Abstract
Cathepsins were originally identified as proteases that act in the lysosome. Recent work has uncovered nontraditional roles for cathepsins in the extracellular space as well as in the cytosol and nucleus. There is strong evidence that subspecialized and compartmentalized cathepsins participate in many physiologic and pathophysiologic cellular processes, in which they can act as both digestive and regulatory proteases. In this review, we discuss the transcriptional and translational control of cathepsin expression, the regulation of intracellular sorting of cathepsins, and the structural basis of cathepsin activation and inhibition. In particular, we highlight the emerging roles of various cathepsin forms in disease, particularly those of the cardiac and renal systems.
The maintenance of a healthy organism largely relies upon controlled biosynthesis, maturation, function, and terminal breakdown of proteins. Proteolytic enzymes contribute to these processes by irreversibly cleaving peptide bonds. This can result in destruction of the substrate protein, its maturation, or modulation of the biological activities of the cleavage products. To accomplish the multitude of selective and well-controlled proteolytic events that keep us healthy, the human genome encodes more than 550 proteases and more than 200 endogenous protease inhibitors (1, 2).
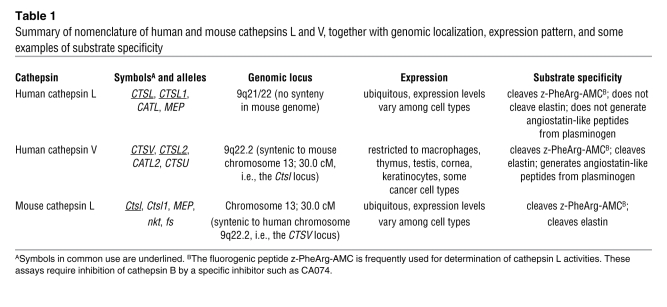
The so-called “catheptic activity” (derived from the Greek word kathépsein, meaning to digest or to boil down) was first described in the gastric juice during the 1920s (3). Today, cathepsins are classified based on their structure and catalytic type into serine (cathepsins A and G), aspartic (cathepsins D and E), and cysteine cathepsins (Figure 1 and Sidebar 1). The latter constitutes the largest cathepsin family, with 11 proteases in humans referred to as clan CA, family C1a: cathepsins B, C (also known as cathepsin J and dipeptidyl-peptidase 1), F, H, K (also known as cathepsin O2), L, O, S, W, V (also known as cathepsin L2), and Z (also known as cathepsin X and cathepsin P) (4). In general, the cysteine cathepsins are stable in acidic cellular compartments, i.e., in lysosomes and endosomes, and capable of efficiently cleaving a wide variety of substrates. Despite this, during the past decade, important and specific functions of cathepsins have been discovered to occur extracellularly and in other locations inside cells, such as secretory vesicles (5, 6), the cytosol (7–9), and the nucleus (10, 11). When studying the function of cathepsins, one needs to consider species differences between humans and mice (Table 1 and Sidebar 2).
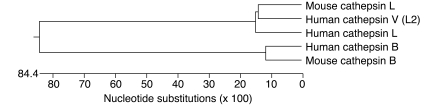
Figure 1. Phylogeny and nomenclature of cathepsins.
Phylogenic tree of mouse and human cathepsin proenzymes. Mouse cathepsin L, human cathepsin V, and human cathepsin L are compared with each other and with human and mouse cathepsin B in order to demonstrate the phylogenic distance to other prototypic members of the cysteine cathepsin family.
Table 1 .
Summary of nomenclature of human and mouse cathepsins L and V, together with genomic localization, expression pattern, and some examples of substrate specificity

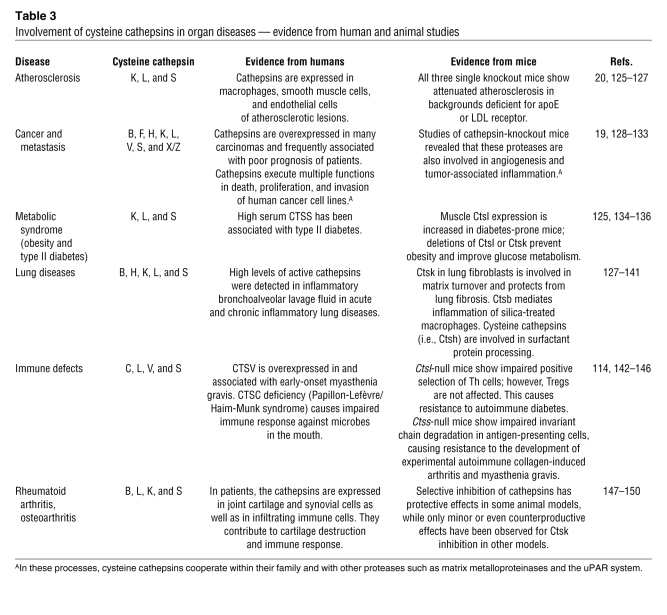
Novel insight into cathepsin function was made possible by various novel genomic, proteomic, and imaging tools as well as the generation and in-depth analysis of knockout and transgenic mice (12). These studies established that cathepsins are not simply redundant, homeostatic enzymes involved in the turnover of proteins delivered to the lysosome by endocytosis or autophagocytosis but are critically involved in the proteolytic processing of specific substrates. Thus, cathepsins contribute to distinct physiologic processes, such as antigen presentation in the immune system (13), collagen turnover in bone and cartilage (14, 15), and neuropeptide and hormone processing (16, 17). Various diseases or phenotypes develop when cathepsins are lost in humans or mice, respectively (Tables 2 and 3). In addition, ectopic or excessive expression and activity of cathepsins promotes the development of several common diseases in humans and mice, including cancer and arthritis (Table 3).
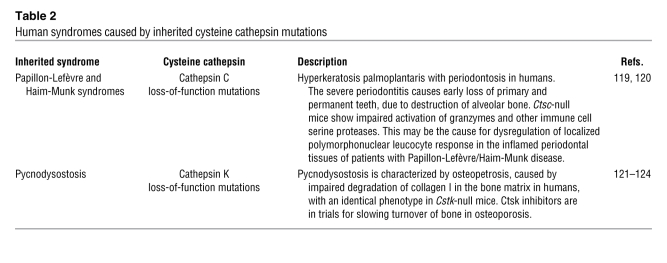
Table 2 .
Human syndromes caused by inherited cysteine cathepsin mutations
Table 3 .
Involvement of cysteine cathepsins in organ diseases — evidence from human and animal studies

The roles of cathepsins in many physiologic and disease processes have been covered by recent comprehensive reviews (17–20). Here, we focus on the recently discovered roles of cathepsins in organ diseases, with a special emphasis on the ubiquitously expressed cysteine endopeptidase cathepsin L. In particular, we discuss how the different functions of a cysteine cathepsin depend on the cell type in which it is expressed and the cellular compartment in which the protease is localized. We address the homeostatic function of cathepsin L in the heart and its potential role in cardiac regeneration, the reciprocal processing function of cathepsins B and L in ectopic trypsinogen activation during the onset of acute pancreatitis, and the emerging roles of cytosolic and nuclear cathepsin L variants in proteinuric kidney disease and stem cell physiology, respectively.
A primer on cathepsin biology
Cathepsin L transcription and translation.
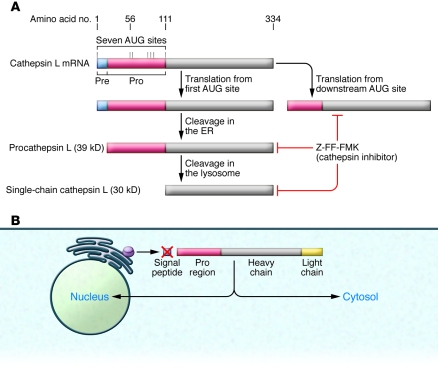
Substantial work has been done to analyze the promoter regions of the human cathepsin L gene (CTSL) promoter as well as to understand the regulation of different splice variants within the 5′ untranslated region of the transcript (21, 22). Of note, one of the splice variants contains a functional internal ribosomal entry site that enables ongoing translation of human cathepsin L under stress conditions, and hypoxia can shut down cap-dependent translation initiation (23). More recent work has focused on the regulation of cathepsin L alternative translation. According to the presence of different forms of cathepsin L in distinct subcellular and extracellular compartments, cathepsin L proteins can be initiated from downstream AUG sites (10), omitting the signal peptide that is normally present at the N terminus of lysosomal cathepsin L that routes the protein to the ER during its synthesis (Figure 2) (10, 24–26).
