Telomeres are essential for the integrity of linear eukaryotic chromosomes. This study provides evidence that GTBP1, a single-strand telomere binding protein, is an essential component for telomere structure and function in tobacco by preventing aberrant interchromosomal telomeric homologous recombination.
Abstract
Telomeres are nucleoprotein complexes essential for the integrity of eukaryotic chromosomes. Cellular roles of single-stranded telomeric DNA binding proteins have been extensively described in yeast and animals, but our knowledge about plant single-strand telomeric factors is rudimentary. Here, we investigated Nicotiana tabacum G-strand-specific single-stranded telomere binding proteins (GTBPs), homologs of a human heterogeneous nuclear ribonucleoprotein. GTBPs bound specifically to the plant single-stranded (TTTAGGG)4 telomeric repeat element in vitro and were associated with telomeric sequences in tobacco BY-2 suspension cells. Transgenic plants (35S:RNAi-GTBP1), in which GTBP1 was suppressed, exhibited severe developmental anomalies. In addition, the chromosomes of 35S:RNAi-GTBP1 cells displayed elongated telomeres, frequent formation of extrachromosomal telomeric circles, and numerous abnormal anaphase bridges, indicating that GTBP1 knockdown tobacco plants experienced genome instability. GTBP1 inhibited strand invasion, an initial step in interchromosomal homologous recombination. We propose that GTBP1 plays a critical role in telomere structure and function by preventing aberrant interchromosomal telomeric homologous recombination in tobacco.
INTRODUCTION
Telomeres are unique nucleoprotein structures that protect the extreme termini of linear eukaryotic chromosomes. They are composed of tandemly repeated G-rich DNA sequence elements (TTAGGG in vertebrates and TTTAGGG in higher plants) along with nonhistone telomere binding proteins (Blackburn, 1991; Collins, 2000; Shore, 2001). Telomere binding proteins play an essential role in telomere architecture. Therefore, without properly functioning telomere binding proteins, telomeres are destabilized and cells undergo senescence, apoptosis, or the ageing process (Blackburn, 2001; Blasco, 2005). Telomere binding proteins are classified into two groups based on their binding modes. Human TRF1/PIN2 and TRF2 and yeast Rap1 and Taz1 are double-stranded telomere binding proteins (Chong et al., 1995; Bilaud et al., 1997; Smogorzewska and de Lange, 2004), whereas yeast Cdc13p and human POT1 are single-stranded specific telomeric binding factors (Nugent et al., 1996; Baumann and Cech, 2001). Cdc13p and POT1 are typified by their association with telomeric DNA through an oligonucleotide-oligosaccharide binding fold (OB-fold). In humans, POT1, TRF1, and TRF2, together with TIN2, TPP1, and Rap1, form a telomere-protein complex sheltrin (de Lange, 2005). In addition, heterogeneous nuclear ribonucleoproteins (HnRNPs) A1 and D can bind the single-stranded telomere sequences (Ishikawa et al., 1993; LaBranche et al., 1998). HnRNP A1 is a positive telomere length regulator, as HnRNP1 A1-deficient mouse cells harbor shorter telomeres than do normal cells, and complementation of HnRNP1 A1 expression in mutant cells restores normal telomere length (LaBranche et al., 1998).
Although single-strand-specific telomere binding factors are largely unstudied in higher plants relative to those in humans and yeast, identification and some cellular aspects of these proteins have recently been elucidated. POT1a and POT1b were identified in Arabidopsis thaliana by their sequence homology with Schizosaccharomyces pombe POT1 (Shakirov et al., 2005). Transgenic Arabidopsis plants, which overexpressed the truncated N-terminal region of POT1b, exhibited extensive erosion of telomeres, nonspecific chromosome fusions, and severe morphological defects (Shakirov et al., 2005). This indicated that POT1b contributes to telomere stability by capping the ends of chromosomes. POT1a interacted with the active telomerase complex, and a knockout mutation of POT1a caused a massive decrease in telomerase activity and gradual shortening of telomeres over generations, suggesting its positive role in telomere length homeostasis (Surovtseva et al., 2007). However, unlike yeast and vertebrate POT1 proteins, recombinant At POT1 proteins have no detectable single-strand telomere binding activity in vitro (Shakirov et al., 2009). In addition, nuclear extracts prepared from pot1a and pot1b T-DNA insertion mutants displayed no changes in single-strand-specific telomere binding activity. These results raise the possibility that POT1 proteins are not major single-stranded telomeric binding proteins in Arabidopsis (Shakirov et al., 2009).
In addition to POT1-like proteins, several putative single-stranded telomeric binding proteins were identified in higher plants. From tobacco (Nicotiana tabacum) bright yellow-2 (BY-2) suspension-cultured cells, Hirata et al. (2004) detected a DNA-protein complex in gel retardation assays using a 32P-(TTTAGGG)4 repeat single-stranded probe. The binding protein was purified, and its partial amino acid sequence was determined. RT-PCR with degenerate primers resulted in the isolation of a tobacco cDNA encoding a 36-kD protein (G-strand-specific single-stranded telomere binding protein 1 [GTBP1]) containing two RNA recognition motifs (RRMs) (Hirata et al., 2004). With the aid of affinity chromatography followed by matrix-assisted laser-desorption ionization time of flight mass spectrometry analysis, two single-strand telomere binding proteins (STEP1 and WHY1) were isolated from Arabidopsis (Kwon and Chung, 2004; Yoo et al., 2007). The binding of STEP1 to telomeric DNA inhibited telomerase-mediated telomere elongation in vitro (Kwon and Chung, 2004). Although a T-DNA insertional mutation of WHY1 did not result in detectable abnormal phenotypes, why1 mutant plants contained longer telomeres, whereas WHY1-overexpressing plants showed shortened telomeres and decreased telomerase activity. Thus, both STEP1 and WHY1 might be involved in the regulation of telomere length in Arabidopsis. However, physiological roles of these putative tobacco and Arabidopsis telomeric proteins remain to be elucidated.
In this study, we isolated two additional GTBP paralogs (GTBP2 and GTBP3) from tobacco. In vitro gel retardation assays indicated that the three GTBP paralogs interacted specifically with the single-stranded TTTAGGG element. RT-PCR with gene-specific primers revealed that the mRNA level of GTBP1 was much higher than those of GTBP2 and GTBP3. Chromatin immunoprecipitation (ChIP) analysis showed that GTBP1 was associated with telomeric sequences in BY-2 cells. We investigated a possible physiological role of GTBP1 via RNA interference (RNAi)-mediated gene knockdown. The T0 and T1 35S:RNAi-GTBP1 transgenic tobacco plants showed severe developmental abnormalities. In addition, chromosomes of the transgenic cells displayed longer telomeres, frequent formation of extrachromosomal telomeric circles (t-circles), and one or more abnormal anaphase bridges, indicating that GTBP1 knockdown plants experienced genome instability. GTBP1 inhibited strand invasion, an initial step for interchromosomal recombination. Based on these results, we propose that GTBPs play important roles in telomere structure and function in tobacco plants.
RESULTS
Isolation and Characterization of Three GTBP Paralogs in Tobacco
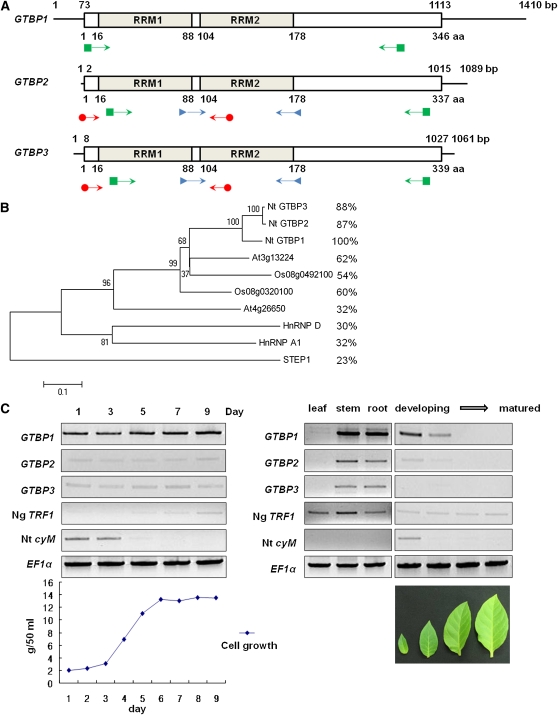
The HnRNP homolog GTBP1 was previously identified in tobacco BY-2 cells (Hirata et al., 2004) and encodes a 36-kD protein with two RRMs. With total RNA from BY-2 cells, we performed RT-PCR using degenerate oligonucleotides designed from the conserved region of the RRMs (Figure 1A; see Supplemental Table 1 online). The PCR products of ~300 bp encoded three homologous partial GTBPs, one of which corresponded to GTBP1. Thus, the products were referred to as GTBP1, GTBP2, and GTBP3. Total recombinant λ DNA was prepared from the λ-uni-Zap II tobacco flower cDNA library. The complete coding regions of GTBP2 and GTBP3 were obtained through 5′- and 3′-rapid amplification of cDNA ends (RACE) using the λ DNA as a template with primers corresponding to the 5′- and 3′-ends of the library vector sequence and the internal regions of the partial GTBP cDNA clones (Figure 1A). The coding regions of GTBP1 (Hirata et al., 2004), GTBP2, and GTBP3 encode 346 (36.2 kD), 337 (35.8 kD), and 339 (36.0 kD) amino acid proteins, respectively. Each GTBP contains two RRMs in the N-terminal region similar to human HnRNP A1. GTBP2 and GTBP3 are 98% identical to each other, and 87 and 88% identical to GTBP1, respectively (Figure 1B; see Supplemental Figure 1 online). Thus, GTBP2 and GTBP3 are more closely related to each other than either is to GTBP1. The three GTBPs are 58 to 63% identical to the homologous dicot Arabidopsis (At3g13224) and monocot rice (Oryza sativa; Os08g0492100) proteins, whose cellular functions are unknown (Figure 1B). The Nt GTBP paralogs are relatively less identical (30 to 32%) to human hnRNP A1 and hnRNP D.
