derivates were eluted (1.1 mL·min−1) at 35°C by a nonlinear solvent gradient: 100% A (8.8 mM sodium hydrogenphosphate, pH 7.5, and 0.2% tetrahydrofuran) for 0 to 2 min, 15% B (19 mM 8.8 mM sodium hydrogenphosphate, pH 7.5, 33% methanol, and 21% acetonitrile) for 2 to 8 min, 18% B for 8 to 10 min, 50% B for 10 to 14 min, 52% B for 14 to 18 min, 60% B for 18 to 25 min, 100% for 25 to 26 min, 100% B for 26 to 28 min, and 0% B for 28 to 30 min.
Fatty Acid Analysis
Total fatty acids in seeds were quantified by gas chromatography after derivatization to fatty acid methyl esters using pentadecanoic acid (15:0) as internal standard (Browse et al., 1986). The desaturation index (ID) was calculated as follows:
Analysis of Cuticular Wax
Wax analysis was conducted as described in detail by Kurdyukov et al. (2006). Arabidopsis shoots were immersed for 10 s in chloroform (CHCl3; Merck) for wax extraction. An internal standard (20 μL of tetracosane [Fluka]; chloroform solution 10 mg into 50 mL) was added to each sample directly after wax extraction. Hydroxyl and carboxylic groups were transformed into corresponding trimethylsilyl derivatives by reaction with N,N-bis-trimethylsilyl-trifluoroacetamide (Macherey-Nagal) in pyridine (30 min at 70°C). A sample of 1 μL was analyzed by gas chromatography with flame detection (CG-Hewlett Packard 5890 series H; Hewlett-Packard) with on-column injection (30 m DB-1 i.d. 0.32 mm, film 0.2 μm; JandW Scientific). Wax compounds were identified by gas chromatography–mass spectrometry (quadrupole mass selective detector HP 5971; Hewlett-Packard).
Determination of Phytohormones
The determination of the phytohormones IAA, ABA, oPDA, JA, SA, and SAG was performed by liquid chromatography–tandem mass spectrometry as previously described (Luo et al., 2009). For analysis of SAG, mass transitions were as follows: 299/137 (declustering potential −45 V, entrance potential −7 V, and collision energy −22 V).
For determination of the relative amount of zeatin, samples were extracted using the method described by Matyash et al. (2008). The polar phases were analyzed by an Ultra Performance LC (ACQUITY UPLC system; Waters) combined with a time-of-flight mass spectrometer (LCT Premier; Waters). For liquid chromatography, an ACQUITY UPLC BEH SHIELD RP18 column (1 × 100 mm, 1.7-μm particle size; Waters) was used at a temperature of 40°C, a flow rate of 0.2 mL/min, and with the following gradient: 0 to 0.5 min 100% A, 0.5 to 3 min from 0% B to 20% B, 3 to 8 min from 20% B to 100% B, 8 to 10 min 100% B, and 10 to 14 min 100% A (solvent system A, water/methanol/acetonitrile/formic acid [90:5:5:0.1, v/v/v/v]; B, acetonitrile/formic acid [100:0.1, v/v]).
The TOF-MS was operated in positive electrospray ionization mode in W optics with a mass resolution larger than 10,000. Data were acquired by MassLynx software in centroided format over a mass range of m/z 50 to 1200 with a scan duration of 0.5 s and an interscan delay of 0.1 s The capillary and the cone voltage were maintained at 2700 and 30 V and the desolvation and source temperature at 250 and 80°C, respectively. Nitrogen was used as cone (30 L/h) and desolvation gas (600 L/h). For accurate mass measurement of > 5 ppm root mean squared, all analyses were monitored using Leu-enkephaline ([M+H+] 556.2771 m/z; Sigma-Aldrich) as lock spray reference compound at a concentration of 0.5 μg/mL in acetonitrile/water (50:50, v/v) and a flow rate of 30 μL/min.