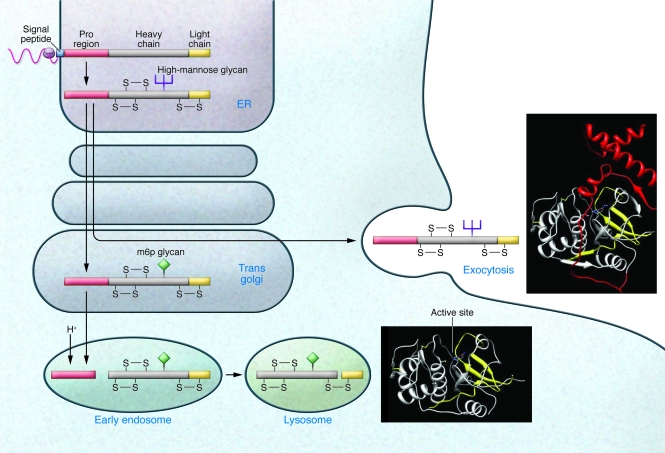
Figure 2. Cathepsin structures and traditional and nontraditional protease trafficking.
Cathepsins contain a signal peptide (blue) that directs insertion of the nascent polypeptide chain into the ER. Within the ER, the signal peptide is cleaved and the protein folds with the assistance of the proregion (red). Disulfide bond formation (indicated by S-S) and N-linked glycosylation with high-mannose glycans subsequently occurs in the ER. Within the Golgi, mannose residues are phosphorylated to form m6p, which is used to route the protein into the endosomal/lysosomal compartment via the m6p receptor. Upon initial acidification of the endosome, cathepsins are activated, which leads to cleavage of the proregion and further activation of the cathepsin, resulting in further proteolytic processing in the lysosome into heavy and light chains (yellow). A portion of the cathepsins is not converted to the m6p form and as a result is shunted into the exocytosis pathway. Conversion to m6p appears to be rate limiting, as overexpression of a given cathepsin greatly increases the proportion of the enzyme in this pathway. Ribbon diagrams depict the structure of mature cathepsin L in the extracellular matrix and in the lysosome. The ribbon colors correspond to the colors used in the diagram on the left.
Cathepsin biosynthesis, processing, and trafficking.
As secreted proteins, cathepsins are synthesized with an N-terminal signal peptide that targets the protein to the lumen of the ER (Figure 2). The signal peptide is cotranslationally cleaved and N-linked glycosylation occurs within the ER. Similar to other proteases, cysteine cathepsins are synthesized as inactive proenzymes and require proteolytic processing for activity. The immature protein possesses an N-terminal proregion, which is removed to activate the enzyme, suggesting that the proregion acts as an autoinhibitor. Indeed, synthetic peptides corresponding to proregions do function as specific inhibitors of the parent cathepsin in the case of cathepsin L and cathepsin-like cysteine proteases (27, 28). The understanding of the nature of cathepsin propeptide inhibition has been advanced by the structural determination of a number of proenzyme forms of cathepsins, including cathepsins B (29, 30), L (29), K (31, 32), X (33), and S (34). The structures are broadly similar, with a small “mini-domain” at the N terminus, which forms a small compact structure, and an extended peptide, which is bound over the active site cleft, occluding it. The inhibitory peptide is not cleaved, as it is held in an inverse, nonproductive orientation over the active site. The folded mini-domain interacts noncovalently with the enzyme to tether the inhibitory peptide over the active site at one end. The tether at the other end is provided by the peptide bond, connecting the enzyme and proregion. Binding within the active site is noncovalent, with the exception of cathepsin X, which has a disulfide bond between a cysteine in the proregion and the active site cysteine (33). In addition to its role in autoinhibition, the N-terminal proregion of cathepsin L appears to be necessary for the correct folding of the protein (35), a finding that is also true for papain (36). Mutations destabilizing the interface between the helices prevent correct folding of the protein (37, 38). Folding of the proregion may precede folding of the full protein, thereby providing a scaffold that then directs the folding of the remaining domains. The full structure would be stabilized by the formation of disulfide bonds that prevent unfolding once the proregion has been removed (Figure 2).
Initial glycosylation generates high mannose glycans within the ER. Cathepsins destined for the lysosome are further processed in the Golgi apparatus by modification of mannose residues to mannose-6-phosphate (m6p). For cathepsin D, recognition of the cathepsin by the initial enzyme in m6p formation requires interactions with surface loops in the structure (39). Two to three lysine residues separated by 34 Å appear to be critical in the recognition motif (40). Modified cathepsins bind to the m6p receptor for targeting to the lysosome. Upon acidification in late endosomes, cathepsins become active and begin proteolytic processing, with cleavage within the proregion allowing proregion dissociation from the enzyme. This process may occur autocatalytically for both cathepsin B (41) and cathepsin L (42). Due to the distance between the active site and the identified cleavage sites, the process is believed to be intermolecular rather than intramolecular (42). This results in an active, single chain form of the protein. Upon arrival at the lysosome, further processing cleaves the protein into two chains. Active cathepsins may also be recruited from late endosomes or lysosomes for secretion into the extracellular space via Ca2+-mediated fusion of these organelles with the plasma membrane (43, 44). In addition, a minor population of cathepsins (approximately 5%) does not travel to the lysosome but is instead secreted as a proenzyme. Furthermore, alternative splicing and exon skipping can lead to cathepsin forms that lack the signal peptide, and these can subsequently localize to the nucleus and mitochondrial matrix (Figure 3) (21, 45). Recent data suggest that truncated forms of cathepsin L are important in regulating the cytoskeleton of kidney podocytes (7, 8), whereas others have described mature cathepsin L outside lysosomes, e.g., during histone processing in embryonic stem cells (25). These cathepsin L variants have been previously shown to arise by translation from an alternate downstream AUG site (10) and to be located in the nucleus of fibroblasts, in which they can cleave the transcription factor cut-like homeobox 1 (Cux1) (10). Cathepsin L also processes histone H3 during mouse embryonic stem cell differentiation (25). Although conventional cathepsin L cleaves various proteins very efficiently, due to the denaturing conditions and low pH of the lysosome, cytosolic and nuclear cathepsin L exhibit remarkable substrate specificity that allows a very specific enzymatic activity at cytosolic or nuclear pH (46). So far, two substrates of cytosolic cathepsin L have been described in podocytes: dynamin (7) and synaptopodin (8). In addition, CD2-associated protein (CD2AP) in podocytes is protected from puromycin-induced degradation in the absence of cathepsin L, suggesting the possibility that CD2AP is an additional cathepsin L substrate (9).
Figure 3. Alternative translation from downstream AUG sites produces cytoplasmic and nuclear cathepsin L.
(A) Alternative translation from downstream AUG sites produces cathepsin L that is devoid of the leader sequence and that can be present outside the lysosome, including in the cytosol and the nucleus. (B) Alternative translation leads to peptides that lack the leader sequence and thus are targeted to the cytosol or nucleus. The initial folding of the protein requires an intact proregion, and the mature protein is stabilized by three disulfide bonds. Thus, this process may require the presence of yet to be identified chaperones to generate a functional enzyme.
Opposing roles of cathepsins B and L in acute pancreatitis
Physiological activation of digestive proteases.
The acinar cells of the exocrine pancreas produce and secrete a wide variety of potent proteolytic enzymes essential for intestinal digestion of nutrient proteins. However, these digestive enzymes are potentially harmful. Therefore, these proteases are produced as precursors (i.e., zymogens) within pancreatic acinar cells and are only activated in the duodenum. The key step in this activation process is the conversion of inactive trypsinogen to active trypsin by limited proteolysis by enteropeptidase, a highly selective trypsinogen-cleaving protease located at the luminal site of duodenal cells (26). The proteolytically active trypsin initiates an activation cascade of proteolytic enzymes within the duodenum, thereby ensuring the high proteolytic capacity needed for food digestion (26).
Cathepsin B: a major trypsin activator in acute pancreatitis.
Because of the vital role of trypsin in protease activation, the ectopic intrapancreatic cleavage of trypsinogen to proteolytically active trypsin has long been considered as a key event in the pathogenesis of acute and chronic pancreatitis (47). The most direct evidence for this pathogenic principle comes from recent work on hereditary chronic pancreatitis: most affected families have heritable mutations that either increase the autoactivation of trypsinogen or impair the inhibition/degradation of active trypsin within pancreatic acinar cells (48). Yet, cathepsin mutations have not been found in hereditary pancreatitis. However, cathepsin B has long been considered a promising candidate for the trypsinogen-activating protease in pancreatic acinar cells. Support for this notion stems from in vitro experiments showing that cathepsin B can remove the N-terminal hexapeptide, the so called “trypsinogen-activation peptide”, from the human, rodent, and bovine trypsinogen zymogens. Notably, this cleavage mimics that of the physiological trypsinogen activator enteropeptidase (49, 50). The isoleucine residue that is exposed and represents the N terminus of trypsin interacts via its free amino group with the side-chain carboxy group of asparagine 194 (chymotrypsin numbering), thereby enabling and stabilizing the proteolytic active conformation of trypsin (51). The functional relevance of cathepsin B for trypsinogen activation in acute pancreatitis has been analyzed in various rodent models. In these studies, both genetic deficiency for cathepsin B and treatment of animals with a cathepsin B inhibitor resulted in reduced trypsin activity and amelioration of pancreatitis (52–54).
