Abstract
Phosphorylation of proteins is an essential signalling mechanism in eukaryotic and prokaryotic cells. Although N-phosphorylation of basic amino acid is known for its importance in biological systems, it is still poorly explored in terms of products and mechanisms. In the present study, two MS fragmentation methods, ECD (electron-capture dissociation) and CID (collision-induced dissociation), were tested as tools for analysis of N-phosphorylation of three model peptides, RKRSRAE, RKRARKE and PLSRTLSVAAKK. The peptides were phosphorylated by reaction with monopotassium phosphoramidate. The results were confirmed by 1H NMR and 31P NMR studies. The ECD method was found useful for the localization of phosphorylation sites in unstable lysine-phosphorylated peptides. Its main advantage is a significant reduction of the neutral losses related to the phosphoramidate moiety. Moreover, the results indicate that the ECD–MS may be useful for analysis of regioselectivity of the N-phosphorylation reaction. Stabilities of the obtained lysine-phosphorylated peptides under various conditions were also tested.
Keywords: electron capture dissociation (ECD), phosphorylation, phospholysine, post-translational modification
Abbreviations: CID, collision-induced dissociation; ECD, electron capture dissociation; ESI–MS, electrospray ionization MS; ETD, electron transfer dissociation; HMBC, heteronuclear multiple bond correlation; PKG, protein kinase G
INTRODUCTION
Protein phosphorylation belongs to the most important post-translational modifications, is involved in many signal transduction pathways and plays a significant role in mechanisms responsible for the regulation of cellular functions. The O-phosphorylation of serine, threonine and tyrosine residues has been extensively studied, since the high stability of O-phosphorylated products has permitted the analysis of this modification with many different techniques [1], especially MS [2,3].
However, other amino acid residues also undergo phosphorylation, among them the basic amino acids histidine, lysine and arginine. With the latter amino acid residues, the modification results in the formation of a phosphoramidate bond (P–N bond), which is highly unstable under acidic conditions and easily undergoes hydrolysis. Therefore, N-phosphorylated proteins are difficult to analyse, and detection of N-phosphorylated peptides using MS creates problems due to low abundance [4] or spontaneous gas-phase dephosphorylation [5].
Protein N-phosphorylation, compared with O-phosphorylation, is still poorly explored in terms of products and mechanisms. A considerable interest has been focused on protein histidine phosphorylation, recognized as an important modification in prokaryotes and eukaryotes, specific histidine kinases being involved in signalling systems called the two-component regulatory systems [6–8]. Histidine was also the first synthetically N-phosphorylated basic amino acid whose chemical properties have been well studied [9]. The histidine modification, in spite of susceptibility to acid hydrolysis, could be analysed using MS both in peptides and proteins, using different analytical techniques such as LC–MS (liquid chromatography MS), ESI–MS (electrospray ionization MS) and MALDI–TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight MS), and different fragmentation methods, mainly CID (collision-induced dissociation), ETD (electron transfer dissociation), EDD (electron detachment dissociation) and ECD (electron capture dissociation) [10–13].
Protein N-phosphorylation may also occur on arginine and lysine residues (reviewed recently by Besant et al. [14]). The presence of protein arginine and lysine kinases in cells has been reported [15]. While several reports concerned arginine kinases involved in cellular metabolic pathways [16,17], N-phosphorylation of the lysine side chain has been much less investigated. Specific protein lysine kinases and the corresponding phosphatases (phosphoramidases) were found in eukaryotic cells [14], e.g. rat liver nuclear protein arginine kinase with an additional lysine kinase activity [18], protein lysine kinases involved in histone H1 phosphorylation [19] and bovine liver protein lysine phosphatases [20]. The involvement of lysine phosphorylation in cellular signalling or metabolic pathways, albeit possible, has not been shown yet.
Although lysine phosphorylation is an important protein modification, it is difficult to detect by ESI–MS. The main reason is the low abundance of the phosphopeptide ions as compared with their non-modified counterparts. Usually, formic acid or acetic acid is added to aid protonation of the analyte molecules during electrospray. Therefore, relatively rapid hydrolysis of the phosphoramidate group under acidic conditions may also complicate the application of ESI–MS to the analysis of phospholysine-containing peptides.
MS has become an alternative to the more traditional methods, e.g. NMR, two-dimensional gel electrophoresis, HPLC analysis and proteolytic digestion using Edman degradation of 32P-labelled peptides, for analysis of phosphorylated peptides [21]. The ESI–MS method may be particularly useful to evaluate the extent of phosphorylation in a protein molecule. In contrast with other analytical methods, MS experiments can be performed successfully even on sub-picomolar amounts of a mixture of proteins, as long as the analytes possess different molecular mosses. Moreover, MS fragmentation techniques may be used to characterize the modification sites with the amino acid residue resolution.
Although the most common method of fragmentation, CID, is very useful for the sequencing of O-phosphorylated peptides, its application to N-phosphorylation is limited as a consequence of considerable losses of the phosphate group HPO3− from the phosphoramidate bond even at relatively low collision energy conditions. Therefore, the resulting CID spectra are dominated by the dephosphorylated forms of the fragments.
