Abstract
Oncogenic human papillomavirus (HPV) infection is the most important risk factor in cervical carcinogenesis cases; high viral loads, viral integration into the host genome, and gain of the telomerase-related genes, TERT and TERC, are all factors associated with progression to cancer. A recently developed multiparameter HPV 16/18 multiplex ligation-dependent probe amplification (MLPA) assay, which allows the simultaneous assessment of these factors, was applied to a series of 67 normal and (pre)malignant frozen uterine cervical samples, as well as to 91 cytological preparations, to test the ability of the MLPA assay to identify high-risk lesions on the basis of these factors. Validation was performed using quantitative PCR, the PapilloCheck and fluorescence in situ hybridization. Only 5 out of 37 normal tissue samples or low-grade cervical lesions (ie, CIN1 and condyloma) showed either an HPV16 viral load higher than 25 copies per cell, viral integration, and/or gain of one of the telomerase-related genes, whereas for the high-grade cervical lesions, one or more of these risk factors was found in 25 of 30 cases. The HPV MLPA assay showed a sensitivity of 83% and a specificity of 86% in frozen cervical specimens. Furthermore, the feasibility of the MLPA assay was shown for cytological samples, where in 57% of high-grade squamous intraepithelial lesion cases, the high-risk factors were detected using this assay.
Currently preventive screening for (pre)malignant lesions of the uterine cervix is based on the analysis of Papanicolaou stained cytological samples, combined with identification of high risk human papillomavirus (HPV) types.1,2 Although the specificity of cytology in the detection of high grade lesions is high, its sensitivity can be as low as 50%.3,4 On the contrary, the sensitivity for HPV testing is very high, while its specificity is low.5 Obviously, there is a need for a screening approach that is both highly specific and highly sensitive. We recently developed an HPV multiplex ligation-dependent probe amplification (MLPA)-assay6 which not only identifies HPV16 and 18, but simultaneously assesses viral load and viral integration for these HPV types, as well as gain of the telomerase genes, since these markers were described to be associated with progression to cancer (see below). In our previous study the MLPA-assay was used to determine these parameters in cervical cancer cell lines and a very small subset of clinically derived tissue.6 Here, the study has been expanded to 67 fresh frozen samples of normal cervix and different stages of CIN, as well as, 91 cytological samples.
MLPA is a molecular technique initially developed by Schouten et al7 for quantification of up to 40 genomic targets. For each target a pair of probes is designed. Each probe contains a universal PCR primer sequence and a sequence complementary to the target. When the probes hybridize immediately adjacent to each other they can be ligated and subsequently amplified using universal primers. Because one of the primers is labeled with a fluorescent dye the amplified products can be visualized using capillary electrophoresis. Furthermore, the products can be discerned based on length because of the variable stuffer sequences.8 We modified the assay as such that a simultaneous quantification of both human and viral targets is possible.
Of the 15 to 18 oncogenic HPV-types described9 HPV16 and 18 account for 70% of the cervical cancer cases.10 In addition, it has been described that the viral load for HPV16 increases during cervical carcinogenesis,11 while also viral integration into the host genome is described to be associated with progression to cancer.12
For HPV18 a high viral load is not related to cervical malignancy,13–15 but integration into the host genome is seen more often for this HPV type.16,17
The telomerase genes are included in our MLPA-assay because gain of chromosome 3q, with telomerase RNA component (TERC) as one of the suggested candidate genes18,19 is frequently identified during carcinogenesis of the cervix. Furthermore, gain of chromosome 5p, containing the telomerase reverse transcriptase (TERT) gene, is also reported to be associated with progression to cancer.20,21
Although all these viral and genomic markers are predominantly detected in high grade lesions they do not identify all these lesions, when assessed separately.11,19,20 As a result the specificity of these individual markers is very high but their sensitivity is low. Through the combination of these markers in the HPV 16/18 MLPA-assay we show that the sensitivity is considerably increased, while the specificity of the assay remains high when applied to frozen cervical specimens. Also we demonstrate the feasibility of the test for cytological preparations to obtain a risk classification based on these MLPA parameters with increasing cytological grading.
Materials and Methods
Uterine Cervical and Cytological Tissue Samples
Sixty-seven frozen cervical specimens including 7 normal ectocervical tissues, 20 normal ectocervical epithelia adjacent to (pre)neoplastic lesions, 10 CIN1 lesions/condylomas, 6 CIN2 lesions, 7 CIN3 lesions, and 17 squamous cell carcinomas were obtained from the Tissue Bank of the University of Liège (Belgium). The samples contained between 30% and 95% (pre)malignant cells. DNA was extracted from each sample by using the NucleoSpin Tissue kit (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions. The project protocol was approved by the Medical Ethics Committee of the University Hospital of Liège.
DNA was also isolated from a series of 84 liquid based cytological samples (Surepath, BD, Franklin Lakes, HJ) from the Department of Pathology (Maastricht University Medical Centre, The Netherlands) which were preselected based on aberrations detected on microscopic inspection. These were also analyzed for HPV triage by GP5+/6+ testing.22 This was also the case for 7 normal samples. DNA was previously isolated using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
MLPA-Assay
The MLPA-assay was performed as previously described6 and illustrated in Figure 1. A multiplex pre-amplification PCR was performed using the Qiagen Multiplex PCR kit. A 20 μl reaction mixture contained Qiagen Master Mix (3 mmol/L MgCl2, dNTPs, and HotstarTaq DNA polymerase), the previously described multiplex primer mix targeting the viral E2, E6, and E7 genes for HPV16 and 18, as well as the human β-globin, MSH2, TERT, and TERC genes6 (final concentration 20 nmol/L for each forward primer and 200 nmol/L for each reverse primer), and 10 ng of sample DNA. Amplification was performed in a Biometra T1 Thermocycler (Biometra, Göttingen, Germany) as follows: 15 minutes at 95°C, followed by 20 cycles of each 30 seconds at 94°C, 90 seconds at 55°C, and 90 seconds at 72°C, and a final extended elongation step for 10 minutes at 72°C.
Figure 1.
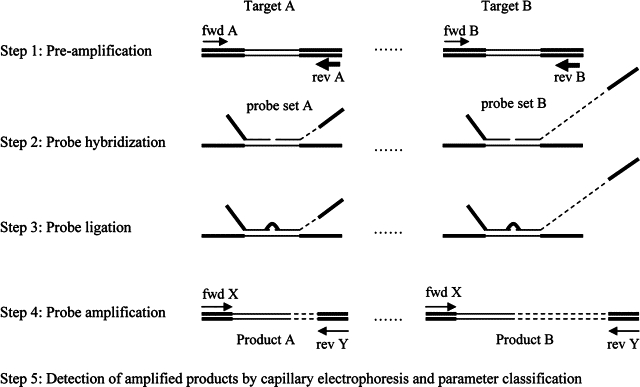
Schematic overview of the different steps in the HPV MLPA-assay. A pre-amplification step is performed with a mixture of specific HPV or human target primers (fwd = forward primer; rev = reverse primer) for each target (step 1). Subsequently, an MLPA reaction is initiated by hybridizing a mixture of probe sets specific for all targets to the pre-amplification products (step 2). A probe set consists of two oligonucleotides: one synthetic oligonucleotide (left, probe set A) and one M13-derived oligonucleotide (right, probe set B). The synthetic oligonucleotide contains a universal forward priming sequence (bold line in probe) next to the target specific sequence (thin line in probe). The M13-derived oligonucleotide contains a universal reverse priming sequence (bold line in probe) and a unique stuffer sequence (discontinuous line in probe) next to the target specific sequence (thin line in probe). The length of the stuffer sequence varies between the different probes. Subsequently, the probe sets are joined by ligation (step 3) and all ligated oligonucleotides are simultaneously amplified by one universal primer set (fwd X and rev Y; step 4). The length of the MLPA products is the combined length of both oligonucleotides, which is unique for each probe set due to the specific length of the stuffer sequence. After these reactions, the products are analyzed by capillary electrophoresis (step 5), and the parameters classified as shown in f2.
