Abstract
A proteomic analysis of the secretome of cultured dermal fibroblasts from patients with systemic sclerosis (SSc) and nephrogenic systemic fibrosis (NSF) was performed to identify proteins that reflect the fibrotic process. Confluent culture supernatants from three cell strains each of normal, SSc, and NSF dermal fibroblasts were pooled separately, and each pool was labeled with a specific fluorochrome. The three pools were electrophoresed together on two-dimension SDS gels, and protein differential expression was evaluated by quantitative fluorescence analysis. The secretome analysis identified 1694 spots per sample, among which 890 spots (52%) were differentially increased or decreased (more than twofold) in SSc fibroblasts, and 985 spots (58%) were differentially increased or decreased in NSF fibroblasts compared with normal fibroblasts. Mass spectrometry analysis was then used to identify the proteins that had increased by the greatest extent in both NSF and SSc secretomes. Three reticulocalbin family members were among the 10 most up-regulated proteins. Confocal microscopy results validated the differential increase of reticulocalbin-1 in affected SSc and NSF skin, and Western blot findings demonstrated its presence in SSc sera. The secretomes of both SSc and NSF fibroblasts display a pattern of shared changes compared with the normal fibroblast secretome. The differentially increased proteins reflect an activated fibroblast phenotype and may represent a specific “fibrosis signature” that can be used as a biomarker for fibrotic diseases.
Fibrotic disorders, which include systemic sclerosis (SSc), idiopathic pulmonary fibrosis, cirrhosis of the liver, and the newly recognized nephrogenic systemic fibrosis (NSF), are characterized by abnormal and excessive deposition of collagen and other extracellular matrix components in various tissues.1–5 Although their etiologies are probably quite diverse, the presence of extracellular matrix-producing fibroblasts displaying an activated phenotype in the affected tissues is typical of all fibrotic diseases. Fibroblast activation is characterized by a marked increase in the transcriptional activity of the genes encoding type I and type III collagens and fibronectin, initiation of the expression of α-smooth muscle actin, and the reduction of extracellular matrix degradative activities.6–9 Activated fibroblasts display contractile properties resulting from the expression of stress fibers containing α-smooth muscle actin, and their profibrotic activation is part of a complex set of molecular and biochemical changes that are conserved for multiple sequential passages in vitro.6,7,10
The most frequent systemic fibrotic disorder is SSc, a disease characterized by excessive deposition of collagen and other connective tissue macromolecules in skin and multiple internal organs, prominent and often severe alterations in the microvasculature, and humoral and cellular immunological abnormalities. The most apparent and almost universal clinical features of SSc are related to the severe fibrotic changes occurring in multiple tissues and very prominently in the microvasculature.1,2 The extent and rate of progression of tissue fibrosis is of paramount importance in determining the clinical features and the prognosis of SSc. Indeed, fibrosis of the skin correlates with both survival and functional limitations.11–14
Although there has been substantial interest in the identification of biomarkers that allow early diagnosis and assessment of disease activity or that carry a predictive prognostic in SSc15–17 including global gene expression and microarray studies,18,19 fully validated biomarkers reflecting the fibrotic process are not available. The clinical semiquantitative assessment of skin thickness by palpation (modified Rodnan skin score) is considered the gold standard and the only primary outcome measure used in clinical trials of SSc disease-modifying agents. This subjective and highly variable assessment is fraught with inaccuracies as pointed out recently.20 It is, therefore, generally accepted that the development of objective and reliable markers reflecting the severity of tissue fibrosis would be of great value for improving the performance of clinical trials and the accurate assessment of the efficacy of a given treatment. Furthermore, such markers would allow a reduction in the number of patients needed for clinical trials to achieve statistical power and would offer an objective and quantitative method independent of the subjective assessment of the investigators involved in the study.
NSF is a recently recognized fibrotic disorder occurring in patients with renal insufficiency after exposure to Gd-containing contrast agents used for magnetic resonance imaging.5,21–23 Affected tissues from patients with NSF display a remarkable fibrotic process and, like SSc fibroblasts, fibroblasts cultured from patients with NSF produce increased levels of collagens and other extracellular matrix proteins, which are maintained in vitro for several passages.24,25
Here, we describe the results of a proteomic analysis of the secretome of fibroblasts from patients with SSc and NSF, which allowed the identification of several shared proteins that were substantially elevated in comparison with the secretome of normal fibroblasts and, thus, they may reflect the increased fibrogenesis of these cells. These proteins should therefore be considered putative biomarkers that may be useful to assess the extent and severity of the fibrotic process in fibrotic diseases.
Materials and Methods
Cell Culture
Dermal fibroblasts were isolated from punch or excisional biopsy samples of affected forearms obtained from patients with SSc fulfilling the American Rheumatism Association criteria for disease classification26 and from patients with a typical clinical presentation of NSF associated with recent exposure to Gd-containing magnetic resonance imaging contrast agents described previously.21,22 All patients with SSc had the diffuse cutaneous subset as defined by LeRoy et al,27 and in all patients the disease was of recent onset (<18 months) and rapidly progressive. The patients with SSc and NSF from whom the biopsy samples were obtained were matched for age and sex and had not received corticosteroids, antifibrotic therapy, or immunosuppressive therapy. Punch biopsy samples from age- and sex-matched normal subjects were used as controls. The biopsy samples, processed within 1 hour of excision, were split in two halves: one half was formalin-fixed and paraffin-embedded for histopathological and immunohistochemical analysis, and the other half was processed for establishment of dermal fibroblast cell strains. For this purpose, the skin biopsy samples were minced with a scalpel, and small pieces of tissue were placed on plastic culture dishes and then covered with tissue culture medium, which was changed every 3 to 5 days until visible outgrowth of cells was obtained in approximately 2 to 3 weeks. The cells were enzymatically disassociated with 1 mg/ml trypsin at 37°C for 5 to 30 minutes and then subcultured exactly as described by LeRoy.6 These culture conditions allowed the expansion of pure fibroblast populations without any contamination with epithelial or endothelial cells or cells of hematopoietic origin. Dermal fibroblasts from patients with SSc or NSF were subcultured and used between passages 5 and 6 to avoid loss of the fibroblast overproducer phenotype, which is preserved for at least 12 serial passages under these in vitro culture conditions.6,17 Normal fibroblasts were also matched for passage number. Cell strains were established from three different patients with SSc, three different patients with NSF, and three normal individuals. All cultures were grown to confluence in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with vitamins (Cellgro, Manassas, VA), 10% fetal bovine serum (Invitrogen), and antibiotics (Cellgro). Once the cultures reached confluence, they were washed twice and incubated for 18 hours in serum-free Dulbecco's modified Eagle's medium.