ECD performed on Fourier transform mass spectrometers was shown to be useful for the characterization of labile post-translational protein modification [22], including non-enzymatic modifications of basic side chains [11,23]. No fragmentation of phosphorylated histidine residues was observed during ECD experiments [24,25]. This suggests that ECD may be a useful method for analysis of peptides containing the N-phosphorylated lysine side chain, although such an analysis has not been reported to date. Recently, we used the ECD method for analysis of distribution of deuterium along the sequence of a protein molecule undergoing the hydrogen exchange under conditions of a high-pressure denaturation [26]. Although fragmentation of deuterium-labelled compounds using the CID method is known to indicate the migration of deuterons (hydrogen scrambling) [27], which makes the analysis impractical, we found that the ECD fragmentation allows for ambiguous recognition of deuterated peptide bonds in a protein molecule. The current study presents an analysis using ESI–MS of the lysine phosphorylation products obtained by the reaction of peptides with monopotassium phosphoramidate. Stabilities of peptides containing N-phosphorylated lysine in solution were also analysed. The comparison between the two MS fragmentation types, CID and ECD, pointed to ECD as a better method for localization of N-phosphorylation on the lysine residue. In addition, the applicability of ECD–MS was tested to study the regioselectivity of peptide N-phosphorylation.
MATERIALS AND METHODS
Materials
Peptides
Three peptides, known to be the substrates or inhibitors of specific protein kinases (Table 1), were chosen for the investigation. Each peptide contained one or two lysine residues localized at different positions in the sequence. All analysed peptides were purchased from Sigma–Aldrich.
Table 1. Model peptides used in experiments and their biological activity.
| Peptide sequence | Biological activity | References |
|---|---|---|
| RKRSRAE | Selective substrate of PKG (protein kinase G) with a strong preference for PKG Iα over PKG II | [30] |
| RKRARKE | An inhibitor of the cGMP (cyclic guanosine monophosphate)dependent protein kinase | [31] |
| PLSRTLSVAAKK | A part of a glycogen synthase sequence; an effective substrate of CaM (calmodulin) kinase II and PKC (protein kinase C) | [32,33] |
Peptide N-phosphorylation
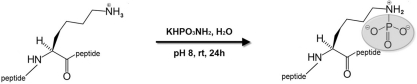
Peptide N-phosphorylation was performed using a protocol previously described by Wei and Matthews [28]. The phosphorylation agent, monopotassium phosphoramidate, was prepared by the classical Stokes' method [29]. In the standard procedure, peptide and monopotassium phosphoramidate were used at a ratio of 1:40 (w/w). The peptide (0.5 mg) sample was dissolved in 10 mM ammonium bicarbonate and the pH of the solution was adjusted to 8 with 0.1 M NaOH. After the addition of 20 mg of monopotassium phosphoramidate, the mixture sample was stirred for 24 h at room temperature (22°C) (Figure 1).
Figure 1. Scheme of the peptide phosphorylation on the ϵ-amino group of lysine.
rt, room temperature.
Sample preparation
For the MS experiments, the phosphorylated sample was desalted using Sep-Pak Plus C18 Cartridges (Waters Corporation). The column was prepared by washing alternately with acetonitrile (POCH, Poland, HPLC grade) and deionized water. The sample was loaded directly on a Sep-Pak column and washed five times with small portions of the deionized water. The phosphorylated peptides were eluted with 60% aqueous solution of acetonitrile (1 ml).
Methods
ESI–MS
All MS experiments were performed on an Apex-Qe Ultra 7T instrument (Bruker Daltonics, Bremen, Germany) equipped with a dual ESI source and a heated hollow cathode dispenser. The instrument was operated in the positive-ion mode and calibrated with the Tunemix™ mixture (Bruker Daltonics). The mass accuracy was better than 5 ppm. Analysis of the obtained mass spectra was carried out using a Biotools (Bruker Daltonics) software. The instrumental parameters were as follows: scan range, 100–1600 m/z; dry gas, nitrogen; temperature, 200°C; potential between the spray needle and the orifice, 4.2 kV.
CID
The precursor ions were selected on the quadrupole and subsequently fragmented in the hexapole collision cell. Argon was used as a collision gas. The obtained fragments were directed to the ICR mass analyser and registered as an MS/MS (tandem MS) spectrum. The collision energy of 20 V was applied in the hexapole collision cell.
ECD
The mass spectrometer was equipped with a heated hollow cathode dispenser, which was operated at 1.7 A for the ECD experiments. The precursor ions were selected on the quadrupole and directed to the ICR cell where they were fragmented. The parameters were set as 150 ms for the ECD pulse length, and ECD bias was 0.8 V.
NMR analysis
All NMR spectra were obtained with a Bruker Avance spectrometer operating in the quadrature mode at 500.13 MHz for 1H and 202.46 MHz for 31P nuclei. The residual peaks of deuterated solvents were used as internal standards in the 1H NMR method. 31P NMR spectra were recorded at 277 K, both with and without proton decoupling. The internal standard used in 31P NMR was inorganic phosphate (Pi), showing resonance at 2.15 ppm (pH 7.8), 2.05 ppm (pH 7.5), 1.65 ppm (pH 5.0) and 0.0 ppm (pH 1.5). Additionally, chemical shifts were checked with external reference (85% H3PO4). All samples were analysed using the gradient-enhanced 1H-31P HMBC (heteronuclear multiple bond correlation) method, with the HMBC experiments optimized for long-range couplings using different 3JP-H values (1–20 Hz). The 1H NMR spectra were obtained with and without the use of the HDO suppression method. All buffer solutions used for NMR spectroscopy were based on deuterium oxide of 100% 2H purity (Armar Chemicals AG).