Cathepsin L: keeping trypsin in check.
Because of their partially overlapping substrate repertoires, it had been widely assumed that cysteine cathepsins other than cathepsin B might also contribute to trypsinogen activation in pancreatitis. Biochemical data obtained from bovine and human proteins revealed a specific cathepsin L cleavage site in trypsinogen at a position 3 amino acids C-terminal from the normal enteropeptidase/cathepsin B cleavage site (5). This cleavage prevents the generation of the N-terminal isoleucine that is essential for the active conformation of trypsin. Hence, in contrast to cathepsin B, cleavage of trypsinogen by cathepsin L results in an inactive trypsin variant. Active trypsin is not cleaved by cathepsin L at this position but at a second cathepsin L cleavage site between amino acids E82 and G83 that inactivates trypsin (5). These data imply that cathepsin L can prevent ectopic trypsinogen activation. In keeping with this notion, Ctsl–/– mice show increased intrapancreatic trypsin activity upon pancreatitis induction (5). Of note, Ctsl–/– mice also show less severe pancreatitis, because, as a result of the less severe local and systemic inflammation, acinar cells undergo apoptosis instead of necrosis. Thus, cysteine cathepsins serve complex roles in the pancreas beyond activating and inactivating trypsinogen.
Cathepsins and protease zymogens: how do they meet?
Despite compelling evidence for selective cleavage of trypsinogen by cathepsins B and L, an important cell biological question remains to be resolved: how can endolysosomal cathepsins meet trypsinogen, which is located in the secretory vesicles of the pancreatic acinar cell that are known as zymogen granules? Of note, colocalization of cathepsin B and trypsinogen has been shown by subcellular fractionation and immunogold labeling of EM sections in pancreata from patients and rodents with pancreatitis (reviewed in ref. 54).
The activation of trypsinogen begins in vesicular organelles that are acidic (55), raising the possibility that the colocalization of trypsinogen and cathepsins could result from the missorting of lysosomal cathepsins, due to inefficient trafficking from the trans-Golgi network to the endosomal/lysosomal compartment via the m6p/m6p receptor pathway (56). It has been estimated that approximately 5%–10% of lysosomal proteins are missorted and enter the constitutive and regulated secretory pathways, and it is clear that cathepsin L and other cysteine cathepsins can selectively effect their protease function in secretory vesicles (18, 57). For example, cathepsin L has been identified as a major enzyme involved in the generation of secreted pituitary neuropeptides (e.g., enkephalin, ACTH, α-MSH, β-endorphin) from their large precursor proteins, such as preproenkephalin and proopiomelanocortin, in human cells and mice (17, 58, 59). However, as it stands, it is not clear how the selective sorting of cathepsins into zymogen granules can be induced during the onset of acute pancreatitis.
In the search for alternative mechanisms, it was recently proposed that incompletely executed autophagy leads to the observed enzyme colocalization in early pancreatitis (60). Both starvation and pancreatitis induce an autophagic response in pancreatic acinar cells that engulfs cytoplasmic proteins and organelles, including zymogen granula (60). After fusion of autophagosomes with lysosomes, the contents of the resulting autophagolysosomes are digested by lysosomal hydrolases, including cathepsins (61). Eventually, the autophagolysosomes break down and disappear in starvation-induced autophagy. In contrast, during pancreatitis the autophagic process is impaired at the level of autophagolysosome degradation, thereby providing an acidic compartment for colocalization of trypsinogen and cathepsins (60). Furthermore, an imbalance consisting of low levels of cathepsin L, which degrades trypsinogen and trypsin, and high levels of cathepsin B, which activates trypsinogen, has been found in autophagic vesicles during pancreatitis (60). This, in turn, results in intracellular accumulation of active trypsin that activates further digestive enzymes, thereby causing damage to pancreatic cells, a hallmark event in the pathogenesis of pancreatitis.
Cysteine cathepsins in the heart
Cathepsin L: a homeostatic protease within the myocardium.
Cardiomyopathies represent a heterogeneous group of heart diseases that are characterized by progressive myocardial remodeling, leading to impaired pump function of the heart (62, 63). Among many other etiologies, defects in lysosomes and lysosomal hydrolases have been shown to cause myocardial heart disease (64, 65). Cardiomyopathies have also been described as a component of inherited disorders caused by deficiency of lysosomal glycosidases; for example, Pompe disease is caused by abnormal accumulation of glycogen. Deficiency of lysosome-associated membrane protein 2 (LAMP-2) induces the accumulation of autophagic vacuoles and causes Danon disease, which is characterized by severe myopathy of cardiac and skeletal muscles (64, 65). Of note, LAMP-2–deficient mice display a vacuolar cardioskeletal myopathy that is similar to that observed in individuals with Danon disease (66). Increased activity of lysosomal enzymes also has been found in patients with hypertensive heart failure (67, 68).
In contrast to those of these well-established lysosomal storage diseases and their causative molecules, the role of lysosomal proteases in the heart remained elusive for a long time. However, recent findings with 1-year-old Ctsl–/– mice shed light on this issue (69, 70). These aging animals develop a cardiac phenotype that displays key features of human dilated cardiomyopathy (69). As such, complete deficiency of cathepsin L causes interstitial myocardial fibrosis and the appearance of pleomorphic nuclei in cardiomyocytes, both characteristics of human cardiomyopathies. It also causes cardiac chamber dilation and impaired cardiac contraction. Moreover, at 1 year of age, Ctsl–/– mice develop supraventricular tachycardia, ventricular extrasystoles, and first-degree atrioventricular blockage (69). Deficiency of cathepsin L in mice affects the endolysosomal system of cardiomyocytes in newborn mice (69). In particular, it increases the number of acidic organelles, although these vesicles lack the accumulation of typical lysosomal storage materials and have altered morphology (70). Subsequently, the defects in the acidic cellular compartment are accompanied by complex biochemical and cellular alterations, with loss of cytoskeletal proteins and mitochondrial impairment (70). These findings raise the question of how cathepsin L deficiency and the observed alteration of the acidic cellular compartment change intracellular signaling toward induction of a hypertrophic response with subsequent dilation of the heart.
Cathepsin L involvement in cardiac signal transduction.
In a gain-of-function approach, human cathepsin L was transgenically overexpressed in the cardiomyocytes of mice (71). The transgenic mice had a decreased hypertrophic response and exhibited reduced cardiomyocyte apoptosis in two models of hypertensive heart failure, i.e., aortic banding and angiotensin II infusion. The observed cardioprotective effect of human cathepsin L overexpression in the mouse heart was associated with inhibition of Akt signaling (71). As of yet, it has not been fully resolved how cathepsin L blocks Akt signaling in the heart, but some information on how cathepsin L affects intracellular signal transduction processes is available from data obtained in mouse epidermis. Cathepsin L–deficient mouse keratinocytes show enhanced recycling of nondegraded plasma membrane receptors and their ligands to the cell surface and sustained growth factor signaling (72). This impaired termination of growth factor signaling within the endolysosomal compartment results in increased Ras, Akt, and MAPK activation (73). As a consequence, the proliferation of basal epidermal keratinocytes is increased. This, in turn, results in the epidermal hyperproliferation and increased susceptibility to squamous carcinogenesis in the skin of Ctsl–/– mice (74). Thus, the overexpression of cathepsin L in the endolysosomal compartment of cardiomyocytes is likely to result in immediate proteolysis of endocytosed receptors and their ligands. Therefore, both the time span available for signaling from the receptors and the rate of receptor recycling are reduced. This premature signal termination lowers the activation state of cytosolic kinases like Akt and therefore reduces the hypertrophic response of the challenged mouse myocardium (71).
Extracellular cathepsins in cardiac remodeling and repair.
It is also worth noting that dilative cardiomyopathy–associated interstitial fibrosis in Ctsl–/– mice is the only defect in the heart that cannot be rescued by transgenic reexpression of mouse cathepsin L in cardiomyocytes of otherwise cathepsin L–deficient mice (75). These results imply that the observed cardiac fibrosis in Ctsl–/– mice is caused by the absence of cathepsin L from cardiac fibroblasts and not from cardiomyocytes. Since collagen I (Col1a1) mRNA expression is not enhanced in cathepsin L–deficient myocardium, the accumulation of collagen in the ECM is most likely due to impaired collagen turnover.
Cathepsin L is mainly located in the endosomal/lysosomal compartment, but a fraction of the proenzyme can be secreted and activated by other proteases such as matrix metalloproteinases. Activated extracellular cathepsin L is capable of processing ECM proteins, such as fibronectin, laminin, and type I, IV, and XVIII collagen, even at neutral pH (76–78). Cysteine cathepsins, such as cathepsin S and cathepsin B, are highly abundant in the left ventricular myocardium of patients with hypertensive heart failure and therefore have been implicated in turnover of the ECM and cardiac remodeling in this disease (68, 79).