The pre-amplified product was diluted five times using sterile water, after which 2 μl was mixed with 1.5 μl MLPA-buffer (1.5 M KCl, 300 mmol/L Tris-HCl pH 8.5, 1 mmol/L EDTA; MRC-Holland, Amsterdam, the Netherlands), 1.5 μl of the previously described probe mix containing probes against the described viral and human targets6 (3 fmol of each synthetic probe oligonucleotide and 1.5 fmol of each M13-derived oligonucleotide in TE buffer) and 3 μl sterile water. After a 5 minutes denaturation step at 98°C in a Biometra T1 Thermocycler with a heated lid, the mixture was incubated for 16 hours at 60°C. For ligation this mixture was diluted to 40 μl with ligation buffer (2.6 mmol/L MgCl2, 5 mmol/L Tris-HCl pH 8.5, 0.013% non-ionic detergents, 0.2 mmol/L nicotinamide adenine dinucleotide) containing 1U heat-stable Ligase-65 enzyme (MRC-Holland, Amsterdam, the Netherlands) and incubated at 54°C for 15 minutes, followed by ligase inactivation at 98°C for 5 minutes. Four μl of this mixture was added to 16 μl of PCR mixture containing dNTPs (2 mmol/L each, Fermentas, St. Leon-Rot, Germany), 1 U Taq-polymerase (MRC-Holland, Amsterdam, the Netherlands), 1× PCR buffer (50 mmol/L KCl, 10 mmol/L Tris-HCl pH 8.5, 1.6 mmol/L MgCl2) and 4 pmol of the two MLPA-PCR primers each, with the forward primer 5′-GTGGCAGGGCGCTACGAACAA-3′ labeled with carboxyfluorescein, and the reverse primer 5′-GGACGCGCCAGCAAGATCCAATCTAGA-3′. Amplification was performed on a Biometra T1 Thermocycler as follows: an initial cycle of 2 minutes at 95°C, followed by 33 cycles of 30 seconds at 94°C, 30 seconds at 60°C, and 1 minute at 72°C, and a final extended elongation step for 10 minutes at 72°C. MLPA buffers and enzymes were obtained from MRC-Holland (Amsterdam, the Netherlands).
Analysis of MLPA Products
Amplified carboxyfluorescein-labeled MLPA products were analyzed by electrophoresis on an ABI3730 genetic analyzer (Applied Biosystems, Foster City, CA). One μl of 20× diluted amplified MLPA products was mixed with 8.5 μl of deionized formamide and 0.5 μl of GeneScan-600 LIZ size standard (Applied Biosystems, Foster City, CA) and run in GeneScan mode. All analyses were done according to the manufacturers' instructions. Electropherograms were analyzed by GeneMarker software (Softgenetics, State College, PA), peak height data were exported to Excel files for calculation of ratios and analyzed according to the flow-chart described in Figure 2. HPV positivity was determined based on the presence of both E6 and E7 peaks (Figure 2, step A). E6 and E7 viral loads were estimated by determining the ratio between E6 or E7 and the seven human targets, ie, β-globin (2×), MSH2, TERT (2×), and TERC (2×), and using this ratio in the following specific equations: L = 55.9r11.1, L = 12.5r11.3, L = 16.8r11.3, and L = 43.2r11.4 for 16E6, 16E7, 18E6, and 18E7, respectively (Figure 2, step B). With L being the viral load estimated as HPV copies per human gene copy and r1 being the ratio between target signal intensity and the total signal intensity of all human targets. The average load of E6 and E7 was multiplied by 2 to obtain the viral load in a diploid cell.
Figure 2.
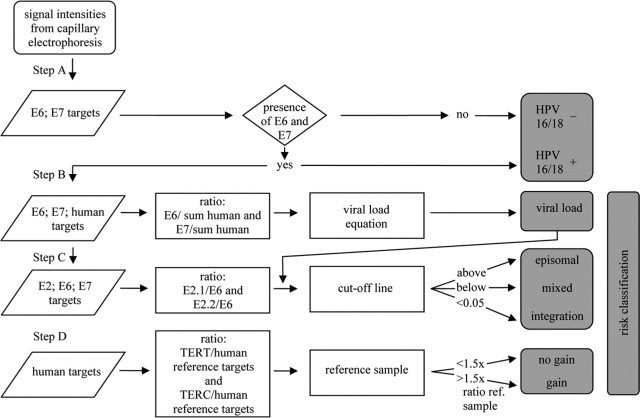
Flow-chart for HPV MLPA parameter classification, ie, HPV 16 and 18 positivity, viral load, physical status (episomal, mixed, integration) and gain for TERT and TERC. For details see Materials and Methods.
All fresh frozen samples were analyzed in duplicate and when the viral load in a sample was determined to be less than 1 copy per cell the assay was repeated with a primer and probe mix containing only HPV targets, thus without primers and probes for the human targets, to confirm HPV positivity. When the viral load in a sample was more than 50 copies per cell the sample was diluted with normal human DNA to decrease the viral load and the diluted sample was analyzed again.
Due to experimental variability a reference series was included in each experiment. This series consisted of 5 mixtures of plasmids and human DNA, mimicking HPV16 samples with viral loads of 2, 5, 10, 20, and 40 copies per cell of which 30% was integrated. In each experiment the reference series was run in duplicate and the subsequent 16E2.1/E6 or 16E2.2/E6 ratio was plotted against the viral load to determine the viral integration cut-off value for different viral loads. Patient samples with a ratio above the cut-off line were scored as episomal and samples with a ratio below the cut-off line were scored as mixed or integrated (Figure 2, step C). This results in a classification of episomal HPV when less than 30% of the virus in a sample shows integration, mixed (ie, episomal and integrated) HPV when between 30% and 95% of the virus shows integration, and integrated HPV when more than 95% of the virus shows integration.
The specificity of the primer and probe mix for HPV16 and 18 was evaluated by tests on HPV16 or 18 plasmid model systems, cell lines containing HPV16 or 18, and clinical samples containing closely related HPV types, ie, HPV31, 33 and 45 (data not shown), as determined with the PapilloCheck (Greiner Bio-One GmbH, Frickenhausen, Germany).
Similar to the fresh frozen tissue samples the cytological samples were analyzed in duplicate and reference samples were included in each experiment. For these experiments the reference samples were mixtures of plasmids and human DNA, mimicking an episomal HPV16 and HPV18 sample with a viral load of 2.
To determine gain for TERT the ratio between the combined signal intensity of TERTa and TERTc on the one hand and the combined signal intensity of β-globin-a, β-globin-b, and MSH2 on the other hand was calculated. This signal intensity ratio was compared to the ratio as measured for these targets in the normal human DNA sample. When the signal intensity ratio was 1.5× higher than that of the reference sample, TERT was classified as gained. To determine gain for TERC the ratio between the combined signal intensity of TERCa and TERCb on the one hand and the combined signal intensity of β-globin-a, β-globin-b, and MSH2 on the other hand was calculated. This signal intensity ratio was also compared to the ratio as measured for these targets in the normal human DNA sample and classified as gain when it was 1.5× higher than that of the reference sample (Figure 2; step D).
Quantitative PCR
All samples were tested for the presence of HPV16, 18, 31, 33 and 45 using a previously described quantitative (qPCR) by Lindh et al23 targeting sequences in the E6/E7 genes, and were scored positive when the threshold cycle (Ct) value was below 35 for HPV16, 18, 31, and 45 and below 30 for HPV33. Furthermore, for the majority of samples viral load and physical status for HPV16 were estimated by the qPCR described by Peitsaro et al24 using primers and probes for HPV16 E6 and HPV16 E2 (Biolegio, Malden, The Netherlands) in a 20 μl PCR mixture containing dNTPs (2 mmol/L each, Fermentas, St. Leon-Rot, Germany), 1 U Taq-polymerase (MRC-Holland, Amsterdam, the Netherlands), 1× PCR buffer (50 mmol/L KCl, 10 mmol/L Tris-HCl pH 8.5, 1.6 mmol/L MgCl2), 3 pmol of each primer, 1 pmol of each probe, and 25 ng of sample DNA. The qPCR reactions were performed in a Rotor-Gene 6000 real-time system (Corbett Life Science, Sydney, Australia) as follows: 2 minutes at 95°C, followed by 50 cycles of 30 seconds at 94°C, 90 seconds at 55°C, and 90 seconds at 72°C, and a final extended elongation step for 10 minutes at 72°C. The standard curves were obtained by amplification of a 10-fold dilution series of 5 × 106 to 500 copies of a HPV16 plasmid.6
The threshold cycle values for both HPV16 E6 and E2 standard curves were plotted against the log of the copy number over the entire range of dilutions and revealed a linear relationship. These standard curves were used to estimate the average HPV copy number in the individual samples based on duplicate measurements. Viral loads were calculated based on the assumption that the samples have a diploid DNA content and a DNA mass of 7.8 pg per diploid cell.25 The physical status of the virus was determined on basis of the copy number ratio between E2 and E6. Samples were classified as integrated when no fluorescence was detected for E2 and the ratio was 0, mixed for ratios between 0 and 0.5, and episomal for ratios above 0.5 as previously described.26
Fluorescence in Situ Hybridization
FISH analysis on a small selection of 4-μm thick, fresh-frozen tissue sections samples was performed as previously described19 using digoxigenin-labeled probes for HPV16 or 18 (PanPath, Budel, The Netherlands) or a Vysis probe set (Vysis, Abbott Molecular, Des Plaines, IL) consisting of: a DNA probe for chromosome 3 centromere labeled with the fluorescent dye Spectrum Green (SG), a 3q26-specific BAC clone containing the TERC gene labeled with Spectrum Orange (SO), and a probe for chromosome 7 centromere labeled with Spectrum Aqua (SA).