RESULTS AND DISCUSSION
Three model peptides were phosphorylated with monopotassium phosphoramidate and analysed using MS, the ESI–MS spectra confirming that the reaction yielded N-phosphorylated products. The peptides are known substrates or inhibitors of protein kinases (Table 1). All lysine-phosphorylated peptides were stable in 60% aqueous acetonitrile solution (pH 7) at room temperature (22°C). No degradation product was observed in the ESI–MS spectra if the samples were incubated at 4°C for 1 week. On the other hand, the phosphorylated peptides were unstable under acidic conditions. At pH 2–3 (in 10% formic acid), the half-time for phosphopeptide at room temperature was approx. 20 min. Thus, the lysine-phosphorylated peptide stability was similar to that described previously for phosphohistidine [12].
The possibility of arginine side-chain phosphorylation under the conditions employed was excluded by various NMR and ESI–MS experiments (not described in the Materials and methods section). We performed the NMR-controlled pH-dependent phosphorylation experiments under conditions similar to those described in the present paper. Our results clearly showed that the 31P NMR spectrum of arginine-phosphoramidate post-reaction mixture contains no phosphoarginine resonance (singlet resonance) over a pH range from 4.0 to 9.0. The traces of phosphoarginine product were found only for reaction mixtures incubated at pH 10 and above (we analysed the pH range 3–12), whereas experiments conducted for lysine proved that phospholysine (triplet resonance) reacts with phosphoramidate ions over the pH range 5–12. A similar conclusion was also obtained on the basis of our ESI–MS experiments. Therefore, the chemical phosphorylation employing monopotassium phosphoramidate in pH 8 takes place exclusively at amino groups, even if arginine residues are present in the peptide chain.
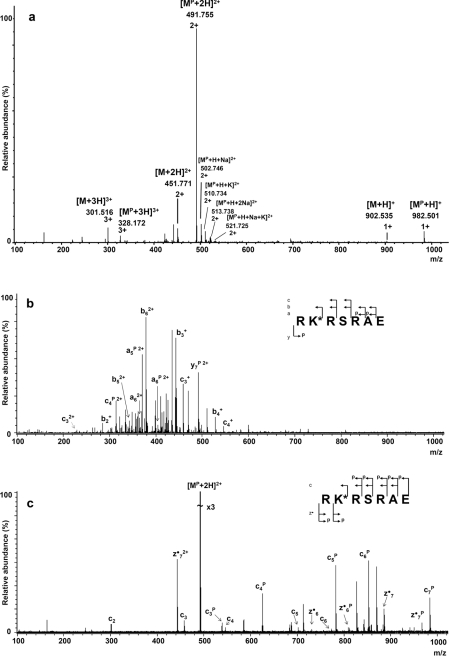
The ESI–MS spectra (Figure 2a) of the phosphorylated RKPRSRAE peptide (where KP is the phosphorylated lysine residue), recorded in the positive-ion mode, present the main peak at m/z 491.755 corresponding to the monophosphorylated compound ([MP+2H]2+). In these spectra, the abundant peak of the phosphorylated peptide is accompanied by its unphosphorylated form. There are additional peaks at m/z: 502.746, 510.734, 513.738 and 521.725 that correspond to 2+ ions of the phosphopeptide containing metal cations: [MP+H+Na]2+, [MP+H+K]2+, [MP+2Na]2+ and [MP+K+Na]2+ respectively. Interestingly, no metal adducts were apparent in the unphosphorylated peptide spectrum. This may suggest that the presence of the phosphoramidate group increases the affinity of the peptide towards the metal ions. The latter effect is observed also in the ESI–MS spectra of the other analysed model peptides.
Figure 2. ESI–MS spectra of the phosphorylated RKPRSRAE peptide.
(a) ESI–MS spectrum recorded in the positive-ion mode, (b) CID fragmentation spectrum (precursor ion m/z 491.7) and (c) ECD fragmentation spectrum (precursor ion m/z 491.7). K* represents possible phosphorylation on the lysine ε-amino group. The fragmentation pattern is shown in each spectrum. cn- and z•n-fragmentation ions with the index P present the phosphorylated fragment of the peptide.
The most abundant peak at 491.7 m/z, corresponding to the [MP+2H]2+ ion, was chosen as a precursor for the fragmentation both by CID and ECD. Figure 2(b) shows the CID spectrum of the products obtained by phosphorylation of RKRSRAE peptide. The number of identified fragments is not sufficient to establish the phosphorylation site. This may be due to the extensive neutral losses of HPO3 (80 Da) and water molecule (18 Da). The most abundant peak corresponds to the loss of 98 Da. Although phosphoramidates cannot directly eliminate H3PO4 molecules because of merely three oxygen atoms being available in the amidate, a concerted loss of one H3PO4 molecule from N-phosphorylated peptides cannot be excluded. Recently, Kleinnijenhuis et al. [12] observed also that collisional activation of the histidine-phosphorylated peptide results in the extensive loss of H3PO4. The authors proved the origin of the lost water to be an aspartic acid residue adjacent to the phosphorylated histidine residue. Therefore, although the phosphorylated lysine residue in RKPRSRAE is surrounded by arginine residues, the eliminated water molecule may derive from the Ser4 or Glu7 residues.
The spectrum in Figure 2(b) is dominated by ions corresponding to the elimination of HPO3 and the water molecule, whereas the abundances of the backbone fragments are relatively low, with many unidentified fragments observed. The CID spectrum is difficult to analyse and the localization of the peptide phosphorylation site is complicated.
