Ycf3 is a plastid-encoded gene known to function in the assembly of photosystem I (PSI). This study revealed that Ycf3-interacting protein 1, a nucleus-encoded thylakoid protein, interacts with Ycf3 to promote the assembly of PSI units in the thylakoid membrane.
Abstract
The intricate assembly of photosystem I (PSI), a large multiprotein complex in the thylakoid membrane, depends on auxiliary protein factors. One of the essential assembly factors for PSI is encoded by ycf3 (hypothetical chloroplast reading frame number 3) in the chloroplast genome of algae and higher plants. To identify novel factors involved in PSI assembly, we constructed an epitope-tagged version of ycf3 from tobacco (Nicotiana tabacum) and introduced it into the tobacco chloroplast genome by genetic transformation. Immunoaffinity purification of Ycf3 complexes from the transplastomic plants identified a novel nucleus-encoded thylakoid protein, Y3IP1 (for Ycf3-interacting protein 1), that specifically interacts with the Ycf3 protein. Subsequent reverse genetics analysis of Y3IP1 function in tobacco and Arabidopsis thaliana revealed that knockdown of Y3IP1 leads to a specific deficiency in PSI but does not result in loss of Ycf3. Our data indicate that Y3IP1 represents a novel factor for PSI biogenesis that cooperates with the plastid genome-encoded Ycf3 in the assembly of stable PSI units in the thylakoid membrane.
INTRODUCTION
Photosystem I (PSI) is a large pigment-protein complex in the thylakoid membrane of plants and photosynthetically active bacteria (Jordan et al., 2001; Nelson and Ben-Shem, 2004; Amunts et al., 2007; Amunts and Nelson, 2009). It acts as a light-driven plastocyanin-ferredoxin oxidoreductase by mediating electron transfer from reduced plastocyanin (or cytochrome c6) to oxidized ferredoxin. PSI is one of the largest and most complex membrane protein assemblies known (Nelson and Ben-Shem, 2004; Amunts and Nelson, 2008). In higher plants, PSI consists of at least 14 core subunits, four different light-harvesting complex proteins (LHCI), and a large number of cofactors. The recently determined crystal structure of PSI from pea (Pisum sativum) at 3.3-Å resolution revealed the presence of 173 chlorophyll molecules, three iron-sulfur clusters, two phylloquinones, and 15 carotenoid molecules (Amunts et al., 2010). The complex architecture of PSI requires a highly ordered and spatially as well as temporally coordinated assembly process to ensure proper subunit incorporation and cofactor attachment (Wollman et al., 1999; Hippler et al., 2002; Dühring et al., 2007; Schöttler and Bock, 2008).
Even small structural perturbations can have significant effects on the stability and function of PSI (Hippler et al., 1998; Rochaix et al., 2000; Schöttler et al., 2007b). Unfortunately, little is known about the mechanisms involved in PSI assembly and about the assembly of large membrane protein complexes in general. A set of protein factors involved in PSI assembly has been identified in both cyanobacteria (Bartsevich and Pakrasi, 1997; Wilde et al., 2001; Shen et al., 2002a, 2002b) and plants (Boudreau et al., 1997; Ruf et al., 1997; Amann et al., 2004; Yabe et al., 2004; Stöckel et al., 2006; Schwenkert et al., 2010). Two of these assembly factors, Ycf3 (hypothetical chloroplast reading frame number 3) and Ycf4 (hypothetical chloroplast reading frame number 4), are encoded by chloroplast genes and were identified by reverse genetics (Boudreau et al., 1997; Ruf et al., 1997). Tobacco (Nicotiana tabacum) and Chlamydomonas reinhardtii ycf3 knockout mutants and Chlamydomonas ycf4 knockout strains completely lack PSI complexes, demonstrating that both factors are essential for the assembly of stable PSI units (Boudreau et al., 1997; Ruf et al., 1997). The Ycf3 protein is conserved in photosynthetically active organisms and contains three tetratricopeptide repeat domains (each formed by two short amphipathic α-helices), which are thought to mediate protein–protein interactions. Indeed, protein interaction studies have provided evidence that Ycf3 interacts with at least two PSI core subunits, PsaA and PsaD, providing additional support for a critical role of Ycf3 in PSI assembly and possibly suggesting that the Ycf3 protein is a chaperone-like factor that may help guide newly synthesized PSI subunits to their target position in the thylakoid membrane (Naver et al., 2001). Rapid insertion of hydrophobic membrane proteins into the thylakoid membrane is believed to be crucial for efficient complex assembly because unintegrated subunits of membrane protein complexes are usually unstable and subjected to rapid proteolytic degradation (Wollman et al., 1999; Choquet and Vallon, 2000).
How Ycf3 regulates PSI assembly and whether it acts alone or together with partner proteins in an assembly complex are currently unknown. To study the functions of Ycf3 in greater detail, we constructed a tagged ycf3 gene version and introduced it into the tobacco plastid genome to replace the wild-type ycf3 allele. Employing an affinity purification approach, we used these transplastomic plants to isolate an interaction partner of the Ycf3 protein, which we have termed Ycf3-interacting protein 1 (Y3IP1). Reverse genetics analysis of Y3IP1 function in tobacco and Arabidopsis thaliana demonstrate that Y3IP1 represents a novel factor that is required for the assembly of stable PSI units in higher plants.
RESULTS
Generation of Transplastomic Tobacco Plants with a FLAG-Tagged ycf3 Gene
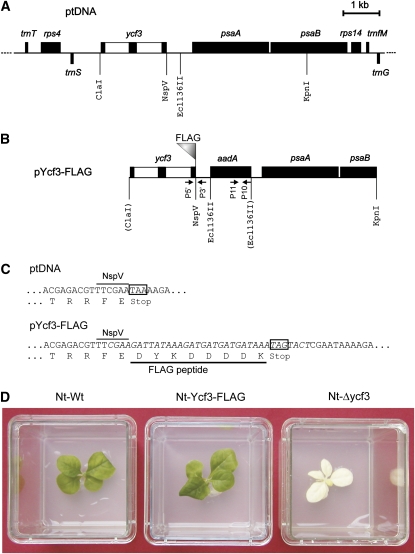
Our previously generated ycf3 knockout tobacco plants were photosynthetically incompetent and failed to accumulate detectable amounts of PSI (Ruf et al., 1997). Unfortunately, all our attempts to detect the Ycf3 protein in wild-type plants by generating specific antibodies had failed and, therefore, a detailed molecular analysis of PSI assembly by Ycf3 was not possible. We thus decided to construct a modified version of the ycf3 gene that encodes an epitope tag sequence that is detectable with a specific antibody. To identify a suitable insertion site for the tag, which does not interfere with Ycf3 function, we analyzed an amino acid sequence alignment of Ycf3 proteins from various plants and cyanobacteria. This analysis revealed that, while the N terminus of the protein is rather conserved, the C terminus displays considerable interspecific sequence variation (see Supplemental Figure 1 online), suggesting that it may provide a suitable site for tag addition. We therefore constructed a ycf3 gene version that encodes a translational fusion with the FLAG tag at the C terminus of Ycf3 (Figures 1A to 1C). The FLAG tag is a strong epitope tag of eight amino acids against which sensitive antibodies are available.
Figure 1.
Construction of a Plastid Transformation Vector with a FLAG-Tagged ycf3.
(A) Physical map of the region of the tobacco chloroplast genome (ptDNA) containing the ycf3 gene. Genes presented above the line are transcribed from left to right, and genes below the line are transcribed in the opposite direction. The ycf3 gene contains two introns, which are shown as open boxes. Relevant restriction sites used for cloning are indicated.
(B) Map of the plastid targeting fragment in transformation vector pYcf3-FLAG. The aadA selectable marker gene is integrated into a unique Ecl136II restriction site within the intergenic spacer between the ycf3 gene and the psaA/B operon. Restriction sites lost during vector construction due to ligation to heterologous ends are shown in parentheses. The binding sites and orientations of primers used for PCR analysis of plastid transformants are also marked.
(C) Introduction of the FLAG-encoding sequence at the end of the ycf3 reading frame. The sequence of the synthetic DNA ligated into the NspV restriction site is shown in italics. The encoded amino acid sequences are shown in the one letter code below the nucleotide sequences.
(D) Phenotypic comparison of Nt-Ycf3-FLAG plants with the wild type (Nt-Wt) and the ycf3 knockout (Nt-Δycf3). The wild-type-like phenotype of plants expressing FLAG-tagged Ycf3 suggests that the C-terminal FLAG tag does not interfere with Ycf3 function (see also Figure 8B).
Once we had cloned FLAG-tagged ycf3 into a tobacco plastid DNA fragment, we constructed a plastid transformation vector, pYcf3-FLAG, by integrating a chimeric selectable marker gene (aadA) into the intergenic spacer between the ycf3 gene and the downstream psaA/B operon (Figures 1A and 1B). The aadA gene product confers resistance to the aminoglycoside antibiotics spectinomycin and streptomycin and, when fused to plastid-specific expression signals, can be used for the selection of plants with transgenic chloroplasts (transplastomic plants; Svab and Maliga, 1993). The chosen aadA insertion site in plastid transformation vector pYcf3-FLAG (Figure 1B) is in a safe distance from the mapped psaA/B promoter (Meng et al., 1988) and hence should be neutral in that no negative effects on expression of the psaA/B operon are to be expected.
Chloroplast transformation experiments by biolistic bombardment of aseptically grown tobacco leaves followed by selection for spectinomycin-resistant cell lines yielded several transplastomic lines (designated Nt-Ycf3-FLAG; Figure 1D), five of which were further characterized (lines 2, 6, 13, 14, and 16; Figure 2A). The lines were subjected to three to four additional rounds of regeneration under stringent antibiotic selection to eliminate residual wild-type genome copies and generate homoplasmic transplastomic cell lines (Bock, 2001). Homoplasmy was verified by sensitive PCR assays that confirmed the absence of untagged wild-type ycf3 alleles (Figure 2A) as well as by genetic crosses and inheritance studies that confirmed the uniparental maternal transmission of aadA-encoded antibiotic resistance, as expected for a plastid genome–encoded trait.
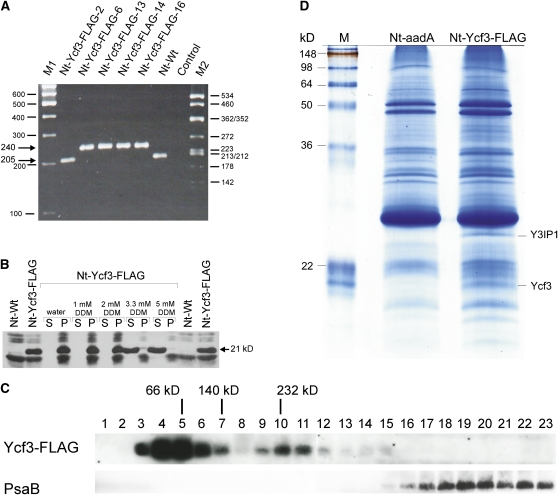
Figure 2.
Molecular Analysis of Nt-Ycf3-FLAG Plants.
(A) Homoplasmy of plastid transformants expressing a FLAG-tagged version of the ycf3 gene. PCR amplification with primer pair P5′/P3′ (Figure 1B) yields a 205-bp product for the wild type and a 240-bp product for the transplastome. Absence of the 205-bp fragment from lines Nt-Ycf3-FLAG-6, 13, 14, and 16 suggests homoplasmy for the FLAG-tagged ycf3 gene. Line Nt-Ycf3-FLAG-2 is transplastomic but carries only the aadA and not the FLAG mutation due to homologous recombination between the FLAG sequence and the aadA insertion site (Figure 1; Bock et al., 1994; Hager et al., 2002). This line was subsequently used as control (and referred to as Nt-aadA; see [C]). Homoplasmy for the aadA selectable marker gene was additionally confirmed by genetic crosses and inheritance assays. M1, M2, molecular mass markers (sizes of bands indicated in base pairs); control, buffer control.
(B) Association of the Ycf3 protein with thylakoid membranes. In Nt-Ycf3-FLAG plants but not in the wild type (Nt-Wt), the monoclonal anti-FLAG antibody recognizes a protein of 21 kD, which corresponds to the theoretical molecular mass of FLAG-tagged Ycf3. DDM treatment of purified thylakoids releases the protein from the membranes at concentrations of 3.3 mM and above (S, supernatant; P, thylakoid pellet). Cross-reacting bands present in thylakoid preparations from both the wild-type and the transplastomic plant are likely to represent LHC proteins. Note that they remain membrane bound at both 3.3 and 5 mM DDM, as expected for integral membrane proteins.
