Abstract
C4 photosynthesis drives productivity in several major food crops and bioenergy grasses, including maize (Zea mays), sugarcane (Saccharum officinarum), sorghum (Sorghum bicolor), Miscanthus x giganteus, and switchgrass (Panicum virgatum). Gains in productivity associated with C4 photosynthesis include improved water and nitrogen use efficiencies. Thus, engineering C4 traits into C3 crops is an attractive target for crop improvement. However, the lack of a small, rapid cycling genetic model system to study C4 photosynthesis has limited progress in dissecting the regulatory networks underlying the C4 syndrome. Setaria viridis is a member of the Panicoideae clade and is a close relative of several major feed, fuel, and bioenergy grasses. It is a true diploid with a relatively small genome of ~510 Mb. Its short stature, simple growth requirements, and rapid life cycle will greatly facilitate genetic studies of the C4 grasses. Importantly, S. viridis uses an NADP-malic enzyme subtype C4 photosynthetic system to fix carbon and therefore is a potentially powerful model system for dissecting C4 photosynthesis. Here, we summarize some of the recent advances that promise greatly to accelerate the use of S. viridis as a genetic system. These include our recent successful efforts at regenerating plants from seed callus, establishing a transient transformation system, and developing stable transformation.
Why Study C4?
C4 photosynthesis is the primary mode of carbon capture for some of the world's most important food, feed, and fuel crops, including maize (Zea mays), sorghum (Sorghum bicolor), sugarcane (Saccharum officinarum), millets (e.g. Panicum miliaceum, Pennisetum glaucum, and Setaria italica), Miscanthus x giganteus, and switchgrass (Panicum virgatum). In contrast with C3 plants, C4 plants first fix CO2 into a C4 acid before delivering the CO2 to the Calvin cycle (Hatch and Slack, 1966; Hatch, 1971). For example, in maize and sorghum leaves, CO2 entering mesophyll (M) cells is first fixed into oxaloacetate, which is then reduced to malate in the M chloroplasts. The malate then diffuses into the inner bundle sheath (BS) cells and is transported into the BS chloroplast. There, malate is decarboxylated by NADP-malic enzyme, releasing CO2 close to ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco). This carbon shuttle greatly lowers rates of photorespiration as Rubisco is both isolated from the site of O2 evolution (oxygen evolving complex of photosystem II) and also maintained in a CO2-rich environment. Indeed, in mature maize or sorghum leaves, rates of photorespiration are at the limits of detection under conditions where C3 plants lose up to 30% of their photosynthetic capacity due to photorespiration (Zhu et al., 2008). Accompanying this partitioning of photosynthetic activities are several anatomical adaptations. This includes close vein spacing and large numerous plastids of the inner BS. Together, these characters enable C4 plants to thrive in environments that induce high rates of photorespiration in C3 plants, such as the tropics or grassland savannas (Sage et al., 1999; Sage and Pearcy, 2000). An added benefit of the C4 syndrome is improved nitrogen and water use efficiencies that have likely contributed to their global distribution and high rates of productivity (Sage, 2004; Tilman et al., 2006; Edwards et al., 2010; Edwards and Smith, 2010).
What Is the Grass?
“A child said, What is the grass? fetching it to me with full hands; How could I answer the child?. . . .I do not know what it is any more than he.”
Walt Whitman wrote these words over one and half centuries ago (Whitman, 1855), and in many ways they are as true today as they were then. Despite the agronomic and ecological importance of C4 grasses, little is known of the molecular mechanisms that underpin C4 differentiation. Maize is perhaps the best-studied C4 grass, and yet not a single transcription factor, kinase, phosphatase, or receptor has been identified that can be linked directly to C4 differentiation. Why is this? Part of the reason is that large-scale genetic screens for C4 mutants have not been conducted. For instance, it would be informative to screen large mutagenized populations for mutants that grow well under ambient conditions, but fail to grow under low CO2 conditions. This could potentially identify mutants with defects in CO2 concentration pathways. Similarly, mutants that are rescued under high CO2 but fail to grow in ambient conditions may identify mutants with leaky BS cells. However, these screens are not trivial to conduct with large plants like maize as it is difficult to scrub CO2 from large chamber volumes or keep concentrations of CO2 elevated to very high levels (>1000 ppm).