This turns attention to another emerging aspect of extracellular cathepsin L — its involvement in cardiac repair. Endothelial progenitor cells home to ischemic areas, differentiate, and build the basis for new blood vessels, a process known as neovascularization (80). Improvement of vascularization and function of ischemic areas in the heart may represent a physiologic function of endothelial progenitor cells. The infusion of excess numbers of in vitro–differentiated progenitors is currently being evaluated in clinical trials aimed at improving the outcome of postinfarction congestive heart failure (81). Bone marrow–derived endothelial progenitor cells show high cathepsin L expression and activity, and neovascularization after experimental hind limb ischemia is substantially impaired in Ctsl–/– mice (82). Furthermore, infusion of cathepsin L–deficient mononuclear cells, but not that of mononuclear cells from cathepsin D– or MMP9-deficient mice, results in a failure of these cells to home efficiently. Mechanistically, cathepsin L is required for the invasion and the proteolytic matrix-degrading activity of the endothelial progenitors (82). Interestingly, diabetes, a typical risk factor for ischemic heart disease, impairs human and mouse cathepsin L activity (but not that of other major proteases) in endothelial progenitor cells and also reduces invasion of these cells in a glucose concentration–dependent manner (83). Hence, the specific impairment of cathepsin L function by hyperglycemia may explain the poor neovascularization and regeneration capacities of ischemic tissues in diabetic patients.
The role of truncated forms of cathepsin L in murine and human renal disease
The role of podocyte cathepsin L in proteinuric kidney diseases.
The kidney glomerulus is a highly specialized structure that ensures the selective ultrafiltration of plasma, so that essential proteins are retained in the blood (84). Podocytes are unique cells with a complex cellular organization, consisting of a cell body, major processes, and foot processes (FPs). The FPs cover the outer aspect of the glomerular basement membrane. They form a characteristic interdigitating pattern with FPs of neighboring podocytes, leaving in between the filtration slits that are bridged by the glomerular slit diaphragm (SD) (84). Human genetic studies have revealed that mutations affecting several podocyte proteins, including α-actinin-4 (85), nephrin (86), PLCε1 (87), podocin (88), transient receptor potential cation channel, subfamily C, member 6 (TRPC6) (89, 90), and inverted forming gene 2 (INF2) (91), lead to renal disease, owing to disruption of the filtration barrier and rearrangement of the podocyte actin cytoskeleton. Cell biologic and mouse genetic studies revealed that additional proteins regulate the plasticity of the podocyte actin cytoskeleton, such as Rho GDIa, podocalyxin, FAT tumor suppressor homolog 1 (FAT1), NCK adaptor protein 1 (Nck1), Nck2, synaptopodin, and dynamin, and are also of critical importance for sustained function of the kidney glomerular filtration barrier (reviewed in ref. 92).
The onset of proteinuria, induced by either LPS (7, 8) or puromycin aminonucleoside (PAN) (9), is associated with the induction of cathepsin L expression and activity in podocytes. The latter study also introduced the emerging concept that the onset of proteinuria represents a migratory event in podocyte FP that is associated by the activation of cathepsin L (9). This study also showed that the SD-associated CD2AP (93) was protected from puromycin-induced degradation in primary podocyte cultures derived from Ctsl–/– mice (9). Subsequently, it was found that a cytoplasmic variant of cathepsin L in podocytes is required for the development of proteinuria in mice through a mechanism that involves the cleavage of the large GTPase dynamin (7) and synaptopodin (8). The clinical relevance of these findings was underscored by the observation that podocyte cathepsin L expression is increased in a variety of human proteinuric kidney diseases, ranging from minimal change disease (MCD) to diabetic nephropathy (7). Together, these results support the notion that cathepsin L–mediated proteolysis plays a critical role in the development of various forms of proteinuria (94).
Cathepsin L–mediated degradation of dynamin leads to proteinuria in mice.
Dynamin is essential for the formation of clathrin-coated vesicles at the plasma membrane during endocytosis (95). Dynamin has also been implicated in the regulation of actin dynamics in certain cell types (96). Using the Prediction of Endopeptidase Proteolytic Sites computer algorithm (PEPS) for predicting putative cathepsin L substrates (46), dynamin was identified as a target of cathepsin L (7). In mouse podocytes, dynamin is cleaved by cytoplasmic cathepsin L during LPS- or PAN-induced experimental proteinuria, and gene delivery of cathepsin L–resistant dynamin protected mice against LPS-induced proteinuria (7). The notion that dynamin is required for proper podocyte structure and function is further supported by the observation that overexpression of dominant-negative dynamin leads to a loss of podocyte stress fibers in vitro and development of proteinuria in mice (7).
Cyclosporine A prevents proteinuria by blocking cathepsin L–mediated degradation of synaptopodin in podocytes.
Calcineurin is a ubiquitously expressed serine/threonine phosphatase (97). Its best-characterized function is the regulation of nuclear factor of activated T cells (NFAT) signaling. The immunosuppressive action of the calcineurin inhibitor cyclosporine A (CsA) stems from its inhibition of NFAT signaling in T cells (98). CsA can also induce remission of the proteinuria associated with diseases such as MCD and focal segmental glomerulosclerosis (FSGS) (99). Although T cell dysfunction is associated with some forms of proteinuria (100), including a subset of children with MCD (101), the salutary action of CsA in MCD and FSGS led to the suspicion that CsA might exert its effect, at least in part, independently of its effects on T cells, a hypothesis also supported by reports of CsA effectiveness in nonimmunological human (102) and experimental (103) Alport syndrome.
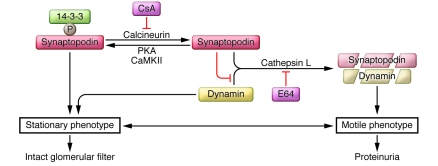
Recently, a mechanism was identified wherein CsA blocks calcineurin-mediated dephosphorylation of synaptopodin in mouse podocytes, thereby preserving the phosphorylation-dependent synaptopodin/14-3-3β interaction (8). This interaction, in turn, protects synaptopodin from cathepsin L–mediated degradation and preserves a stable filtration barrier. Moreover, inducible expression of dominant-active calcineurin in podocytes is sufficient to cause the degradation of synaptopodin and dynamin, thereby inducing proteinuria (8). These data describe a calcineurin/cathepsin L signaling pathway in podocytes that contributes to the regulation of kidney filter function (Figure 4). In contrast to most other calcineurin-NFAT controlled signaling events (97, 98, 104, 105), the antiproteinuric effect of CsA stems, at least in part, from its inhibition of cathepsin L–mediated degradation of synaptopodin in podocytes (Figure 4 and ref. 8).
Figure 4. Cytosolic cathepsin L and its function in proteinuric kidney disease.
Phosphorylation of synaptopodin by PKA or CaMKII promotes 14-3-3 binding, which protects synaptopodin and dynamin against cathepsin L–mediated cleavage, thereby contributing to a stationary podocyte phenotype and an intact glomerular filtration barrier. Dephosphorylation of synaptopodin by calcineurin abrogates the interaction with 14-3-3. This renders the cathepsin L cleavage sites of synaptopodin accessible and promotes the degradation of synaptopodin and dynamin, thereby promoting a motile phenotype and the development of proteinuria. The calcineurin inhibitor CsA and the cathepsin inhibitor E64 safeguard against proteinuria by stabilizing synaptopodin and dynamin protein levels in podocytes.
The emerging role of nuclear cathepsin L in polycystic kidney disease.
Polycystic kidney disease (PKD) represents the most common genetic renal disease in the world. PKD is inherited as an autosomal dominant (ADPKD) or autosomal recessive (ARPKD) trait and characterized by progressive enlargement of renal cysts (106). Cux1 is a homeobox gene that represses the cyclin kinase inhibitors p21 and p27, and transgenic mice ectopically expressing Cux1 develop renal hyperplasia (107). A 246–amino acid deletion in Cux1 accelerates PKD progression in the cpk model of ARPKD (11), and the ensuing phenotype was explained by a missing cathepsin L cleavage site in the truncated Cux1 mutant, which thereby maintains increased tubular cell proliferation and apoptosis. Cux1 is proteolytically processed by a nuclear isoform of cathepsin L (10). In both human ADPKD cells and in kidneys of mice with a targeted deletion in Pkd1, a murine model of PKD, decreased nuclear cathepsin L levels are associated with increased levels of Cux1 protein in the cystic cells in vitro and the cysts in vivo (11). These results suggest a mechanism by which reduced Cux1 processing by nuclear cathepsin L results in the accumulation of Cux1, downregulation of p21/p27, and increased cell proliferation in PKD (11). Furthermore, they provide proof of principle of the hypothesis that nuclear cathepsin L is capable of processing transcription factors that control important cellular programs, such as growth.