(C) Identification of Ycf3 complexes by sucrose gradient centrifugation. Sucrose gradients were fractionated into 23 fractions (numbered from top to bottom) and, following electrophoretic separation, probed with the anti-FLAG antibody to identify Ycf3-containing fractions (top panel) or with an anti-PsaB antibody to identify PSI-containing fractions (bottom panel). Peaks of relevant marker proteins from the molecular mass marker gradient are indicated above the blots.
(D) Detection of a putative interaction partner of Ycf3. Eluates from immunoprecipitation experiments with Nt-Ycf3-FLAG plants and Nt-aadA control plants were separated in a 15% polyacrylamide gel and stained with colloidal Coomassie blue. In addition to the band representing the FLAG-tagged Ycf3, another band, labeled Y3IP1, was specifically and reproducibly detected only in the Nt-Ycf3-FLAG samples, qualifying this protein as a potential interaction partner of Ycf3. M, molecular mass marker.
Since there is no absolute linkage between the aadA selectable marker gene and the FLAG mutation to be cointroduced, homologous recombination can separate the two. Thus, two different types of transplastomic plants were obtained: (1) plants that contained both the aadA marker and the FLAG insertion and (2) plants that harbored aadA but lacked the FLAG sequence. These two types of transplastomic lines have been observed earlier in transformation experiments designed to introduce mutations in chloroplast genes (Bock et al., 1994; Hager et al., 2002) and, as expected, were also obtained with the pYcf3-FLAG vector. Of the five transplastomic lines selected for detailed characterization, one (Nt-Ycf3-FLAG-2) carried aadA but lacked the FLAG sequence and four had both the aadA marker and the FLAG sequence (Figure 2A). Line Nt-Ycf3-FLAG-2 (carrying aadA, but lacking the FLAG tag) served as control in subsequent experiments and, for simplicity, will be referred to as Nt-aadA.
Having generated homoplasmic transplastomic tobacco plants carrying a tagged ycf3 instead of the wild-type ycf3 allele, we next wanted to test whether tethering of the FLAG epitope tag to the C terminus of the Ycf3 protein was phenotypically neutral. Our earlier work had established that loss of Ycf3 function results in a nonphotosynthetic phenotype due to the absence of PSI (Ruf et al., 1997). This phenotype is already recognizable in sterile culture on sucrose-containing medium and is characterized by severe pigment deficiency and leaf bleaching due to the accumulation of photooxidative damage (Figure 1D). When we compared homoplasmic Nt-Ycf3-FLAG plants with wild-type plants and the ycf3 knockout mutant, no visible difference between the wild-type and the FLAG-tagged plants was seen, suggesting that the FLAG tag does not significantly impair Ycf3 function (Figure 1D). As growth on tissue culture medium supplemented with sucrose may cover up more subtle phenotypes and, moreover, because impaired function of a photosynthesis-related protein can produce a mutant phenotype in a light-dependent manner, we also assayed wild-type and mutant plants grown in soil under widely different light intensities (ranging from 4 to 400 μE m−2 s−1). In no case did the transplastomic plants expressing the FLAG-tagged Ycf3 protein show any visible phenotype or growth retardation compared with wild-type plants, suggesting that the FLAG-tagged Ycf3 protein is functional.
Loose Association of the Ycf3 Protein with Tobacco Thylakoid Membranes
In the unicellular green alga C. reinhardtii, the Ycf3 protein is associated with thylakoid membranes (Boudreau et al., 1997), a finding that is compatible with its proposed assembly function for PSI (Naver et al., 2001). Having tagged the Ycf3 protein with the FLAG epitope, we were interested in analyzing the association of the Ycf3 protein with thylakoids in tobacco using anti-FLAG antibodies. The antibodies recognized a protein of the expected molecular mass (Ycf3 + FLAG = 21 kD) in Nt-Ycf3-FLAG plants, but not in the wild type, suggesting that this protein is FLAG-tagged Ycf3 (Figure 2B). The protein was only detectable in isolated thylakoids, demonstrating that, as expected, Ycf3 is also bound to the thylakoid membrane in higher plants. We next sought to determine how tightly Ycf3 is associated with the membrane. To this end, we treated isolated thylakoids with different concentrations of the mild nonionic detergent n-dodecyl maltoside (DDM). Relatively low DDM concentrations of 3.3 to 5 mM were sufficient to release the Ycf3 protein nearly quantitatively from the thylakoid membrane, whereas integral membrane proteins, such as photosystem core subunits and LHC proteins, remain associated with the thylakoid under these conditions (Figure 2B). These data confirm that Ycf3 is only peripherally associated with the thylakoid membrane, as expected for a photosystem assembly factor (Boudreau et al., 1997; Naver et al., 2001).
Presence of Ycf3 in Higher Molecular Mass Complexes
The successful in vivo tagging of Ycf3 allowed us to test if Ycf3 is present in protein complexes. To this end, we prepared thylakoids from Nt-Ycf3-FLAG plants, solubilized them with DDM, and separated the released multiprotein complexes in sucrose gradients by analytical ultracentrifugation. Probing of gradient fractions with anti-FLAG antibodies revealed the presence of Ycf3 in two distinct protein complexes. Whereas the bulk of the Ycf3 protein is present in a complex peaking at ~60 to 70 kD, a substantial fraction is associated with much larger complexes, peaking at ~230 to 250 kD (Figure 2C). Whether the two complexes are present in vivo or, alternatively, the 60- to 70-kD complex arises from partial dissociation of the 230- to 250-kD complex upon solubilization and/or gradient centrifugation remains to be investigated. Nonetheless, detection of Ycf3 in gradient fractions corresponding to much higher molecular masses than free Ycf3 is compatible with the idea that Ycf3 does not act alone but is part of a larger assembly complex for PSI.
Probing of the sucrose gradient fractions with an anti-PsaB antibody demonstrated that neither of the two Ycf3-containing protein complexes is stably associated with PSI (Figure 2C), consistent with the proposed function of Ycf3 as an assembly factor that is not an integral part of PSI complexes.
Isolation of a Protein Interacting with Ycf3
The possibility of detecting the Ycf3-FLAG protein with an anti-FLAG antibody in our transplastomic Nt-Ycf3-FLAG plants allowed us to take a FLAG affinity purification approach toward isolating interactors of Ycf3 that potentially could represent constituents of the Ycf3 complex and, thus, novel components of the PSI assembly machinery. In view of the ease with which Ycf3 could be released from the thylakoid membrane by mild detergent treatment (Figure 2B) and the fact that high molecular mass complexes containing Ycf3 could be detected by sucrose gradient ultracentrifugation (Figure 2C), we reasoned that stable protein–protein interactions should be preserved in solubilized Ycf3 complexes.
Therefore, we subjected detergent-solubilized protein fractions prepared from isolated thylakoid membranes to a coimmunoprecipitation procedure (see Methods). Coimmunoprecipitation experiments are known to suffer from a background of nonspecific protein associations, mainly due to cross-reactivity of the antibodies involved (Rigaut et al., 1999). To distinguish between specific and nonspecific proteins bound to the FLAG antibody matrix, the purification was performed in parallel for the transplastomic Nt-Ycf3-FLAG plants and the transplastomic Nt-aadA control plants (Figure 2D). Proteins binding nonspecifically to the matrix appear in both samples, whereas specific interactors of Ycf3 are expected to be present only in the Nt-Ycf3-FLAG sample. When we compared the protein patterns from these experiments, we indeed reproducibly observed two protein bands that were only detectable in the Nt-Ycf3-FLAG samples (Figure 2D). These proteins had molecular masses of ~21 and 24 kD, respectively, and were stained with colloidal Coomassie blue to approximately equal intensity, possibly suggesting that they are present in about equimolar amounts.
The two bands and the corresponding gel regions from the Nt-aadA lane (Figure 2D) were excised and in-gel digested with trypsin, and the resulting peptides were subjected to mass spectrometric identification by de novo sequencing. Database searches assigned the identified peptides from the 21-kD band to the Ycf3 protein (Table 1). The corresponding gel region from the Nt-aadA control did not contain any peptides matching the Ycf3 amino acid sequence. This was expected because the Ycf3 protein in Nt-aadA plants is not tagged and therefore is not isolated by our FLAG affinity purification. In three independent experiments, two specific peptides were consistently identified from the 24-kD band in Nt-Ycf3-FLAG plants (Figures 2D and 3, Table 1). Both peptides were absent from the Nt-aadA control and could be assigned to two highly similar tentative consensus (TC) sequences, TC40639 and TC52264, from N. tabacum in the TIGR Gene Indices EST database (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=tobacco). Copurification of Ycf3 and the 24-kD protein from Nt-Ycf3-FLAG plants and complete absence of both proteins from the Nt-aadA control strongly suggested that the 24-kD protein represents an interaction partner of Ycf3. Therefore, we tentatively named this protein Ycf3-interacting protein 1 (Y3IP1).
Table 1.
Peptides Identified from the Two Proteins (21 and 24 kD) Specifically Present in Coimmunoprecipitation Experiments with Nt-Ycf3-FLAG Transplastomic Plants
| Exp. | Band | m/z | Sequence | Homology |
| 1 | 24 kD | 535 | LTIMoxIEDPRDVER | TC40639/TC52264 |
| 552 | LTIMoxIEDPR | TC40639/TC52264 | ||
| 657 | LLGIDDENAPTR | TC40639/TC52264 | ||
| 928 | LLGIDDENAPTRDDLAAALEEINEGK | TC40639/TC52264 | ||
| 21 kD | 510 | LEIDPYDR | Ycf3 | |
| 721 | SYILYNIGLIHTSNGEHTK | Ycf3 | ||
| 877 | NYAEALQNYYEAMoxR | Ycf3 | ||
| 957 | AEAWFDQAAEYWK | Ycf3 | ||
| 2 | 24 kD | 657 | LLGIDDENAPTR | TC40639/TC52264 |
| 21 kD | 510 | LEIDPYDR | Ycf3 | |
| 523 | TFSIVANILLR | Ycf3 | ||
| 877 | YAEALQNYYEAMoxR | Ycf3 | ||
| 3 | 24 kD | 552 | LTIMoxIEDPR | TC40639/TC52264 |
| 657 | LLGIDDENAPTR | TC40639/TC52264 | ||
| 21 kD | 510 | LEIDPYDR | Ycf3 | |
| 623 | VANILLR | Ycf3 | ||
| 799 | KALEYYFR | Ycf3 |
Peptide sequences were determined by mass spectrometry–based de novo sequencing in three independent experiments. Exp., experiment; Mox, oxidized Met; m/z, mass-to-charge ratio.
Figure 3.
Amino Acid Sequence Alignment of the Ycf3-Interacting Protein Y3IP1 from Selected Plant Species.
The alignment includes the Y3IP1 sequences from the dicots Arabidopsis, Vitis vinifera, and N. tabacum (TC52264), the monocot Oryza sativa, the moss Physcomitrella patens, and the green algae C. reinhardtii and Ostreococcus lucimarinus. The alignment was produced using the MAFFT program version 6 (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/). Peptides identified by mass spectrometry during isolation of Y3IP1 from tobacco are underlined. A putative transmembrane region identified in silico (using the InterProScan algorithm; http://www.ebi.ac.uk/Tools/InterProScan/) is shaded in gray in the Arabidopsis sequence. Amino acid residues conserved in all species shown here are marked with asterisks, colons denote conserved amino acids exchanges, and single dots indicate semiconserved substitutions. Note that all sequences include the putative transit peptide for protein import into chloroplasts, but no conserved cleavage site is recognizable.
The 702-bp open reading frame of the tobacco EST contig TC40639 encodes a protein of 234 amino acids, whereas the 789-bp open reading frame of EST contig TC52264 encodes a protein of 263 amino acids (see Supplemental Figures 2A and 2B online). The amino acid sequences encoded by the two EST contigs are highly similar but not completely identical (sequence identity, 82.5%; sequence similarity, 84.0%; http://www.ebi.ac.uk/Tools/emboss/align/; see Supplemental Figure 2B online), suggesting that TC40639 does not just represent a C-terminally truncated version of TC52264 and raising the possibility that two Y3IP1 genes are present in the nuclear genome of tobacco. To confirm this supposition, we obtained complete sequence information for both cDNAs using an RT-PCR strategy (see Methods; database accession numbers for the cDNA sequences: HQ015439 for TC40639 and HQ015440 for TC52264). We then performed PCR analyses with tobacco genomic DNA and cDNA as template and sequenced the amplification products. The data revealed that, indeed, Y3IP1 is encoded by two nuclear genes in tobacco and ultimately confirmed that the two EST contigs are not the product of alternative RNA processing (see Supplemental Figure 2C online).