Preparation of Supernatants for Two-Dimensional Gel Electrophoresis
Supernatants from the three SSc, three normal, or three NSF cultures were pooled separately, and each pool was concentrated 40-fold by 18 hours of centrifugation on Centri-Sep 3000-kDa (Applied Biosystems, Carlsbad, CA) columns at 4°C, according to the manufacturer's instructions. To avoid any differences in the starting sample, the same volume of tissue culture media (20 ml) obtained when the cells reached overconfluence for at least 24 hours was used in the final step. Protein concentration in the culture supernatants was determined using a Quant Kit (GE Healthcare, Piscataway, NJ). The samples were then brought to pH 8 to 8.5 with 1 mol/L NaOH to optimize fluorescent tagging. For each gel, 50 μg of protein for each category was added to 400 pmol/L concentrations of Cy2, Cy3, or Cy5 fluorescent tags and allowed to incubate on ice for 30 minutes. The labeling reaction was quenched by addition of 1 μl of 10 mmol/L lysine and subsequent incubation on ice for 15 minutes. For two-dimensional (2D) electrophoresis, the three samples (normal, SSc, and NSF) were pooled, brought up to 350 μl in 8 mol/L urea, 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid, and supplemented with dithiothreitol (final concentration 13 mmol/L) and IPG buffer 3-10 (final concentration 2%) before 2D electrophoresis as described previously.28,29
Two-Dimensional Difference in Gel Electrophoresis Labeling and 2D Gel Electrophoresis
For each gel, 18-cm, pH 3 to 10 immobilized pH gradient (IPG) strips were rehydrated at 30 V for 12 hours in 350 μl of sample, using an IPGphor (GE Healthcare). Once rehydration was complete, samples were focused at 500 V for 1 hour, 1000 V for 1 hour, and finally 8000 V for 6 hours. Immediately after completion, IPG strips were processed for separation by SDS-polyacrylamide gel electrophoresis. To reduce the disulfide bonds in the focused proteins in preparation for the second dimension, IPG strips were incubated for 15 minutes in equilibration buffer I consisting of 0.375 mol/L Tris-HCl, pH 8.8, 6 mol/l urea, 2% SDS, 20% glycerol, and 13 mmol/L dithiothreitol. The IPG strips were soaked in equilibration buffer II for an additional 15 minutes to alkylate the sulfhydryl groups. Buffer II is identical to buffer I with the exception that 2.5% (w/v) iodoacetamide is used instead of dithiothreitol. The strips were embedded in 0.7% w/v agarose on top of 12.5% acrylamide slab gels. Second-dimension separations were performed on a DALT6 platform (GE Healthcare). IPG strips were electrophoresed at 2 W/gel for 30 minutes, followed by 20 W/gel until the dye front reached the bottom of the gel. The gel was rinsed in deionized water and scanned using the difference in gel electrophoresis (DIGE)-enabled Typhoon Scanner (GE Healthcare). After scanning of the images, DeCyder 5.01 software (GE Healthcare) was used for differential gel analysis. The 2D images from the different samples were then compared using the DIA module of DeCyder with a value of 1000 as the initial estimate of protein spots present. DIA analysis allows for the direct comparison of intensities of specific protein spots between different samples within the same gel.30
Tryptic Digestion of Isolated Protein Spots
The 2D gel was poststained with SYPRO Ruby and rescanned using the Typhoon Scanner. The resulting gel images were matched back to the master DIGE image and spots of interest were designated for picking from the SYPRO image. Protein spots of interest were prepared for mass spectral analysis by the Spot Handling Workstation (GE Healthcare). SYPRO-stained spots of interest were automatically cut from the gel and washed twice with 50 mmol/L ammonium bicarbonate in 50% methanol. The plugs were then dehydrated in 75% acetonitrile for 10 minutes and dried under a stream of air. Trypsin (10 μl of 20 μg/ml in 20 mmol/L ammonium bicarbonate) was added to each plug and incubated for 2 hours at 37°C. After digestion, the resulting tryptic peptides were extracted twice into 50% acetonitrile, 0.1% formic acid and then dried completely under a stream of air.
Mass Spectrometry Protein Identification
Proteins were identified via liquid chromatography-mass spectrometry using a Thermo Scientific ProteomeX Workstation consisting of a Surveyor high-performance liquid chromatograph front end, followed by an LCQ DecaXP Plus ion-trap mass spectrometer. The dried-down peptide extracts were resuspended in 15 μl of 1% formic acid, and 10 μl was loaded onto a Thermo Hypersil-Keystone BioBasic C18 column (0.18 × 100 mm). The peptides were separated at a flow rate of 4 μl/min using a linear gradient of 2 to 50% acetonitrile in 0.1% formic acid over 45 minutes. As the peptides were eluted from the column, they were subjected to a full mass spectrometry scan, followed by tandem mass spectrometry of the three largest peaks. The resulting tandem mass spectrometry spectra were analyzed using SEQUEST. Protein identifications were considered valid if they met the Human Proteome Organisation (HUPO) protein identification filter (Xcorr ≥ 1.9, 2.2, or 3.75 for z = 1, 2, and 3, respectively; ΔCN ≥ 0.1; and Rsp ≤ 4) as described previously in the Human Plasma Proteome Project.31
Confocal Microscopy Validation
To validate the results obtained by the proteomic analyses, reticulocalbin (RCN)-1, one of the proteins that displayed the highest differential expression, was selected for semiquantitative assessment of its abundance in normal, SSc, and NSF skin. RCN-1 was analyzed by immunofluorescence using a RCN-1 rabbit polyclonal antibody (Bethyl Laboratories, Montgomery, TX). Isotype control staining was performed using rabbit IgG (Sigma-Aldrich, St. Louis, MO). Secondary antibodies were affinity-purified sheep (FAb)2 anti-rabbit IgG-Cy3 conjugated (Sigma-Aldrich). Paraffin-embedded sections from skin were deparaffinized with two changes of xylene for 10 minutes each and then two changes of ethanol for 5 minutes each. Antigen retrieval was performed by boiling the tissue sections in 10 mmol/L citrate buffer, pH 6.0, for 20 minutes. The sections were then rinsed in PBS for 2 minutes and incubated with 5% normal sheep serum for 20 minutes at room temperature to block nonspecific binding sites. The primary antibody incubation step was performed overnight at 4°C followed by incubation with the sheep polyclonal antibody (1:200). The unbound antibodies were removed from the sections after each incubation with three changes of PBS for 2 minutes each. Tissue sections were counterstained with 4,6-diamidino-2-phenylindole and analyzed using a Zeiss LSM 510 META confocal laser scanning microscope system. Zeiss META confocal software was used in balancing signal strength. The breakthrough of the 4,6-diamidino-2-phenylindole signal into the red and the green channel was recorded separately and subtracted from the 4,6-diamidino-2-phenylindole blue channel. Zeiss META enhancement software was used in balancing the signal strength, and the image was scanned eightfold to separate signal from noise. Panels were assembled using Photoshop software without any RGB modification.
