Abstract
The homeodomain transcription factor pancreatic duodenal homeobox 1 (Pdx1) is a major mediator of insulin transcription and a key regulator of the β cell phenotype. Heterozygous mutations in PDX1 are associated with the development of diabetes in humans. Understanding how Pdx1 expression levels are controlled is therefore of intense interest in the study and treatment of diabetes. Pdx1 C terminus–interacting factor-1 (Pcif1, also known as SPOP) is a nuclear protein that inhibits Pdx1 transactivation. Here, we show that Pcif1 targets Pdx1 for ubiquitination and proteasomal degradation. Silencing of Pcif1 increased Pdx1 protein levels in cultured mouse β cells, and Pcif1 heterozygosity normalized Pdx1 protein levels in Pdx1+/– mouse islets, thereby increasing expression of key Pdx1 transcriptional targets. Remarkably, Pcif1 heterozygosity improved glucose homeostasis and β cell function and normalized β cell mass in Pdx1+/– mice by modulating β cell survival. These findings indicate that in adult mouse β cells, Pcif1 limits Pdx1 protein accumulation and thus the expression of insulin and other gene targets important in the maintenance of β cell mass and function. They also provide evidence that targeting the turnover of a pancreatic transcription factor in vivo can improve glucose homeostasis.
Introduction
The homeodomain transcription factor pancreatic duodenal homeobox 1 (Pdx1) is required for pancreas development in mice and in humans (1–4). Pdx1 is required for the specification of pancreatic progenitors after which its expression becomes restricted primarily to the insulin-producing β cell, where it plays critical roles in insulin gene transcription and insulin secretion as well as β cell survival (reviewed in ref. 5). Tight regulation of Pdx1 protein levels is necessary for Pdx1 to play its developmental and adult physiological roles. Genetic studies in which Pdx1 is conditionally inactivated in mice suggest that Pdx1 gene dosage is critical both for development of the endocrine and exocrine pancreas and for the maintenance of adult β cells (6–8). Heterozygous loss of Pdx1 resulting in approximately 30% reduction in protein levels is sufficient to induce impaired glucose tolerance and insulin secretion in mice, and heterozygous mutations are associated with the development of diabetes in humans (9–11). Furthermore, Pdx1 haploinsufficiency limits the β cell compensatory mechanisms that occur in response to genetic and diet-induced insulin resistance (12–14).
Much of the regulation of Pdx1 dosage occurs at the transcriptional level. Upstream regulatory elements have been described that temporally and spatially control Pdx1 expression and are required for outgrowth and differentiation of pancreatic progenitors (8, 15–20); however, posttranslational regulation of Pdx1 via phosphorylation has also been proposed to have diverse functional effects, including regulation of Pdx1 transactivation and protein stability (13, 21–25). We previously described Pdx1 C terminus–interacting factor 1 (Pcif1, also known as SPOP), a broad complex, tramtrack, bric-a-brac (BTB) domain–containing protein that interacts directly with the C terminus of Pdx1 (26, 27). BTB-domain containing proteins have recently been identified as substrate-specific adaptors for ubiquitin ligase complexes that contain the scaffolding protein Cul3 (28, 29). We demonstrate here that the Pcif1-Cul3 complex targets Pdx1 protein for ubiquitination and proteasomal degradation. Further, we derived and characterized mice with a mutant allele of Pcif1 (Pcif1gt) and found that reduction of Pcif1 gene dosage improves glucose tolerance, normalizes β cell mass, and rescues β cell apoptosis rates in Pdx1+/– mice while normalizing Pdx1 protein levels and the expression of Pdx1 transcriptional targets.
Results
Pcif1 targets Pdx1 for ubiquitination by a Cul3-based E3 ubiquitin ligase.
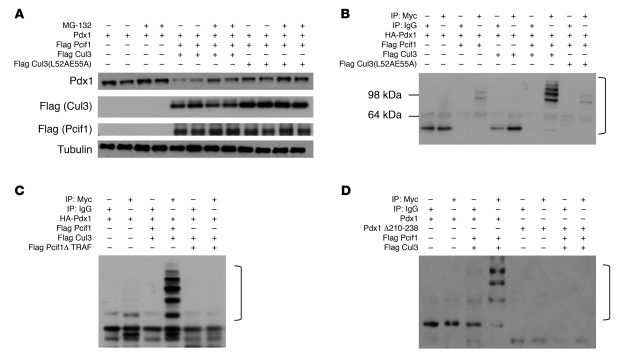
BTB domain–containing proteins have been identified as substrate-specific adaptors for ubiquitin ligase complexes that include the scaffold protein Cul3 (30). The Drosophila ortholog of Pcif1 targets cubitus interruptus for ubiquitination, while the human ortholog SPOP has been linked to multiple targets, including the polycomb protein Bmi1 (31, 32). To determine whether a similar mechanism underlies Pcif1-mediated downregulation of Pdx1 transactivation, we utilized heterologous HEK293T cells overexpressing Pdx1 with or without coexpression of Pcif1 and Cul3. In cells overexpressing Pdx1, treatment with the proteasome inhibitor MG-132 induced a slight increase in Pdx1 protein levels, suggesting that Pdx1 protein is proteasomally degraded in this system (Figure 1A). Addition of Pcif1 and Cul3 resulted in a dramatic decrease in Pdx1 protein accumulation that was partially reversed by proteasome inhibition. Notably, the ability of Cul3 to decrease Pdx1 accumulation was attenuated by alanine substitutions for key leucine and glutamic acid residues required for interactions between Cul3 and BTB domain–containing proteins (Cul3L52AE55A) (30, 33) (Figure 1A).
Figure 1. Pcif1 and Cul3 target Pdx1 for ubiquitination.
(A) HEK293T cells were transfected with the indicated plasmids and treated with vehicle or MG-132 for 4 hours before harvest. Western blots probed for Pdx1, Flag (Cul3 and Pcif1), and Tubulin (loading control). (B–D) HEK293T cells transfected with the indicated plasmids prior to immunoprecipitation with IgG or anti-Myc–coupled agarose. Western blots probed for HA (Pdx1). Brackets indicate ubiquitinated Pdx1.
