In the fungus Ustilago maydis, the ability to infect plants and the coordination of the cell cycle are closely interconnected. This work shows that a protein (Clp1) physically interacts with two master regulatory proteins (bW and Rbf1) that are both essential for pathogenic development and alters their function to coordinate infection and the cell cycle.
Abstract
Regulation of the cell cycle and morphogenetic switching during pathogenic and sexual development in Ustilago maydis is orchestrated by a concerted action of the a and b mating-type loci. Activation of either mating-type locus triggers the G2 cell cycle arrest that is a prerequisite for the formation of the infectious dikaryon; this cell cycle arrest is released only after penetration of the host plant. Here, we show that bW, one of the two homeodomain transcription factors encoded by the b mating-type locus, and the zinc-finger transcription factor Rbf1, a master regulator for pathogenic development, interact with Clp1 (clampless 1), a protein required for the distribution of nuclei during cell division of the dikaryon. In addition, we identify Cib1, a previously undiscovered bZIP transcription factor required for pathogenic development, as a Clp1-interacting protein. Clp1 interaction with bW blocks b-dependent functions, such as the b-dependent G2 cell cycle arrest and dimorphic switching. The interaction of Clp1 with Rbf1 results in the repression of the a-dependent pheromone pathway, conjugation tube formation, and the a-induced G2 cell cycle arrest. The concerted interaction of Clp1 with Rbf1 and bW coordinates a- and b-dependent cell cycle control and ensures cell cycle release and progression at the onset of biotrophic development.
INTRODUCTION
Biotrophic fungi are capable of infecting plants without killing their hosts. The establishment of the biotrophic interface during infection involves complex signaling between the pathogen and the host cells to suppress plant defense pathways or to guarantee the supply of nutrients provided by the host. The infection process requires precise timing and tight control of gene expression not only to ensure the correct temporal and spatial expression of various pathogenicity factors but also to adapt fungal development to the changing environment during disease progression (Skibbe et al., 2010). In the smut fungus Ustilago maydis, pathogenic and sexual development is tightly linked to cell cycle control (Perez-Martin et al., 2006). The sexual cycle of U. maydis is initiated by the fusion of two compatible haploid cells, called sporidia. Recognition of the two mating partners is achieved via a pheromone/receptor system encoded by the a mating-type locus (Bölker et al., 1992). Upon binding of the pheromone Mfa to the cognate receptor Pra, a double-tracked signaling cascade is induced that culminates in differential phosphorylation of the HMG transcription factor Prf1 by Protein Kinase A and the mitogen-activated protein kinase Kpp2 (Kaffarnik et al., 2003; Müller et al., 2003, 2004). In parallel, activation of the pheromone pathway triggers conjugation tube formation and G2 cell cycle arrest, thereby securing a synchronous stage of the cell cycle of the mating partners already before cytogamy (Spellig et al., 1994). After fusion of the conjugation tubes, maintenance of cell cycle arrest, sustained polar growth, and all subsequent steps of pathogenic and sexual development are realized via the action of the heterodimeric transcription factor bE/bW, which is encoded by the multiallelic b mating-type locus (reviewed in Brefort et al., 2009). The functional bE/bW heterodimeric complex is formed only when the bE and bW proteins originate from different alleles. The bE/bW heterodimer is sufficient to initiate the switch from budding to filamentous growth and to promote pathogenic development, as conclusively shown by the generation of haploid strains expressing the bE1/bW2 heterodimer. These so-called solopathogenic strains can infect plants without a mating partner (Bölker et al., 1995a; Kämper et al., 2006). Concomitantly, the formation of the bE/bW complex triggers a cell cycle arrest that is released only after penetration of the host plant. Recently, we identified a set of 345 genes regulated in response to the activation of the b signaling pathway (Heimel et al., 2010). These genes encode, for example, proteins involved in cell wall remodeling or potential secreted effectors, as well as proteins that function in cell cycle control. However, only a few of the 345 b-responsive genes identified are regulated via direct binding of the bE/bW heterodimer. The majority of the genes are regulated via Rbf1, a b-dependently expressed C2H2 zinc-finger transcription factor that serves as master regulator of the b-dependent transcriptional cascade. Rbf1 is required and sufficient for b-induced filament formation and G2 cell cycle arrest (Heimel et al., 2010). The cell cycle arrest is achieved via the Rbf1-dependent regulation of an array of transcription factors, for example, the zinc-finger protein Biz1 that was shown to downregulate the b-type cyclin Clb1, resulting in G2 cell cycle arrest (Flor-Parra et al., 2006). One of the few genes regulated by bE/bW and independently from Rbf1 is clp1 (Scherer et al., 2006; Heimel et al., 2010). Clp1 has been shown to be required for pathogenic development of U. maydis: strains in which clp1 has been deleted are not affected during saprophytic growth and are still able to penetrate the plant cell surface; however, development is stalled before the first cell division, and clamp cell formation, a process that ensures the proper distribution of the two different nuclei during dikaryotic growth, does not occur (Scherer et al., 2006). Although the clp1 gene is induced immediately after formation of the active bE/bW heterodimer, the Clp1 protein can be detected only when the hyphae penetrate the plant surface, which coincides with the release of the bE/bW- and Rbf1-mediated cell cycle block. In addition, ectopic expression of clp1 was shown to prevent b-dependent filament formation and cell cycle arrest (Scherer et al., 2006), indicating that the protein plays a decisive role during cell cycle release. Clp1-related proteins in the Basidiomycetes Coprinopsis cinerea and Cryptococcus neoformans have also been shown to be required for sexual development (Inada et al., 2001; Ekena et al., 2008); however, the molecular function of the protein is still unknown.
Here, we identify the central regulators of pathogenic development, bW and Rbf1, as well as the previously undiscovered pathogenicity factor Cib1 as Clp1-interacting proteins. Whereas the interaction of Clp1 with bW blocks b-function, the interaction of Clp1 with Rbf1 results in repression of the pheromone pathway. We further demonstrate that expression of Rbf1 is sufficient for the early phase of pathogenic development in the absence of the bE/bW heterodimer. The reinitiation of the cell cycle and nuclear division require the additional expression of Clp1. Based on these observations, we provide a model for the control of mating-type-dependent gene regulation and the cell cycle to facilitate pathogenic and sexual development.
RESULTS
We have shown previously that Clp1 is crucial for fungal proliferation after penetration of the host plant and that ectopic clp1 expression counteracts b-dependent filament formation and the b-dependent cell cycle block (Scherer et al., 2006). To gain insight into the molecular function of Clp1, we employed the yeast two-hybrid system to identify Clp1-interacting proteins, using the entire Clp1 open reading frame as bait. Four out of the 37 proteins identified as potential Clp1 interacting proteins (see Supplemental Table 1 online) encode transcription factors: (1) bW, a component of the bE/bW heterodimeric complex encoded by the b mating-type locus; (2) Rbf1, a C2H2 zinc-finger transcription factor that is the central regulator in the bE/bW-mediated regulatory cascade (Heimel et al., 2010); (3) Um02664, a putative GATA-type zinc-finger transcription factor with homology to the Neurospora crassa white collar 2 protein (wc2); and (4) Um11782, a putative bZIP transcription factor that was termed Cib1 (Clp1 interacting bZip1). Since the deletion of um02664 (wc2) did not result in any observable phenotype with respect to pathogenicity (Table 1), we refrained from further characterization of this protein. Quantitative β-galactosidase assays and sensitivity to 3-aminotriazole were used to measure the strength of the interaction of Clp1 with bW, Rbf1, and Cib1 in yeast. Binding of Clp1 to bW (24.51 ± 0.92) or Cib1 (79.52 ± 2.07) was found to be approximately twofold to sixfold stronger than Clp1 binding to Rbf1 (see Supplemental Figure 1 online). The physical interaction of Clp1 with bW, Rbf1, and Cib1 was further verified by coimmunoprecipitation. A full-length Clp1-Myc-tag fusion and HA-tagged versions of full-length Cib1 as well as of the bW and Rbf1 fragments identified in the yeast two-hybrid screen were generated by in vitro translation. Clp1-Myc was specifically coprecipitated from mixtures of Cib1-HA, bW-HA, or Rbf1-HA with an anti-HA antibody (Figure 1A), demonstrating that Clp1 physically interacts with all three proteins in vitro.
Table 1.
Pathogenicity of um02664 and cib1 Deletion Strains
| Strains Inoculated | No. of Plants Infected | Plants with Tumors |
| SG200 | 60 | 57 (95%) |
| SG200Δum02664 | 74 | 70 (95%) |
| SG200Δcib1 | 130 | 0 (0%) |
| FB1 × FB2 | 55 | 53 (96%) |
| FB1Δum02664 × FB2Δum02664 | 96 | 90 (94%) |
| FB1Δcib1 × FB2Δcib1 | 157 | 0 (0%) |
Seven-day-old maize plants were inoculated with the strains indicated, and tumor formation was scored 7 DAI.
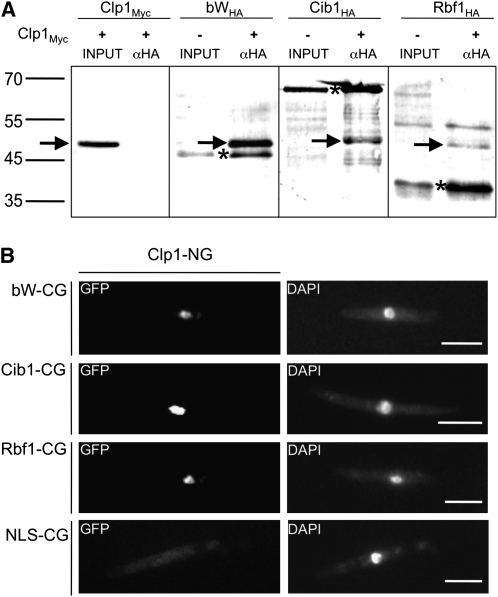
Figure 1.
Clp1 Interacts with bW and Rbf1 in Vitro and in Vivo.
(A) In vitro expression and coimmunoprecipitation of Myc-tagged Clp1 (~48 kD) and HA-tagged Cib1 (~61 kD), bW (~40 kD), and Rbf1 (~35 kD). In the negative control (Clp1Myc), no unspecific binding of Myc-tagged Clp1 to the αHA-antibody was detected after coincubation with lysate. By contrast, after coincubation with HA-tagged Cib1, bW, or Rbf1, the Myc-tagged Clp1 protein was coimmunoprecipitated, demonstrating that the Clp1 protein physically interacts with Cib1, bW, and Rbf1 in vitro. Proteins were labeled with biotin, detected with streptatividin-conjugated alkaline phosphatase, and visualized with NBT/BCIP. Asterisks indicate HA-tagged proteins, and arrows indicate the Myc-tagged Clp1 protein.