Instead, in the most comprehensive genetic screen for C4 mutants, Langdale and colleagues (Hall et al., 1998b) first identified mutants with photosynthetic defects and then examined the accumulation of carbon shuttle enzymes as a secondary screen. These screens have led to the identification of a few genes that affect the accumulation of C4 shuttle enzymes and Rubisco. Examples include Golden 2 (Hall et al., 1998a; Fitter et al., 2002; Waters et al., 2009) and Bundle Sheath Defective 2 (Roth et al., 1996; Brutnell et al., 1999; Wostrikoff and Stern, 2007). These mutants display BS cell-specific defects, but this is because the process or protein that is regulated by the gene products is localized to that cell type; therefore, these genes should not be regarded as genes regulating C4 differentiation. For instance, BSD2 is required for the assembly of Rubisco holoenzyme (Brutnell et al., 1999) and displays a BS-specific defect because Rubisco is localized to the BS chloroplast. However, the Bsd2 gene is expressed in both BS and M cells and plays an essential role in regulating Rubisco accumulation in both C3 and C4 plants (Roth et al., 1996; Brutnell et al., 1999; Wostrikoff and Stern, 2007). This localization of a general C3 function to either BS or M cells is likely to be prevalent in C4 grasses, as over 18% of the maize transcriptome is differentially expressed between these two cell types (Sawers et al., 2007), including a sizable fraction of the plastid proteome (Majeran et al., 2005, 2008; Friso et al., 2010). Thus, screens for BS or M cell-specific defects are likely to identify many activities that are strictly compartmentalized but are not directly promoting C4 differentiation per se.
As alluded to above, the slow progress in dissecting C4 traits is due, at least in part, to the relative recalcitrance of the current C4 models to high-throughput genetic screens. The most extensively characterized C4 plants include maize, sorghum, Flaveria sp (Asteraceae), Amaranthus (Chenopodiaceae), and Cleome (Cleomaceae); all lack efficient transformation systems, are large in stature, and have relatively long generation times, often of several months (Brown et al., 2005). Thus, the forward genetic screens that have been the foundation of Arabidopsis thaliana research have yielded little in defining the networks underlying C4 differentiation.
The Time for a New C4 Model Has Arrived
The critical need for major gains in crop productivity (http://www.fao.org/docrep/011/i0100e/i0100e00.htm) and the burgeoning biofuels industry have together refocused attention on understanding C4 photosynthesis. A long-standing goal of many members of the C4 community has been to engineer C4 traits into rice (Oryza sativa) as a way potentially to increase rice yields by 50% with reduced fertilizer inputs (Mitchell and Sheehy, 2007; Sheehy et al., 2007). This ambitious project is now being driven by the generous support of the Bill and Melinda Gates Foundation and taps the expertise of an international consortium of scientists (http://beta.irri.org/projects15/c4rice). However, these engineering efforts will require a more sophisticated understanding of both a C3 grass (rice) and its C4 relatives to identify the genes and networks that can be manipulated to transform a C3 into a C4 photosynthetic system (Hibberd et al., 2008). This will include restructuring the rice leaf to place veins closer together, increasing the cross-sectional area of the BS and engineering cell walls to facilitate the metabolic flux of sugars, amino acids, and C4 acids between the BS and M cells. Major biochemical hurdles must also be overcome, including the engineering of a C4 shuttle to pump CO2 into the BS cells and driving cell-specific accumulation of transporter proteins. It is accepted that this is a high-risk project that likely will not be realized within the next decade. However, if successful, C4 rice has the potential to form the foundation of a second green revolution.
Over the past 5 years, several billion dollars have been invested in bioenergy research as a means to offset the US dependence on oil and reduce greenhouse gas emissions (Ohlrogge et al., 2009). In a highly cited report by the U.S. Department of Energy (Perlack et al., 2005), several grasses were identified as potential feedstocks for a lignocellulosic fuel industry. Interestingly, of the top seven grasses named (maize, sorghum, M. x giganteus, switchgrass, big bluestem, Arundo donax, and reed canary grass), five use C4 photosynthesis. Thus, the primary targets of the biofuels feedstocks community (cellulose, lignin, and hemicellulose) are the products of C4 photosynthesis.
In summary, there is a critical need to understand the networks underlying C4 photosynthetic differentiation as a foundation for engineering these traits into rice and for manipulating existing C4 systems to yield more.