Images were acquired using a Leica DMRXA microscope (Leica, Wetzlar, Germany) equipped with custom optical filters for 4,6-diamidino-2-phenylindole, SA, SO, and SG (Chroma Technologies, Brattleboro, VT) with a ×40 Plan Apo (NA 1.20) objective. The microscope was connected to a digital black and white CCD camera (Metasystems Image Pro System, Sandhausen, Germany). To determine gain for the TERC target the copy number for chromosome 7 was used as a control for the ploidy of cells. For chromosome 3 and target 3q26 (TERC) the maximum number of signals per nucleus was determined and used as an indicator of copy number, as described previously.19
PapilloCheck Assay
The PapilloCheck HPV-Screen DNA-chip (Greiner, Frickenhausen, Germany) was used for the qualitative detection and differentiation between 24 types of genital HPV (18 high-risk and 6 low-risk). HPV genotyping was performed as previously described by Jones et al27 Briefly, for each test at least 40 ng of DNA was used and a 350 bp fragment of the HPV E1 gene was amplified using a multiplex PCR with type-specific primers. An internal PCR control targeting a fragment of the human housekeeping gene ADAT1 was included in each run to avoid false negative results. PCR fragments were fluorescence-labeled with Cy5 and hybridized to specific probes on the PapilloCheck DNA chip. The amplification level was determined by the binding of PCR products to five control spots and their subsequent signal intensity on the chip. Following hybridization and subsequent washing steps, the chip was scanned at excitation wavelengths of 532 and 635 nm.
Results
Fresh Frozen Tissue Samples
HPV-Typing
In order for the MLPA-assay to be useful in cervical screening the sensitivity in HPV-typing has to be high. As summarized in Table 1, 52 of the 67 patient samples were found to be positive for HPV16, including 4 of the 7 (57%) normal samples, 14 of the 20 (70%) normal samples adjacent to a lesion, 7 of the 10 (70%) CIN1/condyloma samples, 12 of the 13 (92%) CIN2/3 samples and 15 of the 17 (88%) carcinoma samples. Furthermore, one CIN2/3 and one carcinoma sample were found to be positive for HPV18.
Table 1.
Results of the HPV MLPA-Assay in Frozen Uterine Cervical Tissue Samples, Validated by qPCR, PapilloCheck, and FISH
| Diag. | Case | Age | % Cells | Typing |
Viral load |
Physical status |
Telomerase genes gain |
High risk class. MLPA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MLPA | qPCR | PapilloCheck | MLPA | qPCR* Ct HPV16/18 | qPCR† HPV16 | MLPA | qPCR† E2/E6 | MLPA TERT | MLPA TERC | |||||
| Normal no lesion (n = 7) | 1 | 64 | − | − | − | − | 35/37 | − | − | − | − | − | − | |
| 2‡ | 47 | − | − | − | − | 36/37 | − | − | − | − | − | − | ||
| 3 | 52 | − | − | − | − | 35/− | − | − | − | − | − | − | ||
| 4 | 52 | 16 | − | − | 0.11 | 36/− | − | epi | − | − | − | − | ||
| 5 | 73 | 16 | − | − | 0.14 | 35/40 | − | epi | − | − | − | − | ||
| 6 | 38 | 16 | 16 | 16 | 0.22 | 34/40 | − | epi | − | − | − | − | ||
| 7 | 60 | 16 | 16 | 16 | 2.10 | 34/38 | 0.16 | epi | int | − | − | − | ||
| Normal adj. to lesion (n = 20) | 8 | 75 | − | − | − | − | 38/− | n.d. | − | − | − | − | − | |
| 9 | 35 | − | 16 | − | − | 34/− | n.d. | − | − | − | − | − | ||
| 10 | − | − | − | − | − | 38/38 | n.d. | − | − | − | − | − | ||
| 11 | 47 | − | − | − | − | 35/− | n.d. | − | − | − | − | − | ||
| 12 | 36 | − | − | − | − | 37/− | n.d. | − | − | − | − | − | ||
| 13 | 43 | − | − | 45 | − | −/− | n.d. | − | − | − | − | − | ||
| 14 | 50 | 16 | − | 16 | 0.004 | 37/− | − | epi | − | − | − | − | ||
| 15 | 34 | 16 | 16 | − | 0.05 | 34/39 | 0.05 | epi | mix | − | − | − | ||
| 16 | − | 16 | 16 | 16 | 0.07 | 33/− | 0.02 | epi | mix | − | − | − | ||
| 17 | 32 | 16 | 16 | 58 | 0.13 | 32/− | 0.1 | epi | mix | − | − | − | ||
| 18 | − | 16 | 16 | 16; 59 | 0.18 | 34/− | 0.04 | epi | mix | − | − | − | ||
| 19 | 52 | 16 | 16; 31 | n.c. | 0.18 | 34/39 | 0.05 | epi | epi | − | − | − | ||
| 20 | − | 16 | 16 | 16 | 0.33 | 31/40 | 0.33 | mix (a)§ | mix | − | − | + | ||
| 21 | 64 | 16 | 16 | n.c. | 0.48 | 31/− | 0.22 | epi | mix | − | − | − | ||
| 22 | − | 16 | 16 | 16; 59 | 0.51 | 31/38 | 0.17 | epi | mix | − | − | − | ||
| 23 | 49 | 16 | 16 | 16 | 0.91 | 32/− | 0.34 | epi | n.c. | − | − | − | ||
| 24 | 32 | 16 | 16 | 16 | 2.5 | 30/− | 0.83 | epi | mix | − | − | − | ||
| 25 | 51 | 16 | 16 | n.c. | 6.8 | 26/38 | 1.6 | epi | epi | − | − | − | ||
| 26 | − | 16 | 16 | 16 | 13 | 26/− | 8 | epi | epi | − | − | − | ||
| 27 | − | 16 | 16 | 16 | 31 | 26/36 | 4.5 | epi¶ | mix | − | − | + | ||
| CIN1/condyloma (n = 10) | 28 | − | 60 | − | − | − | − | 37/− | n.d. | − | − | − | − | − |
| 29 | − | 60 | − | − | 52 | − | 39/40 | n.d. | − | − | − | − | − | |
| 30 | − | 95 | − | − | − | − | −/− | n.d. | − | − | − | − | − | |
| 31 | 22 | 40 | 16 | − | 56; 59 | 0.02 | 35/39 | − | epi | − | − | − | − | |
| 32 | − | 70 | 16 | 16 | 16 | 0.06 | 33/39 | − | epi | − | − | − | − | |
| 33 | 58 | 50 | 16 | 16 | 16; 59 | 0.09 | 33/37 | 0.09 | epi | epi | − | − | − | |
| 34 | 23 | 40 | 16 | 16 | 56; 59 | 0.14 | 33/39 | − | epi | − | − | − | − | |
| 35 | 32 | 90 | 16 | 16 | n.c. | 6.4 | 27/− | 2.4 | int (b)§ | int | − | − | + | |
| 36 | − | 60 | 16 | 16 | 16; 59 | 90 | 23/37 | 54 | epi | epi | − | − | + | |
| 37 | 21 | 90 | 16 | 16 | 16; 59 | 1008 | 19/− | 568 | epi¶ | epi | + | + | + | |
| CIN2 (n = 6) | 38 | 38 | 50 | 16 | 16; 45 | 16; 45 | 0.01 | 34/− | 0.03 | epi | mix | − | − | − |
| 39‡ | 43 | 60 | 16 | 16 | 16 | 1.4 | 29/38 | 0.76 | mix (c)¶ | mix | − | −¶ | + | |
| 40 | − | 70 | 18 | 18 | 16; 18; 59 | 1.7 | 35/27 | − | mix (b)§ | − | + | + | + | |
| 41 | − | 90 | 16 | 16 | 16; 33 | 50 | 23/− | 45 | epi | epi | + | + | + | |
| 42 | 59 | 70 | 16 | 16 | 16; 59 | 94 | 23/38 | 73 | epi | epi | + | + | + | |