Western Blots
Sera from 10 patients with SSc and two healthy normal subjects (volunteer laboratory personnel) were assayed for RCN-1. All SSc sera were from patients who fulfilled the American Rheumatism Association classification criteria for SSc26 and with the diffuse subset of the disease classified according to LeRoy et al.27 Sera were diluted 1:10 with saline solution and electrophoresed on a 10% Tris-glycine SDS gel. Primary antibody against human RCN-1 (Bethyl Laboratories) was used at a 1:1000 dilution, incubated overnight at 4°C. Anti-rabbit horseradish peroxidase-conjugated antibodies at 1:5000 dilution were used as secondary antibody.
Results
2D DIGE
We analyzed by 2D DIGE the supernatants from three SSc, three NSF, and three normal fibroblast cultures. The secreted proteins from normal fibroblast cell strains were pooled and conjugated with Cy2 fluorochrome, the secreted proteins from the three SSc fibroblast cell strains were pooled and conjugated with Cy3, and the secreted proteins from the three NSF cell strains were pooled and conjugated with Cy5. The images detected for each fluorochrome are shown in Figure 1. A total of 1694 spots were detected on the 2D electrophoresis. DIGE software analysis allowed quantitative determination of each fluorescent signal in every detected spot. The differential analysis of SSc versus NSF fibroblast secretomes revealed that 1403 spots (85%) had similar levels, whereas 65 spots were increased in the SSc sample and 226 spots were increased in the NSF sample (Figure 2A). In contrast, the volume ratio analysis comparison of the secretome of normal fibroblasts with that of SSc fibroblasts indicated that 450 spots were increased (more than twofold) in the SSc fibroblast secretome, whereas 440 were decreased (<50%) and 804 had similar levels (Figure 2B). The differential analysis of NSF versus normal fibroblast secretome identified 391 spots increased in NSF, 590 decreased, and 713 with similar expression (Figure 2C).
Figure 1.
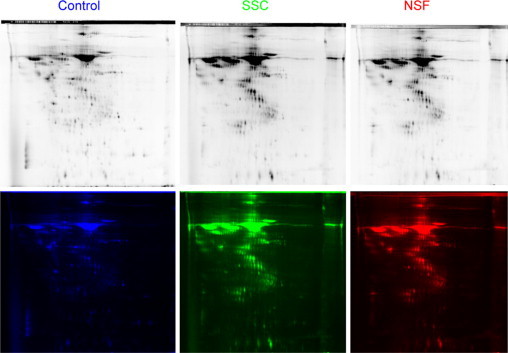
2D DIGE. The supernatants from three SSc, three NSF and three normal fibroblast cultures were pooled separately and analyzed. The secretome from normal fibroblasts was conjugated with Cy2 fluorochrome (blue), the secretome from SSc fibroblasts was conjugated with Cy3 (green), and the NSF secretome was conjugated with Cy5 (red). The top panels reflect the fluorescence intensity in each channel. The bottom panels represent the color assigned to each channel for visual differential analysis.
Figure 2.
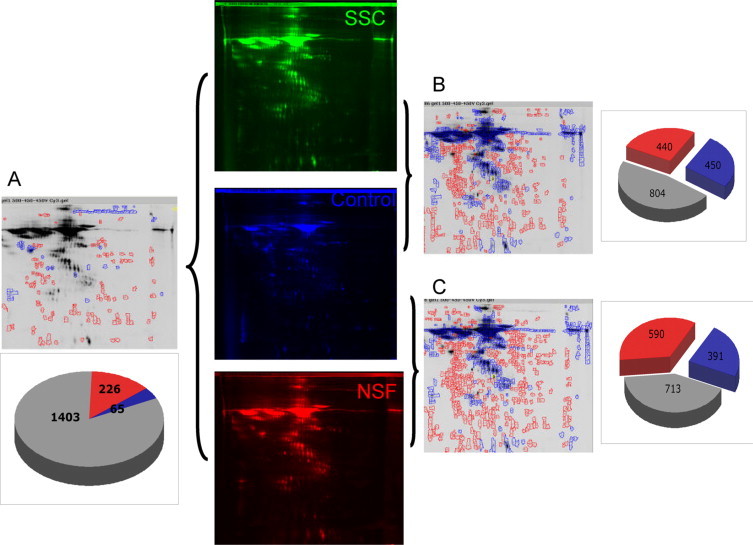
Differential analysis of fluorescence intensity. A: Differential analysis of SSc versus NSF fibroblast secretomes, showing 1403 spots (85%) with similar levels with 65 spots increased (green) in the SSc sample and 226 spots increased in the NSF sample (red). B: Volume ratio analysis comparison of the secretomes of normal fibroblasts and SSc fibroblasts, showing 450 spots increased (more than twofold) in the SSc fibroblast secretome, 440 spots decreased (<50%), and 804 spots with similar levels. C: Differential analysis of NSF versus normal fibroblast secretomes showing 391 spots increased in NSF, 590 spots decreased, and 713 spots with similar levels.