Setaria viridis: Redefining the Model System
Recent advances in sequencing technologies are transforming the field of plant science (Wang et al., 2010). Rather than decades, it will soon be possible to obtain draft genome sequences in a matter of months or weeks (Eid et al., 2009; Pushkarev et al., 2009). Thus, the availability of genome sequence soon will not be a limiting factor in developing a model genetic system. Instead, more mundane characteristics, such as generation time, ease of growth, crossing, size, and self-fertility, likely will dictate the choice of a model system. A case in point is Brachypodium distachyon. Despite the availability of the rice and sorghum genomes, B. distachyon is gaining momentum as a model system because it requires less space and flowers faster than sorghum or rice and, perhaps most importantly, is readily transformable (Vogel and Hill, 2008). With an extremely small and well-annotated genome, B. distachyon is poised to become a powerful model system for understanding grasses (Vogel et al., 2010). However, as a C3 grass, its utility in dissecting C4 photosynthesis is limited.
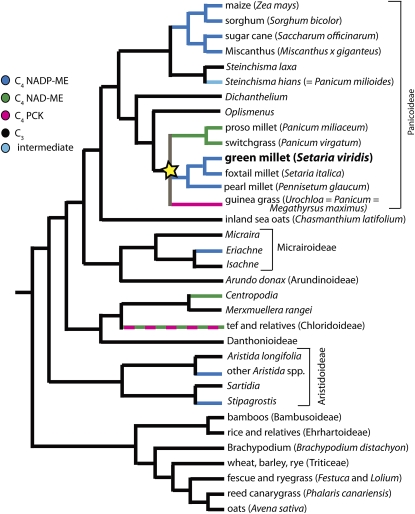
The C4 photosynthetic pathway has originated multiple times within the grasses (Kellogg, 1999; Christin et al., 2008; Vicentini et al., 2008), having evolved, apparently independently, in subfamilies Aristidoideae, Chloridoideae, Micrairoideae, and Panicoideae (Sinha and Kellogg, 1996; Grass Phylogeny Working Group, 2001). The number of origins has been estimated to be as high as 17, although the precise number depends on whether reversal to C3 is considered a possibility (Vicentini et al., 2008). C4 species are divided into subtypes, named for the primary decarboxylating enzyme that is localized to the BS. Maize, sorghum, and sugarcane use an NADP-dependent malic enzyme (NADP-ME subtype), whereas switchgrass and tef (Eragrostis tef) use an NAD-malic enzyme (NAD-ME subtype). A few taxa, such as guinea grass and some species of sand dropseed, use PEP carboxykinase (PCK) to generate a CO2 pump in the BS cells. Most C4 lineages are of the NADP-ME subtype, but there is a minimum of three independent origins of the NAD-ME subtype and two (and probably more) of the PCK subtype (Christin et al., 2009). Thus, C4 lineages have likely exploited existing diversity in C3 ancestors as a foundation for the C4 syndrome (Hibberd and Quick, 2002; Sawers et al., 2007). Most current work has focused on maize and sorghum, which together represent only one of the C4 origins and a single subtype. A summary of the origins and diversity of C4 subtypes in the grasses is shown in Figure 1. This is a pruned tree based on relationships described by Vicentini et al. (2008) and Christin et al. (2008).
Figure 1.
Phylogeny of the Grass Family.
Relationships based largely on Vicentini et al. (2008) and Christin et al. (2009), showing multiple origins of C4 photosystems. S. viridis is an NADP-ME subtype C4 grass that is closely related to the bioenergy feedstock switchgrass (NAD-ME subtype), the grain crop foxtail millet, and the agricultural weed guinea grass (PCK). The C4 photosynthetic systems in this Setaria/Urochloea/Panicum (SUPa clade, indicated with a yellow star) arose independently from the NADP-ME family members of the Andropononeae (maize, sorghum, sugarcane, and M. x giganteus). Dashed lines show clades with multiple subtypes.