Determination of Flavonoids
For flavonoid measurements, six flowers were pulverized using liquid nitrogen and extracted as described previously (Pinto et al., 1999) in 200 μL of methanol:water:HCl (70:20:1 by volume) at 60°C for 1 h (99.8 methanol and 35.4% HCl), and the absorption at 310 nm was determined after cooling down the extracts to room temperature. As a standard, naringinin (Sigma-Aldrich) was used.
Determination of Protein, Soluble Sugars, and Starch in Seeds
For total protein analysis, 10 seeds were homogenized in 100 μL of extraction buffer (25 mM Tris-HCl, pH 8.0, 125 mM NaCl, 0.5 mM EDTA, and 0.5% [w/v] SDS) (Ruuska et al., 2002). The total protein content was measured in 100 μL of crude homogenate with a Bio-Rad detergent-compatible protein assay system using γ-globulin as standard according to the manufacturer’s instructions. Starch and soluble sugars were essentially extracted and determined enzymatically as described by Lin et al. (1988) starting from 35 seeds per sample.
Photosynthesis Measurements
Modulated chlorophyll a fluorescence emission from the upper leaf surface was measured with a pulse amplitude modulation fluorometer (IMAGING PAM, Maxi version; Walz) according to the principles of Schreiber et al. (1986). Photosynthetic ETRs were assessed according to Genty et al. (1989).
Statistical Evaluation of Experimental Data
The data are expressed as mean values ± se of the indicated number of independent measurements. Significant differences between two data sets were analyzed using the Welch test, which allows for unequal relative errors between two groups of measurements assuming that a Gauss distribution is applicable (Welch, 1947). For data plotting, SigmaPlot8.0 for Windows (SPSS) was used.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Actin2 (At5g09810; NM_121018), BDG (At1g64670; NM_105142), CER1 (At1g02205; NM_180601), CER10 (At3g55360; NM_115394), ENO1 (At1g74030; NM_106062), KCR1 (At1g67730; NM_105441), KCR2 (At1g24470; NM_102292), PPT1 (At5g33320; NM_122856), PPT2 (At3g01550; NM_111021), and WAX2 (At5g57800; NM_125164).
Supplemental Data
The following materials are available in the online version of this article
Supplemental Figure 1. Comparison of the Shoot and Root Phenotypes of Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe) with cue1 Single Mutants.
Supplemental Figure 2. Confocal Microscopy Images of Pollen Sacs from Col-0 Wild-Type Plants and Heterozygous ccEe Double Mutants.
Supplemental Figure 3. Autofluorescence of Pollen from a Wild-Type Plant and Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe).
Supplemental Figure 4. Comparison of Pollen Germination Rates between Arabidopsis Wild-Type (Col-0) and ccEe Plants with Alleles cue1-1/eno1-2(+/−) and cue1-6/eno1-2(+/−).
Supplemental Figure 5. Relative Contents of Amino Acids Extracted from Flower Buds or Rosette Leaves of the Wild Type (Col-0), cue1-6, and eno1 Single Mutants as well as the Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe).
Supplemental Figure 6. Cross Sections of the Inflorescence Stem of Heterozygous eno1 Mutants in the Homozygous cue1 Background (ccEe) Compared with Wild-Type and cue1 Plants.
Supplemental Figure 7. Typical Toluidine Blue Staining for Cuticle Integrity of Aerial Parts of the Heterozygous eno1 Mutant in the Homozygous cue1 Background (ccEe) Compared with the Wild Type (pOCA = Ecotype Bensheim, Col-0) and the cue1 and eno1 Single Mutants.
Supplemental Figure 8. Scanning Electron Microscopy Images of Epiculticular Wax Crystals of Inflorescence Stems of Arabidopsis Wild Type (Col-0, pOCA = Ecotype Bensheim) as well as Mutant Alleles of cue1, eno1, and ccEe.
Supplemental Figure 9. Epicuticular Wax Analysis of Inflorescence Stems of Arabidopsis Wild Type (pOCA = Ecotype Bensheim, Col-0) as well as Alleles of cue1, eno1, and cue1-1/eno1-2(+/−).
Supplemental Figure 10. Chl Leaching Experiment with Cut Leaves of 6-Week-Old Col-0, eno1-2, cue1-6, and cue1-6/eno1-2(+/−).