(B) BiFC analysis of Clp1-protein interactions. Clp1 fused to NG (N-terminal part of eGFP) was coexpressed with bW1, Cib1, Rbf1, or a nuclear localization sequence (NLS) from VP16 (negative control) fused to CG (C-terminal part of eGFP). A nuclear localized GFP signal was detected in strains coexpressing Clp1 with bW1, Cib1, or Rbf1. By contrast, only background fluorescence was observed in the negative control. For visualization of nuclei, cells were stained with 4’,6-diamidino-2-phenylindole (DAPI). Bars = 10 μm.
To confirm the physical interactions in vivo, we used the bimolecular fluorescence complementation assay (BiFC) (Hu et al., 2002; Kerppola, 2006). Full-length Clp1, Cib1, bW1, or Rbf1 proteins were fused to the N- (NG) or C-terminal part (CG) of enhanced green fluorescent protein (eGFP) and were coexpressed in the U. maydis strain FB2 under the control of the arabinose-inducible crg1 promoter. Coexpression of Clp1-NG either with Cib1-CG, bW1-CG, or Rbf1-CG led to a focused fluorescent signal in the nucleus, indicating protein–protein interactions. By contrast, when Clp1-NG was coexpressed with a nuclear-localized NLS-CG as control, only background fluorescence was observed (Figure 1B). Similarly, expression of bW-CG, Cib1-CG and Rbf1-CG with NLS-NG resulted in only background fluorescence (see Supplemental Figure 2 online). From our results, we conclude that Clp1 interacts with bW, Cib1, and Rbf1.
Cib1 Encodes a Putative bZIP Transcription Factor That Is Required for Pathogenic Development
cib1 encodes a predicted protein of 574 amino acids, containing a bZIP domain (amino acids 97 to 161), as predicted by SMART (Schultz et al., 1998). bZIP domains are commonly found in transcription factors and consist of a basic region that facilitates DNA binding and a leucine-zipper domain that is required for homo- or heterodimerization (Ellenberger et al., 1994; Hurst, 1995). To test whether Cib1 is required for pathogenic development of U. maydis, we deleted the cib1 open reading frame in the solopathogenic strain SG200 (a1 mfa2 bE1 bW2) and in the compatible haploid strains FB1 (a1 b1) and FB2 (a2 b2). Mutant strains were not impaired during growth in axenic culture, filament formation, or mating (data not shown). However, Δcib1 strains were blocked in pathogenic development. Maize (Zea mays) plants inoculated with a mixture of the strains FB1Δcib1 and FB2Δcib1 still showed chlorosis, but formation of tumors was not observed (Table 1). Similar results were obtained for the cib1 deletion in the solopathogenic strain SG200, indicating that loss of pathogenicity cannot be attributed to a defect in cell recognition or cell/cell fusion, since this strain infects plants without the requirement for a compatible mating partner.
To determine at which stage of pathogenic development the cib1 mutant strains were blocked, fungal cells in infected leaves were visualized by staining with Chlorazole Black E. Strain SG200 was used as control for the infection experiments. Three days after inoculation, SG200 had entered the plant via appressorium-like structures, and extensive proliferation of fungal material was observed in the leaf tissue (Figures 2A and 2B). Clamp-like structures with Y-shaped septa were frequently found (Figure 2C). cib1 deletion strains were not impaired in appressorium formation and were capable of penetrating the plant surface (Figure 2D). However, further proliferation of fungal cells was not observed. Invading hyphae were short and restricted to single plant cells of the outer epidermal layer (Figure 2E), and clamp-like structures were absent in the cib1 mutant strain (Figure 2F). Taken together, the phenotype is of striking similarity to that observed for clp1 deletion strains (Figures 2G to 2I) and infers a key function for the Clp1-Cib1 protein interaction during pathogenic development.
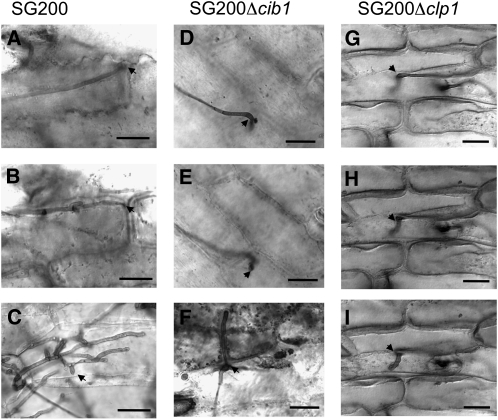
Figure 2.
cib1 Is Required for Proliferation in Planta.
Infected leaves were stained with Chlorazole Black E 3 DAI. The solopathogenic strain SG200 (a1 mfa2; bE1/bW2) penetrates the leaf surface via appressorium-like structures (A) and continues to proliferate in planta (B). Clamp cells of proliferating SG200 hyphae (C), indicative of active mitosis. Deletion of cib1 (D) or clp1 (G) in SG200 does not interfere with formation of appressoria, but proliferation of fungal hyphae is blocked ([E] and [H]) and clamp cells are not observed ([F] and [I]). Arrows mark appressorium-like structures ([A], [B], and [D] to [I]) or clamp cells (C). Bars = 20 μm.
Cib1 Is Posttranscriptionally Regulated and Localizes to the Nucleus
To analyze cib1 gene expression in vivo, we replaced the cib1 open reading frame with eGFP, generating strain UKH140 (a1 b1 Pcib1:eGFP). We detected strong GFP fluorescence in haploid sporidia during axenic growth (Figure 3A), indicating that the clp1 promoter mediates high expression under these conditions. We next aimed to determine the subcellular localization of Cib1 by fusing the C terminus of Cib1 to triple eGFP. The fusion construct was integrated into the cib1 locus of the solopathogenic strain SG200, replacing the endogenous cib1 gene. The resulting strain, USA1 (a1 mfa2 bE1 bW2 cib1:3xeGFP), infected maize plants with rates similar to that of the respective wild-type strain SG200, indicating that the Cib1:3xeGFP fusion protein was functional (see Supplemental Table 2 online). Surprisingly, despite the high expression of the cib1 gene during growth in axenic culture, expression of the Cib1:3xeGFP fusion protein was not detectable. However, when grown on the leaf surface of maize plants, the Cib1:3xeGFP fusion protein was found to localize in the nucleus of appressorium-like structures (Figure 3B). We were not able to detect a Cib1:3xGFP signal in cells prior to penetration of the leaf surface. Apparently, Cib1 protein expression is subject to posttranscriptional regulation.
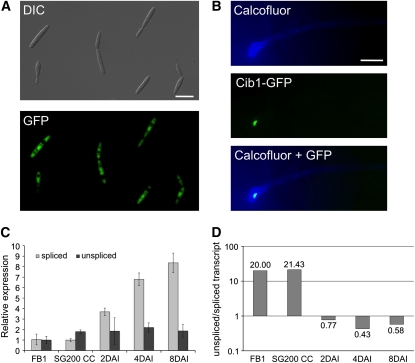
Figure 3.
Developmental Regulation of Cib1 Protein Expression and cib1 Splicing.
(A) cib1 gene expression was monitored by replacing the cib1 ORF with 3xeGFP (UKH140, a1 b1; Pcib1::3xeGFP). Shown is the morphology (differential interference contrast [DIC]) and eGFP expression of cells grown in axenic culture. Bar = 10 μm.
(B) A functional Cib1:3eGFP fusion protein was expressed under the control of the native promoter in strain USA1 (a1 mfa2; bE1/bW2; cib1:3xeGFP). Fungal hyphae were stained 24 h after inoculation with calcofluor to visualize appressorium-like structures. Nuclear localized GFP fluorescence was observed in fungal hyphae only after penetration of the leaf surface. Bar = 10 μm.
(C) Gene expression analysis of cib1 mRNA derivatives using qRT-PCR. Gene expression is shown relative to the lowest expression value. RNA samples were isolated from strain FB1 (a1b1) during growth in liquid MM-NG, from strain SG200 (a1 mfa2 bE1/bW2) during filamentous growth on solid Potato Dextrose medium containing charcoal (CC), and from leaf tissue infected with strain SG200 2, 4, and 8 DAI. Actin and eIF2b were used for normalization. Shown are the mean values of two technical replicates. Error bars represent the sd. The experiment was repeated three times with similar results.
(D) Molecular ratio of unspliced to spliced cib1 mRNA derivatives. The ratio of unspliced and spliced cib1 mRNA was obtained by calculating the absolute number of mRNA molecules by RT-PCR, using serial dilutions with standardized concentrations of 500-bp cib1 mRNA fragments with (unspliced) or without intron (spliced) for calibration. Calibration curves were calculated using the Bio-Rad Icycler software (unspliced r2 = 0.969; spliced r2 = 0.975); the absolute concentration of unspliced und spliced cib1 mRNA molecules from U. maydis strains grown in axenic culture or during pathogenic development were calculated by plotting the CT values of the RT-PCR analysis against the calibration curves. Samples for calibration were done in triplicate and samples for absolute quantification of gene expression in duplicates. The experiment was repeated three times with similar results.
Sequence analysis of cib1 cDNA obtained by 5′- and 3′-rapid amplification of cDNA ends revealed a 45-nucleotide 5′ untranslated region (UTR), a 196-nucleotide 3′ UTR, and a 65-nucleotide intron (see Supplemental Figure 3 online) that contained unusual splice sites not following the GU/AG rule. However, six out of eight cDNA clones still contained the intron sequences. Thus, we reasoned that the expression of the Cib1 protein may be achieved via developmentally regulated alternative splicing. Translation of the unspliced cib1 mRNA would result in a 436–amino acid protein identical to Cib1 up to amino acid 273, but dissimilar in the remaining 163 amino acid residues. Notably, the domain interacting with Clp1 (amino acids 374 to 535), as deduced from the common region in the three independent Clp1 interacting Cib1 fragments identified in the yeast two-hybrid screen (see Supplemental Table 1 online), would not be present in the protein resulting from unspliced cib1 mRNA. To address the alternative splicing, we performed quantitative RT-PCR (qRT-PCR) with primer sets specific for (1) the spliced or (2) the putative unspliced cib1 mRNA derivative (see Supplemental Figure 3 online) using RNA isolated from different developmental stages. The constitutively expressed genes for actin and eIF2b were used for normalization. Expression of both cib1 mRNA derivatives was similar in FB1 cells grown (by budding) in liquid medium (minimal medium containing nitrate and glucose [MM-NG]) as well as in SG200 cells growing filamentously on solid Potato Dextrose medium plates containing charcoal (Figure 2C). However, during pathogenic development, expression of the spliced derivative increased 8.3-fold (8 d after inoculation [DAI]), whereas the expression levels of the unspliced mRNA remained constant (Figure 3C). Since the relative expression values obtained by normalization to constitutively expressed genes does not allow a direct comparison of the abundance of different mRNA molecules, we determined the ratio of unspliced to spliced cib1 mRNA by calculating the absolute number of mRNA molecules by RT-PCR using serial dilutions with standardized concentrations of 500-bp cib1 mRNA fragments with (unspliced) or without intron (spliced) for calibration. During axenic culture of strain FB1 in liquid MM-NG and during filamentous growth of SG200 on solid Potato Dextrose medium plates containing charcoal, the ratio of unspliced to spliced derivative was determined to be 20:1 and 21.4:1, respectively. By contrast, during biotrophic development, the unspliced derivative was approximately twofold less abundant than the spliced derivative (Figure 3D). These data are in line with the upregulation of the spliced transcript during biotrophic development, suggesting that the restriction of Cib1 expression to the biotrophic growth phase is mediated via an alternative splicing mechanism.