The clade within the Panicoideae indicated by the star in Figure 1 is of particular note as it includes representatives of all three C4 subtypes. This clade includes a grain crop, Setaria italica (foxtail millet); a promising biofuel feedstock, Panicum virgatum (switchgrass); and a major agricultural weed, Urochloa maxima (guinea grass), each representing a different subtype of C4. S. viridis (green millet) is closely related to pearl millet (Pennisetum glaucum) and foxtail millet (Setaria italica) and is likely the weedy relative of the domesticated foxtail millet (Dekker, 2003). As recently discussed, the crop S. italica offers a number of advantages as a model for bioenergy grasses (Doust et al., 2009), but as a model for forward and reverse genetics, S. viridis has some distinct advantages. Perhaps most importantly, S. viridis is significantly shorter than most S. italica accessions (see Supplemental Figure 1 online). We grow S. viridis using the same growth conditions, soil, and flats as Arabidopsis and typically grow 50 plants/flat to maturity in the growth chamber. Under short-day treatments, plants can be <10 cm in height at flowering. Second, S. viridis has a much shorter generation time. In short days, some accessions of S. viridis begin flowering within 2 weeks of planting, and mature seed can be harvested within 6 weeks of planting, approximately half the time of S. italica. One of the advantages of S. italica as a model system is that a draft genome assembly will soon be available from JGI-DOE as a major output of a recent DOE-USDA grant (D. Rokhsar and J. Bennetzen, personal communication; http://genomicscience.energy.gov/research/DOEUSDA/abstracts/2008bennetzen_abstract.shtml). However, this likely will not be limiting for long as efforts to sequence S. viridis are now underway as well at DOE and the Beijing Genomics Institute in China (D. Rokhsar and S. Huang, personal communication).
Although performing crosses in Setaria is challenging, it can be done. Because S. viridis flowers are bisexual, they must be emasculated or the pollen destroyed before crossing. The flowers can be emasculated by waiting until just before anthesis, cutting off the top of the anthecium with fine scissors, and then removing the anthers with forceps. This procedure is described in detail by Siles et al. (2001). Alternatively, the entire inflorescence can be dipped in boiling water for a few seconds to kill the pollen. In either case, because of the large number of flowers and the possibility of accidental self-pollination, the identity of seedlings must be verified using molecular markers. Seed propagation generally is not problematic with S. viridis. Even under conditions of rapid cycling (i.e., short days) a single plant can generate upwards of several hundred to several thousand seed. Although the fruits disarticulate readily, they are all caught within bags (we typically use maize ear shoot bags) that are placed over the inflorescence after flowering is complete. We have also found that placing the dried seed in a −80°C freezer overnight greatly improves the frequency of germination.
A recombinant inbred population derived from a cross between S. viridis × S. italica has been generated and a number of quantitative trait loci mapped using this population (Doust et al., 2005; Doust and Kellogg, 2006). In addition, some S. viridis accessions are available at GRIN (http://www.ars-grin.gov/), though little molecular characterization of these materials has been performed. Thus, while S. viridis is suitable for classic genetic analysis, the genetics resources for this organism are clearly lagging the primary C4 model grass, maize.
As an NADP-ME subtype C4 plant, with a small genome of 510 Mb that will likely be available within the year, a rapid generation time, and small size, S. viridis is poised to be an excellent model system. The one major hurdle yet to overcome, however, is transformation. To the best of our knowledge, there are no published reports on tissue culture or transformation methods for S. viridis.