Figure 1.
Structure and Expression of Nt GTBPs.
(A) Schematic representation of three tobacco GTBP cDNAs and their predicted proteins. The solid lines show 5′- and 3′-untranslated regions. The bars depict coding regions. The RRMs (RRM1 and RRM2) are indicated. PCR primers for partial cDNA cloning, 5′- and 3′-RACE, and RT-PCR are indicated by arrows with triangles, circles, and squares, respectively.
(B) Phylogenetic relationship of 10 GTBP homologs from tobacco, Arabidopsis, rice, and human. The tree was constructed using MEGA4 software with the neighbor-joining method and the summed branch length of the tree is 3.21911554. Amino acid identities of these proteins to GTBP1 are shown in right side. A text file of the alignment used to generate this tree is available as Supplemental Data Set 1 online.
(C) Expression profiles of three GTBP paralogs. Total RNA was isolated from BY-2 cells over the course of 9 d (left gel) and from leaves, stems, and roots of 1-month-old tobacco plants (right gel) and analyzed by RT-PCR using gene-specific primers. Growth curve of BY-2 cells is shown in the bottom of left panel. Tobacco leaves that are in four different developmental stages are shown in the bottom of the right panel. Ng TRF1 and Nt cyM were used as G-phase and M-phase markers, respectively. The level of elongation factor 1α (EF1α) transcript is shown as a loading control.
[See online article for color version of this figure.]
Genomic DNA was isolated from mature tobacco leaves, digested by EcoRV and HindIII, and hybridized with 32P-labeled GTBP1, GTBP2, or GTBP3 probes. These hybridizations detected two to four bands with each enzyme digestion (see Supplemental Figure 2 online). To examine the expression of each GTBP mRNA, RT-PCR was conducted with gene-specific primers (Figure 1A; see Supplemental Figure 3 online). In BY-2 suspension-cultured cells, all three GTBP genes were ubiquitously expressed during a 9-d culture period (Figure 1C). The GTBP genes were highly expressed in mature tobacco stems and roots, while their expression gradually decreased as leaf tissues matured. Notably, the level of GTBP1 mRNA was markedly higher than those of GTBP2 and GTBP3 regardless of the different cell and tissue types, indicating that GTBP1 was the predominantly expressed paralog in tobacco (Figure 1C). Taken together, the three GTBP paralogs do not represent a cloning artifact, but rather are encoded by a gene family.
Nt GTBPs Bind Plant Single-Stranded Telomeric Repeats with Sequence Specificity
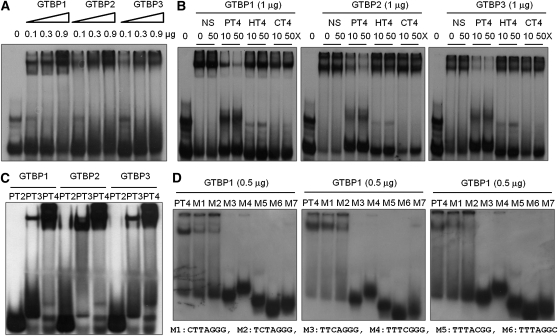
GTBP1, GTBP2, and GTBP3 were expressed in Escherichia coli as fusion proteins with maltose binding protein (MBP). The purified proteins were incubated with a 32P-labeled single-stranded (TTTAGGG)4 repeat in the presence or absence of various competitors and analyzed by gel retardation assays. The GTBP proteins produced a single DNA-protein complex. Increasing amounts of the proteins (0.1 to 0.9 μg) generated more intense shifted bands (Figure 2A). The binding affinities of all three GTBPs were largely disrupted when a 50-fold excess of unlabeled plant telomeric element (PT4) was included in the reaction mixtures. By contrast, nonspecific DNA or telomeric repeats from other species, including human (HT4) and Caenorhabditis elegans (CT4), did not exert any inhibitory effects on the interaction of Nt GTBPs with PT4 (Figure 2B). GTBPs did not bind double-stranded telomeric elements at detectable levels. Thus, GTBPs bound specifically to single-stranded plant telomeric repeats in vitro.
Figure 2.
Nt GTBPs Bind Plant Single-Stranded Telomeric Repeats with Sequence Specificity.
(A) GTBPs bind plant single-stranded telomeric sequences. Various quantities (0, 0.1, 0.3, and 0.9 μg) of three Nt GTBP paralogs were incubated with radiolabeled single-stranded (TTTAGGG)4 (PT4) repeats and analyzed in a gel retardation assay.
(B) Sequence-specific binding activity of GTBPs. Nt GTBPs (1 μg) were incubated with radiolabeled PT4 in the presence of a 10- or 50-fold excess of unlabeled plant (PT4), human (HT4), C. elegans (CT4), or nonspecific (NS) telomeric DNA as competitors. The first “0” lane of each gel contained radiolabeled PT4 without protein.
(C) Gel retardation assays with various telomere repeats. GTBPs (1 μg) were analyzed with radiolabeled PT2, PT3, or PT4 (containing two, three, or four repeats, respectively).
(D) Gel retardation assays with a series of single nucleotide mutants of PT4. Each mutant (M1 to M7) contained a single nucleotide substitution in the repeated sequence, as listed below the gels.
Figure 2C also shows that all three GTBPs were able to form shifted bands only with the three-repeat or longer TTTAGGG single-stranded telomeric element. The binding activities of GTBPs were markedly increased with four telomeric repeats (PT4) relative to those with three repeats (PT3), which suggested that, under our experimental conditions, three repeats were minimal telomeric sequences. In addition, substitutions of the first and second thymine nucleotides (T1 to C1 and T2 to C2) reduced the binding signals by ~50%, whereas single mutations introduced to nucleotides between the third and seventh positions (T3→C3, A4→C4, G5→C5, G6→C6, or G7→C7) completely abrogated the DNA binding capacity (Figure 2D). Overall, these results indicate that all three Nt GTBP paralogs have similar binding specificities for plant single-stranded telomeric repeats in vitro.
GTBP1 Is Associated with Telomeric Sequences in Tobacco BY-2 Cells
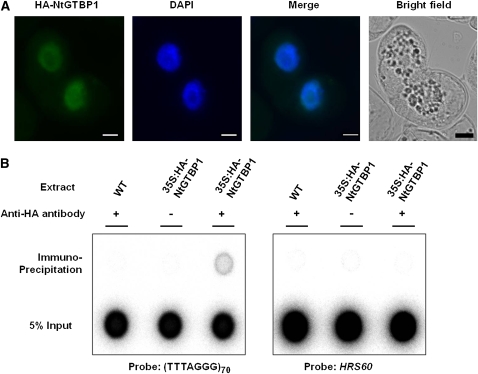
ChIP assays were performed to provide in vivo evidence for the interaction of GTBPs with telomeric sequences. The hemagglutinin (HA)-tagged GTBP1 fusion gene was transiently expressed in BY-2 suspension cells under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The cellular localization of HA-GTBP1 was examined by immunostaining using primary anti-HA antibody and secondary Cy3-conjugated anti-mouse antibody. The HA-GTBP1 localization signal was detected exclusively in the nucleus, indicating that the HA-GTBP1 fusion protein was effectively expressed in BY-2 cells (Figure 3A). The genomic DNA-protein complex was prepared from wild-type and 35S:HA-GTBP1 BY-2 cells, fragmented by sonication, and subjected to immunoprecipitation using an anti-HA antibody. The coimmunoprecipitated DNA was then detected by hybridization with a 32P-labeled (TTTAGGG)70 repeat element. As shown in Figure 3B, telomeric fragments were pulled down by the anti-HA antibody in 35S:HA-GTBP1 BY-2 cells, while HRS60, a tandemly organized highly repeated DNA sequence in tobacco (Koukalova et al., 1993), was not pulled down by the antibody. In addition, there was no hybridization signal in wild-type cell-free extracts. These results support the notion that GTBP1 is associated with telomeric sequences in vivo.