Clp1 Blocks the Function of a bE-bW Fusion Protein
Induced expression of either the active bE/bW heterodimer or Rbf1 triggers the dimorphic switch from budding yeast-like growth to filamentous tip growth, accompanied by a G2 cell cycle arrest (Brachmann et al., 2001; Mielnichuk et al., 2009). We have previously shown that simultaneous induction of clp1 and bE1/bW2 blocks the b-dependent developmental switch as well as the expression of b-dependent genes (Scherer et al., 2006). It is conceivable that the clp1-dependent inhibition of b function is achieved via physical interaction of Clp1 and bW.
To test whether the inhibition of the bE/bW function by Clp1 may be realized via the hindrance of bE/bW complex formation, we tested whether Clp1 would also alter the function of Kon8. Kon8 is a fusion protein in which the bE1 and bW2 proteins, both lacking the N-terminal dimerization domains, are tethered together via a kinker region, thus bypassing the requirement for heterodimerization (Romeis et al., 1997; Grandel et al., 2000). Although the Kon8 protein harbors a deletion within the C-terminal part of bW, the remaining part of bW is sufficient for interaction with Clp1 (see Supplemental Figure 4 online). To address whether Clp1 alters the function of Kon8, we used strain UKH155 (a2 Pnar1:kon8), which harbors a nitrate-inducible kon8 allele integrated into the ip locus, and strain UKH125 (a2 Δb::Pcrg1:clp1; Pnar1:kon8), a UKH155 derivative in which the b mating-type locus was replaced by an arabinose-inducible clp1 allele. When spotted on charcoal-containing solid minimal media supplemented with nitrate and glucose, induced expression of kon8 triggered extensive filament formation in both strains, indicative of an active b pathway. By contrast, on charcoal-containing solid minimal media supplemented with nitrate and arabinose, strain UKH155 (expressing kon8) exhibited filamentous growth, whereas no filament formation was observed in strain UKH125 (expressing kon8 and clp1) (see Supplemental Figure 4 online). Accordingly, these data indicate that induced expression of clp1 blocks kon8-dependent filament formation and that the clp1-mediated counteraction of b function is unlikely to occur via disruption of bE/bW dimerization.
Clp1 Counteracts rbf1-Dependent Filament Formation and Blocks Expression of the Pheromone Precursor and Receptor
To determine whether Clp1 would also affect Rbf1 function, we constructed the U. maydis strains UKH156 (a1 Δb::Pnar1:rbf1), in which the b locus was replaced by a nitrate-inducible allele of rbf1, and UKH164 (a1 Δb::Pnar1:rbf1; Pcrg1:clp1), a UKH156 derivative harboring an arabinose-inducible allele of clp1 integrated in the ip locus. Induction of rbf1 in MM-NG led in both strains to identical filament formation (Figures 4A and 4B). However, the simultaneous induction of clp1 and rbf1 in minimal medium containing nitrate and arabinose (MM-NA) led to a fourfold reduction of filamentous cells in strain UKH164 compared with UKH156 (Figures 4A and 4B). The effect of Clp1 on rbf1-dependent gene expression was assessed by microarray analysis comparing strains UKH156 (induction of rbf1) and UKH164 (induction of clp1 and rbf1) after 12 h of growth under inducing conditions in MM-NA. We noted a 37.5-fold reduction of the expression of the pheromone gene mfa1 and a 4-fold reduced expression of the pheromone receptor gene pra1 upon clp1 expression (see Supplemental Table 3 online). To verify the microarray results, we performed qRT-PCR analysis. mfa1 and pra1 expression was comparable in strain UKH156 (induced expression of rbf1) and the control strain JB1 (a1 Δb), demonstrating that rbf1 expression itself does not affect mfa1 and pra1 expression. However, in strain UKH164, the coexpression of rbf1 and clp1 led to a drastic repression of the mfa1 and pra1 genes (Figure 4C), confirming the results obtained by microarray analyses.
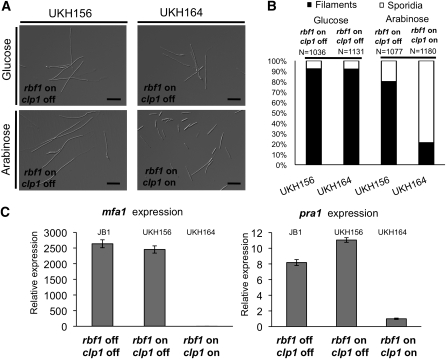
Figure 4.
Induced Coexpression of clp1 and rbf1 Results in Reduced rbf1-Dependent Filament Formation and Repression of the a Mating-Type Genes.
(A) Strains UKH156 (a1 Δb::Pnar1:rbf1) and UKH164 (a1 Δb::Pnar1:rbf1; Pcrg1:clp1) were grown in nitrate minimal medium supplemented with glucose (clp1 not expressed) or arabinose (clp1 expressed) as sole the carbon source. In UKH156, the nitrate-induced expression of rbf1 leads to filament formation; the arabinose-induced coexpression of clp1 in UKH164 results in reduced filament formation.
(B) Quantification of filament formation in strains UKH156 and UKH164 12 h after induction of rbf1 with or without clp1 as shown in (A). N = number of cells counted.
(C) qRT-PCR analysis of mfa1 and pra1 gene expression in response to rbf1 or clp1 induction. Strains UKH156 and UKH164 and, as a control, JB1 (a1 Δb) were grown for 12 h in nitrate minimal medium supplemented with arabinose. Shown are the mean values of two technical replicates. The experiment was repeated twice with similar results. Actin and eIF2b were used for normalization. Error bars represent the sd. Expression was calculated relative to the lowest expression value.
Clp1 Inhibits the Morphological and Regulatory Pheromone Response in an rbf1-Dependent Manner
The expression of both mfa1 and pra1 is highly induced after activation of the pheromone pathway (Urban et al., 1996). Thus, the expression monitored in UKH156 and JB1 reflects only the noninduced, basal transcription level of the genes, since the pheromone pathway is not induced. Consequently, we tested whether pheromone-dependent induction of mfa1 and pra1 would also be influenced by coexpression of clp1 and rbf1.
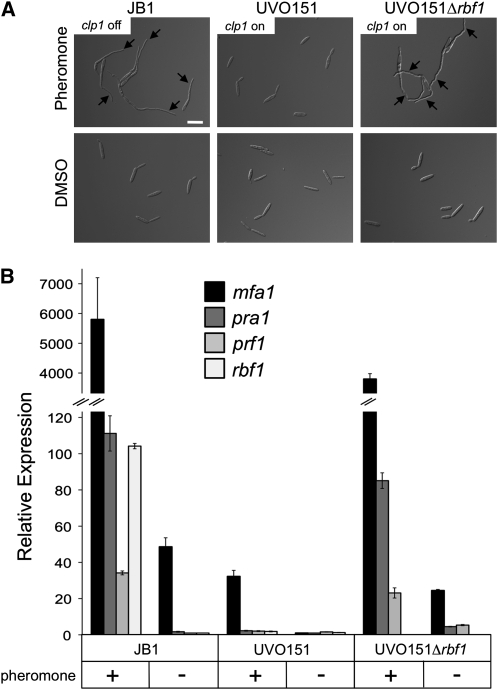
Since rbf1 expression itself is induced via the pheromone pathway (Zarnack et al., 2008), we used strain UVO151 (a1 Δb Pcrg1:clp1) (Scherer et al., 2006) that allows pheromone-mediated rbf1 induction and arabinose-dependent clp1 expression. Strains JB1 and a UVO151 derivative deleted in rbf1 (UVO151Δrbf1; a1 Δb Δrbf1; Pcrg1:clp1) were used as controls. To induce the pheromone pathway, cells were treated for 6 h with synthetic a2 pheromone in minimal medium containing nitrate and arabinose (MM-NA); under these conditions, both rbf1 and clp1 are induced. Upon pheromone treatment of JB1, >80% of the cells formed filamentous conjugation tubes (Figure 5A). The simultaneous expression of rbf1 and clp1 in UVO151 led to a drastically reduced conjugation tube formation of <2%. Cells were not cell cycle arrested but continued to grow by budding (Figure 5A). Pheromone treatment of UVO151Δrbf1, however, led to the formation of conjugation tubes comparable to that of JB1 (Figure 5A).
Figure 5.
Induced Expression of clp1 Blocks Pheromone-Induced Conjugation Tube Formation and Cell Cycle Arrest in an rbf1-Dependent Manner.
(A) Strains JB1 (a1 Δb), UVO151 (a1 Δb; Pcrg1:clp1), and UVO151Δrbf1 (a1 Δb; Δrbf1 Pcrg1:clp1) were treated with synthetic a2 pheromone (2.5 μg/mL) for 6 h in MM-NA. DMSO, which was used as solvent for the a2 pheromone, served as negative control. Strains JB1 and UVO151Δrbf1 respond to pheromone treatment with the formation of conjugation tubes (arrows). By contrast, cells of strain UVO151 do not form conjugation tubes, but continue to grow by budding, indicating that the cell cycle is not arrested upon pheromone treatment in that strain. Bar = 15 μm.
(B) Pheromone-dependent gene expression in strains JB1 (a1 Δb), UVO151 (a1 Δb; Pcrg1:clp1), and UVO151Δrbf1 (a1 Δb; Δrbf1; Pcrg1:clp1). Cells were treated as described above, and gene expression of the a mating-type locus genes mfa1 and pra1 and of the pheromone response factors prf1 and rbf1 was determined using qRT-PCR. Shown are mean values of two technical replicates. Error bars represent the sd. Actin and eIF2b were used for normalization. The experiment was repeated three times with similar results.
Expression analysis by qRT-PCR revealed that mfa1 and pra1, as well as prf1, encoding the central transcriptional regulator of pheromone signaling (Hartmann et al., 1996; Urban et al., 1996), and rbf1 were drastically induced upon pheromone treatment in JB1 and UVO151Δrbf1. By contrast, no induction was observed in strain UVO151 under identical conditions (Figure 5B). These data demonstrate that induced expression of clp1 blocks pheromone-dependent gene regulation, conjugation tube formation, and the pheromone-induced cell cycle arrest. All three inhibitory effects of Clp1 depend on Rbf1.