To determine whether ubiquitination of Pdx1 is promoted by Pcif1 and Cul3, we employed an in vivo ubiquitination assay in HEK293T cells. Cells expressing Myc-tagged ubiquitin were subjected to immunoprecipitation with anti-Myc or isotype-matched control IgG and the products immunoblotted for Pdx1. Ubiquitination of Pdx1 was detectable as a ladder of high-molecular-weight bands that were immunoprecipitated in cells that expressed Pcif1 and Pdx1, and ubiquitination was further increased by the addition of Cul3 (Figure 1B and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI40440DS1). Cul3L52AE55A supported a minimal level of ubiquitination, consistent with the proposed role of Cul3 interaction with Pcif1 in promoting Pdx1 ubiquitination.
We previously determined that the TRAF domain of Pcif1 is required for the physical and functional interaction with Pdx1 in vitro (27). To determine whether the interaction between Pcif1 and Pdx1 was required for Pdx1 ubiquitination, we replaced full-length Pcif1 with Pcif1 lacking the TRAF domain (Pcif1Δ TRAF) in the in vivo ubiquitination system and found that the TRAF domain was required to promote Pdx1 ubiquitination (Figure 1C). Further, deletion of a conserved 28-amino-acid motif in the C terminus of Pdx1 (aa 210–238) that is required for the physical and functional interaction with Pcif1 (26, 27) abolished Pcif1-mediated ubiquitination of Pdx1 (Figure 1D). Taken together, these data strongly suggest that physical interactions among Cul3, Pcif1, and Pdx1 are required to promote Pdx1 ubiquitination.
Pcif1 regulates Pdx1 protein accumulation in Min6 β cells.
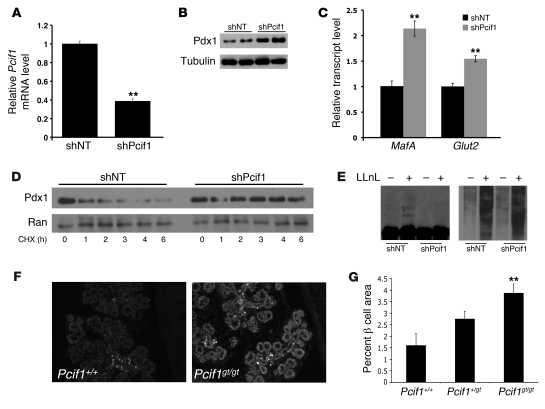
The experiments described above are in agreement with those of Bunce et al, who demonstrated Pcif1-mediated ubiquitination of Pdx1 in an overexpression system (34). To determine whether endogenous Pcif1 regulates Pdx1 protein levels in β cells, we next examined the impact of Pcif1 loss of function on Pdx1 protein accumulation in Min6 mouse insulinoma cells. shRNA-mediated silencing of Pcif1 resulted in a greater than 50% reduction in Pcif1 transcript (Figure 2A). Compared with cells transfected with a nontargeting shRNA, endogenous Pdx1 protein levels were substantially increased in cells after Pcif1 silencing (Figure 2B). Pdx1 protein levels are critical for maintaining β cell function and mediate these effects by regulating the transcription of β cell genes such as MafA and the glucose transporter Glut2 (35, 36). Downregulation of Pcif1 in Min6 cells was sufficient to induce a greater than 2-fold upregulation of MafA and 1.5-fold upregulation of Glut2 transcript (Figure 2C). Insulin mRNA was not regulated under these conditions, likely due to its long half-life. Downregulation of Pcif1 in Min6 cells also extended the half-life of endogenous Pdx1, dramatically increasing protein stability over the course of a 6-hour treatment with cycloheximide (Figure 2D). Treatment of Cul3-overexpressing Min6 cells with the proteasome inhibitor LLnL induced the accumulation of high-molecular-weight ubiquitinated proteins (Figure 2E), including bands detected by Pdx1 Western blot that may represent polyubiquitinated Pdx1. Accumulation of this species was abolished upon Pcif1 knockdown (Figure 2E, left panel). Probing Pdx1 immunoprecipitates from LLnL-treated Min6 cells with an anti-ubiquitin antibody revealed the presence of a high-molecular-weight (~98 kDa) band that was substantially diminished upon Pcif1 knockdown (Supplemental Figure 2). These data suggest that Pcif1-mediated regulation of endogenous Pdx1 protein levels directly impacts Pdx1 protein ubiquitination, accumulation, and stability and the expression of key Pdx1 target genes.
Figure 2. Increased Pdx1 accumulation and stability in Pcif1-deficient Min6 β cells and embryonic pancreas.
(A–C) Min6 cells nucleofected with control nontargeting shRNA (shNT) or shPcif1. (A) Pcif1 transcript measured by QPCR and normalized to Hprt. n = 3, **P < 0.01. (B) Western blot probed for Pdx1 and tubulin (loading control). (C) Transcript levels of Pdx1 transcriptional targets MafA and Glut2. n = 3, **P < 0.01. (D) Min6 cells expressing shNT or shPcif1, treated with the translation inhibitor cyclohexamide (CHX) and harvested at the time points indicated. Western blots are probed for Pdx1 and Ran (loading control). (E) Min6 cells overexpressing Cul3 and nontargeting shRNA or shPcif1 were treated with vehicle or 20 μM LLnL for 8 hours. Western blots are probed for Pdx1 (left) and ubiquitin (right). (F) Fluorescence staining of E16.5 embryos with dilute Pdx1 antibody. Original magnification ×10. (G) β Cell area of E18.5 embryos. n = 6, **P < 0.01 compared with Pcif1+/+.
Derivation of a Pcif1 gene trap allele.
To investigate the role of Pcif1 in vivo, we generated mice from an ES cell clone containing a gene trap insertion in the first intron of the Pcif1 locus. Mice heterozygous for the gene trap allele (Pcif1+/gt) were viable and fertile. In heterozygous crosses, Pcif1gt/gt embryos harvested at E18.5 were found at Mendelian ratios (n = 372); however, no viable Pcif1gt/gt mice were found after postnatal day 1, indicating that homozygous loss of Pcif1 results in postnatal lethality. At E18.5, Pcif1gt/gt embryos displayed normal body and pancreas weight and normal blood sugar (Supplemental Figure 3).