As the tobacco nuclear genome is not yet sequenced, we also searched for sequences encoding homologs of Y3IP1 in fully sequenced plant genomes. Putative Y3IP1 homologs were identified in photosynthetic green algae, mosses, and higher plants (Figure 3). In all fully sequenced plant genomes included in the alignment presented in Figure 3 as well as in the fully sequenced poplar (Populus trichocarpa) genome, Y3IP1 appears to be encoded by a single-copy nuclear gene. In Arabidopsis, the Y3IP1 gene is split by five introns. Whether or not the presence of two Y3IP1 gene copies in tobacco is due to the amphidiploid genome structure of N. tabacum (with one gene copy coming from Nicotiana sylvestris and the other from Nicotiana tomentosiformis) remains to be analyzed once the full genome sequence of tobacco becomes available. Searches for structural and functional motifs in the Y3IP1 sequence revealed the presence of a C-terminal transmembrane domain (Figure 3) but no further domains or motifs of known function.
Y3IP1 Is Targeted to Chloroplasts
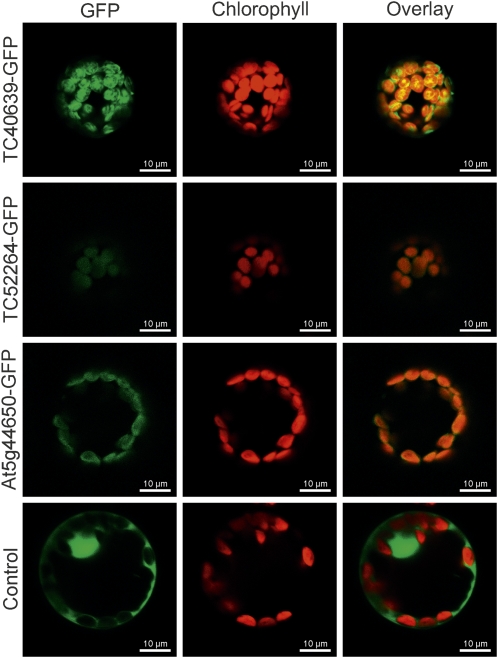
The isolation of Y3IP1 as an interaction partner of the plastid genome-encoded PSI assembly factor Ycf3 suggests that Y3IP1 is a chloroplast protein. In silico predictions of subcellular localization using the iPSORT (http://hc.ims.u-tokyo.ac.jp/iPSORT/), TargetP V1.1 (http://www.cbs.dtu.dk/services/TargetP/), and ChloroP 1.1 (http://www.cbs.dtu.dk/services/ChloroP/) algorithms also suggested chloroplast localization of all Y3IP1 homologs analyzed. To experimentally confirm the chloroplast localization of Y3IP1, we constructed green fluorescent protein (GFP) fusions with three full-length Y3IP1 gene versions: the two tobacco EST contigs (TC40639 and TC52264) and the single-copy Arabidopsis Y3IP1 gene (At5g44650). The fusion constructs (driven by the strong constitutively expressed cauliflower mosaic virus [CaMV] 35S promoter) were introduced into tobacco protoplasts by transient transformation. Analysis of the subcellular localization of the Y3IP1-GFP fusion proteins by confocal laser scanning microscopy revealed that all three proteins localized to the chloroplast (Figure 4), thus confirming that Y3IP1 is a chloroplast protein in both tobacco and Arabidopsis.
Figure 4.
In Vivo Analysis of Subcellular Localization of Y3IP1.
Fusion constructs of the full-length Y3IP1 coding sequences from N. tabacum (TC40639 and TC52264) and Arabidopsis (At5g44650) to GFP were transiently expressed in tobacco protoplasts by polyethylene glycol–mediated DNA uptake. GFP fluorescence, chlorophyll fluorescence, and the overlay of the two signals are shown for all three constructs. As an empty vector control, a fusion of the CaMV 35S promoter to GFP was included, which shows the typical nucleocytosolic localization of GFP fluorescence. Colocalization of GFP fluorescence with chlorophyll fluorescence in all three Y3IP1-GFP fusions demonstrates that Y3IP1 is targeted to chloroplasts. Single plane micrographs were taken with a confocal laser scanning microscope 48 h after protoplast transformation.
Generation of Y3IP1 Mutants in Tobacco and Arabidopsis
Y3IP1 was isolated as an interaction partner of the PSI-specific assembly factor Ycf3. This suggests a possible role for Y3IP1 in the PSI assembly process. To test this hypothesis, we sought to generate mutants with reduced levels of Y3IP1 in N. tabacum and Arabidopsis. From a large collection of tobacco antisense and cosuppression lines (Lein et al., 2008), two cosuppression lines were obtained. The lines (subsequently referred to as Nt-cs-Y3IP1-1 and Nt-cs-Y3IP1-2) carry a 560-bp Y3IP1 cDNA fragment (overlapping by 511 bp with the TC40639 sequence; see Supplemental Figure 2A online) in the sense orientation under the control of the strong CaMV 35S promoter and were therefore considered as cosuppression lines.
Arabidopsis RNA interference (RNAi) lines for Y3IP1 (At-RNAi-Y3IP1) were generated by Agrobacterium tumefaciens–mediated transformation with a hairpin-type RNAi construct against Y3IP1 (obtained from the AGRIKOLA clone collection; Hilson et al., 2004). To track the fate of Ycf3 upon downregulation of Y3IP1, we also produced RNAi lines in our transplastomic tobacco background expressing the FLAG-tagged Ycf3 from the chloroplast genome (referred to as Nt-RNAi-Y3IP1). To this end, transplastomic Nt-Ycf3-FLAG plants were supertransformed by Agrobacterium-mediated transformation with a hairpin RNAi construct directed against both TC40639 and TC52264.
From all three gene silencing approaches, mutants with clear phenotypes were obtained (Figure 5). The typical mutant phenotype was characterized by growth retardation, delayed development, and light-green leaf color. Most of the Nt-RNAi-Y3IP1 lines were so severely affected that they did not grow photoautotrophically and therefore could only be propagated in sterile culture on sucrose-containing medium (Figure 5C). Collectively, the phenotypes of Arabidopsis and tobacco Y3IP1 mutants suggested that photosynthetic performance was reduced upon downregulation of Y3IP1 expression.
Figure 5.
Phenotype of Mutants with Reduced Y3IP1 Expression.
(A) An N. tabacum var Samsun NN wild-type plant and two cosuppression lines (Nt-cs-Y3IP1-1 and Nt-cs-Y3IP1-2). The picture was taken after 90 d of growth in soil.
(B) An Arabidopsis Col-0 wild-type plant and six independently generated RNAi lines (At-RNAi-Y3IP1 plants). The plants were photographed 35 d after transfer from sucrose-containing selection medium to soil.
(C) An N. tabacum var Petit Havana wild-type plant, a transplastomic plant expressing the in vivo–tagged Ycf3 (Nt-Ycf3-FLAG), and three independent RNAi lines (Nt-RNAi-Y3IP1 plants). Plants had to be grown on sucrose-containing medium in sterile culture because the severity of the mutant phenotypes in some of the RNAi lines did not permit autotrophic growth when transferred to soil.
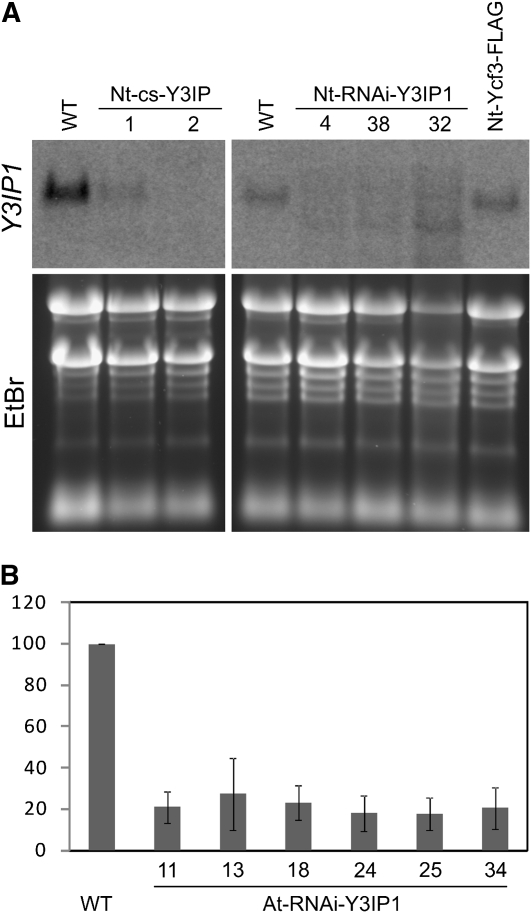
Downregulation of Y3IP1 transcript levels in N. tabacum cosuppression lines and RNAi lines was analyzed by RNA gel blot experiments using a Y3IP1-specific probe (Figure 6A). As expected, the signal for the Y3IP1 mRNA was strongly reduced in both the cosuppression lines and the RNAi lines. Hybridizing RNA species of lower molecular mass accumulated especially in the strong Nt-RNAi-Y3IP1 lines (Figure 6A). These RNAs most likely represent degradation products of the Y3IP1 transcript and result from RNAi-induced cleavage of the mRNA. Downregulation of Y3IP1 in six isolated At-RNAi-Y3IP1 mutants was analyzed by real-time quantitative RT-PCR (Figure 6B). All six lines showed a strong reduction in Y3IP1 mRNA, of up to <20% of the transcript levels in the wild type.
Figure 6.
Accumulation of Y3IP1 Transcripts in Tobacco and Arabidopsis Mutants.
(A) Transcript accumulation in N. tabacum cosuppression lines (left panel) and RNAi lines (right panel). mRNA accumulation was analyzed by RNA gel blot analysis using a Y3IP1-specific probe. To confirm equal loading and integrity of all RNA samples, the ethidium bromide–stained agarose gel prior to blotting is shown below.
(B) Downregulation of Y3IP1 expression in six independently generated Arabidopsis RNAi lines as determined by quantitative real-time PCR. Data comprise four technical replicates (using three independent RNA isolations from the same plant), and error bars indicate sd.
Y3IP1 Mutants Are Specifically Impaired in PSI Accumulation
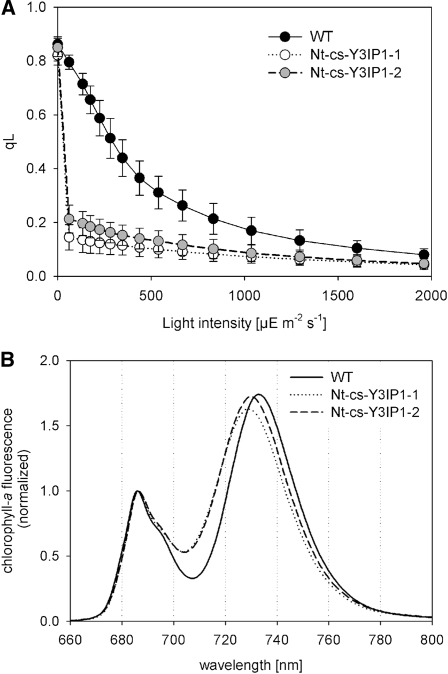
The growth phenotypes and the pigment deficiency of the tobacco and Arabidopsis Y3IP1 mutants (Figure 5) were compatible with a function of Y3IP1 in photosynthesis. To characterize the physiological basis of the mutant phenotype in greater detail, the chlorophyll content, the efficiency of photosynthetic electron transport, and the contents of the multiprotein complexes of the photosynthetic electron transport chain were determined using spectroscopic methods. To circumvent limitations in the available leaf material (At-RNAi-Y3IP1 plants) and possible artifacts from heterotrophic growth (Nt-RNAi-Y3IP1 lines), these analyses were performed with soil-grown Nt-cs-Y3IP1 mutant plants. As suggested by the light-green phenotype of the Y3IP1 mutants, the chlorophyll content per leaf area was found to be reduced in Nt-cs-Y3IP1 plants (Table 2). The mutants also have a strongly decreased chlorophyll a:chlorophyll b ratio, indicating that the primary defect may reside in one of the photosystem cores rather than in the light-harvesting antennae (Table 2). The maximum quantum efficiency of photosystem II (PSII; FV/FM), a standard measure of PSII integrity, was significantly reduced in the mutants (Table 2). To preliminarily distinguish between a defective PSII and a defect in one of the downstream components of the linear electron transport chain, we determined the chlorophyll fluorescence parameter qL (Kramer et al., 2004) in relation to light intensity. qL correlates with the fraction of PSII reaction centers that are open (with the primary quinone-type acceptor QA in the oxidized state). Already at low light intensities, the qL values measured in the Y3IP1 mutants were greatly reduced compared with those in the wild type (Figure 7A), indicating an extreme reduction of the PSII acceptor side. This suggests that the mutants have a basically intact PSII photochemistry but are incapable of efficiently transferring electrons to the downstream components of the electron transport chain.