Transformation Methods for S. viridis
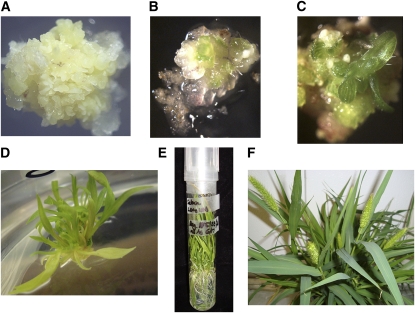
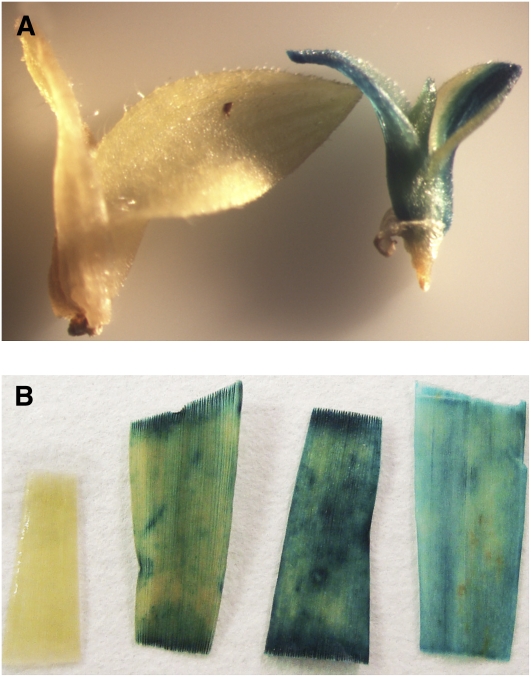
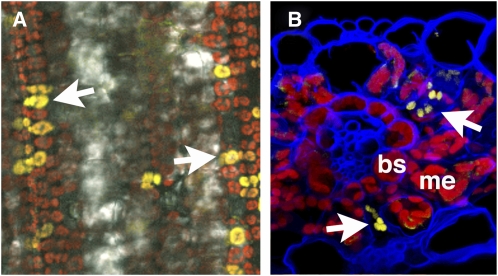
Often in monocot transformation, regeneration of plants from tissue culture is rate limiting. Thus, we first attempted to induce callus formation and regenerate plants from seed of S. viridis using published protocols for B. distachyon and S. italica (Rao et al., 1988; Rout et al., 1998; Vogel and Hill, 2008). As shown in Figure 2, we were able to successfully regenerate mature plants, which flowered and set seed, from seed callus. Although we have only propagated a small number of plants from seed callus through flowering, we have not observed any instances of somaclonal variation, and seed set in all regenerants is very high (>1000/plant). This is likely due to the relatively brief callus phase before plants are moved to regeneration media. A detailed protocol is provided in Supplemental Methods online. Most recently we have been able to transform S. viridis callus with a construct expressing a β-glucuronidase (GUS) transgene (the transgene pOL001 described in Vogel and Hill [2008]). We have been able to generate transformed shoots and several T0 plants (as shown in Figure 3) and are now optimizing our transformation procedure. We have also developed a transient expression system using Agrobacterium tumefaciens–mediated transformation (Janssen and Gardner, 1990) and have been able to introduce a plastid-localized yellow fluorescent protein (YFP) construct into leaf cells of S. viridis (Figure 4). We are now testing a series of constructs for subcellular localization patterns. Together, these studies are extremely encouraging and suggest that routine stable and transient transformation of S. viridis is now in reach.
Figure 2.
Regeneration of S. viridis.
Several steps in the regeneration process are shown, including the following.
(A) Callus formation induced from germinating S. viridis seeds.
(B) Initial shoot regeneration from callus.
(C) Young shoot after regeneration.
(D) Plantlets after transfer to rooting media.
(E) Rooted plant.
(F) Regenerated plants at flowering.
Figure 3.
A. tumefaciens–Mediated Transformation of S. viridis.
(A) Developing shoots grown on selective media following cocultivation with A. tumefaciens. Image shows nontransformed (left) and GUS-positive (right) shoot.
(B) Mature leaf tissue from three independent GUS-positive transformants and a nontransgenic control (leftmost sample). A detailed protocol for S. viridis transformation is provided in Supplemental Methods online, and constructs used are shown in Supplemental Figure 2 online.
Figure 4.
Transient A. tumefaciens–Mediated Transformation of S. viridis.
S. viridis (accession A10) seedlings were grown on MetroMix 360 soil mix under a 12-h light/dark cycle with high relative humidity (75%) at a constant temperature of 23°C. The plants were watered as needed and every 3rd day with 20-10-20 fertilizer. Twelve days after germination, leaf 4 of a healthy S. viridis seedling was inoculated with A. tumefaciens strain AGL1 carrying the pPTN469 vector (Sattarzadeh et al., 2010) at a concentration of 0.05 (OD600). The images show transient expression of a plastid-targeted YFP fusion protein. A detailed protocol for transient transformation is provided in the Supplemental Methods online, and constructs used are shown in Supplemental Figure 2 online.
(A) Low magnification fluorescence image showing abundance of transformed cells (arrows). The picture was taken using a Leica TCS SP5 laser scanning confocal microscope (Leica Microsystems) using ×63 oil submerged objective lens. For the YFP signal (yellow), leaf tissue was excited with a 514-nm laser, and emitted light was collected from 525 to 575 nm. Autofluorescence (red) was captured from 650 to 789 nm. Image was compiled using Leica image software LAS-AF (version 1.8.2.) and Adobe Photoshop CS3 version 9.0.2 (Adobe Systems).