The gene trap insertion cassette contains a strong splice acceptor sequence followed by the βGeo reporter and ends with a polyadenylation signal. The transcript generated by the gene trap should prevent splicing between exons 1 and 2, thereby blocking the generation of the normal Pcif1 transcript. Using primers that span exons 1 and 2, Pcif1 transcript levels were reduced by greater than 50% in E18.5 pancreas and lung from heterozygous animals and undetectable in Pcif1gt/gt tissues (Supplemental Figure 4, A and B). Similar reductions in Pcif1 protein were observed in Western blots of embryonic kidney lysates (Supplemental Figure 4C). No Pcif1 transcript was detected in Pcif1gt/gt tissues using primers spanning exons 7 and 8 (data not shown), indicating that the gene trap efficiently prevents the generation of a protein-coding Pcif1 transcript.
Transcript analysis indicated that Pcif1 is expressed during embryonic pancreas development (data not shown). To determine whether Pcif1 modulates Pdx1 protein accumulation during development, we assessed staining intensity in sections from E16.5 and E18.5 embryos using an anti-Pdx1 antibody at a dilution that enabled semiquantitative evaluation of Pdx1 protein levels. Pdx1 staining intensity was markedly increased in all pancreatic lineages in Pcif1gt/gt mice at these stages (Figure 2F and data not shown). Further, mice homozygous for the Pcif1 gene trap allele demonstrated an approximately 3-fold increase in relative β cell area at E18.5 (Figure 2G), consistent with previously established roles of Pdx1 protein level in regulating endocrine progenitor specification and lineage allocation during pancreatic development (7, 8, 37, 38). There was no significant change in β cell area in the Pcif1+/gt mice at this stage of development (Figure 2E).
Increased Pdx1 protein levels in Pcif1+/gtPdx1+/– mouse islets.
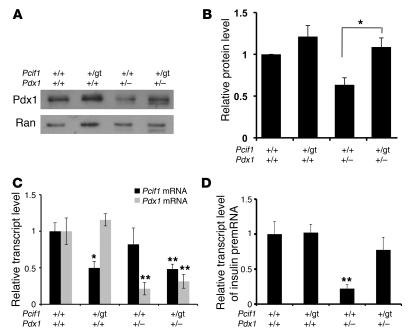
To determine whether Pcif1 regulates Pdx1 protein levels in adult β cells in vivo, we intercrossed Pcif1+/gt and Pdx1+/– mice and analyzed islets isolated from wild-type, Pcif1+/gt, Pdx1+/–, and Pcif1+/gtPdx1+/– littermates (Figure 3, A and B). In agreement with a study by Brissova et al. (10), Pdx1 protein levels were reduced in Pdx1+/– islets compared with those of wild-type littermates, whereas Pdx1 protein levels were normalized in Pcif1+/gtPdx1+/– islets. Pdx1 transcript levels were not affected by Pcif1 genotype, consistent with the hypothesis that Pcif1-mediated regulation of Pdx1 occurs posttranscriptionally (Figure 3C). Further, the normalization of Pdx1 protein levels in Pcif1+/gtPdx1+/– islets was associated with normalization of pre-mRNA level for insulin1, a key Pdx1 transcriptional target (Figure 3D). These data demonstrate that Pcif1 regulates the accumulation of Pdx1 protein in the adult β cell and may thus modulate the actions of Pdx1 as a critical regulator of glucose homeostasis.
Figure 3. Pcif1 regulates Pdx1 protein accumulation and Pdx1 target gene expression in primary mouse islets.
(A) Western blot of islets isolated from individual mice, probed for Pdx1. (B) Quantification of Western blots of islet protein extracts, normalized to loading control. n = 4, *P < 0.05. (C) Pdx1 and Pcif1 transcript levels in isolated mouse islets. n = 5–8, *P < 0.05, **P < 0.01 compared with wild-type. (D) Insulin transcript level in mouse islets. n = 5–8, **P < 0.01.
Pcif1 heterozygosity normalizes β cell mass and improves β cell survival in Pdx1+/– mice.
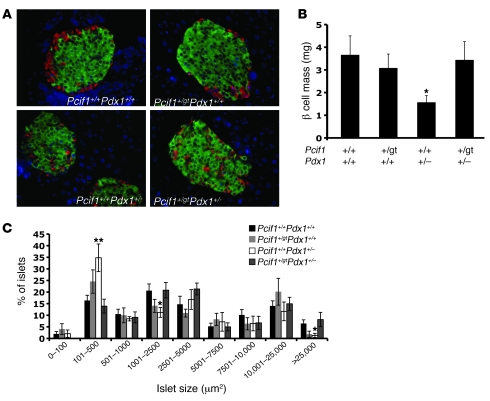
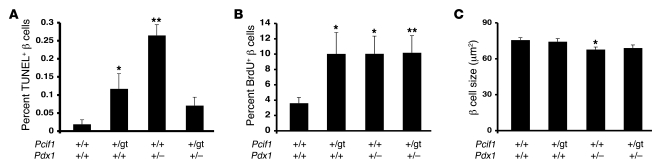
Pdx1+/– mice were previously reported to exhibit an age-related decline in β cell mass due to impaired β cell survival (39). Examination of islet architecture in Pdx1+/– pancreata revealed the previously described “mixed islet” phenotype, with glucagon-positive α cells scattered throughout the core of the islet; this defect was not normalized in Pcif1+/gtPdx1+/– mice (Figure 4A). We quantified β cell mass by determining the relative area occupied by insulin staining of pancreatic sections and observed a marked decrease in the cross-sectional area stained by insulin in Pdx1+/– pancreas compared with all other genotypes. Upon quantification, Pcif1+/gt mice were found to have β cell mass similar to that of wild-type littermates, whereas Pdx1+/– mice displayed a greater than 50% decrease in β cell mass compared with wild-type and Pcif1+/gt mice (Figure 4B). Notably, Pcif1 heterozygosity normalized the reduced β cell mass of Pdx1+/– mice. This was accompanied by a reduction in the rate of apoptotic β cells in Pcif1+/gtPdx1+/– mice as compared with Pdx1+/– mice (Figure 5A).
Figure 4. Pcif1 heterozygosity normalizes β cell mass in Pdx1+/– mice.