| 43 | − | 60 | 16 | 16 | 16 | 130 | 22/− | 91 | epi§ | epi | − | − | + | |
| CIN3 (n = 7) | 44 | 53 | 50 | 16 | 16 | 16 | 0.65 | 31/38 | 0.14 | epi | mix | − | − | − |
| 45 | 30 | 70 | 16 | 16 | − | 3.1 | 28/− | 3.8 | int (b) | mix | − | − | + | |
| 46 | − | 30 | 16 | 16 | 16 | 5.9 | 27/40 | 15 | int (a) | int | − | + | + | |
| 47 | 49 | 50 | 16 | 16 | 16 | 8.4 | 27/− | 3.8 | epi | epi | − | − | − | |
| 48 | 36 | 50 | 16 | 16 | 16; 59 | 9.5 | 26/36 | 6.1 | mix (c) | epi | − | + | + | |
| 49 | − | 80 | 16 | 16 | 16 | 10 | 25/38 | 25 | epi | epi | − | + | + | |
| 50‡ | 27 | 60 | 16 | 16 | 16 | 249 | 24/− | 112 | epi¶ | epi | − | −¶ | + | |
| SCC (n = 17) | 51 | 30 | 95 | − | 45 | 45; 11 | − | 38/− | n.d. | − | n.d. | + | − | + |
| 52 | 44 | 30 | 16 | 33 | 16; 59; 33 | 0.01 | 35/− | − | epi | − | + | + | + | |
| 53 | − | 60 | 16 | 31 | 16 | 0.04 | 35/− | − | epi | − | − | − | − | |
| 54 | − | 80 | 18 | 18 | 16; 59 | 3.7 | 35/28 | − | epi§ | − | − | − | − | |
| 55 | 46 | 90 | 16 | 16 | − | 4.5 | 27/36 | 4.4 | int (b) | int | − | − | + | |
| 56 | − | 15 | 16 | 16 | 16 | 4.8 | 26/38 | 17 | int (b) | int | + | + | + | |
| 57 | 45 | 50 | 16 | 16 | 16 | 5.3 | 27/− | 12 | epi | n.c. | − | + | + | |
| 58 | − | 70 | 16 | 16 | 16 | 5.6 | 26/36 | 13 | epi | epi | − | + | + | |
| 59‡ | 51 | 80 | 16 | 16 | 16 | 9.4 | 27/40 | 3.7 | int (a) | int | − | − | + | |
| 60 | 70 | 95 | 16 | 16 | 16 | 25 | 24/− | 15 | epi | epi | − | − | + | |
| 61 | 50 | 60 | 16 | 16 | 16 | 62 | 23/39 | 81 | epi | epi | − | − | + | |
| 62 | 62 | 90 | 16 | 16 | 16 | 70 | 25/− | 23 | epi | epi | − | − | + | |
| 63 | 45 | 60 | 16 | 16 | 16 | 83 | 22/− | 153 | epi | epi | + | + | + | |
| 64 | 55 | 90 | 16 | 16 | 16 | 126 | 20/38 | 504 | mix (a) | mix | − | + | + | |
| 65 | 48 | 60 | 16 | 16 | 16; 59 | 134 | 22/37 | 38 | mix (b) | mix | − | + | + | |
| 66‡ | 58 | 80 | 16 | 16 | 16 | 149 | 20/37 | 198 | mix (b)¶ | mix | − | +¶ | + | |
| 67 | − | 70 | 16 | 16 | 16 | 166 | 20/37 | 404 | mix (a) | mix | − | + | + | |
% Cells, percentage (pre)malignant cells; HPV, human papillomavirus; MLPA, multiplex ligation-dependent probe amplification; qPCR, quantitative PCR; FISH, fluorescence in situ hybridization; +, detected; −, not detected; n.c., not conclusive; n.d., not determined; a, (partial) loss of target sequence 16E2.1; b, (partial) loss of target sequence 16/18E2.1 and (partial) loss of target sequence 16/18E2.2; c, (partial) loss of target sequence 16E2.2); epi, episomal; mix, episomal and integrated; int, integrated; TERC, telomerase RNA component; TERT, telomerase reverse transcriptase; high risk class; MLPA, high risk classification based on viral load of 25 HPV16 copies or more and/or viral integration and/or gain of the telomerase genes.
according to Lindh et al.23
according to Peitsaro et al.24
previously described by Theelen et al.6
FISH inconclusive.
Confirmed by FISH.
To determine the validity of the MLPA results all samples were analyzed by two independent qPCR protocols for HPV16, a qPCR for HPV18, 31, 33, and 45, and the PapilloCheck. The MLPA results were confirmed by qPCR in 48 of the 54 HPV16/18 positive cases and 12 of the 13 HPV16/18 negative cases based on a threshold cycle (Ct) value below 35. Furthermore, using the PapilloCheck HPV16 was detected in 40 of the 52 cases that were determined to be HPV16 positive by the MLPA-assay. In 4 samples the PapilloCheck was inconclusive, while 6 of the 8 samples in which no HPV16 was detected with the PapilloCheck, were determined to have a viral load of less than 1 copy per 5 cells, and the remaining 2 samples had a viral load of 3 and 5 copies per cell. In two of these HPV16 negative samples a double infection with HPV56 and 59 was detected, one sample had an infection with HPV58 and the other 5 samples were negative for all HPV types detected by the PapilloCheck (Table 2).
Table 2.
MLPA Typing versus qPCR and PapilloCheck in Frozen Tissue Samples
| MLPA |
qPCR |
PapilloCheck |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HPV | n | 16+ | 18+ | 16− 18− | 16+ | 18+ | 16+ 18+ | Others | Negative | n.c. |
| 16+ | 52 | 46 | − | 6 | 40 | − | − | 3 | 5 | 4 |
| 18+ | 2 | − | 2 | − | 1 | − | 1 | − | − | − |
| 16− 18− | 13 | 1 | − | 12 | − | − | − | 3 | 10 | − |
n, number of cases; −, not detected; n.c., not conclusive.
All 13 samples that were determined to be HPV16/18 negative by the MLPA-assay were validated using the PapilloCheck, which was also used to identify infections with other HPV types. There were 3 samples that were positive for one or more of the other HPV types. This concerned an HPV45 infection in a normal sample which was adjacent to a lesion, an HPV52 infection in a CIN1/condyloma sample and a co-infection with HPV11 and 45 in a carcinoma sample. This latter HPV45 infection as well as a co-infection of HPV16 and 45 was also confirmed by the qPCR by Lindh et al23 The 10 remaining samples were negative for all HPV types detected by the PapilloCheck.
Viral Load
For HPV16 a high viral load is described to be associated with progression to cancer. Overall the viral load detected by the MLPA-assay ranged from less than 1 copy per 10 cells to approximately 1000 copies per cell. As depicted in Figure 3A the reproducibility of the MLPA procedure for viral load detection between the duplicate analyses, starting from the same DNA sample, is very high. A median viral load of 1 copy per 5 cells was detected for the normal samples, 2 copies per 5 cells for the normal samples adjacent to a lesion, 1 copy per 7 cells for the CIN1/condyloma samples, 26 copies per cell for the CIN2 samples, 8 copies per cell for the CIN3 samples, and 17 copies per cell for the carcinoma samples.
Figure 3.