(B) Confocal reconstruction of leaf section showing transformed mesophyll cells (arrows). Chlorophyll autofluorescence is red and cell walls counterstained blue. bs, bundle sheath cell; m, mesophyll cells. Leaves were fixed in 2.5% paraformaldehyde, embedded in 7% low melting point agarose, cryosectioned, and counterstained with calcofluor white as described (Goldshmidt et al., 2008). The image was taken with a Carl Zeiss LSM 710 laser scanning microscope. A 514-nm laser excitation and 520- to 550-nm prism filter set was used to detect YFP emission (yellow), a 405-nm laser excitation and 475- to 500-nm prism filter set used for calcofluor white emission (blue), and a 488-nm laser excitation and 650- to 700-nm prism filter set used for detection of the chlorophyll emission (red). Subsequently, the confocal z-stack was reconstructed using Bitplane Imaris 7 software.
Given the many attractive features of S. viridis as a genetic system, the time is right to begin detailed histological, physiological, and molecular characterizations of this plant. Systems modeling, which has shown its capacity in identifying limitations of C3 photosynthesis (Lefebvre et al., 2005; Zhu et al., 2007), can be used to identify features critical for the high efficiency of C4 photosynthesis. To do this, a systems model of C4 photosynthesis must faithfully describe not only the biophysical and biochemical processes involved in C4 photosynthesis, but also the diffusion process of major metabolites and gases through the system. Once such a model is available, combining it with measurements of the biochemical and anatomical properties of C4 leaves will in turn allow sensitivity analysis of the CO2 fixation efficiency to various parameters and environmental factors. Such an analysis will help identify the critical biochemical and anatomical features controlling efficiencies of C4 photosynthesis. These features can then be focused on engineering C4 photosynthesis into C3 plants. Thus, the C4 community could greatly benefit from a concerted effort to characterize the growth, development, and physiology of S. viridis.
Given the rapid progress that has been made in developing tools for S. viridis, the C4 community may soon be able to rally around a common C4 model. With any luck, we may be able to answer that outstanding question, “What is the [C4] grass?”, by fetching a handful of S. viridis.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mature S. viridis and S. italica at Flowering.
Supplemental Figure 2. Schematic of Expression Constructs.
Supplemental Methods. Plant Regeneration and Agrobacterium-Mediated Transformation of S. viridis.
Supplementary Material
Acknowledgments
We thank the Boyce Thompson Institute imaging center for training and use of the confocal microscope (supported through National Science Foundation DBI-0618969 and the Triad foundation). We thank Pinghua Li and Ying Rong for the photographs of the Setaria plants.
References
- Brown N.J., Parsley K., Hibberd J.M. (2005). The future of C4 research–Maize, Flaveria or Cleome? Trends Plant Sci. 10: 215–221 [DOI] [PubMed] [Google Scholar]
- Brutnell T.P., Sawers R.J., Mant A., Langdale J.A. (1999). BUNDLE SHEATH DEFECTIVE2, a novel protein required for posttranslational regulation of the rbcL gene of maize. Plant Cell 11: 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin P.A., Besnard G., Samaritani E., Duvall M.R., Hodkinson T.R., Savolainen V., Salamin N. (2008). Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr. Biol. 18: 37–43 [DOI] [PubMed] [Google Scholar]
- Christin P.A., Salamin N., Kellogg E.A., Vicentini A., Besnard G. (2009). Integrating phylogeny into studies of c4 variation in the grasses. Plant Physiol. 149: 82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J. (2003). Evolutionary biology of the foxtail (Setaria) species-group. Weed Biology and Management, Inderjit, ed (Dordrecht, The Netherlands: Kluwer; ), pp. 65–114 [Google Scholar]
- Doust A.N., Devos K.M., Gadberry M.D., Gale M.D., Kellogg E.A. (2005). The genetic basis for inflorescence variation between foxtail and green millet (poaceae). Genetics 169: 1659–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust A.N., Kellogg E.A. (2006). Effect of genotype and environment on branching in weedy green millet (Setaria viridis) and domesticated foxtail millet (Setaria italica) (Poaceae). Mol. Ecol. 15: 1335–1349 [DOI] [PubMed] [Google Scholar]