(A) Representative images of insulin- and glucagon-stained adult pancreas sections. Original magnification ×20. (B) Quantification of β cell mass. n = 7–10 mice per group; *P < 0.05 compared with wild-type. (C) Islet size distribution. n = 7–9 mice per group;*P < 0.05, **P < 0.01 compared with wild-type.
Figure 5. Normalized β cell mass is mediated by improved survival in Pcif1+/gtPdx1+/– mice.
(A) Apoptosis measured by quantifying insulin- and TUNEL-copositive cells. (B) β Cell replication measured by quantifying insulin and BrdU double positive cells. (C) β Cell size calculated by dividing insulin-positive islet area by the number of DAPI-stained nuclei counted within that area. All morphometric analyses performed on pancreas sections from 16-week-old male mice. n = 5–8 per group; *P < 0.05, **P < 0.01 compared with wild-type.
To determine whether the rescue of β cell area in Pcif1+/gtPdx1+/– was associated with changes in islet size, we determined the distribution of islet size in all genotypes. Although average islet size was not different among genotypes (data not shown), Pdx1+/– mice had a dramatic increase in the number of small (101–250 μm2) islets and a corresponding decrease in the percentage of medium-sized (1,001–2,500 μm2) and large (>25,000 μm2) islets (Figure 4C). Notably, each of these differences in islet size distribution was normalized in Pcif1+/gtPdx1+/– mice (Figure 4C).
In Pcif1+/gt mice, the finding of increased β cell apoptosis in the setting of normal β cell mass (Figure 4B and Figure 5A) suggested that β cell replication may also be influenced by Pcif1 gene dosage. Indeed, Pcif1+/gt mice displayed an increase in the rate of β cell replication compared with wild-type littermates; however, the rates of BrdU incorporation were not different among Pcif1+/gt, Pdx1+/–, and Pcif1+/gtPdx1+/– mice (Figure 5B). The finding of increased β cell replication in Pcif1+/gt mice was confirmed by quantification of Ki67 and phospho–histone H3–positive β cells (Supplemental Figure 5, A and B). We also observed a modest but significant decrease in average β cell size in Pdx1+/– mice that was not altered in the context of Pcif1 heterozygosity (Figure 5C). Taken together, these data indicate that the normalization of β cell mass observed in Pcif1+/gtPdx1+/– mice is due primarily to a reduction in the rate of apoptosis.
To determine whether the effect of Pcif1 heterozygosity on β cell turnover was age dependent, we assessed the β cell replication rate of each genotype in 5-week-old mice. Notably, we observed no differences in BrdU incorporation among genotypes at this age (Supplemental Figure 6). In these adolescent animals, pancreas size and β cell apoptosis rates as assessed by TUNEL staining were extremely low, precluding rigorous evaluation; preliminary analysis did not reveal differences based on genotype (data not shown). Taken together, these data suggest that the effect of Pcif1 heterozygosity on β cell replication rates is age dependent and that Pcif1 deficiency improves the age-related decline in β cell mass previously observed in Pdx1+/– mice (39).
Pcif1 deficiency reduces ER stress–associated apoptosis in Pdx1-deficient β cells.
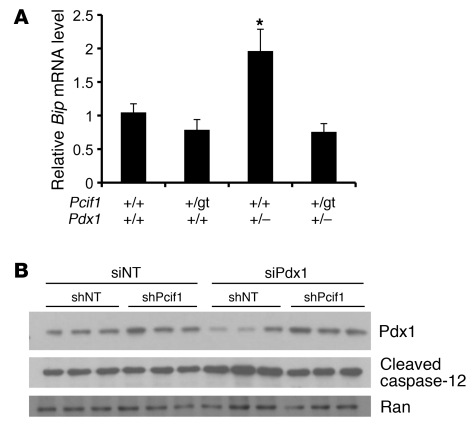
Pdx1 deficiency was recently shown to result in increased susceptibility to ER stress–induced apoptosis in β cells (14). To determine whether improved ER homeostasis contributed to the improvement in β cell survival in Pcif1+/gtPdx1+/– mice, we measured mRNA levels of the ER chaperone protein Bip in islets isolated from each genotype. As recently described (14), we observed an increase in Bip mRNA levels in Pdx1+/– islets compared with those of wild-type littermates (Figure 6A). Bip mRNA levels were normalized in Pcif1+/gtPdx1+/– compared with Pdx1+/– islets, suggesting that improved ER homeostasis may underlie the improved β cell survival in transheterozygous animals. In support of this finding, knockdown of endogenous Pdx1 protein in Min6 cells resulted in increased cleavage of the ER-resident caspase-12, while simultaneous knockdown of Pcif1 restored Pdx1 protein to control levels and prevented caspase-12 cleavage (Figure 6B).
Figure 6. Pcif1 deficiency improves ER homeostasis in Pdx1-deficient β cells.
(A) Bip mRNA levels as a measure of ER stress in isolated islets, normalized to Hprt. n = 7 per group; *P < 0.05 compared with wild-type. (B) Western blot for Pdx1 and the ER-resident caspase-12 after Pcif1 or Pdx1 knockdown in Min6 cells.
Pcif1 heterozygosity improves glucose homeostasis in Pdx1+/– mice.