Table 2.
Quantification of Chlorophyll Contents, the Chlorophyll a:Chlorophyll b Ratio, the Maximum Quantum Efficiency of PSII Photochemistry (FV/FM), and the Amounts of Photosynthetic Complexes in Thylakoid Membranes of Wild-Type Plants and Y3IP1 Cosuppression Lines
| Parameter | Wild Type | Nt-cs-Y3IP1-1 | Nt-cs-Y3IP1-2 |
| Chlorophyll [mg·m−2] | 343.5 ± 19.5 | 224.4 ± 43.2 | 249.9 ± 36.4 |
| Chlorophyll a/b | 3.71 ± 0.11 | 2.99 ± 0.12 | 3.09 ± 0.13 |
| FV/FM | 0.81 ± 0.01 | 0.51 ± 0.06 | 0.58 ± 0.07 |
| PSII [mmol·mol chlorophyll−1] | 2.56 ± 0.10 | 3.67 ± 0.47 | 3.44 ± 0.48 |
| Cyt-bf [mmol·mol chlorophyll−1] | 1.05 ± 0.20 | 1.27 ± 0.27 | 1.11 ± 0.32 |
| PSI [mmol·mol chlorophyll−1] | 2.26 ± 0.09 | 0.49 ± 0.06 | 0.72 ± 0.14 |
| PSII [μmol·m−2] | 0.88 ± 0.08 | 0.83 ± 0.24 | 0.87 ± 0.22 |
| Cyt-bf [μmol·m−2] | 0.36 ± 0.08 | 0.29 ± 0.10 | 0.29 ± 0.11 |
| PSI [μmol·m−2] | 0.78 ± 0.06 | 0.11 ± 0.03 | 0.18 ± 0.05 |
The contents of photosynthetic complexes were determined by difference absorption spectroscopy measurements performed with isolated thylakoids and normalized on a chlorophyll basis (mmol·mol chlorophyll−1) or leaf area basis (μmol·m−2). The values represent averages from several measurements ± sd (n = 5 for the wild type, n = 7 for Nt-cs-Y3IP1-1, and n = 11 for Nt-cs-Y3IP1-2).
Figure 7.
Analysis of the Chlorophyll Fluorescence Parameter qL and the 77K Chlorophyll a Fluorescence Emission in Wild-Type and Nt-cs-Y3IP1 Plants.
(A) Determination of qL, the fraction of PSII reaction centers that are open (with the primary quinone acceptor QA oxidized) in relation to light intensity. Low qL values already at low light intensities in the Y3IP1 mutants indicate an extreme reduction of the PSII acceptor side. The values represent averages from several measurements (n = 10 for the wild type, n = 14 for Nt-cs-Y3IP1-1, and n = 22 for Nt-cs-Y3IP1-2), and error bars indicate sd.
(B) Chlorophyll a fluorescence emission measurements at 77K. The fluorescence emission signals were normalized to the PSII emission maximum at 685 nm. The PSI emission signal peaks at 733 nm in the wild type. The blue-shifted wavelength of the PSI emission maximum in the Y3IP1 mutants indicates that PSI antenna organization is altered and suggests that a significant proportion of PSI antenna proteins are not attached to PSI.
While these data indicated a deficiency in photosynthetic electron transport in the Y3IP1 mutants, none of the above measurements can be used to distinguish between primary defects in PSI and primary defects in PSII or the cytochrome b6f complex. Therefore, we directly determined the contents of the protein complexes participating in photosynthetic electron transport (PSII, cytochrome b6f, and PSI) by difference absorption spectroscopy (Schöttler et al., 2007a). Interestingly, the Nt-cs-Y3IP1 mutants displayed a reduction in PSI contents (to 24 and 31%, respectively, of the wild-type level), while the PSII contents and, to a lesser extent, also the cytochrome b6f contents were elevated (Table 2). Elevated PSII and cytochrome b6f contents have also been observed in other PSI-deficient mutants (Schöttler et al., 2007a). This is largely due to data normalization to chlorophyll, which results in an apparent increase in the amounts of the unaffected complexes. To exclude this effect, we also normalized the data on a leaf area basis (Table 2). Comparison of the amounts of the three complexes per leaf area revealed that the Y3IP1 mutants are specifically deficient in PSI, while the amounts of PSII and the cytochrome b6f complex are not significantly altered (Table 2).
To further analyze the PSI-deficient phenotype of the Y3IP1 mutants, chlorophyll a fluorescence emission measurements at 77K were conducted. At this low temperature, the characteristic long-wavelength emission from PSI can be readily analyzed (Krause and Weis, 1991). Comparison of 77K chlorophyll a fluorescence emission spectra of wild-type plants and the Nt-cs-Y3IP1 mutants revealed that the maximum fluorescence emission from PSI (peaking at 733 nm in the wild type) was blue shifted by ~5 nm in the mutants (Figure 7B). This indicates an altered functional organization of PSI (Jensen et al., 2004; Schöttler et al., 2007b) and suggests that a substantial proportion of the PSI antenna is not connected to PSI reaction centers. This is in line with the earlier finding that LHCI proteins stably accumulate in mutants with reduced amounts of PSI reaction center subunits (Stöckel et al., 2006).
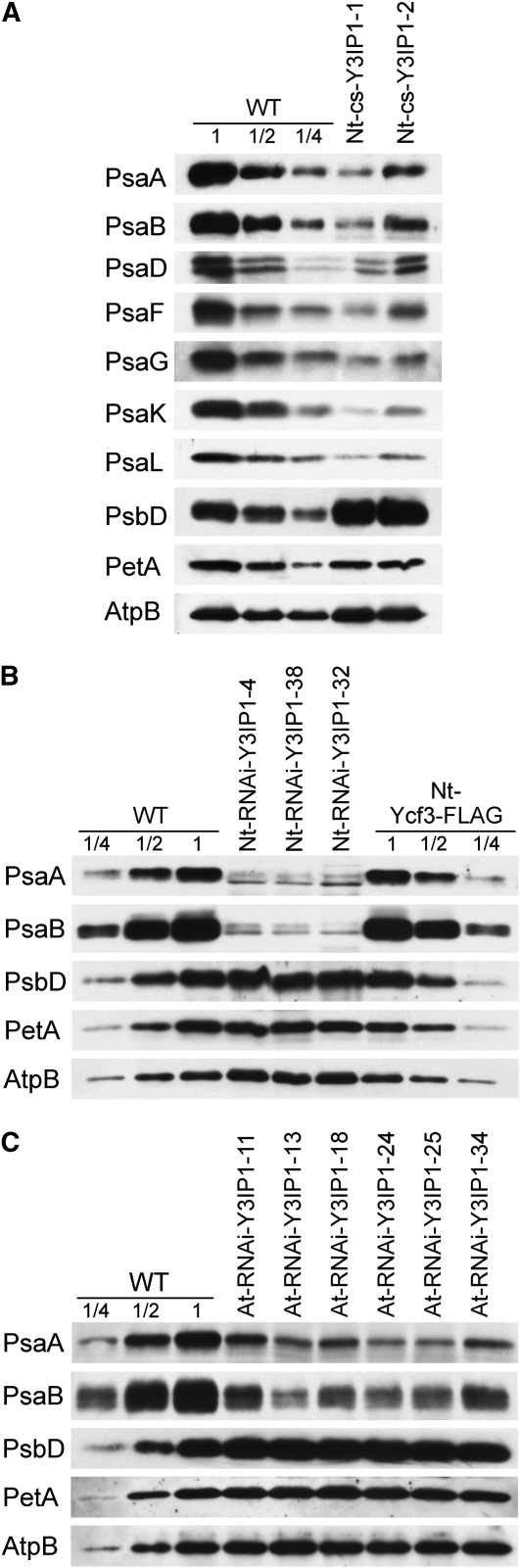
We next wanted to confirm the impaired PSI accumulation in Y3IP1 mutants at the molecular level. To this end, thylakoids were isolated from all three sets of mutants (the tobacco cosuppression lines, the tobacco RNAi lines, and the Arabidopsis RNAi lines) and analyzed immunobiochemically using a set of specific antibodies raised against diagnostic subunits of all four multiprotein complexes in the thylakoid membrane (PSII, cytochrome b6f complex, PSI, and ATP synthase). Altogether, seven subunits of PSI were investigated: the two reaction center proteins, PsaA and PsaB, the stromal ridge subunit PsaD, and the four small subunits PsaF, PsaG, PsaK, and PsaL. At least the two reaction center proteins, PsaA and PsaB, are diagnostic of PSI complex accumulation in that no PSI can assemble in thylakoid membranes of knockout mutants for either psaA or psaB (Rochaix, 1997; Redding et al., 1999). The same holds true for the tested subunits of the other thylakoidal protein complexes: PsbD (diagnostic of PSII), PetA (diagnostic of the cytochrome b6f complex), and AtpB (diagnostic of ATP synthase). Probing of thylakoid proteins from Nt-cs-Y3IP1 lines revealed a strong reduction in PSI amounts compared with the wild type. The reduction was similarly strong for all PSI subunits tested (Figure 8A) and was in the same range as our spectroscopy measurements had suggested (Table 2). The tobacco RNAi lines showed an even stronger reduction in PSI (Figure 8B), which is in good agreement with their more severe phenotypes and their inability to grow photoautotrophically (Figure 5). As the tobacco RNAi lines were generated in the transplastomic Ycf3-FLAG background, we additionally compared them with the Nt-Ycf3-FLAG plants. No difference in thylakoid protein accumulation was detectable between wild-type plants and the Nt-Ycf3-FLAG plants, thus confirming that the tethering of the FLAG tag to the C terminus of Ycf3 (and downstream integration of the aadA marker) is phenotypically neutral and does not affect protein function in any way (Figure 8B). Finally, we also confirmed the specific downregulation of PSI in six independent Arabidopsis RNAi lines (Figure 8C).
Figure 8.
Immunoblot Analysis of Diagnostic Components of the Multiprotein Complexes in the Thylakoid Membrane in Tobacco and Arabidopsis Mutants with Reduced Y3IP1 Expression.
Isolated thylakoid proteins were separated by SDS-PAGE (in 12 or 15% gels), blotted, and probed with antibodies against the proteins indicated on the left side of each panel. Equal amounts of chlorophyll were loaded. For quantitative assessment of protein accumulation in the mutants, a dilution series of the wild-type sample (100, 50, and 25%) was loaded.
(A) Immunoblot analysis of thylakoid proteins in the tobacco cosuppression lines. Note that the PsaD subunit is detected as a double band because it is present in two different isoforms with slightly different molecular masses (Obokata et al., 1993).
(B) Analysis of thylakoid proteins in the tobacco RNAi lines. To confirm wild-type levels of thylakoid protein complexes in the Nt-Ycf3-FLAG lines, an additional dilution series (100, 50, and 25%) of the Nt-Ycf3-FLAG sample was loaded.
(C) Analysis of Arabidopsis RNAi mutants.
In all three sets of mutants, the diagnostic subunits of PSII (PsbD), the cytochrome b6f complex (PetA = cytochrome f), and ATP synthase (AtpB) accumulated to identical or slightly elevated levels compared with the wild type (Figure 8). This confirms that Y3IP1 is specifically involved in PSI biogenesis and is not required for accumulation of the other components of the photosynthetic electron transport chain.
Y3IP1 Regulates PSI Accumulation at the Posttranslational Level
The identification of Y3IP1 as a protein–protein interaction partner of Ycf3 suggests that Y3IP1 is involved in PSI accumulation at the level of complex assembly. To confirm this supposition, we wanted to exclude the alternative possibilities that Y3IP1 is involved in the transcription of PSI genes, accumulation of stable PSI mRNAs, and/or translation of PSI subunits. As Y3IP1 is localized to plastids (Figure 4), only the PSI genes in the plastid genome are relevant to any possible function of Y3IP1 prior to PSI assembly (i.e., in gene expression). The plastid genome of higher plants encodes three essential subunits of PSI: PsaA, PsaB (the two reaction center proteins of PSI encoded by the psaA/B operon and translated from a polycistronic mRNA; Meng et al., 1988), and PsaC (an essential iron-sulfur protein; Takahashi et al., 1991). As Ycf3 interacts with Y3IP1 and is also essential for PSI accumulation (Ruf et al., 1997), we additionally included the ycf3 gene in our analysis of the expression of plastid PSI genes in Y3IP1 mutants. Finally, the plastid-encoded ycf4 gene shown to be required for PSI accumulation in C. reinhardtii (Boudreau et al., 1997) was also included.