Supplemental Figure 11. Light Microscopy Images Showing the Distribution of Stomata as well as the Phenotypes of Stomatal Guard Cells on the Adaxial Surface of Rosette Leaves of the Wild Type (Col-0), the cue1-6 and eno1-2 Single Mutants, as well as in the Heteozygous eno1-2 Mutant in the Homozygous cue1-6 Background [cue1-6/eno1-2(+/−)].
Supplemental Figure 12. Relative Transcipt Levels of Genes Involved in Cuticular Wax Biosynthesis in cue1-6/eno1-2(+/−) Compared with the Wild Type.
Supplemental Figure 13. Relative Composition of Saturated and Unsaturated Fatty Acids Determined by Gas Chromatography after Derivatization to Fatty Acid Methyl Esters.
Supplemental Figure 14. Spatial and Developmental Expression Profiles of Genes Involved in PEP Delivery to Plastids.
Supplemental Table 1. Genotype Analysis of the F2 Generation of Crosses between cue1 and eno1 Mutants.
Supplemental Table 2. Content of Flavonoids in Flowers of Arabidopsis Wild-Type Plants, cue1 and eno1 Mutant Alleles, and Heterozygous eno1-2 Mutants in the Homozygous cue1-6 Background.
Supplemental Table 3. Detailed Tissue and Development-Specific Expression Profiles of Genes Involved in PEP Provision to Plastids as well as Pyruvate Synthesis in Plastids and the Cytosol.
Supplemental Table 4. Primer Pairs Used for qRT-PCR.
Supplementary Material
Acknowledgments
We thank the Deutsche Forschungsgemeinschaft and the International Graduate School in Genetics and Functional Genomics (Cologne) for funding. We thank Tanja Fuchs (University of Bonn) for help with fatty acid measurements. We also thank K.H. Linne von Berg (University of Cologne) for help with the scanning electron microscopy and Mojgan Shahriari (University of Cologne) for the introduction into confocal laser scanning microscopy.
References
- Aarts M.G., Keijzer C.J., Stiekema W.J., Pereira A. (1995). Molecular characterization of the CER1 gene of arabidopsis Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7: 2115–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A., Dixit S., Jetter R., Thoenes E., van Arkel G., Pereira A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Andre C., Froehlich J.E., Moll M.R., Benning C. (2007). A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 19: 2006–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T., Hatakeyama K., Hinata K., Inatsugi R., Nishida I., Sato S., Kato T., Tabata S., Toriyama K. (2004). Disruption of the novel plant protein NEF1 affects lipid accumulation in plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 39: 170–181 [DOI] [PubMed] [Google Scholar]
- Bagge P., Larsson C. (1986). Biosynthesis of aromatic amino acids by highly purified spinach chloroplasts - Compartmentation and regulation of the reactions. Physiol. Plant. 68: 641–647 [Google Scholar]
- Bartel B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. Mol. Biol. 48: 51–66 [DOI] [PubMed] [Google Scholar]
- Baud S., Graham I.A. (2006). A spatiotemporal analysis of enzymatic activities associated with carbon metabolism in wild-type and mutant embryos of Arabidopsis using in situ histochemistry. Plant J. 46: 155–169 [DOI] [PubMed] [Google Scholar]
- Baud S., Mendoza M.S., To A., Harscoët E., Lepiniec L., Dubreucq B. (2007a). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J. 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Baud S., Wuillème S., Dubreucq B., de Almeida A., Vuagnat C., Lepiniec L., Miquel M., Rochat C. (2007b). Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J. 52: 405–419 [DOI] [PubMed] [Google Scholar]
- Beaudoin F., Wu X., Li F., Haslam R.P., Markham J.E., Zheng H., Napier J.A., Kunst L. (2009). Functional characterization of the Arabidopsis thaliana β-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 150: 1174–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanvillain R., Boavida L.C., McCormick S., Ow D.W. (2008). Exportin1 genes are essential for development and function of the gametophytes in Arabidopsis thaliana. Genetics 180: 1493–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Borchert S., Harborth J., Schünemann D., Hoferichter P., Heldt H.W. (1993). Studies of the enzymatic capacities and transport properties of pea root plastids. Plant Physiol. 101: 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J., McCourt P.J., Somerville C.R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal. Biochem. 152: 141–145 [DOI] [PubMed] [Google Scholar]