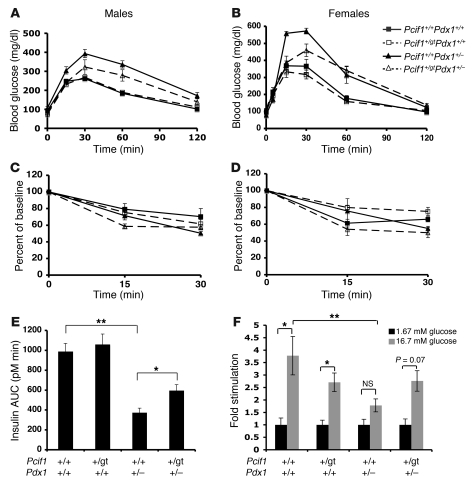
To determine whether the observed normalization of β cell mass in Pcif1+/gtPdx1+/– mice is associated with improved β cell function, we assessed glucose tolerance and insulin secretion. Male and female Pcif1+/gt mice displayed normal glucose tolerance upon intraperitoneal glucose tolerance testing, and Pdx1+/– mice were glucose intolerant as previously described (Figures 7, A and B). The glucose intolerance phenotype of Pdx1+/– mice was significantly improved in both male and female Pcif1+/gtPdx1+/– transheterozygous mice. All genotypes displayed similar insulin sensitivity, determined by insulin tolerance testing (Figures 7, C and D), suggesting that the observed alterations in glucose tolerance were due to differences in insulin secretion; therefore, acute glucose-stimulated insulin secretion was assessed in vivo. As expected, Pdx1+/– mice displayed a blunted insulin secretory response to glucose. Notably, the defect in Pdx1+/– mice was attenuated, though not fully rescued, in Pcif1+/gtPdx1+/– mice (insulin AUC: Pdx1+/– vs. Pcif1+/gtPdx1+/–, 373 ± 44 vs. 595 ± 58 pM min; P < 0.05) (Figure 7E).
Figure 7. Pcif1 heterozygosity improves glucose homeostasis and β cell function in Pdx1+/– mice.
(A) Intraperitoneal glucose tolerance of male mice (7–8 weeks old). n = 6–9 per group; P < 0.05, Pdx1+/– versus Pcif1+/gtPdx1+/–, by ANOVA. (B) Intraperitoneal glucose tolerance of female mice (10–12 weeks old). n = 6–11 per group; P < 0.001, Pdx1+/– versus Pcif1+/gtPdx1+/– by ANOVA. (C) Insulin tolerance of male mice (8–9 weeks old). P = NS for all groups. (D) Insulin tolerance of female mice (11–13 weeks old). P = NS for all groups. (E) Acute glucose-stimulated insulin secretion calculated as AUC for 15 minutes after glucose bolus (12-week-old male mice). n = 7–12 per group, *P < 0.05, **P < 0.01. (F) Glucose-stimulated insulin secretion during static incubations of isolated islets from 6- to 8-week-old male mice. Insulin secretion values were normalized to islet insulin content and expressed as fold stimulation. n = 3–6 mice per group, *P < 0.05.
To determine whether Pcif1 heterozygosity directly influences islet function, we further analyzed insulin secretion in isolated islets. Wild-type and Pcif1+/gt islets displayed a significant glucose stimulation of insulin secretion in static incubations (Figure 7F). Although Pdx1+/– islets were previously reported to display normal insulin secretion ex vivo, we observed a blunted response after static incubation (1.76 ± 0.26–fold over basal secretion; P = NS). Glucose-stimulated insulin secretion was restored in Pcif1+/gtPdx1+/– islets (2.76 ± 0.4–fold over basal secretion, P = 0.07) (Figure 7F). These data suggest that Pcif1 haploinsufficiency improves glucose responsiveness in Pdx1+/– β cells.
Pcif1 regulation of Pdx1 protein levels is regulated by glucose.
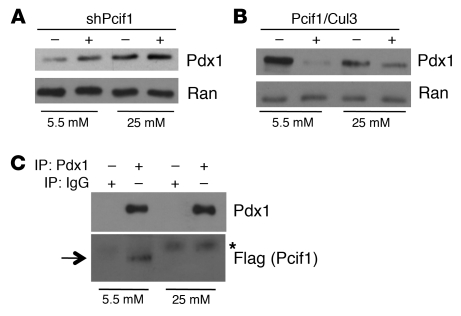
The increase in Pdx1 protein level that followed shRNA-mediated Pcif1 silencing in Min6 cells (Figure 2B and Figure 8A) was observed in cells cultured in low-glucose-containing medium (5.5 mM). Although Min6 cells chronically maintained in high glucose exhibit levels of Pcif1 similar to those maintained in low glucose (Supplemental Figure 7), Pcif1 silencing in Min6 cells cultured in high-glucose medium (25 mM) did not significantly alter endogenous Pdx1 protein levels (Figure 8A), suggesting that Pcif1 modulation of Pdx1 protein accumulation is regulated by glucose. In addition, the level of Pdx1 protein was increased in high-glucose- compared with low-glucose-cultured cells, in agreement with recent findings by Humphrey et al. demonstrating that Pdx1 protein is stabilized in Min6 cells and primary islets cultured under high-glucose conditions (25). Similarly, we found that in the HEK293T overexpression system, the previously observed effect of Pcif1 and Cul3 on Pdx1 protein accumulation (Figure 1A) was amplified in cells cultured in 5.5 mM glucose as compared with standard (25 mM glucose) conditions (Figure 8B). In agreement with these findings, overexpressed Pdx1 more readily coimmunoprecipitated with Flag-tagged Pcif1 in cells cultured in low glucose (Figure 8C), suggesting that decreased interaction between Pcif1 and Pdx1 in high-glucose conditions may underlie the increase in Pdx1 protein accumulation in those cells.
Figure 8. Posttranscriptional regulation of Pdx1 protein level by glucose.
(A) Western blot of lysates from Min6 cells maintained in 5.5 or 25 mM glucose, expressing nontargeting shRNA or shPCIF1. Blots are probed for Pdx1 and Ran (loading control). (B) Western blot of lysates from HEK293T cells maintained in 5.5 or 25 mM glucose, expressing Pdx1 in the presence or absence of overexpressed Pcif1 and Cul3. Blots are probed for Pdx1 and Ran. (C) HEK293T cells maintained in 5.5 or 25 mM glucose, transfected with plasmids expressing Pdx1 and Flag-Pcif1, and subjected to immunoprecipitation with IgG or Pdx1 antisera. Western blots are probed for Pdx1 or Flag (Pcif1). Arrow indicates migration of Flag-Pcif1; asterisk indicates heavy chain IgG signal.
In primary mouse islets, overnight culture in high glucose (30 mM) resulted in an approximately 50% decrease in Pcif1 mRNA, which corresponded to a posttranscriptional increase in Pdx1 protein (Supplemental Figure 8, A and B). This increase in Pdx1 protein was associated with a greater than 10-fold increase in insulin mRNA level (Supplemental Figure 8C), suggesting that acute glucose regulation of Pcif1 transcription could contribute to the posttranscriptional increase in Pdx1 protein levels and increased expression of Pdx1 transcriptional targets. Taken together, these data suggest that glucose modulation of Pdx1 protein accumulation involves multiple mechanisms including regulation of levels of and interaction with Pcif1.