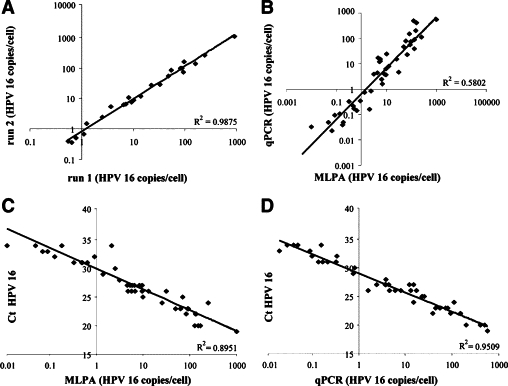
A: Reproducibility of viral load estimations (HPV copies per cell) when comparing duplicate MLPA measurements. B–D: Comparison of the viral load estimations by MLPA and qPCR for fresh frozen tissue samples. B: Comparison of viral load as determined by MLPA and qPCR as applied by Peitsaro et al.24C: Correlation between the Ct values determined by qPCR using the method of Lindh et al23 and the viral loads as determined by the MLPA. D: Correlation between the Ct values as determined by qPCR using the method of Lindh et al23 and qPCR using the method of Peitsaro et al.24
To validate the viral load as determined by the MLPA-assay, all HPV16 positive samples were analyzed by qPCR according to Peitsaro et al.24 Viral load could be determined in 43 of these 52 samples analyzed using qPCR. For the 9 qPCR negative samples the viral load was determined to be less than 1 copy per 4 cells using the MLPA-assay. When comparing the viral loads as determined by the MLPA-assay or qPCR for the individual cases it became evident that both procedures detect a similar range of HPV copies per cell (Figure 3B).23–24 Overall, the viral load as detected by the qPCR according to Lindh et al23 using Ct values correlated well with the copy numbers as detected by the MLPA (Figure 3C)23–24 and the qPCR according to Peitsaro et al24 (Figure 3D).23–24 The detected HPV copies per cell ranged from less than 1 copy per 10 cells to approximately 570 copies per cell, with a viral load of 0.16 copies per cell for the normal sample, and a median viral load of 1 copy per 5 cells for the normal samples adjacent to a lesion, 2 copies per 7 cells for the CIN1/condyloma samples, 11 copies per cell for the CIN2/3 samples, and 23 copies per cell for the carcinoma samples.
Viral Integration
Viral integration into the host genome is described to be associated with progression to cancer. As a consequence of integration (part of) the HPV E2 gene is almost always deleted, while the HPV E6 gene is retained (see Figure 4, A–C). Quantification of the E2/E6 ratio is therefore used for the estimation of viral integration. The reproducibility of both the E2.1/E6 and the E2.2/E6 signal intensity ratio is depicted in Figure 5, A and B, and found to be high. Integrated HPV was detected in 1 of the 16 HPV16 positive normal samples adjacent to a lesion, 1 of the 7 HPV16 positive CIN1/condyloma samples, 4 of the 12 HPV16 positive CIN2/3 samples, as well as the HPV18 positive CIN2/3 sample and 7 of the 15 HPV16 positive carcinoma samples.
Figure 4.
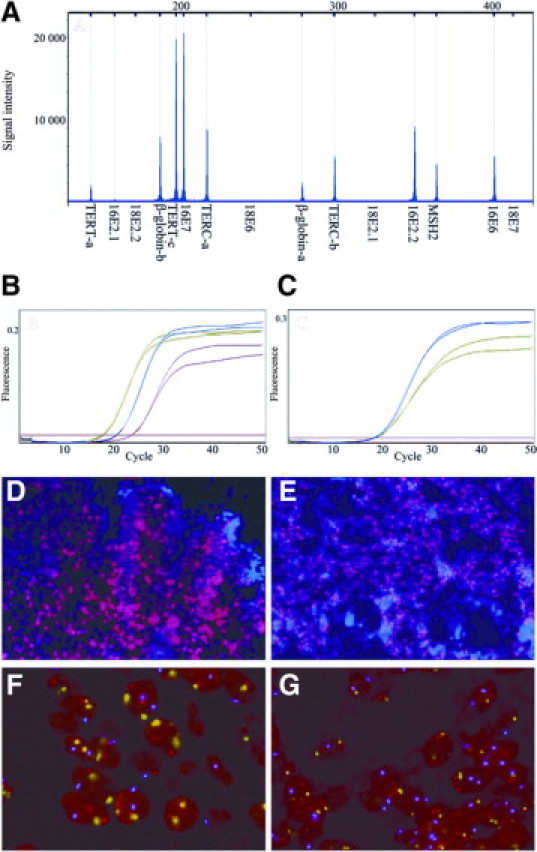
A–C: Examples of the different techniques for fresh frozen tissue samples: an MLPA capillary electrophoresis peak profile (A; case 59); qPCR curves for E6 (B) and E2 (C) for a sample with exclusively episomal HPV in blue (case 61); a sample with both episomal and integrated HPV in green (case 64); and a sample with fully integrated HPV in red (case 43). D–G: FISH patterns for episomal HPV (D; case 37), integrated HPV (E; case 66), and the absence (F; case 50) or presence (G; case 66) of gain for TERC (red) as compared to copy numbers of chromosome 7 (blue) and chromosome 3 (green).
Figure 5.
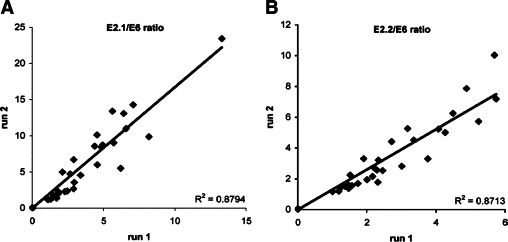
Reproducibility of detection of viral integration using E2/E6 ratios for fresh frozen tissue samples. Comparison of the duplicate MLPA measurements for the E2.1/E6 (A) and E2.2/E6 (B) signal intensity ratios.
For the validation of the physical status as determined by the MLPA-assay we used the E2/E6 duplex qPCR. Thirteen of the 14 samples that were determined to contain integrated HPV by the MLPA-assay were confirmed by qPCR. One of these samples (case 45) was determined to contain predominantly integrated HPV by the MLPA-assay whereas qPCR determined the sample to contain both integrated and episomal HPV. In addition, 17 of the 39 samples that were determined to be episomal by MLPA were confirmed by qPCR. Nine of these 39 samples could not be confirmed because no HPV16 was detected by qPCR and in 2 of these 39 samples qPCR was inconclusive because of inconsistency between duplicates. Strikingly, in the remaining 11 samples in which the MLPA-assay detected episomal HPV the qPCR indicated the presence of integrated HPV (mixed or integrated; summarized in Table 3). These 11 discordant cases comprised 9 normal samples (of which 8 adjacent to a lesion), and 2 CIN2/3.
Table 3.
Comparison between MLPA and qPCR for Physical Status Analysis in Frozen Tissue Samples
| MLPA Physical status | qPCR |
|||||
|---|---|---|---|---|---|---|
| n | epi | mix | int | n.c. | − | |
| Epi | 39 | 17 | 10 | 1 | 2 | 9 |
| mix 2.1 | 3 | − | 3 | − | − | − |
| mix 2.2 | 2 | 1 | 1 | − | − | − |
| mix 2.1 and 2.2 | 2 | − | 2 | − | − | − |
| int 2.1 | 2 | − | − | 2 | − | − |
| int 2.1 and 2.2 | 4 | − | 1 | 3 | − | − |
n, number of cases; epi, episomal; mix, episomal and integrated; int, integrated; 2.1, (partial) loss of target sequence 16E2.1; 2.2, (partial) loss of target sequence 16E2.2; n.c., not conclusive; −, not detected.
Of the 10 samples that were analyzed for the physical status of HPV16 and HPV18 by FISH episomal HPV16 was confirmed in three cases (cases 27, 37, and 50 in Table 1; Figure 4D) and integrated HPV in two cases (cases 40, and 66 in Table 1; Figure 4E). In the remaining 3 HPV16 samples (cases 20, 35, and 43) FISH analysis was inconclusive, which was also the case for the 2 samples that were analyzed for HPV18 (cases 40 and 54). All these latter 5 cases had a relatively low viral load.