Volume Ratio Overlay
Each protein spot fluorescence intensity was calculated by DIGE software and the ratios between SSc and normal and NSF and normal were calculated (Figure 3A). To analyze the differentially expressed proteins with common quantitative changes among the NSF and SSc fibroblast secretomes, each spot was assigned a progressive number, and the fluorescence intensity ratio between SSc fibroblast secretome and normal fibroblast secretome was plotted against the spot number. The same analysis was performed for the volume ratio between the NSF and normal fibroblast secretomes and the two series of dot plots were overlaid in the same graph (Figure 3B). For the purpose of biomarker identification, we elected to perform mass spectrometry analysis of the spots that were most elevated in both SSc and NSF samples (Figure 3B, dotted line). Each of the spots with the highest level of increase compared with the normal secretome was digested and analyzed by mass spectrometry.
Figure 3.
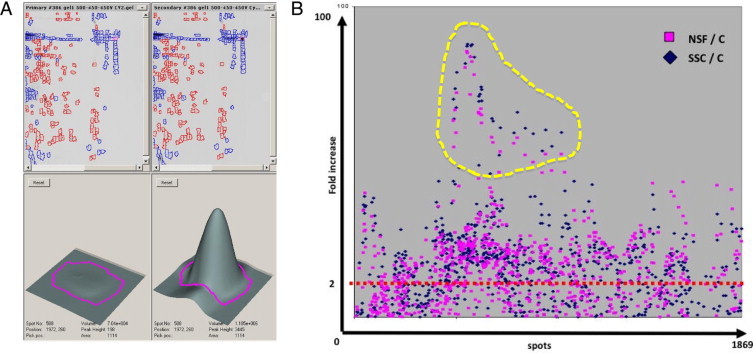
Identification of proteins increased in both SSc and NSF secretomes. A: Three-dimensional view displaying the intensity of fluorescence of a representative spot in two different channels and their fluorescence intensities calculated by DIGE software. The magenta lines in the bottom panels indicate the spot boundaries obtained using the pixel data from the images. The spot boundaries are used for spot volume calculations using the DeCyder software.30B: Dot plot of fluorescence intensity ratio of the SSc (blue diamonds) and NSF (pink squares) secretomes, both calculated against the fluorescence values of normal secretome protein spots. The yellow dotted area indicates the spots with the highest increase in both SSc and NSF secretomes compared with the normal secretome.
Mass Spectrometry Identification of the Shared Changes in SSc and NSF Fibroblast Secretomes
The list of the most up-regulated proteins identified by mass spectrometry is shown in Table 1, and the complete list of all differentially expressed proteins that were identified by mass spectrometry is provided in a Supplemental Table (see http://ajp.amjpathol.org). Of note, the most up-regulated protein, RCN-3 (accession number 28626510), and the fourth most up-regulated protein, RCN-1(accession number 4506455), belong to the CREC (Cab45, reticulocalbin, ERC-45, and calumenin) family of calcium-binding proteins residing in the endoplasmic reticulum, which are probably involved in trafficking and transport of newly synthesized proteins.32–34 Remarkably, calumenin (accession number 4502551), identified to be up-regulated on average 2.5-fold, also belongs to the same family of proteins. Of further interest was the fact that the second and third most up-regulated proteins identified were SPARC or osteonectin (accession number 4507171) and the α2 chain of type I collagen (accession number 48762934). Both proteins have been shown to be increased in SSc tissues by several independent, hypothesis-driven studies.35–37
Table 1.
List of the Most Highly and Differentially Increased Proteins in Both SSc and NSF Secretomes Identified by Mass Spectrometry
| Protein | Accession no. | Maximum fold change |
|---|---|---|
| Reticulocalbin-3 | 28626510 | 26.18 |
| Osteonectin | 4507171 | 18.28 |
| α2 chain of type I collagen | 48762934 | 17.69 |
| Reticulocalbin-1 | 4506455 | 17.69 |
| Tropomyosin 4 | 4507651 | 14.32 |
| Enolase 1 | 450371 | 10.28 |
| Calreticulin precursor | 4757900 | 7.08 |
| Actin, alpha 1 | 4501881 | 5.56 |
| Pigment epithelium-derived factor | 39725934 | 2.95 |
RCN-1 Expression Is Increased in Vivo in SSc and NSF
To validate the proteomic results, we examined the tissue levels in affected SSc and NSF skin of RCN-1, a protein found to be among the most increased in the secretomes of both SSc and NSF fibroblasts. For this purpose we analyzed by immunofluorescence followed by confocal microscopy the tissue abundance of RCN-1 in three skin biopsy samples from SSc patients, three skin biopsy samples from an affected area of skin of a patient with NSF, and three normal skin biopsy samples. Figure 4A shows representative images demonstrating substantially increased levels of RCN-1 in the dermis of the SSc and the NSF skin samples analyzed. The distribution of RCN was both cellular and pericellular, with a remarkable accumulation surrounding small vessels. Quantitative analysis of fluorescence performed in the three SSc, three NSF, and three normal skin samples showed that the overall levels of RCN-1 in SSc skin biopsies were 2.5-fold higher than those in normal skin, whereas the levels in the NSF sample were 3.1 fold higher than those in normal skin (Figure 4B). To further examine the presence of RCN-1 in the sera from patients with SSc we performed Western blots and showed that a protein band recognized by the specific anti-RCN-1 antibody and of the appropriate molecular size was clearly detectable in sera from the 10 patients with SSc examined (Figure 4C) as well as in the sera from the two healthy volunteers.
Figure 4.