- Doust A.N., Kellogg E.A., Devos K.M., Bennetzen J.L. (2009). Foxtail millet: A sequence-driven grass model system. Plant Physiol. 149: 137–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards E.J., Smith S.A. (2010). Phylogenetic analyses reveal the shady history of C4 grasses. Proc. Natl. Acad. Sci. USA 107: 2532–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards E.J., et al. (2010). The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science 328: 587–591 [DOI] [PubMed] [Google Scholar]
- Eid J., et al. (2009). Real-time DNA sequencing from single polymerase molecules. Science 323: 133–138 [DOI] [PubMed] [Google Scholar]
- Fitter D.W., Martin D.J., Copley M.J., Scotland R.W., Langdale J.A. (2002). GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 31: 713–727 [DOI] [PubMed] [Google Scholar]
- Friso G., Majeran W., Huang M., Sun Q., van Wijk K.J. (2010). Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol. 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y. (2008). Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 20: 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group (2001). Phylogeny and subfamilial classification of the Poaceae. Ann. Mo. Bot. Gard. 88: 373–457 [Google Scholar]
- Hall L.N., Rossini L., Cribb L., Langdale J.A. (1998a). GOLDEN 2: A novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 10: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L.N., Roth R., Brutnell T.P., Langdale J.A. (1998b). Cellular differentiation in the maize leaf is disrupted by bundle sheath defective mutations. Symp. Soc. Exp. Biol. 51: 27–31 [PubMed] [Google Scholar]
- Hatch M.D. (1971). The C4-pathway of photosynthesis: Evidence for an intermediate pool of carbon dioxide of the donor C4-dicarboxylic acid. Biochem. J. 125: 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M.D., Slack C.R. (1966). Photosynthesis by sugar-cane leaves: A new carboxylation reaction and the pathway of sugar formation. Biochem. J. 101: 103–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd J.M., Quick W.P. (2002). Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415: 451–454 [DOI] [PubMed] [Google Scholar]
- Hibberd J.M., Sheehy J.E., Langdale J.A. (2008). Using C4 photosynthesis to increase the yield of rice-rationale and feasibility. Curr. Opin. Plant Biol. 11: 228–231 [DOI] [PubMed] [Google Scholar]
- Janssen B.J., Gardner R.C. (1990). Localized transient expression of GUS in leaf discs following cocultivation with Agrobacterium. Plant Mol. Biol. 14: 61–72 [DOI] [PubMed] [Google Scholar]
- Kellogg E.A. (1999). Phylogenetic aspects of the evolution of C4 photosynthesis. C4 Plant Biology, Sage R.F., Monson R.K., eds (San Diego, CA: Academic Press; ), pp. 411–444 [Google Scholar]
- Lefebvre S., Lawson T., Zakhleniuk O.V., Lloyd J.C., Raines C.A. (2005). Increased sedoheptulose-1,7-bisphosphatase activity in transgenic tobacco plants stimulates photosynthesis and growth from an early stage in development. Plant Physiol. 138: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Cai Y., Sun Q., van Wijk K.J. (2005). Functional differentiation of bundle sheath and mesophyll maize chloroplasts determined by comparative proteomics. Plant Cell 17: 3111–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran W., Zybailov B., Ytterberg A.J., Dunsmore J., Sun Q., van Wijk K.J. (2008). Consequences of C4 differentiation for chloroplast membrane proteomes in maize mesophyll and bundle sheath cells. Mol. Cell. Proteomics 7: 1609–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P.L., Sheehy J.E. (2007). The case for C4 rice. Charting New Pathways to C4 Rice J.E., Sheehy P.L., Mitchell, Hardy B., eds (Los Banos, Philippines: International Rice Research Institute; ), pp. 27–36 [Google Scholar]
- Ohlrogge J., Allen D., Berguson B., Dellapenna D., Shachar-Hill Y., Stymne S. (2009). Energy. Driving on biomass. Science 324: 1019–1020 [DOI] [PubMed] [Google Scholar]
- Perlack R.D., Wright L.L., Turhollow A.F., Graham R.L. (2005). Biomass as a feedstock for a bioenergy and bioproducts industry: The technical feasibility of a billion-ton annual supply. (Oak Ridge, TN: Oak Ridge National Laboratory Report TM-2005, under contract DOE/GO-102005-2135; ). [Google Scholar]