Discussion
Here, we demonstrate that Pcif1, a BTB domain protein partner of Pdx1, directly contributes to the accumulation of Pdx1 protein in the adult β cell and thus to the actions of Pdx1 as a critical regulator of glucose homeostasis. Our results demonstrate that Pcif1 and Cul3 cooperatively target Pdx1 for ubiquitination and proteasomal degradation and that Pcif1 deficiency increases Pdx1 protein levels and stability in Min6 cells and in the developing pancreas and adult islets in vivo. Normalization of Pdx1 protein levels in Pcif1+/gtPdx1+/– transheterozygous mice was associated with improved β cell function and mass, leading to improved glucose tolerance and insulin secretion in Pdx1+/– mice. The improvement in β cell mass was due to an attenuation of the survival defect caused by Pdx1 deficiency and associated with an improvement in ER homeostasis. These results provide genetic evidence of posttranscriptional regulation of Pdx1 as a mediator of glucose homeostasis and of β cell survival in vivo.
Pcif1 heterozygosity completely rescued β cell mass and islet insulin transcript levels in Pdx1+/– mice but mediated only a partial rescue of insulin secretion and thus glucose tolerance. Further, the defect in islet architecture observed in Pdx1+/– mice was not normalized. These discrepancies may reflect different thresholds of Pdx1 required for optimal activation of Pdx1 transcriptional targets involved in islet development, insulin secretion, and β cell growth and survival. Our analysis of Pcif1gt/gt embryos demonstrated that although Pcif1 is not required for pancreatic organogenesis, it does influence the formation of developing β cells; however, the spatial and temporal sequence of Pcif1 expression in the pancreas has not yet been fully elucidated, and thus the relative importance of Pcif1 in regulation of the multiple roles of Pdx1 in the developing pancreas merits further investigation.
Although our findings suggest that the improved glucose homeostasis in Pcif1+/gtPdx1+/– mice is due to rescue of islet Pdx1 protein levels and thus improved β cell mass and function, the data also suggest a complex role for Pcif1 in the maintenance of β cell mass. Pcif1+/gt mice displayed normal β cell mass, but also balanced increases in both β cell replication and apoptosis rates. Furthermore, Pcif1 heterozygosity induced an increased rate of β cell replication that was not affected in the setting of Pdx1 heterozygosity, indicating roles of Pcif1 in β cell replication, and perhaps apoptosis, that may be independent of its regulation of Pdx1 protein accumulation. In addition, Pcif1 heterozygosity had no effect on β cell replication in younger (5-week-old) mice, pointing to a role in age-dependent replication rates. The human ortholog of Pcif1, SPOP, has been implicated in the ubiquitination of multiple targets, including the polycomb group protein Bmi1 (32), recently implicated in pancreatic β cell proliferation via its control of the Ink4a/Arf locus (40, 41). SPOP has also been described to ubiquitinate the proapoptotic protein Daxx (42). Thus, the contribution of Pcif1 to the β cell–replicative and apoptotic rates may be due to regulation of Bmi1, Daxx, or other, as-yet-undiscovered ubiquitination targets.
Our findings indicate that in adult β cells, Pcif1 limits Pdx1 protein accumulation and thus the expression of insulin and other gene targets critical to the maintenance of β cell mass and function. The identification of this new Pdx1 regulatory mechanism and validation of its role in vivo suggest that the therapeutic targeting of Pcif1-Cul3–mediated turnover of Pdx1 could raise Pdx1 protein levels to improve β cell function and viability in the treatment of diabetes.
Methods
Animals and physiological experiments.
Mouse ES cells containing a gene trap insertion in the first intron of Pcif1 were purchased from BayGenomics (clone Bgb118). Mice were derived by injection of ES cells into blastocysts by the University of Pennsylvania Transgenic and Chimeric Mouse Facility. Animals were housed in the animal care facility at the University of Pennsylvania and maintained on a 12-hour light/12-hour dark cycle with ad libitum access to food and water. All experiments described were performed on mice of a mixed 129/Sv × C57BL/6 background. For embryonic studies, the date of vaginal plug was considered E0.5. For glucose tolerance tests, animals were fasted for 16 hours before delivery of a 2-g/kg glucose bolus. Blood glucose was measured by handheld glucometers (Freestyle/One Touch). Serum was collected during the glucose tolerance test and insulin assessed by ELISA (Chemicon). For insulin tolerance tests, animals were fasted for 6 hours prior to an i.p. injection of 0.75 U/kg insulin. All animal procedures were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Pancreas morphometry.
BrdU was administered in the drinking water (1 g/l) for 1 week prior to sacrifice. Five- and 16-week-old male mice were anesthetized and the pancreas dissected rapidly, weighed, and fixed overnight in 4% paraformaldehyde. Paraffin sections were stained for insulin (Linco), BrdU (US Biologicals), and TUNEL (Chemicon Apoptag Peroxidase In Situ Kit). For β cell mass measurements, whole slide images of insulin-stained sections were captured on an Aperio slide scanner and the stained area quantified using Image-Pro software (Media Cybernetics). The percentage of insulin-stained area was then multiplied by pancreas weight to obtain an estimate of β cell mass. For replication measurements, an average of 1,119 and 1,463 β cells were counted per animal in the 5- and 16-week-old cohorts, respectively. For apoptosis measurements, an average of 2,398 β cells were counted per animal. For Ki67 and pHH3 confirmation of replication measurements, an average of 2,381 and 2,715 cells were counted per animal, respectively.
Islet isolation.
Islets were isolated from 8- to 12-week-old mice by collagenase digestion followed by 3–4 rounds of hand picking. For RNA and protein analysis, samples were matched for islet purity by measuring amylase and insulin transcript levels. For static incubations, 5 islets were analyzed in triplicate from each animal. Islets were incubated in glucose-free KRBB buffer for 1 hour before the addition of 1.67 mM glucose. Samples were taken after 1 hour of incubation, and glucose was added to a final concentration of 16.7 mM. After 1 hour of stimulation, secretion samples were collected and islets were harvested in islet lysis buffer and analyzed for insulin content. Insulin secretion and content were measured by ELISA (Chemicon).