Outlook
Recent studies have uncovered multiple divergent roles for different cathepsins in a variety of physiologic and pathophysiologic processes. From the findings in the different organs discussed above, it has become clear that cathepsins serve as regulatory enzymes beyond acting as simple housekeeping proteases and harbor important functions outside the lysosome. Future studies are required to delineate the translational mechanisms leading to the generation of the truncated forms of cathepsin L. Structural insights should aid drug development of cathepsin inhibitors that act in an allosteric manner and, therefore, may be more specific for individual cathepsin forms than currently available inhibitors. The understanding of cathepsins and their recently identified substrates continues to be an expanding area in biology. Most importantly, they provide starting points for the development of novel selective therapeutic modalities for various human diseases.
Acknowledgments
This work was supported in part by a US NIH grant (DK073495) to J. Reiser, the American Society of Nephrology Carl W. Gottschalk Research Scholar grant to B.D. Adair, and the Deutsche Forschungsgemeinschaft SFB 850 Project B7, the Centre of Chronic Immunodeficiency Freiburg grant (TP8), the Excellence Initiative of the German Federal and State Governments (EXC 294), and the European Union Framework Program (FP7 “MICROENVIMET” no. 201279) to T. Reinheckel.
Footnotes
Authorship note: Jochen Reiser, Brian Adair, and Thomas Reinheckel contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(10):3421–3431. doi:10.1172/JCI42918.
References
- 1.Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4(7):544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 2.Barrett AJ, Rawlings ND. ‘Species’ of peptidases. Biol Chem. 2007;388(11):1151–1157. doi: 10.1515/BC.2007.151. [DOI] [PubMed] [Google Scholar]
- 3.Willstätter R, Bamann E. Über die Proteasen der Magenschleimhaut. Erste Abhandlung über die Enzyme der Leukozyten. Hoppe-Seyler’s Z Physiol Chem. 1929;180(1–3):127–143. [Google Scholar]
- 4.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36(Database issue):D320–D325. doi: 10.1093/nar/gkn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wartmann T, et al. Cathepsin L inactivates human trypsinogen, whereas cathepsin L-deletion reduces the severity of pancreatitis in mice. Gastroenterology. 2010;138(2):726–737. doi: 10.1053/j.gastro.2009.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puri N, Roche PA. Mast cells possess distinct secretory granule subsets whose exocytosis is regulated by different SNARE isoforms. Proc Natl Acad Sci U S A. 2008;105(7):2580–2585. doi: 10.1073/pnas.0707854105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sever S, et al. Proteolytic processing of dynamin by cytoplasmic cathepsin L is a mechanism for proteinuric kidney disease. J Clin Invest. 2007;117(8):2095–2104. doi: 10.1172/JCI32022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faul C, et al. The actin cytoskeleton of kidney podocytes is a direct target of the antiproteinuric effect of cyclosporine A. Nat Med. 2008;14(9):931–938. doi: 10.1038/nm.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiser J, et al. Podocyte migration during nephrotic syndrome requires a coordinated interplay between cathepsin L and alpha3 integrin. J Biol Chem. 2004;279(33):34827–34832. doi: 10.1074/jbc.M401973200. [DOI] [PubMed] [Google Scholar]
- 10.Goulet B, et al. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol Cell. 2004;14(2):207–219. doi: 10.1016/S1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- 11.Alcalay NI, et al. Acceleration of polycystic kidney disease progression in cpk mice carrying a deletion in the homeodomain protein Cux1. Am J Physiol Renal Physiol. 2008;295(6):F1725–F1734. doi: 10.1152/ajprenal.90420.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasiljeva O, Reinheckel T, Peters C, Turk D, Turk V, Turk B. Emerging roles of cysteine cathepsins in disease and their potential as drug targets. Curr Pharm Des. 2007;13(4):387–403. doi: 10.2174/138161207780162962. [DOI] [PubMed] [Google Scholar]
- 13.Honey K, Rudensky AY. Lysosomal cysteine proteases regulate antigen presentation. Nat Rev Immunol. 2003;3(6):472–482. doi: 10.1038/nri1110. [DOI] [PubMed] [Google Scholar]
- 14.Stoch SA, Wagner JA. Cathepsin K inhibitors: a novel target for osteoporosis therapy. Clin Pharmacol Ther. 2008;83(1):172–176. doi: 10.1038/sj.clpt.6100450. [DOI] [PubMed] [Google Scholar]
- 15.Deal C. Potential new drug targets for osteoporosis. Nat Clin Pract Rheumatol. 2009;5(1):20–27. doi: 10.1038/ncprheum0977. [DOI] [PubMed] [Google Scholar]
- 16.Friedrichs B, et al. Thyroid functions of mouse cathepsins B, K, and L. J Clin Invest. 2003;111(11):1733–1745. doi: 10.1172/JCI15990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funkelstein L, et al. Major role of cathepsin L for producing the peptide hormones ACTH, beta-endorphin, and alpha-MSH, illustrated by protease gene knockout and expression. J Biol Chem. 2008;283(51):35652–35659. doi: 10.1074/jbc.M709010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brix K, Dunkhorst A, Mayer K, Jordans S. Cysteine cathepsins: cellular roadmap to different functions. . Biochimie. 2008;90(2):194–207. doi: 10.1016/j.biochi.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 19.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6(10):764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 20.Lutgens SP, Cleutjens KB, Daemen MJ, Heeneman S. Cathepsin cysteine proteases in cardiovascular disease. FASEB J. 2007;21(12):3029–3041. doi: 10.1096/fj.06-7924com. [DOI] [PubMed] [Google Scholar]
- 21.Abudula A, Rommerskirch W, Weber E, Gunther D, Wiederanders B. Splice variants of human cathepsin L mRNA show different expression rates. Biol Chem. 2001;382(11):1583–1591. doi: 10.1515/BC.2001.193. [DOI] [PubMed] [Google Scholar]
- 22.Jean D, Guillaume N, Frade R. Characterization of human cathepsin L promoter and identification of binding sites for NF-Y, Sp1 and Sp3 that are essential for its activity. Biochem J. 2002;361(pt 1):173–184. doi: 10.1042/0264-6021:3610173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jean D, Rousselet N, Frade R. Cathepsin L expression is up-regulated by hypoxia in human melanoma cells: role of its 5′-untranslated region. Biochem J. 2008;413(1):125–134. doi: 10.1042/BJ20071255. [DOI] [PubMed] [Google Scholar]
- 24.Bulynko YA, Hsing LC, Mason RW, Tremethick DJ, Grigoryev SA. Cathepsin L stabilizes the histone modification landscape on the Y chromosome and pericentromeric heterochromatin. Mol Cell Biol. 2006;26(11):4172–4184. doi: 10.1128/MCB.00135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duncan EM, et al. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell. 2008;135(2):284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng XL, Kitamoto Y, Sadler JE. Enteropeptidase, a type II transmembrane serine protease. Front Biosci (Elite Ed). 2009;1:242–249. doi: 10.2741/E23. [DOI] [PubMed] [Google Scholar]
- 27.Carmona E, et al. Potency and selectivity of the cathepsin L propeptide as an inhibitor of cysteine proteases. Biochemistry. 1996;35(25):8149–8157. doi: 10.1021/bi952736s. [DOI] [PubMed] [Google Scholar]
- 28.Schilling K, et al. Selectivity of propeptide-enzyme interaction in cathepsin L-like cysteine proteases. Biol Chem. 2009;390(2):167–174. doi: 10.1515/BC.2009.023. [DOI] [PubMed] [Google Scholar]
- 29.Coulombe R, Grochulski P, Sivaraman J, Menard R, Mort JS, Cygler M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15(20):5492–5503. [PMC free article] [PubMed] [Google Scholar]
- 30.Turk V, Turk B, Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20(17):4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quraishi O, et al. The occluding loop in cathepsin B defines the pH dependence of inhibition by its propeptide. Biochemistry. 1999;38(16):5017–5023. doi: 10.1021/bi981950o. [DOI] [PubMed] [Google Scholar]
- 32.LaLonde JM, et al. The crystal structure of human procathepsin K. Biochemistry. 1999;38(3):862–869. doi: 10.1021/bi9822271. [DOI] [PubMed] [Google Scholar]
- 33.Sivaraman J, Nagler DK, Zhang R, Menard R, Cygler M. Crystal structure of human procathepsin X: a cysteine protease with the proregion covalently linked to the active site cysteine. J Mol Biol. 2000;295(4):939–951. doi: 10.1006/jmbi.1999.3410. [DOI] [PubMed] [Google Scholar]