- Buer C.S., Muday G.K., Djordjevic M.A. (2007). Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol. 145: 478–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A., Andre C., Hoffmann-Benning S., Benning C. (2006). WRI1 is required for seed germination and seedling establishment. Plant Physiol. 141: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A., Benning C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chalker-Scott L. (1999). Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 70: 1–9 [Google Scholar]
- Chen M., Mooney B.P., Hajduch M., Joshi T., Zhou M., Xu D., Thelen J.J. (2009). System analysis of an Arabidopsis mutant altered in de novo fatty acid synthesis reveals diverse changes in seed composition and metabolism. Plant Physiol. 150: 27–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Goodwin S.M., Boroff V.L., Liu X., Jenks M.A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- del Rio L.A., Corpasa F.J., Barroso J.B. (2004). Nitric oxide and nitric oxide synthase activity in plants. Phytochemistry 65: 783–792 [DOI] [PubMed] [Google Scholar]
- Dennis D.T. (1989). Fatty acid biosynthesis in plastids. Physiology, Biochemistry, and Genetics of Non-Green Plastids, Boyer C.D., Shannon J.C., Hardison R.C., (Rockville, MD: American Society of Plant Physiologists; ), pp. 120–129 [Google Scholar]
- Dinkova-Kostova A.T. (2008). Phytochemicals as protectors against ultraviolet radiation: Versatility of effects and mechanisms. Planta Med. 74: 1548–1559 [DOI] [PubMed] [Google Scholar]
- Dobritsa A.A., Shrestha J., Morant M., Pinot F., Matsuno M., Swanson R., Moller B.L., Preuss D. (2009). CYP704B1 is a long-chain fatty acid ω-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis thaliana. Plant Physiol. 151: 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P.J., Rawsthorne S. (2000). Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol. 122: 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias B.A., Givan C.V. (1979). Localization of pyruvate dehydrogenase complex in Pisum sativum chloroplasts. Plant Sci. Lett. 17: 115–122 [Google Scholar]
- Ellis C., Karafyllidis I., Wasternack C., Turner J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzold H. (2002). Simultanfärbung von Pflanzenschnitten mit Fuchsin, Chrysoidin und Astrablau. Mikrokosmos 91: 316–318 [Google Scholar]
- Fan L.M., Wang Y.F., Wang H., Wu W.H. (2001). In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. J. Exp. Bot. 52: 1603–1614 [PubMed] [Google Scholar]
- Fernie A.S., Carrari F., Sweetlove L.J. (2004). Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7: 254–261 [DOI] [PubMed] [Google Scholar]
- Fischer K., Kammerer B., Gutensohn M., Arbinger B., Weber A., Häusler R.E., Flügge U.I. (1997). A new class of plastidic phosphate translocators: A putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell 9: 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N., Benning C. (1998). wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genty B., Briantais J.M., Baker N.R. (1989). The relationship between the quantum yield of photosynthetic transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta 990: 87–92 [Google Scholar]
- Givan C.V. (1999). Evolving concepts in plant glycolysis: Two centuries of progress. Biol. Rev. Camb. Philos. Soc. 74: 277–309 [Google Scholar]
- Guilford W.J., Schneider D.M., Labovitz J., Opella S.J. (1988). High resolution solid state 13C NMR spectroscopy of sporopollenins from different plant taxa. Plant Physiol. 86: 134–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F.-Q., Okamoto M., Crawford N.M. (2003). Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302: 100–103 [DOI] [PubMed] [Google Scholar]
- He Y., et al. (2004). Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Herrmann K.M. (1995). The shikimate pathway: Early steps in the biosynthesis of aromatic compounds. Plant Cell 7: 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann K.M., Weaver L.M. (1999). The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 473–503 [DOI] [PubMed] [Google Scholar]