Ectopic Expression of Rbf1 Is Sufficient for the Initial Stages of Pathogenic Development Independent from the b Mating-Type Locus
Although the b locus is considered to be the major control instance for pathogenic development, 90% of the b-responsive genes depend on Rbf1 for their regulation. Rbf1 is required for the b-induced cell cycle arrest; U. maydis cells deleted in rbf1 do not arrest the cell cycle upon b induction (Heimel et al., 2010). The cell cycle arrest is released only when the U. maydis hyphae penetrate the plant surface (Snetselaar and Mims, 1992), which coincides with the nuclear localization of the Clp1 protein (Scherer et al., 2006). To release the b-mediated cell cycle arrest and to facilitate further fungal development within the plant, the function of either bE/bW or Rbf1 (or both) has to be altered. To elucidate whether clp1 is required for the cell cycle release, we attempted to dissect the bE/bW-Rbf1 regulatory cascade during pathogenic development. Since plant-derived signals/factors influence this regulatory network (Wahl et al., 2010) and are crucial for the reinitiation of growth after appressorium formation, analysis of fungal proliferation is restricted to the host plant.
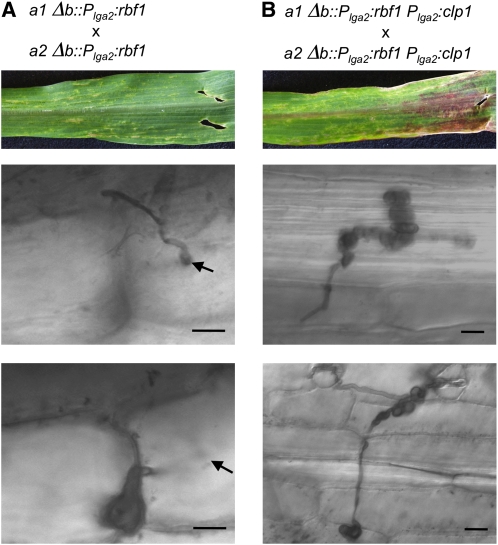
Our initial question was whether rbf1 would be sufficient to initiate pathogenic development independently from b. Since expression of rbf1 results in G2 cell cycle arrest, our attempts to use constitutively active promoters to drive rbf1 expression failed. We thus replaced the b mating-type locus with the rbf1 gene under the control of the lga2 promoter. lga2 expression can be activated both via pheromone and via the ectopic expression of rbf1 (Urban et al., 1996; Heimel et al., 2010). Thus, pheromone treatment should initiate expression of the Plga2:rbf1 gene, and the resulting Rbf1 expression then should maintain the Plga2-driven expression of rbf1. This self-maintaining positive feedback loop should ensure high expression of rbf1 after pheromone stimulation. In infection experiments, we used a mixture of the strains UKH184 (a1Δb::Plga2:rbf1) and UKH186 (a2Δb::Plga2:rbf1). Sensing of the pheromone from the compatible mating partner should trigger the positive rbf1 feedback loop. Infected maize plants displayed disease symptoms, such as the chlorosis that typically spread throughout the leaf. Fungal hyphae developed appressoria and penetrated the leaf surface. However, similar to infection experiments with U. maydis strains deleted in clp1, most hyphae were retained in the first epidermal cell layer, and only in very rare cases progressed to a neighboring cell. In all cases, hyphae developed bulbous structures at the tips (Figure 6A). Our data clearly show that expression of rbf1 is sufficient for appressorium formation and penetration of the leaf surface, the initial steps of pathogenic development, and that this development is independent from the b mating-type locus.
Figure 6.
Ectopic Expression of rbf1 and clp1 Is Sufficient for the Initial Steps of Pathogenic Development.
Mixtures of strains UKH184 (a1 Δb::Plga2:rbf1) and UKH186 (a2 Δb::Plga2:rbf1) (A) or UKH178 (a1 Δb::Plga2:rbf1; Plga2:clp1) and UKH180 (a2 Δb::Plga2:rbf1; Plga2:clp1) (B) were coinoculated into 7-d-old maize seedlings. Pictures of leaves were taken 6 DAI; fungal development was investigated by Chlorazole Black E staining of infected leaf samples 4 DAI. Infection of maize plants with a mixture of strains UKH184 and UKH186 expressing rbf1 under the control of the lga2 promoter resulted in local chlorosis. Strains were able to form appressoria and to penetrate the host plant; however, development was arrested immediately or shortly after penetration by formation of enlarged bulbous cells. By contrast, infection with strains UKH178 and UKH180, expressing both rbf1 and clp1 under the control of the lga2 promoter, led to more severe disease symptoms, as enhanced formation and spreading of chlorotic areas and anthocyanin production. Fungal progression after plant penetration was not restricted to single plant cells, but hyphae continued to spread throughout the leaf. The formation of bulbous cells was observed as well; however, these structures did not terminate hyphal growth. Arrows indicate the point of plant penetration. Bars = 20 μm.
Cooperative Action of Rbf1 and Clp1 Facilitates Plant Infection and Hyphal Development Independent from the b Mating-Type Locus
We next asked whether further proliferation of the rbf1-expressing strains would be facilitated via additional Clp1 expression. Strains UKH178 (a1Δb::Plga2:rbf1 Plga2:clp1) and UKH180 (a2Δb::Plga2:rbf1 Plga2:clp1) are isogenic to UKH184 (a1Δb::Plga2:rbf1) and UKH186 (a2Δb::Plga2:rbf1) but harbor an additional clp1 allele under the control of the lga2 promoter integrated into the ip locus. We used a mixture of strains UKH178 and UKH180 for infection experiments and monitored fungal development as described before. Similar to the strains UKH184 and UKH186, UKH178 and UKH180 were able to induce appressoria and to penetrate the leaf surface. However, in contrast with the strains expressing only rbf1, the simultaneous expression of rbf1 and clp1 in strains UKH178 and UKH180 promoted fungal proliferation inside the plant. Development proceeded up to 3 to 4 DAI, although the hyphae showed abnormal phenotypes (Figure 6B). The hyphae developed bulbous structures, but in contrast with strains UKH184 and UKH186, the bulbous cells did not ultimately terminate fungal growth, as we observed in several cases that filamentous hyphae emanated from these structures.
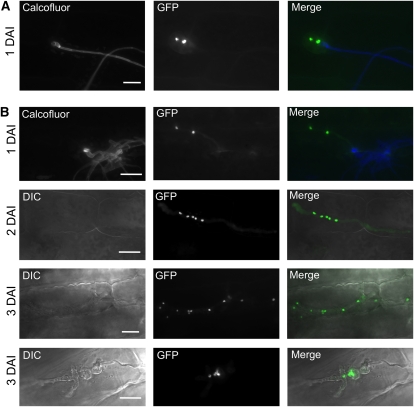
To determine whether hyphal growth in planta is associated with nuclear division, we visualized fungal nuclei during in planta development by expression of a nuclear localized eGFP under the control of the plant-induced mig2-5 promoter (Scherer et al., 2006; Wahl et al., 2010), yielding strains UKH184GN (green nuclei), UKH186GN, UKH178GN, and UKH180GN. In line with our previous observations, we observed that the strains expressing only rbf1 were unable to proliferate inside the plant. The bulbous cells formed immediately after plant penetration contained only two nuclei (Figure 7A), indicating that nuclear division is stalled and strains are unable to release the rbf1-induced cell cycle arrest. Microscopy observations at later time points revealed no alteration at 2 DAI, while at 3 DAI, most hyhae were collapsed. By contrast, we observed multiple nuclei in strains that simultaneously express both rbf1 and clp1. Over time, the number of nuclei in the hyphae increased (Figure 7B). In addition, we found that the number of nuclei in hyphal compartments was often uneven and that no clamps were formed, indicating that nuclear division was not synchronous.
Figure 7.
Nuclear Distribution during Pathogenic Development of U. maydis Strains Expressing rbf1 and rbf1/clp1.
(A) Nuclear distribution in infection structures of a mixture of UKH184GN (a1 Δb::Plga2:rbf1 Pmig2_5:NLS-3eGFP) and UKH186GN (a2 Δb::Plga2:rbf1 Pmig2_5:NLS-3eGFP) expressing nuclear-localized GFP under the control of the plant-induced mig2-5 promoter. Calcofluor staining was used to visualize fungal growth on the leaf surface. Bulbous structures were formed directly after penetration of the host plant and invariably contained only the two nuclei of the dikaryotic hyphae formed after cell fusion. No progression of fungal development was observed at later time points of infection.
(B) Mixtures of UKH178GN (a1 Δb::Plga2:rbf1; Plga2:clp1 Pmig2_5:NLS-3eGFP) and UKH180GN (a2 Δb::Plga2:rbf1; Plga2:clp1 Pmig2_5:NLS-3eGFP), expressing nuclear-localized GFP during in planta growth. Hyphae infected the plant via appressorium-like structures, continued to proliferate inside the plant, and contained multiple nuclei. Hyphae were often found to contain uneven numbers and irregularly spaced nuclei. At 3 DAI, morphologically abnormal fungal hyphae (top panel, 3 DAI) or bulbous structures (bottom panel, 3 DAI) were frequently found that accumulated an increasing number of nuclei over time.
Bars = 20 μm.
In essence, our data demonstrate that simultaneous Clp1 and Rbf1 expression is required and sufficient for fungal proliferation inside the host plant. For coordinated development of fungal hyphae and the establishment of a biotrophic interface, however, additional factors are required that most probably demand the action of bE/bW itself or of other b-dependent regulatory factors.
DISCUSSION
Sexual and pathogenic development in U. maydis are interdependent and tightly connected to the establishment of the biotrophic stage. Development prior to and after fusion of compatible haploid sporidia is controlled by the two mating-type loci a and b (Rowell and DeVay, 1954; Rowell, 1955; Holliday, 1961; Puhalla, 1970; Day et al., 1971). Besides their distinctive functions in cell fusion and pathogenic development, both loci also ensure the synchronization of the cell cycle: the a-mediated pheromone response leads to G2 cell cycle arrest prior to cell fusion (Garcia-Muse et al., 2003), and the arrest is maintained by the activity of the b locus after cell fusion until the hyphae penetrate their host plant. The b-mediated cell cycle arrest is achieved by the action of different transcriptional regulators, of which the bE/bW heterodimer and Rbf1 are of particular importance. Induction of either bE/bW or Rbf1 leads to G2 cell cycle arrest (Heimel et al., 2010). In this study, we show that the Clp1 protein binds to both bW and Rbf1. Clp1 binding alters both b and Rbf1 function, leads to release of the cell cycle arrest, and allows the consecutive developmental progression.
In addition, we identified the putative bZIP transcription factor Cib1 as a Clp1 interacting protein. Deletion of cib1 resulted in a phenotype that is of striking similarity to that of Δclp1 strains. Mutant strains are not impaired during growth in axenic culture, in filamentous growth, or in appressorium formation. However, after penetration of the leaf surface, development of the fungus was blocked prior the first mitotic division. The conserved domain structure and the nuclear localization of a functional Cib1-GFP fusion protein suggest that Cib1 functions as a transcriptional regulator. However, despite intense efforts, including genome-wide expression profiling of cib1 deletion strains during the early infection stage (i.e., when the cib1 deletion blocks further development), we were unable to identify Cib1 target genes. Thus, the elucidation of the molecular function of Cib1 requires further investigation. Since deletion of cib1 did not affect the Clp1-mediated inactivation of bW function (see Supplemental Figure 5 online), it is possible that cib1 regulates specific processes after plant penetration that are involved in, for example, clamp cell formation.