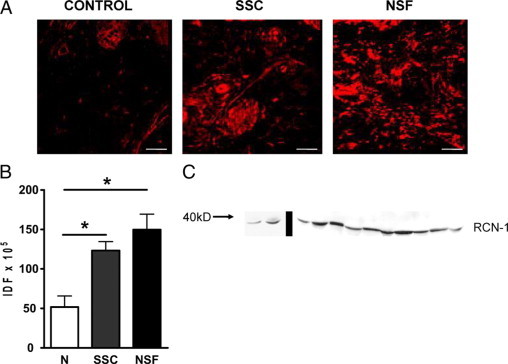
In vivo validation of RCN-1 in SSc and NSF tissues and sera. A: Tissue expression of RCN-1 in representative biopsy samples of affected skin from one patient with SSc, of affected skin from one patient with NSF, and in a sample of normal skin analyzed by immunofluorescence followed by confocal microscopy. Note the substantially increased abundance of RCN-1 in the dermis of the SSc and NSF skin samples analyzed with a cellular, pericellular, and perivascular distribution. The samples shown are representative of similar analyses of three SSc, three NSF, and three normal skin samples. Original magnification, ×200. B: Quantitative analysis of fluorescence in the three SSc, three NSF, and three normal (N) skin biopsy samples, showing overall RCN-1 abundance levels to be 2.5-fold higher in SSc skin biopsy samples and 3.1-fold higher in NSF skin biopsy samples than those in normal skin. *P < 0.05 in a two-tailed, unpaired t-test. IDF, integrated density of fluorescence. C: Western blot showing that a band of protein recognized by the specific anti-RCN-1 antibody and of the appropriate molecular size was clearly detectable in sera from the 10 patients with SSc examined (lanes to the right of the bar) as well as in sera from two normal healthy volunteers (lanes to the left of the bar).
Discussion
The excessive deposition of collagen and other extracellular matrix components in SSc and NSF tissue is due to overproduction of these proteins by fibroblasts.6–9,38–40 A key feature of SSc dermal fibroblasts as well as of fibroblasts from patients with NSF is that the profibrotic phenotype of these cells is conserved in vitro for several passages.6,8,24,25 This unique feature is likely to result from selective epigenetic regulation of a complex set of genes that is maintained for several passages in vitro as shown for SSc fibroblasts.41
Although activated fibroblasts play the central role in the development of tissue and organ fibrosis in SSc and other fibrotic diseases there is currently a lack of reliable fibroblast activity markers.17 To identify possible markers that directly reflect the level of fibroblast activity in various fibrotic diseases and that may be detectable in the sera of affected individuals we analyzed the differences in the secretome of normal versus activated or profibrotic fibroblasts using a proteomic approach. Because of the complexity and heterogeneity of skin pathology in SSc, we considered likely the possibility that some of the genes/proteins detected in the SSc fibroblast secretome may reflect other concomitant pathophysiological events such as immune activation or vasculopathy. Therefore, because the target of the studies was the identification of markers that specifically reflected tissue fibrosis and fibroblast activation, we performed in parallel differential analysis of the secretome of cultured fibroblasts derived from affected skin of patients with NSF, another fibrotic condition in which vasculopathy and autoimmunity are not considered to be present.
The rationale for studying fibroblast culture supernatants for biomarker identification relies on the assumption that a soluble secreted or shed cellular product released and present in the tissue culture media could be detected in vivo in serum and, therefore, be easily accessible for assay by venipuncture. The approach has been extremely informative for a variety of cancer-associated molecules.42,43 Another important advantage of the proteomic approach is that any biomarker identified using proteomics is completely unbiased and only depends on the sensitivity of the differential analysis of two-dimensional gel electrophoresis with the accuracy of mass spectrometry identification. The use of proteomic approaches for biomarker identification and for the study of disease pathogenesis for numerous diseases including SSc44–46 has been rapidly growing and is highly promising.
The comparative analysis of the differentially expressed proteins from SSc and NSF fibroblast secretomes revealed a pattern of shared alterations in these two clinically and etiologically different fibrotic conditions. Of interest, the mass spectrometry identification of the most increased spots revealed proteins that are already known to be part of the profibrotic phenotype of SSc fibroblasts such as α2 chain of type I collagen, α-smooth muscle actin, and osteonectin. The identification of these proteins by the comprehensive proteomic approach described here is in agreement with the numerous studies that identified their increase as a typical feature of the profibrotic phenotype of SSc fibroblasts and, most importantly, is a very strong internal validation of the functional relevance of the procedure we used here. However, we acknowledge that the data as acquired only suggest that the proteins identified are putative biomarkers. Although the sample pooling strategy we used decreases the sensitivity and does not allow statistical analyses, we believe this approach substantially increased the specificity of the study by selecting only those proteins that were overexpressed in at least six different fibroblast strains from two different fibrotic disorders. We also acknowledge that it is true that proteins lacking lysine will not be observed using this labeling method. However, such proteins account for only a small percentage of the proteome (∼8% depending on species). Therefore, despite this limitation we are observing the vast majority of the proteome. Because the DIGE step of this work is being used primarily to detect candidate proteins that might be changing in abundance, the fact that we are not observing the entire proteome, whereas certainly not ideal, is acceptable. We set out to find proteins that changed, and we were able to find them, even though we recognize that we were examining only ∼92% of the total proteome. For future work, there is a second class of Cy dyes that label cysteine rather than lysine. We plan to use these new Cy dyes in the future to cover the ∼8% of proteins that do not contain lysine.
Of remarkable interest was the identification of several proteins that were not previously known to be associated with the profibrotic phenotype. Three of the 10 most up-regulated proteins identified belonged to the CREC family of proteins, which are calcium-binding proteins residing in the endoplasmic reticulum that appear to participate in trafficking and transport of newly synthesized proteins.32–34 Furthermore, given the colocalization of the CREC proteins with protein disulfide isomerases in the endoplasmic reticulum and their opposite regulation during endoplasmic reticulum stress responses,47 they may also be involved in the regulation of the proper folding of procollagen molecules and play a role in the complex pathway of procollagen assembly, processing, and secretion mediated by protein disulfide isomerase.48–51 The presence of intracellular proteins in the concentrated supernatants may reflect cellular death, as well as increased cell turnover or the increased secretion of cell microparticles as has been suggested.52,53
We subsequently focused on the in vivo validation of the proteomic results, a necessary initial point to support the contention that these proteins may represent potential biomarkers of the fibrotic process. The most abundant member of the family of reticulocalbins is RCN-1; therefore, it represented the most important candidate for our study. Before we could consider RCN-1 as a potential biomarker of fibrosis, it was necessary to demonstrate whether it was actually overexpressed in vivo in SSc- and NSF-affected skin and whether it was detectable in the sera of affected individuals. Therefore, we performed immunofluorescence studies of skin samples from three patients with SSc and three patients with NSF to compare the amount of expression of RCN-1 with the levels detectable in normal skin biopsy samples. We observed that RCN-1 epitopes were detectable in skin, and their abundance in the dermis was significantly higher in both SSc and NSF skin samples than in normal skin samples. To determine whether RCN-1 was detectable in the serum we analyzed by Western blots sera from 10 patients with SSc, and the results indicated that a protein band recognized by specific RCN-1 antibodies and of a correct molecular size was indeed present and detectable in sera from these patients. Further validation of the “fibrosis signature” in a larger number of serum samples from patients with SSc or NSF classified by extent of fibrosis and the duration and severity of disease activity will be necessary to conclusively demonstrate whether RCN-1 and/or the other proteins in the fibrosis signature identified here represent a reliable biomarker for early diagnosis and assessment of activity of the fibrotic process in SSc and other fibrotic conditions.