- 34.Kaulmann G, Palm GJ, Schilling K, Hilgenfeld R, Wiederanders B. The crystal structure of a Cys25 -> Ala mutant of human procathepsin S elucidates enzyme-prosequence interactions. Protein Sci. 2006;15(11):2619–2629. doi: 10.1110/ps.062401806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao K, Stearns NA, Dong J, Wu QL, Sahagian GG. The proregion of cathepsin L is required for proper folding, stability, and ER exit. Arch Biochem Biophys. 1994;311(1):19–27. doi: 10.1006/abbi.1994.1203. [DOI] [PubMed] [Google Scholar]
- 36.Vernet T, et al. Processing of the papain precursor. The ionization state of a conserved amino acid motif within the Pro region participates in the regulation of intramolecular processing. J Biol Chem. 1995;270(18):10838–10846. doi: 10.1074/jbc.270.18.10838. [DOI] [PubMed] [Google Scholar]
- 37.Kreusch S, et al. An evolutionarily conserved tripartite tryptophan motif stabilizes the prodomains of cathepsin L-like cysteine proteases. Eur J Biochem. 2000;267(10):2965–2972. doi: 10.1046/j.1432-1033.2000.01312.x. [DOI] [PubMed] [Google Scholar]
- 38.Schilling K, Pietschmann S, Fehn M, Wenz I, Wiederanders B. Folding incompetence of cathepsin L-like cysteine proteases may be compensated by the highly conserved, domain-building N-terminal extension of the proregion. Biol Chem. 2001;382(5):859–865. doi: 10.1515/BC.2001.105. [DOI] [PubMed] [Google Scholar]
- 39.Baranski TJ, Faust PL, Kornfeld S. Generation of a lysosomal enzyme targeting signal in the secretory protein pepsinogen. Cell. 1990;63(2):281–291. doi: 10.1016/0092-8674(90)90161-7. [DOI] [PubMed] [Google Scholar]
- 40.Braulke T, Bonifacino JS. Sorting of lysosomal proteins. Biochim Biophys Acta. 2009;1793(4):605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Mach L, Mort JS, Glossl J. Noncovalent complexes between the lysosomal proteinase cathepsin B and its propeptide account for stable, extracellular, high molecular mass forms of the enzyme. J Biol Chem. 1994;269(17):13036–13040. [PubMed] [Google Scholar]
- 42.Menard R, et al. Autocatalytic processing of recombinant human procathepsin L. Contribution of both intermolecular and unimolecular events in the processing of procathepsin L in vitro. J Biol Chem. 1998;273(8):4478–4484. doi: 10.1074/jbc.273.8.4478. [DOI] [PubMed] [Google Scholar]
- 43.Jaiswal JK, Andrews NW, Simon SM. Membrane proximal lysosomes are the major vesicles responsible for calcium-dependent exocytosis in nonsecretory cells. J Cell Biol. 2002;159(4):625–635. doi: 10.1083/jcb.200208154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001;106(2):157–169. doi: 10.1016/S0092-8674(01)00421-4. [DOI] [PubMed] [Google Scholar]
- 45.Muntener K, Zwicky R, Csucs G, Rohrer J, Baici A. Exon skipping of cathepsin B: mitochondrial targeting of a lysosomal peptidase provokes cell death. J Biol Chem. 2004;279(39):41012–41017. doi: 10.1074/jbc.M405333200. [DOI] [PubMed] [Google Scholar]
- 46.Lohmuller T, et al. Toward computer-based cleavage site prediction of cysteine endopeptidases. Biol Chem. 2003;384(6):899–909. doi: 10.1515/BC.2003.101. [DOI] [PubMed] [Google Scholar]
- 47.Chiari H. Über die Selbstverdauung des menschlichen Pankreas. Zeitschrift für Heilkunde. 1896;17:69–96. [Google Scholar]
- 48.Chen JM, Ferec C. Chronic pancreatitis: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2009;10:63–87. doi: 10.1146/annurev-genom-082908-150009. [DOI] [PubMed] [Google Scholar]
- 49.Greenbaum LM, Hirshkowitz A, Shoichet I. The activation of trypsinogen by cathepsin B. J Biol Chem. 1959;234:2885–2890. [PubMed] [Google Scholar]
- 50.Figarella C, Miszczuk-Jamska B, Barrett AJ. Possible lysosomal activation of pancreatic zymogens. Activation of both human trypsinogens by cathepsin B and spontaneous acid. Activation of human trypsinogen 1. Biol Chem Hoppe Seyler. 1988;369 suppl:293–298. [PubMed] [Google Scholar]
- 51.Bode W, Huber R. Induction of the bovine trypsinogen-trypsin transition by peptides sequentially similar to the N-terminus of trypsin. FEBS Lett. 1976;68(2):231–236. doi: 10.1016/0014-5793(76)80443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Acker GJ, Saluja AK, Bhagat L, Singh VP, Song AM, Steer ML. Cathepsin B inhibition prevents trypsinogen activation and reduces pancreatitis severity. Am J Physiol Gastrointest Liver Physiol. 2002;283(3):G794–G800. doi: 10.1152/ajpgi.00363.2001. [DOI] [PubMed] [Google Scholar]
- 53.Halangk W, et al. Role of cathepsin B in intracellular trypsinogen activation and the onset of acute pancreatitis. J Clin Invest. 2000;106(6):773–781. doi: 10.1172/JCI9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Acker GJ, Perides G, Steer ML. Co-localization hypothesis: a mechanism for the intrapancreatic activation of digestive enzymes during the early phases of acute pancreatitis. World J Gastroenterol. 2006;12(13):1985–1990. doi: 10.3748/wjg.v12.i13.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lerch MM, Saluja AK, Runzi M, Dawra R, Steer ML. Luminal endocytosis and intracellular targeting by acinar cells during early biliary pancreatitis in the opossum. J Clin Invest. 1995;95(5):2222–2231. doi: 10.1172/JCI117912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roshy S, Sloane BF, Moin K. Pericellular cathepsin B and malignant progression. Cancer Metastasis Rev. 2003;22(2–3):271–286. doi: 10.1023/A:1023007717757. [DOI] [PubMed] [Google Scholar]
- 57.Hook V, Funkelstein L, Lu D, Bark S, Wegrzyn J, Hwang SR. Proteases for processing proneuropeptides into peptide neurotransmitters and hormones. Annu Rev Pharmacol Toxicol. 2008;48:393–423. doi: 10.1146/annurev.pharmtox.48.113006.094812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasothornsrikul S, et al. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc Natl Acad Sci U S A. 2003;100(16):9590–9595. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hwang SR, et al. Cathepsin L expression is directed to secretory vesicles for enkephalin neuropeptide biosynthesis and secretion. J Biol Chem. 2007;282(13):9556–9563. doi: 10.1074/jbc.M605510200. [DOI] [PubMed] [Google Scholar]
- 60.Mareninova OA, et al. Impaired autophagic flux mediates acinar cell vacuole formation and trypsinogen activation in rodent models of acute pancreatitis. J Clin Invest. 2009;119(11):3340–3355. doi: 10.1172/JCI38674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ravikumar B, et al. Mammalian macroautophagy at a glance. J Cell Sci. 2009;122(pt 11):1707–1711. doi: 10.1242/jcs.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7(8):589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 63.Hill JA, Olson EN. Cardiac plasticity. N Engl J Med. 2008;358(13):1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 64.Nishino I, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature. 2000;406(6798):906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 65.Maron BJ, et al. Clinical outcome and phenotypic expression in LAMP2 cardiomyopathy. JAMA. 2009;301(12):1253–1259. doi: 10.1001/jama.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanaka Y, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406(6798):902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 67.Akbar H, Cancelas J, Williams DA, Zheng J, Zheng Y. Rational design and applications of a Rac GTPase-specific small molecule inhibitor. Methods Enzymol. 2006;406:554–565. doi: 10.1016/S0076-6879(06)06043-5. [DOI] [PubMed] [Google Scholar]
- 68.Cheng XW, et al. Elastolytic cathepsin induction/activation system exists in myocardium and is upregulated in hypertensive heart failure. Hypertension. 2006;48(5):979–987. doi: 10.1161/01.HYP.0000242331.99369.2f. [DOI] [PubMed] [Google Scholar]
- 69.Stypmann J, et al. Dilated cardiomyopathy in mice deficient for the lysosomal cysteine peptidase cathepsin L. Proc Natl Acad Sci U S A. 2002;99(9):6234–6239. doi: 10.1073/pnas.092637699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petermann I, et al. Lysosomal, cytoskeletal, and metabolic alterations in cardiomyopathy of cathepsin L knockout mice. FASEB J. 2006;20(8):1266–1268. doi: 10.1096/fj.05-5517fje. [DOI] [PubMed] [Google Scholar]
- 71.Tang Q, et al. Lysosomal cysteine peptidase cathepsin L protects against cardiac hypertrophy through blocking AKT/GSK3beta signaling. J Mol Med. 2009;87(3):249–260. doi: 10.1007/s00109-008-0423-2. [DOI] [PubMed] [Google Scholar]