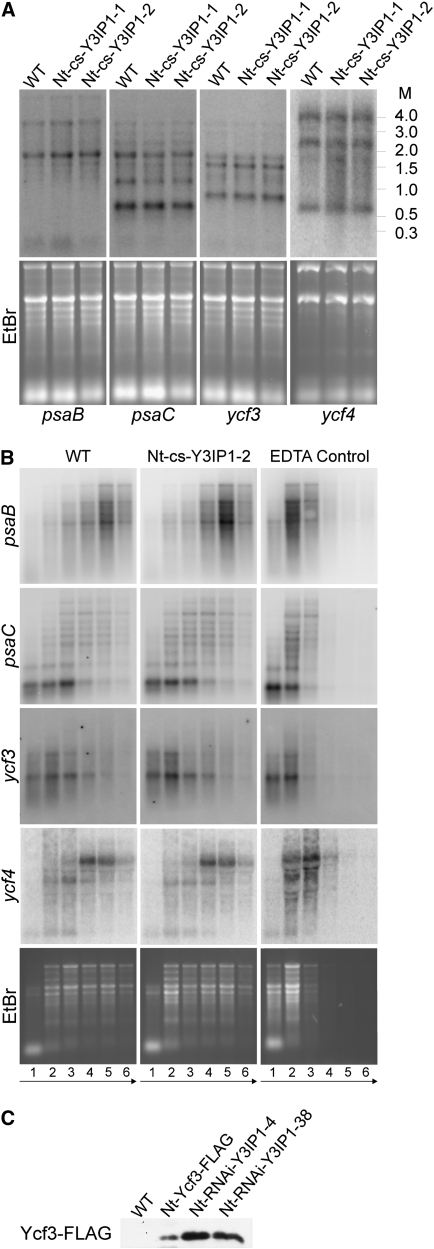
We first compared PSI transcript patterns and accumulation levels in Nt-cs-Y3IP1 lines and wild-type plants. None of the transcripts (psaA/B, psaC, ycf3, and ycf4) showed any difference in RNA abundance or RNA processing patterns (Figure 9A), indicating that Y3IP1 acts neither at the transcriptional level nor at the level of mRNA maturation or stability.
Figure 9.
Unaltered PSI Gene Expression in Y3IP1 Mutants and Y3IP1-Independent Accumulation and Thylakoid Targeting of Ycf3.
(A) Analysis of PSI gene transcription in wild-type tobacco plants and two cosuppression lines. Transcript patterns and accumulation levels were determined for the three essential plastid genome-encoded PSI subunits (psaA/B and psaC) and the ycf3 and ycf4 genes encoding essential PSI assembly factors (Boudreau et al., 1997; Ruf et al., 1997). To confirm equal loading and integrity of all RNA samples, the ethidium bromide–stained agarose gels prior to blotting are shown below. M, molecular mass marker (sizes in kilobases).
(B) Analysis of translational efficiency for essential plastid genome-encoded PSI genes. Polysome loading was analyzed for the psaA/B, psaC, ycf3, and ycf4 transcripts. Collected fractions from the sucrose density gradients are numbered from the top to the bottom. Equal aliquots of extracted RNAs from all fractions were separated by denaturing agarose gel electrophoresis, blotted, and hybridized to gene-specific radiolabeled probes. The arrows at the bottom indicate the gradient in sucrose density (from low to high). As a control, a sample was treated with EDTA to cause dissociation of ribosomes from the mRNAs. Ribosome distribution in the gradients is revealed by ethidium bromide (EtBr) staining of agarose gels prior to blotting.
(C) Accumulation of the Ycf3 protein in tobacco Y3IP1 RNAi mutants. The FLAG-tagged Ycf3 protein in the transplastomic Nt-Ycf3-FLAG line and two Y3IP1 RNAi lines (generated in the transplastomic background) was detected with an anti-FLAG antibody. Slightly higher Ycf3 levels in the RNAi lines are due to loading of thylakoid proteins on an equal chlorophyll basis (and the reduced chlorophyll content in the RNAi lines).
To test for the possible involvement of Y3IP1 in the translation of PSI mRNAs, the association of psaA/B, psaC, ycf3, and ycf4 mRNAs with ribosome in Nt-cs-Y3IP1 plants and wild-type plants was compared in polysome-loading experiments. Polysomes are complexes of mRNAs covered with translating ribosomes, and their migration into continuous sucrose gradients upon ultracentrifugation correlates with the number of ribosomes bound to the mRNA molecule. Thus, heavily translated mRNAs are loaded with many ribosomes and migrate deeply into the gradient, whereas poorly translated mRNAs accumulate in the upper fractions of the gradient. For all mRNAs analyzed, the polysome profiles in wild-type and Y3IP1 mutant plants were indistinguishable (Figure 9B), indicating that PSI mRNA translation proceeds at comparable efficiency and suggesting that Y3IP1 is not a translation factor for plastid genome-encoded PSI mRNAs.
Together, the analysis of PSI gene transcription and translation suggests that the deficiency in PSI in Y3IP1 mutants is not caused by a defect in gene expression. This supports a function of Y3IP1 at the posttranslational level and is in line with the isolation of Y3IP1 as an interaction partner of the PSI assembly factor Ycf3.
Y3IP1 Is Not Required for Stable Accumulation of Ycf3
The posttranslational function of Y3IP1 in PSI accumulation and the interaction of Y3IP1 with the PSI assembly factor Ycf3 suggest two possible roles of Y3IP1 in PSI biogenesis. First, Y3IP1 could cooperate with Ycf3 in an assembly complex and thus be directly involved in PSI assembly. Alternatively, Y3IP1 could be solely required for stability and/or the membrane association of Ycf3 and, in this way, be indirectly involved in PSI assembly. To distinguish between these possibilities, we analyzed the fate of Ycf3 in the Y3IP1 knockdown mutants. This was possible because we had generated the tobacco Y3IP1 RNAi lines in the transplastomic Ycf3-FLAG–tagged background. We therefore could employ anti-FLAG antibodies to test if Ycf3 accumulates to normal levels and is associated with thylakoid membranes in Y3IP1 mutants. This was indeed the case (Figure 9C), indicating that Y3IP1 does not merely exert its function in PSI biogenesis via Ycf3 stabilization or Ycf3 membrane targeting. Instead, this finding supports a direct involvement of Y3IP1 in PSI assembly, presumably as a component of a Ycf3-Y3IP1 assembly complex.
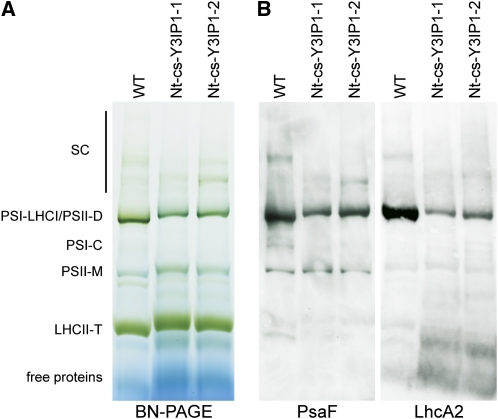
We also tested the possibility that Y3IP1 is required for coupling of the antenna proteins to the PSI core. To this end, thylakoidal protein complexes were separated by blue-native PAGE (BN-PAGE), and the blotted protein complexes were probed with an antibody against a diagnostic subunit of PSI and a subunit of LHCI. These experiments confirmed the strongly reduced PSI accumulation in the Y3IP1 mutants and revealed that the residual PSI is associated with LHCI (Figure 10).
Figure 10.
Undisturbed Association of LHCI with PSI in Y3IP1 Mutants.
(A) Separation of thylakoidal protein complexes by BN-PAGE. Profiles for the wild type and two tobacco cosuppression lines are shown. LHCII-T, light-harvesting complex II (LHCII) trimer; PSII-M, PSII monomer; PSI-C, PSI core; PSI-LHCI, PSI complex with LCHI attached; PSII-D, PSII dimer; SC, various supercomplexes.
(B) Identification of PSI and LHCI by immunoblotting. The BN gel was blotted and probed with diagnostic antibodies for PSI (PsaF) and LHCI (LhcA2). Colocalization of PsaF and LHCI suggests association of LHCI with PSI.
DISCUSSION
In the course of this work, we identified a nuclear-encoded protein factor involved in PSI assembly in higher plants. The protein was isolated using a combination of approaches, in which we fused the known PSI assembly factor Ycf3 (Boudreau et al., 1997; Ruf et al., 1997) with an epitope tag in vivo using stable transformation of the tobacco chloroplast genome. Subsequent affinity purification identified a specific protein interaction partner of Ycf3, referred to as Y3IP1. The Y3IP1 protein is conserved in higher plants and green algae (Figure 3), but we were unable to identify a putative homolog in cyanobacteria. This may suggest that Y3IP1 represents a eukaryotic acquisition and is not required for the assembly of PSI in photosynthetic prokaryotes. This is remarkable because its interaction partner, Ycf3, is conserved in cyanobacteria and known to be essential for PSI assembly also in Synechocystis (Schwabe et al., 2003). However, it remains possible that a cyanobacterial Y3IP1-like protein does exist but has escaped bioinformatic identification due to very low sequence conservation.
Our reverse genetics analyses of the Y3IP1 genes in tobacco and Arabidopsis uncovered a specific function of Y3IP1 in PSI biogenesis. In the absence of T-DNA insertion lines for the Y3IP1 gene, cosuppression and RNAi were the methods of choice to generate mutants with downregulated Y3IP1 expression. Spectroscopy measurements (Table 2, Figure 7) and biochemical analyses (Figure 8) consistently revealed a strong and specific reduction in PSI accumulation in all Y3IP1 mutants, thus providing a straightforward explanation for their pigment-deficient and growth-retarded phenotype (Figure 5).
Together with the specific impairment of Y3IP1 mutants in PSI accumulation, identification of Y3IP1 as an interaction partner of the essential PSI assembly factor Ycf3 suggested that Y3IP1 also participates in PSI assembly. However, formally, two alternative possibilities needed to be considered: (1) involvement in PSI gene expression at the transcriptional and/or posttranscriptional levels or (2) function of Y3IP1 as a PSI subunit that is assembled into the PSI complex by Ycf3 and therefore identified as a protein interaction partner of Ycf3. The latter possibility can largely be excluded on theoretical grounds because the three-dimensional structure of higher plant PSI has been resolved to very high resolution (Ben-Shem et al., 2003; Amunts et al., 2007, 2010; Amunts and Nelson, 2009), and, clearly, the structure cannot accommodate an additional protein subunit as large as 24 kD. Although a loose peripheral association cannot be definitively excluded, our finding that the Ycf3-containing PSI assembly complex is not stably bound to PSI complexes (Figure 2C) tentatively argues against this possibility. The possible involvement of Y3IP1 in PSI gene expression was tested experimentally. As Y3IP1 is exclusively localized to plastids (Figure 4), it cannot be involved in the transcription or translation of nuclear PSI genes; therefore, only the expression of plastid PSI genes needed to be investigated. Our analyses of transcript patterns, RNA accumulation levels, and translation of plastid PSI genes excluded the possibility that Y3IP1 represents a specific transcription or translation factor for plastid-encoded PSI genes (Figures 9A and 9B). Therefore, we propose that Y3IP1 acts at the posttranslational level and is a novel assembly factor for PSI that, together with its interaction partner Ycf3, mediates incorporation of PSI subunits into the thylakoid membrane.