A different fragmentation pattern was observed in the spectrum obtained following the ECD fragmentation of RKPRSRAE (Figure 2c). In contrast with CID, the ECD fragmentation of the [MP+2H]2+ ion retains the phosphorylation of the majority of fragments containing Lys2. On the other hand, in the obtained cn- and zn•-series of ions, phosphorylated and unphosphorylated species are observed, indicating that even the ECD fragmentation causes certain level of dephosphorylation. In spite of a higher abundance of the fragments containing phospholysine and domination of the corresponding peaks in the spectrum, a partial dephosphorylation and the lack of certain ions make the spectrum interpretation difficult. The ECD fragmentation pattern suggests that the phosphoramidate group is located mainly on the lysine side chain, although a partial phosphorylation of the N-terminal amino group cannot be excluded.
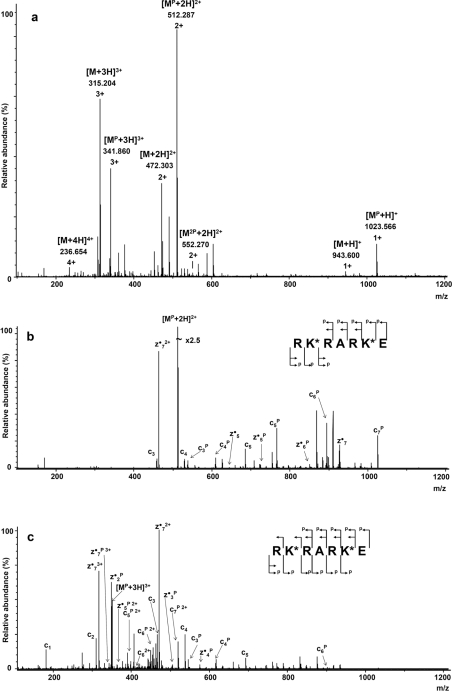
The analysed peptide of the RKRARKE sequence encompasses two lysine residues – Lys2 and Lys6. Although the peptide may be expected to yield a doubly phosphorylated product, following the reaction with an excess of the phosphorylating agent, the ESI–MS spectrum (presented in Figure 3a) shows that the predominant peak corresponds to a singly phosphorylated product. To simplify interpretation of the spectrum, the ESI–MS analysis was performed using 2% formic acid as a solvent preventing metal ion co-ordination to the phosphopeptide, observed at higher pH values. Although the phosphoramidate bond is known to hydrolyse quickly under acidic conditions, we found the phosphopeptides to be stable for long enough to perform the MS experiment (t1/2~30 min at room temperature).
Figure 3. ESI–MS spectrum of RKPRARKPE.
(a) Mass spectrum of the phosphorylation products of RKRARKE, (b) ECD fragmentation spectrum (precursor ion m/z 512.3, [MP+2H]2+) and (c) ECD fragmentation spectrum (precursor ion m/z 341.9, [MP+3H]3+). K* represents a possible phosphorylated lysine residue.
The mass spectrum (Figure 3a) does not show any peaks coming from the metal-ion adducts. The main peaks result from the ionization of the unphosphorylated [m/z 472.303 (+2)], monophosphorylated [m/z 512.287 (+2)] and diphosphorylated [m/z 552.270 (+2)] peptide. The unphosphorylated peptide ion dominates (m/z 315.20) in its triply protonated form. The intensity of the diphosphorylated peptide ions [represented by m/z 552.27 (+2) and 369.24 (+3)] is low. The latter phenomenon may be a result of either the phosphorylation procedure used being capable of modifying only one lysine residue in RKRARKE or the neutralization of the molecule's overall charge by the phosphorylation, which makes the ionization more difficult.
The ECD experiment allowed us to localize the phosphorylation site in doubly and triply charged precursor ions (Figures 3b and 3c respectively) of the monophosphopeptide. The ECD analysis performed on the doubly charged ion ([MP+2H]2+, Figure 3b) is represented predominantly by three series of fragment ions: a longer series of singly phosphorylated ions zn•P cnP, and shorter series of non-phosphorylated cn and zn ions. The mass peaks, corresponding to the species containing the phosphoramidate group, are characterized by a higher abundance.
The ECD fragmentation, performed on the triply charged, singly phosphorylated peptide ion ([MP+3H]3+) (Figure 3c), resulted in a better sequence coverage. The cn- and zn•-series are almost complete and the number of identified phosphorylated fragment ions is sufficient to sequence the peptide and to localize the phosphorylation site. The presence of a series of peaks in the spectrum, corresponding to z2•P, z3•P, z4•P and z5•P, suggests that the phosphorylation site is located on Lys6, whereas the presence of peaks corresponding to c3P, c4P and c5P, indicates that the phosphoramidate may be also present on Lys2. Thus, the ECD fragmentation spectrum indicates both lysine residues to undergo N-phosphorylation. The peptide's chemical modification does not appear regioselective, albeit the phosphorylation levels of Lys2 and Lys6 are different and the relative intensity of fragment ions suggests the modification of Lys6 to be preferred.
The third model peptide, PLSRTLSVAAKK, contains a sequence of two lysine residues located at the C-terminus (Lys11 and Lys12). The positions of lysine moieties might be expected to influence the N-phosphorylation regioselectivity. The ESI–MS spectrum presents the peak masses derived from the monophosphorylated PLSRTLSVAAKK peptide, with the measurement done in 2% formic acid to aid the peptide protonation. Because of a relatively rapid hydrolysis of the phosphoramidate, the mass experiment was performed without delay just after the addition of the acid.