- Holloway P.J. (1983). Some variations in the composition of suberin from the cork layers of higher plants. Phytochemistry 22: 495–502 [Google Scholar]
- Journet E.P., Douce R. (1985). Enzymic capacities of purified cauliflower bud plastids for lipid-synthesis and carbohydrate-metabolism. Plant Physiol. 79: 458–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer B., Fischer K., Hilpert B., Schubert S., Gutensohn M., Weber A., Flügge U.I. (1998). Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6-phosphate/phosphate antiporter. Plant Cell 10: 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F., Rawsthorne S. (1994). Starch and fatty acid biosynthesis in plastids from developing embryos of oil seed rape. Plant J. 6: 795–805 [Google Scholar]
- Kasahara H., Takei K., Ueda N., Hishiyama S., Yamaya T., Kamiya Y., Yamaguchi S., Sakakibara H. (2004). Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J. Biol. Chem. 279: 14049–14054 [DOI] [PubMed] [Google Scholar]
- Kinsman E.A., Pyke K.A. (1998). Bundle sheath cells and cell-specific plastid development in Arabidopsis leaves. Development 125: 1815–1822 [DOI] [PubMed] [Google Scholar]
- Knappe S., Löttgert T., Schneider A., Voll L., Flügge U.I., Fischer K. (2003). Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis-AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J. 36: 411–420 [DOI] [PubMed] [Google Scholar]
- Koornneef M. (1990). Mutations affecting the testa color in Arabidopsis. Arabidopsis Inf. Serv. 27: 1–4 [Google Scholar]
- Kruger N.J., von Schaewen A. (2003). The oxidative pentose phosphate pathway: Structure and organisation. Curr. Opin. Plant Biol. 6: 236–246 [DOI] [PubMed] [Google Scholar]
- Kubis S.E., Pike M.J., Everett C.J., Hill L.M., Rawsthorne S. (2004). The import of phosphoenolpyruvate by plastids from developing embryos of oilseed rape, Brassica napus (L.), and its potential as a substrate for fatty acid synthesis. J. Exp. Bot. 55: 1455–1462 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S., Faust A., Nawrath C., Bär S., Voisin D., Efremova N., Franke R., Schreiber L., Saedler H., Métraux J.P., Yephremov A. (2006). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lernmark U., Gardestrom P. (1994). Distribution of pyruvate dehydrogenase complex activities between chloroplasts and mitochondria from leaves of different species. Plant Physiol. 106: 1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Culligan K., Dixon R.A., Chory J. (1995). CUE1: A mesophyll cell specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell 7: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Beisson F., Pollard M., Ohlrogge J. (2006). Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1999). The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 47–65 [DOI] [PubMed] [Google Scholar]
- Lin T.P., Caspar T., Somerville C., Preiss J. (1988). Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) lacking ADPglucose pyrophosphorylase activity. Plant Physiol. 86: 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth P., Mopper K. (1979). High-performance liquid-chromatographic determination of subpicomole amounts of amino-acids by precolumn fluorescence derivatization with ortho-phthaldialdehyde. Anal. Chem. 51: 1667–1674 [Google Scholar]
- Lonien J., Schwender J. (2009). Analysis of metabolic flux phenotypes for two Arabidopsis mutants with severe impairment in seed storage lipid synthesis. Plant Physiol. 151: 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z.-B., Janz D., Jiang Y., Göbel C., Wildhagen H., Tan H., Rennenberg H., Feussner I., Polle A. (2009). Upgrading root physiology for stress tolerance by ectomycorrhizas: insights from metabolite and transcriptional profiling into reprogramming for stress anticipation. Plant Physiol. 151: 1902–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyash V., Liebisch G., Kurzchalia T., Shevchenko A., Schwudkte D. (2008). Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49: 1137–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk J.A., Dennis D.T. (1992). A developmental analysis of the enolase isozymes from Ricinus communis. Plant Physiol. 99: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S., Mandel T., Rogachev I., Feldberg L., Lotan O., Yativ M., Wang Z., Jetter R., Venger I., , Adato A., Aharoni A. (2008). Gene expression and metabolism in tomato fruit surface tissues. Plant Physiol. 