Studies in the unicellular green alga C. reinhardtii revealed that Ycf3 interacts with the PSI subunits PsaA and PsaD (Naver et al., 2001), suggesting that Ycf3 functions as an assembly chaperone that possibly helps inserting the A and D subunits into the PSI core. Ycf3 is loosely attached to the stromal side of the thylakoid membrane (Boudreau et al., 1997; Figure 2B) and therefore may be predominantly involved in the assembly of the reducing side of PSI. It seems reasonable to speculate that Y3IP1, in a complex with Ycf3, also participates in assembly of the stromal side of PSI and, perhaps, interacts with additional subunits of PSI, although a role in cofactor insertion into PSI also seems possible. It is noteworthy that our Ycf3-FLAG affinity purification approach identified Y3IP1 but did not result in the detection of PsaA, PsaD, or any other PSI subunit. The likely reason for this is that the molecular interactions between assembly factors and the proteins they chaperone are usually transient in nature and therefore are unlikely to be preserved during our rigorous affinity purification procedures. By contrast, the interaction between Ycf3 and Y3IP1 appears to be rather stable, and the two proteins were isolated in seemingly 1:1 stoichiometric amounts (Figure 2D). As expected for an assembly factor, Ycf3 is present in substoichiometric amounts relative to the structural subunits of PSI (Boudreau et al., 1997), and the same may be the case for Y3IP1. However, at present, this conclusion is based solely on similar staining intensities, and the actual stoichiometry of the Ycf3-Y3IP1 complexes remains to be determined more precisely.
Our analysis of Ycf3 accumulation in Y3IP1 mutants revealed that Y3IP1 is not required for the stability of Ycf3 or for its association with thylakoids (Figure 9C). Whether or not, conversely, Y3IP1 is stable in the absence of Ycf3 is currently unknown. Our ycf3 knockout mutants (Ruf et al., 1997) provide a suitable tool to address this question, but thus far, we have not been successful in obtaining Y3IP1-specific antibodies that could be used to analyze Y3IP1 accumulation in a Ycf3-free mutant background.
PSI represents one of the largest and most complex macromolecular assemblies in nature. To dissect how its many protein subunits are inserted into the thylakoid membrane in a concerted manner and how nearly 200 cofactors are built into the complex represents a daunting task. Although a set of factors involved in PSI assembly (Boudreau et al., 1997; Ruf et al., 1997; Stöckel et al., 2006; Ozawa et al., 2009), cofactor biogenesis, and cofactor attachment (Shen et al., 2002a, 2002b; Amann et al., 2004; Yabe et al., 2004; Xu et al., 2005; Lohmann et al., 2006) has been identified, we are still far from understanding the molecular mechanisms underlying the assembly of PSI and other multiprotein complexes in energy-transducing membranes. Systematic studies of the molecular interactions between photosystem subunits and their assembly chaperones will be key in deciphering the mechanistic details of the assembly process. At the same time, the development of novel untargeted approaches that are able to identify protein interaction partners of known players in photosystem assembly will be of the utmost importance in identifying novel assembly chaperones and other accessory factors involved in photosystem biogenesis. The combination of in vivo tagging and immunoaffinity purification developed here for Ycf3 is generally applicable and thus should be instrumental in identifying additional factors that participate in the assembly of protein complexes in the thylakoid membrane.
METHODS
Plant Material
Sterile tobacco plants (Nicotiana tabacum var Petit Havana) were grown on agar-solidified MS medium containing 30 g/L sucrose (Murashige and Skoog, 1962). Homoplasmic transplastomic lines were rooted and propagated on the same medium. RNAi plants were generated in the transplastomic Petit Havana background. Tobacco cosuppression lines were produced in N. tabacum var SNN (Lein et al., 2008). For generation of transgenic Arabidopsis thaliana plants, the standard ecotype Columbia-0 (Col-0) was used. Tobacco plants used for physiological analyses were grown under long-day conditions (16 h light) with an average growth light intensity of 300 μE m−2 s−1.
Construction of a Plastid Transformation Vector with a FLAG-Tagged ycf3 Allele
The region of the tobacco plastid genome containing the ycf3 reading frame was cloned as a ClaI/KpnI restriction fragment into a pBluescript vector cut with KpnI and NaeI. The ClaI end of the insert was blunted by a fill-in reaction with the Klenow fragment of the Escherichia coli DNA polymerase I (Roche Applied Science). The FLAG-encoding DNA sequence was integrated upstream of the ycf3 stop codon by linearizing the resulting plasmid with the restriction enzyme NspV and ligating it to a double-stranded FLAG fragment prepared by annealing two synthetic oligonucleotides (Figure 1C). As a selectable marker for chloroplast transformation, a chimeric aadA gene (Svab and Maliga, 1993) was integrated into a unique Ecl136II restriction site within the intergenic spacer between the ycf3 gene and the psaA/B operon (Figure 1B). A plasmid clone carrying the aadA marker gene in the same transcriptional orientation as ycf3 and psaA/B was identified by diagnostic restriction digests and designated pYcf3-FLAG.
Plastid Transformation and Selection of Homoplasmic Transplastomic Tobacco Lines
Young leaves from sterile tobacco plants were bombarded with plasmid pYcf3-FLAG-coated 0.6-μm gold particles using a biolistic gun (PDS1000He; Bio-Rad). Primary spectinomycin-resistant lines were selected on regeneration medium containing 500 mg/L spectinomycin (Svab et al., 1990; Svab and Maliga, 1993). Plastid transformation was confirmed by double resistance tests on medium with both spectinomycin and streptomycin (500 mg/L each; Bock, 2001) and by PCR amplification using the primer pair P10 and P11 (Bock et al., 1996; Figure 1B). Five independently generated transplastomic lines were subjected to three additional rounds of regeneration on medium with spectinomycin to obtain homoplasmic tissue.
Crosses and Inheritance Tests
Wild-type and Nt-Ycf3-FLAG plants were transferred to soil and grown to maturity under greenhouse conditions. Seedpods were collected from selfed plants and from reciprocal crosses of the Nt-Ycf3-FLAG transplastomic lines with wild-type plants. Surface-sterilized seeds were germinated on MS medium with spectinomycin (500 mg/L) and analyzed for uniparental inheritance of the chloroplast genome-localized spectinomycin resistance gene aadA. Selfed homoplasmic transformants and crosses with a Nt-Ycf3-FLAG transplastomic line as the maternal parent gave rise to uniformly resistant (green) progeny, whereas seeds collected from wild-type plants yielded exclusively sensitive (white) seedlings.
Construction of RNAi Vectors and Plant Nuclear Transformation
The two cosuppression lines in N. tabacum var SNN (Nt-cs-Y3IP1-1 and Nt-cs-Y3IP1-2) carry a 560-bp tobacco cDNA fragment spanning 511 bp of EST contig TC40639 in sense orientation under the control of the CaMV 35S promoter (Lein et al., 2008). Insertion of the cassette was tested by PCR with genomic DNA as template and primers 35S (5′-GTGGATTGATGTGATATCTCC-3′) and OCS (5′-GTAAGGATCTGAGCTACACAT-3′) followed by direct sequencing of the PCR products. Transgenic plants were grown in soil under standard glasshouse conditions (16 h light at 25°C/8 h dark at 20°C).
For the generation of RNAi lines in transplastomic N. tabacum var Petit Havana, a 196-bp sequence in the 3′ region of EST contig TC40639 (sharing >97% similarity with EST contig TC52264; see Supplemental Figure 2A online) was chosen and amplified from tobacco cDNA with the primer pairs C7 (5′-TTGAATTCATGGCCTAATTTGG-3′)/C8 (5′-TTGGTACCCTGACCCAAAGCTGGAGGAA-3′), introducing the restriction sites EcoRI/KpnI (underlined), and C9 (5′-TTTCTAGAGAATTCATGGCCTAATTTGG-3′)/C10 (5′-TTGGATCCCTGACCCAAAGCTGGAGGAA-3′), introducing the restriction sites XbaI and BamHI (underlined). The C7/C8 PCR product was digested with EcoRI and KpnI and cloned (in sense orientation) into multiple cloning site 1 (MCS1) of the similarly cut RNAi vector pHANNIBAL (Wesley et al., 2001). Subsequently, the C9/C10 PCR product was digested with BamHI and XbaI and inserted (in antisense orientation) into MCS2. The resulting vector contains a Y3IP1-specific hairpin construct under the control of the CaMV 35S promoter and the ocs terminator. For Agrobacterium tumefaciens–mediated nuclear transformation of tobacco plants, the expression cassette was excised with SacI and SpeI and cloned into the similarly cut binary plant transformation vector pART27 carrying the nptII selection marker (Gleave, 1992). The resulting final vector was transformed into transplastomic tobacco plants expressing an in vivo FLAG-tagged Ycf3 by Agrobacterium-mediated transformation using established protocols (Rosahl et al., 1987). Successful integration of the RNAi construct into the nuclear genome was confirmed by PCR with genomic DNA as template and the primers 35S and OCS. The RNAi lines were subsequently grown in sterile culture on agar-solidified medium containing 30 g/L sucrose.
For the generation of Arabidopsis RNAi lines, the plasmid CATMA5a40480, which carries a 166-bp PCR product corresponding to positions 587 to 752 of the Y3IP1 coding sequence, was obtained from the AGRIKOLA collection (Hilson et al., 2004). The vector was transformed into Agrobacterium (strain GV3101::pMP90::pSOUP) by electroporation. Agrobacterium-mediated transformation of Arabidopsis ecotype Col-0 was performed using the floral dip method (Clough and Bent, 1998). Selection of transgenic plants was performed by germination on agar-solidified 0.5× MS medium (Duchefa) containing 6 μg/mL phosphinotricine (Duchefa). Resistant seedlings were transferred to soil and grown to maturity in a growth chamber under short-day conditions (12 h light/12 h dark, light intensity 145 μE m−2 s−1). Insertion of the RNAi cassette was verified by PCR (Hilson et al., 2004).
Extraction of Nucleic Acids, Isolation of Polysomes, and Hybridization Procedures
Total plant DNA was isolated from fresh leaf material by a rapid miniprep procedure (Doyle and Doyle, 1990). Total cellular RNA was extracted using the NucleoSpin RNA II kit (Macherey-Nagel) or the peqGOLD TriFast reagent (Peqlab). Isolation of polysomes and RNA extraction from polysomal fractions was performed as described previously (Kahlau and Bock, 2008; Rogalski et al., 2008). Gradient fractionation was performed using the Auto Densi-Flow (Labconco) and the Pharmacia LKB RediFrac fraction collector (GE Healthcare). RNA pellets were dissolved in 30 μL sterile water, and 7-μL aliquots were used for RNA gel blot analyses.
Total cellular RNA (5 μg for detection of plastid transcripts and 40 μg for detection of nuclear transcripts) or polysomal RNA (samples of 7 μL per fraction) were denatured at 95°C and separated in a 1% (w/v) denaturing agarose gel containing 1 mL 37% formaldehyde/100 mL gel (PerfectBlue Gelsystem; PeqLab). The RNA was then transferred onto Hybond nylon membranes (Hybond-XL; GE Healthcare) by capillary blotting using standard procedures. Hybridization was performed at 65°C in a standard buffer (Church and Gilbert, 1984). Probes were produced by PCR amplification from cDNA using gene-specific primers described previously (Ruf et al., 1997). In addition, a ycf4-specific probe was generated by amplification with the primers Pycf4_5′ (5′-CCCTTCTATGACAAATTTGA-3′) and Pycf4_3′ (5′-CGAGTCAAGGGAATGGACCCC-3′). For hybridization, [α-32P]dCTP-labeled probes were generated by random priming (Multiprime DNA labeling system; GE Healthcare). Hybridization signals were analyzed using a Typhoon Trio+ variable mode imager (GE Healthcare).
PCR, cDNA Synthesis, and DNA Sequencing
PCR amplifications were performed according to standard protocols (30 cycles) in an Eppendorf thermocycler using Taq DNA polymerase (Fermentas), Pfu DNA polymerase (Fermentas), or Phusion DNA polymerase (Finnzymes). Primer pair P10 (5′-AACCTCCTATAGACTAGGC-3′; complementary to the psbA 3′-untranslated region of the chimeric aadA gene) and P11 (5′-AGCGAAATGTAGTGCTTACG-3′; derived from the 3′ portion of the aadA coding region; Figure 1B) were used to confirm successful plastid transformation by amplifying a portion of the selectable marker gene aadA. Primers P5′ (5′-GAAGCGCATAATTGGTTGAA-3′) and P3′ (5′-GATTGGGTCTTCCAAAAGC-3′) flank the FLAG insertion site (Figure 1B) and were used to assay for the presence of the FLAG insertion and to test for homoplasmy of the transplastomic FLAG-tagged ycf3 lines. Amplification products were separated by electrophoresis in 2.5% agarose gels according to standard procedures.