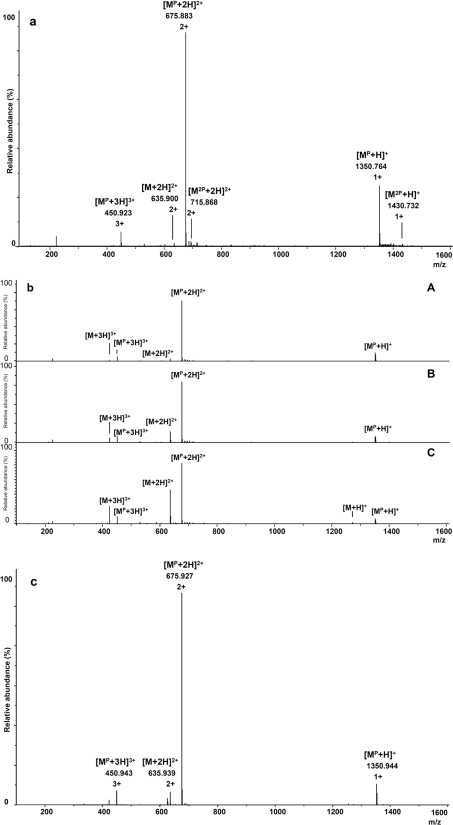
In the ESI–MS spectrum (Figure 4a), only peaks corresponding to the singly phosphorylated peptide are observed, with the abundance of non-phosphorylated and diphosphorylated peaks being low. The main peak in the spectrum at m/z 675.883 represents a doubly charged, singly phosphorylated peptide [MP+2H]2+. Two other forms, singly and triply charged, are apparent at m/z 1350.754 and 450.923 respectively. It is of note that the phosphorylated peptide is relatively resistant towards hydrolysis and does not lose the phosphoramidate group in 2% formic acid even after 20 min.
Figure 4. ESI–MS spectra of the products of phosphorylation of the PLSRTLSVAAKK peptide.
(a) ESI–MS spectrum in positive-ion mode in 2% formic acid. (b) ESI–MS spectra in 10% formic acid after: A, 2 min; B, 10 min; and C, 20 min. (c) ESI–MS spectrum of the phosphorylated peptide sample after 6 months storage at −20°C.
To check the resistance of the modification in phosphorylated PLSRTLSVAAKK, the sample was incubated with 10% formic acid. The ESI–MS spectra, recorded after 2, 10 and 20 min (Figure 4b), show evolution with time pointing to an apparent slow dephosphorylation of the peptides. While the singly phosphorylated form is still predominant after 2 min, the following 20 min incubation period results in an increase in the peaks corresponding to the unphosphorylated species. We also examined the stability of the phosphorylated lysine residues in the phosphopeptide solution (60% aqueous acetonitrile) stored at −20°C. Surprisingly, the ESI–MS spectrum, recorded for the lysine-phosphorylated peptides tested, remains unchanged even after a 6-month incubation period (Figure 4c), indicating that phospholysine containing peptides can be stored at low temperature and neutral pH.
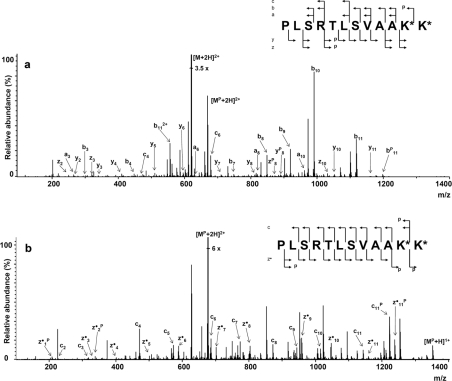
To check whether the peptide was N-phosphorylated regioselectively, fragmentation was performed using the CID and ECD methods (Figures 5a and 5b). The doubly charged, singly phosphorylated ion (MP+2H)2+ of the PLSRTLSVAAKK peptide was chosen (m/z 675.883) for the fragmentations. The CID spectrum shows, beside a majority of all ions derived from the nonphosphorylated form, only three peaks corresponding to the phosphorylated peptide fragments and several unidentified peaks. Although the number of fragment ions is sufficient for the peptide sequencing, it is difficult to establish the phosphorylation site. The ECD spectrum provides much more information. The resulting cn- and zn•-fragment ions cover 100% of the peptide sequence. Furthermore, there are four identified fragment ions, bearing the phosphoramidate moiety. The intensity of certain ions belonging to the zn•P series was very low and, therefore, the corresponding peaks were not pointed in Figure 5(b). The presence of the phosphorylated c11P ion proves that Lys11 is phosphorylated, at least partially. On the other hand, the occurrence of the z1•P phosphorylated ion can be explained only assuming a partial phosphorylation of Lys12. Thus, the analysis of the ECD spectrum reveals that both Lys11 and Lys12 moieties are phosphorylated to some extent. Fragmentation of the PLSRTLSVAAKK peptide phosphorylation products, performed by the ECD method, results in a better sequence coverage and significantly higher retention of the phosphoramidate group, as compared with the CID method. On the other hand, the fragmentation behaviour of the lysine-phosphorylated peptide reflects a high lability of the phosphoramidate moieties. Even with the ECD technique, significant phosphate-related losses are observed. This observation correlates well with the recently reported electron-based dissociation (ECD and ETD) of phosphorylated histidine in polypeptides [11]. Our results demonstrate that, although in the process of electron-based dissociation the phosphorylated lysine residue shows a lability similar to that of the phosphorylated histidine residue, it may be sequenced using ECD. On the other hand, the dephosphorylation level observed during the ECD fragmentation is surprisingly high. The electron-based fragmentation methods are believed to be extremely selective with respect to peptide bonds [34], and consequently, the ECD should not influence the modifications of peptides. Our results suggest a relatively low stability of the phosphoramidate bond in the gas phase, compared with other known post-translational modifications.