147: 823–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X., Zhu Q., Li X., Li J., Zeng Q., Rong H., Zhang H., Wu P. (2006). The hpa1 mutant of Arabidopsis reveals a crucial role of histidine homeostasis in root meristem maintenance. Plant Physiol. 141: 1425–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P., Knappe S., Geimer S., Fischer K., Schulz B., Unte U.S., Rosso M.G., Ache P., Flügge U.I., Schneider A. (2005). The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17: 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J.B., Jaworski J.G. (1997). Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Mol. Biol. 48: 109–136 [DOI] [PubMed] [Google Scholar]
- Ohlrogge J.B., Kuhn D.N., Stumpf P.K. (1979). Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proc. Natl. Acad. Sci. USA 76: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat G.C., Alandete-Saez M., Bowman J.L., Sundaresan V. (2009). Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324: 1684–1689 [DOI] [PubMed] [Google Scholar]
- Panikashvili D., Aharoni A. (2008). ABC-type transporters and cuticle assembly: Linking function to polarity in epidermis cells. Plant Signal. Behav. 3: 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.K., Howden R., Twell D. (1998). The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125: 3789–3799 [DOI] [PubMed] [Google Scholar]
- Parsley K., Hibberd J.M. (2006). The Arabidopsis PPDK gene is transcribed from two promoters to produce differentially expressed transcripts responsible for cytosolic and plastidic proteins. Plant Mol. Biol. 62: 339–349 [DOI] [PubMed] [Google Scholar]
- Pinto M.E., Casati P., Hsu T.P., Ku M.S., Edwards G.E. (1999). Effects of UV-B radiation on growth, photosynthesis, UV-B-absorbing compounds and NADP-malic enzyme in bean (Phaseolus vulgaris L.) grown under different nitrogen conditions. J. Photochem. Photobiol. 48: 200–209 [DOI] [PubMed] [Google Scholar]
- Plaxton W.C. (1996). The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 185–214 [DOI] [PubMed] [Google Scholar]
- Prabhakar V., Löttgert T., Gigolashvili T., Bell K., Flügge U.I., Häusler R.E. (2009). Molecular and functional characterization of the plastid-localized phosphoenolpyruvate enolase ENO1 from Arabidopsis thaliana. FEBS Lett. 583: 983–991 [DOI] [PubMed] [Google Scholar]
- Qi Q.G., Kleppingersparace K.F., Sparace S.A. (1995). The utilization of glycolytic intermediates as precursors for fatty acid biosynthesis by pea root plastids. Plant Physiol. 107: 413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawsthorne S. (2002). Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 41: 182–196 [DOI] [PubMed] [Google Scholar]
- Reid E.E., Thompson P., Lyttle C.R., Dennis D.T. (1977). Pyruvate dehydrogenase complex from higher plant mitochondria and proplastids. Plant Physiol. 59: 842–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds E.S. (1963). The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17: 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez V.M., Chételat A., Majcherczyk P., Farmer E.E. (2010). Chloroplastic phosphoadenosine phosphosulfate metabolism regulates basal levels of the prohormone jasmonic acid in Arabidopsis leaves. Plant Physiol. 152: 1335–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M.G., Li Y., Strizhov N., Reiss B., Dekker K., Weisshaar B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Ruuska S.A., Girke T., Benning C., Ohlrogge J.B. (2002). Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruuska S.A., Schwender J., Ohlrogge J.B. (2004). The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol. 136: 2700–2709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid J., Amrhein N. (1995). Molecular organization of the shikimate pathway in higher plants. Phytochemistry 39: 737–749 [Google Scholar]
- Schnurr J., Shockey J., Browse J. (2004). The Acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber U., Schliwa U., Bilger B. (1986). Continuous recording of photochemical and non-photochemical chlorophyll fluorescence quenching with a new type of modulation fluorometer. Photosynth. Res. 10: 51–62 [DOI] [PubMed] [Google Scholar]