Telomerase Genes
Gain for the telomerase related genes TERT and TERC is described to be associated with progression to cancer, and was therefore included in our HPV MLPA-assay. Gain for TERT was detected in 8 samples, ie, 1 CIN1, 3 CIN2/3, and 4 carcinoma samples. Gain for TERC was detected in 16 samples, ie, 1 CIN1, 6 CIN2/3, and 9 carcinoma samples. Seven of the 8 cases with gain for TERT overlapped with the TERC gain cases. Only in case 51 TERT gain was seen to be independent of gain for TERC. The FISH analyses for TERC performed on tissue sections from three patients (cases 39, 50, and 66 in Table 1; Figure 4, F and G) gave confirmatory results.
Sensitivity and Specificity of the MLPA-Assay
To quantify the diagnostic capacity of the MLPA-assay, sensitivity and specificity were calculated for each of the four parameters, ie, HPV type 16/18, viral load, viral integration, and gain of the telomerase related genes. For this purpose the ability of the assay to distinguish high grade CIN2/3 and carcinoma lesions on the one hand, from normal samples, low grade CIN1 and condyloma lesions on the other hand, was determined. For the presence of HPV type 16 and 18 the sensitivity is calculated to be 97% but the specificity to be only 32%. For a viral load of more than 25 copies per cell the sensitivity and specificity are calculated to be 40% and 92%, respectively. For viral integration this is 40% and 95%, for gain of TERT 23% and 97%, and for gain of TERC 50% and 97%, respectively (Table 4). By combining 2 parameters the sensitivity can be increased up to 63 to 70%, with a small reduction in specificity to 86 to 92%. When all parameters are combined the sensitivity increases up to 83%, while retaining a specificity of 86%.
Table 4.
Sensitivity and Specificity of the HPV MLPA-Assay when Applied to Frozen Tissue Samples
| Histology | MLPA single parameter |
MLPA multiparameter |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type 16 and/or 18 (%) | Viral load >25 (%) | Integration (%) | TERT gain (%) | TERC gain (%) | Telom gain (%) | Viral load >25 and/or integration (%) | Viral load >25 and/or telom gain (%) | Integration and/or telom gain (%) | Viral load >25 and/or integration and/or telom gain (%) | |
| Normal | 4/7 (57) | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) | 0/7 (0) |
| Normal adjacent to lesion | 14/20 (70) | 1/20 (5) | 1/20 (5) | 0/20 (0) | 0/20 (0) | 0/20 (0) | 2/20 (10) | 1/20 (5) | 1/20 (5) | 2/20 (10) |
| CIN1/condyloma | 7/10 (70) | 2/10 (20) | 1/10 (10) | 1/10 (10) | 1/10 (10) | 1/10 (10) | 3/10 (30) | 2/10 (20) | 2/10 (20) | 3/10 (30) |
| CIN2/3 | 13/13 (100) | 4/13 (31) | 5/13 (38) | 3/13 (23) | 6/13 (46) | 6/13 (46) | 8/13 (62) | 8/13 (62) | 8/13 (62) | 10/13 (77) |
| SCC | 16/17 (94) | 8/17 (47) | 7/17 (41) | 4/17 (24) | 9/17 (53) | 10/17 (59) | 11/17 (65) | 13/17 (76) | 12/17 (71) | 15/17 (88) |
| Sensitivity (95% CI) | 97% (80.9–99.8%) | 40% (23.2–59.2%) | 40% (23.2–59.2%) | 23% (10.6–42.7%) | 50% (31.7–68.3%) | 53% (34.6–71.2%) | 63% (43.9–79.5%) | 70% (50.4–84.6%) | 67% (47.1–82.1%) | 83% (64.5–93.6%) |
| Specificity (95% CI) | 32% (18.6–49.9%) | 92% (77.0–97.9%) | 95% (80.5–99.1%) | 97% (84.2–99.9%) | 97% (84.2–99.9%) | 97% (84.2–99.9%) | 86% (70.4–94.9%) | 92% (77.0–97.9%) | 92% (77.0–97.9%) | 86% (70.4–94.9%) |
CI, confidence interval; CIN, cervical interaepithelial neoplasia; SCC, squamous cell carcinoma; telom, telomerase gene; TERC, telomerase RNA component; TERT, telomerase reverse transcriptase; >25, more than 25 copies per cell.
Cytological Samples
Cytological samples of 91 preselected cases (Table 5), ranging from normal to HSIL (Pap classification 1 to 3A28), for which the presence of HPV was known based on GP5+/6+ testing,22 were analyzed by the MLPA-assay and verified by the two qPCR protocols. The MLPA-assay determined a total of 32 samples to be positive for HPV16/18 of which 27 were positive for HPV16 and 5 were positive for HPV18. Among the HPV16 samples 11 had a viral load higher than 25 copies per cell and 6 samples contained integrated HPV whereas among the HPV18 samples 2 contained integrated HPV. Of these 32 cases, 31 were confirmed by qPCR. The remaining sample was determined to have a HPV16 viral load of 1 copy per 10 cells by the MLPA-assay and a Ct-value of 28 for HPV31 by qPCR. Additionally, of the 59 HPV16/18 negative samples qPCR found 4 samples (2 normal and 2 LSIL) to be infected with HPV45, 9 samples (2 ASCUS and 7 LSIL) with HPV31 and 1 LSIL sample with HPV33. Furthermore, a gain for TERT and/or TERC was detected in 10 of the 32 HPV 16/18 positive samples, as well as in 5 of the 59 HPV16/18 negative samples. As summarized in Table 5, 23 of the 91 samples were determined to be high risk according to the MLPA-assay. This concerned 1 of the 7 (14%) normal, 4 of the 28 (14%) ASCUS, 14 of the 49 (29%) LSIL, and 4 of the 7 (57%) HSIL samples.
Table 5.
Summary of Results of the MLPA Assay in Cytological Samples Compared with qPCR and GP5+/6+
| Cytology | n | HR HPV (%) | GP5+/6+ HPV 16/18 (%) | qPCR HPV 16/18 (%) | MLPA |
High-risk class. MLPA (%) | ||
|---|---|---|---|---|---|---|---|---|
| High load (%) | Integration (%) | Telom. gain (%) | ||||||
| Normal | 7 | 3 (43) | 1 (14) | 1 (14) | 0 | 0 | 1 (14) | 1 (14) |
| ASCUS | 28 | 13 (46) | 9 (32) | 9 (32) | 0 | 1 (4) | 3 (11) | 4 (14) |
| LSIL | 49 | 38 (78) | 14 (29) | 15 (31) | 8 (16) | 6 (12) | 8 (16) | 14 (29) |
| HSIL | 7 | 7 (100) | 7 (100) | 7 (100) | 3 (43) | 1 (14) | 3 (43) | 4 (57) |
| Total | 91 | 61 | 31 | 32 | 11 | 8 | 15 | 23 |
n, number of cases; HPV, human papillomavirus; MLPA, multiplex ligation-dependent probe amplification; qPCR, quantitative PCR; Telom., telomerase genes; high-risk class; MLPA, high-risk classification based on viral load of 25 HPV16 copies or more and/or viral integration and/or gain of the telomerase genes.
Discussion
HPV-related parameters, such as oncogenic type, viral load, and viral integration are prognosticators in cervical (pre)neoplasia.9,11,19 Since these individual markers have been shown to exhibit either a low sensitivity or a low specificity,5,11 we have developed a multiparameter assay for the simultaneous assessment of these indicators in combination with the detection of increased copy numbers of the telomerase related genes. The predictive value of the individual markers in this HPV MLPA-assay was similar to that described in the literature,11,19,20 but when combining the results of the individual parameters in the MLPA-assay a balance between high specificity and high sensitivity was reached which is more acceptable for a screening test.
For typing of HPV16 and 18 all of the fresh frozen tissue samples that were determined to be negative by the MLPA-assay were confirmed by the PapilloCheck and/or qPCR. This was also the case for 48 of the 52 fresh frozen tissue samples that were found to be HPV16 positive. The discrepant samples were characterized by a low viral load, which implies that the MLPA-assay is at least as sensitive as qPCR and the PapilloCheck in the detection of these HPV types. This is particularly evident since for the two latter assays higher amounts of DNA were used as compared to the MLPA-assay. Also the number of cycles used for the qPCR as described by Peitsaro et al24 was 50 instead of 40.