The cib1 gene was found to be expressed constitutively, but, similar to Clp1, expression of the Cib1 protein was restricted to cells that had penetrated the leaf surface. The posttranscriptional regulation of Cib1 expression can be attributed to development-specific alternative splicing via intron retaining. EST analyses provided the first evidence for alternative splicing in U. maydis, and intron retaining was found to be the most prominent mode of action (Ho et al., 2007). However, no biological significance of alternative splicing has been demonstrated yet in U. maydis (Feldbrügge et al., 2008). Hence, the regulation of Cib1 provides important evidence for the involvement of alternative splicing mechanisms in the regulation of developmental processes. Since translation of both the nonspliced and the spliced cib1 mRNA would result in proteins harboring all characteristic domains of a bZIP transcription factor, it is tempting to speculate that the dynamics of homo- and heterodimerization of the putative Cib1 isoforms could account for additional regulatory cues. A similar mode of action has been observed for the cAMP-responsive element modulator that is involved in the regulation of spermatogenesis in mammalia (Foulkes et al., 1992). Dimerization dynamics of the different interacting isoforms generated via alternative splicing determine whether the homo- or heterodimeric complex acts as an activator or repressor on a common subset of regulated genes. Since expression of the various isoforms varies in different cell types, target gene regulation by cAMP-responsive element modulator homo- or heterodimers is cell type specific (Foulkes et al., 1992; Don and Stelzer, 2002). Another example is provided by the bZIP transcription factor TCF11 that is involved in the antioxidant response during murine embryonic development. TCF11-mediated transactivation requires the homodimerization of full-length TCF11, whereas heterodimerization with a shorter isoform represses transactivation and blocks the interaction with cell-type-specific interaction partners that are required to gain full activity as a transcription factor (Husberg et al., 2001).
Impact of Clp1-bW Interaction on Pathogenic Development
The central role of the bE/bW heterodimer for the regulation of pathogenic development has been conclusively demonstrated (Bölker et al., 1995a). We have shown that b function is blocked by Clp1 expression (Scherer et al., 2006), and we now demonstrate that this block is achieved via the physical interaction of Clp1 with bW. The interaction domain is located in the C-terminal part of the bW protein that is conserved between different bW alleles. Inhibition of b function by the Clp1-bW interaction might by facilitated by blocking of DNA binding, by masking of transcription factor transactivation domains, or by a Clp1-mediated disruption of the bE/bW heterodimer. Since Clp1 expression also led to functional inhibition of Kon8, a bE-bW fusion protein, we can exclude that disruption of the bE/bW heterodimer is required for Clp1-mediated inhibition of b function. The Kon8 protein harbors a stretch of 75 amino acids that is sufficient for the Clp1 interaction. Interestingly, deletion of this domain abolished the ability of bE-bW fusion proteins to trigger pathogenic development, while the ability to initiate filamentous growth remained unaltered (Grandel et al., 2000). We postulate that this domain is required for the Clp1-mediated cell cycle release after plant penetration and to promote biotrophic development. For the initiation of filamentous growth, Clp1 is not required; thus, the domain can be abolished.
Similar to U. maydis, the Clp1 protein in C. neoformans was also found to be required for dikaryotic growth. Cells lacking clp1 were incapable of reinitiating growth after mating (Ekena et al., 2008), which is in line with our observation that Clp1 counteracts mating-type-induced cell cycle arrest.
Role of the Clp1-Rbf1 Interaction in Pathogenic Development
Rbf1 is a master regulator of b-dependent development that is expressed in response to both pheromone stimulation (Zarnack et al., 2008) and b induction (Heimel et al., 2010). Rbf1 is not required for pheromone-induced conjugation tube formation and pheromone-induced G2 cell cycle arrest but is essential for b-dependent morphogenesis, b-dependent cell cycle arrest, and b-dependent gene regulation (Heimel et al., 2010). Whereas induction of rbf1 by itself did not alter the expression of mfa1, the simultaneous expression of rbf1 and clp1 led to a drastic reduction in both basal and pheromone-induced expression of mfa1 and a less pronounced reduction of pra1 expression. Currently, we can only speculate about the mechanism for Rbf1/Clp1-mediated repression of mfa1. One possibility is that the Clp1/Rbf1 complex leads to the activation of a repressor for mfa1. Alternatively, the Clp1/Rbf1 complex could bind directly to the mfa1 promoter to act as a repressor. In this context, it is interesting to note that the Saccharomyces cerevisiae Rme1 protein and the human YY1 protein, two of the small number of transcription factors that have been shown to function both as transcriptional activators and repressors, are, similar to Rbf1, also C2H2 zinc-finger proteins (Toone et al., 1995; Thomas and Seto, 1999). It is possible that Rbf1 acts as a transcriptional activator and in collaboration with Clp1 as repressor.
But what is the biological function of a protein interfering with the activity of both mating types? Ectopic clp1 expression inhibits activation of the pheromone pathway and thereby pheromone-dependent conjugation tube formation and gene regulation in an rbf1-dependent manner. However, since Clp1 protein expression is restricted to the biotrophic phase, it appears unlikely that the inhibition of conjugation tube formation via Clp1 is of biological significance. The Clp1-mediated repression of mfa1, and thus of the pheromone pathway, confers additional consequences on gene regulation during pathogenic development. For the initiation of the developmental program upon plant infection, it is necessary to release both the a- and b-dependent cell cycle blocks. Clp1 expression leads to the downregulation of the pheromone pathway, and, as a consequence, releases the a-dependent cell cycle block. To release the b-mediated cell cycle arrest, Clp1 interacts with, and inhibits, bW. In addition, the Clp1/Rbf1-dependent repression of mfa1 results in reduced gene expression of bE and bW, since the a pathway is required for expression of both b genes (Figure 8). The ultimate result of this regulatory circuit is the downregulation of rbf1 expression after plant penetration, which is most likely a prerequisite for the reinitiation of growth in planta. Because clp1 itself is regulated by the bE/bW heterodimer (Scherer et al., 2006), the feedback regulation could establish an oscillatory self-perpetuating regulatory network that couples cell cycle control to pathogenic development of U. maydis. Similarly, in S. cerevisiae, the periodic expression of 70% of the cell cycle–dependent expressed genes continued in the absence of mitotic and S-phase cyclins, and a network of transcription factors was proposed to drive the periodic expression pattern of these genes independently of, and in tandem with, the CDK oscillator (Orlando et al., 2008).
Figure 8.
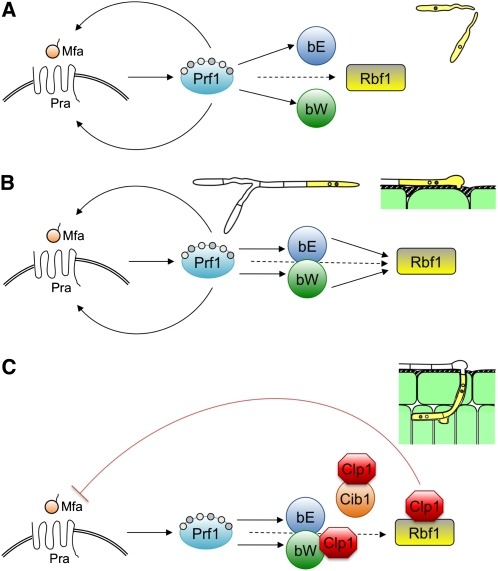
Model for the Coordinated Action of the Mating Types to Regulate Different Stages of Pathogenic Development of U. maydis.
(A) Recognition of a compatible mating partner (Mfa) by the a-encoded pheromone/receptor (Pra) triggers signaling cascades that culminate in the activation of the high mobility group transcription factor Prf1 via differential phosphorylation by Protein Kinase A and the mitogen-activated protein kinase Kpp2 (not shown). Activated Prf1 in turn induces the expression of the a locus genes mfa and pra, the b locus genes bE and bW, as well as rbf1. Activation of the a pathway leads to formation of conjugation tubes and G2 cell cycle arrest.
(B) After cell fusion, compatible bE/bW proteins dimerize and further induce the expression of rbf1. High levels of rbf1 ensure the maintenance of cell cycle arrest, induce filamentous growth, and facilitate the initial stages of plant infection.
(C) After penetration of the host plant, Clp1 expression interferes with a- and b-dependent gene regulation via interaction with bW and Rbf1. The Clp1/Rbf1 interaction effectively represses the a-dependent pathway, whereas Clp1/bW interaction blocks b-dependent gene regulation. Since Clp1 is regulated by the bE/bW heterodimer, a negative autoregulatory feedback loop is established. The consequences of these regulatory cross-connections are (1) the downregulation of rbf1 after plant penetration and (2) the counteraction of the a- and b-induced cell cycle arrest. Further proliferation requires in addition the expression of the Cib1 protein, which is subject to developmentally regulated splicing.
Although the bE/bW heterodimer is required and sufficient to promote pathogenic development, the active pheromone pathway is necessary for full virulence of U. maydis. The haploid strain CL13 (a1 bE1/bW2) is solopathogenic due to the presence of compatible b alleles. However, the introduction of the mfa2 gene to activate the pheromone pathway (resulting in strain SG200) increased virulence significantly, demonstrating that the interplay between the a and b pathway is required for full virulence (Di Stasio et al., 2009). As outlined, the activation of the a pathway leads to induction of bE, bW, and rbf1 (Urban et al., 1996; Zarnack et al., 2008), the key regulators for pathogenic development, and thus primes the cells for the biotrophic phase. However, there must be a principal difference between the a-dependent rbf1 induction and the subsequent b-dependent rbf1 induction. We have conclusively shown that ectopic induction of rbf1 leads to the initiation of pathogenic development (i.e., the cells switch to filamentous growth, form appressoria, and penetrate the plant surface). Since these developmental steps are initiated in strains from which the b locus has been deleted, we can exclude that they are caused by interplay with other b-dependent factors. We favor the explanation that fungal development during different stages of pathogenic development is controlled via different expression levels of rbf1. Whereas filamentous growth, G2 cell cycle arrest, and appressorium formation require high levels of rbf1, reinitiation of growth after penetration of the leaf surface is likely achieved via Clp1-mediated downregulation of a- and b-dependent pathways, leading to reduced rbf1 expression and the release of the G2 cell cycle arrest. The pheromone-induced rbf1 expression level is not sufficient to facilitate plant infection, as strains with compatible a and incompatible b alleles are apathogenic (Banuett and Herskowitz, 1989). Indeed, the measurement of expression levels on the plant surface via Affymetrix arrays revealed that rbf1 expression in a strain with compatible a and b genes (SG200: 794 ± 67) was 2.9-fold increased when compared with a strain in which only the a pathway is activated (SG200Δb: 275 ± 40; M. Vranes, personal communication). Further support for this model is provided by the observation that expression levels of rbf1 during in planta development are drastically reduced. Expression levels were not detectable by array analysis of tumor material 5, 9, and 13 DAI (M. Vranes, personal communication); furthermore, qRT-PCR analysis revealed a 140-fold lower expression in planta when compared with the rbf1 expression values achieved upon b induction in axenic culture (see Supplemental Figure 6 online).