- Pushkarev D., Neff N.F., Quake S.R. (2009). Single-molecule sequencing of an individual human genome. Nat. Biotechnol. 27: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A.M., Kavi Kishor P.B., Ananda Reddy L., Vaidvanath K. (1988). Callus induction and high frequency plant regeneration in Italian millet (Setaria italica). Plant Cell Rep. 7: 557–559 [DOI] [PubMed] [Google Scholar]
- Roth R., Hall L.N., Brutnell T.P., Langdale J.A. (1996). bundle sheath defective2, a mutation that disrupts the coordinated development of bundle sheath and mesophyll cells in the maize leaf. Plant Cell 8: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout G.R., Samataray S., Das D. (1998). In vitro selection and characterization of Ni-tolerant callus lines of Setaria italica L. Acta Physiol. Plant. 20: 269–275 [Google Scholar]
- Sage R.F. (2004). The evolution of C4 photosynthesis. New Phytol. 161: 341–370 [DOI] [PubMed] [Google Scholar]
- Sage R.F., Li M., Monson R.K. (1999). The taxonomic distribution of C4 photosynthesis. C4 Plant Biology, Sage R.F., Monson R.K., eds (San Diego, CA: Academic Press; ), pp. 551–584 [Google Scholar]
- Sage R.F., Pearcy R.W. (2000). The physiological ecology of C4 photosynthesis. Photosynthesis: Physiology and Metabolism, Sharkey T.D., von Caemmerer S., eds (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 497–532 [Google Scholar]
- Sattarzadeh A., Fuller J., Moguel S., Wostrikoff K., Sato S., Covshoff S., Clemente T., Hanson M., Stern D.B. (2010). Transgenic maize lines with cell-type specific expression of fluorescent proteins in plastids. Plant Biotechnol. J. 8: 112–125 [DOI] [PubMed] [Google Scholar]
- Sawers R.J., Liu P., Anufrikova K., Hwang J.T., Brutnell T.P. (2007). A multi-treatment experimental system to examine photosynthetic differentiation in the maize leaf. BMC Genomics 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy J.E., Ferrer A.B., Mitchell P.L., Elmido-Mabilangan A., Pablico P., Dionara M.J.A. (2007). How the rice crop works and why it needs a new engine. Charting New Pathways to C4 Rice, Sheehy J.E., Mitchell P.L., Hardy B., eds (Los Banos, Philippines: International Rice Research Institute; ), pp. 3–26 [Google Scholar]
- Siles M.M., Baltensperger D.D., Nelson L.A. (2001). Technique for artificial hybridization of foxtail millet. Setaria italica (L.) Beauv. Crop Sci. 41: 1408–1412 [Google Scholar]
- Sinha N.R., Kellogg E.A. (1996). Parallelism and diversity in multiple origins of C4 photosynthesis in the grass family. Am. J. Bot. 83: 1458–1470 [Google Scholar]
- Tilman D., Hill J., Lehman C. (2006). Carbon-negative biofuels from low-input high-diversity grassland biomass. Science 314: 1598–1600 [DOI] [PubMed] [Google Scholar]
- Vicentini A., Barber J.C., Aliscioni S.A., Giussani L.M., Kellogg E.A. (2008). The age of the grasses and clusters of origins of C4 photosynthesis. Glob. Change Biol. 14: 1–15 [Google Scholar]
- Vogel J., Hill T. (2008). High-efficiency Agrobacterium-mediated transformation of Brachypodium distachyon inbred line Bd21–3. Plant Cell Rep. 27: 471–478 [DOI] [PubMed] [Google Scholar]
- Vogel J.P., et al. (2010). Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463: 763–768 [DOI] [PubMed] [Google Scholar]
- Wang L., Li P., Brutnell T.P. (2010). Exploring plant transcriptomes using ultra high-throughput sequencing. Brief Funct. Genomics 9: 118–128 [DOI] [PubMed] [Google Scholar]
- Waters M.T., Wang P., Korkaric M., Capper R.G., Saunders N.J., Langdale J.A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. (1855). Leaves of Grass. (New York: self; ). [Google Scholar]
- Wostrikoff K., Stern D. (2007). Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 104: 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.-G., de Sturler E., Long S.P. (2007). Optimizing the distribution of resources between enzymes of carbon metabolism can dramatically increase photosynthetic rate: A numerical simulation using an evolutionary algorithm. Plant Physiol. 145: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.G., Long S.P., Ort D.R. (2008). What is the maximum efficiency with which photosynthesis can convert solar energy into biomass? Curr. Opin. Biotechnol. 19: 153–159 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.