Figure 3.
GTBP1 Is Associated with Telomeric Sequences in Tobacco BY-2 Cells.
(A) Nuclear localization of HA-GTBP1. 35S:HA-GTBP1 was transformed into BY-2 cells, and localization of HA-GTBP1 was examined by immunohistochemistry using an anti-HA antibody. The BY-2 cells were viewed by fluorescence microscopy under dark field and light field. The nuclei were stained with DAPI. Bars = 10 μm.
(B) ChIP assay. The genomic DNA-protein complex from wild-type and 35S:HA-GTBP1 BY-2 cells was fragmented by sonication and subjected to immunoprecipitation using an anti-HA antibody. The coimmunoprecipitated DNA was hybridized with 32P-labeled (TTTAGGG)70 or HRS60 repeated tobacco DNA sequences.
Generation and Characterization of 35S:RNAi-GTBP1 Transgenic Tobacco Plants
Because GTBP1 was predominantly expressed in all examined tobacco tissues (Figure 1C), GTBP1 was chosen for further examination. To address the cellular functions of GTBP1, we employed an RNAi-mediated gene knockdown transgenic approach. Transgenic tobacco plants (35S:RNAi-GTBP1) with suppressed GTBP1 were established by introducing a complementary hairpin-forming DNA fragment of the N-terminal (122 to 498 bp) or C-terminal (726 to 1070 bp) region of GTBP1 cDNA under the control of the CaMV 35S promoter (Figure 4A). Several independent primary transformants were selected due to their kanamycin resistance. The presence of the transgene was confirmed by genomic DNA gel blot analysis (see Supplemental Figure 4 online). RT-PCR analysis showed that these independent RNAi transgenic lines contained markedly reduced levels of GTBP1 mRNA compared with wild-type tobacco plants (Figure 4B).
Figure 4.
Construction and Characterization of 35S:RNAi-GTBP1 Knockdown Transgenic Tobacco Plants.
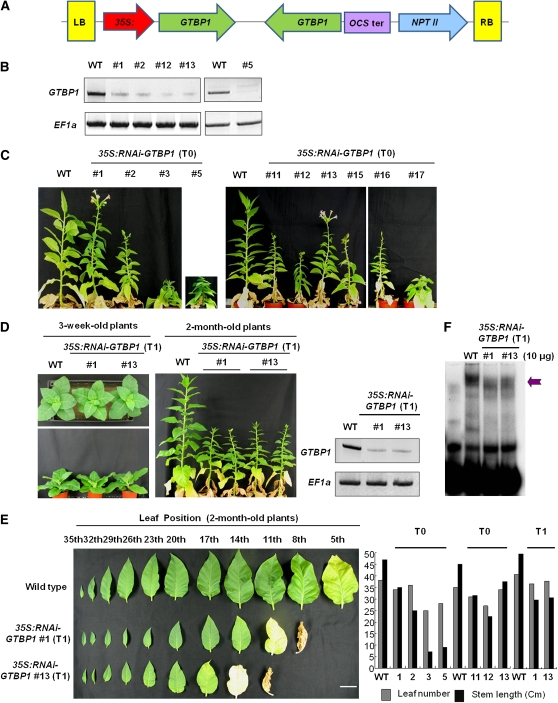
(A) Schematic structure of GTBP1 RNAi binary vector construct. The 35S:RNAi-GTBP1 vector includes the inverted-repeat sequence of the N-terminal (122 to 498 bp) or C-terminal (726 to 1070 bp) regions of GTBP1 cDNA. LB, left border; OCS ter, octopine synthase terminator; NPTII, neomycin phosphotransferase II; RB, right border.
(B) Repression of GTBP1 mRNA in transgenic plants. Total leaf RNA from wild-type and independent T0 35S:RNAi-GTBP1 lines was analyzed by RT-PCR. EF1α was used as a loading control.
(C) Gross morphology of 2-month-old wild-type and independent T0 transgenic lines grown under greenhouse conditions.
(D) Morphology of 3-week-old and 2-month-old wild-type and T1 RNAi transgenic lines (#1 and #13). GTBP1 transcript levels were determined by RT-PCR in T1 generation (shown at the right with EF1α as a control).
(E) Morphological comparison of leaves from 2-month-old wild-type and T1 transgenic (#1 and #13) plants. Leaf number and stem length of wild-type and 35S:RNAi-GTBP1 (T0 and T1) tobacco plants are shown in the right panel. Bar = 3 cm.
(F) Single-strand telomere binding activities of wild-type and T1 RNAi transgenic seedlings. The cell-free nuclear extracts containing 10 μg of protein were prepared from wild-type and T1 transgenic seedlings (#1 and #13), incubated with PT4, and analyzed by gel retardation assays as described in Figure 2. Arrow indicates protein-PT4 complex.
[See online article for color version of this figure.]
As shown in Figure 4C, knockdown of GTBP1 resulted in abnormal pleiotropic phenotypes in tobacco plants. The independent T0 35S:RNAi-GTBP1 transgenic plants showed severe growth retardation, failed to produce healthy leaves and shoots, and exhibited premature senescence. Floral organs emerged earlier in 35S:RNAi-GTBP1 plants, but they usually fell off before maturation; thus, most transgenic plants were unable to produce seeds. In general, abnormal phenotypes of the 35S:RNAi-GTBP1 plants were less apparent during the earlier stages of development but became gradually more serious as plants matured. Among the various independent RNAi transgenic plants, lines #3, #5, #11, #12, #15, #16, and #17, which all contained multiple copies of the transgene (see Supplemental Figure 4 online), displayed more severe phenotypes and were sterile. Thus, we obtained subsequent T1 progeny from transgenic lines #1 and #13 that contained a single copy of the transgene with less severe phenotypes. The T1 #1 and #13 plants also displayed retarded growth (Figure 4D). Although leaf number in wild-type and T1 transgenic plants was similar, transgenic leaves were smaller and became senescent earlier than did wild-type leaves (Figure 4E). In addition, internode lengths in the RNAi lines were shorter relative to those in wild-type tobacco plants; hence, T1 35S:RNAi-GTBP1 plants were ~50% as tall as wild-type tobacco plants (Figure 4D). Cell-free nuclear extracts were prepared from 1-week-old wild-type and T1 transgenic seedlings (#1 and #13), incubated with single-stranded four telomeric repeats (PT4), and subjected to gel retardation assays. The results revealed that the RNAi transgenic seedlings possessed decreased binding activities for the telomeric repeats compared with wild-type plants (Figure 4F). Collectively, the results in Figure 4 suggest that downregulation of GTBP1 caused severe developmental anomalies in transgenic tobacco, which correlated with the reduction of binding activity to single-stranded telomeric repeats.
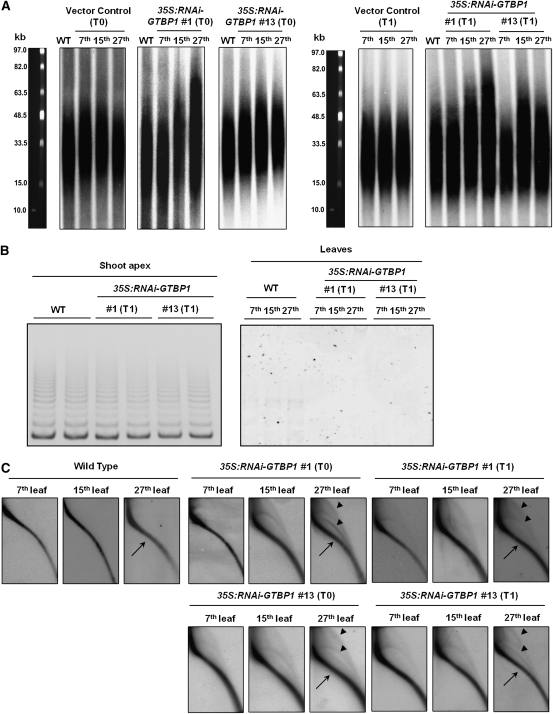
Telomere Lengthening and Formation of Extrachromosomal Telomeric Circles in 35S:RNAi-GTBP1 Transgenic Tobacco Plants
The massive abnormalities of the 35S:RNAi-GTBP1 transgenic tobacco plants may be due to defects of telomere metabolism. To investigate whether GTBP1 participates in telomere regulation, telomere lengths were determined in wild-type, vector control, and 35S:RNAi-GTBP1 (lines #1 and #13) leaves. Telomeres in wild-type and vector control leaves were ~15 to 50 kb, which were consistent with previous studies (Fajkus et al., 1995; Yang et al., 2004). On the other hand, both #1 and #13 T0 transgenic lines displayed varied lengths of telomeres depending on the developmental stages of the leaf tissues. For example, in line #1, the seventh leaf contained normal telomeres (15 to 50 kb), whereas the 15th leaf contained telomeres ranging from 15 to 65 kb. Telomeres in the 27th leaf were even longer (15 to 75 kb) (Figure 5A, left panel). Similarly, the 15th and 27th leaves in the RNAi transgenic line #13 also had longer telomeres (15 to 60 kb) than did control plants. These results parallel our observation that aberrant morphology of the transgenic plants becomes progressively more apparent as the plants mature (Figure 4), suggesting that the abnormal phenotypes are associated with altered telomere lengths.