- 72.Reinheckel T, et al. The lysosomal cysteine protease cathepsin L regulates keratinocyte proliferation by control of growth factor recycling. J Cell Sci. 2005;118(pt 15):3387–3395. doi: 10.1242/jcs.02469. [DOI] [PubMed] [Google Scholar]
- 73.Dennemarker J, et al. Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol Chem. 2010;391(8):913–922. doi: 10.1515/BC.2010.097. [DOI] [PubMed] [Google Scholar]
- 74.Dennemarker J, et al. Deficiency for the cysteine protease cathepsin L promotes tumor progression in mouse epidermis. Oncogene. 2010;29(11):1611–1621. doi: 10.1038/onc.2009.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spira D, et al. Cell type-specific functions of the lysosomal protease cathepsin L in the heart. J Biol Chem. 2007;282(51):37045–37052. doi: 10.1074/jbc.M703447200. [DOI] [PubMed] [Google Scholar]
- 76.Everts V, et al. Osteoclastic bone degradation and the role of different cysteine proteinases and matrix metalloproteinases: differences between calvaria and long bone. J Bone Miner Res. 2006;21(9):1399–1408. doi: 10.1359/jbmr.060614. [DOI] [PubMed] [Google Scholar]
- 77.Felbor U, Dreier L, Bryant RA, Ploegh HL, Olsen BR, Mothes W. Secreted cathepsin L generates endostatin from collagen XVIII. EMBO J. 2000;19(6):1187–1194. doi: 10.1093/emboj/19.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maciewicz RA, Etherington DJ. A comparison of four cathepsins (B, L, N and S) with collagenolytic activity from rabbit spleen. Biochem J. 1988;256(2):433–440. doi: 10.1042/bj2560433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schenke-Layland K, et al. Cardiomyopathy is associated with structural remodelling of heart valve extracellular matrix. Eur Heart J. 2009;30(18):2254–2265. doi: 10.1093/eurheartj/ehp267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chavakis E, Urbich C, Dimmeler S. Homing and engraftment of progenitor cells: a prerequisite for cell therapy. J Mol Cell Cardiol. 2008;45(4):514–522. doi: 10.1016/j.yjmcc.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 81.Chavakis E, Koyanagi M, Dimmeler S. Enhancing the outcome of cell therapy for cardiac repair: progress from bench to bedside and back. Circulation. 2010;121(2):325–335. doi: 10.1161/CIRCULATIONAHA.109.901405. [DOI] [PubMed] [Google Scholar]
- 82.Urbich C, et al. Cathepsin L is required for endothelial progenitor cell-induced neovascularization. Nat Med. 2005;11(2):206–213. doi: 10.1038/nm1182. [DOI] [PubMed] [Google Scholar]
- 83.Urbich C, Dernbach E, Rossig L, Zeiher AM, Dimmeler S. High glucose reduces cathepsin L activity and impairs invasion of circulating progenitor cells. J Mol Cell Cardiol. 2008;45(3):429–436. doi: 10.1016/j.yjmcc.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 84.Somlo S, Mundel P. Getting a foothold in nephrotic syndrome. Nat Genet. 2000;24(4):333–335. doi: 10.1038/74139. [DOI] [PubMed] [Google Scholar]
- 85.Kaplan JM, et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet. 2000;24(3):251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- 86.Kestila M, et al. Positionally cloned gene for a novel glomerular protein--nephrin--is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1(4):575–582. doi: 10.1016/S1097-2765(00)80057-X. [DOI] [PubMed] [Google Scholar]
- 87.Hinkes B, et al. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38(12):1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 88.Boute N, et al. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet. 2000;24(4):349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- 89.Winn MP, et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science. 2005;308(5729):1801–1804. doi: 10.1126/science.1106215. [DOI] [PubMed] [Google Scholar]
- 90.Reiser J, et al. TRPC6 is a glomerular slit diaphragm-associated channel required for normal renal function. Nat Genet. 2005;37(7):739–744. doi: 10.1038/ng1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brown EJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010;42(1):72–76. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mundel P, Reiser J. Proteinuria: an enzymatic disease of the podocyte? Kidney Int. 2010;77(7):571–580. doi: 10.1038/ki.2009.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shih NY, et al. Congenital nephrotic syndrome in mice lacking CD2-associated protein. Science. 1999;286(5438):312–315. doi: 10.1126/science.286.5438.312. [DOI] [PubMed] [Google Scholar]
- 94.Defilippi P, et al. p125FAK tyrosine phosphorylation and focal adhesion assembly: studies with phosphotyrosine phosphatase inhibitors. Exp Cell Res. 1995;221(1):141–152. doi: 10.1006/excr.1995.1361. [DOI] [PubMed] [Google Scholar]
- 95.Kirchhausen T. Three ways to make a vesicle. Nat Rev Mol Cell Biol. 2000;1(3):187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- 96.Schafer DA. Regulating actin dynamics at membranes: a focus on dynamin. Traffic. 2004;5(7):463–469. doi: 10.1111/j.1600-0854.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 97.Aramburu J, Heitman J, Crabtree GR. Calcineurin: a central controller of signalling in eukaryotes. EMBO Rep. 2004;5(4):343–348. doi: 10.1038/sj.embor.7400133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109 suppl:S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 99.Meyrier A. Treatment of focal segmental glomerulosclerosis. Expert Opin Pharmacother. 2005;6(9):1539–1549. doi: 10.1517/14656566.6.9.1539. [DOI] [PubMed] [Google Scholar]
- 100.Shalhoub RJ. Pathogenesis of lipoid nephrosis: a disorder of T-cell function. Lancet. 1974;2(7880):556–560. doi: 10.1016/s0140-6736(74)91880-7. [DOI] [PubMed] [Google Scholar]
- 101.Audard V, et al. Minimal change nephrotic syndrome and classical Hodgkin’s lymphoma: report of 21 cases and review of the literature. Kidney Int. 2006;69(12):2251–2260. doi: 10.1038/sj.ki.5000341. [DOI] [PubMed] [Google Scholar]
- 102.Charbit M, Gubler MC, Dechaux M, Gagnadoux MF, Grunfeld JP, Niaudet P. Cyclosporin therapy in patients with Alport syndrome. Pediatr Nephrol. 2007;22(1):57–63. doi: 10.1007/s00467-006-0227-y. [DOI] [PubMed] [Google Scholar]
- 103.Chen D, et al. Cyclosporine a slows the progressive renal disease of alport syndrome (X-linked hereditary nephritis): results from a canine model. J Am Soc Nephrol. 2003;14(3):690–698. doi: 10.1097/01.ASN.0000046964.15831.16. [DOI] [PubMed] [Google Scholar]
- 104.Heit JJ, et al. Calcineurin/NFAT signalling regulates pancreatic beta-cell growth and function. Nature. 2006;443(7109):345–349. doi: 10.1038/nature05097. [DOI] [PubMed] [Google Scholar]
- 105.Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E. NFATc1 Balances Quiescence and Proliferation of Skin Stem Cells. Cell. 2008;132(2):299–310. doi: 10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Igarashi P, Somlo S. Polycystic kidney disease. J Am Soc Nephrol. 2007;18(5):1371–1373. doi: 10.1681/ASN.2007030299. [DOI] [PubMed] [Google Scholar]
- 107.Sansregret L, Nepveu A. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene. 2008;412(1–2):84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 108.McGrath ME. The lysosomal cysteine proteases. Annu Rev Biophys Biomol Struct. 1999;28:181–204. doi: 10.1146/annurev.biophys.28.1.181. [DOI] [PubMed] [Google Scholar]
- 109.Turk D, et al. Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 2001;20(23):6570–6582. doi: 10.1093/emboj/20.23.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Erickson AH. Biosynthesis of lysosomal endopeptidases. J Cell Biochem. 1989;40(1):31–41. doi: 10.1002/jcb.240400104. [DOI] [PubMed] [Google Scholar]
- 111.Garnero P, et al. The collagenolytic activity of cathepsin K is unique among mammalian proteinases. . J Biol Chem. 1998;273(48):32347–32352. doi: 10.1074/jbc.273.48.32347. [DOI] [PubMed] [Google Scholar]
- 112.Mason RW. Emerging functions of placental cathepsins. Placenta. 2008;29(5):385–390. doi: 10.1016/j.placenta.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 113.Bethel PA, et al. Design of selective Cathepsin inhibitors. Bioorg Med Chem Lett. 2009;19(16):4622–4625. doi: 10.1016/j.bmcl.2009.06.090. [DOI] [PubMed] [Google Scholar]