Figure 5. Fragmentation spectra of the phosphorylated PLSRTLSVAAKK peptide.
(a) CID fragmentation spectrum (precursor ion m/z 675.9; [MP+2H]2+) and (b) ECD fragmentation spectrum (precursor ion m/z 675.9; [MP+2H]2+). K* represents a possible phosphorylated lysine residue.
NMR analysis was performed on unphosphorylated and phosphorylated peptides, RKRARKE and PLSRTLSVAAKK, to confirm correctness of the ECD–MS analysis. The 1H NMR chemical shift of the RKRARKE lysine ε-CH2 moiety found in the non-phosphorylated peptide spectrum is approx. 2.93 ppm. The 1H NMR spectrum of the phosphorylated RKRARKE peptide, compared with non-phosphorylated RKRARKE peptide, shows clearly a new multiplet resonance at 3.08 ppm, strongly correlated with that of ε-CH2 of the phosphorylated lysine moiety. The observed downfield shift of the ε-CH2 moiety of the phosphorylated form (0.15–0.25 ppm) is typical of phosphorylated/non-phosphorylated aliphatic amino acid side-chain systems [35,36]. The 31P NMR spectrum of the phosphorylated peptide shows a resonance at approx. 7.0 ppm.
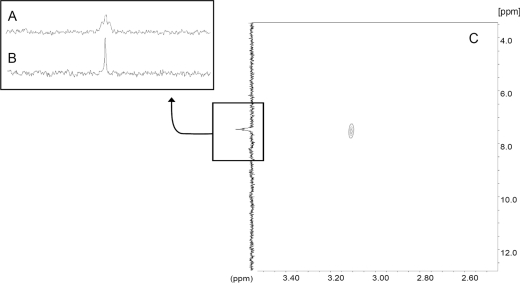
The 1H NMR spectrum of phosphorylated peptide PLSRTLSVAAKK contains a multiplet resonance at 3.06 ppm, corresponding to the phospholysine ε-CH2 moiety, and the resonances of the non-phosphorylated lysine ε-CH2 moiety visible at 2.90 ppm, with relative integrals of 1:1 respectively. The 31P NMR spectrum of the phosphorylated peptide clearly shows a new triplet resonance at 7.6 ppm (3JP-H=6.8 Hz, Figure 6), with the coupling constants in the range typical of the three-bond P–H couplings [37,38]. It is of note that the latter resonance appears to correspond to a single form of phospholysine, as with proton-decoupling applied. The 31P NMR spectrum shows a single, narrow (half-width 2.67 Hz), symmetrical singlet resonance at 7.6 ppm (Figure 6). The previously mentioned triplet form of the resonance in the 31P NMR spectrum results apparently from the heteronuclear coupling of phosphate phosphorus with two magnetically equivalent hydrogen atoms of the ε-CH2 methylene group of the N-phosphorylated lysine moiety of the PLSRTLSVAAKK peptide.
Figure 6. 31P NMR spectrum (A), 31P NMR spectrum with proton decoupling (B) and 1H-31P HMBC spectrum (C) of the PLSRTLSVAAKK peptide KP moiety.
The gradient-enhanced 1H-31P HMBC (see Figure 6c) proved that the above-mentioned P-coupled methylene hydrogen atoms show 1H NMR shifts of approx. 3.1 ppm, in agreement with the 1H NMR spectrum of the phosphorylated PLSRTLSVAAKK peptide. In view of the analysis of COSY and 1H-13C HSQC (heteronuclear single-quantum coherence) spectra of the phosphorylated peptide, as well as the data on several homopeptide derivatives [39], the α-CH proton of the lysine moiety with a free carboxylic group (Lys12, δα-CH=3.74 ppm) should not have the phosphate group at the side chain. Consequently, the other lysine moiety (Lys11, δα-CH=3.80 ppm) must have the phosphoramidate group in the ε-CH2 region. In view of the analysis of the NMR data, the presence of the O-phosphoserine moiety in the analysed peptides is not apparent. The three-bond coupling between serine CH2 and phosphorus would result in a two-doublets resonance pattern. Moreover, the 1H-31P HMBC spectra indicate the phosphoserine-, and also thiophosphoserine-CH2, group to show 1H chemical shifts in the 3.6–4.2 ppm range [35,36,40]. Furthermore, the observed resonances do not belong to the phosphorylated arginine or glutamate moieties, as would be expected with those singlet resonances in 31P NMR. Finally, the triplet resonances could not originate from the phosphorylated N-terminal end of a peptide, as a doublet 31P NMR resonance would have to result from the proximity of the α-CH moiety.
It is of note that, while in view of the NMR results the reaction appears rather regiospecific, the ECD spectra suggest modification of both Lys11 and Lys12. In our opinion, the discrepancy results from different characteristics of the two methods, with the former providing quantitative results, but not always able to detect minor constituents, and the latter allowing a higher sensitivity, but not necessarily a quantitative evaluation.
CONCLUSIONS
Comparative MS analyses are presented, accompanied by 1H NMR and 31P NMR studies, of model peptides non-phosphorylated and N-phosphorylated on lysine residue(s). Although the modified peptides, monitored by MS, proved labile under acidic conditions in accordance with the well-known phosphoramidate acid-lability, neutral solutions of the peptides could be stored for a long time at −20°C without losing the phosphoramide group.