Figure 5.
Telomere Deregulation in 35S:RNAi-GTBP1 Tobacco Plants.
(A) Telomere elongation in T0 and T1 transgenic plants. Genomic DNA was purified from the 27th leaves of wild-type plants and the 7th, 15th, and 27th leaves of T0 and T1 transgenic (#1 and #13) plants. After digestion with TaqI, DNA was subjected to TRF analysis by pulse-field gel electrophoresis.
(B) Telomerase activity in wild-type and T1 transgenic (#1 and #13) plants. The level of telomerase activity was examined in shoot apexes and various leaves by PCR-based TRAP assays.
(C) Formation of extrachromosomal t-circles in T0 and T1 transgenic (#1 and #13) plants. Genomic DNA from the 7th, 15th, and 27th leaves of wild-type and T0 and T1 35S:RNAi-GTBP1 (lines #1 and #13) tobacco plants were subjected to TRF analysis by 2D pulse-field gel electrophoresis. Linear telomeric DNA and extrachromosomal t-circles are indicated by arrows and arrowheads, respectively.
We then measured telomere length in T1 transgenic leaves (lines #1 and #13). Intriguingly, telomeres of the seventh leaves in the T1 generation were similar to the wild-type telomere ranges (15 to 50 kb) (Figure 5A, right panel). Subsequently, as the T1 transgenic tobacco plants matured, telomeres elongated to 15 to 75 kb in the 15th and 27th leaves, in which the elongation pattern was comparable to that of T0 transgenic plants. In addition, we noticed that, in both T0 and T1 transgenic lines, the lengths of the longer telomeres were more drastically changed than were the shorter telomeres; 50-kb telomeres were lengthened to 60 to 75 kb, while 15-kb telomeres were fairly constant in the transgenic plants (Figure 5A). Overall, these results suggest that knockdown of GTBP1 resulted in increased telomere length, which was coupled with anomalous morphology, and that telomere length homeostasis is subject to developmental regulation. Furthermore, it is likely that unusually elongated telomeres in 35S:RNAi-GTBP1 transgenic leaves are reset to normal lengths after each generation.
Telomerase enzymatic activities are closely tied with the cell division program and are generally low in differentiated vegetative tissues (Fitzgerald et al., 1996; Riha et al., 1998; Yang et al., 2002). Because 35S:RNAi-GTBP1 leaves possess longer telomeres than do control leaves, we monitored telomerase activity in wild-type and RNAi transgenic leaves by telomere repeat amplification (TRAP) assays (Yang et al., 2002). In spite of elongated telomeres in 35S:RNAi-GTBP1 lines, the levels of telomerase activity were indistinguishable between wild-type and transgenic shoot apexes and leaves (Figure 5B). Thus, we concluded that the longer telomeres found in the transgenic leaves are not due to increased telomerase activity.
In mammalian cells, a lack of functional TRF2 and Pot1 caused generation of extrachromosomal telomeric circles (t-circles) that arose from aberrant homologous recombination (HR) of the telomeres (Wang et al., 2004; Wu et al., 2006). Recently, Zellinger et al. (2007) detected increased formation of t-circles in Arabidopsis ku70 and ku80 mutant cells. Because 35S:RNAi-GTBP1 lines contained longer telomeres without enhanced telomerase activity, we considered the possibility that the elongated telomeres were subject to HR. To examine this possibility, we performed two-dimensional (2D) pulse-field gel electrophoresis to detect possible formations of extrachromosomal t-circles. Telomeres from both T0 and T1 35S:RNAi-GTBP1 plants showed extrachromosomal arcs of open-circular and super-coiled double-stranded circles in addition to the linear telomeric arcs (Figure 5C). These t-circles were slightly detectable in the seventh leaves but became increasingly apparent in the 15th to 27th leaves, a pattern reminiscent of telomere elongation (Figure 5A). Thus, t-circle generation appeared to coincide temporally and spatially with telomeric elongation and also with progressively worsening phenotypes observed in both T0 and T1 35S:RNAi-GTBP1 plants. We did not detect such additional t-circles in wild-type tobacco leaves regardless of their developmental stages. These results indicate that knockdown of GTBP1 resulted not only in telomere elongation but also in t-circle formation that may be due to the aberrant HR of the telomeres. Both of these events, in turn, caused impaired development of 35S:RNAi-GTBP1 tobacco plants.
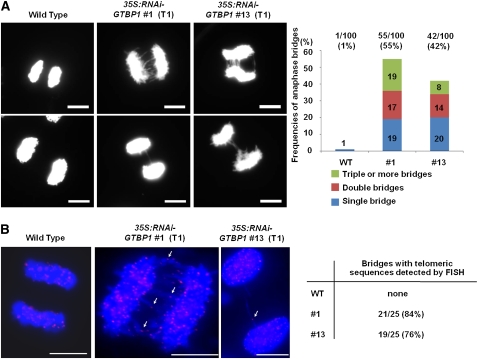
GTBP1 Knockdown Plants Exhibit a High Frequency of Anaphase Bridges in Anther Cells
Most mutants that have impaired telomere stability inevitably suffer cytogenetic abnormalities that arise from end-to-end chromosome fusions (Shakirov et al., 2005; Vannier et al., 2006; Hong et al., 2007, 2010; Song et al., 2008; Surovtseva et al., 2009). To investigate whether repression of GTBP1 also caused chromosome instability in tobacco, we examined wild-type and transgenic chromosomes during meiotic progression of anther cells. As expected, there was negligible anaphase bridge formation in wild-type tobacco meiotic cells (1 of 100) (Figure 6A). By contrast, T1 35S:RNAi-GTBP1 meiotic cells presented abnormal anaphase bridges with frequencies of 55% (55 of 100) and 42% (42 of 100) in lines #1 and #13, respectively. Moreover, anaphase chromosomes in the transgenic cells contained double (14 to 17%) and multiple (8 to 19%) bridges in addition to single bridges (19 to 20%) (Figure 6A).
Figure 6.
Formation of Anaphase Bridges in T1 35S:RNAi-GTBP1 Transgenic Cells.
(A) Anaphase chromosomes from wild-type and T1 35S:RNAi-GTBP1 (lines #1 and #13) anther cells. Anaphase chromosome spreads were obtained from anthers of wild-type and 35S:RNAi-GTBP1 transgenic lines (#1 and #13), stained with DAPI, and observed by fluorescence microscopy. Abnormal anaphase bridges were detected in transgenic meiotic cells. Frequencies of single, double, and multiple anaphase bridges are indicated in the right panel. Bars = 10 μm.
(B) FISH analysis of anaphase chromosomes in wild-type and T1 35S:RNAi-GTBP1 (lines #1 and #13) meiotic cells using a (TTTAGGG)70 repeat telomeric fluorescent probe. Chromosomal DNA was denatured and incubated with Texas red-dUTP–incorporated (TTTAGGG)70 telomeric probe. The chromosomes were counterstained with DAPI and observed using fluorescence microscopy. Arrows indicate telomeric signals at the fusion points of anaphase bridges. Percentages of anaphase bridges with telomeric sequences detected by FISH are indicated in the right panel. Bars = 10 μm.