The induced expression of rbf1 enabled the cells to form appressoria and to penetrate the plant surface; however, no nuclear division was observed, and hyphal progression was blocked early upon plant penetration. Simultaneous expression of Clp1 resulted in the release of the cell cycle arrest, which emphasizes the central role for Clp1 in cell cycle control. The Clp1-mediated cell cycle release in strains UKH178 and UKH180 (and derivatives UKH178GN and UKH180GN) is independent from b, which may be on first sight contradictory to our model. However, since the Clp1/Rbf1 interaction leads to the repression of the a pathway, Clp1 induction results in a downregulation of the lga2 promoter driving the rbf1 expression, thereby reducing rbf1 levels.
Coinduction of rbf1 and clp1 in strains UKH178GN and UKH180GN led to severe morphological alterations: nuclear division occurred in a nonsynchronous manner, and no clamp cell formation was observed. Interestingly, the bulbous structures observed in strains UKH178GN and UKH180GN are similar to the structures formed after the inactivation of a temperature-sensitive bE2 allele during pathogenic development, which also leads to loss of polarity and results in multinucleated, unseptated cells (Wahl et al., 2010).
Notably, as Clp1-interacting proteins (see Supplemental Table 1 online), we identified Cdc15, a protein required for cytokinesis and nuclear distribution in U. maydis (Böhmer et al., 2009) and, in addition, the orthologs of BZZ1, Mob1, and CDC36, which were shown to be involved in septum formation, polar growth, and cell cycle control in various fungal species (Hartwell et al., 1970; Reed, 1980; Breter et al., 1983; Luca and Winey, 1998; Luca et al., 2001; Soulard et al., 2002; Weiss et al., 2002; Song et al., 2008; Maerz et al., 2009). A possible role for these proteins during in planta development of U. maydis has not been investigated yet and thus remains speculative. However, it is possible that Clp1 function is not exclusively restricted to the modulation of transcription factors and that the function of the interacting proteins contributes to the observed phenotype.
In basidiomycetes such as C. cinerea and Schizophyllum commune, clamp cell formation is a highly regulated process that ensures correct nuclear distribution during dikaryotic growth. The coordinated activation of the A (homeodomain proteins) and B (pheromone/receptor) pathways is required to execute this complex developmental program, as distinct steps are regulated by the individual mating-type loci (Swiezynski and Day, 1960b, 1960a; Raper, 1966). Whereas nuclear migration, induction of the clamp primordium, and formation of the septum are A-dependent processes, fusion of the clamp cell with the subapical compartment is regulated via the B pathway (Kües, 2000; Brown and Casselton, 2001; Kothe, 2001). Similarly, in U. maydis, clamp cells participate in nuclear distribution during dikaryotic growth, and clamp formation depends on the bE/bW heterodimer; however, in contrast with the other basidiomycetes, clamp cell formation is independent from the a pathway (Bölker et al., 1995b; Scherer et al., 2006). The Clp1-mediated cross connection of the a and b pathway might therefore represent an evolutionary remnant of the mating-type-dependent clamp cell regulation in tetrapolar fungi that has been adapted by U. maydis for its biotrophic development: in both cases, the coordination of the cell cycle with developmental processes may be instrumental. The requirement for tight cell cycle regulation during infection has recently been demonstrated for the hemibiotrophic pathogen Magnaporthe grisea in which the formation of appressoria is regulated via a DNA replication–dependent checkpoint (Saunders et al., 2010).
In summary, our results indicate that Clp1 functions as an internode, connecting multiple layers of regulation, to synchronize and regulate cell cycle and hyphal proliferation during biotrophic growth of U. maydis. We believe that this is of importance not only for fungal biology but represents a paradigm for how cell cycle control, morphogenesis, and pathogenicity are interdependently connected to mediate complex developmental processes.
METHODS
Yeast Two-Hybrid Analysis
Screening for Clp1-interacting proteins was performed essentially as described by Mendoza-Mendoza et al. (2009), using the Matchmaker III system (Clontech). Plasmid pGBKT7-Clp1 was generated by PCR amplification of the clp1 open reading frame (ORF), introducing an NdeI site at the 5′ and a BamHI site at the 3′ end and subsequent ligation into the respective sites of pGBKT7 (Clontech). Plasmid pGADbW2_567-645 was generated by PCR amplification of the bW2 fragment encompassing amino acids 567 to 645, introducing a 3′ and a 5′ SfiI site and subsequent ligation into the respective SfiI sites of plasmid pGAD-DS (Dualsystems Biotech). Primer sequences are given in Supplemental Table 4 online.
Strains and Growth Conditions
Escherichia coli strain TOP10 (Invitrogen) was used for cloning purposes. Growth conditions for E. coli and Ustilago maydis were as described by Scherer et al. (2006). Growth conditions and media used to induce nar1 promoter or crg1 promoter-driven gene expression follow the protocol of Brachmann et al. (2001). U. maydis strains with relevance for this study are listed in Supplemental Table 5 online. Mating assays and plant infections were described in Brachmann et al. (2001). The maize (Zea mays) cultivar Early Golden Bantam was used for infection experiments under controlled greenhouse conditions (16 h light, daylight, supplemented with 20 kLux artificial light, 28 to 32°C; 8 h dark, 20°C) or in a CLF Plant Climatics GroBank with a 16 h (28°C)/8 h (22°C) day/night rhythm. For pheromone stimulation of U. maydis cells, we followed the protocol of Müller et al. (2003) with the exception that minimal medium containing nitrate and arabinose was used instead of complete medium.
DNA and RNA Procedures
Molecular methods followed described protocols (Sambrook et al., 1989). DNA isolation and transformation procedures for U. maydis were performed as described (Schulz et al., 1990). Sequences of primers used for PCR are given in Supplemental Table 4 online.
For all gene deletions, we used a PCR-based approach (Kämper, 2004). For the cib1-3xeGFP fusion, 1 kb of the 3′ end of the cib1 ORF and 1 kb of the 3′ UTR were PCR amplified, introducing two SfiI sites and removing the stop codon of cib1; both fragments were ligated to an SfiI 3xeGFP-HygR fragment of pUMA647 (Scherer et al., 2006) in pCR2.1 TOPO (Invitrogen) as backbone, yielding pCib3eGFP. For the Pcib1:eGFP fusion, the cib1 ORF of plasmid pCib3eGFP was replaced by a 1-kb cib1 promoter fragment to yield pPcib1:eGFP. To replace the b mating-type locus with the arabinose-inducible clp1 allele, a 1.3-kb NdeI-NotI fragment of pRU-ATG1 (Scherer et al., 2006), a 1.4-kb BstXI(blunt)-NdeI crg1 promotor fragment, and a 0.3-kb NotI-EcoRI(blunt) nos terminator fragment from pRU12 were integrated into the StuI site of pCRΔb (Kämper, 2004) to generate pCRΔb-crg:clp1. To replace the b mating-type locus with the nitrate-inducible rbf1 allele, the rbf1 ORF was PCR amplified, creating an NdeI site at the start and a NotI site following the stop codon, and cloned into pCR2.1 TOPO. The NdeI-NotI ORF fragment, a 1-kb HpaI(blunt)-NdeI nar1 promotor fragment, and a 0.3-kb NotI-EcoRI(blunt) nos terminator fragment from pRU12 (Brachmann et al., 2001) were integrated into the StuI site of pCRΔb to generate pCRΔb-nar:rbf1.
To replace the b mating-type locus with the rbf1 gene under control of the lga2 promoter, the procedure was identical, with the exception that a 1-kb PvuII(blunt)/NdeI lga2 promoter fragment was used instead of the nar1 promoter and transformed into strains FB1 and FB2. For expression of the clp1 allele under the control of the lga2 promoter, the crg1 promoter from pRU-ATG1 (Scherer et al., 2006) was replaced with a 1-kb PvuII(blunt)/NdeI lga2 promoter fragment. The resulting plasmid plga2-clp1 was linearized with SspI and integrated into the ip locus of strains UKH184 and UKH186 via homologous recombination. Visualization of fungal nuclei during biotrophic growth in strains UKH178, UKH180, UKH184, and UKH186 was performed essentially as described by Wahl et al. (2010). The strains were generated by ectopic integration of the plasmid pRWnlsGfp (linearized with SspI) harboring triple eGFP fused to a VP16-NLS under control of the plant-induced mig2-5 promoter.
For expression of the N-terminal GFP (NG) fragment (amino acids 1 to 155) fused to the C terminus of Clp1, plasmid pNG-Clp1 was used, a pRU12 derivative harboring the arabinose-inducible crg1 promoter (Brachmann et al., 2001). For construction of pNG-Clp1, a 1-kb fragment upstream of the leu2 ORF was PCR amplified and integrated into pRU12 as a NotI/HindIII fragment into the respective sites, replacing the carboxin resistance marker. Subsequently, a 1-kb PCR fragment downstream to the leu2 ORF was inserted into the NotI(blunt) site by partial digestion, followed by the integration of a PCR-amplified 0.5-kb SfiI/AscI NG fragment and a PCR-amplified 1.4-kb NdeI/SfiI Clp1 fragment together with a 0.3-kb AscI/NotI fragment into the NdeI/NotI opened vector backbone. Finally, a 1.9-kb NotI fragment with the hygromycin resistance cassette from pNEBUH (Müller et al., 1999) was integrated into the respective site. The plasmid was linearized with BamHI and integrated into the leu2 locus of strain FB2 via homologous recombination. For expression of the C-terminal GFP (CG) fragment (amino acids 155 to 238) fused to the C termini of Cib1, bW1, and Rbf1 plasmids pCG-Cib1, pCG-bW1, and pCG-Rbf1 were used, respectively. All plasmids are derivatives of plasmid pRU11 harboring the arabinose-inducible crg1 promoter (Brachmann et al., 2001). Corresponding ORFs were PCR amplified and integrated as NdeI/SfiI fragments together with the PCR-amplified 0.3-kb SfiI/NotI CG fragment into the NdeI/NotI opened pRU11 vector. Subsequently, linkers (Hu et al., 2006) between the ORFs and the C-GFP domain were inserted by cloning of complementary oligonucleotides into SfiI sites. Plasmid pCG-NLS was constructed by integration of a 0.3-kb NdeI/NotI CG fragment into the respective sites of pRU11. The NLS from VP16 (Quadbeck-Seeger et al., 2000) was inserted in frame via a linker consisting of two annealed oligonucleotides into the SfiI site of the CG fragment. For construction of plasmid pNG-NLS, the 46-bp NdeI/SfiI fragment of pCG-NLS was used to replace the clp1 ORF in plasmid pNG-Clp1. Plasmids were linearized with SspI and integrated into the ip locus via homologous integration (Brachmann et al., 2001) in strain UKH50.