For cDNA synthesis, total cellular RNA extracted with the peqGOLD TriFast reagent was treated with RNase-free DNase I (Epicentre Biotechnologies) to eliminate residual DNA followed by reverse transcription with SuperScript III reverse transcriptase (Invitrogen) using random hexanucleotide primers. cDNA synthesis for quantitative real-time PCR was performed with SuperScript III reverse transcriptase (Invitrogen) using RNA extracted with the NucleoSpin RNA II kit (Macherey-Nagel) as template and oligo-dT18 primers. Quantitative RT-PCR was performed with the StepOnePlus real-time PCR system (Applied Biosystems) and Absolute SYBR Green ROX mix (Thermo Scientific) as described previously (Karcher and Bock, 2009) using the same housekeeping genes for normalization and the Y3IP1-specific primers C87 (5′-TCCGTTGGCCAAATTTAGAG-3′) and C77 (5′-TGGCTGCTTCTTTTGGATCT-3′).
To confirm the presence of two genes for Y3IP1 in tobacco, complete cDNA sequence information was obtained for both TC sequences. To this end, PCR was performed with primer C9, which binds to both TCs (see Supplemental Figure 2A online), and an oligo-dT18 primer using N. tabacum cDNA as template. The PCR products were cloned employing the TOPO TA Cloning vector system (Invitrogen) and subsequently sequenced with the M13 forward primer. The additional sequence information was used to design a primer pair that results in PCR products of two different sizes for TC40639 (187 bp) and TC52264 (300 bp). PCR amplifications were then performed with genomic DNA and cDNA using the primer pair C98/C101 (see Supplemental Figure 2A online). The PCR products were separated in a 1.5% agarose gel.
Transient Expression of GFP Fusion Proteins in Protoplasts
The complete coding sequences of the two tobacco EST contig sequences existing for Y3IP1 (TC40639 and TC52264) were amplified from N. tabacum cDNA with the primer pairs C78 (5′-TTCTCGAGATGGCCTCGAACATGCTTCA-3′)/C90 (5′-TTCCATGGCCTGACCCAAAGCTGGAGGAA-3′) and C78/C91 (5′-TTCCATGGCCTGTAGAGAATTATAGAAGAGA-3′), respectively. The coding sequence of Y3IP1 from Arabidopsis (At5g44650) was amplified from Arabidopsis cDNA using the primer pair C81 (5′-TTCTCGAGATGACGACGCAGATATTTCAG-3′)/C92 (5′-TTCCATGGCCTGTAGGGAATTGTAAAACA-3′). The restriction sites XhoI and NcoI (underlined) were introduced in all PCR products via the primer sequences. The PCR products were digested with XhoI and NcoI and cloned in frame into the respective restriction sites of vector pA7-GFP (Voelker et al., 2006), generating a fusion of the Y3IP1 coding region with GFP (TC40639-GFP, TC52264-GFP, and At5g44650-GFP) under the control of the CaMV 35S promoter. For transient expression of the Y3IP1-GFP fusions, protoplasts were prepared from aseptically grown N. tabacum leaves and transformed according to a published protocol (Huang et al., 2002). Analysis of subcellular localization of GFP fluorescence was performed as described previously (Karcher and Bock, 2009).
Isolation of Thylakoids, Immunoprecipitation, and Immunoblotting
To detect the FLAG-tagged Ycf3 protein, thylakoid proteins from wild-type and transplastomic plants were isolated from total leaf material using published procedures (Machold et al., 1979). Equal amounts of thylakoid proteins were then electrophoretically separated on tricine-SDS polyacrylamide gels (Schägger and von Jagow, 1987) and transferred to Hybond-P polyvinylidene difluoride membranes (GE Healthcare) using the Trans-Blot Cell (Bio-Rad) and a standard transfer buffer (192 mM glycine and 25 mM Tris, pH 8.3). Immunoblot detection was performed with specific antibodies using the enhanced chemiluminescence system (ECL PLUS system; GE Healthcare) and anti-FLAG M2 monoclonal antibodies (Sigma-Aldrich) or affinity-purified anti-FLAG polyclonal antibodies (Rockland Immunochemicals). To determine the strength of Ycf3 binding to the thylakoid membrane, purified thylakoids were incubated for 20 min at room temperature with different concentrations of the nonionic detergent DDM. Following centrifugation for 20 min at 40,000g, pelleted thylakoids and supernatants were analyzed by SDS-PAGE.
The size of Ycf3-containing protein complexes was estimated by analytical ultracentrifugation. To this end, leaf material from Nt-Ycf3-FLAG plants and control plants (Nt-aadA) was homogenized in extraction buffer (350 mM sucrose, 50 mM HEPES, pH 7.5, 2 mM EDTA, 50 mM NaCl, and 5 mM MgCl2). Chloroplasts were pelleted by centrifugation at 1800g for 8 min, washed with extraction buffer, and lysed in lysis buffer (20 mM Tricine, pH 8.0, 5 mM EDTA, and 0.5× Complete Protease Inhibitor Cocktail; Roche). Thylakoid membranes were then pelleted by centrifugation at 2500g for 5 min, and membrane proteins were solubilized in lysis buffer containing 0.6% (w/v) DDM at a chlorophyll concentration of 1 mg/mL. Solubilized proteins corresponding to 0.8 mg chlorophyll were separated in a linear sucrose gradient by ultracentrifugation for 17 h at 160,000g (SW41Ti; Beckmann). Linear sucrose gradients were prepared by freezing and thawing a 10% (w/v) sucrose solution in 20 mM Tricine/NaOH, pH 8.0, 0.125× Complete Protease Inhibitor Cocktail, and 0.05% (w/v) DDM. Five-hundred–microliter fractions were collected from the gradients, and proteins were extracted by methanol-chloroform precipitation (Wessel and Flügge, 1984). Precipitated proteins were redissolved in loading buffer for subsequent SDS-PAGE separation in 15% polyacrylamide gels and immunoblot analysis. For size estimation of protein complexes, the distribution of marker proteins (66 kD, BSA; 140 kD, lactate dehydrogenase; 232 kD, catalase; High Molecular Weight Marker from GE Healthcare) treated with 0.6% (w/v) DDM in lysis buffer was analyzed in a parallel gradient.
For immunoprecipitation experiments, chloroplasts were isolated from fresh leaf material homogenized in extraction buffer (330 mM sucrose, 50 mM HEPES, 2 mM EDTA, and 50 mM NaCl, pH 7.5) and filtered through four layers of gauze and one layer of Miracloth (Calbiochem). Cell debris was pelleted at 100g for 5 min, and chloroplasts were then collected by centrifugation (10 min, 2000g) and washed twice in extraction buffer. Chloroplasts representing 2.5 mg chlorophyll were mixed with 1 mL lysis buffer (25 mM HEPES/KOH, pH 7.5, 10 mM EDTA, 5 mM MgCl2, and 0.2× Complete Protease Inhibitor Cocktail) and incubated for 10 min on ice. Subsequently, membranes were pelleted for 5 min at 2500g and washed with lysis buffer. For release of Ycf3 complexes from the membranes, thylakoids were mixed with 1 mL TBS buffer (50 mM Tris/HCl, pH 7.4, 50 mM NaCl, 1 mM EDTA, and 0.05× Complete Protease Inhibitor Cocktail) containing 0.6% DDM and incubated for 20 min at 4°C. Membranes were pelleted at 25,000g, and the supernatant was incubated with 20 μL EZview Red ANTI-FLAG M2 Affinity gel (Sigma-Aldrich) for 1.5 h at 4°C. After washing the Affinity gel four times with TBS buffer, the bound proteins were eluted with SDS-PAGE sample buffer (Laemmli, 1970) and separated in a 15% polyacrylamide gel followed by staining with the NOVEX Colloidal Blue Stain Kit (Invitrogen) and destaining in distilled water.
Immunoprecipitates and thylakoid proteins (isolated according to Schöttler et al., 2004) were analyzed by separation in 12 or 15% SDS-polyacrylamide gels (Laemmli, 1970) using a Perfect Blue twin gel system (PeqLab) and blotted as described above. For immunochemical detection with the ECL PLUS system, specific antibodies against PsaA, PsaB, PsaD, PsaF, PsaG, PsaK, PsaL, PsbD, PetA, and AtpB were used. Antibodies were kindly provided by Reinhold G. Herrmann (University of Munich, Germany), Ralf Oelmüller, and Thomas Pfannschmidt (University of Jena, Germany) or purchased from Agrisera. The chemiluminescence signal was visualized by exposition to Kodak Biomax XAR film (Sigma-Aldrich).
For BN-PAGE, isolated thylakoid membranes (Schöttler et al., 2004) equivalent to 20 μg chlorophyll from tobacco wild-type plants and Y3IP1 cosuppression lines (Nt-cs-Y3IP1) were solubilized with 1% DDM at a chlorophyll concentration of 1 mg chlorophyll/mL and analyzed by BN-PAGE using published protocols (D'Amici et al., 2009). BN gradient gels (5 to 12%) were used for protein complex separation (PROTEAN II xi cell; Bio-Rad).
In-Gel Tryptic Digestion and Mass Spectrometry
Tryptic digestion and mass spectrometry were performed as described previously (Walz et al., 2002) with minor modifications. Bands of interest were excised, transferred to Protein LoBind tubes (Eppendorf), destained (in 40% [v/v] acetonitrile and 60% [v/v] 50 mM NH4HCO3) twice for 1 h and dehydrated in 100% acetonitrile for 5 min. Proteins were then digested in gel with Trypsin Proteomics Grade (Sigma-Aldrich) at 37°C overnight. The residual digestion solution was collected and combined with supernatants of the following extraction steps. Tryptic peptides were extracted from the gel by adding 50 μL 5% (v/v) trifluoroacetic acid (TFA) to the gel slice and incubating it for 20 min, and then incubating the gel slice three times in 50 μL 5% (v/v) TFA in 50% (v/v) acetonitrile for 20 min. After vacuum drying of the collected supernatants, peptides were solubilized in 7 μL 0.1% TFA and 3 μL 8 M guanidinum chloride in 2.5% (v/v) TFA. After desalting (ZipTIPs C18; Millipore), samples were eluted in 4 μL 75% (v/v) acetonitrile containing 0.1% acetic acid and subjected to tandem mass spectrometry in a Q-TOF instrument (Micromass) equipped with a z-spray source as described (Walz et al., 2002).
Chlorophyll Fluorescence Measurements and Difference Absorption Spectroscopy
Chlorophyll fluorescence was recorded with a pulse amplitude modulated fluorimeter (Dual-PAM-100; Heinz Walz) on intact plants at room temperature. Plants were dark adapted for 1 h prior to determination of the maximum PSII quantum efficiency (FV/FM). Subsequently, the light response curves of the redox state of the PSII acceptor side were measured and the chlorophyll a fluorescence parameter qL was calculated based on a lake model for PSII-LHCII (Kramer et al., 2004). The contents of PSII, the cytochrome b6f complex, and PSI were determined in thylakoids prepared as described previously (Schöttler et al., 2004). PSI was quantified from P700 difference absorption signals at 830 to 870 nm in solubilized thylakoids using the Dual-PAM instrument (Schöttler et al., 2007a). Contents of PSII and the cytochrome b6f complex were determined from difference absorption measurements of cytochrome b559 (PSII) and the cytochromes f and b6. Measurement procedures and deconvolution methods were described in detail previously (Kirchhoff et al., 2002; Schöttler et al., 2007b).
Chlorophyll contents were determined in 80% (v/v) acetone (Porra et al., 1989). The 77K chlorophyll a fluorescence emission spectra of isolated thylakoids (equivalent to 10 μg chlorophyll mL−1) were recorded using a Jasco F-6500 fluorimeter (Jasco). Chlorophyll a fluorescence was excited at 430-nm wavelength (10-nm spectral bandwidth), and fluorescence emission was determined with a spectral bandwidth of 1 nm in a wavelength range from 660 to 800 nm.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: N. tabacum TC40639 (HQ015439), N. tabacum TC52264 (HQ015440), and Arabidopsis Y3IP1 (At5g44650).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Ycf3 Amino Acid Sequences from Various Photosynthetically Active Organisms.
Supplemental Figure 2. Alignment of Two Tobacco EST Contig Sequences (TC40639 and TC52264) Potentially Encoding the Ycf3-Interacting Protein Y3IP1 and Confirmation of the Presence of Two Y3IP1-Encoding Genes in the Tobacco Nuclear Genome.