Fluorescence in situ hybridization (FISH) analysis revealed that anaphase bridges in Arabidopsis tert mutant and rad50 tert double mutant cells contained telomeric repeats, suggesting that the unusual chromosome-end fusions included telomeric repeats at the fusion points (Vannier et al., 2006). Similarly, Hong et al. (2007) showed that the anaphase bridges in rice telomere binding protein1 (rtbp1) loss-of-function mutant meiotic cells contained telomeric DNA. To examine whether the anaphase bridges in 35S:RNAi-GTBP1 chromosomes were due to telomere dysfunction, we conducted FISH using a (TTTAGGG)70 telomeric DNA repeat probe (see Supplemental Figure 5 online). Single and multiple hybridization signals of the telomeric probe were detected with anaphase bridges in the 35S:RNAi-GTBP1 chromosomes (Figure 6B). The frequencies of positive FISH signals with anaphase bridges were 84% (21 of 25) and 76% (19 of 25) in lines #1 and #13, respectively (Figure 6B). Collectively, these results suggest that suppression of GTBP1 indeed resulted in telomere malfunction, which is closely associated with the chromosome instability and abnormal phenotypes of 35S:RNAi-GTBP1 plants.
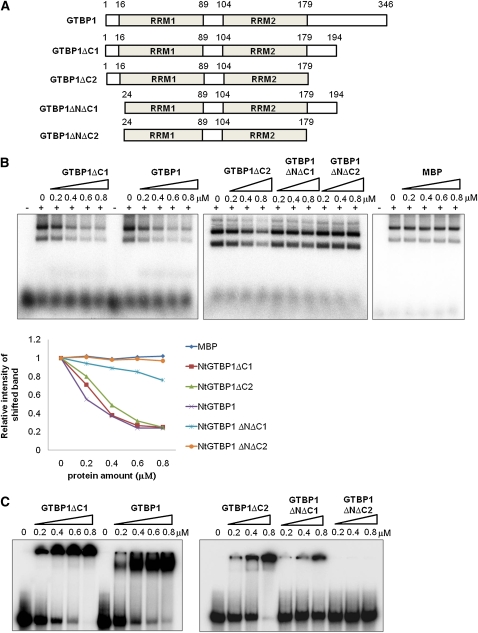
GTBP1 Inhibits Telomeric Strand Invasion in Vitro
In the process of interchromosomal telomeric HR, invasion of a 3′-overhang single-stranded telomeric sequence into the double-stranded telomeric region is considered to be a prerequisite step (Cesare and Reddel, 2010). In view of the single-strand-specific telomere binding activity of GTBP1 and the high frequency of anaphase telomeric fusion in GTBP1 knockdown plants, we postulated that telomere binding of GTBP1 affects the strand invasion process. To test this hypothesis, we performed a recently developed strand invasion assay for telomeric DNA repeats (Amiard et al., 2007; Zhang et al., 2007). The recombinant full-length (MBP-GTBP11-346) or deletion mutants (MBP-GTBP1ΔC11-194, MBP-GRP1ΔC21-179, MBP-GRP1ΔNΔC124-194, and MBP-GRP1ΔNΔC224-179) of GTBP1 were incubated with a 32P-labeled single-stranded (TTTAGGG)8 repeat probe with T-vector plasmid containing double-stranded (TTTAGGG)70 telomeric repeats (Figure 7A). The protein-DNA complexes were treated with proteinase K and then analyzed by gel electrophoresis. The degree of invasion of the single-stranded telomeric probe into the plasmid was determined by the intensity of the shifted band. MBP-GTBP11-346 inhibited the invasion of single-stranded telomeric probe into the plasmid in a dose-dependent manner (Figure 7B). Approximately 80% of the single-strand invasion was inhibited by a saturating amount of MBP-GTBP11-346. Similar concentration-dependent inhibitory profiles were detected when the MBP-GTBP1ΔC11-194 or MBP-GRP1ΔC21-179 mutants that retained high binding activities for single-strand telomere (Figure 7C) were included in the assay mixture. Notably, the MBP-GRP1ΔNΔC124-194 mutant protein, which had reduced telomere binding activity, partially inhibited the strand invasion process. Finally, MBP-GTBP1ΔNΔC224-179 and MBP, which lack telomere binding activity (Figure 7C), both failed to inhibit strand invasion (Figure 7B). Therefore, binding activity of GTBP1 was proportionally correlated with the degree of strand invasion inhibitory activity in vitro. These results argue that GTBP1 may prevent aberrant interchromosomal telomeric HR by inhibiting telomeric strand invasion.
Figure 7.
Nt GTBP1 Inhibits Telomeric Strand Invasion in Vitro.
(A) Schematic representation of full-length and deletion mutants of GTBP1.
(B) Strand invasion assay. Increasing concentrations of GTBP1 derivatives (0, 0.2, 0.4, 0.6, and 0.8 μM) were incubated with a 32P-labeled single-stranded (TTTAGGG)8 repeat probe with T-vector plasmid containing double-stranded (TTTAGGG)70 telomere repeats. The degree of invasion of the single-stranded telomeric probe into the plasmid was determined by the shifted band intensity. The first “−” lane of each gel contained radiolabeled PT8 without protein. The graph in the bottom panel shows relative intensities of shifted bands.
(C) Binding activities of full-length and deletion mutants of GTBP1 to single-strand telomere sequences. Various concentrations (0, 0.2, 0.4, 0.6, and 0.8 μM) of GTBP1 derivatives were analyzed with radiolabeled single-stranded (TTTAGGG)8 (PT8) repeats.
[See online article for color version of this figure.]
DISCUSSION
Telomeres shield the extreme ends of linear eukaryotic chromosomes. Both animal and plant telomeres form a stable t-loop, in which the 3′ single-strand overhang is inserted into a double-stranded telomeric region so that telomeres can be protected and distinguished from double-strand break repair mechanisms (Griffith et al., 1999; Cesare et al., 2003). Single-strand telomere binding proteins, along with double-strand-specific factors, play critical roles in telomere maintenance. In mammals, POT1 is an essential factor for telomere protection as loss-of-function of POT1 caused severe genomic anomalies due to telomere malfunction (Hockemeyer et al., 2006; Wu et al., 2006). In addition, animal POT1 participates in the negative regulation of telomere length homeostasis by inhibiting access of telomerase to telomeric ends (Kelleher et al., 2005). Higher plants also have homologs of animal POT1. Arabidopsis POT1b was shown to function in telomere protection (Shakirov et al., 2005), whereas POT1a, contrary to its animal homolog, appeared to work as a positive regulator for telomerase and telomere length regulation (Surovtseva et al., 2007). However, neither telomeric protein exhibited single-strand telomere binding activities. Therefore, it was postulated that POT1s may not be major single-stranded telomeric factors in Arabidopsis (Shakirov et al., 2009). These results prompted us to examine the cellular role for GTBP paralogs in tobacco plants.
While three homologous tobacco GTBPs displayed similar modes of binding to single-stranded telomeric sequences in vitro, GTBP1 was the predominantly expressed gene in tobacco (Figures 1 and 2). We noticed that the expression profile of Nt GTBP1 was quite different from that of the double-stranded telomeric factor Ng TRF1 in tobacco. Nt GTBP1 was constitutively expressed in BY-2 suspension cells during the 9-d culture period, as opposed to Ng TRF1, whose expression was inversely correlated with cell division activity (Figure 1C; Yang et al., 2003). In addition, Nt GTBP1, but not Ng TRF1, was developmentally regulated during leaf development, suggesting that Nt GTBP1 and Ng TRF1 may have different roles in telomeres. Nt GTBP1 was associated with telomeric sequences as evidenced by ChIP assay (Figure 3). The independent T0 and T1 35S:RNAi-GTBP1 plants displayed severely aberrant phenotypes with retarded vegetative growth, premature senescence, and infertility, which were coupled with reduced binding activity to single-stranded telomeric sequences (Figure 4). Also, the GTBP1 knockdown leaves contained longer telomeres with frequent formation of extrachromosomal t-circles (Figure 5). Thus, Nt GTBP1 is a negative regulator of telomere length, which is opposite of the role of At POT1a, but, at least in part, similar to mammalian POT1. This raises the tantalizing possibility that telomere length homeostasis could be regulated by the functional balance between two different single-stranded telomeric factors, POT1 and GTBP, in plants. Furthermore, given that the GTBP1 knockdown meiotic cells suffered frequent anaphase bridges that contained telomeric sequences at the fusion points, it is highly plausible that GTBP1 plays a positive role in telomere stability (Figure 6). This view is further supported by in vitro results that GTBP1 inhibited the single-strand invasion into double-stranded telomeric DNA that is a prerequisite step for interchromosomal telomeric HR (Figure 7). Therefore, it is reasonable to consider that GTBP1 suppression-induced abnormal phenotypes are closely correlated with genomic instability due to the dysfunctional telomeres in the transgenic tobacco. With this in mind, we conclude that GTBP1 is an essential component for both structure and function of tobacco telomeres.