The prevalence for HPV16 among the CIN2/3 and carcinoma cases in this study is as high as 92% and 88%, respectively, while the literature reports a prevalence of about 60% in carcinoma cases.9 Based on the abnormally high prevalence for HPV16 in this study it is clear that the sensitivity of this assay is overestimated. The prevalence for HPV16 is also high in the histologically normal samples, particularly when combined with a (pre) neoplastic lesion. The risk of false positive results is, however, unlikely in the MLPA-assay because of the use of four probes per HPV type. HPV infection is only scored as being positive when a product is detected for both the E6 and E7 probe, while in a majority of cases also products will be detected for one or both of the E2 probes, providing additional proof for the reliability of the outcome. Furthermore, the samples in our study were randomly selected without prior knowledge of HPV status, and as described above mostly confirmed by qPCR or the PapilloCheck providing additional arguments against false positive MLPA-results. The presence of HPV in the normal samples, either without a lesion or adjacent to a premalignancy, indicates that the sole detection of HPV is not a very specific prognosticator for the presence of a high grade lesion.
In the literature, the cut-off value for HPV16 viral load used to identify with high specificity women with prevalent high grade lesions is described to be between 22 and 35 copies per cell.11,29 Therefore, the cut-off value used in this study was set at 25 viral copies per cell. Based on this value 40 of the 43 samples were identically classified using either the MLPA-assay or qPCR. However, in this study, the viral load as detected using the MLPA-assay was generally somewhat higher than the viral load as detected using qPCR. As a result, 2 fresh frozen tissue samples, ie, 1 normal sample adjacent to a lesion and 1 carcinoma sample, were classified as high grade based on the viral load determined by the MLPA-assay and as low grade based on qPCR, which might be an indication for the use of a higher cut-off value for the MLPA-assay. On the other hand, one CIN2/3 sample was classified as low-grade based on the viral load as determined by the MLPA-assay and as high grade based on the viral load as determined by qPCR, arguing against such an increase of the cut-off value.
The incidence for viral integration based on qPCR was very high in the samples that were classified as normal. Although this observation is in agreement with previous qPCR studies on viral integration24 various other studies using APOT, FISH, or 2D-agarose gel electrophoresis techniques showed that integration occurs late in the carcinogenic process.12,30,31 To explain this apparent discrepancy it should be noted that 10 of our samples had a viral load of less than five copies per cell. The error in qPCR threshold cycle values and E2/E6 ratios is possibly higher in samples with a low viral load than in samples with a high viral load.32,33 Furthermore, there was one case that was determined to contain integrated HPV using the MLPA-assay, but exclusively episomal HPV using qPCR. In this case the HPV16E2.2 target sequence was deleted while the HPV16E2.1 target sequence was retained. Integration was therefore missed by qPCR because the probe used recognizes a sequence close to HPV16E2.1. Finally, in two cases the qPCR results were inconclusive.
With respect to the discriminating power of the MLPA-assay in the frozen tissue samples a sensitivity and specificity of 63% and 86%, respectively, were obtained by combining the three HPV parameters. Inclusion of the results for the telomerase related genes in these analyses leads to a significant increase of the sensitivity to 83%, while retaining the specificity of 86%. In addition to four high-grade cases with an episomal HPV16 infection and a low viral load, the carcinoma case that was not infected with HPV16 or 18 was also classified as high grade based on a gain for one of the telomerase related genes. Furthermore, the absence or presence of gain for TERC was confirmed by FISH in selected cases, while the incidence was in agreement with our previous findings.19 Similarly, Heselmeyer et al,18 using comparative genomic hybridization in cases of severe cervical dysplasia and invasive carcinoma, demonstrated an increasing incidence for gain of TERC with increasing severity of the lesion. We conclude that the assessment of gain of the telomerase genes contributes significantly to the proper classification of high risk patients.
In 10 of the 67 frozen tissue samples discordance between the morphological and molecular characteristics was observed. Four of the five cases among the CIN1 and normal samples with either integration or a high viral load were confirmed by qPCR. These cases therefore deserve closer monitoring than the other low grade cases. In five CIN2/3 and carcinoma cases the MLPA-assay resulted in an underestimation of the severity of the lesion. This can be partially explained by the presence of other oncogenic HPV types, indicating the necessity to include additional HPV types in the assay. An expansion of the number of probes targeting the E2 gene could also lead to an increased detection of integration and subsequently to a higher sensitivity of the assay.
Application of the HPV MLPA-assay to a series of cytological samples revealed an evident increase of high risk classification with the increase of cytological grading. Overall, 25% of these preselected samples were classified as high risk cases using our MLPA assay. Again, inclusion of additional HPV types in this test will increase its sensitivity when applied to cytological samples. Our MLPA analysis of cytological samples showed that high risk HPV16 and 18 is more often associated with high grade cytological outcomes as compared to any other high risk HPV type. This confirms the results as described earlier by Munoz et al9 Furthermore, a high frequency of integration was seen in the LSIL cases in our series, which is in agreement with a previous study by Gallo et al34 who detected that viral integration was particularly evident in LSIL of a younger population of women. Finally, gain for TERT and TERC was detected in 8 of the 49 LSIL samples, which is in agreement with a study by Heselmeyer et al35 where gain for TERC was demonstrated in 7% of these low-grade samples. Our results thus show that the MLPA-assay is able to detect gain for TERT and TERC in cytological samples, despite the fact that these preparations may contain a high proportion of normal cells. Apparently, in these samples a sufficient fraction of (pre)malignant cells is present to reach and exceed the detection threshold for gain of the telomerase associated genes.
In conclusion, we showed that the HPV 16/18 MLPA-assay is capable of distinguishing high grade from low grade cervical lesions, both with a high sensitivity and high specificity. In biopsy material the combination of three parameters results in risk classification which can be applied as a potential indicator for cervical (pre)neoplasia. Furthermore, the potential of the MLPA assay as a complementary technique for cytological samples has been illustrated.
Acknowledgements
We thank Nathalie Krusy (Laboratory of Experimental Pathology, University of Liège, Belgium) for the DNA isolation and PapilloCheck analysis.
Footnotes
Supported by the Transnational University Limburg (Maastricht, The Netherlands), a grant provided by LIOF (Limburg Development and Investment Company, Maastricht, The Netherlands) and a grant from OP-Zuid (Operationeel Programma Zuid; 31R263, The Netherlands) through PathoFinder BV.
P.D., M.H., and J.S. are Research Associates of the Belgian National Fund for Scientific Research.