Two fragmentation methods, ECD and CID, were compared as tools for the analysis of the N-phosphorylation products. The results pointed to the ECD fragmentation as an advantageous method, allowing a significant reduction of the neutral losses related to the phosphoramidate moiety and permitting, in most cases, the localization of phosphorylation sites. On the other hand, the lysine-phosphorylated peptides are relatively unstable in a vacuum, and localization of the phosphorylation site, even using the ECD method, should be done with caution.
FUNDING
This work was supported by the Ministry of Science and Higher Education [grant number N401 2334 34].
References
- 1.Raggiaschi R., Gotta S., Terstappen G. C. Phosphoproteome analysis. Biosci. Rep. 2005;25:33–44. doi: 10.1007/s10540-005-2846-0. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P. The origins of protein phosphorylation. Nature Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 3.Marks F. Protein Phosphorylation. York: Wiley-VCH, New; 1996. [Google Scholar]
- 4.Smith J. R., Olivier M., Greene A. S. Relative quantification of peptide phosphorylation in a complex mixture using 18O labeling. Physiol. Genomics. 2007;31:357–363. doi: 10.1152/physiolgenomics.00096.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLachlin D. T., Chait B. T. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr. Opin. Chem. Biol. 2001;5:591–602. doi: 10.1016/s1367-5931(00)00250-7. [DOI] [PubMed] [Google Scholar]
- 6.Besant P. G., Attwood P. V. Mammalian histidine kinases. Biochim. Biophys. Acta. 2005;1754:281–290. doi: 10.1016/j.bbapap.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 7.Hoch J. A. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 8.Thomason P., Kay R. Eukaryotic signal transduction via histidine-aspartate phosphorelay. J. Cell Sci. 2000;113:3141–3150. doi: 10.1242/jcs.113.18.3141. [DOI] [PubMed] [Google Scholar]
- 9.Attwood P. V., Piggott M. J., Zu X. L., Besant P. G. Focus on phosphohistidine. Amino Acids. 2007;32:145–156. doi: 10.1007/s00726-006-0443-6. [DOI] [PubMed] [Google Scholar]
- 10.Zu X. L., Besant P. G., Imhof A., Attwood P. V. Mass spectrometric analysis of protein histidine phosphorylation. Amino Acids. 2007;32:347–357. doi: 10.1007/s00726-007-0493-4. [DOI] [PubMed] [Google Scholar]
- 11.Wind M., Wegener A., Kellner R., Lehmann W. D. Analysis of CheA histidine phosphorylation and its influence on protein stability by high-resolution element and electrospray mass spectrometry. Anal. Chem. 2005;77:1957–1962. doi: 10.1021/ac040140h. [DOI] [PubMed] [Google Scholar]
- 12.Kleinnijenhuis A. J., Kjeldsen F., Kallipolitis B., Haselmann K. F., Jensen O. N. Analysis of histidine phosphorylation using tandem MS and ion-electron reactions. Anal. Chem. 2007;79:7450–7456. doi: 10.1021/ac0707838. [DOI] [PubMed] [Google Scholar]
- 13.Besant P. G., Attwood P. V. Detection and analysis of protein histidine phosphorylation. Mol. Cell. Biochem. 2009;329:93–106. doi: 10.1007/s11010-009-0117-2. [DOI] [PubMed] [Google Scholar]
- 14.Besant P. G., Attwood P. V., Piggott M. J. Focus on phosphoarginine and phospholysine. Curr. Protein Pept. Sci. 2009;10:536–550. doi: 10.2174/138920309789630598. [DOI] [PubMed] [Google Scholar]
- 15.Matthews H. R., Huebner V. D. Nuclear protein kinases. Mol. Cell. Biochem. 1984;59:81–99. doi: 10.1007/BF00231306. [DOI] [PubMed] [Google Scholar]
- 16.Wakim B. T., Aswad G. D. Ca2+-calmodulin-dependent phosphorylation of arginine in histone 3 by a nuclear kinase from mouse leukemia cells. J. Biol. Chem. 1994;269:2722–2727. [PubMed] [Google Scholar]
- 17.Uda K., Fujimoto N., Akiyama Y., Mizuta K., Tanaka K., Ellington W. R., Suzuki T. Evolution of the arginine kinase gene family. Comp. Biochem. Physiol. D. 2006;1:209–218. doi: 10.1016/j.cbd.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Sikorska M., Whitfield J. F. Isolation and purification of a new 105 kDa protein kinase from rat liver nuclei. Biochim. Biophys. Acta. 1982;703:171–179. doi: 10.1016/0167-4838(82)90045-0. [DOI] [PubMed] [Google Scholar]
- 19.Matthews H. R. Protein kinases and phosphatases that act on histidine, lysine or arginine residues in eukaryotic proteins: a possible regulator of the mitogen-activated protein kinase cascade. Pharmacol. Ther. 1995;67:323–350. doi: 10.1016/0163-7258(95)00020-8. [DOI] [PubMed] [Google Scholar]
- 20.Hiraishi H., Yokoi F., Kumon A. 3-Phosphohistidine and 6-phospholysine are substrates of a 56-kDa inorganic pyrophosphatase from bovine liver. Arch. Biochem. Biophys. 1998;349:381–387. doi: 10.1006/abbi.1997.0480. [DOI] [PubMed] [Google Scholar]