Cell culture and transfections.
Min6 β cells were cultured in DMEM with 5.5 mM or 25 mM glucose (Invitrogen) supplemented with 10% FBS and used between passages 25 and 35. For knockdown experiments, cells were nucleofected by AMAXA with predesigned shRNA plasmids directed against Pcif1 or a nontargeting control (Sigma-Aldrich shPcif1 TRCN0000124643: CCGGGTAAACCCGAAAGGGCTAGATCTCGAGATCTAGCCCTTTCGGGTTTACTTTTTG). After 72 hours, cells were harvested for protein and RNA. HEK293T cells were cultured in high-glucose DMEM under standard conditions. For expression assays, cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions, and protein was harvested after 48 hours. For proteasome inhibition, 10 μM MG-132 (Calbiochem) or vehicle (DMSO) was added to the medium 4 hours before harvest. For proteasome inhibition in Min6 cells, 20 μM LLnL (Sigma-Aldrich) or vehicle (DMSO) was added to medium 8 hours before harvest.
In vivo ubiquitination assay and coimmunoprecipitation.
HEK293T cells were transfected as described above. After 48 hours, cells were harvested in RIPA buffer and subjected to immunoprecipitation with mouse α-Myc–coupled agarose beads or mouse IgG–coupled control beads (Santa Cruz Biotechnology Inc.). To assess Pcif1-Pdx1 interaction, HEK293T cells were transfected as described above and subjected to immunoprecipitation with goat α-Pdx1 antibody or goat IgG (Santa Cruz Biotechnology Inc.) 48 hours later.
Western blots and antisera.
Proteins were separated by SDS-PAGE and immunoblotted with the following antibodies: rabbit anit-Pcif1 (26), mouse anti-tubulin (Sigma-Aldrich), rabbit anti-Pdx1 (43), mouse anti-Flag epitope (Sigma-Aldrich), rabbit anti-HA epitope (Santa Cruz Biotechnology Inc.), rabbit anti-ubiquitin (Dako), rat anti–caspase-12 (Sigma-Aldrich), and mouse anti-Ran (BD Biosciences).
RNA isolation and transcript analysis.
Embryonic tissues were harvested and stored in RNAlater (Ambion), then homogenized in TRIzol (Invitrogen) and processed according to the manufacturer’s instructions. Islet RNA was extracted using the RNeasy Mini Kit (QIAGEN). All samples were reverse transcribed using SuperScript (Invitrogen) and oligo(dT) for priming. Transcript was analyzed by quantitative PCR and normalized to the Hprt transcript as an internal control. For isolated islets, samples were purity matched by comparing insulin and amylase transcripts.
Statistics.
Data are presented as mean ± SEM. Differences between groups were compared by 2-tailed Student’s t tests and considered significant when P values were less than 0.05. Group measurements of glucose and insulin tolerance were compared by factorial ANOVA.
Supplementary Material
Acknowledgments
We thank K.H. Kaestner for critical reading of the manuscript and for assistance with ES cell culture, C.V. Wright (Vanderbilt University) for providing us with the Pdx1+/– mice, and Jean Richa of the University of Pennsylvania Chimeric and Transgenic Mouse Facility for blastocyst injection. We received technical assistance in image capture and analysis from G. Swain of the Morphology Core (P30-DK050306) and D. Martinez of the Pathology Core at the Children’s Hospital of Philadelphia. This work was supported by NIH NIDDK grants R01 DK068157 and P01 DK049210 (to D.A. Stoffers) and NIH grant T32-GM07229 (to K.C. Claiborn).
Footnotes
Conflict of interest: Doris A. Stoffers is a co-inventor on U.S. patent nos. 6,274,310, 2001, and PCT/US08/02792, 2008. Doris A. Stoffers received an honorarium from Merck Research Labs for participation in a Global Diabetes and Obesity Expert Forum.
Citation for this article: J Clin Invest. 2010;120(10):3713–3721. doi:10.1172/JCI40440.
References
- 1.Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122(5):1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 2.Offield MF, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122(3):983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 3.Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15(1):106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 4.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371(6498):606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 5.Babu DA, Deering TG, Mirmira RG. A feat of metabolic proportions: Pdx1 orchestrates islet development and function in the maintenance of glucose homeostasis. Mol Genet Metab. 2007;92(1–2):43–55. doi: 10.1016/j.ymgme.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale MA, et al. The homeodomain protein PDX1 is required at mid-pancreatic development for the formation of the exocrine pancreas. Dev Biol. 2005;286(1):225–237. doi: 10.1016/j.ydbio.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 7.Holland AM, Gonez LJ, Naselli G, Macdonald RJ, Harrison LC. Conditional expression demonstrates the role of the homeodomain transcription factor Pdx1 in maintenance and regeneration of beta-cells in the adult pancreas. Diabetes. 2005;54(9):2586–2595. doi: 10.2337/diabetes.54.9.2586. [DOI] [PubMed] [Google Scholar]
- 8.Fujitani Y, et al. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes Dev. 2006;20(2):253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hani EH, et al. Defective mutations in the insulin promoter factor-1 (IPF-1) gene in late-onset type 2 diabetes mellitus. J Clin Invest. 1999;104(9):R41–R48. doi: 10.1172/JCI7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brissova M, et al. Reduction in pancreatic transcription factor PDX-1 impairs glucose-stimulated insulin secretion. J Biol Chem. 2002;277(13):11225–11232. doi: 10.1074/jbc.M111272200. [DOI] [PubMed] [Google Scholar]
- 11.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17(2):138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 12.Kulkarni RN, Jhala US, Winnay JN, Krajewski S, Montminy M, Kahn CR. PDX-1 haploinsufficiency limits the compensatory islet hyperplasia that occurs in response to insulin resistance. J Clin Invest. 2004;114(6):828–836. doi: 10.1172/JCI21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brissova M, et al. Reduced PDX-1 expression impairs islet response to insulin resistance and worsens glucose homeostasis. Am J Physiol Endocrinol Metab. 2005;288(4):E707–E714. doi: 10.1152/ajpendo.00252.2004. [DOI] [PubMed] [Google Scholar]