References
- 1.Jordan J, Arbyn M, Martin-Hirsch P, Schenck U, Baldauf JJ, Da Silva D, Anttila A, Nieminen P, Prendiville W. European guidelines for quality assurance in cervical cancer screening: recommendations for clinical management of abnormal cervical cytology, part 1. Cytopathology. 2008;19:342–354. doi: 10.1111/j.1365-2303.2008.00623.x. [DOI] [PubMed] [Google Scholar]
- 2.Spitzer M, Apgar BS, Brotzman GL. Management of histologic abnormalities of the cervix. Am Fam Physician. 2006;73:105–112. [PubMed] [Google Scholar]
- 3.ACOG Practice Bulletin No. 99: management of abnormal cervical cytology and histology. Obstet Gynecol. 2008;112:1419–1444. doi: 10.1097/AOG.0b013e318192497c. [DOI] [PubMed] [Google Scholar]
- 4.Cuzick J, Clavel C, Petry KU, Meijer CJ, Hoyer H, Ratnam S, Szarewski A, Birembaut P, Kulasingam S, Sasieni P, Iftner T. Overview of the European and North American studies on HPV testing in primary cervical cancer screening. Int J Cancer. 2006;119:1095–1101. doi: 10.1002/ijc.21955. [DOI] [PubMed] [Google Scholar]
- 5.Rao A, Pather S, Dalrymple C, Mackie A, Deans R, Carter J. The role of HPV testing in patients with possible high-grade cervical cytology. J Obstet Gynaecol Res. 2009;35:503–506. doi: 10.1111/j.1447-0756.2008.00944.x. [DOI] [PubMed] [Google Scholar]
- 6.Theelen W, Reijans M, Simons G, Ramaekers FCS, Speel EJM, Hopman AHN. A new multiparameter assay to assess HPV 16/18 viral load and physical status together with gain of telomerase genes in HPV-related cancers. Int J Cancer. 2010;126:959–975. doi: 10.1002/ijc.24844. [DOI] [PubMed] [Google Scholar]
- 7.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Y, Wu BL. Designing a simple multiplex ligation-dependent probe amplification (MLPA) assay for rapid detection of copy number variants in the genome. J Genet Genomics. 2009;36:257–265. doi: 10.1016/S1673-8527(08)60113-7. [DOI] [PubMed] [Google Scholar]
- 9.Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 10.Munoz N, Castellsague X, de Gonzalez AB, Gissmann L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006;24(Suppl 3):1–10. doi: 10.1016/j.vaccine.2006.05.115. [DOI] [PubMed] [Google Scholar]
- 11.Saunier M, Monnier-Benoit S, Mauny F, Dalstein V, Briolat J, Riethmuller D, Kantelip B, Schwarz E, Mougin C, Pretet JL. Analysis of human papillomavirus type 16 (HPV16) DNA load and physical state for identification of HPV16-infected women with high-grade lesions or cervical carcinoma. J Clin Microbiol. 2008;46:3678–3685. doi: 10.1128/JCM.01212-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klaes R, Woerner SM, Ridder R, Wentzensen N, Duerst M, Schneider A, Lotz B, Melsheimer P, von Knebel Doeberitz M. Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Res. 1999;59:6132–6136. [PubMed] [Google Scholar]
- 13.Cheung JL, Cheung TH, Ng CW, Yu MY, Wong MC, Siu SS, Yim SF, Chan PK. Analysis of human papillomavirus type 18 load and integration status from low-grade cervical lesion to invasive cervical cancer. J Clin Microbiol. 2009;47:287–293. doi: 10.1128/JCM.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hesselink AT, Berkhof J, Heideman DA, Bulkmans NW, van Tellingen JE, Meijer CJ, Snijders PJ. High-risk human papillomavirus DNA load in a population-based cervical screening cohort in relation to the detection of high-grade cervical intraepithelial neoplasia and cervical cancer. Int J Cancer. 2009;124:381–386. doi: 10.1002/ijc.23940. [DOI] [PubMed] [Google Scholar]
- 15.Xi LF, Koutsky LA, Castle PE, Wheeler CM, Galloway DA, Mao C, Ho J, Kiviat NB. Human papillomavirus type 18 DNA load and 2-year cumulative diagnoses of cervical intraepithelial neoplasia grades 2–3. J Natl Cancer Inst. 2009;101:153–161. doi: 10.1093/jnci/djn461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pirami L, Giache V, Becciolini A. Analysis of HPV16, 18, 31, and 35 DNA in pre-invasive and invasive lesions of the uterine cervix. J Clin Pathol. 1997;50:600–604. doi: 10.1136/jcp.50.7.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinokurova S, Wentzensen N, Kraus I, Klaes R, Driesch C, Melsheimer P, Kisseljov F, Durst M, Schneider A, von Knebel Doeberitz M. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008;68:307–313. doi: 10.1158/0008-5472.CAN-07-2754. [DOI] [PubMed] [Google Scholar]
- 18.Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hopman AHN, Theelen W, Hommelberg PP, Kamps MA, Herrington CS, Morrison LE, Speel EJM, Smedts F, Ramaekers FCS. Genomic integration of oncogenic HPV and gain of the human telomerase gene TERC at 3q26 are strongly associated events in the progression of uterine cervical dysplasia to invasive cancer. J Pathol. 2006;210:412–419. doi: 10.1002/path.2070. [DOI] [PubMed] [Google Scholar]
- 20.Huang FY, Kwok YK, Lau ET, Tang MH, Ng TY, Ngan HY. Genetic abnormalities and HPV status in cervical and vulvar squamous cell carcinomas. Cancer Genet Cytogenet. 2005;157:42–48. doi: 10.1016/j.cancergencyto.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Matthews CP, Shera KA, McDougall JK. Genomic changes and HPV type in cervical carcinoma. Proc Soc Exp Biol Med. 2000;223:316–321. doi: 10.1046/j.1525-1373.2000.22345.x. [DOI] [PubMed] [Google Scholar]
- 22.van den Brule AJ, Pol R, Fransen-Daalmeijer N, Schouls LM, Meijer CJ, Snijders PJ. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J Clin Microbiol. 2002;40:779–787. doi: 10.1128/JCM.40.3.779-787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindh M, Gorander S, Andersson E, Horal P, Mattsby-Balzer I, Ryd W. Real-time taqman PCR targeting 14 human papilloma virus types. J Clin Virol. 2007;40:321–324. doi: 10.1016/j.jcv.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Peitsaro P, Johansson B, Syrjanen S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J Clin Microbiol. 2002;40:886–891. doi: 10.1128/JCM.40.3.886-891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee GM, Thornthwaite JT, Rasch EM. Picogram per cell determination of DNA by flow cytofluorometry. Anal Biochem. 1984;137:221–226. doi: 10.1016/0003-2697(84)90374-9. [DOI] [PubMed] [Google Scholar]
- 26.Kulmala SM, Shabalova IP, Petrovitchev N, Syrjanen KJ, Gyllensten UB, Johansson BC, Syrjanen SM. Type-specific persistence of high-risk human papillomavirus infections in the New Independent States of the former Soviet Union Cohort Study. Cancer Epidemiol Biomarkers Prev. 2007;16:17–22. doi: 10.1158/1055-9965.EPI-06-0649. [DOI] [PubMed] [Google Scholar]
- 27.Jones J, Powell NG, Tristram A, Fiander AN, Hibbitts S. Comparison of the PapilloCheck DNA micro-array Human Papillomavirus detection assay with Hybrid Capture II and PCR-enzyme immunoassay using the GP5/6+ primer set. J Clin Virol. 2009;45:100–104. doi: 10.1016/j.jcv.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 28.Rebolj M, van Ballegooijen M, Berkers LM, Habbema D. Monitoring a national cancer prevention program: successful changes in cervical cancer screening in the Netherlands. Int J Cancer. 2007;120:806–812. doi: 10.1002/ijc.22167. [DOI] [PubMed] [Google Scholar]
- 29.Cricca M, Morselli-Labate AM, Venturoli S, Ambretti S, Gentilomi GA, Gallinella G, Costa S, Musiani M, Zerbini M. Viral DNA load, physical status and E2/E6 ratio as markers to grade HPV16 positive women for high-grade cervical lesions. Gynecol Oncol. 2007;106:549–557. doi: 10.1016/j.ygyno.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 30.Hudelist G, Manavi M, Pischinger KI, Watkins-Riedel T, Singer CF, Kubista E, Czerwenka KF. Physical state and expression of HPV DNA in benign and dysplastic cervical tissue: different levels of viral integration are correlated with lesion grade. Gynecol Oncol. 2004;92:873–880. doi: 10.1016/j.ygyno.2003.11.035. [DOI] [PubMed] [Google Scholar]
- 31.Hopman AHN, Smedts F, Dignef W, Ummelen M, Sonke G, Mravunac M, Vooijs GP, Speel EJM, Ramaekers FCS. Transition of high-grade cervical intraepithelial neoplasia to micro-invasive carcinoma is characterized by integration of HPV 16/18 and numerical chromosome abnormalities. J Pathol. 2004;202:23–33. doi: 10.1002/path.1490. [DOI] [PubMed] [Google Scholar]
- 32.Roberts I, Ng G, Foster N, Stanley M, Herdman MT, Pett MR, Teschendorff A, Coleman N. Critical evaluation of HPV16 gene copy number quantification by SYBR green PCR. BMC Biotechnol. 2008;8:57. doi: 10.1186/1472-6750-8-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruutu MP, Kulmala SM, Peitsaro P, Syrjanen SM. The performance of the HPV16 real-time PCR integration assay. Clin Biochem. 2008;41:423–428. doi: 10.1016/j.clinbiochem.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 34.Gallo G, Bibbo M, Bagella L, Zamparelli A, Sanseverino F, Giovagnoli MR, Vecchione A, Giordano A. Study of viral integration of HPV-16 in young patients with LSIL. J Clin Pathol. 2003;56:532–536. doi: 10.1136/jcp.56.7.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heselmeyer-Haddad K, Janz V, Castle PE, Chaudhri N, White N, Wilber K, Morrison LE, Auer G, Burroughs FH, Sherman ME, Ried T. Detection of genomic amplification of the human telomerase gene (TERC) in cytologic specimens as a genetic test for the diagnosis of cervical dysplasia. Am J Pathol. 2003;163:1405–1416. doi: 10.1016/S0002-9440(10)63498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