- 21.Yan J. X., Packer N. H., Gooley A. A., Williams K. L. Protein phosphorylation: technologies for the identification of phosphoamino acids. J. Chromatogr. A. 1998;808:23–41. doi: 10.1016/s0021-9673(98)00115-0. [DOI] [PubMed] [Google Scholar]
- 22.Syrstad E. A., Turecek F. Toward a general mechanism of electron capture dissociation. J. Am. Soc. Mass. Spectrom. 2005;16:208–224. doi: 10.1016/j.jasms.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Stefanowicz P., Kijewska M., Szewczuk Z. Sequencing of peptide-derived Amadori products by the electron induced dissociation method. J. Mass. Spectrom. 2009;44:1047–1052. doi: 10.1002/jms.1580. [DOI] [PubMed] [Google Scholar]
- 24.Shi S. D., Hemling M. E., Carr S. A., Horn D. M., Lindh I., McLafferty F. W. Phosphopeptide/phosphoprotein mapping by electron capture dissociation mass spectrometry. Anal. Chem. 2001;73:19–22. doi: 10.1021/ac000703z. [DOI] [PubMed] [Google Scholar]
- 25.Sweet S. M., Bailey C. M., Cunningham D. L., Heath J. K., Cooper H. J. Large scale localization of protein phosphorylation by use of electron capture dissociation mass spectrometry. Mol. Cell. Proteomics. 2009;8:904–912. doi: 10.1074/mcp.M800451-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stefanowicz P., Petry-Podgorska I., Kowalewska K., Jaremko L., Jaremko M., Szewczuk Z. Electrospray ionization mass spectrometry as a method for studying the high-pressure denaturation of proteins. Biosci. Rep. 2010;30:91–99. doi: 10.1042/BSR20090015. [DOI] [PubMed] [Google Scholar]
- 27.Rand K. D., Jørgensen T. J. D. Development of a peptide probe for the occurrence of hydrogen (H/H) scrambling upon gas phase fragmentation. Anal. Chem. 2007;79:8686–8693. doi: 10.1021/ac0710782. [DOI] [PubMed] [Google Scholar]
- 28.Wei Y. F., Matthews H. R. Identification of phosphohistidine in proteins and purification of protein-histidine kinases. Methods Enzymol. 1991;200:388–414. doi: 10.1016/0076-6879(91)00156-q. [DOI] [PubMed] [Google Scholar]
- 29.Stokes H. N. On diamidoorthophosphoric and diamidotrihydroxylphosphoric acids. Am. Chem. J. 1893;15:198–214. [Google Scholar]
- 30.Hall K. U., Collins S. P., Gamm D. M., Massa E., DePaoli-Roach A. A., Uhle M. D. Phosphorylation-dependent inhibition of protein phosphatase-1 by G-substrate. J. Biol. Chem. 1999;274:3485–3495. doi: 10.1074/jbc.274.6.3485. [DOI] [PubMed] [Google Scholar]
- 31.Glass D. B. Differential responses of cyclic GMP-dependent and cyclic AMP-dependent protein kinases to synthetic peptide inhibitors. Biochem. J. 1983;213:159–164. doi: 10.1042/bj2130159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.House C., Kemp B. E. Protein kinase C contains a pseudosubstrate prototope in its regulatory domain. Science. 1987;238:1726–1728. doi: 10.1126/science.3686012. [DOI] [PubMed] [Google Scholar]
- 33.Alexander D. R., Graves J. D., Lucas S. C., Cantrell D. A., Crumpton M. J. A method for measuring protein kinase C activity in permeabilized T lymphocytes by using peptide substrates. Evidence for multiple pathways of kinase activation. Biochem. J. 1990;268:303–308. doi: 10.1042/bj2680303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelleher N. L., Zubarev R. A., Bush K., Furie B., Furie B. C., McLafferty F. W., Walsh C. T. Localization of labile posttranslational modifications by electron capture dissociation: the case of γ-carboxyglutamic acid. Anal. Chem. 1999;71:4250–4253. doi: 10.1021/ac990684x. [DOI] [PubMed] [Google Scholar]
- 35.Pogliani L., Ziessow D., Krüger Ch. Conformational study of phosphoserine in aqueous solutions. II - 1H n.m.r. results. Org. Magn. Res. 1977;10:26–30. [Google Scholar]
- 36.Raeck C., Berger S. A 2D NMR method to study peptide phosphorylation. Anal. Bioanal. Chem. 2007;389:2161–2165. doi: 10.1007/s00216-007-1653-9. [DOI] [PubMed] [Google Scholar]
- 37.Isab A. A., Hussain M. S., Akhtar M. N., Wazeer M. I. M., Al-Arfaj A. R. 13C, 15N and 31P NMR studies of the disproportination of cyanogold(I) complexes: [R3PAu13C15N] Polyhedron. 1999;18:1401–1409. [Google Scholar]
- 38.Lindon J. C., Baker D. J., Farrant R. D., Williams J. M. 1H, 13C and 31P n.m.r. spectra and molecular conformation of myo-inositol 1,4,5-trisphosphate. Biochem. J. 1986;233:275–277. doi: 10.1042/bj2330275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varga-Defterdarović L., Hrlec G. Synthesis and intramolecular reactions of Tyr-Gly and Tyr-Gly-Gly related 6-O-glucopyranose esters. Carbohydr. Res. 2004;339:67–75. doi: 10.1016/j.carres.2003.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Ruman T., Długopolska K., Jurkiewicz A., Rut D., Frączyk T., Cieśla J., Leś A., Szewczuk Z., Rode W. Thiophosphorylation of free amino acids and enzyme protein by thiophosphoramidate ions. Bioorg. Chem. 2010;38:74–80. doi: 10.1016/j.bioorg.2009.11.002. [DOI] [PubMed] [Google Scholar]