It is worth noting that phenotypic severity, telomere elongation, and the frequency of t-circle formation in T0 RNAi transgenic plants became increasingly apparent as the plants grew to maturity (Figures 4 and 5). Intriguingly, these abnormalities were restored to normal levels in the early stages of the T1 generation but then concomitantly reappeared when T1 transgenic plants were further grown. Thus, phenotypic properties were subject to developmental regulation and influenced by telomere length and architecture, with its severity being more serious in the later stage of development. In Arabidopsis ku70 mutant cells, abnormally elongated telomeres were reset to a new length after three generations through the telomere rapid deletion process (Watson and Shippen, 2007). It was previously proposed that individual telomere tracts on homologous chromosomes were coordinately regulated through development in Arabidopsis (Shakirov and Shippen, 2004). Based on these results, we speculate that elongated telomeres were reset during the T0 to T1 generation of the 35S:RNAi-GTBP1 transgenic plants. However, due to the lack of properly functioning GTBP1, telomeres were again gradually lengthened and destabilized, which in turn caused gradual phenotypic defects.
Higher plants usually exhibit flexible phenotypes in response to changes in telomere length. For example, although the last five generations show multiple phenotypic problems, the telomerase-deficient Arabidopsis tert mutants could survive for up to 10 generations (Riha et al., 2001). Transgenic tobacco 35S:TRF1 cells, in which the double-strand binding protein Ng TRF1 was overexpressed, showed progressive reduction in cell viability with shortened telomeres over successive rounds of batch subculture (Yang et al., 2004). A rice rtbp1 mutant that suppressed telomere binding protein 1 already contained markedly elongated telomeres in the G1 generation but showed gradually more defective phenotypes during the consecutive four generations (Hong et al., 2007). By contrast, knockdown of GTBP1 resulted in immediate defective phenotypes, and most independent T0 plants were seriously impaired and sterile (Figure 4). The rapid onset of defective phenotypes was also identified in Arabidopsis ctc1 (for conserved telomere maintenance component1), in which telomere length deregulation, recombination, and end-to-end fusion were strongly present in the first generation of loss-of-function mutant plants (Surovtseva et al., 2009). Thus, suppression of At CTC1 and Nt GTBP1 had more immediate effects on telomere stability than did suppression of Ng TRF1 or Os RTBP1. This may be due to the existence of a gene family for Ng TRF1 and Os RTBP1 and/or to the more direct roles of Nt GTBP1 and At CTC1. In this respect, it is of interest to note that knockdown of Nt GTBP1 induced longer telomeres, and Arabidopsis ctc1 mutants, by contrast, exhibited shorter telomeres. Therefore, defects in the 35S:RNAi-GTBP1 and ctc1 plants might not be due to telomere length deregulation per se but to the structural instability that resulted from the absence of capping proteins. This notion is supported by the results that telomeres in the seventh leaves of T0 and T1 35S:RNAi-GTBP1 plants displayed normal lengths but showed extrachromosomal t-circles (Figure 5C).
Approximately 10% of human cancer cells maintain their telomeres under telomerase negative conditions by alternative lengthening of telomeres (ALT) (Cesare and Reddel, 2010). ALT cancer cells are typified by enhanced t-circle formation, as ALT is initiated by telomeric HR (Wang et al., 2004). Likewise, Arabidopsis tert ku70 double mutant callus cells displayed a large number of t-circles, which is typical of ALT (Zellinger et al., 2007). Given that t-circles are frequently found in 35S:RNAi-GTBP1 lines and GTBP1 inhibits strand invasion in vitro, it is of keen interest to determine whether GTBP1 is a negative regulator of the ALT initiation process in tobacco. In conclusion, our data provide evidence that GTBP1 is an essential factor for telomere integrity and normal growth and development in tobacco, a dicot model crop plant.
METHODS
Plant Materials
Wild-type and 35S:RNAi-GTBP1 transgenic tobacco (Nicotiana tabacum cv Samsun NN) plants were grown on 0.8% agar for 7 d or on Murashige and Skoog (MS) medium containing 3% sucrose, vitamin B5 (12 mg/L), and 0.8% agar, pH 5.8, for 14 d in a growth chamber under a 16-h-light/8-h-dark photoperiod at 25°C. Tobacco seedlings were transferred to pots and moved to a growth room for further growth. The tobacco BY-2 suspension cell line was maintained in MS medium on a rotary shaker (150 rpm) at 27°C in the dark. Every 9 d, stationary phase cells (5 mL) were transferred to 45 mL of fresh medium and cultured (Yang et al., 2002).
Isolation of Three Full-Length Nt GTBP Paralogs
To isolate the cDNAs for tobacco GTBP paralogs, degenerate oligonucleotides corresponding to the amino acid sequence VEIKRTIP for the upstream primer and VEIKKA for the downstream primer were produced (see Supplemental Table 1 online). These amino acid sequences are conserved in RRMs (Figure 1A). RT-PCR was performed with oligonucelotide primers and first-strand cDNA prepared from total RNA (2.0 μg) of log-phase BY-2 suspension-cultured cells using high-fidelity Ex-Taq DNA polymerase (Takara). PCR comprised 35 amplification cycles with an annealing temperature of 52°C for 45 s, an elongation temperature of 72°C for 1 min, and a denaturing temperature of 95°C for 30 s. PCR products were separated via agarose gel electrophoresis, and ~300-bp-long cDNA fragments were eluted from the gel and cloned into the pGEM-T Easy vector (Promega) for sequencing. Three homologous Nt GTBP cDNAs were identified and named GTBP1 (GenBank accession number HM049166; Hirata et al., 2004), GTBP2 (GenBank accession number HM049167), and GTBP3 (GenBank accession number HM049168). To obtain full-length cDNAs, total recombinant λ DNA was prepared from the λ-uni-Zap II tobacco flower cDNA library (Kim et al., 2000) and used for RACE (Frohman, 1994) with primers corresponding to the 5′- and 3′-ends of the library vector sequence (T7 and T3 promoters) and the internal regions of the partial GTBP cDNA clones (see Supplemental Table 1 online).
Phylogenetic Analysis
Protein sequences were aligned (available as Supplemental Data Set 1 online) using ClustalW in Mega4 software (Tamura et al., 2007). The evolutionary relationship was analyzed by the neighbor-joining method (Saitou and Nei, 1987). The optimal tree that shows the sum of branch length was 4.37412444. Boostrapping was performed with 20,000 replicates. The evolutionary distances were calculated by the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site.
Genomic DNA Isolation and DNA Gel Blot Analysis
Leaf genomic DNA was isolated from tobacco plants as described previously (Yang et al., 2003). Genomic DNA (10 μg) was digested with indicated restriction enzymes, blotted onto Hybond-N nylon membranes (Amersham), and hybridized with 32P-labeled probes (Nt GTBP1, Nt GTBP2, and Nt GTBP3 full-length cDNAs [Figure 1A], respectively, in Supplemental Figure 2 online and NPTII in Supplemental Figure 4 online).
RNA Extraction and RT-PCR
Total RNAs were extracted from BY-2 cells and various tobacco tissues as described previously (Yang et al., 2003). Total RNA (2 μg) was used for first-strand cDNA synthesis with MMLV reverse transcriptase (Promega). To investigate gene expression profiles, RT-PCR was performed with gene-specific primers (see Supplemental Figure 2 and Supplemental Table 1 online) using Ex-Taq polymerase (Takara). Amplification comprised 25 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C in an automatic thermal cycler (Perkin-Elmer/Cetus). PCR products were separated on 1.0% agarose gels and visualized with ethidium bromide under UV light.
Gel Retardation Assays
The full-length or various deletion mutants of GTBP cDNAs were generated using primers listed in Supplemental Table 1 online and inserted into the pMAL-c2x vector (New England Biolabs) using BamHI and SalI sites. MBP-GTBP fusion proteins were expressed in Escherichia coli and purified by affinity chromatography using amylose resin (New England Biolabs) as described by Yang et al. (2003). The cell-free nuclear extracts were prepared from 1-week-old wild-type and 35S:RNAi-GTBP1 seedlings as described previously (Lee et al., 2000). Gel retardation assays were performed with bacterially expressed recombinant proteins or nuclear extracts containing 10 μg proteins as described by Byun et al. (2008). Various telomeric DNA probes and competitors used in this study are listed in Supplemental Table 1 online.