Acknowledgments
We thank Brigitte Buchwald (Max-Planck-Institut für Molekulare Pflanzenphysiologie [MPI-MP]) for help with plant transformation, the MPI-MP Green Team for plant care, Marita Hermann (University of Freiburg, Germany) and Marita Hermann (University of Münster, Germany) for excellent technical assistance during the initial phase of this work, Wolfram Thiele (MPI-MP) for help with photosynthesis measurements, Yvonne Weber (MPI-MP) for help with protoplast transformation, Katrin Piepenburg (MPI-MP) for help with microscopy, Frederik Börnke (University of Erlangen, Germany) for help with generating the cosuppression lines, and Nadine Tiller (MPI-MP) for helpful discussion. We also thank Reinhold G. Herrmann (University of Munich, Germany), Ralf Oelmüller, and Thomas Pfannschmidt (University of Jena, Germany) for providing antibodies. This research was supported by the Deutsche Forschungsgemeinschaft (SFB 429) and the Max Planck Society.
References
- Amann K., Lezhneva L., Wanner G., Herrmann R.G., Meurer J. (2004). ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16: 3084–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A., Drory O., Nelson N. (2007). The structure of a plant photosystem I supercomplex at 3.4A resolution. Nature 447: 58–63 [DOI] [PubMed] [Google Scholar]
- Amunts A., Nelson N. (2008). Functional organization of a plant photosystem I: Evolution of a highly efficient photochemical machine. Plant Physiol. Biochem. 46: 228–237 [DOI] [PubMed] [Google Scholar]
- Amunts A., Nelson N. (2009). Plant photosystem I design in the light of evolution. Structure 17: 637–650 [DOI] [PubMed] [Google Scholar]
- Amunts A., Toporik H., Borovikova A., Nelson N. (2010). Structure determination and improved model of plant photosystem I. J. Biol. Chem. 285: 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsevich V.V., Pakrasi H.B. (1997). Molecular identification of a novel protein that regulates biogenesis of photosystem I, a membrane protein complex. J. Biol. Chem. 272: 6382–6387 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A., Frolow F., Nelson N. (2003). Crystal structure of plant photosystem I. Nature 426: 630–635 [DOI] [PubMed] [Google Scholar]
- Bock R. (2001). Transgenic chloroplasts in basic research and plant biotechnology. J. Mol. Biol. 312: 425–438 [DOI] [PubMed] [Google Scholar]
- Bock R., Hermann M., Kössel H. (1996). In vivo dissection of cis-acting determinants for plastid RNA editing. EMBO J. 15: 5052–5059 [PMC free article] [PubMed] [Google Scholar]
- Bock R., Kössel H., Maliga P. (1994). Introduction of a heterologous editing site into the tobacco plastid genome: The lack of RNA editing leads to a mutant phenotype. EMBO J. 13: 4623–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E., Takahashi Y., Lemieux C., Turmel M., Rochaix J.-D. (1997). The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 16: 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Vallon O. (2000). Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82: 615–634 [DOI] [PubMed] [Google Scholar]
- Church G.M., Gilbert W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- D'Amici G.M., Huber C.G., Zolla L. (2009). Separation of thylakoid membrane proteins by sucrose gradient ultracentrifugation or blue native-SDS-PAGE two-dimensional electrophoresis. Methods Mol. Biol. 528: 61–70 [DOI] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. (1990). Isolation of plant DNA from fresh tissue. Focus 12: 13–15 [Google Scholar]
- Dühring U., Ossenbühl F., Wilde A. (2007). Late assembly steps and dynamics of the cyanobacterial photosystem I. J. Biol. Chem. 282: 10915–10921 [DOI] [PubMed] [Google Scholar]
- Gleave A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20: 1203–1207 [DOI] [PubMed] [Google Scholar]
- Hager M., Hermann M., Biehler K., Krieger-Liszkay A., Bock R. (2002). Lack of the small plastid-encoded PsbJ polypeptide results in a defective water-splitting apparatus of photosystem II, reduced photosystem I levels, and hypersensitivity to light. J. Biol. Chem. 277: 14031–14039 [DOI] [PubMed] [Google Scholar]
- Hilson P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res. 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippler M., Redding K., Rochaix J.-D. (1998). Chlamydomonas genetics, a tool for the study of bioenergetic pathways. Biochim. Biophys. Acta 1367: 1–62 [DOI] [PubMed] [Google Scholar]
- Hippler M., Rimbault B., Takahashi Y. (2002). Photosynthetic complex assembly in Chalmydomonas reinhardtii. Protist 153: 197–220 [DOI] [PubMed] [Google Scholar]
- Huang F.-C., Klaus S.M.J., Herz S., Zou Z., Koop H.-U., Golds T.J. (2002). Efficient plastid transformation in tobacco using the aphA-6 gene and kanamycin selection. Mol. Genet. Genomics 268: 19–27 [DOI] [PubMed] [Google Scholar]
- Jensen P.E., Haldrup A., Zhang S., Scheller H.V. (2004). The PSI-O subunit of plant photosystem I is involved in balancing the excitation pressure between the two photosystems. J. Biol. Chem. 279: 24212–24217 [DOI] [PubMed] [Google Scholar]
- Jordan P., Fromme P., Witt H.T., Klukas O., Saenger W., Krauß N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Kahlau S., Bock R. (2008). Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: Chromoplast gene expression largely serves the production of a single protein. Plant Cell 20: 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher D., Bock R. (2009). Identification of the chloroplast adenosine-to-inosine tRNA editing enzyme. RNA 15: 1251–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff H., Mukherjee U., Galla H.J. (2002). Molecular architecture of the thylakoid membrane: Lipid diffusion space for plastoquinone. Biochemistry 41: 4872–4882 [DOI] [PubMed] [Google Scholar]
- Kramer D.M., Johnson G., Kiirats O., Edwards G.E. (2004). New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynth. Res. 79: 209–218 [DOI] [PubMed] [Google Scholar]
- Krause G.H., Weis E. (1991). Chlorophyll fluorescence and photosynthesis: The basics. Annu. Rev. Plant Physiol. Plant Mol. Biol. 42: 313–349 [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lein W., Usadel B., Stitt M., Reindl A., Ehrhardt T., Sonnwald U., Börnke F. (2008). Large-scale phenotyping of transgenic tobacco plants (Nicotiana tabacum) to identify essential leaf functions. Plant Biotechnol. J. 6: 246–263 [DOI] [PubMed] [Google Scholar]
- Lohmann A., Schöttler M.A., Bréhélin C., Kessler F., Bock R., Cahoon E.B., Dörmann P. (2006). Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J. Biol. Chem. 281: 40461–40472 [DOI] [PubMed] [Google Scholar]
- Machold O., Simpson D.J., Moller B.L. (1979). Chlorophyll-proteins of thylakoids from wild-type and mutants of barley (Hordeum vulgare L.). Carlsberg Res. Commun. 44: 235–254 [Google Scholar]
- Meng B.Y., Tanaka M., Wakasugi T., Ohme M., Shinozaki K., Sugiura M. (1988). Cotranscription of the genes encoding two P700 chlorophyll a apoproteins with the gene for ribosomal protein CS14: Determination of the transcriptional initiation site by in vitro capping. Curr. Genet. 14: 395–400 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue culture. Physiol. Plant. 15: 473–497 [Google Scholar]
- Naver H., Boudreau E., Rochaix J.-D. (2001). Functional studies of Ycf3: Its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 13: 2731–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N., Ben-Shem A. (2004). The complex architecture of oxygenic photosynthesis. Nat. Rev. Mol. Cell Biol. 5: 1–12 [DOI] [PubMed] [Google Scholar]
- Obokata J., Mikami K., Hayashida N., Nakamura M., Sugiura M. (1993). Molecular heterogeneity of photosystem I. Plant Physiol. 102: 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa S.-I., Nield J., Terao A., Stauber E.J., Hippler M., Koike H., Rochaix J.-D., Takahashi Y. (2009). Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell 21: 2424–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394 [Google Scholar]
- Redding K., Cournac L., Vassiliev I.R., Golbeck J.H., Peltier G., Rochaix J.-D. (1999). Photosystem I is indispensable for photoautotrophic growth, CO2 fixation, and H2 photoproduction in Chlamydomonas reinhardtii. J. Biol. Chem. 274: 10466–10473 [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Rochaix J.-D. (1997). Chloroplast reverse genetics: New insights into the function of plastid genes. Trends Plant Sci. 2: 419–425 [Google Scholar]
- Rochaix J.-D., Fischer N., Hippler M. (2000). Chloroplast site-directed mutagenesis of photosystem I in Chlamydomonas: Electron transfer reactions and light sensitivity. Biochimie 82: 635–645 [DOI] [PubMed] [Google Scholar]
- Rogalski M., Karcher D., Bock R. (2008). Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 15: 192–198 [DOI] [PubMed] [Google Scholar]
- Rosahl S., Schell J., Willmitzer L. (1987). Expression of a tuber-specific storage protein in transgenic tobacco plants: Demonstration of an esterase activity. EMBO J. 6: 1155–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S., Kössel H., Bock R. (1997). Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 139: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Schöttler M.A., Bock R. (2008). Extranuclear inheritance: Plastid-nuclear cooperation in photosystem I assembly in photosynthetic eukaryotes. Prog. Bot. 69: 89–115 [Google Scholar]
- Schöttler M.A., Flügel C., Thiele W., Bock R. (2007a). Knock -out of the plastid-encoded PetL subunit results in reduced stability and accelerated leaf age-dependent loss of the cytochrome b6f complex. J. Biol. Chem. 282: 976–984 [DOI] [PubMed] [Google Scholar]
- Schöttler M.A., Flügel C., Thiele W., Stegemann S., Bock R. (2007b). The plastome-encoded PsaJ subunit is required for efficient photosystem I excitation, but not for plastocyanin oxidation in tobacco. Biochem. J. 403: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöttler M.A., Kirchhoff H., Weis E. (2004). The role of plastocyanin in the adjustment of the photosynthetic electron transport to the carbon metabolism in tobacco. Plant Physiol. 136: 4265–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabe T.M., Gloddek K., Schluesener D., Kruip J. (2003). Purification of recombinant BtpA and Ycf3, proteins involved in membrane protein biogenesis in Synechocystis PCC 6803. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 786: 45–59 [DOI] [PubMed] [Google Scholar]
- Schwenkert S., Netz D.J., Frazzon J., Pierik A.J., Bill E., Gross J., Lill R., Meurer J. (2010). Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 425: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen G., Antonkine M.L., von der Est A., Vassiliev I.R., Brettel K., Bittl R., Zech S.G., Zhao J., Stehlik D., Bryant D.A., Golbeck J.H. (2002a). Assembly of photosynthesis I. II. Rubredoxin is required for the in vivo assembly of Fx in Synechococcus sp. PCC 7002 as shown by optical and EPR spectroscopy. J. Biol. Chem. 277: 20355–20366 [DOI] [PubMed] [Google Scholar]
- Shen G., Zhao J., Reimer S.K., Antonkine M.L., Cai Q., Weiland S.M., Golbeck J.H., Bryant D.A. (2002b). Assembly of potosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J. Biol. Chem. 277: 20343–20354 [DOI] [PubMed] [Google Scholar]
- Stöckel J., Bennewitz S., Oelmüller R. (2006). The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol. 141: 870–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z., Hajdukiewicz P., Maliga P. (1990). Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA 87: 8526–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z., Maliga P. (1993). High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90: 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Goldschmidt-Clermont M., Soen S.-Y., Franzen L.G., Rochaix J.-D. (1991). Direct chloroplast transformation in Chlamydomonas reinhardtii: Insertional inactivation of the psaC gene encoding the iron-sulfur protein destabilizes photosystem I. EMBO J. 10: 2033–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker C., Schmidt D., Mueller-Roeber B., Czempinski K. (2006). Members of the Arabidopsis AtTPK/KCO family form homomeric vacuolar channels in planta. Plant J. 48: 296–306 [DOI] [PubMed] [Google Scholar]
- Walz C., Juenger M., Schad M., Kehr J. (2002). Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. Plant J. 31: 189–197 [DOI] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wessel D., Flügge U.-I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Wilde A., Lünser K., Ossenbühl F., Nickelsen J., Börner T. (2001). Characterization of the cyanobacterial ycf37: Mutation decreases the photosystem I content. Biochem. J. 357: 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman F.-A., Minai L., Nechushtai R. (1999). The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta 1411: 21–85 [DOI] [PubMed] [Google Scholar]
- Xu X.M., Adams S., Chua N.-H., Moller S.G. (2005). AtNAP1 represents an atypical SufB protein in Arabidopsis plastids. J. Biol. Chem. 280: 6648–6654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T., Morimoto K., Kikuchi S., Nishio K., Terashima I., Nakai M. (2004). The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16: 993–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]