- 14.Sachdeva MM, et al. Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proc Natl Acad Sci U S A. 2009;106(45):19090–19095. doi: 10.1073/pnas.0904849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma S, Leonard J, Lee S, Chapman HD, Leiter EH, Montminy MR. Pancreatic islet expression of the homeobox factor STF-1 relies on an E-box motif that binds USF. J Biol Chem. 1996;271(4):2294–2299. doi: 10.1074/jbc.271.4.2294. [DOI] [PubMed] [Google Scholar]
- 16.Sharma S, Jhala US, Johnson T, Ferreri K, Leonard J, Montminy M. Hormonal regulation of an islet-specific enhancer in the pancreatic homeobox gene STF-1. Mol Cell Biol. 1997;17(5):2598–2604. doi: 10.1128/mcb.17.5.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerrish K, Van Velkinburgh JC, Stein R. Conserved transcriptional regulatory domains of the pdx-1 gene. Mol Endocrinol. 2004;18(3):533–548. doi: 10.1210/me.2003-0371. [DOI] [PubMed] [Google Scholar]
- 18.Boyer DF, Fujitani Y, Gannon M, Powers AC, Stein RW, Wright CV. Complementation rescue of Pdx1 null phenotype demonstrates distinct roles of proximal and distal cis-regulatory sequences in pancreatic and duodenal expression. Dev Biol. 2006;298(2):616–631. doi: 10.1016/j.ydbio.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22(24):3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerrish K, et al. Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J Biol Chem. 2000;275(5):3485–3492. doi: 10.1074/jbc.275.5.3485. [DOI] [PubMed] [Google Scholar]
- 21.Petersen HV, et al. Glucose stimulates the activation domain potential of the PDX-1 homeodomain transcription factor. FEBS Lett. 1998;431(3):362–366. doi: 10.1016/S0014-5793(98)00776-5. [DOI] [PubMed] [Google Scholar]
- 22.Lebrun P, Montminy MR, Van Obberghen E. Regulation of the pancreatic duodenal homeobox-1 protein by DNA-dependent protein kinase. J Biol Chem. 2005;280(46):38203–38210. doi: 10.1074/jbc.M504842200. [DOI] [PubMed] [Google Scholar]
- 23.Boucher MJ, Selander L, Carlsson L, Edlund H. Phosphorylation marks IPF1/PDX1 protein for degradation by glycogen synthase kinase 3-dependent mechanisms. J Biol Chem. 2006;281(10):6395–6403. doi: 10.1074/jbc.M511597200. [DOI] [PubMed] [Google Scholar]
- 24.Kishi A, Nakamura T, Nishio Y, Maegawa H, Kashiwagi A. Sumoylation of Pdx1 is associated with its nuclear localization and insulin gene activation. Am J Physiol Endocrinol Metab. 2003;284(4):E830–E840. doi: 10.1152/ajpendo.00390.2002. [DOI] [PubMed] [Google Scholar]
- 25.Humphrey RK, Yu SM, Flores LE, Jhala US. Glucose regulates steady-state levels of PDX1 via the reciprocal actions of GSK3 and AKT kinases. J Biol Chem. 2010;285(5):3406–3416. doi: 10.1074/jbc.M109.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu A, Desai BM, Stoffers DA. Identification of PCIF1, a POZ domain protein that inhibits PDX-1 (MODY4) transcriptional activity. Mol Cell Biol. 2004;24(10):4372–4383. doi: 10.1128/MCB.24.10.4372-4383.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu A, Oliver-Krasinski J, Stoffers DA. Two conserved domains in PCIF1 mediate interaction with pancreatic transcription factor PDX-1. FEBS Lett. 2006;580(28–29):6701–6706. doi: 10.1016/j.febslet.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 28.Pintard L, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425(6955):311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- 29.Krek W. BTB proteins as henchmen of Cul3-based ubiquitin ligases. Nat Cell Biol. 2003;5(11):950–951. doi: 10.1038/ncb1103-950. [DOI] [PubMed] [Google Scholar]
- 30.Xu L, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425(6955):316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10(6):719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Hernandez-Munoz I, et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci U S A. 2005;102(21):7635–7640. doi: 10.1073/pnas.0408918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 34.Bunce MW, Boronenkov IV, Anderson RA. Coordinated activation of the nuclear ubiquitin ligase Cul3-SPOP by the generation of phosphatidylinositol 5-phosphate. J Biol Chem. 2008;283(13):8678–8686. doi: 10.1074/jbc.M710222200. [DOI] [PubMed] [Google Scholar]
- 35.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. β-Cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12(12):1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raum JC, et al. FoxA2, Nkx2.2, and PDX-1 regulate islet beta-cell-specific mafA expression through conserved sequences located between base pairs -8118 and -7750 upstream from the transcription start site. Mol Cell Biol. 2006;26(15):5735–5743. doi: 10.1128/MCB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holland AM, Hale MA, Kagami H, Hammer RE, MacDonald RJ. Experimental control of pancreatic development and maintenance. Proc Natl Acad Sci U S A. 2002;99(19):12236–12241. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliver-Krasinski JM, et al. The diabetes gene Pdx1 regulates the transcriptional network of pancreatic endocrine progenitor cells in mice. J Clin Invest. 2009;119(7):1888–1898. doi: 10.1172/JCI37028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson JD, et al. Increased islet apoptosis in Pdx1+/– mice. . J Clin Invest. 2003;111(8):1147–1160. doi: 10.1172/JCI16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23(8):975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dhawan S, Tschen SI, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes Dev. 2009;23(8):906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon JE, et al. BTB domain-containing speckle-type POZ protein (SPOP) serves as an adaptor of Daxx for ubiquitination by Cul3-based ubiquitin ligase. J Biol Chem. 2006;281(18):12664–12672. doi: 10.1074/jbc.M600204200. [DOI] [PubMed] [Google Scholar]
- 43.Stoffers DA, Stanojevic V, Habener JF. Insulin promoter factor-1 gene mutation linked to early-onset type 2 diabetes mellitus directs expression of a dominant negative isoprotein. J Clin Invest. 1998;102(1):232–241. doi: 10.1172/JCI2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.