All PCR amplified fragments were verified by sequencing. For transformation, either linearized plasmid DNA or PCR-generated linear DNA was used; homologous integration was verified by DNA gel blots. RNA extraction and qRT-PCR analysis for cib1, mfa1, pra1, prf1, rbf1, ppi, eIf2b, and actin was performed as described (Scherer et al., 2006) with the following changes: MESA GREEN qPCR Master mix (Eurogentech) was used; cycling was 7 min 95°C followed by 45 cycles of 95°C for 30s, 60°C for 20 s, and 72°C for 40s; specificity of PCR products was checked by melting curve analysis from 60 to 95°C. For absolute quantification of spliced/unspliced cib1 mRNAs, RT-PCR was performed on serial dilutions of calibrated solutions of 500-bp cib1 mRNA fragment with (unspliced) or without the intron (spliced) to generate calibration curves (Bio-Rad Icycler software). Absolute numbers of unspliced and spliced cib1 mRNA molecules from U. maydis strains grown in axenic culture or during pathogenic development were then calculated by plotting the respective CT values of the RT-PCR analysis against the calibration curves (Leong et al., 2007). Calibration samples were processed in triplicates and all other samples in duplicates. Full-length cib1 cDNA was isolated using the GeneRacer kit (Invitrogen), cloned in pCR2.1 TOPO (Invitrogen), and sequenced.
In Vitro Protein Expression and Coimmunoprecipitation
The TNT quick coupled transcription/translation system (Promega) was used for in vitro expression of HA- or Myc-tagged proteins according to the manufacturer’s protocol. For expression of the HA- or Myc-tagged proteins, plasmids isolated in the yeast two-hybrid screen were used in the case of bW and Rbf1. For expression of HA-tagged Cib1, full-length cDNA was PCR amplified, introducing a 3′ and a 5′ SfiI site and subsequently ligated into the respective SfiI sites of plasmid pGAD-DS (Dualsystems Biotech). For expression of Myc-tagged Clp1, plasmid pGBKT7-Clp1 was used. After in vitro expression of the proteins, 15 μL of Clp1 protein lysate were mixed with equal volumes of bW-, Rbf1-, Cib1-, or unprogrammed rabbit reticulocyte lysate (no addition of vector DNA, as a negative control) and diluted with coimmunoprecipitation buffer (25 mM HEPES, 100 mM NaCl, 12.5 mM MgCl, 1 mM EDTA, 10% [w/v] glycerol, 0.1% [w/v] Nonidet P-40, and 1 mM DTT, pH 7.5) to a final volume of 150 μL. After 30 min at room temperature, 10 μL α-HA-coupled agarose beads (Sigma-Aldrich) were added, and the reactions were incubated at 4°C overnight on a rotary wheel. After washing four times with 1 mL coimmunoprecipitation buffer, beads were resuspended in 20 μL Laemmli buffer, boiled for 5 min at 95°C, and subjected to SDS-PAGE. Following semidry blotting, biotinylated proteins were detected with the Transcend colorimetric nonradioactive translation detection system (Promega) according to the manufacturer’s protocol.
BiFC Analysis
Protein interaction studies were performed according to Hu et al. (2006). The N-terminal eGFP fragment (NG) encompassing amino acids 1 to 154 was fused to the C terminus of Clp1 and integrated into the leu2 locus of strain FB2 generating strain UKH50. The C-terminal eGFP (CG) encompassing amino acids 155 to 238 was fused to the C terminus of bW1, Rbf1, and Cib1 or to a nuclear localization sequence from VP16 (VP16-NLS) as negative control and subsequently integrated into the ip locus as described by Brachmann et al. (2001). Expression of fusion proteins was under the control of the arabinose-inducible crg1 promoter (Brachmann et al., 2001). Construction of strains UKH214, UKH215, and UKH216 was performed as described above, with the exception that pNG-NLS was used instead of pNG-Clp1. For BiFC analysis, cells were grown to an OD600 = 0.25 in complete medium and shifted to complete medium containing arabinose instead of glucose to induce protein expression. Fluorescence microscopy was performed 20 h after induction. Exposure time for the GFP channel was 250 ms and for the DAPI channel 90 ms.
DNA Array and Data Analysis
Custom-designed Affymetrix chips (UstilagoA) were used for DNA array analysis. Probe sets for the individual genes are visualized at http://mips.gsf.de/genre/proj/ustilago/. Target preparation, hybridization, and data acquisition were performed essentially as described before (Eichhorn et al., 2006), with the following alterations: 5 μg RNA were used for first-strand cDNA synthesis at 50°C with Superscript II (Invitrogen). Software dChip1.3 (Li and Wong, 2003) was used to calculate mean expression values and fold changes based on a 90% confidence interval (lower bound of fold change), allowing a conservative evaluation of fold change (Li and Wong, 2001). Induced and repressed genes were filtered according to the following filter criteria: a change in expression of ≥2 and a difference of mean expression values >100 were considered significant.
Microscopy
Microscopy analysis was performed using an Axioimager equipped with an Axiocam MRm camera or a Lumar V12 equipped with an Axiocam HRc camera (Zeiss). Nuclei were stained with DAPI Vectashield H-1200 (Vector Laboratories) and fungal cell walls with 2 μg/mL Calcofluor white (Sigma-Aldrich) in PBS. All images were processed with Axiovision (Zeiss). Chlorazole Black E staining was performed according to Brachmann et al. (2001).
Accession Numbers
Sequence data from this article can be found at the Munich Information Center for Protein Sequences Ustilago maydis database (http://mips.helmholtz-muenchen.de/genre/proj/ustilago/) and the National Center for Biotechnology Information database under the following accession numbers: actin (um11232), XP_762364; eIf2b (um04869), XP_761016; clp1 (um02438), XP_758585; cib1 (um11782), XP_759656; leu2 (um01245), XP_757392; rbf1 (um03172), XP_759319; bW1 (um00578), XP_756725; bW2 (M84182); and wcl2 (um02664) XP_758811. All array data have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE21121.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Quantification of Clp1 Binding to bW, Rbf1, and Cib1.
Supplemental Figure 2. bW-CG, Cib1-CG, and Rbf1-CG Do Not Display Nonspecific Interactions in BiFC Analysis.
Supplemental Figure 3. Schematic Overview of the cib1 Locus and Transcribed mRNAs.
Supplemental Figure 4. Kon8-Dependent Filament Formation Is Suppressed by Induced Expression of clp1.
Supplemental Figure 5. Cib1 Is Not Required for Clp1-Mediated Inhibition of b-Dependent Filament Formation.
Supplemental Figure 6. qRT-PCR Analysis of rbf1 Expression after b Induction in Vitro and during Pathogenic Development.
Supplemental Table 1. Clp1-Interacting Proteins Identified by Yeast Two-Hybrid Screening.
Supplemental Table 2. Pathogenicity of Strain USA1.
Supplemental Table 3. Differential Gene Expression upon Induced Coexpression of clp1 and rbf1 in U. maydis.
Supplemental Table 4. Primers Used in This Study.
Supplemental Table 5. Strains Used in This Study.
Supplementary Material
Acknowledgments
We thank M. Feldbrügge for various plasmid constructs, S. Adler for construction and initial characterization of strain USA1, and M. Vranes for providing DNA array results. This work was supported by grants from the German Ministry of Education and Science (BMBF) for the DNA array setup. K.H. was supported by grants from the International Max Planck Research School for Environmental, Cellular, and Molecular Microbiology and the International DFG Graduate School GRK 767 “Transcriptional Control of Developmental Processes.”