Transformation of 35S:HA-GTBP1 Construct into BY-2 Cells
The 3xHA-tagged GTBP1 cDNA was generated using HA tag-PstI sense, HA tag-PstI antisense, GTBP1-SalI sense, and GTBP1-BamHI antisense primers (see Supplemental Table 1 online) and cloned into the pCAMBIA 1390 vector (CAMBIA), which contained the CaMV 35S promoter. The 35S:HA-GTBP1 construct was transformed into Agrobacterium tumefaciens strain LBA4404 by electroporation. Agrobacterium was then coincubated with actively dividing tobacco BY-2 suspension cultured cells for 3 d at 25°C with gently shaking (150 rpm). Transgenic BY-2 cells were selected on MS medium supplemented with 100 μg/mL hygromycin and 100 μg/mL carbenicillin. Expression and localization of HA-GTBP1 fusion protein were confirmed by immunostaining using primary anti-HA antibody (New England Biolabs) and secondary Cy3-conjugated anti-mouse antibody (Millipore) as described by Lee et al. (2009). Nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI) in mounting solution (Vector Laboratories).
ChIP Assays
An established ChIP assay protocol (http://www.epigenome-noe.net/researchtools/protocol.php?protid=13) was followed with some modifications. To cross-link genomic DNA with bound proteins, wild-type and 35S:HA-GTBP1 transgenic BY-2 cells were incubated with a 1% formaldehyde solution for 10 min. Cross-linking was quenched by the addition of glycine up to 150 mM. After harvesting cells by vacuum filtration through filter paper, chromatin was fractionated by sonication into fragments 0.2 to 1.0 kb in length in ChIP dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.0, and 167 mM NaCl). ChIP was performed by incubating sonicated cell extracts without (as a negative control) or with 1:100 diluted anti-HA antibody (New England Biolabs). Bound chromatin fragments were eluted with protein G agarose beads (Santa Cruz Biotechnology). Elution products were dot blotted onto Hybond-N nylon membranes (Amersham) and subjected to hybridization with 32P-labeled (TTTAGGG)70 repeat or HRS 60 (Koukalova et al., 1993) probes.
Generation of 35S:RNAi-GTBP1 Knockdown Transgenic Tobacco Plants
GTBP1 cDNA encompassing 377 bp of the 5′ end (from 122 to 498 bp) (RNAi lines #11, #12, #13, #15, #16, and #17) or 345 bp of the 3′ end (from 726 to 1070 bp) (RNAi lines #1, #2, #3, and #5) was generated by primers listed in Supplemental Table 1 online and inserted into the pHANNIBAL vector (CSIRO) using XhoI, EcoRI, BamHI, and HindIII sites in inverted orientations. The hairpin-producing cassette was then cloned into NotI site of binary vector pART (http://www.pi.csiro.au/RNAi/vectors.htm). The produced Nt GTBP1 RNAi constructs were transferred to Agrobacterium strain LBA4404 by electroporation. Leaf discs of N. tabacum cv Samsun NN were cocultivated with Agrobacterium as described by Bae et al. (2009), and transgenic tobacco plants were generated on medium containing 200 mg/L kanamycin. The regenerated T0 plants were grown in a growth room under a 16-h-light/8-h-dark photoperiod at 25°C. The T0 transformants (lines #1 and #13) were self-fertilized, and seeds were collected. T1 plants were selected on MS medium containing 200 mg/L kanamycin. T0 and T1 transgenic plants were used for phenotypic analysis.
TRAP Assay and Telomere Repeat Fragment Analysis
TRAP assays, which measure telomerase enzyme activity, were performed as described by Yang et al. (2002). TRAP assays were performed in 50 μL of reaction mixture composed of 50 mM Tris-acetate, pH. 8.3, 50 mM potassium glutamate, 0.1% Triton X-100, 1 mM spermidine, 1 mM DTT, 50 μM each deoxynucleotide triphosphate, 5 mM MgCl2, 10 mM EGTA, 100 ng/μL BSA, and 0.5 unit Taq polymerase (Takara). For telomere repeat fragment (TRF) analysis, tobacco leaf genomic DNA (5 μg) was digested by TaqI restriction enzyme, separated by pulse-field gel electrophoresis using a CHEF-DRIII system (Bio-Rad), transferred to nylon membrane, and hybridized with a 32P-labeled (TTTAGGG)70 telomeric probe.
2D Pulse-Field Gel Electrophoresis
The TaqI-digested tobacco leaf genomic DNA (5 μg) was separated in a 0.5% agarose gel by pulse-field gel electrophoresis using a CHEF-DRIII system (Bio-Rad) at 1 V/cm for 25.5 h running time at an angle of 120° with switching times ramped from 1 to 6 s at 14°C. The second dimension electrophoresis was performed in a 1.1% agarose gel at 6 V/cm for 8.5 h with the same conditions. The gel was blotted onto a Hybond-N nylon membrane (Amersham) and hybridized to a 32P-labeled (TTTAGGG)70 telomeric repeat probe under highly stringent conditions. The blots were visualized by autoradiography at −80°C.
Cytogenetic Analysis and FISH
Anaphase chromosome spreads were obtained from anthers of wild-type and 35S:RNAi-GTBP1 transgenic lines (#1 and #13) and stained with DAPI as described previously (Hong et al., 2007) with minor modifications. Anthers were fixed in 1:3 (v/v) acetic acid:ethanol for 2 h, soaked in distilled water, and digested with 2% cellulose, 1.5% macerozyme, 0.3% pectolyase, and 1 mM EDTA, pH 4.2, for 1 h. After being squashed on slide glass with 60% acetic acid, digested anthers were dried on hot plate at 45°C. Chromosomal DNAs were stained with DAPI and observed by fluorescence microscopy (Axio imager; Zeiss).
Chromosomal DNA was denatured with 70% formamide at 70°C for 3 min. Samples were then dehydrated in a 70, 85, 95, and 100% ethanol series at −20°C for 3 min each as described by Hong et al. (2007). Texas red-dUTP (Invitrogen) incorporated (TTTAGGG)70 telomeric probe was applied to the denatured chromosomal DNA in hybridization solution (50% [v/v] formamide, 10% [w/v] dextran sulfate, 5 ng/μL salmon sperm DNA, and 2× SSC), covered with a plastic cover slip, and incubated in a humid chamber at 37°C for 36 h. The chromosomes were counterstained with DAPI and observed using fluorescence microscopy with Qmax Blue and XF 102-2 filters (Omega Optical).
Strand Invasion Assays
Strand invasion assays were performed with a 32P-labeled single stranded (TTTAGGG)8 (PT8) repeat probe and pGEM-T Easy plasmid containing double-stranded (TTTAGGG)70 telomeric repeats according to Amiard et al. (2007). The full-length or various deletion mutants of GTBP1 were constructed using primers listed in Supplemental Table 1 online and incubated with PT8 (0.25 ng) in invasion buffer (50 mM HEPES, pH 8.0, 0.1 mg/mL BSA, 1 mM DTT, 100 mM NaCl, and 2% [v/v] glycerol] for 15 min at 4°C. The plasmid (100 ng) was added to the reaction mixture and incubated for 15 min at room temperature. Reactions were stopped by the addition of stop buffer [10% (w/v) SDS, 6 μg proteinase K, and 25 ng (CCCTAAA)4]. Samples were separated on 1.1% agarose gels in 0.5× TBE at 85 V at room temperature. Gels were dried and visualized by autoradiography. Strand invasion signals were quantified with a phosphor imager (Fuji). The degree of invasion of the single-stranded telomeric probes into the plasmids was determined by the shifted band intensity.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: tobacco GTBP1 (HM049166), GTBP2 (HM049167), and GTBP3 (HM049168); Arabidopsis STEP1 (NP194208); Arabidopsis proteins (At3g13224 and At4g26650); rice proteins (Os08g0320100 and Os08g0492100); and human HnRNP D (BAA09522) and HnRNP A1 (P09651).
Supplemental Data
The following materials are available in the online version of the article.
Supplemental Figure 1. Sequence Analysis of Tobacco GTBP Paralogs.
Supplemental Figure 2. DNA Gel Blot Analysis of Tobacco DNA.
Supplemental Figure 3. Specificity of the Gene-Specific Primers.
Supplemental Figure 4. DNA Gel Blot Analysis of Wild-Type and 10 T0 35S:RNAi-GTBP1 Tobacco Plants.
Supplemental Figure 5. FISH Analysis of Meiotic Chromosomes in Wild-Type Tobacco Anther Cells.
Supplemental Table 1. Oligonucleotide Sequences Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Figure 1B.
Supplementary Material
Acknowledgments
This work was supported by grants from the Technology Development Program for Agriculture and Forestry (Project No. 309017-5 funded by the Ministry for Agriculture, Forestry, and Fisheries, Republic of Korea), the National Research Foundation (Project No. 2009-0078317 funded by the Ministry of Education, Science, and Technology, Republic of Korea), and the BioGreen 21 Program (funded by the Rural Development Administration, Republic of Korea) to W.T.K.