- Schulze-Siebert D., Heineke D., Scharf H., Schultz G. (1984). Pyruvate-derived amino acids in spinach chloroplasts - Synthesis and regulation during photosynthetic carbon metabolism. Plant Physiol. 76: 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R., Flügge U.I., Kunze R. (2004). Plant membrane protein databases. Plant Physiol. Biochem. 42: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Schwender J., Ohlrogge J.B. (2002). Probing in vivo metabolism by stable isotope labeling of storage lipids and proteins in developing Brassica napus embryos. Plant Physiol. 130: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J., Ohlrogge J.B., Shachar-Hill Y. (2003). A flux model of glycolysis and the oxidative pentosephosphate pathway in developing Brassica napus embryos. J. Biol. Chem. 278: 29442–29453 [DOI] [PubMed] [Google Scholar]
- Schwender J., Shachar-Hill Y., Ohlrogge J.B. (2006). Mitochondrial metabolism in developing embryos of Brassica napus. J. Biol. Chem. 281: 34040–34047 [DOI] [PubMed] [Google Scholar]
- Smith R.G., Gauthier D.A., Dennis D.T., Turpin D.H. (1992). Malate dependent and pyruvate dependent fatty acid synthesis in leukoplastids from developing castor endosperm. Plant Physiol. 98: 1233–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M., Ap Rees T. (1979). Capacities of pea chloroplasts to catalyze the oxidative pentose phosphate pathway and glycolysis. Phytochemistry 18: 1905–1911 [Google Scholar]
- Streatfield S.J., Weber A., Kinsman E.A., Häusler R.E., Li J., Post-Beittenmiller D., Kaiser W.M., Pyke K.A., Flügge U.I., Chory J. (1999). The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid dependent nuclear gene expression. Plant Cell 11: 1609–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Tanaka H., Machida C., Watanabe M., Machida Y. (2004). A new method for rapid visualization of defects in leaf cuticule reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Turner S., Sieburth L.E. (March 22, 2003). Vascular patterning. The Arabidopsis Book, Somerville C.R., Meyerowitz E.M., (Rockville, MD: American Society of Plant Biologists; ), doi/10.1199/tab.0073, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Straeten D., Rodrigues-Pousada R.A., Goodman H.M., Van Montagu M. (1991). Plant enolase - Gene structure, expression, and evolution. Plant Cell 3: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2009). Auxin: A trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Voelker T., Kinney A.J. (2001). Variations in the biosynthesis of seed-storage lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 335–361 [DOI] [PubMed] [Google Scholar]
- Voll L., Häusler R.E., Hecker R., Weber A., Weissenbock G., Fiene G., Waffenschmidt S., Flügge U.I. (2003). The phenotype of the Arabidopsis cue1 mutant is not simply caused by a general restriction of the shikimate pathway. Plant J. 36: 301–317 [DOI] [PubMed] [Google Scholar]
- Voll L.M., Allaire E.E., Fiene G., Weber A.P. (2004). The Arabidopsis phenylalanine insensitive growth mutant exhibits a deregulated amino acid metabolism. Plant Physiol. 136: 3058–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll L.M., Hajirezaei M.R., Czogalla-Peter C., Lein W., Stitt M., Sonnewald U., Börnke F. (2009). Antisense inhibition of enolase strongly limits the metabolism of aromatic amino acids, but has only minor effects on respiration in leaves of transgenic tobacco plants. New Phytol. 184: 607–618 [DOI] [PubMed] [Google Scholar]
- Welch B.L. (1947). The generalization of “student’s” problem when several different population variances are involved. Biometrika 34: 28–35 [DOI] [PubMed] [Google Scholar]
- Wheeler M.C., Tronconi M.A., Drincovich M.F., Andreo C.S., Flügge U.I., Maurino V.G. (2005). A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol. 139: 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermann R., Ahlers F., Schmitz-Thom I. (2001). Sporopollenin. Biopolymers, Vol. 1, Stenbüchel A., Hofrichter M., (Weinheim, Germany: Wiley-VCH Verlag; ), pp. 209–227 [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An ‘‘Electronic Fluorescent Pictograph’’ browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Turner J.G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Rowland O., Kunst L. (2005). Disruptions of the Arabidopsis enoylenoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.