Acknowledgements
We thank Megan Music and Susan V. Castro, Ph.D., for their assistance.
Footnotes
Supported by National Institutes of Health (grant R01-AR019616 to S.A.J.) and the generous support of the John Murray Foundation, the Lindsay's Hope Foundation and the Scleroderma Research Foundation of New Jersey.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
A guest editor acted as editor-in-chief for this manuscript. No person at Thomas Jefferson University was involved in the peer review process or final disposition for this article.
Current address of F.D.G.: Scleroderma Programme, Section of Musculoskeletal Diseases, Leeds Institute of Molecular Medicine, Leeds University, Leeds, UK; of M.A.S.: Clinical Pharmacology Consultant, ClinPharm Consulting, LLC, Research Triangle Park, North Carolina.
Web Extra Material
List of all protein spots that had a positive identification by mass spectrometry with their relative ratio in the two conditions. C/S ratio: Normal fibroblast secretome/SSc fibroblast secretome. C/NF ratio: Normal fibroblast secretome/NSF fibroblast secretome.
References
- 1.Varga J, Abraham D. Systemic sclerosis: a prototypic multisystem fibrotic disorder. J Clin Invest. 2007;117:557–567. doi: 10.1172/JCI31139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenbloom J, Castro SV, Jimenez SA. Fibrotic diseases: cellular and molecular mechanisms and novel therapies (Physiology in Medicine Series) Ann Intern Med. 2010;152:159–167. doi: 10.7326/0003-4819-152-3-201002020-00007. [DOI] [PubMed] [Google Scholar]
- 3.Gharaee-Kermani M, Phan SH. Molecular mechanisms of and possible treatment strategies for idiopathic pulmonary fibrosis. Curr Pharm Des. 2005;11:3943–3971. doi: 10.2174/138161205774580561. [DOI] [PubMed] [Google Scholar]
- 4.Tsukada S, Parson CJ, Rieppe RA. Mechanisms of liver fibrosis. Clin Chim Acta. 2006;364:33–60. doi: 10.1016/j.cca.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 5.Cowper SE, Su LD, Bhawan J, Robin HS, LeBoit PE. Nephrogenic fibrosing dermopathy. Am J Dermatopathol. 2001;23:383–393. doi: 10.1097/00000372-200110000-00001. [DOI] [PubMed] [Google Scholar]
- 6.LeRoy EC. Connective tissue synthesis by scleroderma skin fibroblasts in cell culture. J Exp Med. 1972;135:1351–1362. doi: 10.1084/jem.135.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jelaska A, Arakawa M, Broketa G, Korn JH. Heterogeneity of collagen synthesis in normal and systemic sclerosis skin fibroblasts. Increased proportion of high collagen-producing cells in systemic sclerosis fibroblasts. Arthritis Rheum. 1996;39:1338–1346. doi: 10.1002/art.1780390811. [DOI] [PubMed] [Google Scholar]
- 8.Jiménez SA, Feldman G, Bashey RI, Bienkowski R, Rosenbloom J. Co-ordinate increase in the expression of type I and type III collagen genes in progressive systemic sclerosis fibroblasts. Biochem J. 1986;237:837–843. doi: 10.1042/bj2370837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiménez SA, Saitta B. Alterations in the regulation of expression of the α 1(I) collagen gene (COL1A1) in systemic sclerosis (scleroderma) Springer Semin Immunopathol. 1999;21:397–414. doi: 10.1007/BF00870302. [DOI] [PubMed] [Google Scholar]
- 10.Kirk TZ, Mark ME, Chua CC, Chua BH, Mayes MD. Myofibroblasts from scleroderma skin synthesize elevated levels of collagen and tissue inhibitor of metalloproteinase (TIMP-1) with two forms of TIMP-1. J Biol Chem. 1995;270:3423–3428. doi: 10.1074/jbc.270.7.3423. [DOI] [PubMed] [Google Scholar]
- 11.Steen VD, Medsger TA., Jr Improvement in skin thickening in systemic sclerosis associated with improved survival. Arthritis Rheum. 2001;44:2828–2835. doi: 10.1002/1529-0131(200112)44:12<2828::aid-art470>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 12.Clements PJ, Hurwitz EL, Wong WK, Siebold JR, Mayes M, White B, Wigley F, Weisman M, Barr W, Moreland L, Medsger TA, Jr, Steen VD, Martin RW, Collier D, Weinstein A, Lally E, Varga J, Weiner SR, Andrews B, Abeles M, Furst DE. Skin thickness score as a predictor and correlate of outcome in systemic sclerosis: high-dose versus low-dose penicillamine trial. Arthritis Rheum. 2000;43:2445–2454. doi: 10.1002/1529-0131(200011)43:11<2445::AID-ANR11>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 13.Seibold JR, McCloskey DA. Skin involvement as a relevant outcome measure in clinical trials of systemic sclerosis. Curr Opin Rheumatol. 1997;9:571–575. doi: 10.1097/00002281-199711000-00014. [DOI] [PubMed] [Google Scholar]
- 14.Denton CP, Black CM, Abraham DJ. Mechanisms and consequences of fibrosis in systemic sclerosis. Nat Clin Pract Rheumatol. 2006;2:134–144. doi: 10.1038/ncprheum0115. [DOI] [PubMed] [Google Scholar]
- 15.Doran JP, Veale DJ. Biomarkers in systemic sclerosis. Rheumatology (Oxford) 2008;47:v36–v38. doi: 10.1093/rheumatology/ken270. [DOI] [PubMed] [Google Scholar]
- 16.Hummers LK. Biomarkers of vascular disease in scleroderma. Rheumatology (Oxford) 2008;47:v21–v22. doi: 10.1093/rheumatology/ken281. [DOI] [PubMed] [Google Scholar]