References
- Amiard S., et al. (2007). A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol. 14: 147–154 [DOI] [PubMed] [Google Scholar]
- Bae H., Choi S.M., Yang S.W., Pai H.S., Kim W.T. (2009). Suppression of the ER-localized AAA ATPase NgCDC48 inhibits tobacco growth and development. Mol. Cells 28: 57–65 [DOI] [PubMed] [Google Scholar]
- Baumann P., Cech T.R. (2001). Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Bilaud T., Brun C., Ancelin K., Koering C.E., Laroche T., Gilson E. (1997). Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet. 17: 236–239 [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. (1991). Structure and function of telomeres. Nature 350: 569–573 [DOI] [PubMed] [Google Scholar]
- Blackburn E.H. (2001). Switching and signaling at the telomere. Cell 106: 661–673 [DOI] [PubMed] [Google Scholar]
- Blasco M.A. (2005). Telomeres and human disease: Ageing, cancer and beyond. Nat. Rev. Genet. 6: 611–622 [DOI] [PubMed] [Google Scholar]
- Byun M.Y., Hong J.-P., Kim W.T. (2008). Identification and characterization of three telomere repeat-binding factors in rice. Biochem. Biophys. Res. Commun. 372: 85–90 [DOI] [PubMed] [Google Scholar]
- Cesare A.J., Quinney N., Willcox S., Subramanian D., Griffith J.D. (2003). Telomere looping in P. sativum (common garden pea). Plant J. 36: 271–279 [DOI] [PubMed] [Google Scholar]
- Cesare A.J., Reddel R.R. (2010). Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11: 319–330 [DOI] [PubMed] [Google Scholar]
- Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T. (1995). A human telomeric protein. Science 270: 1663–1667 [DOI] [PubMed] [Google Scholar]
- Collins K. (2000). Mammalian telomeres and telomerase. Curr. Opin. Cell Biol. 12: 378–383 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2005). Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Fajkus J., Kovarik A., Kralovics R., Bezdek M. (1995). Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol. Gen. Genet. 247: 633–638 [DOI] [PubMed] [Google Scholar]
- Fitzgerald M.S., McKnight T.D., Shippen D.E. (1996). Characterization and developmental patterns of telomerase expression in plants. Proc. Natl. Acad. Sci. USA 93: 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman M.A. (1994). On beyond classic RACE (rapid amplification of cDNA ends). Genome Res. 4: S40–S58 [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97: 503–514 [DOI] [PubMed] [Google Scholar]
- Hirata Y., Suzuki C., Sakai S. (2004). Characterization and gene cloning of telomere-binding protein from tobacco BY-2 cells. Plant Physiol. Biochem. 42: 7–14 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Danieis J.-P., Takai H., de Lange T. (2006). Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell 126: 63–77 [DOI] [PubMed] [Google Scholar]
- Hong J.-P., Byun M.Y., An K., Yang S.-J., An G., Kim W.T. (2010). OsKu70 is associated with developmental growth and genome stability in rice (Oryza sativa L.). Plant Physiol. 152: 374–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.-P., Byun M.Y., Koo D., An K., Bang J., Chung I.K., An G., Kim W.T. (2007). Suppression of RICE TELOMERE BINDING PROTEIN1 results in severe and gradual developmental defects accompanied by genome instability in rice. Plant Cell 19: 1770–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F., Matunis M.J., Dreyfuss G., Cech T.R. (1993). Nuclear proteins that bind the pre-mRNA 30 splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell. Biol. 13: 4301–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher C., Kurth I., Lingner J. (2005). Human protection of telomeres 1 (POT1) is a negative regulator of telomerase activity in vitro. Mol. Cell. Biol. 25: 808–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.S., Lee J.H., Yoon G.M., Cho H.S., Park S.W., Suh M.C., Choi D., Ha H.J., Liu J.R., Pai H.S. (2000). CHRK1, a chitinase-related receptor-like kinase in tobacco. Plant Physiol. 123: 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukalova B., Komarnitsky I.K., Kuhrova V. (1993). The distribution of tobacco HRS60 DNA repeated sequences in species of the genus Nicotiana. Plant Sci. 88: 39–44 [Google Scholar]
- Kwon C., Chung I.K. (2004). Interaction of an Arabidopsis RNA-binding protein with plant single-stranded telomeric DNA modulates telomerase activity. J. Biol. Chem. 279: 12812–12818 [DOI] [PubMed] [Google Scholar]
- LaBranche H., Dupuis S., Ben-David Y., Bani M.R., Wellinger R.J., Chabot B. (1998). Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 19: 199–202 [DOI] [PubMed] [Google Scholar]
- Lee H.K., Cho S.K., Son O., Xu Z., Hwang I., Kim W.T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Kim J.H., Kim W.T., Kang B.G., Chung I.K. (2000). Characterization and developmental expression of single-stranded telomeric DNA-binding proteins from mung bean (Vigna radiata). Plant Mol. Biol. 42: 547–557 [DOI] [PubMed] [Google Scholar]
- Nugent C.I., Hughes T.R., Lue N.F., Lundblad V. (1996). Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274: 249–252 [DOI] [PubMed] [Google Scholar]
- Riha K., Fajkus J., Siroky J., Vyskot B. (1998). Developmental control of telomere lengths and telomerase activity in plants. Plant Cell 10: 1691–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K., McKnight T.D., Griffing L.R., Shippen D.E. (2001). Living with genome instability: Plant response to telomere dysfunction. Science 291: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Shakirov E.V., McKnight T.D., Shippen D.E. (2009). POT1-independent single-strand telomeric DNA binding activities in Brassicaceae. Plant J. 58: 1004–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov E.V., Shippen D.E. (2004). Length regulation and dynamics of individual telomere tracks in wild-type Arabidopsis. Plant Cell 16: 1959–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov E.V., Surovtseva Y.V., Osbun N., Shippen D.E. (2005). The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 25: 7725–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D. (2001). Telomeric chromatin: Replicating and wrapping up chromosome ends. Curr. Opin. Genet. Dev. 11: 189–198 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., de Lange T. (2004). Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73: 177–208 [DOI] [PubMed] [Google Scholar]
- Song X., Leehy K., Warrington R.T., Lamb J.C., Surovtseva Y.V., Shippen D.E. (2008). STN1 protects chromosome ends in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 19815–19820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y.V., Churikov D., Boltz K.A., Song X., Lamb J.C., Warrington R., Leehy K., Heacock M., Price C.M., Shippen D.E. (2009). Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva Y.V., Shakirov E.V., Vespa L., Osbun N., Song X., Shippen D.E. (2007). Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 26: 3653–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Vannier J.-B., Depeiges A., White C., Gallego M.E. (2006). Two roles for Rad50 in telomere maintenance. EMBO J. 25: 4577–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.W., Jin E.S., Chung I.K., Kim W.T. (2002). Cell cycle-dependent regulation of telomerase activity by auxin, ABA and protein phosphorylation in tobacco BY-2 suspension culture cells. Plant J. 29: 617–626 [DOI] [PubMed] [Google Scholar]
- Yang S.W., Kim D.H., Lee J.J., Chun Y.J., Lee J.-H., Kim Y.J., Chung I.K., Kim W.T. (2003). Expression of the telomeric repeat binding factor gene NgTRF1 is closely coordinated with the cell division program in tobacco BY-2 suspension culture cells. J. Biol. Chem. 278: 21395–21407 [DOI] [PubMed] [Google Scholar]
- Yang S.W., Kim S.K., Kim W.T. (2004). Perturbation of NgTRF1 expression induces apoptosis-like cell death in tobacco BY-2 cells and implicates NgTRF1 in the control of telomere length and stability. Plant Cell 16: 3370–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H.H., Kwon C., Lee M.M., Chung I.K. (2007). Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 49: 442–451 [DOI] [PubMed] [Google Scholar]
- Wang R.C., Smogorzewska A., de Lange T. (2004). Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119: 355–368 [DOI] [PubMed] [Google Scholar]
- Watson J.M., Shippen D.E. (2007). Telomere rapid deletion regulates telomere length in Arabidopsis thaliana. Mol. Cell. Biol. 27: 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., et al. (2006). Pot1 deficiency initiates DNA damage chenkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62 [DOI] [PubMed] [Google Scholar]
- Zellinger B., Akimcheva S., Puizina J., Schirato M., Riha K. (2007). Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol. Cell 27: 163–169 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Fan H.Y., Goldman J.A., Kingston R.E. (2007). Homology-driven chromatin remodeling by human RAD54. Nat. Struct. Mol. Biol. 14: 397–405 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Evolutionary divergence and convergence in proteins. Evolving Genes and Proteins, Bryson V., Vogel H.J., (New York: Academic Press; ), pp. 97–166 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.