- 114.Viken MK, et al. Polymorphisms in the cathepsin L2 (CTSL2) gene show association with type 1 diabetes and early-onset myasthenia gravis. Hum Immunol. 2007;68(9):748–755. doi: 10.1016/j.humimm.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 115.Zeeuwen PL, et al. Colocalization of cystatin M/E and cathepsin V in lamellar granules and corneodesmosomes suggests a functional role in epidermal differentiation. J Invest Dermatol. 2007;127(1):120–128. doi: 10.1038/sj.jid.5700480. [DOI] [PubMed] [Google Scholar]
- 116.Haider AS, et al. Genomic analysis defines a cancer-specific gene expression signature for human squamous cell carcinoma and distinguishes malignant hyperproliferation from benign hyperplasia. J Invest Dermatol. 2006;126(4):869–881. doi: 10.1038/sj.jid.5700157. [DOI] [PubMed] [Google Scholar]
- 117.Hagemann S, et al. The human cysteine protease cathepsin V can compensate for murine cathepsin L in mouse epidermis and hair follicles. Eur J Cell Biol. 2004;83(11–12):775–780. doi: 10.1078/0171-9335-00404. [DOI] [PubMed] [Google Scholar]
- 118.Sevenich L, Pennacchio LA, Peters C, Reinheckel T. Human cathepsin L rescues the neurodegeneration and lethality in cathepsin B/L double-deficient mice. Biol Chem. 2006;387(7):885–891. doi: 10.1515/BC.2006.112. [DOI] [PubMed] [Google Scholar]
- 119.Toomes C, et al. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat Genet. 1999;23(4):421–424. doi: 10.1038/70525. [DOI] [PubMed] [Google Scholar]
- 120.Pham CT, Ley TJ. Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc Natl Acad Sci U S A. 1999;96(15):8627–8632. doi: 10.1073/pnas.96.15.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Saftig P, et al. Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proc Natl Acad Sci U S A. 1998;95(23):13453–13458. doi: 10.1073/pnas.95.23.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gowen M, et al. Cathepsin K knockout mice develop osteopetrosis due to a deficit in matrix degradation but not demineralization. J Bone Miner Res. 1999;14(10):1654–1663. doi: 10.1359/jbmr.1999.14.10.1654. [DOI] [PubMed] [Google Scholar]
- 123.Gelb BD, Shi GP, Chapman HA, Desnick RJ. Pycnodysostosis, a lysosomal disease caused by cathepsin K deficiency. Science. 1996;273(5279):1236–1238. doi: 10.1126/science.273.5279.1236. [DOI] [PubMed] [Google Scholar]
- 124.Potts W, et al. Cathepsin L-deficient mice exhibit abnormal skin and bone development and show increased resistance to osteoporosis following ovariectomy. Int J Exp Pathol. 2004;85(2):85–96. doi: 10.1111/j.0959-9673.2004.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kitamoto S, et al. Cathepsin L deficiency reduces diet-induced atherosclerosis in low-density lipoprotein receptor-knockout mice. Circulation. 2007;115(15):2065–2075. doi: 10.1161/CIRCULATIONAHA.107.688523. [DOI] [PubMed] [Google Scholar]
- 126.Lutgens E, et al. Disruption of the cathepsin K gene reduces atherosclerosis progression and induces plaque fibrosis but accelerates macrophage foam cell formation. Circulation. 2006;113(1):98–107. doi: 10.1161/CIRCULATIONAHA.105.561449. [DOI] [PubMed] [Google Scholar]
- 127.Sukhova GK, et al. Deficiency of cathepsin S reduces atherosclerosis in LDL receptor-deficient mice. . J Clin Invest. 2003;111(6):897–906. doi: 10.1172/JCI14915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gocheva V, et al. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;20(5):543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vasiljeva O, et al. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006;66(10):5242–5250. doi: 10.1158/0008-5472.CAN-05-4463. [DOI] [PubMed] [Google Scholar]
- 130.Gondi CS, et al. Adenovirus-mediated expression of antisense urokinase plasminogen activator receptor and antisense cathepsin B inhibits tumor growth, invasion, and angiogenesis in gliomas. Cancer Res. 2004;64(12):4069–4077. doi: 10.1158/0008-5472.CAN-04-1243. [DOI] [PubMed] [Google Scholar]
- 131.Tummalapalli P, et al. RNAi-mediated abrogation of cathepsin B and MMP-9 gene expression in a malignant meningioma cell line leads to decreased tumor growth, invasion and angiogenesis. Int J Oncol. 2007;31(5):1039–1050. [PMC free article] [PubMed] [Google Scholar]
- 132.Sevenich L, et al. Synergistic antitumor effects of combined cathepsin B and cathepsin Z deficiencies on breast cancer progression and metastasis in mice. Proc Natl Acad Sci U S A. 2010;107(6):2497–2502. doi: 10.1073/pnas.0907240107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gocheva V, et al. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24(3):241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Maehr R, et al. Cathepsin L is essential for onset of autoimmune diabetes in NOD mice. J Clin Invest. 2005;115(10):2934–2943. doi: 10.1172/JCI25485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang M, et al. Deficiency and inhibition of cathepsin K reduce body weight gain and increase glucose metabolism in mice. Arterioscler Thromb Vasc Biol. 2008;28(12):2202–2208. doi: 10.1161/ATVBAHA.108.172320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang X, Vaag A, Carlsson E, Hansson M, Ahren B, Groop L. Impaired cathepsin L gene expression in skeletal muscle is associated with type 2 diabetes. Diabetes. 2003;52(9):2411–2418. doi: 10.2337/diabetes.52.9.2411. [DOI] [PubMed] [Google Scholar]
- 137.Perdereau C, Godat E, Maurel MC, Hazouard E, Diot E, Lalmanach G. Cysteine cathepsins in human silicotic bronchoalveolar lavage fluids. . Biochim Biophys Acta. 2006;1762(3):351–356. doi: 10.1016/j.bbadis.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 138.Hornung V, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9(8):847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Srivastava M, et al. Overexpression of cathepsin K in mice decreases collagen deposition and lung resistance in response to bleomycin-induced pulmonary fibrosis. Respir Res. 2008;9:54. doi: 10.1186/1465-9921-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Buhling F, et al. Pivotal role of cathepsin K in lung fibrosis. Am J Pathol. 2004;164(6):2203–2216. doi: 10.1016/S0002-9440(10)63777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Guttentag S, Robinson L, Zhang P, Brasch F, Buhling F, Beers M. Cysteine protease activity is required for surfactant protein B processing and lamellar body genesis. Am J Respir Cell Mol Biol. 2003;28(1):69–79. doi: 10.1165/rcmb.2002-0111OC. [DOI] [PubMed] [Google Scholar]
- 142.Honey K, et al. Thymocyte expression of cathepsin L is essential for NKT cell development. Nat Immunol. 2002;3(11):1069–1074. doi: 10.1038/ni844. [DOI] [PubMed] [Google Scholar]
- 143.Hsieh CS, deRoos P, Honey K, Beers C, Rudensky AY. A role for cathepsin L and cathepsin S in peptide generation for MHC class II presentation. . J Immunol. 2002;168(6):2618–2625. doi: 10.4049/jimmunol.168.6.2618. [DOI] [PubMed] [Google Scholar]
- 144.Nakagawa T, et al. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. . Science. 1998;280(5362):450–453. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- 145.Nakagawa TY, et al. Impaired invariant chain degradation and antigen presentation and diminished collagen-induced arthritis in cathepsin S null mice. Immunity. 1999;10(2):207–217. doi: 10.1016/S1074-7613(00)80021-7. [DOI] [PubMed] [Google Scholar]
- 146.Tolosa E, et al. Cathepsin V is involved in the degradation of invariant chain in human thymus and is overexpressed in myasthenia gravis. J Clin Invest. 2003;112(4):517–526. doi: 10.1172/JCI18028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dejica VM, et al. Cleavage of type II collagen by cathepsin K in human osteoarthritic cartilage. Am J Pathol. 2008;173(1):161–169. doi: 10.2353/ajpath.2008.070494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schurigt U, et al. Cathepsin K deficiency partially inhibits, but does not prevent, bone destruction in human tumor necrosis factor-transgenic mice. Arthritis Rheum. 2008;58(2):422–434. doi: 10.1002/art.23224. [DOI] [PubMed] [Google Scholar]
- 149.Svelander L, et al. Inhibition of cathepsin K reduces bone erosion, cartilage degradation and inflammation evoked by collagen-induced arthritis in mice. Eur J Pharmacol. 2009;613(1–3):155–162. doi: 10.1016/j.ejphar.2009.03.074. [DOI] [PubMed] [Google Scholar]
- 150.Asagiri M, et al. Cathepsin K-dependent toll-like receptor 9 signaling revealed in experimental arthritis. Science. 2008;319(5863):624–627. doi: 10.1126/science.1150110. [DOI] [PubMed] [Google Scholar]