- 17.Castro SV, Jimenez SA. Biomarkers in systemic sclerosis. Biomarkers Med. 2010;4:133–147. doi: 10.2217/bmm.09.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sargent JL, Milano A, Connolly MK, Whitfield ML. Scleroderma gene expression and pathway signatures. Curr Rheumatol Rep. 2008;10:205–211. doi: 10.1007/s11926-008-0034-5. [DOI] [PubMed] [Google Scholar]
- 19.Farina G, Lafyatis D, Lemaire R, Lafyatis R. A four-gene biomarker predicts skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheum. 2010;62:580–588. doi: 10.1002/art.27220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clements PJ, Lachenbruch PA, Seibold JR, Zee B, Steen VD, Brennan P, Silman AJ, Allegar N, Varga J, Massa M, Wigley FM, Ingenito F, Weisman M, White B, Martin RW, McCloskey D, Moreland LW, Mayes M, Lally EV, Unanue M, Collier DH, Weiner S, Weinstein A, Medsger TA, Jr, Andrews B, Dixon M, Furst DE. Skin thickness score in systemic sclerosis: an assessment of interobserver variability in 3 independent studies. J Rheumatol. 1993;20:1892–1896. [PubMed] [Google Scholar]
- 21.Jiménez SA, Artlett CM, Sandorfi N, Derk C, Latinis K, Sawaya H, Haddad R, Shanahan JC. Dialysis-associated systemic fibrosis (nephrogenic fibrosing dermopathy): study of inflammatory cells and transforming growth factor beta1 expression in affected skin. Arthritis Rheum. 2004;50:2660–2666. doi: 10.1002/art.20362. [DOI] [PubMed] [Google Scholar]
- 22.Mendoza FA, Artlett CM, Sandorfi N, Latinis K, Piera-Velazquez S, Jiménez SA. Description of 12 cases of nephrogenic fibrosing dermopathy and review of the literature. Semin Arthritis Rheum. 2006;35:238–249. doi: 10.1016/j.semarthrit.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grobner T, Prischl FC. Gadolinium and nephrogenic systemic fibrosis. Kidney Int. 2007;72:260–264. doi: 10.1038/sj.ki.5002338. [DOI] [PubMed] [Google Scholar]
- 24.Piera-Velazquez S, Louneva N, Wermuth PJ, Fertala J, Del Galdo F, Jimenez SA. Persistent activation of dermal fibroblasts from patients with gadolinium-associated Nephrogenic Systemic Fibrosis. Ann Rheum Dis. 2010 doi: 10.1136/ard.2009.127761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edward M, Fitzgerald L, Thind C, Leman J, Burden AD. Cutaneous mucinosis associated with dermatomyositis and nephrogenic fibrosing dermopathy: fibroblast hyaluronan synthesis and the effect of patient serum. Br J Dermatol. 2007;156:473–479. doi: 10.1111/j.1365-2133.2006.07652.x. [DOI] [PubMed] [Google Scholar]
- 26.Subcommittee for Scleroderma Criteria of the American Rheumatism Association Preliminary criteria for the classification of systemic sclerosis (scleroderma) Arthritis Rheum. 1980;23:581–590. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- 27.LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA, Jr, Rowell N, Wollheim F. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988;15:202–205. [PubMed] [Google Scholar]
- 28.Sharma K, Lee S, Han S, Francos B, McCue P, Wassell R, Shaw MA, RamachandraRao SP. Two-dimensional fluorescence difference gel electrophoresis analysis of the urine proteome in human diabetic nephropathy. Proteomics. 2005;5:2648–2655. doi: 10.1002/pmic.200401288. [DOI] [PubMed] [Google Scholar]
- 29.Ramachandra Rao SP, Wassell R, Shaw MA, Sharma K. Profiling of human mesangial cell subproteomes reveals a role for calmodulin in glucose uptake. Am J Physiol Renal Physiol. 2007;292:F1182–F1189. doi: 10.1152/ajprenal.00268.2006. [DOI] [PubMed] [Google Scholar]
- 30.Karp NA, Kreil DP, Lilley KS. Determining a significant change in protein expression with DeCyder™ during a pair-wise comparison using two-dimensional difference gel electrophoresis. Proteomics. 2004;4:1421–1432. doi: 10.1002/pmic.200300681. [DOI] [PubMed] [Google Scholar]
- 31.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 32.Honoré B, Vorum H. The CREC family, a novel family of multiple EF-hand, low-affinity Ca2+-binding proteins localised to the secretory pathway of mammalian cells. FEBS Lett. 2000;466:11–18. doi: 10.1016/s0014-5793(99)01780-9. [DOI] [PubMed] [Google Scholar]
- 33.Vorum H, Hager H, Christensen BM, Nielsen S, Honoré B. Human calumenin localizes to the secretory pathway and is secreted to the medium. Exp Cell Res. 1999;248:473–481. doi: 10.1006/excr.1999.4431. [DOI] [PubMed] [Google Scholar]
- 34.Fukuda T, Oyamada H, Isshiki T, Maeda M, Kusakabe T, Hozumi A, Yamaguchi T, Igarashi T, Hasegawa H, Seidoh T, Suzuki T. Distribution and variable expression of secretory pathway protein reticulocalbin in normal human organs and non-neoplastic pathological conditions. J Histochem Cytochem. 2007;55:335–345. doi: 10.1369/jhc.6A6943.2006. [DOI] [PubMed] [Google Scholar]
- 35.Verrecchia F, Laboureau J, Verola O, Roos N, Porcher R, Bruneval P, Ertault M, Tiev K, Michel L, Mauviel A, Farge D. Skin involvement in scleroderma—where histological and clinical scores meet. Rheumatology (Oxford) 2007;46:833–841. doi: 10.1093/rheumatology/kel451. [DOI] [PubMed] [Google Scholar]