References
- Banuett F., Herskowitz I. (1989). Different a alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86: 5878–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhmer C., Ripp C., Bölker M. (2009). The germinal centre kinase Don3 triggers the dynamic rearrangement of higher-order septin structures during cytokinesis in Ustilago maydis. Mol. Microbiol. 74: 1484–1496 [DOI] [PubMed] [Google Scholar]
- Bölker M., Böhnert H.U., Braun K.H., Görl J., Kahmann R. (1995b). Tagging pathogenicity genes in Ustilago maydis by restriction enzyme-mediated integration (REMI). Mol. Gen. Genet. 248: 547–552 [DOI] [PubMed] [Google Scholar]
- Bölker M., Genin S., Lehmler C., Kahmann R. (1995a). Genetic regulation of mating, and dimorphism in Ustilago maydis. Can. J. Bot. 73: 320–325 [Google Scholar]
- Bölker M., Urban M., Kahmann R. (1992). The a mating type locus of U. maydis specifies cell signaling components. Cell 68: 441–450 [DOI] [PubMed] [Google Scholar]
- Brachmann A., Weinzierl G., Kämper J., Kahmann R. (2001). Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42: 1047–1063 [DOI] [PubMed] [Google Scholar]
- Brefort T., Doehlemann G., Mendoza-Mendoza A., Reissmann S., Djamei A., Kahmann R. (2009). Ustilago maydis as a pathogen. Annu. Rev. Phytopathol. 47: 423–445 [DOI] [PubMed] [Google Scholar]
- Breter H.J., Ferguson J., Peterson T.A., Reed S.I. (1983). Isolation and transcriptional characterization of three genes which function at start, the controlling event of the Saccharomyces cerevisiae cell division cycle: CDC36, CDC37, and CDC39. Mol. Cell. Biol. 3: 881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.J., Casselton L.A. (2001). Mating in mushrooms: Increasing the chances but prolonging the affair. Trends Genet. 17: 393–400 [DOI] [PubMed] [Google Scholar]
- Day P.R., Anagnostakis S.L., Puhalla J.E. (1971). Pathogenicity resulting from mutation at the b locus of Ustilago maydis. Proc. Natl. Acad. Sci. USA 68: 533–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stasio M., Brefort T., Mendoza-Mendoza A., Munch K., Kahmann R. (2009). The dual specificity phosphatase Rok1 negatively regulates mating and pathogenicity in Ustilago maydis. Mol. Microbiol. 73: 73–88 [DOI] [PubMed] [Google Scholar]
- Don J., Stelzer G. (2002). The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis. Mol. Cell. Endocrinol. 187: 115–124 [DOI] [PubMed] [Google Scholar]
- Eichhorn H., Lessing F., Winterberg B., Schirawski J., Kämper J., Müller P., Kahmann R. (2006). A ferroxidation/permeation iron uptake system is required for virulence in Ustilago maydis. Plant Cell 18: 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekena J.L., Stanton B.C., Schiebe-Owens J.A., Hull C.M. (2008). Sexual development in Cryptococcus neoformans requires CLP1, a target of the homeodomain transcription factors Sxi1alpha and Sxi2a. Eukaryot. Cell 7: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T., Fass D., Arnaud M., Harrison S.C. (1994). Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8: 970–980 [DOI] [PubMed] [Google Scholar]
- Feldbrügge M., Zarnack K., Vollmeister E., Baumann S., Koepke J., König J., Münsterkötter M., Mannhaupt G. (2008). The posttranscriptional machinery of Ustilago maydis. Fungal Genet. Biol. 45(Suppl 1): S40–S46 [DOI] [PubMed] [Google Scholar]
- Flor-Parra I., Vranes M., Kämper J., Perez-Martin J. (2006). Biz1, a zinc finger protein required for plant invasion by Ustilago maydis, regulates the levels of a mitotic cyclin. Plant Cell 18: 2369–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes N.S., Mellstrom B., Benusiglio E., Sassone-Corsi P. (1992). Developmental switch of CREM function during spermatogenesis: from antagonist to activator. Nature 355: 80–84 [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T., Steinberg G., Perez-Martin J. (2003). Pheromone-induced G2 arrest in the phytopathogenic fungus Ustilago maydis. Eukaryot. Cell 2: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandel A., Romeis T., Kämper J. (2000). Regulation of pathogenic development in the corn smut fungus Ustilago maydis. Mol. Plant Pathol. 1: 61–66 [DOI] [PubMed] [Google Scholar]
- Hartmann H.A., Kahmann R., Bölker M. (1996). The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J. 15: 1632–1641 [PMC free article] [PubMed] [Google Scholar]
- Hartwell L.H., Culotti J., Reid B. (1970). Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA 66: 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimel K., Scherer M., Vranes M., Wahl R., Pothiratana C., Schuler D., Vincon V., Finkernagel V., Flor-Parra I., Kämper J. (2010). The transcription factor Rbf1 is the master regulator for b-mating type controlled pathogenic development in Ustilago maydis. PLoS Pathog. 6: e1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho E.C., Cahill M.J., Saville B.J. (2007). Gene discovery and transcript analyses in the corn smut pathogen Ustilago maydis: Expressed sequence tag and genome sequence comparison. BMC Genomics 8: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. (1961). The genetics of Ustilago maydis. Genet. Res. Camb. 2: 204–230 [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. (2002). Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Hu C.D., Grinberg A.V., Kerppola T.K. (2006). Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis. Current Protocols of Cell Biology, Bonifacino J.S., Dasso M., Harford J.B., Lippincott-Schwartz J., Yamada K.M., (New York: Wiley; ), Unit 21.3 [DOI] [PubMed] [Google Scholar]
- Hurst H.C. (1995). Transcription factors 1: bZIP proteins. Protein Profile 2: 101–168 [PubMed] [Google Scholar]
- Husberg C., Murphy P., Martin E., Kolsto A.B. (2001). Two domains of the human bZIP transcription factor TCF11 are necessary for transactivation. J. Biol. Chem. 276: 17641–17652 [DOI] [PubMed] [Google Scholar]
- Inada K., Morimoto Y., Arima T., Murata Y., Kamada T. (2001). The clp1 gene of the mushroom Coprinus cinereus is essential for A-regulated sexual development. Genetics 157: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffarnik F., Müller P., Leibundgut M., Kahmann R., Feldbrügge M. (2003). PKA and MAPK phosphorylation of Prf1 allows promoter discrimination in Ustilago maydis. EMBO J. 22: 5817–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper J. (2004). A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271: 103–110 [DOI] [PubMed] [Google Scholar]
- Kämper J., et al. (2006). Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444: 97–101 [DOI] [PubMed] [Google Scholar]
- Kerppola T.K. (2006). Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1: 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe E. (2001). Mating-type genes for basidiomycete strain improvement in mushroom farming. Appl. Microbiol. Biotechnol. 56: 602–612 [DOI] [PubMed] [Google Scholar]
- Kües U. (2000). Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64: 316–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong D.T., Gupta A., Bai H.F., Wan G., Yoong L.F., Too H.P., Chew F.T., Hutmacher D.W. (2007). Absolute quantification of gene expression in biomaterials research using real-time PCR. Biomaterials 28: 203–210 [DOI] [PubMed] [Google Scholar]
- Li C., Wong W.H. (2001). Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wong W.H. (2003). DNA-chip analyzer (dChip). The Analysis of Gene Expression Data: Methods and Software, Parmigiani G., Garrett E.S., Irizarry R., Zeger S.L., (New York: Springer; ), pp. 120–141 [Google Scholar]
- Luca F.C., Mody M., Kurischko C., Roof D.M., Giddings T.H., Winey M. (2001). Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol. Cell. Biol. 21: 6972–6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F.C., Winey M. (1998). MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell 9: 29–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerz S., Dettmann A., Ziv C., Liu Y., Valerius O., Yarden O., Seiler S. (2009). Two NDR kinase-MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa. Mol. Microbiol. 74: 707–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Mendoza A., Eskova A., Weise C., Czajkowski R., Kahmann R. (2009). Hap2 regulates the pheromone response transcription factor Prf1 in Ustilago maydis. Mol. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- Mielnichuk N., Sgarlata C., Perez-Martin J. (2009). A role for the DNA-damage checkpoint kinase Chk1 in the virulence program of the fungus Ustilago maydis. J. Cell Sci. 122: 4130–4140 [DOI] [PubMed] [Google Scholar]
- Müller P., Aichinger C., Feldbrügge M., Kahmann R. (1999). The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34: 1007–1017 [DOI] [PubMed] [Google Scholar]
- Müller P., Leibbrandt A., Teunissen H., Cubasch S., Aichinger C., Kahmann R. (2004). The Gbeta-subunit-encoding gene bpp1 controls cyclic-AMP signaling in Ustilago maydis. Eukaryot. Cell 3: 806–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P., Weinzierl G., Brachmann A., Feldbrügge M., Kahmann R. (2003). Mating and pathogenic development of the Smut fungus Ustilago maydis are regulated by one mitogen-activated protein kinase cascade. Eukaryot. Cell 2: 1187–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando D.A., Lin C.Y., Bernard A., Wang J.Y., Socolar J.E., Iversen E.S., Hartemink A.J., Haase S.B. (2008). Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature 453: 944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martin J., Castillo-Lluva S., Sgarlata C., Flor-Parra I., Mielnichuk N., Torreblanca J., Carbo N. (2006). Pathocycles: Ustilago maydis as a model to study the relationships between cell cycle and virulence in pathogenic fungi. Mol. Genet. Genomics 276: 211–229 [DOI] [PubMed] [Google Scholar]
- Puhalla J.E. (1970). Genetic studies on the b incompatibility locus of Ustilago maydis. Genet. Res. Camb. 16: 229–232 [Google Scholar]
- Quadbeck-Seeger C., Wanner G., Huber S., Kahmann R., Kämper J. (2000). A protein with similarity to the human retinoblastoma binding protein 2 acts specifically as a repressor for genes regulated by the b mating type locus in Ustilago maydis. Mol. Microbiol. 38: 154–166 [DOI] [PubMed] [Google Scholar]
- Raper J.R. (1966). Genetics of Sexuality in Higher Fungi. (New York: The Ronald Press; ). [Google Scholar]
- Reed S.I. (1980). The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics 95: 561–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T., Kämper J., Kahmann R. (1997). Single-chain fusions of two unrelated homeodomain proteins trigger pathogenicity in Ustilago maydis. Proc. Natl. Acad. Sci. USA 94: 1230–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowell J.B. (1955). Functional role of compatibility factors and an in vitro test for sexual incompatibility with haploid lines of Ustilago zea. Phytopathology 45: 370–374 [Google Scholar]
- Rowell J.B., DeVay J.E. (1954). Genetics of Ustilago zea in relation to basic problems of its pathogenicity. Phytopathology 44: 356–362 [Google Scholar]
- Sambrook J., Frisch E.F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Saunders D.G., Aves S.J., Talbot N.J. (2010). Cell cycle-mediated regulation of plant infection by the rice blast fungus. Plant Cell 22: 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M., Heimel K., Starke V., Kämper J. (2006). The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell 18: 2388–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P. (1998). SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz B., Banuett F., Dahl M., Schlesinger R., Schäfer W., Martin T., Herskowitz I., Kahmann R. (1990). The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60: 295–306 [DOI] [PubMed] [Google Scholar]
- Skibbe D.S., Doehlemann G., Fernandes J., Walbot V. (2010). Maize tumors caused by Ustilago maydis require organ-specific genes in host and pathogen. Science 328: 89–92 [DOI] [PubMed] [Google Scholar]
- Snetselaar K.M., Mims C.W. (1992). Sporidial fusion and infection of maize seedlings by the smut fungus Ustilago maydis. Mycologia 84: 193–203 [Google Scholar]
- Song Y., Cheon S.A., Lee K.E., Lee S.Y., Lee B.K., Oh D.B., Kang H.A., Kim J.Y. (2008). Role of the RAM network in cell polarity and hyphal morphogenesis in Candida albicans. Mol. Biol. Cell 19: 5456–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A., Lechler T., Spiridonov V., Shevchenko A., Li R., Winsor B. (2002). Saccharomyces cerevisiae Bzz1p is implicated with type I myosins in actin patch polarization and is able to recruit actin-polymerizing machinery in vitro. Mol. Cell. Biol. 22: 7889–7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellig T., Bölker M., Lottspeich F., Frank R.W., Kahmann R. (1994). Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 13: 1620–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezynski K.M., Day P.R. (1960a). Migration of nuclei in Coprinus lagopus. Genet. Res. 1: 129–139 [Google Scholar]
- Swiezynski K.M., Day P.R. (1960b). Heterokaryon formation in Coprinus lagopus. Genet. Res. 1: 114–128 [Google Scholar]
- Thomas M.J., Seto E. (1999). Unlocking the mechanisms of transcription factor YY1: Are chromatin modifying enzymes the key? Gene 236: 197–208 [DOI] [PubMed] [Google Scholar]
- Toone W.M., Johnson A.L., Banks G.R., Toyn J.H., Stuart D., Wittenberg C., Johnston L.H. (1995). Rme1, a negative regulator of meiosis, is also a positive activator of G1 cyclin gene expression. EMBO J. 14: 5824–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban M., Kahmann R., Bölker M. (1996). Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251: 31–37 [DOI] [PubMed] [Google Scholar]
- Wahl R., Zahiri A., Kamper J. (2010). The Ustilago maydis b mating type locus controls hyphal proliferation and expression of secreted virulence factors in planta. Mol. Microbiol. 75: 208–220 [DOI] [PubMed] [Google Scholar]
- Weiss E.L., Kurischko C., Zhang C., Shokat K., Drubin D.G., Luca F.C. (2002). The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158: 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnack K., Eichhorn H., Kahmann R., Feldbrügge M. (2008). Pheromone-regulated target genes respond differentially to MAPK phosphorylation of transcription factor Prf1. Mol. Microbiol. 69: 1041–1053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.