- 36.Davies CA, Jeziorska M, Freemont AJ, Herrick AL. Expression of osteonectin and matrix Gla protein in scleroderma patients with and without calcinosis. Rheumatology (Oxford) 2006;45:1349–1355. doi: 10.1093/rheumatology/kei277. [DOI] [PubMed] [Google Scholar]
- 37.Macko RF, Gelber AC, Young BA, Lowitt MH, White B, Wigley FM, Goldblum SE. Increased circulating concentrations of the counteradhesive proteins SPARC and thrombospondin-1 in systemic sclerosis (scleroderma). Relationship to platelet and endothelial cell activation. J Rheumatol. 2002;29:2565–2570. [PubMed] [Google Scholar]
- 38.LeRoy EC. Increased collagen synthesis by scleroderma skin fibroblasts in vitro: a possible defect in the regulation or activation of the scleroderma fibroblast. J Clin Invest. 1974;54:880–889. doi: 10.1172/JCI107827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buckingham RB, Prince RK, Rodnan GP, Taylor F. Increased collagen accumulation in dermal fibroblast cultures from patients with progressive systemic sclerosis (scleroderma) J Lab Clin Med. 1978;92:5–21. [PubMed] [Google Scholar]
- 40.Hitraya EG, Jiménez SA. Transcriptional activation of the α(I) procollagen gene in systemic sclerosis dermal fibroblasts: role of intronic sequences. Arthritis Rheum. 1996;39:1347–1354. doi: 10.1002/art.1780390812. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54:2271–2279. doi: 10.1002/art.21948. [DOI] [PubMed] [Google Scholar]
- 42.Kulasingam V, Diamandis EP. Tissue culture-based breast cancer biomarker discovery platform. Int J Cancer. 2008;123:2007–2012. doi: 10.1002/ijc.23844. [DOI] [PubMed] [Google Scholar]
- 43.Sardana G, Marshall J, Diamandis EP. Discovery of candidate tumor markers for prostate cancer via proteomic analysis of cell culture-conditioned medium. Clin Chem. 2007;53:429–437. doi: 10.1373/clinchem.2006.077370. [DOI] [PubMed] [Google Scholar]
- 44.Rottoli P, Magi B, Perari MG, Liberatori S, Nikiforakis N, Bargagli E, Cianti R, Bini L, Pallini V. Cytokine profile and proteome analysis in bronchoalveolar lavage of patients with sarcoidosis, pulmonary fibrosis associated with systemic sclerosis and idiopathic pulmonary fibrosis. Proteomics. 2005;5:1423–1430. doi: 10.1002/pmic.200301007. [DOI] [PubMed] [Google Scholar]
- 45.Bogatkevich GS, Ludwicka-Bradley A, Singleton CB, Bethard JR, Silver RM. Proteomic analysis of CTGF-activated lung fibroblasts: identification of IQGAP1 as a key player in lung fibroblast migration. Am J Physiol Lung Cell Mol Physiol. 2008;295:L603–L611. doi: 10.1152/ajplung.00530.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aden N, Shiwen X, Aden D, Black C, Nuttall A, Denton CP, Leask A, Abraham D, Stratton R. Proteomic analysis of scleroderma lesional skin reveals activated wound healing phenotype of epidermal cell layer. Rheumatology (Oxford) 2008;47:1754–1760. doi: 10.1093/rheumatology/ken370. [DOI] [PubMed] [Google Scholar]
- 47.Mintz M, Vanderver A, Brown KJ, Lin J, Wang Z, Kaneski C, Schiffmann R, Nagaraju K, Hoffman EP, Hathout Y. Time series proteome profiling to study endoplasmic reticulum stress response. J Proteome Res. 2008;7:2435–2444. doi: 10.1021/pr700842m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bassuk JA, Berg RA. Protein disulphide isomerase, a multifunctional endoplasmic reticulum protein. Matrix. 1989;9:244–258. doi: 10.1016/s0934-8832(89)80057-5. [DOI] [PubMed] [Google Scholar]
- 49.Kellokumpu S, Suokas M, Risteli L, Myllylä R. Protein disulfide isomerase and newly synthesized procollagen chains form higher-order structures in the lumen of the endoplasmic reticulum. J Biol Chem. 1997;272:2770–2777. doi: 10.1074/jbc.272.5.2770. [DOI] [PubMed] [Google Scholar]
- 50.Wilson R, Lees JF, Bulleid NJ. Protein disulfide isomerase acts as a molecular chaperone during the assembly of procollagen. J Biol Chem. 1998;273:9637–9643. doi: 10.1074/jbc.273.16.9637. [DOI] [PubMed] [Google Scholar]
- 51.Ko MK, Key EP. PDI-mediated ER retention and proteasomal degradation of procollagen I in corneal endothelial cells. Exp Cell Res. 2004;295:25–35. doi: 10.1016/j.yexcr.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 52.Guiducci S, Distler JHW, Jüngel A, Huscher C, Huber LC, Michel BA, Gay RE, Pisetsky DS, Gay S, Matucci-Cerinic M, Distler O. The relationship between plasma microparticles and disease manifestations in patients with systemic sclerosis. Arthritis Rheum. 2008;58:2845–2853. doi: 10.1002/art.23735. [DOI] [PubMed] [Google Scholar]
- 53.Jüngel A, Distler O, Schulze-Horsel U, Huber LC, Ha HR, Simmen B, Kalden JR, Pisetsky DS, Gay S, Distler JH. Microparticles stimulate the synthesis of prostaglandin E2 via induction of cyclooxygenase 2 and microsomal prostaglandin E synthase 1. Arthritis Rheum. 2007;56:3564–3574. doi: 10.1002/art.22980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all protein spots that had a positive identification by mass spectrometry with their relative ratio in the two conditions. C/S ratio: Normal fibroblast secretome/SSc fibroblast secretome. C/NF ratio: Normal fibroblast secretome/NSF fibroblast secretome.


